Abstract
Different structures and cell types of the periodontium respond to orthodontic tooth movement (OTM) individually. Cementoblasts (OC/CM) located in the immediate vicinity of the fibroblasts on the cement have found way to the centre of actual research. Here, we identify and validate possible reference genes for OC/CM cells by RT-qPCR with and without static compressive loading. We investigated the suitability of 3 reference genes in an in vitro model of cementoblast cells using four different algorithms (Normfinder, geNorm, comparative delta-Ct method and BestKeeper) under different confluences and time. Comparable to our previous publications about reference genes in OTM in rats and human periodontal ligament fibroblasts (hPDLF), Rpl22 in murine OC/CM cells appears as the least regulated gene so that it represents the most appropriate reference gene. Furthermore, unlike to the expression of our recommended reference genes, the expression of additionally investigated target genes changes with confluence and under loading compression. Based on our findings for future RT-qPCR analyses in OC/CM cells, Rpl22 or the combination Rpl22/Tbp should be favored as reference gene. According to our results, although many publications propose the use of Gapdh, it does not seem to be the most suitable approach.
Subject terms: Cell biology, Molecular biology, Rheumatology
Introduction
The periodontal ligament (PDL), a component of the periodontal apparatus, represents a microenvironment essential for anchoring teeth, fulfils functions of proprioception and buffering of acting forces and it enables periodontal remodeling allowing for orthodontic tooth movement. During orthodontic treatment, teeth are moved through the alveolar bone with the help of mechanical force. This force initially deflects the tooth inside the alveolar cavity1. As a result, the periodontium can be divided into tension and compression areas. An inflammatory reaction associated with bone loss and risk of root resorptions on the compression side2,3 and bone formation on the tension side has been detected4.
The individual role of different cell types involved in orthodontic tooth movement must be identified and understood in order to gain a better insight into the mechanisms of orthodontic tooth movement for research purposes.
Apart from its important role in anchoring teeth, the PDL contributes to tooth nutrition, homoeostasis and damaged tissue repair5. PDL fibroblasts (PDLF), PDL stem cells (PDLSC), osteoblasts, osteoclasts and cementoblasts are important cells in this area. PDLFs are the most common and best studied cell type in investigations of tooth movement1.
Cementoblasts are located on the cementum covered root surface. Cementum, produced by cementoblasts all lifelong, plays an important role in anchoring the tooth to the surrounding alveolar bone6,7. It represents the key structure for attaching Sharpey’s fibers on the tooth side8. These fibers are the unique connecting link between tooth and alveolar bone. The absence of cementum results in loss of attachment, a major problem in periodontal degeneration and disease. For this reason, new cementum formation and restoration of soft tissue attachment to the cementum is one of the major goals of regenerative periodontal therapy and research9. Furthermore, cementoblasts are assumed to have an essential protective function on the root surface by protecting it from resorption2. It has been shown that root resorption lacunas are regenerated with cementum in absence of the causal stimulus (e.g. orthodontic force)10,11.
First RT-qPCR data regarding murine cementoblasts under compressive force have already been published12–15. Mechanoreceptors, enabling stressed cells to react to compressive stimuli, are less investigated in OC/CM cells. The ion channel Piezo1 seems to be involved in hPDLF16 and in OCCM13 regulation. In addition, in previous work we were able to demonstrate the participation of TLR-4 after compression of hPDLF17. It is also known that compression leads to increased VEGF-a expression on protein level18 as well as to enhanced cytokine expression in hPDLF19. Moreover, there are information that induced oxygen partial pressure affects RANKL mRNA upregulation in murine cementoblasts20 and in hPDLF21. In hPDLF, hypoxia related VEGF mRNA upregulation is published22. Considering these findings we already investigated the influence of hypoxia regarding to loading compression experiments with hPDLF with the result, that hypoxic effects appear to play a minor role in the regulation of osteoclastogenesis23. These findings need to be verified for cementoblasts. However, a systematic validation of reference genes with focus on the in vitro model of mechanical loading in cementoblasts is missing.
The best characterized murine cementoblasts cells were isolated and immortalized by setting SV40 large T antigen under control of the osteocalcin promotor, being the reason for naming these cells osteocalcin cementoblasts (OC/CM)24. In vitro, this procedure only immortalizes cells that express osteocalcin (OCN) and SV40, because preliminary in situ studies showed, that OCN was expressed by cementoblasts during root development, but not by PDL cells. Consequently, when populations are isolated from developing molars using collagenase/trypsin digestion, only cementoblasts, not PDL cells, are immortalized and will survive in culture24,25. A primary murine cell line is currently not available.
To simulate the mechanical forces acting in periodontal remodeling in vitro, a static mechanic compression model has been developed26. For this purpose, a sterile glass cylinder is placed on the monolayer for simulating the stimulus that a cell of the periodontal microenvironment experiences when being compressed between root and alveolar bone during OTM.
Studies of gene expression observing cells under different conditions play an important role in investigating molecular mechanisms. Quantitative real-time PCR (RT-qPCR) is the gold standard for quantification of gene expression due to the method’s merits concerning specificity, sensitivity, accuracy and reproducibility27.
Even with greatest care, variations in the path from sample collection to data acquisition can influence the RT-qPCR statement if normalization is not performed with a suitable reference gene28. The reference gene should meet three criteria: (1) the amplification efficiency should be similar to the target genes; (2) a moderate expression level; (3) stable expression under all test conditions29.
Furthermore, a well working reference gene in condition ‘A’ may not be suitable in condition ‘B’30. There is evidence that a reference gene working for one cell type cannot be adopted for other cell types without checking the requirements above31. In addition, it is known that some classic reference genes such as ß-actin do not perform adequately in mechanically stressed cells32,33. We used these findings as a basis for selecting possible genes for the present investigation.
Possible reference genes were purposely selected based on research already made, and their stability in neighboring cells using comparable experimental conditions. We included 3 potential references in our investigation. First, Gapdh (glyceraldehyde-3-phosphate dehydrogenase), a glycolysis enzyme which is frequent in the normalization of RT-qPCR and has also been used in previous experiments with murine cementoblasts13,15. Second, Rpl22 (the ribosomal 60S protein L22), a component of the ribosomal 60S subunit which is proven to be the most stable reference gene in compression experiments with human periodontal fibroblasts. Third, Tbp (TATA-binding protein), a transcription factor exhibiting stable gene expression under comparable experimental conditions in human periodontal fibroblasts34.
Due to the high proliferation rate of OC/CM cells which significantly increase their confluence, we postulate that gene expression of a suitable reference gene should be independent from different cell densities. We examined the expression of different target genes at 60% and 100% confluence to show the effect of an altered confluence on the gene expression of target genes. The chosen target genes are inflammation and remodeling markers. This allows to investigate their changes during this compression associated inflammatory reaction that is accompanied by bone loss.
In this study, we propose recommendable reference genes for OC/CM cells at different confluences and in an in vitro model of orthodontic tooth movement in order to facilitate reliable research respecting the MIQE guidelines30. Our publication should provide as a basis for future RT-qPCR analyses with murine cementoblasts, investigating their role in periodontal microenvironment, delineation to other cells and regulation under mechanical stimulation. In addition, we show that precise monitoring of cell confluence is fundamental for comparable RT-qPCR results with these cells.
Results
Selection of primer, amplicon quality and specificity, RT-qPCR efficiencies and expression levels
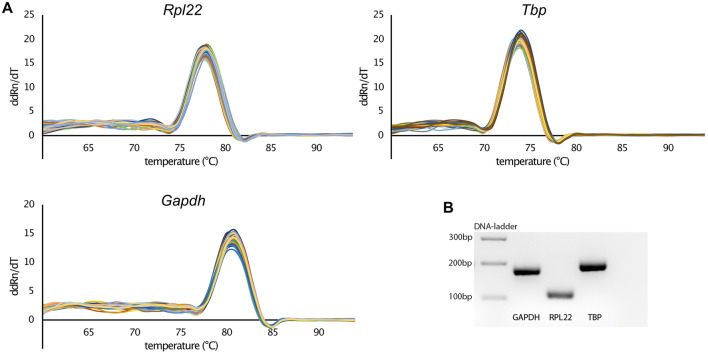
First of all, we were able to design primers for our candidate reference genes that meet our criteria to ensure adequate PCR specificity and efficiency. Primer specificity was controlled by performing melting curves and gel electrophoresis of the RT-qPCR products. All melting curves show a single peak that indicates the absence of side products and primer dimers (Fig. 1). This is confirmed by electrophoretic separation. For each gene, a single, sharply bordered fluorescent band shows up (Fig. 1, Supplementary Data 1) at the expected molecular amplicon weight (Table 1).
Figure 1.
Validation of qPCR primers. (A) Melting curve analysis for testing primer specificity. A single peak indicates a single PCR-product. (B) qPCR products of each target gene were run on a 2% agarose gel. Each gene shows a single fluorescent band at the expected amplicon size (Table 1). The gel was detected with ChemiDoc MP Imaging System (BioRad), exported as a TIFF image, inverted and cropped to show the relevant gel area. See Supplementary Data 1 for the unedited image. (bp = base pairs).
Table 1.
RT-qPCR gene, primer and target/amplicon information for the 3 investigated candidate references genes and 10 investigated target genes.
| Gene symbol | Gene name (Mus musculus) | Gene function | Accession number (NCBI Gene Bank) | Chromosoma location (length) | 5ʹ-forward primer-3ʹ (length/Tm/%GC) | 5ʹ reverse primer-3ʹ (length/Tm/%GC) | Primer location | Amplicon length (bp) | Amplicon location (bp of start/stop) | Intron-flanking (length) | Variants targeted (transcript/ splice) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Rpl22 | Ribosomal protein L22 | Translation of mRNA in protein | NM_001277113.1 | 4; 4 E2 (2153 bp) | AAGTTCACCCTGGACTGCAC (20 bp/60.18 °C/55%) | AGGTTGCCAGCTTTCCCATT (20 bp/60.18 °C/50%) | Exon 2/3 | 110 | 166/275 | Yes | Yes |
| Tbp | TATA box binding protein | General transcription factor | NM_013684.3 | 17 A2; 17 8.95 cM (1842 bp) | GACCCACCAGCAGTTCAGTAG (21 bp/60.34 °C/57.14%) | ATGTGGTCTTCCTGAATCCCTTT (23 bp/59.67 °C/43.48) | Exon 5/7 | 194 | 1003/1196 | Yes | – |
| Gapdh | Glyceraldehyde-3-phosphate dehydrogenase | Enzyme in glycolysis and gluconeo-genesis | NM_001289726 | 6 F2; 6 59.32 cM (1296 bp) | CCCCCATGTTTGTGATGGGT (20 bp/60.25 °C/55.25%) | TCTTCTGGGTGGCAGTGATG (20 bp/59.76 °C/55%) | Exon 4/5 | 177 | 469/645 | Yes | Yes |
| IL-6 | Interleukin 6 | Important role in bone metabolism; osteoclastogenesis | NM_031168.2 | 5 B1; 5 15.7 cM (1083 bp) | ACTTCACAAGTCGGAGGCTTA (21 bp/59.03 °C/47.62%) | TTTTCTGCAAGTGCATCATCGT (22 bp/59.45 °C/40.91%) | Exon 2/3 | 116 | 220/335 | YES | Yes |
| IL-1a | Interleukin 1a | Important role in bone metabolism; osteoclastogenesis | NM_010554 | 2 F1; 2 62.9 cM (1974 bp) | GCCATTGACCATCTCTCTCTGA (22 bp/59.57 °C/50%) | TGATACTGTCACCCGGCTCT (20 bp/60.32 °C/55%) | Exon 3/4 | 156 | 130/285 | Yes | Yes |
| Spp1 (OPN) | Osteopontin | Extracellular structural component of bone | NM_001204201.1 | 5 E5; 5 50.68 cM (1475 bp) | TGGACTGAGGTCAAAGTCTAGGA (23 bp/60.18 °C/47.83%) | ACAGGGATGACATCGAGGGA (20 bp/60.03 °C/55%) | - | 126 | 450/575 | – | Yes |
| Bglap (OCN) | Osteocalcin | Marker for bone formation | NM_007541 | 3 F1; 3 38.82 cM (496 bp) | GGTAGTGAACAGACTCCGGC (20 bp/61.11 °C/60%) | GGGCAGCACAGGTCCTAAAT (20 bp/60.03 °C/55%) | Exon 2/3 | 177 | 177/353 | Yes | – |
| Tnfsf11 (Rankl) | Tumor necrosis factor (ligand) superfamily, member 11 | Differentiation and activation of osteoclasts | NM_011613.3 | 14 D3; 14 41.26 cM (2243 bp) | CATTTGCACACCTCACCATCAA (22 bp/59.7 °C/45.45%) | CGTTGCTTAACGTCATGTTAGAGAT (25 bp/59.71 °C/40%) | Exon 4/5 | 120 | 642/761 | Yes | – |
| Tnfrsf11b (Opg) | Tumor necrosis factor receptor superfamily member 11B | Inactivates RANKL | NM_008764.3 | 15; 15 D1 (2818 bp) | AGACCAGGAAATGGTGAAGAAGAT (24 bp/59.71 °C/41.67%) | CAAGAAGCTGCTCTGTGGTGA (21 bp/60.54 °C/52.38%) | Exon 4/5 | 104 | 1023/1126 | Yes | – |
| Runx2 | Runt related transcription factor 2 | Associated with osteoblast differentiation | NM_001145920.2 | 17 B3; 17 21.33 cM (6475 bp) | TCTCAGTAAGAAGAGCCAGGCA (22 bp/60.82 °C/50%) | TTCGTGGGTTGGAGAAGCG (19 bp/60.3 °C/57.89%) | Exon 6/7 | 110 | 2058/2149 | Yes | Yes |
| Ptgs2 (Cox2) | Prostaglandin-endoperoxide synthase 2 | Involved in prostaglandin synthesis | NM_011198.4 | MT (non nuclear) (4460 bp) | TGAGTACCGCAAACGCTTCT (20 bp/59.97 °C/50%) | GCAGGGTACAGTTCCATGACA (21 bp/60 °C/52.38%) | Exon 9/10 | 126 | 1543/1668 | Yes | – |
| Col1a1 | Collagen, type 1, alpha 1 | Subunit of the fibril-forming type I collagen | NM_007742 | 11 59.01 cM 5946 bp | AGCATGACCGATGGATTCCC (20 bp/59.89 °C/55%) | ATTAGGCGCAGGAAGGTCAG (20 bp/59.82 °C/55%) | Exon 48/49 | 89 | 4097/4166 | Yes | – |
Tm melting temperature of primer/specific qPCR product (amplicon), %GC guanine/cytosine content, bp base pairs, MT mitochondrial.
Primer efficiency ranges between 90.2 and 100.2% after linear regression with a minimum coefficient of determination R2 = 0.9974 and between 96.1 and 106.2% in LineReg (Table2).
Table 2.
Primer efficiency obtained from a 4 step log10 dilution series (100/10/1/0,1 ng/µL) with technical duplicates in 4 (Tbp) respectively 5 (Rpl22, Gapdh) independent probes, logarithmic linearization, linear regression and finally calculated with 51.
| Linear regression | LineReg | |||||
|---|---|---|---|---|---|---|
| Primer efficiency Ep [%] (2E/100%) | SD of (2E/100%) | Slope | Coefficient of determination R2 | Amplification efficiency EL [%] (2E/100%) | SD of (2E/100%) | |
| RPL22 | 99.6 (1.995) | 0.027 | − 3.334 | 0.9981 | 106.2 (2.088) | 0.028 |
| TBP | 90.2 (1.902) | 0.046 | − 3.586 | 0.9984 | 96.1 (1.946) | 0.026 |
| GAPDH | 100.2 (2.001) | 0.051 | − 3.323 | 0.9974 | 98.2 (1.975) | 0.032 |
In addition to that, amplification graphs of each well were analyzed with LineReg41 (https://www.medischebiologie.nl/files/) to get well-specific efficiencies (n = 50).
The average expression of our investigated genes ranges from Ct = 11.48 (SD = 0.34; Gapdh) to Ct = 18.27 (SD = 0.34; Tbp) (Fig. 2).
Figure 2.
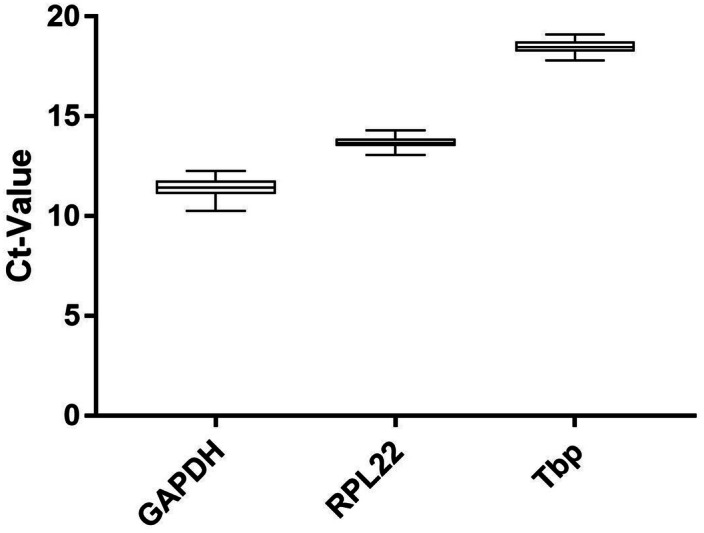
Expression levels of candidate reference genes in all experimental groups (n = 69). Ct values exported with identical threshold settings (mean of two technical replicates). Boxplots show median, interquartile range (box) and data range (whiskers).
Stability analysis of candidate reference genes through mathematical algorithms
To test the stability of reference genes we performed analysis with four different algorithms. All 4 algorithms show that Rpl22 is the least regulated reference gene under loading compression. Although Tbp is not as stable as Rpl22, it scores better than Gapdh in all calculations (Table 3).
Table 3.
Ranking of reference gene stability for OC/CM cells under compressive orthodontic force (compression vs. control) and controls with different confluences (60%, 70%, 100%).
| RefFinder | R | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total of 4 methods | delta-Ct | Bestkeeper | Normfinder | geNorm | Normfinder R | geNorm (NormqPCR) | ||||||||||
| Ranking order | Rank sum | Ranking order | Stability value | Ranking order | Stability value | SD | CV | Ranking order | Stability value | Ranking order | Stability value | Ranking order | Stability value | Ranking order | Stability value | |
| OC/CM control + static compression (n = 24) | ||||||||||||||||
| (1) | Rpl22 | 1 | Rpl22 | 0.49 | Rpl22 | 0.101 | 1.07 | 0.78 | Rpl22 | 0.086 | Tbp/Rpl22 | 0.472 | Rpl22 | 0.14 | Rpl22 | 0.395 |
| (2) | Tbp | 1.682 | Tbp | 0.577 | Tbp | 0.263 | 1.2 | 1.47 | Tbp | 0.464 | Tbp | 0.24 | Tbp | 0.495 | ||
| (3) | Gapdh | 3 | Gapdh | 0.596 | Gapdh | 0.33 | 1.26 | 3.03 | Gapdh | 0.501 | Gapdh | 0.554 | Gapdh | 0.37 | Gapdh | 0.576 |
| OC/CM controls with different confluences (n = 12) | ||||||||||||||||
| (1) | Rpl22 | 1 | Rpl22 | 0.439 | Rpl22 | 0 | 1 | 0 | Rpl22 | 0.199 | Rpl22/Gapdh | 0.41 | Rpl22 | 0.09 | Rpl22 | 0.274 |
| (2) | Gapdh | 1.682 | Gapdh | 0.482 | Gapdh | 0.21 | 1.16 | 1.9 | Gapdh | 0.359 | Tbp | 0.11 | Tbp | 0.326 | ||
| (3) | Tbp | 3 | Tbp | 0.511 | Tbp | 0.358 | 1.28 | 2.01 | Tbp | 0.423 | Tbp | 0.477 | Gapdh | 0.16 | Gapdh | 0.381 |
Stability values are calculated by using RefFinder Web-application (delta-Ct method, Bestkeeper, Normfinder, Genorm) with raw Ct values, all exported with the same threshold. In addition, Normfinder and geNorm were performed in R, where individual primer efficiency was taken into account
SD standard deviation, CV coefficient of variantion, Ct threshold cycle.
Even at different confluences it is shown that Rpl22 reaches the best stability values. In contrast to the loaded samples, Gapdh performs better than Tbp in probes without mechanical loading. Again, Bestkeeper, Normfinder, geNorm and the delta Ct method show the same results (Table 3).
The results of Normfinder and geNorm gained with RefFinder were reviewed with the original Normfinder script for the statistical program R respectively with geNorm as a part of the NormqPCR package for R. In consideration of PCR primer efficiency in R, the results of RefFinder were confirmed for probes with loading compression. In contrast, outcome for probes with different confluences differ. In this case, Rpl22 still wins the comparison, but Tbp is less stable compared to Gapdh.
Influence of different cell confluences with and without loading compression in gene expression
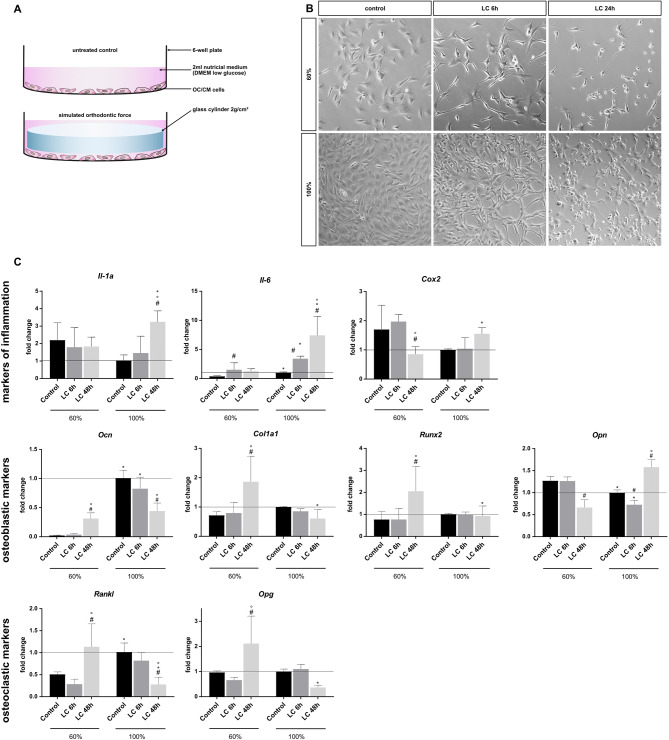
Using loaded and unloaded probes with 60% and 100% confluence, we aimed to analyse the effect of different cell confluences on gene expression of target genes know to be central regulators in periodontal remodeling. The target genes are divided in three groups: Markers of inflammation, osteoblastic markers and osteoclastic markers (Fig. 3).
Figure 3.
Influence of different cell confluences with and without loading compression in gene expression. (A) Illustration of the loading compression (LC) setup. Cell monolayer loaded with a sterile glass cylinder in 6-well plate to apply a static compression of 2 g/cm2. (B) Light microscopy photography of cells before loading (control) and before unloading (LC 6 h and LC 24 h). ×200 magnification, cropped, exposure adjustment and sharpening in Photoshop CC. (C) mRNA expression of selected markers of inflammation, osteoblastic and osteoclastic markers mean ± SD, n = 6 (two independent experiments in triplicate); normalization by ddCt method to Rpl22 and control 100% statistical analysis with GraphPad Prism 7, two-way-ANOVA, p < 0.05, #control vs. LC (loading compression); *60% vs. 100%; °LC 6 h vs. LC 48 h.
According to our data the target gene expression differs under mechanical stress (LC) and at different confluences. Interleukin 6 (Il-6), Osteocalcin (Ocn) and Receptor Activator of NF-κB Ligand (Rankl) show a significant upregulation within the uncompressed controls with increased confluence. For Osteopontin (Opn), a significant downregulation could be observed. Interleukin 1a (Il-1a) and Cyclooxygenase 2 (Cox2) show a non-significant tendency of downregulation, while the basal expression of Collagen type 1a1 (Col1a1), Run-related transcription factor 2 (Runx2) and Osteoprotegerin (Opg) remains unaltered (Fig. 3).
Cells that are mechanically compressed at a confluence of 60% show different regulatory patterns for the investigated target genes. For example Ocn, Col1a1, Rankl and Opg show a significant upregulation after a loading compression time of 48 h. Il-6 was only significantly upregulated after 6 h loading compression. There was a significant downregulation of Cox2 and Opn after 48 h LC while the gene expression of Il-1a at 60% confluence remained unaltered.
At 100% confluence, the target gene expression was changed by the loading compression in a different manner. Il-1a, Il-6, Cox2 and Opn were significantly upregulated after a LC of 48 h, Il-6 even after 6 h LC while Opn was downregulated after 6 h LC time. Ocn showed a significant downregulation after 6 h and 48 h LC time, while Col1a1, Runx22, Rankl und Opg were significantly downregulated only after a LC time of 48 h.
Some markers showed reverse tendencies under loading compression at different cell densities. Il-1a, Cox2 and Opn were downregulated when being compressed at 60% confluence while they are upregulated at 100%. Ocn, Col1a1, Rankl, Opg indicate opposite changes.
Discussion
RT-qPCR is a commonly used method to study changes in gene expression due to its capacity of relative quantification of gene expression. It is always necessary to find at least one or better multiple individually tested reference genes for each cell type through experimental condition considering the error susceptibility and sensitivity of this method35.
Experiments with cells under mechanical compression require special caution in selection of reference genes because well-tried genes such as Gapdh and ß-actin do not perform well under these conditions32,33,36.
We have decided to test three reference genes: Rpl22, Tbp and Gapdh. Gapdh is a commonly known reference gene, that has already been used in first loading compression experiments with OC/CM cells37. For this reason, we included Gapdh in our selection despite its’ well known weakness in our test conditions. Rpl22 performed best and Tbp performed very well as reference gene in loading compression experiments with periodontal ligament fibroblasts34. The selection of potential reference genes was based on the experiences with PDLF cells because of three facts: First, the spatial proximity of cementoblasts to PDLF. Second, we always use the same loading compression method. Third, PDL fibroblasts as well as cementoblasts are differentiated from the same progenitor cells38.
The self-designed primers are working well with satisfying specificity and efficiency. There are no side products detectable. Both methods for determining primer efficiency (dilution series and LineReg) show identical patterns: The Tbp primer has the lowest performance but it is still in the range of acceptable efficiency of 90–110%39.
In our analysis, Gapdh is the most regulated gene under mechanical stimulation. In contrast, Rpl22 and Tbp gene expression seems to be less affected in our experiments. In particular Rpl22 achieved the best results in all four algorithms for the use as a housekeeping gene according to the comparison of all potential reference genes. This matches with our previous publication about hPDLF34. Interestingly, the four algorithms, which show significant differences in their way of calculating stability, deliver identical results. These results were generated with RefFinder web application which offers an easy to use and fast way to analyze gene stability with four different algorithms in just one step. To obtain valid statements, these results have to be verified, because in some cases, RefFinder estimates stabilities differing from the original algorithms. These variation can be traced back to the input of raw Ct values without consideration of primer efficiencies40. For this reason, we compared the RefFinder results of Normfinder with the original Normfinder package and geNorm as part of the NormqPCR package in R. Even this control, where the primer efficiency was taken into account, shows no deviation from the previous results. This is a key argument for the reliability of our reference gene ranking for compression experiments.
Our second analysis in RefFinder with untreated cells at different confluences gives a hint of Gapdhs’ qualities as a classic reference gene. Without compression, Gapdh replaces Tbp at position 2 in the ranking. As before, Rpl22 wins the comparison in all four algorithms and proves its’ usability in experiments without compressive stimulation. We confirmed these results by the calculation in R. However, calculated with primer efficiency, the ranking of Tbp and Gapdh shows up in reverse order.
Confluence plays a crucial role in the regulation of multiple target genes in loading compression experiments of OCCM cell lines. Exemplary markers of inflammation as well as markers of bone remodeling differ significantly in their mRNA expression between 60 and 100% confluence. Especially Il-6 and Ocn show clearly shifted expression levels in the controls. This may correlate with the quantity of cell-junctions to neighboring cells, whose number changes with cell confluence (Fig. 3B). In addition, many markers (e.g. Il-1a, Rankl, Ocn) indicate inverted regulations under compression at variable confluences particularly after 48h strain, that can convert the outcome of a whole experiment. Therefore, a strict observation and documentation of cell confluence is essential for a meaningful and comparable result.
Based on our results, Rpl22 is a recommended RT-qPCR reference gene for experiments with murine cementoblasts both under static compression and in analyses without a compressive stimulus. The combination of Rpl22 and Tbp can improve the normalization, especially in loading experiments where Tbp scores clearly better than Gapdh. The use of Rpl22 as reference gene will help in future investigations to discover the role of cementoblasts in orthodontic tooth movement and their functional distinction from fibroblasts. Cell confluence is a not neglectable aspect for getting reliable results in experiments with cementoblasts. A conscientious monitoring should be a part of experimental quality assurance.
Material and methods
Cell culture
Immortalized murine osteocalcin expressing cementoblasts (OC/CM), friendly provided by Prof. Somerman24, were cultured in DMEM low glucose (1 g/L) (Gibco, USA), 10% FCS (Gibco, USA), 100 units/mL of penicillin and 100 µg/mL of streptomycin (Gibco, USA) in cell culture plates under normal cell culture conditions (37 °C, 5% CO2, water saturated). Cells were trypsinized, centrifuged at 350g, quantified using Counting Chamber “Neubauer improved” and different numbers of cells were seeded in 6-well plates.
Amplification, primer efficiency and validation
Primer specificity was controlled by 2 methods. First, a melting curve analysis was performed after RT-qPCR cycles. Second, the PCR products were analyzed by agarose gel electrophoresis. For this, RT-qPCR products (10 µL) were mixed with 4× loading dye (0.25% (w/v) bromophenol blue, 30% glycerol, 10 mM tris pH7) and loaded on a 2% agarose gel, which was prepared with GelRed Nucleic Acid Gel Stain (Biotium, USA). Amplification products were separated parallel to a 100 bp DNA ladder (Thermo Fisher Scientific, USA) at 120 V for 60 min in TAE buffer. Fluorescent bands were visualized by the ChemiDoc MP Imaging System (BioRad, USA).
Primer efficiency was determined with a 4 step log10 dilution series (100/10/1/0.1 ng/µL cDNA concentration of untreated cells). Two technical replicates per dilution level were used in 5 (Rpl22 & Tbp) respectively 4 (Gapdh) independent experiments. Standard curves were created by linear regression of the resulting Ct values with the relative cDNA dilution. Basing on the slope of each standard curve, primer efficiency was calculated with . In addition, the amplification efficiency of each well (n = 50) was checked up by LineReg41,42 (https://www.medischebiologie.nl/files/).
Compression experiments
The compression method, initially developed by Kanzaki et al.26 has already been used with cementoblasts12,13,37. According to this, cells were compressed with massive glass cylinders (2 g/cm2) in 6-well-plates for 3; 6; 12; 24 and 48 h. The loading times correspond to the time periods established in hPDLF experiments1. Before use, the glass cylinders were washed in deionized water, ethanol (70%) and finally autoclaved.
The pressure level used in the present investigation was 0.02 N/cm2 (similar to 2 g/cm2) and has been adapted from in vivo conditions in the periodontal ligament during orthodontic tooth movement in order to simulate mechanical loading in vitro with periodontal ligament fibroblasts1,43,44 and with cementoblast cells13.
Isolation and purification of RNA
For RNA-isolation cells in each well were first washed with 2 mL phosphate-buffered saline (Gibco) and then harvested with 0.5 mL TRIzol™ Reagent (Thermo Fisher Scientific, USA), two wells were pooled. This leads to biological triplicates for each condition. After isolation, according to the manufacturer’s instructions, the RNA yield of each sample was verified photometrically at 280 nm and 260 nm (Nanodrop One™, Thermo Fisher Scientific, USA). Afterwards RNA purification was performed with RNeasy Mini Kit (Qiagen, Germany) following the producers’ protocol including an on-column DNA digestion (RNase-Free DNase, Qiagen, Germany). In order to control the success of the purification and to ensure a uniform cDNA synthesis, each sample was measured again (Nanodrop One™).
Quantitative realtime-PCR analysis (RT-qPCR)
The RNA was transcribed into cDNA (SuperScript III RT, Thermo Fisher Scientific, USA). Basing upon the measurement after RNA purification, the final concentration was 25 ng/µL. All steps from RNA isolation to cDNA synthesis were performed in parallel for all samples of each experiment in order to avoid experimental variations.
RT-qPCR was performed in technical duplicates using 2.5 ng/µL cDNA in each reaction and a primer concentration of 0.5 µM. The qTower3 (Analytik Jena, Germany), High Green Mastermix (Thermo Fisher Scientific, USA), qPCR-Soft 3 (Analytik Jena, Germany) and self-designed intron spanning primers were used (Eurofins, Luxembourg). Primers were designed by using Primer-BLAST (NCBI, USA) followed by a PCR-Check (Eurofins Oligo Analyse Tool, Luxembourg) to ensure in silico qPCR specificity. Our criteria were length ca 20 bp, annealing temperature 60 °C, max product length 200 bp, intron spanning, covering possible transcript variants. The RT-qPCR protocol included an initial step of 50 °C for 2 min, 95 °C for 10 min followed by 40 cycles of 95 °C/15 s, 60 °C/30 s and 72 °C/30 s. A step of 95 °C for 15 s forms the transition to melting curve analysis (60–95 °C).
Data analysis
Statistical analysis regarding reference gene stability was performed by using RefFinder45 Web-application including the following algorithms: GeNorm46, Normfinder47, Bestkeeper48 and comparative delta-Ct method49. RefFinder analyses raw Ct-values which were exported from qPCRsoft 4.0 (Analytik Jena, Germany) with an automatic threshold. The Ct determines the number of RT-qPCR cycles required for the fluorescence to exceed a certain, defined detection threshold and shows an inverse correlation with the amount of cDNA template of the target gene sequence49. The highest automatically determined threshold was manually applied to all RT-qPCR plates across all experiments to ensure a clean separation between background noise and signal. This guaranteed that comparable Ct values could be generated for further analysis. Next, the mean value was determined from technical duplicates. These mean values from 4 independent experiments were combined into one data set for each gene.
Based on these data sets, two final data sets were generated. One contained exclusively control samples of different confluence while the other contained both loaded and control samples. Consequently, some control samples are part of both final data sets.
GeNorm performs a pairwise expression ratio under all control genes. The calculated stability value M is basing on average pairwise expression variations. As a result, genes with the lowest M value have the most stable expression46.
NormFinder was used to determine and combine expression variations of reference genes between the different groups (loaded/unloaded and 60%/100% confluence) and within each group to obtain a stability value47.
With BestKeeper we were able to determines the ‘optimal’ reference gene employing the pair-wise correlation analysis of all pairs of candidate genes. The geometric mean of the ‘best’ suited ones was calculated48.
The delta Ct method compares the relative expression of 'pairs of genes' within each sample. If the ΔCt value between the two genes remains constant when analysed in different samples, it means either both genes are stably expressed among those samples, or co-regulated (here we assume the stability of both genes). If the delta-Ct changes, one or both genes are variably expressed49. The lowest value always implies the most stable gene expression in all four algorithms. Respecting the fact that RefFinder uses algorithms differ from the original without any possibility to take into account the PCR efficiency40, we also analyzed the dataset with the original algorithms in R. For this, Ct values were brought into linear scale (RQ) by RQ = E−(minCt − sampleCt)40 (E = primer efficiency basing on dilution series; see Table 2). Finally, these RQ values were processed with Normfinder in the statistic software R and with geNorm (part of the NormqPCR package50) in R.
Statistical analysis
Data were analyzed using a two-way analysis of variance (ANOVA) with post hoc Tukey test (Prism version 8.1.0; GraphPad Software), where p < 0.05 was considered statistically significant. Graphs (Fig. 3) show mean values (n = 6) ± standard deviation (SD).
Supplementary information
Acknowledgements
The authors would like to the IZKF Unit of the Medical Faculty of the RWTH Aachen to support this study. The authors would like to thank Dr. Martha Somerman (NIH) and her lab for their kind support and providing cells.
Author contributions
C.N. and R.B.C. conceived the idea of the study and the study design. A.B. designed and C.N. and I.A. validated the used primer pairs. C.N., R. B. C., I.A., J.B. and M.W. contributed to discussion and study design. C.N. and I.A. conducted the experiments. C.K., C.N. and R.B.C. analysed the results. C.N. and I.A. wrote the manuscript and created the figures and tables. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Christian Niederau and Rogerio B. Craveiro.
Change history
2/11/2021
A Correction to this paper has been published: 10.1038/s41598-021-82274-5
Supplementary information
is available for this paper at 10.1038/s41598-020-67449-w.
References
- 1.Janjic M, Docheva D, Trickovic Janjic O, Wichelhaus A, Baumert U. In vitro weight-loaded cell models for understanding mechanodependent molecular pathways involved in orthodontic tooth movement: a systematic review. Stem Cells Int. 2018;2018:1–17. doi: 10.1155/2018/3208285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iglesias-Linares A, Hartsfield JK. Cellular and molecular pathways leading to external root resorption. J. Dent. Res. 2017;96:145–152. doi: 10.1177/0022034516677539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolf M, et al. Reduced orthodontic tooth movement in Enpp1 mutant mice with hypercementosis. J. Dent. Res. 2018;97:937–945. doi: 10.1177/0022034518759295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krishnan V, Davidovitch Z. Cellular, molecular, and tissue-level reactions to orthodontic force. Am. J. Orthod. Dentofac. Orthop. 2006;129:469.e1–469.e32. doi: 10.1016/j.ajodo.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 5.Seo BM, et al. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet. 2004;364:149–155. doi: 10.1016/S0140-6736(04)16627-0. [DOI] [PubMed] [Google Scholar]
- 6.Zweifler LE, et al. Counter-regulatory phosphatases TNAP and NPP1 temporally regulate tooth root cementogenesis. Int. J. Oral Sci. 2015;7:27–41. doi: 10.1038/ijos.2014.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hughes FJ. Periodontium and Periodontal Disease. Stem Cell Biology and Tissue Engineering in Dental Sciences. Amsterdam: Elsevier Inc.; 2015. [Google Scholar]
- 8.Nakashima M, Hayashi Y. Dental stem cells. Encycl. Biomed. Eng. 2018;1–3:554–564. [Google Scholar]
- 9.Gonçalves PF, Sallum EA, Sallum AW. Dental cementum reviewed: Development, structure, composition, regeneration and potential functions. Braz. J. Oral Sci. 2005;4:651–658. [Google Scholar]
- 10.Sawicka M, Bedini R, Wierzbicki PM, Pameijer CH. Interrupted orthodontic force results in less root resorption than continuous force in human premolars as measured by microcomputed tomography. Folia Histochem. Cytobiol. 2014;52:289–296. doi: 10.5603/FHC.a2014.0037. [DOI] [PubMed] [Google Scholar]
- 11.Owman-Moll P, Kurol J. The early reparative process of orthodontically induced root resorption in adolescents—location and type of tissue. Eur. J. Orthod. 1998;20:727–732. doi: 10.1093/ejo/20.6.727. [DOI] [PubMed] [Google Scholar]
- 12.Liu H, et al. Long noncoding RNA expression profile of mouse cementoblasts under compressive force. Angle Orthod. 2019;89:455–463. doi: 10.2319/061118-438.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang YY, et al. Cementogenesis is inhibited under a mechanical static compressive force via Piezo. Angle Orthod. 2017;87:618–624. doi: 10.2319/110616-799.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian Y, et al. Expression of alkaline phosphatase in immortalized murine cementoblasts in response to compression-force. Saudi Med. J. 2011;32:1235–1240. [PubMed] [Google Scholar]
- 15.Rego EB, et al. Effect of PGE 2 induced by compressive and tensile stresses on cementoblast differentiation in vitro. Arch. Oral Biol. 2011;56:1238–1246. doi: 10.1016/j.archoralbio.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Jin Y, et al. Functional role of mechanosensitive ion channel Piezo1 in human periodontal ligament cells. Angle Orthod. 2015;85:87–94. doi: 10.2319/123113-955.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marciniak J, et al. Orthodontic cell stress modifies proinflammatory cytokine expression in human PDL cells and induces immunomodulatory effects via TLR-4 signaling in vitro. Clin. Oral Investig. 2020;24:1411–1419. doi: 10.1007/s00784-019-03111-8. [DOI] [PubMed] [Google Scholar]
- 18.Ichioka H, et al. Biomechanical force induces the growth factor production in human periodontal ligament-derived cells. Odontology. 2016;104:27–34. doi: 10.1007/s10266-015-0206-5. [DOI] [PubMed] [Google Scholar]
- 19.Andrade I, Taddei SRA, Souza PEA. Inflammation and tooth movement: The role of cytokines, chemokines, and growth factors. Semin. Orthod. 2012;18:257–269. [Google Scholar]
- 20.Wu Y, et al. Response of immortalized murine cementoblast cells to hypoxia in vitro. Arch. Oral Biol. 2013;58:1718–1725. doi: 10.1016/j.archoralbio.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 21.Yu XJ, et al. Effect of hypoxia on the expression of RANKL/OPG in human periodontal ligament cells in vitro. Int. J. Clin. Exp. Pathol. 2015;8:12929–12935. [PMC free article] [PubMed] [Google Scholar]
- 22.Kifune T, et al. Hypoxia-induced upregulation of angiogenic factors in immortalized human periodontal ligament fibroblasts. J. Oral Sci. 2018;60:519–525. doi: 10.2334/josnusd.17-0441. [DOI] [PubMed] [Google Scholar]
- 23.Ullrich N, et al. The role of mechanotransduction versus hypoxia during simulated orthodontic compressive strain—an in vitro study of human periodontal ligament fibroblasts. Int. J. Oral Sci. 2019;11:1–10. doi: 10.1038/s41368-019-0066-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Errico JAD, et al. Employing a transgenic animal model to obtain cementoblasts in vitro. J Periodontol.. 2000;71:63–72. doi: 10.1902/jop.2000.71.1.63. [DOI] [PubMed] [Google Scholar]
- 25.MacNeil RL, et al. Isolation of murine cementoblasts: Unique cells or uniquely-positioned osteoblasts? Eur. J. Oral Sci. 1998;106:350–356. doi: 10.1111/j.1600-0722.1998.tb02197.x. [DOI] [PubMed] [Google Scholar]
- 26.Kanzaki H, Chiba M, Shimizu Y, Mitani H. Periodontal ligament cells under mechanical stress induce osteoclastogenesis by receptor activator of nuclear factor κb ligand up-regulation via prostaglandin E2 synthesis. J. Bone Miner. Res. 2002;17:210–220. doi: 10.1359/jbmr.2002.17.2.210. [DOI] [PubMed] [Google Scholar]
- 27.Adamski MG, Gumann P, Baird AE. A method for quantitative analysis of standard and high-throughput qpcr expression data based on input sample quantity. PLoS ONE. 2014;9:103917. doi: 10.1371/journal.pone.0103917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bustin SA, Nolan T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J. Biomol. Tech. 2004;15:155–166. [PMC free article] [PubMed] [Google Scholar]
- 29.Teng X, Zhang Z, He G, Yang L, Li F. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in four lepidopteran insects. J. Insect Sci. 2012;12:1–17. doi: 10.1673/031.012.6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bustin SA, et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 31.Jacob F, et al. Careful selection of reference genes is required for reliable performance of RT-qPCR in human normal and cancer cell lines. PLoS One. 2013;8:e59180. doi: 10.1371/journal.pone.0059180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yurube T, et al. Modified house-keeping gene expression in a rat tail compression loading-induced disc degeneration model. J. Orthop. Res. 2011;29:1284–1290. doi: 10.1002/jor.21406. [DOI] [PubMed] [Google Scholar]
- 33.Lee, C., Grad, S., MacLean, J., Iatridis, J. & Alini, M. Mechanical loading affects endogenous-control gene expression: articular chondrocytes in vitro and intervertebral disc in vivo. 51st Annu. Meet. Orthop. Res. Soc. Poster No: 0410 (2005).
- 34.Kirschneck C, et al. Valid gene expression normalization by RT-qPCR in studies on hPDL fibroblasts with focus on orthodontic tooth movement and periodontitis. Sci. Rep. 2017;7:1–3. doi: 10.1038/s41598-017-15281-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao Y, Gao Y, Huang B, Meng Z, Jia Y. Reference gene validation for quantification of gene expression during ovarian development of turbot (Scophthalmus maximus) Sci. Rep. 2020;10:823. doi: 10.1038/s41598-020-57633-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirschneck C, et al. Reference genes for valid gene expression studies on rat dental, periodontal and alveolar bone tissue by means of RT-qPCR with a focus on orthodontic tooth movement and periodontitis. Ann. Anat. 2016;204:93–105. doi: 10.1016/j.aanat.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 37.Diercke K, Kohl A, Lux CJ, Erber R. IL-1β und Kompression führen zu einer signifikanten Induktion der RANKL-Expression in primären humanen Zementoblasten. J. Orofac. Orthop. 2012;73:397–412. doi: 10.1007/s00056-012-0095-y. [DOI] [PubMed] [Google Scholar]
- 38.Bosshardt DD. Are cementoblasts a subpopulation of osteoblasts or a unique phenotype? J. Dent. Res. 2005;84:390–406. doi: 10.1177/154405910508400501. [DOI] [PubMed] [Google Scholar]
- 39.Taylor SC, et al. The ultimate qPCR experiment: Producing publication quality, reproducible data the first time. Trends Biotechnol. 2019;37:761–774. doi: 10.1016/j.tibtech.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 40.De Spiegelaere W, et al. Reference gene validation for RT-qPCR, a note on different available software packages. PLoS ONE. 2015;10:1–13. doi: 10.1371/journal.pone.0122515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruijter JM, et al. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009;37:45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramakers C, Ruijter JM, Lekanne Deprez RH, Moorman AFM. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 43.Kirschneck C, Proff P, Maurer M, Reicheneder C, Römer P. Kieferorthopädische Kräfte verstärken nikotininduzierten parodontalen Knochenverlust: In-vivo- und In-vitro-Studie. J. Orofac. Orthop. 2015;76:195–212. doi: 10.1007/s00056-015-0283-7. [DOI] [PubMed] [Google Scholar]
- 44.Li M, Zhang C, Yang Y. Effects of mechanical forces on osteogenesis and osteoclastogenesis in human periodontal ligament fibroblasts. Bone Joint Res. 2019;8:19–31. doi: 10.1302/2046-3758.81.BJR-2018-0060.R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie F, Xiao P, Chen D, Xu L, Zhang B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012;80:75–84. doi: 10.1007/s11103-012-9885-2. [DOI] [PubMed] [Google Scholar]
- 46.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe ASF. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012;80:75–84. doi: 10.1007/s11103-012-9885-2. [DOI] [PubMed] [Google Scholar]
- 47.Andersen CL, Jensen JL, Falck Ørntoft T. Normalization of real-Time Quantitative Reverse Transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–50. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 48.Pfaffl MW, Tichopad A, Prgomet C, Neuvians T. Determination of most stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper. Biotechnol. Lett. 2004;26:509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 49.Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006;7:1–9. doi: 10.1186/1471-2199-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perkins JR, et al. ReadqPCR and NormqPCR: R packages for the reading, quality checking and normalisation of RT-qPCR quantification cycle (Cq) data. BMC Genomics. 2012;13:296. doi: 10.1186/1471-2164-13-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Broeders S, et al. Guidelines for validation of qualitative real-time PCR methods. Trends Food Sci. Technol. 2014;37:115–126. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.