Abstract
The identification of disease hotspots is an increasingly important public health problem. While geospatial modeling offers an opportunity to predict the locations of hotspots using suitable environmental and climatological data, little attention has been paid to optimizing the design of surveys used to inform such models. Here we introduce an adaptive sampling scheme optimized to identify hotspot locations where prevalence exceeds a relevant threshold. Our approach incorporates ideas from Bayesian optimization theory to adaptively select sample batches. We present an experimental simulation study based on survey data of schistosomiasis and lymphatic filariasis across four countries. Results across all scenarios explored show that adaptive sampling produces superior results and suggest that similar performance to random sampling can be achieved with a fraction of the sample size.
Subject terms: Medical research, Epidemiology
Introduction
Recent years have seen considerable success towards control and elimination of a range of globally important infectious diseases. For many of these diseases, decisions relating to interventions are made across administrative units. For example, decisions about where to conduct mass drug administration (MDA) campaigns for neglected tropical diseases (NTDs) are made at an implementation unit (IU), typically the district or sub-district level1. A similar approach is typically taken in the control and elimination of malaria, where entire districts or sub-districts may receive insecticide treated nets or indoor residual spraying where others do not.
For NTDs, decisions relating to MDA are based on infection prevalence estimates at the IU level obtained from cross sectional surveys. Where IU level prevalence exceeds a threshold, the entire IU is treated1. Where prevalence does not exceed this threshold, the IU does not qualify for MDA and no individuals in that area are treated. For example, for schistosomiasis, current guidelines recommend that MDA is conducted in areas where prevalence is greater than 10%, whereas for soil-transmitted helminths, this threshold is 20%1.
While operationally straightforward, this approach ignores any within IU heterogeneity. In many instances, districts with prevalence below the threshold that triggers intervention contain a number of villages with active transmission2. Modeling and intuition therefore suggest that as disease transmission declines, moving away from decision making at coarse scales towards a more targeted approach is more cost-effective3. Such targeting is predicated on sufficiently accurate information on the location of sites with an infection prevalence above a policy relevant threshold, from hereon referred to as hotspots.
Missing hotspots could cause setbacks for elimination efforts. Hence, various approaches to identify them have been proposed. Variations of contact tracing, whereby testing is targeted at families and neighbours of individuals found positive during surveys or routine surveillance, have been explored for a number of diseases including schistosomiasis4, lymphatic filariasis5 and malaria6,7. Such approaches can, however, be expensive and can still fail to identify hotspots if positive individuals from those communities are not identified by the initial surveys.
An alternative approach is to use less costly survey methods to sample a higher proportion of locations than would otherwise be possible. Techniques such as lot quality assurance sampling, a method designed to minimize sampling effort in order to categorize outcomes over a given population, is one such approach and has been used to identify hotspot communities for schistosomiasis8,9. Similarly, school-based questionnaires relating to blood in urine and eye worm occurrence, have been used to map urinary schistosomiasis10–12 and loiasis13,14 respectively. These methods are inherently noisy as they only allow measurement of proxies of infection and can suffer from issues of recall.
Another approach to mapping hotspots, which reduces the need to sample a large fraction of the population, is using geospatial modeling. Climatological, environmental and ecological layers can help predict the spatial distribution of many infectious diseases. Furthermore, above and beyond patterns that can be explained by these layers alone, disease outcomes often display some spatial structure, with neighbouring values being correlated due to shared characteristics and transmission. This spatial structure means that information from one site provides information about neighbouring sites. Over the past decade, the ability to predict pathogen infection prevalence across entire regions based on survey data and relationships using geospatial modeling has improved considerably15–17. These advances in geospatial modeling have opened the door to more targeted approaches, potentially allowing decisions about treatment to be made with higher precision and granularity.
Despite these advances, surprisingly little attention has been paid to optimizing the survey design for risk mapping efforts. Evidence from other fields has shown that random sampling is suboptimal for spatial prediction18–21. For lymphatic filariasis, a grid sampling approach has been proposed as a mechanism to allow for more efficient spatial interpolation22,23. Diggle and Lophaven24 propose the use of grid sampling supplemented with clusters of close pairs of points which is useful when estimates of Kriging (covariance) parameters are required24. Simulation studies also suggest that this design provides a cost-effective approach to mapping schistosomiasis3. Similarly, Fronterre et al.25 show that spatially regulated surveys, in combination with spatial modeling, can reduce the sample size required to estimate IU level prevalence.
Recent studies by Chipeta et al.26 and Kabaghe et al.27 propose the use of spatially adaptive designs that leverage information from prior data to inform the locations of future sampling sites to minimize prediction error. Using malaria as an example, results from simulations and field studies show that adaptive spatial designs can be used to produce more precise predictions of infection prevalence using geostatistical modeling26.
Building on the adaptive spatial sampling approach, we incorporate ideas from Bayesian optimization theory28,29 to propose an adaptive spatial sampling approach optimized to identify hotspot communities.
Results
We compared the performance of two approaches for selecting survey sites: random sampling (RS), where sites are chosen randomly; and adaptive sampling (AS), that follows the acquisition function of Eq. (6). The underlying statistical model is the same in both cases (see Eqs. (1)–(2)). The initial dataset is also the same in both cases (see Table 2 lines 7 and 8). Hence, the variations in the performance with respect to the predictions based on depend only on the mechanism of selecting the new survey locations . Adding measurements at new locations improved out-of-sample sites classification under both sampling approaches. However, across the four scenarios tested we observed that adaptive sampling was consistently superior to random sampling in terms of accuracy, positive predictive value and sensitivity.
Table 2.
Experimental procedure.
| Pseudo code for experiments | ||
|---|---|---|
| 1 | for rep in : | |
| 2 | for m in : | m = batch size |
| 3 | random selection: with | all villages |
| 4 | random/adaptive | |
| 5 | + 1 | total number of iterations |
| 6 | for t in | |
| 7 | ||
| 8 | =environmental data | |
| 9 | find | |
| 10 | compute validation statistics on | |
| 11 | random selection | |
| 12 | acquisition function Eq. (6) | |
We repeated each experiment a hundred times (line 1), for batches of size 1, 10 and 50 (line 2). We started with an initial random sample of 100 locations (line 3) for both random and adaptive methods (line 4). We incorporated subsequent samples until 100 additional sampling locations were added (line 5). For the locations selected to be sampled we simulated the observed positive cases according to a Binomial distribution with prevalence (line 7) and incorporated the environmental data (line 8). We then used the accumulated data to find the probability of exceeding the threshold (line 9). Finally we defined a new batch of locations according to a random mechanism (line 11) and to the adaptive sampling method proposed (line 12).
for step .
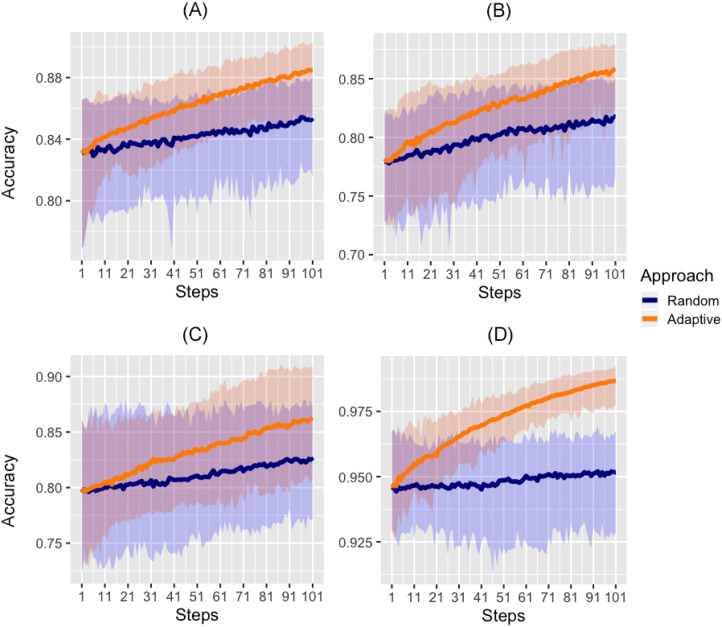
This confirms that under adaptive sampling each new batch of locations leads to a better classification of the unmeasured sites. Figure 1 shows the accuracy computed at each step in the four country scenarios using a batch of size 1. Note that when selecting a batch of size 1, the adaptive design does not take into account the exploration component. In this case the new location suggested is the one that maximizes entropy.
Figure 1.
Out of sample accuracy (batch size = 1). The solid line represents the average value across 50 repetitions. The shaded area represents the 2.5% and 97.5% quantiles of the values observed across all 50 repetitions at each step. Note that step 1 here refers to the initial random sample of 100 sites. (A) Côte d’Ivoire (). (B) Malawi (). (C) Haiti (). (D) Philippines ().
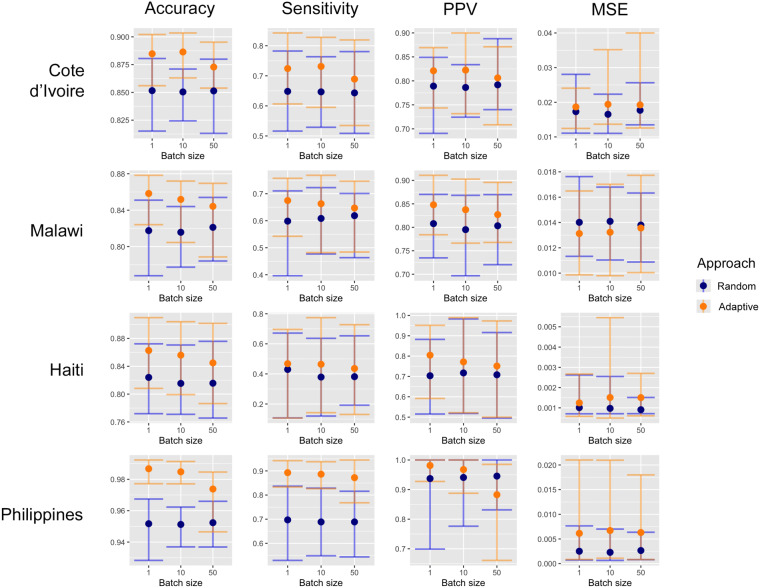
Figure 2 shows a summary of the validation statistics after adding 100 new samples, using different batch sizes (1, 10 and 50), across the four scenarios. The results show that adaptive sampling produces superior accuracy, sensitivity and PPV across every scenario, metric and batch size except in the Philippines where an adaptive approach with a batch size of 50 produced inferior PPV. Better performance across all metrics translates into a smaller number of false positives and a improved identification of hotspots in locations that have not been visited yet. In contrast to the validation statistics discussed above, MSE (bottom row) is lower across all scenarios when random sampling was employed, except in Malawi where adaptive sampling produced lower MSE.
Figure 2.
Summary of validation statistics. Metrics computed after adding 100 new samples in batches of 1, 10 and 50 sites. Dots represent the mean and whiskers represent the the 2.5% and 97.5% quantiles of values observed across all 50 repetitions. The thresholds used to define a hotspot are: in Côte d’Ivoire and Malawi and in Haiti and Philippines.
At larger batch sizes there were smaller differences between random and adaptive sampling in terms of accuracy, PPV and sensitivity (Fig. 2). There are two ways of interpreting this result. One interpretation is that when the batch is large enough, random sampling provides a good coverage of the sampling universe negating the need for a trade-off between exploitation and exploration. The more locations in the batch the more redundant the information they provide, regardless of how they are chosen. A second interpretation is that the adaptive sampling design is more efficient and therefore requires smaller sample sizes to achieve the same results of a larger random sample. Table 1 illustrates this and shows the number of sample points needed when using adaptive sampling to achieve the same accuracy of random sampling with a sample size of 100 locations. For batches of size 50, adaptive sampling produced at least the same level of accuracy with just half the number of additional points across all scenarios. This difference becomes larger for smaller batch sizes (1 or 10). For batch sizes of 10, adaptive sampling required 10–40% of the sample size to achieve the level of accuracy achieved with 100 additional randomly selected sites and for batch sizes of 1, only 7–36% was required. With such sample sizes the adaptive sampling also achieved similar levels of sensitivity and PPV but higher MSE.
Table 1.
For random design RS with sample size of 100, we show the sample size needed to achieve a similar accuracy using an adaptive design AS.
| Country | Num. obsv. | Accuracy (%) | PPV (%) | Sensitivity (%) | MSE () | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| RS | AS | RS | AS | RS | AS | RS | AS | RS | AS | ||
| Côte d’Ivoire | 1 | 100 | 27 | 85.2 | 85.3 | 64.9 | 78.6 | 64.8 | 65.1 | 17.3 | 20.5 |
| 10 | 100 | 30 | 85.0 | 85.3 | 78.6 | 78.7 | 64.7 | 65.2 | 16.5 | 20.4 | |
| 50 | 100 | 50 | 85.1 | 85.5 | 79.2 | 78.6 | 64.3 | 65.5 | 17.7 | 20.0 | |
| Malawi | 1 | 100 | 36 | 81.8 | 81.9 | 80.8 | 80.6 | 59.9 | 59.4 | 14.0 | 14.9 |
| 10 | 100 | 40 | 81.6 | 82.0 | 79.5 | 80.5 | 60.8 | 59.7 | 14.1 | 15.0 | |
| 50 | 100 | 50 | 82.1 | 82.1 | 80.3 | 80.3 | 61.9 | 60.4 | 13.8 | 14.5 | |
| Haiti | 1 | 100 | 31 | 82.4 | 82.6 | 70.3 | 75.7 | 43.0 | 35.0 | 1.0 | 1.4 |
| 10 | 100 | 30 | 81.5 | 81.6 | 71.7 | 71.0 | 38.9 | 36.4 | 1.0 | 1.5 | |
| 50 | 100 | 50 | 81.5 | 82.3 | 70.8 | 70.5 | 38.1 | 39.8 | 0.9 | 1.5 | |
| Philippines | 1 | 100 | 7 | 95.2 | 95.2 | 93.7 | 94.4 | 69.7 | 67.7 | 2.5 | 4.3 |
| 10 | 100 | 10 | 95.1 | 95.2 | 94.1 | 93.6 | 68.9 | 68.6 | 2.3 | 5.1 | |
| 50 | 100 | 50 | 95.2 | 95.6 | 94.6 | 85.0 | 68.9 | 79.8 | 2.7 | 5.5 | |
Additional validation statistics: PPV, sensitivity and MSE are also shown. Along the rows, results are shown per country and batch size .
Discussion
The identification of disease hotspots is an increasingly important public health problem. This is particularly true in disease elimination settings, where transmission is rare and typically focal. Numerous examples illustrate the use of geospatial modeling to predict hotspots, but very little attention has been given to the optimal survey design for such modeling efforts. Here, using simulation studies based on schistosomiasis and lymphatic filariasis survey data, we described a novel, spatially adaptive approach and demonstrate the superiority of this approach at identifying hotspots compared with the standard approach to surveys based on purely random sampling.
Results showed that across all batch sizes investigated, adaptive approaches produced higher levels of accuracy, sensitivity, and PPV compared with random sampling. Yet, the superiority of an adaptive approach declined with larger batch sizes. With a batch size of 1, the adaptive approach has an opportunity to identify the optimal next location to survey in the presence of all available data. In contrast, with larger batch sizes, the impact on predictions of each adaptively sampled location is not known until all locations in the batch are sampled and the model updates.
The use of an adaptive approach only produced marginal gains in accuracy (2–4%) after adding 100 sites to the initial sample, but this could represent hundreds of locations when applied at a country scale. Perhaps more importantly, however, adaptive sampling was more efficient in terms of achieving a given level of accuracy with a far smaller sample size. As outlined in Table 1, across all scenarios explored, adaptive sampling was able to achieve the same level of accuracy and sensitivity to that achieved by adding 100 locations randomly with between 7 and 50% the sample size. These results demonstrate that an adaptive spatial sampling approach has the potential to substantially reduce the resources required to ensure hotspot locations receive treatment, while maintaining similar rates of false positives. In control and elimination settings, an operationalized adaptive spatial sampling approach for several years could render non-negligible improvements in cost-effectiveness. Further simulation studies could be used to help determine the magnitude of such benefits in cost-effectiveness.
It should also be pointed out that in almost all settings the mean squared error estimates were higher for adaptive approach (Fig. 2, Table 1). This illustrates the fact that optimizing a design for one goal, here hotspot classification accuracy, leads to compromising other goals (e.g. precision in the prevalence estimates). Where the goal is to produce the most precise prevalence estimates at any given location, using adaptive approaches based on prediction variance as opposed to entropy would be more appropriate26.
While this approach was demonstrated for two diseases only, it could be used to support the identification of hotspots of any binomial outcome. This includes prevalence of infection of other infectious and non-infectious diseases, particularly those that display strong spatial correlation. Vector-borne diseases, such as malaria, onchocerciasis and loiasis would certainly fall into this category given the association between disease transmission and ecological and environmental conditions. While it is likely that such spatial correlation will be masked following several years of intervention, evidence suggests that residual hotspots still occur2,30. In addition to identifying hotspots of infection, this approach also has potential utility for identifying cold spots in intervention coverage, such as pockets of undervaccination31. While this would likely require use of different covariates related to intervention access, such as distance to roads, population density and poverty, the statistical problem is analogous.
In principle, in the context of schistosomiasis and lymphatic filariasis, including additional relevant covariates could improve the spatial model predictions. Among others, these could include information on intervention coverage, population density, poverty, housing type and soil type. For the purpose of this simulation study, we opted to use WorldClim data as the focus was on the marginal improvement of using an adaptive sampling approach over a random sampling, as opposed to identifying the optimal model and covariates with which to predict infection.
While we used a combination of random forest and model-based geostatistics to produce posterior prevalence estimates, the general adaptive sampling scheme we have proposed would work for any suitable modeling approach that produces posterior estimates with which to estimate exceedance probabilities. Combining random forests with other base learners such as generalized additive models and support vector machines may lead to improvements over using random forest alone. Such a ‘super learner’ approach to ensemble modeling, based on minimizing cross-validation error, may help to address any issues of model misspecification which if not properly addressed could lead the adaptive design to become overly confident about choice of sampling location. Super learning has been used across a range of other statistical problems including causal inference and prediction32–34. Furthermore, the super learner approach can be extended to be ‘online’ whereby models are updated rather than refit from scratch, yielding computational benefits35.
Similarly, an underlying binomial model is not essential to the methodology described here. What is important is the spatial correlation component in which the exploration rule is based. For example, this methodology could work in a Poisson setting, for some definition of hotspot based on a threshold incidence or numbers of cases.
A further potential extension of this work would be to incorporate covariates into the adaptive sampling algorithm. The approach outlined here attempts to reduce redundancy when selecting sampling locations by prioritizing uncertainty across geographic space. However, the uncertainty of the model does not only depend on the geographic space encoded through the Matérn kernel. Uncertainty is also dependent on the remaining features (covariate values). In this application, a good spread of points in geographic space is likely to achieve a good spread of points in the remaining features, as their values are determined by location. In applications where the covariates are less influenced by geography it may be necessary to optimize the design for the uncertainty in the whole feature space. Not doing so may lead to a design that is no better than random. The most straightforward way to implement this extension would be to model the covariates with another kernel (Matérn, linear or any other considered adequate). In such a model the term , in Eq. (2), would be substituted by another Gaussian process that depends on the new kernel. Equation (4) would remain the same, but this time K would in fact be the sum of two kernels: the Matérn that models geographic space and the new kernel that models the remaining features.
Another possible extension of this methodology is applying it to cases where the classification of interest is not binary. For example, for schistosomiasis, MDA is recommended once per year in areas where prevalence is and and twice per year in areas where prevalence is 1. As estimation of entropy is not restricted to binary classification problems, adapting the approach to such a setting is straightforward assuming it is possible to produce probabilistic classifications from the underlying model.
This study had a number of limitations. Firstly, the adaptive sampling approach described requires a georeferenced set of candidate sampling locations. Complete georeferenced lists of settlements are, however, often not available. In the absence of such data, there are several options available. Georeferenced locations could be extracted and compiled from open sources, such as OpenStreetMap, Geonames and openAFRICA. Alternatively, village locations can be derived from gridded population data using the approach described here (see Supplementary Information) or using alternative approaches as suggested by Thomson et al.36.
A second limitation is that we did not consider the temporal aspect of adaptive surveys. In reality, there may be a time lag between the date at which survey data are available and when adaptive surveys take place. Similarly, prior survey data may have been collected over multiple time periods. To address this issue it would be possible to extend the spatial model used, to a spatio-temporal model. Hotspot probabilities could then be forecast from the historic data to the time point at which adaptive surveys are to take place. Additionally, there may be value in using temporally dynamic covariates as opposed to static, long-term averages as used here.
A third limitation was that we defined a site as a hotspot if there was at least a 50% chance that prevalence exceeded the relevant threshold. In some cases, programs may a priori wish to define hotspots more conservatively by classifying sites as hotspots with smaller probabilities (e.g. chance a site is a hotspot). While the methodology would not change, such an approach would have a large impact on the performance of the classifications, increasing sensitivity, but decreasing positive predictive value. In such cases, it may also be useful to modify the acquisition function.
A fourth limitation of this study is that we used a single acquisition function. In the acquisition function we used, the exploration component has an increasing concave weight as more locations are added to the new batch. This assumption, or the specific shape of this weight, could be substituted for an alternative. Also, the utility function, defined here as entropy, could be modified depending on the goal pursued. For example, a program interested in targeting sampling efforts at hotspots, instead of achieving a better binary classification, could use the probability of a location being a hotspot as the utility function. Such an approach would be suitable for situations where testing is required before an intervention/treatment is administered. This approach may also be useful for surveys whose goal is to determine freedom from infection37,38.
A fifth limitation stems from the simulated nature of the experiments. The strength of a simulated approach is that multiple experiments can be conducted without the need for expensive field validation studies. On the basis of these results, a valuable next step would be to conduct field studies comparing random to adaptive designs. Such studies would also allow an exploration of some of the more logistical elements and constraints and using an adaptive approach.
This study has demonstrated the value in adopting an adaptive approach to surveys designed to identify disease hotspots. Results show that a spatially adaptive sampling approach produced consistently superior accuracy in hotspot classification over a random sampling approach, and could dramatically lower the resources requirements to conduct surveys whose goal is to detect disease hotspots.
Methods
Spatial model
To predict the probability that a given site (e.g. a village or other type of settlement) is a hotspot or not, and to guide adaptive sampling schemes, requires fitting a spatial model to observed data. As a reminder, here a hotspot is defined as a location where infection prevalence is greater than a defined threshold. We assume that an initial representative population sample exists to allow a model to be fit. If this is not the case, a randomly sampled set of measurements would be one option, although there may be superior approaches, particularly if data relating to the expected spatial structure or covariate values at candidate survey sites exist24,39,40.
There are a range of different modeling approaches available to predict prevalence at unsurveyed sites. Here, we use a combination of machine learning and model-based geostatistics15,41.
Let be a region (e.g. a country) where we are interested in determining if a set of sites are hotspots or not. As mentioned above, it is assumed that an initial dataset from which we can estimate the overall prevalence exists. Say we have the dataset , where are the GPS coordinates that describe the location of a site of interest, is the number of people tested in such site, are the number of positive cases out of and are other features associated to the site, like elevation, distance to water bodies or average temperature; is the total number of observations. Given these data we can model the prevalence in as a spatially continuous process given by
| 1 |
| 2 |
where are a set of real parameters and f is a spatially correlated random effect using a Matérn correlation function (see Supplementary Information, Eq. (7)) and is a residual independent error term.
Instead of including linear covariate effects, we first fit a random forest model using 20-fold cross validation using all the covariates, excluding latitude and longitude. For each observation, we then have a cross-validated prevalence prediction (from hereon termed out-of-sample predictions). Additionally, we fit a random forest using all observations and use this model to predict to all observation and prediction points (from hereon termed in-sample predictions). Out-of-sample predictions from the random forest are then included as a single covariate in the geostatistical model described by Eq. (2).
When making predictions, in-sample predicted prevalence values from the random forest using all observations were used as the covariate at each prediction point. While this model allows us to predict prevalence across the continuous region , in this case we are only interested in predictions at the location of human settlements. Here, we denote these discrete locations as .
In addition to obtaining estimates of predicted prevalence, the model described above allows us to simulate a posterior distribution of prevalence values at each cluster which can be used to estimate the exceedance probabilities, i.e. the probability that prevalence at location is above a given threshold .
Adaptive sampling
Exploitation
The goal we seek when using adaptive sampling or adaptive design is to leverage the information available and select the optimal sampling locations to improve our statistical inference28,29. The criteria to define what is optimal depends on what quantity is to be estimated. Hence, it is first necessary to define an objective or utility function, i.e. the measure by which we evaluate the performance of any given design. For situations where the goal is to produce as precise predictions as possible over the study region, measures such as average prediction variance is a sensible option26. If, however, the goal is to find hotspots, we are less interested in the precision of our estimates and should be focused on minimizing hotspot classification error from our model. Put another way, we wish to increase our confidence that the prevalence at any given location is above or below the predefined threshold. A measure that fits naturally into this framework is Shannon entropy. Shannon entropy measures the uncertainty of a random variable based on its probability distribution42. Let be the relevant threshold. Given the model described in Eq. (1), for every we can estimate its probability of being a hotspot . Then the entropy value at such location regarding it being a hotspot or not is defined as
| 3 |
Locations with exceedance probabilities of 0.5 (i.e. ) are the most uncertain and have an entropy value of one. On the contrary, the more certainty in the event (i.e. exceedance probabilities close to 0 or 1), the entropy gets closer to 0. By targeting high entropy values, sampling is focused on those sites with highest classification (hotspot or not) uncertainty.
Exploration
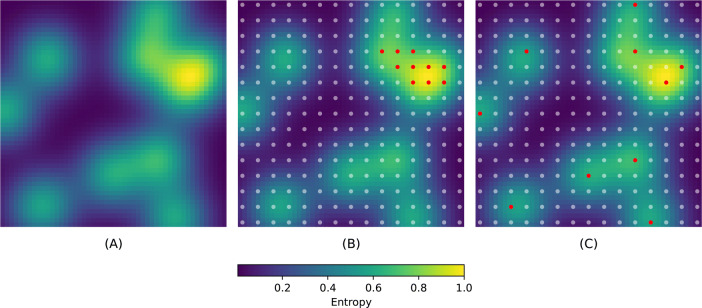
Giving preference to locations with higher uncertainty is intuitively more efficient than a uniform random selection, but choosing the design based only on entropy values (Eq. (3)) may not be efficient because prevalence is usually a spatially correlated process. For example, see Fig. 3 panel A, where we show a simulated field of uncertainty where values are spatially correlated. Since locations with high uncertainty can be expected to be clustered together, by defining a batch of sample points based only on we may end up selecting locations that are very close to each other. However, such an approach leads to redundancy, as taking a measurement at one location also provides information about neighboring locations due to the spatial correlation present. In Fig. 3 panel B, we choose the 10 locations (red dots) with highest uncertainty values from a grid of potential locations (white dots). The Figure demonstrates how this greedy approach can result in poor coverage of the field.
Figure 3.
Exploration-exploitation trade-off. (A) Spatially correlated uncertainty. (B) Batch selected (red dots) by using the greedy approach of targeting the highest values of uncertainty. (C) Batch of locations selected (red dots) using the acquisition function described in Eq. (6).
It would be preferable to sample high entropy points, while ensuring a good spread of points across the study area to avoid redundancy. This allows a balance between exploitation (i.e. targeting high values of ) and exploration (i.e. spread batch locations in )43. If in Eq. (2) we assume that f is a multivariate Gaussian with spatial covariance , then the average amount of information contained in a batch of locations is given by the joint differential entropy
| 4 |
where and .
The differential entropy is the continuous case of the Shannon entropy introduced before42. A low value of implies that the random variable is confined to a small volume, whereas a large value of the differential entropy implies a that the variable is widely dispersed. Given a batch size, by choosing the elements in it that maximize the differential entropy, we would be maximizing the average information content of the batch with respect to the random field f. Finding the batch with highest information content is a problem of combinatorial complexity. However an exact solution is not needed44. A approximate solution can be found through a sequential approach that at step t selects the new element of the batch according to
| 5 |
Trade-off
Once we have a utility function and a rule for exploration, we only need to define a trade-off strategy between exploration and exploitation that helps us select a batch of new survey locations. In Bayesian optimization, this strategy is defined by the acquisition function45,46. Notice, however, that our setting is simpler than the usual setting for Bayesian optimization, where evaluating the utility function is considered to be expensive and the exploration sites could be infinite. In this application we assume a finite set of potential survey locations, as they represent villages or some type of human settlements. Also, in all of these locations we have a measurement of our utility function through the posterior distribution of .
As trade-off strategy we define the step-wise algorithm that combines Eqs. (3) and (5), so that at step t the new element in the batch is chosen according to
| 6 |
In the expression above we are explicitly defining y as a function of to emphasize that we are interested in selecting survey locations. By using this acquisition function we induce batch locations to be spatially scattered and therefore achieve a better exploration. In Fig. 3 panel C, we show a batch of 10 locations (red dots) chosen according to Eq. (6). The locations selected are not the ones with the overall highest uncertainty, but the ones with the highest uncertainty within a neighborhood. This approach allows targeting high entropy values, while reducing information redundancy and exploring the region of interest.
The acquisition function in Eq. (6) is based on the Gaussian process upper confidence bound (GP-UCB) algorithm44. The GP-UBC is used in Bayesian optimization problems with an underlying Gaussian processes regression of the form . The difference between our formulation in Eq. (6) and the original GP-UCB is that the latter uses the mutual information between the observations and the process f42, as opposed to the joint differential entropy of only. The mutual information between and f is theoretically a better approach. However, the assumption of Binomial outcomes that depend on a transformation of f, makes this quantity harder to compute. On the other hand, the use of differential entropy showed satisfactory results in our simulation studies, as shown below.
Experimental simulation
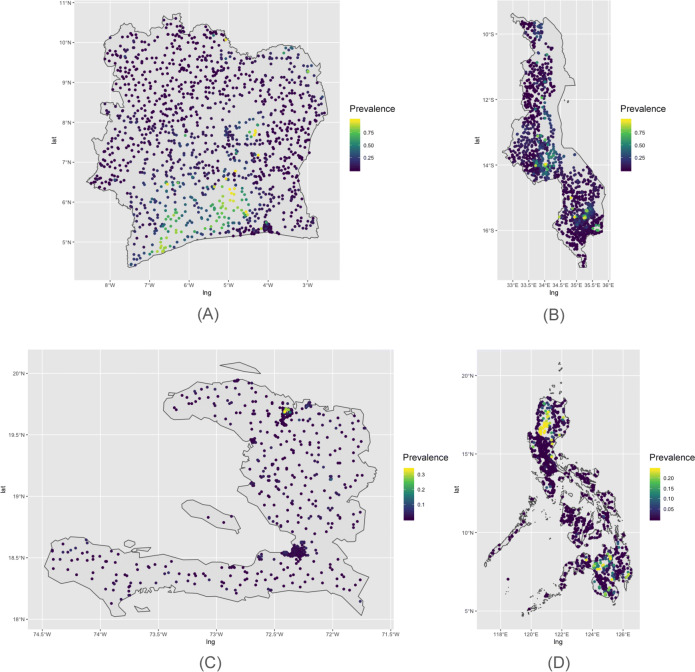
To test the proposed adaptive spatial sampling approach, we conducted a series of experimental simulation studies parameterized using data from NTD surveys across multiple diseases and countries. We created different scenarios in which the task was to adaptively select new sampling locations with the goal of classifying sites as hotspots and not hotspots. In this procedure, our benchmark was the prediction performance when selecting batches of sampling sites randomly without adaptation. We defined four prevalence scenarios based on cross-sectional prevalence survey data of schistosomiasis from Côte d’Ivoire and Malawi and lymphatic filariasis from Haiti and Philippines (see Supplementary Information). In each of the four countries, we used a universe of up to 1500 candidate survey sites identified with the Village Finder algorithm (see Supplementary Information). This algorithm uses gridded population estimates of 2015 from Worldpop to identify clusters of populated places47. For computational reasons, if this resulted in more than 1500 clusters for any given country, we randomly sampled 1500 to obtain a final set of candidate locations. Figure 4 shows the cluster locations in each country and the simulated prevalence used as the truth during these experiments.
Figure 4.
Simulated prevalence scenarios. The locations of the villages is marked by the dots, whose colors represent the hypothetical prevalence of each scenario. (A) Côte d’Ivoire (schistosomiasis). (B) Malawi (schistosomiasis). (C) Haiti (lymphatic filariasis). (D) Philippines (lymphatic filariasis).
In order to have consistency in our results, we repeated our experiments 50 times per country. In each replicate, we randomly selected 100 locations from the universe of clusters and used them as the locations of the initial set . We ran three versions of the experiments, by sequentially selecting batches of size 1, 10 and 50, until we had incorporated 100 new samples. Given a set of initial sampling locations and batch size, we sampled additional locations either completely at random or adaptively following Eq. (6). At each step we fitted the model described in Eqs. (1) and (2). As environmental variables we used: annual mean temperature, temperature seasonality, annual precipitation and precipitation seasonality48, elevation (SRTM) and distance to inland water resampled to the same 1km resolution. After fitting the spatial model on each iteration, we computed four out-of-sample validation statistics to measure performance (see Supplementary Information): accuracy, positive predictive value (PPV), sensitivity and mean squared error (MSE). To compute the validation statistics we fitted the model in Eq. (1) to all the available data at each iteration (i.e. at step t) and made predictions on the villages that had not been visited yet (i.e. at step t). MSE was computed comparing the predicted prevalence vs the simulated prevalence. To compute accuracy, positive predictive value and sensitivity we first classified the villages as hotspots when and compared this classification vs the actual class according to the simulated prevalence. Table 2 shows the algorithm followed to carry on our experiments.
Random forest and geostatistical models were fit using the R packages ranger 0.11.249 and spaMM 3.0.050 respectively. All the simulated datasets and code developed as part of this study, including that used to conduct the simulation experiments, is available at https://github.com/disarm-platform/adaptive_sampling_simulation_r_functions. Internal Review Board approval was granted from the University of California, San Francisco (IRB Number: 18-25235).
We are also in the process of developing a user-friendly web application to allow both the hotspot mapping and adaptive sampling algorithms to be run without code.
Supplementary information
Acknowledgements
This work received financial support from the Coalition for Operational Research on Neglected Tropical Diseases (Grant No. 141G), which is funded at The Task Force for Global Health primarily by the Bill & Melinda Gates Foundation, by the United States Agency for International Development through its Neglected Tropical Diseases Program, and with UK aid from the British people.
Author contributions
H.J.W.S., R.A.P. and B.F.A. conceptualized the study. Formal analyses were performed by R.A.P, F.R. and H.J.W.S., R.A.P. and H.J.W.S. led the writing of the manuscript. J.L., L.H., M.A. and L.J. oversaw collection of the survey data and A.F.B. and M.J.L. provided input on the statistical methodology and reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41598-020-67666-3) contains supplementary material, which is available to authorized users.
References
- 1.World Health Organization. Preventive Chemotherapy in Human Helminthiasis. Coordinated Use of Anthelmintihic Drugs in Control Interventions: A Manual for Health Professionals and Programme Managers. (WHO Press, Geneva, 2006).
- 2.Rao RU, et al. Reassessment of areas with persistent lymphatic filariasis nine years after cessation of mass drug administration in Sri Lanka. PLoS Neglect. Trop. Dis. 2017;11:e0006066. doi: 10.1371/journal.pntd.0006066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sturrock HJW, et al. Planning schistosomiasis control: investigation of alternative sampling strategies for Schistosoma mansoni to target mass drug administration of praziquantel in East Africa. Int. Health. 2011;3:165–175. doi: 10.1016/j.inhe.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Massara CL, et al. Evaluation of an improved approach using residences of schistosomiasis-positive school children to identify carriers in an area of low endemicity. Am. J. Trop. Med. Hyg. 2006;74:495–499. [PubMed] [Google Scholar]
- 5.Harris JR, Wiegand RE. Detecting infection hotspots: modeling the surveillance challenge for elimination of lymphatic filariasis. PLoS Neglect. Trop. Dis. 2017;11:e0005610. doi: 10.1371/journal.pntd.0005610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sturrock H, et al. Reactive case detection for malaria elimination: real-life experience from an ongoing program in Swaziland. PLoS ONE. 2013;8:e63830. doi: 10.1371/journal.pone.0063830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sturrock H, et al. Targeting asymptomatic malaria infections: active surveillance in control and elimination. PLoS Med. 2013;10:e1001467. doi: 10.1371/journal.pmed.1001467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooker S, Kabatereine NB, Myatt M, Stothard RJ, Fenwick A. Rapid assessment of Schistosoma mansoni: the validity, applicability and cost-effectiveness of the lot quality assurance sampling method in Uganda. Trop. Med. Int. Health. 2005;10:647–658. doi: 10.1111/j.1365-3156.2005.01446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rabarijaona LP, et al. Lot quality assurance sampling for screening communities hyperendemic for Schistosoma mansoni. Trop. Med. Int. Health. 2003;8:322–328. doi: 10.1046/j.1365-3156.2003.01019.x. [DOI] [PubMed] [Google Scholar]
- 10.Clements AC, Brooker S, Nyandindi U, Fenwick A, Blair L. Bayesian spatial analysis of a national urinary schistosomiasis questionnaire to assist geographic targeting of schistosomiasis control in Tanzania, East Africa. Int. J. Parasitol. 2008;38:401–15. doi: 10.1016/j.ijpara.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lengeler C, Utzinger J, Tanner M. Questionnaires for rapid screening of schistosomiasis in sub-Saharan Africa. Bull. World Health Organ. 2002;80:235–242. [PMC free article] [PubMed] [Google Scholar]
- 12.Sturrock HJW, Pullan RL, Kihara JH, Mwandawiro C, Brooker SJ. The use of bivariate spatial modeling of questionnaire and parasitology data to predict the distribution of Schistosoma haematobium in Coastal Kenya. PLoS Neglect. Trop. Dis. 2013;7:e2016. doi: 10.1371/journal.pntd.0002016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takougang I, et al. Rapid assessment method for prevalence and intensity of Loa loa infection. Bull. World Health Organ. 2002;80:852–858. [PMC free article] [PubMed] [Google Scholar]
- 14.Diggle PJ, et al. Spatial modelling and the prediction of Loa loa risk: decision making under uncertainty. Ann. Trop. Med. Parasitol. 2007;101:499–509. doi: 10.1179/136485907X229121. [DOI] [PubMed] [Google Scholar]
- 15.Bhatt S, et al. Improved prediction accuracy for disease risk mapping using Gaussian process stacked generalization. J. R. Soc. Interface. 2017;14:20170520. doi: 10.1098/rsif.2017.0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ton J-F, Flaxman S, Sejdinovic D, Bhatt S. Spatial mapping with Gaussian processes and nonstationary Fourier features. Spat. Stat. 2018;28:59–78. doi: 10.1016/j.spasta.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pullan RL, et al. Spatial modelling of soil-transmitted helminth infections in Kenya: a disease control planning tool. PLoS Neglect. Trop. Dis. 2011;5:e958. doi: 10.1371/journal.pntd.0000958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melles SJ, et al. Optimizing the spatial pattern of networks for monitoring radioactive releases. Comput. Geosci. 2011;37:280–288. [Google Scholar]
- 19.Heuvelink G, Brus D, de Gruijter J, et al. Optimization of sample configurations for digital mapping of soil properties with universal kriging. In: Lagacherie P, et al., editors. Digital Soil Mapping: An Introductory Perspective. Amsterdam: Elsevier; 2006. pp. 139–153. [Google Scholar]
- 20.de Gruijter J, Brus D, Bierkens M, Knotters M. Sampling for Natural Resource Monitoring. Berlin: Springer; 2006. [Google Scholar]
- 21.Brus DJ, de Gruijter JJ. Random sampling or geostatistical modelling? Choosing between design-based and model-based sampling strategies for soil (with discussion) Geoderma. 1997;80:1–44. [Google Scholar]
- 22.Gyapong JO, Remme JH. The use of grid sampling methodology for rapid assessment of the distribution of bancroftian filariasis. Trans. R. Soc. Trop. Med. Hyg. 2001;95:681–686. doi: 10.1016/s0035-9203(01)90115-4. [DOI] [PubMed] [Google Scholar]
- 23.Ngwira BM, Tambala P, Perez AM, Bowie C, Molyneux DH. The geographical distribution of lymphatic filariasis infection in Malawi. Filaria J. 2007;6:12. doi: 10.1186/1475-2883-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diggle P, Lophaven S. Bayesian geostatistical design. Scand. J. Stat. 2006;33:53–64. [Google Scholar]
- 25.Fronterre C, Amoah B, Giorgi E, Stanton MC, Diggle PJ. Design and analysis of elimination surveys for neglected tropical diseases. J. Infect. Dis. 2020;221(Supplement_5):S554–S560. doi: 10.1093/infdis/jiz554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chipeta MG, Terlouw DJ, Phiri KS, Diggle PJ. Adaptive geostatistical design and analysis for prevalence surveys. Spat. Stat. 2016;15:70–84. [Google Scholar]
- 27.Kabaghe AN, et al. Adaptive geostatistical sampling enables efficient identification of malaria hotspots in repeated cross-sectional surveys in rural Malawi. PLoS ONE. 2017;12:e0172266. doi: 10.1371/journal.pone.0172266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaloner K, Verdinelli I. Bayesian experimental design: a review. Stat. Sci. 1995;10:273–304. [Google Scholar]
- 29.Sacks J, Welch WJ, Mitchell TJ, Wynn HP. Design and analysis of computer experiments. Stat. Sci. 1989;4:409–423. doi: 10.1214/ss/1177012413. [DOI] [Google Scholar]
- 30.Kittur N, et al. Defining persistent hotspots: areas that fail to decrease meaningfully in prevalence after multiple years of mass drug administration with praziquantel for control of schistosomiasis. Am. J. Trop. Med. Hyg. 2017;97:1810–1817. doi: 10.4269/ajtmh.17-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi S, Metcalf CJE, Ferrari MJ, Tatem AJ, Lessler J. The geography of measles vaccination in the African Great Lakes region. Nat. Commun. 2017;8:15585. doi: 10.1038/ncomms15585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davies MM, Van Der Laan MJ. Optimal spatial prediction using ensemble machine learning. Int. J. Biostat. 2016;12:179–201. doi: 10.1515/ijb-2014-0060. [DOI] [PubMed] [Google Scholar]
- 33.Sturrock HJ, Woolheater K, Bennett AF, Andrade-Pacheco R, Midekisa A. Predicting residential structures from open source remotely enumerated data using machine learning. PLoS ONE. 2018;13:e0204399. doi: 10.1371/journal.pone.0204399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pirracchio R, Petersen ML, van der Laan M. Improving propensity score estimators’ robustness to model misspecification using super learner. Am. J. Epidemiol. 2015;181:108–119. doi: 10.1093/aje/kwu253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benkeser D, Ju C, Lendle S, van der Laan M. Online cross-validation-based ensemble learning. Stat. Med. 2018;37:249–260. doi: 10.1002/sim.7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomson DR, Stevens FR, Ruktanonchai NW, Tatem AJ, Castro MC. Gridsample: an R package to generate household survey primary sampling units (PSUs) from gridded population data. Int. J. Health Geograph. 2017;16:25. doi: 10.1186/s12942-017-0098-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ziller M, Selhorst T, Teuffert J, Kramer M, Schlüter H. Analysis of sampling strategies to substantiate freedom from disease in large areas. Prev. Vet. Med. 2002;52:333–343. doi: 10.1016/s0167-5877(01)00245-8. [DOI] [PubMed] [Google Scholar]
- 38.Michael E, et al. Substantiating freedom from parasitic infection by combining transmission model predictions with disease surveys. Nat. Commun. 2018;9:4324. doi: 10.1038/s41467-018-06657-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lark RM. Optimized spatial sampling of soil for estimation of the variogram by maximum likelihood. Geoderma. 2002;105:49–80. [Google Scholar]
- 40.Chipeta M, Terlouw D, Phiri K, Diggle P. Inhibitory geostatistical designs for spatial prediction taking account of uncertain covariance structure. Environmetrics. 2017;28:e2425. [Google Scholar]
- 41.Bhattacharjee NV, et al. Mapping exclusive breastfeeding in africa between 2000 and 2017. Nat. Med. 2019;25:1205–1212. doi: 10.1038/s41591-019-0525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cover TM, Thomas JA. Elements of Information Theory. Hoboken: Wiley; 1991. [Google Scholar]
- 43.Auer P. Using confidence bounds for exploitation–exploration trade-offs. J. Mach. Learn. Res. 2002;3:397–422. [Google Scholar]
- 44.Srinivas, N., Krause, A., Kakade, S. & Seeger, M. Gaussian process optimization in the bandit setting: no regret and experimental design. In Proceedings of the 27th International Conference on International Conference on Machine Learning, ICML’10, 1015–1022 (Omnipress, USA, 2010).
- 45.Shahriari B, Swersky K, Wang Z, Adams RP, De Freitas N. Taking the human out of the loop: a review of bayesian optimization. Proc. IEEE. 2015;104:148–175. [Google Scholar]
- 46.Snoek, J., Larochelle, H. & Adams, R. P. Practical bayesian optimization of machine learning algorithms. Advances in Neural Information Processing Systems, 2951–2959 (2012).
- 47.Doxsey-Whitfield E, et al. Taking advantage of the improved availability of census data: a first look at the gridded population of the world, version 4. Pap. Appl. Geogr. 2015;1:226–234. [Google Scholar]
- 48.Fick SE, Hijmans RJ. Worldclim 2: new 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017;37:4302–4315. [Google Scholar]
- 49.Wright MN, Ziegler A. ranger: a fast implementation of random forests for high dimensional data in C++ and R. J. Stat. Softw. 2017;77:1–17. doi: 10.18637/jss.v077.i01. [DOI] [Google Scholar]
- 50.Rousset F, Ferdy J-B. Testing environmental and genetic effects in the presence of spatial autocorrelation. Ecography. 2014;37:a781–790. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.