Abstract
Background:
The CXCL subfamily of chemokines (CXCL9, CXCL10, and CXCL11; angiostatic chemokines) plays a key role in many inflammatory diseases. However, the expression of CXCLs in adipose tissue (AT) during obesity and association of these CXCLs with inflammatory markers and insulin resistance are poorly understood. Therefore, this study aimed to investigate the effects of CXCL gene expression on subcutaneous AT inflammatory markers and insulin resistance.
Methods:
Subcutaneous-fat biopsies were collected from 59 nondiabetic (lean/overweight/obese) individuals for RNA isolation. Expression levels of AT CXCL and inflammatory markers were determined by quantitative reverse transcriptase polymerase chain reaction (RT-qPCR). Biomedical parameters in the plasma were measured by enzyme-linked immunosorbent assay (ELISA). Insulin resistance was estimated using homeostatic model assessment (HOMA-IR).
Results:
AT CXCL expression was higher in obese compared with lean individuals (p < 0.05) and positively correlated with body mass index (BMI; r ⩾ 0.269, p < 0.05). Expression of CXCL9, CXCL10, and CXCL11 correlated significantly with various pro-inflammatory markers, including family members of interleukins, chemokines, and their prospective receptors (r ⩾ 0.339, p ⩽ 0.009), but not anti-inflammatory markers. CXCL11 expression correlated specifically with the expression of CCL5, CCL18, TLR3, TLR4, TLR8, IRF5, and NF-κB (r ⩾ 0.279, p ⩽ 0.039). Notably, CXCL11 was correlated with C-reactive protein (CRP), fasting blood glucose (FBG), and HOMA-IR. In multiple regression analysis, CXCL11 was identified as an independent predictor of CCL19, CCL5, IL-6, and TLR3.
Conclusion:
These data suggest that the CXCL family members, specifically CXCL10 and CXCL11, are potential biomarkers for the onset of AT inflammation during obesity.
Keywords: adipose tissue, CXCL9, CXCL10, CXCL11, insulin resistance, metabolic inflammation, obesity
Introduction
The prevalence of obesity continues to surge globally and constitutes a major source of morbidity, mortality, and low quality of life.1 Obesity is a systemic disease caused by abnormal accumulation of body fat, which triggers diverse chronic complications, including diabetes, angiopathy, and cardiovascular diseases.2,3 Despite significant awareness efforts, obesity and its complications continue to increase, reaching epidemic levels.4
Obesity-mediated chronic low-grade inflammation is called meta-inflammation, and is characterized by increased plasma levels of systemic inflammatory C-reactive protein (CRP), adipokines including interleukin (IL)-1 beta (IL-1β), tumor necrosis factor alpha (TNF-α), IL-6, IL-8 (or CXCL8), IL-12, and the hormone leptin.5–7 Immune cells in obese adipose tissue (AT) exacerbate local inflammation by producing inflammatory factors, including chemokines and cytokines. Chemokines (CCL2, CCL3, CCL5, CCL7, CCL8, CCL19, CCL11) as well as chemokine receptors (CCR1, CCR2, CCR3, CCR5) have been associated with obesity and metabolic inflammation.8–10 AT is a plastic and dynamic endocrine organ. Besides functioning as the primary location for triacylglycerol deposit, AT produces hormones and secretes cytokines, chemokines, and angiogenic factors.11 The plasticity of AT is linked closely to angiogenesis, which is crucial for its physiological expansion or regression by a mechanism that has yet to be delineated.12 Aberrant AT vascularization, however, induces pathological inflammatory and metabolic dysfunctions that are often detected in obese individuals.13,14
CXCLs (CXCL9, CXCL10, and CXCL11) play a key role in many inflammatory diseases.15 Like other chemokines, these factors regulate cell trafficking of various types of leukocytes into inflammatory sites and aggravate inflammation.16 Abnormal angiogenesis, inflammation, and immune cell infiltration usually characterize actively expanding AT during obesity.17 CXCL9, CXCL10, and CXCL11 are involved in the recruitment, migration, and activation of immune cells.18,19 AT expression levels of these chemokines and their association with other inflammatory markers are not well defined during obesity. Therefore, in this study, we show, for the first time, that the simultaneous increased expression of CXCL9, CXCL10, and CXCL11 in ATs of overweight/obese individuals correlates with inflammatory and metabolic markers in the same population.
Materials and methods
Study population and anthropometric measurements
The study cohort comprised 59 non-diabetic individuals from both genders who were enrolled at a gymnasium facility or from exercise studies at Dasman Diabetes Institute. Using the standard formula for the body mass index (BMI) = body weight (kg)/height2 (m2), the cohort was divided into 10 lean (BMI < 25 kg/m2), 20 overweight (25 ⩽ BMI < 30 kg/m2), and 29 obese (BMI ⩾ 30 kg/m2) individuals. For each category, the sample size was dependent on the sample availability and each participant’s decision to be engaged in the research study. Exclusion criteria included diseases of the lung, heart, kidney, or liver; hematologic disorders; pregnancy; immune dysfunction; diabetes (types 1 and 2); or malignancy, as previously described.20 Written informed consent was obtained from all study participants following the ethical guidelines of the Declaration of Helsinki and approved by the ethics committee at Dasman Diabetes Institute, Kuwait (grant numbers RA 2010-003; RA AM 2016-007). Heights and weights were measured using calibrated, portable electronic weighing scales and portable, inflexible height-measuring bars; waist circumferences were measured using constant-tension tape. Whole-body compositions including percent body fat, soft lean mass, and total body water were measured using an IOI 353 Body Composition Analyzer (Jawon Medical, Seoul, South Korea). Biochemical and physical characteristics of the participants are summarized in Table 1.
Table 1.
Demographic and clinical characteristics of study population.
| Phenotype | Lean |
Overweight |
Obese |
Lean versus overweight |
Lean versus obese |
|---|---|---|---|---|---|
| (n = 10) (Mean ± SD) | (n = 20) (Mean ± SD) | (n = 29) (Mean ± SD) | (p value) | (p value) | |
| Age (years) | 42.7 ± 8.17 | 43.05 ± 11.19 | 45.41 ± 13.15 | 0.931 | 0.546 |
| Weight (kg) | 62.93 ± 11.90 | 79.49 ± 9.66 | 93.84 ± 13.77 | <0.0001 | <0.0001 |
| Height (cm) | 1.66 ± 0.12 | 1.67 ± 0.11 | 1.64 ± 0.10 | 0.701 | 0.659 |
| BMI (kg/m2) | 22.82 ± 2.35 | 28.30 ± 1.17 | 34.87 ± 3.30 | <0.0001 | <0.0001 |
| Waist (cm) | 81.33 ± 12.44 | 95.19 ± 8.81 | 106.74 ± 12.86 | 0.003 | <0.0001 |
| Body fat (%) | 28.37 ± 6.27 | 32.52 ± 4.87 | 39.56 ± 4.25 | 0.073 | <0.0001 |
| TGL (mmol/l) | 0.63 ± 0.24 | 1.21 ± 0.62 | 1.35 ± 0.86 | 0.001 | <0.0001 |
| FBG (mmol/l) | 4.97 ± 0.64 | 5.24 ± 0.67 | 5.37 ± 0.77 | 0.292 | 0.151 |
| HbA1c (%) | 5.66 ± 0.46 | 5.51 ± 0.44 | 5.69 ± 0.65 | 0.388 | 0.884 |
| Chol (mmol/l) | 5.3 ± 1.11 | 4.97 ± 0.71 | 5.03 ± 1.14 | 0.323 | 0.521 |
| HDL (mmol/l) | 1.69 ± 0.51 | 1.26 ± 0.29 | 1.16 ± 0.27 | 0.006 | 0.009 |
| LDL (mmol/l) | 3.31 ± 0.93 | 3.18 ± 0.65 | 3.28 ± 1.01 | 0.659 | 0.933 |
| WBC | 5.57 ± 1.60 | 6.25 ± 1.52 | 6.58 ± 1.93 | 0.283 | 0.161 |
BMI, body mass index, Chol, cholesterol; FBG, fasting blood glucose; LDL, low-density lipoprotein; HbA1c, glycated hemoglobin; HDL, high-density lipoprotein; SD, standard deviation; TGL, plasma triglycerides.
Collection of subcutaneous AT
Human AT samples (approximately 0.5 g) were collected via abdominal subcutaneous fat pad biopsy lateral to the umbilicus using standard surgical procedure as described previously.9 Briefly, the periumbilical area was sterilized by alcohol swab and then locally anesthetized using 2% lidocaine (2 ml). Fat tissue was collected through a small superficial skin incision (0.5 cm). After removal, biopsy tissue was further incised into smaller pieces of approximately 50–100 mg, rinsed with cold phosphate-buffered saline and preserved in RNAlater and stored at −80°C until use.21
Measurement of metabolic inflammatory markers
Peripheral blood was collected from overnight-fasted individuals and analyzed for fasting blood glucose (FBG), lipid profile, glycated hemoglobin (HbA1c), fasting insulin, and CRP. FBG and lipid profiles [plasma triglycerides (TGL), high-density lipoprotein (HDL), and cholesterol (Chol)] were measured using Siemens Dimension RXL chemistry analyzer (Diamond Diagnostics, Holliston, MA, USA). HbA1c was measured using a Variant™ device (Bio-Rad, Hercules, CA, USA). Homeostatic model assessment (HOMA-IR) as a measure of insulin resistance was calculated from basal (fasting) glucose and insulin concentrations using the following formula: HOMA-IR = fasting insulin (μU/l) × FBG (nmol/l)/22.5. Plasma high-sensitivity CRP levels were measured by enzyme-linked immunosorbent assay (ELISA; BioVendor, Ashville, NC, USA). All assays were performed following instructions of the manufacturers.
RNA extraction, cDNA synthesis, and RT-qPCR reactions
Total RNA was extracted from AT using RNeasy kit (Qiagen, Valencia, CA, USA) following the manufacturer’s protocol. The first-strand cDNA was synthesized from 0.5 µg RNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Waltham, MA, USA). Real-time RT-qPCR was performed as described9; cDNA samples (50 ng) were amplified using TaqMan® Gene Expression Master Mix (Applied Biosystems) and gene-specific 20 × TaqMan gene expression assays (Applied Biosystems) containing forward and reverse primers (listed in Supplemental Table S1) and target-specific TaqMan® MGB probes labeled with FAM dye at the 5′-end and NFQ-MGB at the 3′-end of the probe using 7500 Fast Real-Time PCR System (Applied Biosystems). Each cycle involved denaturation (15 s at 95°C), annealing/extension (1 min at 60°C) after uracil-DNA glycosylases (UDG, 2 min at 50°C) and AmpliTaq gold enzyme (10 min at 95°C) activation. Relative gene expression to the lean AT control was calculated using the comparative cycles to threshold (CT) method as previously described.22 Results were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and means ± standard error of the mean (SEM) are shown expressed as fold changes in expression relative to controls as indicated.23
Statistical analysis
Statistical analysis was performed using GraphPad Prism software (GraphPad, La Jolla, CA, USA) and SPSS for Windows version 19.01 (IBM SPSS Inc., Chicago, IL, USA). Unless otherwise indicated, data were shown as mean ± standard deviation (SD) values. Unpaired Student’s t test was used to compare means between groups. Correlation and stepwise multiple regression analysis were performed to determine associations between different variables. For all analyses, a p value < 0.05 was considered significant. Standard multiple linear regression by the Enter method was used; variables that significantly correlated with CXCL11 were selected as predictor variables and were entered simultaneously to generate the model. The F-test was used to assess whether the set of entered independent variables collectively predicts the dependent variable. R-squared was used to determine how much variance in the dependent variable can be accounted for by the set of independent variables. The t test, (p value) and beta coefficients (β-value) were used to determine the significance and the magnitude of prediction for each independent variable, respectively.
Results
Demographic and clinical characteristics of the study population
The characteristics of the 59 individuals included in this study are detailed in Table 1. All participants were within the age range of 34–58 years old, with no significant differences in height. Relative to the lean individuals, overweight and obese individuals showed a statistically significant increase in BMI, waist circumference, total body fat, and plasma TGL. Mean values for total plasma Chol and low-density lipoprotein (LDL) levels were comparable between all three groups. HDL measurements were significantly higher in lean individuals (Table 1).
CXCL gene expression is associated with obesity
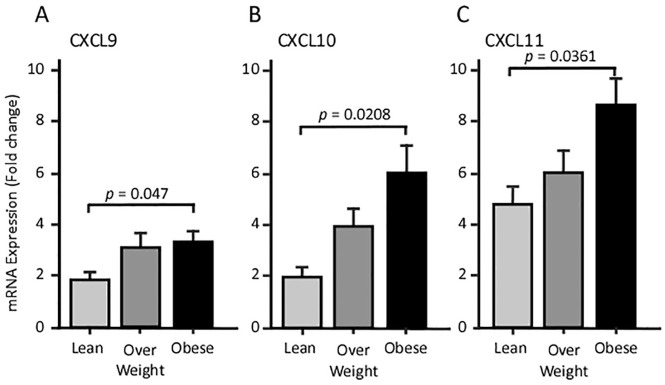
We studied gene expression at the transcriptional (mRNA) level in subcutaneous AT. At a basal level, the expression of CXCL9 and CXCL10 were comparable, whereas CXCL11 showed 2.5-fold higher expression levels (Figure 1A–C). Relative to lean individuals, AT RT-qPCR analysis revealed a marginal surge of CXCL transcripts in overweight participants and a statistically significant increase in its levels in obese subjects (Figure 1).
Figure 1.
The expression levels of CXCLs in AT; RT-qPCR analysis for CXCLs RNA isolated from AT from lean, overweight and obese individuals. A CXCL9, B CXCL10, C CXCL11.
AT, adipose tissue; CXCL, angiostatic chemokines; RT-qPCR, quantitative reverse transcriptase polymerase chain reaction.
In biomedical parameters, CXCL9 and CXCL10 expression exhibited a positive association with the BMI (r ⩾ 0.269, p < 0.05; Table 2). By contrast, CXCL11 transcripts positively correlated with several parameters including BMI (r = 0.286, p = 0.029), FBG (r = 0.292, p = 0.026), body fat percentage (r = 0.402, p = 0.004), and plasma CRP (r = 0.356, p = 0.024) (Table 2). No correlation between the CXCL family members and the lipid profile (plasma TGL, HDL, LDL, and Chol) was noted. Interestingly, CXCL9 and CXCL10 gene expressions were not correlated to each other, yet both were positively correlated with that of CXCL11 (Table 3).
Table 2.
Correlation of CXCLs expression levels in non-diabetic overweight and obese subjects with various clinical and biochemical markers.
| Metabolic markers | CXCL9 |
CXCL10 |
CXCL11 |
|||
|---|---|---|---|---|---|---|
| Pearson correlation (49) |
||||||
| r value | p value | r value | p value | r value | p value | |
| Age | 0.012 | 0.929 | 0.05 | 0.71 | 0.131 | 0.324 |
| Weight | −0.072 | 0.584 | 0.077 | 0.566 | 0.152 | 0.25 |
| Height | −0.091 | 0.488 | −0.121 | 0.367 | −0.048 | 0.716 |
| BMI | 0.339* | 0.012 | 0.281* | 0.037 | 0.290* | 0.03 |
| PBF | 0.082 | 0.562 | 0.193 | 0.175 | 0.286* | 0.042 |
| CHOL | 0.152 | 0.248 | 0.09 | 0.502 | 0.057 | 0.669 |
| HDL | −0.075 | 0.57 | −0.195 | 0.142 | −0.121 | 0.363 |
| LDL | 0.201 | 0.123 | 0.168 | 0.208 | 0.107 | 0.42 |
| TGL | −0.005 | 0.972 | 0.011 | 0.937 | 0.004 | 0.975 |
| GLU | 0.141 | 0.282 | 0.142 | 0.287 | 0.292* | 0.026 |
| HbA1c | 0.009 | 0.944 | 0.135 | 0.318 | 0.189 | 0.154 |
| Insulin | −0.051 | 0.742 | −0.051 | 0.747 | 0.208 | 0.181 |
| HOMA-IR | −0.039 | 0.799 | −0.026 | 0.872 | 0.395* | 0.009 |
| CRP | 0.272 | 0.086 | 0.168 | 0.299 | 0.349* | 0.025 |
BMI, body mass index, CHOL, cholesterol; CXCLs, angiostatic chemokines; FBG, fasting blood glucose; CRP, C-reactive protein; GLU, glucose; HbA1c, glycated hemoglobin; HDL, high-density lipoprotein; HOMA-IR, homeostatic model assessment; LDL, low-density lipoprotein; PBF, percent body fat; SD, standard deviation; TGL, plasma triglycerides.
Bold values with statistically significant correlations as indicated with the prospective p-value column.
Table 3.
Correlation of CXCLs with various cytokines/chemokines in non-diabetic overweight and obese subjects.
| Inflammatory markers | CXCL9 |
CXCL10 |
CXCL11 |
|||
|---|---|---|---|---|---|---|
| Pearson correlation (49) |
||||||
| r value | p value | r value | p value | r value | p value | |
| CXCL9 | 1 | 0.235 | 0.075 | 0.285* | 0.029 | |
| CXCL10 | 0.235 | 0.075 | 1 | 0.739** | 0.0001 | |
| CXCL11 | 0.285* | 0.029 | 0.739** | 0.0001 | 1 | |
| Interleukins | ||||||
| IL-1β | 0.03 | 0.845 | 0.656** | 0.0001 | 0.516** | 0.0001 |
| IL-2 | 0.044 | 0.745 | 0.229 | 0.089 | 0.426** | 0.001 |
| IL-5 | 0.002 | 0.99 | −0.056 | 0.691 | 0.023 | 0.87 |
| IL-6 | 0.008 | 0.956 | 0.504** | 0.0001 | 0.470** | 0.0001 |
| IL-8 | −0.056 | 0.7 | 0.121 | 0.414 | 0.146 | 0.315 |
| IL-12A | 0.034 | 0.826 | 0.199 | 0.211 | 0.285 | 0.068 |
| IL-13 | 0.054 | 0.702 | −0.1 | 0.486 | −0.022 | 0.878 |
| TNF-α | 0.1 | 0.472 | 0.427 ** | 0.002 | 0.450** | 0.001 |
| CC chemokine ligands | ||||||
| CCL2 | 0.132 | 0.327 | 0.546** | 0.0001 | 0.537** | 0.0001 |
| CCL3 | −0.004 | 0.979 | 0.097 | 0.489 | 0.215 | 0.118 |
| CCL5 | 0.121 | 0.417 | 0.264 | 0.076 | 0.468** | 0.001 |
| CCL8 | 0.229 | 0.107 | 0.721 ** | 0.0001 | 0.501** | 0.0001 |
| CCL7 | −0.04 | 0.77 | 0.125 | 0.368 | 0.174 | 0.204 |
| CCL11 | 0.214 | 0.116 | 0.393** | 0.004 | 0.325* | 0.015 |
| CCL15 | 0.211 | 0.108 | −0.03 | 0.822 | −0.106 | 0.428 |
| CCL18 | 0.114 | 0.395 | 0.238 | 0.075 | 0.373** | 0.004 |
| CCL19 | 0.233 | 0.082 | 0.272* | 0.045 | 0.489** | 0.0001 |
| CCL20 | 0.051 | 0.704 | 0.516** | 0.0001 | 0.391** | 0.003 |
| CCR2 | 0.255 | 0.068 | 0.582** | 0.0001 | 0.589** | 0.0001 |
| CCR5 | 0.268* | 0.04 | 0.398** | 0.002 | 0.562** | 0.0001 |
| Macrophage markers | ||||||
| CD86 | 0.397** | 0.002 | 0.337* | 0.012 | 0.362** | 0.007 |
| CD163 | 0.227 | 0.083 | 0.309* | 0.02 | 0.413** | 0.001 |
CXCLs, angiostatic chemokines; IL, interleukin; TNF-α, tumor necrosis factor alpha.
Bold values with statistically significant correlations as indicated with the prospective p-value column.
In AT, increased CXCL gene expression relates to inflammatory signatures
We next correlated the levels of CXCL transcripts to those of inflammatory regulatory markers in AT. As shown in Table 3, our data indicate that, in obese subjects, CXCL10 expression was associated positively with pro-inflammatory markers including IL-1β and IL-6 (r ⩾ 0.504; p = 0.0001). CXCL11 showed correlations with a broader number of interleukins: IL-1β (r = 0.516; p = 0.0001), IL-2 (r = 0.426; p = 0.001) and IL-6 (r = 0.470; p = 0.0001). No correlation between CXCL9 and the studied interleukins was observed (Table 3). Moreover, no statistically significant correlation was observed between CXCLs and the B-cell maturation and differentiation markers IL-5 or IL-13, or with the Th1 cell marker IL-12A, suggesting that these CXCLs are not associated with anti-inflammatory markers. TNF-α transcription levels were correlated with those of CXCL10 and CXCL11 (r = 0.427, p = 0.002; and r = 0.450, p = 0.001; respectively; Table 3).
Interestingly, CXCL expression levels were positively associated with several of the closely related CC motif chemokines observed in macrophages and lymphocytes that mediate obesity-induced chronic inflammation, including CCL2, CCL5, CCL8, CCR2, and CCR5 (0.325 ⩽ r ⩽0.589; p < 0.01). Like the interleukins, the CXCLs associated differentially with CCL family members. All three factors correlated positively with CCR5, whereas CXCL10 and CXCL11 were closely correlated with other cytokines (Table 3). Parameters that showed significant associations with CXCL11 were further studied. As shown in Table 4, multiple regression analysis indicated that IL-6, CCL19, and CCR5 were independently associated with CXCL11.
Table 4.
Multi linear regression: non-diabetic subjects, dependent variable: CXCL11.
| ANOVA (Sig) R2 = 0.56 | ||
|---|---|---|
| Predictor variable | B value | p value |
| IL-6 | 0.182 | 0.001 |
| CCL19 | 0.612 | 0.004 |
| CCR5 | 0.534 | 0.001 |
| TLR3 | 1.198 | 0.002 |
IL, interleukin; TLR, toll-like receptor.
Association between AT CXCL expression and toll-like receptors
We next investigated any prospective correlations between CXCL expression and the toll-like receptors (TLRs). Indeed, we observed a statistically significant positive correlation between the three CXCL transcripts and that of TLR2 and TLR7 (r ⩾ 0.313; p < 0.01). Excluding TLR9, CXCL11 expression was significantly associated with all other TLRs as well (TLR2, TLR3, TLR4, TLR7, TLR8, TLR10; r ⩾ 0.311; p ⩽ 0.03; Table 5).
Table 5.
Correlation of CXCL11 with TLR signaling pathways in non-diabetic overweight and obese subjects.
| TLRs | CXCL9 |
CXCL10 |
CXCL11 |
|||
|---|---|---|---|---|---|---|
| Peasron correlation (49) |
||||||
| r value | p value | r value | p value | r value | p value | |
| TLR2 | 0.594** | 0 | 0.438** | 0.002 | 0.445** | 0.001 |
| TLR3 | 0.191 | 0.17 | 0.25 | 0.071 | 0.343* | 0.013 |
| TLR4 | 0.273 | 0.057 | 0.276 | 0.055 | 0.311* | 0.031 |
| TLR7 | 0.365** | 0.004 | 0.313* | 0.017 | 0.438** | 0.001 |
| TLR8 | 0.343** | 0.009 | 0.238 | 0.081 | 0.437** | 0.001 |
| TLR9 | 0.157 | 0.236 | −0.007 | 0.96 | 0.207 | 0.119 |
| TLR10 | 0.076 | 0.575 | 0.297* | 0.029 | 0.337* | 0.012 |
| MyD88 | 0.395** | 0.002 | 0.322* | 0.015 | 0.339** | 0.009 |
| IRF3 | 0.132 | 0.372 | 0.063 | 0.678 | 0.113 | 0.45 |
| IRF5 | 0.217 | 0.109 | 0.243 | 0.077 | 0.279* | 0.039 |
| NF-κB | 0.178 | 0.175 | 0.154 | 0.248 | 0.283* | 0.03 |
IL, interleukin; IRF, interferon regulatory factor; NF-κB, nuclear factor kappa B.
Bold values with statistically significant correlations as indicated with the prospective p-value column.
TLRs exert their cellular effects through canonical signaling pathways mediated by MyD88 and non-canonical signaling mediated by the interferon regulatory factor (IRF) family members. Our data analysis showed positive correlations with these downstream effectors of TLR signaling. Expression levels of the three CXCLs and MyD88 were found to correlate positively (r ⩾ 0.322; p < 0.01). No correlation was observed between CXCLs and the MyD88-independent signaling molecule IRF3. In addition, CXCL11 expression was correlated positively with that of IRF5 (r = 0.279, p = 0.04) and NF-κB (r = 0.283, p = 0.03) (Table 5).
Association between AT CXCL expression and M1-M2 macrophage transition
Positive correlation with the TLRs suggested a prospective association with macrophage markers. Our data revealed a positive correlation between CXCL transcripts and the M1 macrophage protein CD86 (r ⩾ 0.337; p = 0.01). CXCL10 and CXCL11 were associated positively with the M2 macrophage marker CD163 (r = 0.309; p = 0.02) (Table 3).
Discussion
AT is a lipid depot and an active endocrine organ secreting a variety of adipokines and chemokines.24 Accumulative data suggest that AT dysfunction, inflammation, and inadequate vascularization causes obesity and associated complications.25,26 Angiogenesis is crucial for maintaining normal AT metabolic function and for providing expanding AT with oxygen and nutrients. Insufficient neovascularization causes hypoxia, which then increases inflammation and metabolic dysfunction. CXCLs (CXCL9, CXCL10, and CXCL11; angiostatic chemokines) are involved in the recruitment, migration, and activation of immune cells in inflammatory diseases. Therefore, understanding the role of AT CXCLs in metabolic inflammation may be fundamental to prevent AT abnormalities in obese individuals.
We observed a significant increase in the gene expression of CXCLs in subcutaneous AT from obese subjects relative to that of lean individuals, which positively correlate with BMI, body fat percentage, and CRPs, suggesting a potential association between obesity and metabolic alteration. Similarly, Deiuliis et al. reported elevated transcripts of the other angiostatic proteins, CXCR3 and CXCL10, in omental AT from obese patients compared with corresponding tissue from lean subjects.27 Comparing subcutaneous versus visceral AT from morbidly obese patients, Hueso et al. observed a higher gene expression of CXCR3, CXCL10, and CXCL11 in visceral AT, which is associated with a reduced neovascularization.28 Therefore, impaired angiogenesis is most likely associated with differential expression of the angiostatic chemokines.
Chemokines coordinate leukocyte trafficking to inflammatory sites. In obese individuals, the upregulation of CXCLs was correlated with pro-inflammatory mediators IL-1β, IL-2, IL-6, CCL2, CCL8, CCL20, and TNF-α, but not with anti-inflammatory factors. These pro-inflammatory intermediaries have been implicated in the pathogenesis of various inflammatory disorders including atherosclerosis, coronary heart disease, and diabetes. Using an animal model, the Spiegelman group, in 1993, was the first to associate TNF-α with AT-insulin-resistance-associated obesity.29 Gene expressions of IL-1β, IL-2, and IL-6 are significantly higher in AT isolated from obese individuals, relative to prospective tissues from lean individuals.30,31 Likewise, we recently reported a potent association of CCL19 elevated transcripts with insulin resistance in AT from obese and overweight patients.9 Taken together, these associations between CXCLs and inflammatory markers suggest that, at the onset of obesity, CXCLs and cytokines are produced heavily in inflamed AT, and may subsequently impair insulin sensitivity. The significant correlation of CXCLs with inflammatory factors suggests that they are implicated in the infiltration of immune cells into AT, which substantially contributes to the pathogenesis of obesity. In a similar fashion, CXCL10 is associated with inflammatory factors.32,33 Accordingly, elevated levels of plasma-circulating angiostatic mediators were observed in several inflammation-associated diseases, including chronic ischemic heart disease,34 systematic advanced heart failure,35,36 and hypertension.37
TLRs are well characterized immune receptors that mediate inflammation. Obesity and associated metabolic syndromes are triggered by the activation of TLR2 and TLR4, which is followed by a dynamic cascade leading to the activation of MyD88 and NFκB and the release of pro-inflammatory mediators.38 Several reports from our laboratory show that the AT expression of several TLRs (TLR2, TLR4, TLR8, and TLR10) and downstream signaling molecules (MyD88, IRAK1, IRF3, and NFκB) are higher in obese individuals with or without type 2 diabetes, and these changes were associated directly with BMI, cytokine/chemokine expression, and insulin resistance.21,39–41 Interestingly, our data revealed a significant positive correlation between CXCLs and TLRs and their downstream responsive molecules, suggesting a close interaction between the CXC chemokines and the TLR signaling pathways leading to metabolic inflammation.
A notable biological plausibility was observed in the correlations between CXCLs and inflammatory markers, where some markers with comparable Pearson correlation coefficients showed different statistical significance responses, which can be arbitrated to the complexity of the cell–cell crosstalk between AT cells and resident immune cells. Further, we cannot rule out the implication of other non-studied factors that can specifically affect the significance associations between the studied factors. Moreover, the presence of genetic polymorphisms within the studied population at the effector regions of some markers may cause such variations in correlation studies.
In summary, our findings show that AT CXCL10 and CXCL11 expression levels are associated closely with the expression levels of cytokines, chemokines, and TLRs (Figure 2), which are characteristics of the development of inflammation and progression of insulin resistance in obese individuals. Our results suggest that CXCL10 and CXCL11 are potential predictor molecules for the onset of obesity, and prospective therapeutic targets to maintain normal glucose homeostasis.
Figure 2.
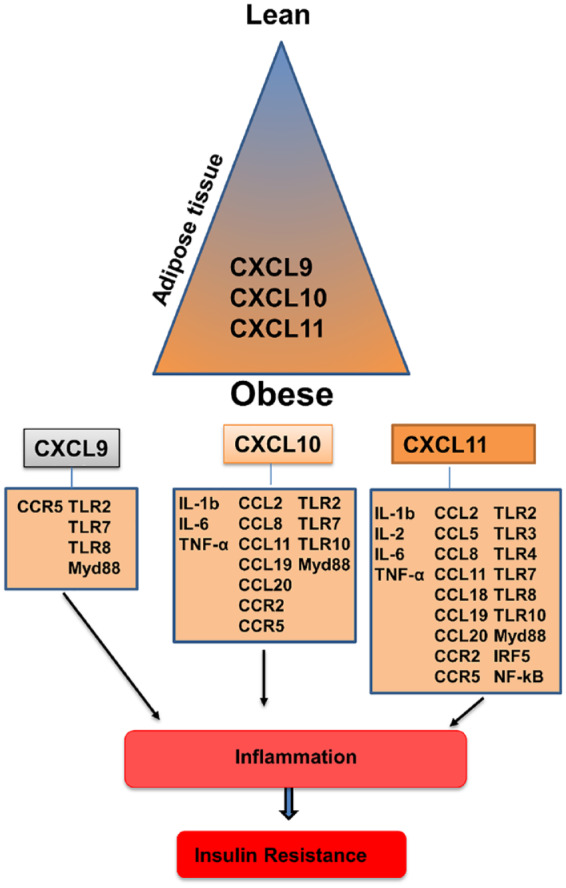
Schematic representation of CXCLs and their association with metabolic inflammation.
CXCL, angiostatic chemokines; IL, interleukin; NF-κB, nuclear factor kappa B; TLR, toll-like receptor; TNF-α, tumor necrosis factor alpha.
Supplemental Material
Supplemental material, sj-pdf-1-tae-10.1177_2042018820930902 for Adipose tissue gene expression of CXCL10 and CXCL11 modulates inflammatory markers in obesity: implications for metabolic inflammation and insulin resistance by Shihab Kochumon, Ashraf Al Madhoun, Fatema Al-Rashed, Rafaat Azim, Ebaa Al-Ozairi, Fahd Al-Mulla and Rasheed Ahmad in Therapeutic Advances in Endocrinology and Metabolism
Acknowledgments
The authors also thank Fahad Al-Ghamlas for help with patient recruitment. The authors would like to thank the editors at Enago company for the academic English editing service (www.enago.com).
Footnotes
Author contribution(s): Shihab Kochumon: Data curation; Formal analysis; Methodology; Writing-original draft.
Ashraf Al Madhoun: Formal analysis; Investigation; Writing-original draft.
Fatema Al-Rashed: Data curation; Formal analysis; Writing-review & editing.
Rafaat azim: Data curation; Formal analysis; Methodology; Writing-review & editing.
Ebaa Al-Ozairi: Conceptualization; Writing-review & editing.
Fahd Al-Mulla: Conceptualization; Writing-review & editing.
Rasheed Ahmad: Conceptualization; Funding acquisition; Supervision; Validation; Writing-review & editing.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by Kuwait Foundation for Advancement of Sciences (KFAS) (Grant #: RA 2010-003; RA AM 2016-007).
Conflict of interest statement: The authors declare that there is no conflict of interest.
Research ethics and patient consent: Written informed consent was obtained from all individual participants included in the study. The study was approved by the ethics committee at Dasman Diabetes Institute, Kuwait (grant numbers RA 2010-003; RA AM 2016-007).
ORCID iD: Rasheed Ahmad  https://orcid.org/0000-0001-5746-0743
https://orcid.org/0000-0001-5746-0743
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Shihab Kochumon, Immunology and Microbiology Department, Dasman Diabetes Institute, Dasman, Kuwait.
Ashraf Al Madhoun, Animal and Imaging Core Facilities, Dasman Diabetes Institute, Dasman, Kuwait; Genetics and Bioinformatics, Dasman Diabetes Institute, Kuwait, Dasman, Kuwait.
Fatema Al-Rashed, Immunology and Microbiology Department, Dasman Diabetes Institute, Dasman, Kuwait.
Rafaat Azim, School of Medicine, Royal College of Surgeons in Ireland, Medical University of Bahrain, Busaiteen, Bahrain.
Ebaa Al-Ozairi, Medical Division, Dasman Diabetes Institute, Kuwait.
Fahd Al-Mulla, Genetics and Bioinformatics, Dasman Diabetes Institute, Kuwait, Dasman, Kuwait.
Rasheed Ahmad, Immunology & Microbiology Department, Dasman Diabetes Institute, AL-Soor Street, P.O. Box 1180 Dasman, Kuwait 15462, Kuwait.
Reference
- 1. Arroyo-Johnson C, Mincey KD. Obesity epidemiology worldwide. Gastroenterol Clin North Am 2016; 45: 571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abdelaal M, le Roux CW, Docherty NG. Morbidity and mortality associated with obesity. Ann Transl Med 2017; 5: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Phelan SM, Burgess DJ, Yeazel MW, et al. Impact of weight bias and stigma on quality of care and outcomes for patients with obesity. Obes Rev 2015; 16: 319–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Report of a WHO Consultation. Obesity: preventing and managing the global epidemic. World Health Organ Tech Rep Ser 2000; 894: i–xii, 1–253. [PubMed] [Google Scholar]
- 5. de Luca C, Olefsky JM. Inflammation and insulin resistance. FEBS Lett 2008; 582: 97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sindhu S, Thomas R, Shihab P, et al. Obesity is a positive modulator of IL-6R and IL-6 expression in the subcutaneous adipose tissue: significance for metabolic inflammation. PLoS One 2015; 10: e0133494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hasan AU, Rahman A, Kobori H. Interactions between Host PPARs and gut microbiota in health and disease. Int J Mol Sci 2019; 20: 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ahmad R, Al-Roub A, Kochumon S, et al. The synergy between palmitate and TNF-alpha for CCL2 production is dependent on the TRIF/IRF3 pathway: implications for metabolic inflammation. J Immunol 2018; 200: 3599–3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kochumon S, Al-Rashed F, Abu-Farha M, et al. Adipose tissue expression of CCL19 chemokine is positively associated with insulin resistance. Diabetes Metab Res Rev 2019; 35: e3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huber J, Kiefer FW, Zeyda M, et al. CC chemokine and CC chemokine receptor profiles in visceral and subcutaneous adipose tissue are altered in human obesity. J Clin Endocrinol Metab 2008; 93: 3215–3221. [DOI] [PubMed] [Google Scholar]
- 11. Waki H, Tontonoz P. Endocrine functions of adipose tissue. Annu Rev Pathol 2007; 2: 31–56. [DOI] [PubMed] [Google Scholar]
- 12. Lemoine AY, Ledoux S, Larger E. Adipose tissue angiogenesis in obesity. Thromb Haemost 2013; 110: 661–668. [DOI] [PubMed] [Google Scholar]
- 13. Pasarica M, Sereda OR, Redman LM, et al. Reduced adipose tissue oxygenation in human obesity: evidence for rarefaction, macrophage chemotaxis, and inflammation without an angiogenic response. Diabetes 2009; 58: 718–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garcia-Martin R, Alexaki VI, Qin N, et al. Adipocyte-specific hypoxia-inducible factor 2α deficiency exacerbates obesity-induced brown adipose tissue dysfunction and metabolic dysregulation. Mol Cell Biol 2016; 36: 376–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Turner MD, Nedjai B, Hurst T, et al. Cytokines and chemokines: at the crossroads of cell signalling and inflammatory disease. Biochim Biophys Acta 2014; 1843: 2563–2582. [DOI] [PubMed] [Google Scholar]
- 16. Hughes CE, Nibbs RJB. A guide to chemokines and their receptors. FEBS J 2018; 285: 2944–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fuster JJ, Ouchi N, Gokce N, et al. Obesity-induced changes in adipose tissue microenvironment and their impact on cardiovascular disease. Circ Res 2016; 118: 1786–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tannenbaum CS, Tubbs R, Armstrong D, et al. The CXC chemokines IP-10 and MIG are necessary for IL-12-mediated regression of the mouse RENCA tumor. J Immunol 1998; 161: 927–932. [PubMed] [Google Scholar]
- 19. Tokunaga R, Zhang W, Naseem M, et al. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation - a target for novel cancer therapy. Cancer Treat Rev 2018; 63: 40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sindhu S, Thomas R, Kochumon S, et al. Increased adipose tissue expression of interferon regulatory factor (IRF)-5 in obesity: association with metabolic inflammation. Cells 2019; 8: 1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ahmad R, Shihab PK, Thomas R, et al. Increased expression of the interleukin-1 receptor-associated kinase (IRAK)-1 is associated with adipose tissue inflammatory state in obesity. Diabetol Metab Syndr 2015; 7: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Khadir A, Tiss A, Abubaker J, et al. MAP kinase phosphatase DUSP1 is overexpressed in obese humans and modulated by physical exercise. Am J Physiol Endocrinol Metab 2015; 308: E71–E83. [DOI] [PubMed] [Google Scholar]
- 23. Ahmad R, Al-Mass A, Al-Ghawas D, et al. Interaction of osteopontin with IL-18 in obese individuals: implications for insulin resistance. PLoS One 2013; 8: e63944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wieser V, Moschen AR, Tilg H. Inflammation, cytokines and insulin resistance: a clinical perspective. Arch Immunol Ther Exp (Warsz) 2013; 61: 119–125. [DOI] [PubMed] [Google Scholar]
- 25. Qatanani M, Lazar MA. Mechanisms of obesity-associated insulin resistance: many choices on the menu. Genes Dev 2007; 21: 1443–1455. [DOI] [PubMed] [Google Scholar]
- 26. Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. J Clin Invest 2006; 116: 1793–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Deiuliis JA, Oghumu S, Duggineni D, et al. CXCR3 modulates obesity-induced visceral adipose inflammation and systemic insulin resistance. Obesity (Silver Spring) 2014; 22: 1264–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hueso L, Ortega R, Selles F, et al. Upregulation of angiostatic chemokines IP-10/CXCL10 and I-TAC/CXCL11 in human obesity and their implication for adipose tissue angiogenesis. Int J Obes (Lond) 2018; 42: 1406–1417. [DOI] [PubMed] [Google Scholar]
- 29. Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science 1993; 259: 87–91. [DOI] [PubMed] [Google Scholar]
- 30. Moschen AR, Molnar C, Enrich B, et al. Adipose and liver expression of interleukin (IL)-1 family members in morbid obesity and effects of weight loss. Mol Med 2011; 17: 840–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hoene M, Weigert C. The role of interleukin-6 in insulin resistance, body fat distribution and energy balance. Obes Rev 2008; 9: 20–29. [DOI] [PubMed] [Google Scholar]
- 32. Xu L, Kitade H, Ni Y, et al. Roles of chemokines and chemokine receptors in obesity-associated insulin resistance and nonalcoholic fatty liver disease. Biomolecules 2015; 5: 1563–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Singh UP, Venkataraman C, Singh R, et al. CXCR3 axis: role in inflammatory bowel disease and its therapeutic implication. Endocr Metab Immune Disord Drug Targets 2007; 7: 111–123. [DOI] [PubMed] [Google Scholar]
- 34. Keeley EC, Moorman JR, Liu L, et al. Plasma chemokine levels are associated with the presence and extent of angiographic coronary collaterals in chronic ischemic heart disease. PLoS One 2011; 6: e21174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Altara R, Mallat Z, Booz GW, et al. The CXCL10/CXCR3 axis and cardiac inflammation: implications for immunotherapy to treat infectious and noninfectious diseases of the heart. J Immunol Res 2016; 2016: 4396368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Altara R, Manca M, Hessel MH, et al. CXCL10 Is a circulating inflammatory marker in patients with advanced heart failure: a pilot study. J Cardiovasc Transl Res 2016; 9: 302–314. [DOI] [PubMed] [Google Scholar]
- 37. Youn JC, Yu HT, Lim BJ, et al. Immunosenescent CD8+ T cells and C-X-C chemokine receptor type 3 chemokines are increased in human hypertension. Hypertension 2013; 62: 126–133. [DOI] [PubMed] [Google Scholar]
- 38. Jialal I, Kaur H, Devaraj S. Toll-like receptor status in obesity and metabolic syndrome: a translational perspective. J Clin Endocrinol Metab 2014; 99: 39–48. [DOI] [PubMed] [Google Scholar]
- 39. Ahmad R, Al-Mass A, Atizado V, et al. Elevated expression of the toll like receptors 2 and 4 in obese individuals: its significance for obesity-induced inflammation. J Inflamm (Lond) 2012; 9: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ahmad R, Kochumon S, Thomas R, et al. Increased adipose tissue expression of TLR8 in obese individuals with or without type-2 diabetes: significance in metabolic inflammation. J Inflamm (Lond) 2016; 13: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sindhu S, Akhter N, Kochumon S, et al. Increased expression of the innate immune receptor TLR10 in obesity and type-2 diabetes: association with ROS-mediated oxidative stress. Cell Physiol Biochem 2018; 45: 572–590. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-tae-10.1177_2042018820930902 for Adipose tissue gene expression of CXCL10 and CXCL11 modulates inflammatory markers in obesity: implications for metabolic inflammation and insulin resistance by Shihab Kochumon, Ashraf Al Madhoun, Fatema Al-Rashed, Rafaat Azim, Ebaa Al-Ozairi, Fahd Al-Mulla and Rasheed Ahmad in Therapeutic Advances in Endocrinology and Metabolism