Multiple levels of PD-L1 regulation in cancer immunotherapy
To date, the PD-1/PD-L1 blockade strategy has been tested in a variety of cancer types. However, only bladder cancer, melanoma, mismatch repair-deficient colorectal cancer, and certain hematopoietic malignancies have been highly responsive in terms of clinical outcomes.1,2 It is presumed that the expression level of the PD-L1 protein largely determines the therapeutic efficacy of PD-1/PD-L1-targeted cancer immunotherapy.3,4 Previous studies have shown that several transcriptional factors are involved in the upregulation of PD-L1 by directly binding to its promoter region.5 For instance, individual activation of STAT1, NF-κB, HIF-1α, c-Myc, or MAPK increased PD-L1 transcription and immune evasion of tumor cells. On the other hand, oncogenic RAS signaling reportedly stabilizes PD-L1 mRNA to promote tumor immunoresistance.6
In addition to transcriptional and translational regulation, the protein stability of PD-L1 is controlled at multiple points at the posttranslational level, which is hypothesized to be one of the leading causes of PD-1/PD-L1-targeted immunotherapy inefficiency and resistance to it. Thus far, all phosphorylation, glycosylation, palmitoylation, ubiquitination, methylation, and acetylation modifications have been found to be involved in the posttranslational modification (PTM) of PD-L1.5,7 The phosphorylation of PD-L1 is frequently observed in multiple cancers. GSK3β can directly bind to the C-terminal domain of PD-L1 and phosphorylate the T180 and S184 residues on PD-L1, which are subsequently recognized by β-TRCP to negatively affect PD-L1 stability and thus enhance PD-1 blockade therapy.8 On the other hand, PD-L1 can be modified by N-linked glycosylation, which stabilizes PD-L1 primarily by preventing GSK3β/β-TRCP-mediated PD-L1 degradation. In addition, Cha et al. found that metformin-activated AMPK directly phosphorylates PD-L1 at the S195 residue to induce abnormal glycosylation, which results in PD-L1 degradation through the ERAD pathway.9
Accumulating evidence suggests that ubiquitin-proteasome system (UPS)-mediated regulation of PD-L1 stability directly influences anti-PD-1/PD-L1 therapeutic efficacy. CMTM6, a type-3 transmembrane protein, and its closest family member, CMTM4, were recently confirmed to be positive regulators of PD-L1 in a wide range of human tumor cells, with both acting in a UPS-dependent manner.10 CMTM6 was found to interact with PD-L1 on the cell surface, interfering with its ubiquitination to prolong its half-life, and it has been confirmed through functional analysis that CMTM6 enhances the immune evasion ability of PD-L1-positive tumor cells. Zhang et al. showed that the Cullin 3-SPOP E3 ligase promotes PD-L1 polyubiquitination and proteasome-mediated degradation to regulate its abundance and cancer immune surveillance.11 In contrast to ubiquitination, deubiquitination of PD-L1 protects the protein from degradation. Recently, CSN5 was found to be required for TNF-α-mediated PD-L1 stabilization in cancer cells and was identified as a crucial protein required for the promotion of PD-L1 deubiquitination.12 In addition, Wu et al. discovered that ubiquitin-specific peptidase USP9X could deubiquitinate and stabilize PD-L1 in oral squamous cell carcinoma cells, thus suppressing USP9X blocks to tumor cell growth.13 Collectively, these results provide a theoretical basis for targeting deubiquitination and stabilization of PD-L1 to activate antitumor immunity.
USP22 is a novel deubiquitinase of PD-L1 in liver cancer
In our recent study,14 we found that USP22 strongly interacts with PD-L1 in vitro and in vivo to induce its deubiquitination and thus prevents the proteasomal degradation of PD-L1. Consistent with this action, the half-life of PD-L1 was markedly shorter in USP22-knockout (KO) cells but largely extended in USP22-overexpressing cells. These results demonstrate that USP22 acts as a deubiquitinase of PD-L1 to enhance its stabilization. Furthermore, we generated PD-L1-overexpressing USP22-KO cells, as well as USP22 and PD-L1 double-KO cells, to investigate the potential pathophysiological significance of the USP22-PD-L1 axis. We found that neither overexpression nor depletion of PD-L1 had any significant enhancing or inhibitory effects on cell proliferation, viability or colony formation, which indicates that PD-L1 was not required for USP22-mediated cancer cell growth in vitro.
In contrast, the therapeutic efficiencies of the anti-PD-L1 antibody and cisplatin were individually enhanced in USP22-KO tumors in immunocompetent C57BL/6 mice but not in immunodeficient NCG mice. These findings, while preliminary, indicate that USP22 depletion significantly promotes the antitumor immune response. Since tumor-infiltrating lymphocytes (TILs) are crucial to the efficacy of cancer immunotherapy, we further evaluated TILs in our mouse models. Indeed, we found that the absolute number of CD3+ TILs and the relative ratio of CD8+GZMB+ cells to CD3+ TILs were both largely increased in the USP22-KO tumors from mice individually inoculated during immunotherapy and chemotherapy.
In addition, we analyzed the correlation between USP22 and PD-L1 in clinical cancer cases from the TCGA and found that USP22 and PD-L1 are associated with a significant tendency for frequent genetic alterations. In particular, USP22 is highly expressed in hepatocellular carcinoma (HCC) but not in other cancer types, which strongly suggests that liver cancer is a potent target for exploring the tumor-suppressing effects of blocking USP22-PD-L1 regulation. Furthermore, to determine whether USP22 is a prognostic marker for liver cancer, we performed Kaplan–Meier survival analyses and found that high USP22 expression is indeed correlated with poor overall survival of untreated liver cancer patients and for those who received sorafenib treatment. To confirm the role of USP22 in the regulation of PD-L1 in human patients, we determined the expression of both genes in HCC and found that the expression level of USP22 was strongly correlated with that of PD-L1. Taken together, data from our study reveal that, in addition to CSN5 and other deubiquitinases, USP22 inhibition may be a potential immune-mediating strategy for the treatment of liver cancer (Fig. 1).
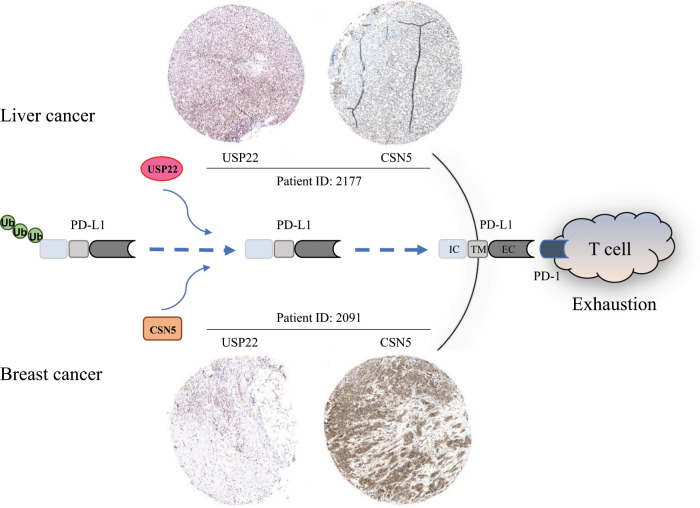
Fig. 1. USP22 dominates PD-L1 deubiquitination in liver cancer to suppress antitumor immunity, while CSN5 contributes to PD-L1 stabilization in breast cancer.
The expression of USP22 and CSN5 in tissue samples was analyzed individually by immunohistochemistry staining and visualized through use of the Protein Atlas program (http://proteinatlas.org). Images of stained USP22 and CSN5 from one liver cancer patient and one breast cancer patient, identified as ID 2177 and ID 2091, respectively, in the Protein Atlas program, were randomly selected and shown as indicated. IC, intracellular domain; TM, transmembrane domain; EC, extracellular domain.
Targeting the USP22-PD-L1 axis in liver cancer immunotherapy
HCC, a kind of liver cancer with both etiologic and geographic diversity, is one of the most prevalent cancers and the fourth leading cause of worldwide cancer deaths, especially in developing countries. In contrast to results from previous studies of primary invasive melanomas or non-small cell lung cancer, some of the findings pertaining to the role of PD-L1 in HCC have been conflicting and disappointing. For instance, PD-L1 expression is a predictive biomarker for cytokine-induced killer cell immunotherapy in HCC patients;15 however, the therapeutic efficacy of targeting PD-L1 using this strategy has been unsatisfactory.
USP22 is an established oncoprotein that has already been found to contribute to tumorigenesis, malignant behavior, and multidrug resistance in HCC.16 The findings from our study revealed that USP22 is a novel deubiquitinase of PD-L1 in liver cancer. USP22 functions to deubiquitinate PD-L1 and stabilize its protein expression level, thereby causing cancer immune resistance. Our findings support the contention that a PD-L1 PTM-targeted strategy has an important role in activating antitumor immunity. We can safely hypothesize that the unsatisfactory clinical efficacy of the PD-L1-targeted strategy is likely caused by USP22-mediated stabilization of PD-L1, which counteracts the effect of anti-PD-L1 drugs.
Of note, the previously well-established functions of USP22 include transcriptional regulation and cell-cycle progression. Given the ability of USP22 to deubiquitinate histones, its function was initially linked with the regulation of gene transcription.17 USP22 has been shown to affect the expression of c-Myc-targeted genes, and its depletion leads to the arrest of cells in the G1 phase of the cell cycle. Moreover, USP22 can also facilitate the progression of the cell cycle by targeting CCNB1.18 In view of such important functions, further research is required to determine whether side effects are induced by inhibiting USP22 expression, as the USP22-PD-L1 axis plays a critical role in the immunotherapeutic efficacy of liver cancer treatment.
Our study identified one critical regulatory mechanism for PD-L1 stabilization and provided supportive evidence for targeting USP22-mediated PD-L1 deubiquitination as a new efficacious treatment for use in cancer immunotherapeutic strategies. Despite these promising results, no drug candidates are currently available for the direct inhibition of USP22, which is an important issue for future research and clinical application. Therefore, USP22-specific inhibitors should be developed as therapeutics for cancer treatment in the near future.
Acknowledgements
X.H. would like to express deepest thanks to Guido Kroemer for the cancer immunity-associated technological training, ideological inspiration and moral edification in his laboratory. This work was supported by grants from the National Natural Science Foundation of China (31970696 and 81502975 to X.H. and 81830089 to T.L.), China Postdoctoral Science Foundation (2016T90413 and 2015M581693 to X.H.), SEU-Alphamab Joint Center (SA2015001 to X.H.), and Zhejiang Provincial Program for the Cultivation of High-level Innovative Health Talents (to X.B.).
Author contributions
T.L., X.B. and X.H. conceived the correspondence. X.H. and X.Z. wrote and revised the paper, and T.L. and X.B. discussed and commented on the paper.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Xing Huang, Xiaozhen Zhang
References
- 1.Le DT, et al. N. Engl. J. Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herbst RS, et al. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel SP, Kurzrock R. Mol. Cancer Therapeutics. 2015;14:847–856. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 4.Li X, Song W, Shao C, Shi Y, Han W. Cell Mol. Immunol. 2019;16:28–39. doi: 10.1038/s41423-018-0086-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J, Dang F, Ren J, Wei W. Trends Biochem. Sci. 2018;43:1014–1032. doi: 10.1016/j.tibs.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coelho MA, et al. Immunity. 2017;47:1083–1099. doi: 10.1016/j.immuni.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y, et al. Cell Res. 2019;29:83–86. doi: 10.1038/s41422-018-0124-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li CW, et al. Nat. Commun. 2016;7:12632. doi: 10.1038/ncomms12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cha JH, et al. Mol. Cell. 2018;71:606–620. doi: 10.1016/j.molcel.2018.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mezzadra R, et al. Nature. 2017;549:106–110. doi: 10.1038/nature23669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, et al. Nature. 2018;553:91–95. doi: 10.1038/nature25015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim SO, et al. Cancer Cell. 2016;30:925–939. doi: 10.1016/j.ccell.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jingjing W, et al. Cancer Med. 2018;7:4004–4011. doi: 10.1002/cam4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang X, et al. Cancer Immunol. Res. 2019;7:1580–1590. doi: 10.1158/2326-6066.CIR-18-0910. [DOI] [PubMed] [Google Scholar]
- 15.Chen C-L, et al. OncoImmunology. 2016;5:e1176653. doi: 10.1080/2162402X.2016.1176653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ling S, et al. Mol. Oncol. 2017;11:682–695. doi: 10.1002/1878-0261.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeusset LM, McManus KJ. Cancers. 2017. p. E167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin Z, et al. Cell Discov. 2015;1:pii: 15028. doi: 10.1038/celldisc.2015.28. [DOI] [PMC free article] [PubMed] [Google Scholar]