Abstract
Mononuclear molybdoenzymes of the dimethyl sulfoxide reductase (DMSOR) family catalyze a number of reactions essential to the carbon, nitrogen, sulfur, arsenic, and selenium biogeochemical cycles. These enzymes are also ancient, with many lineages likely predating the divergence of the last universal common ancestor into the Bacteria and Archaea domains. We have constructed rooted phylogenies for over 1,550 representatives of the DMSOR family using maximum likelihood methods to investigate the evolution of the arsenic biogeochemical cycle. The phylogenetic analysis provides compelling evidence that formylmethanofuran dehydrogenase B subunits, which catalyze the reduction of CO2 to formate during hydrogenotrophic methanogenesis, constitutes the most ancient lineage. Our analysis also provides robust support for selenocysteine as the ancestral ligand for the Mo/W atom. Finally, we demonstrate that anaerobic arsenite oxidase and respiratory arsenate reductase catalytic subunits represent a more ancient lineage of DMSORs compared to aerobic arsenite oxidase catalytic subunits, which evolved from the assimilatory nitrate reductase lineage. This provides substantial support for an active arsenic biogeochemical cycle on the anoxic Archean Earth. Our work emphasizes that the use of chalcophilic elements as substrates as well as the Mo/W ligand in DMSORs has indelibly shaped the diversification of these enzymes through deep time.
Subject terms: Biochemistry, Computational biology and bioinformatics, Evolution, Microbiology, Biogeochemistry
Introduction
Ubiquitous in Archaea and Bacteria, mononuclear molybdoenzymes of the dimethyl sulfoxide reductase (DMSOR) family are believed to have been core components of the first anaerobic respiratory chains, and thus present at life’s origins1-4. Reactions catalyzed by these enzymes are integral components of the carbon, nitrogen, and sulfur biogeochemical cycles, as well as the biogeochemical cycles of arsenic and selenium5, and likely antimony6,7. The family, which has been defined by the presence of a mononuclear molybdopterin or tungstopterin bis(pyranopterin guanine dinucleotide) (Mo/W-bisPGD) co-factor5, was named after DMSO reductases, the first members of the family to be well-characterized8-12.
As more representatives of the family were discovered and characterized, many were found to be heterotrimeric complexes, consisting of the catalytic subunit (the Mo/W-bisPGD-harboring subunit), an electron transfer subunit with as many as four [Fe–S] clusters, and a membrane anchoring subunit that tethers the complex to the membrane and transfers electrons to or from the membrane quinone pool. Thus, members of the family have also been referred to as Complex Iron-Sulfur Molybdoenzymes (CISMs)13. The catalytic subunit may additionally have a twin-arginine translocation motif for export to the periplasm, and a [4Fe–4S] or [3Fe–4S] iron-sulfur cluster13,14. It has been noted, however, that these characteristics are not uniform across the family, as there are numerous examples where one or more of the associated subunits are missing, or the catalytic subunits associate with different subunits altogether5,13,15. One such example is periplasmic nitrate reductase (Nap), which can be as simple as one peptide (e.g., the catalytic subunit NapA) or have additional subunits in a complex (e.g., NapABCGH)16.
Arsenic metabolism has previously been documented in 2.72 billion year old stromatolites, providing conclusive evidence that arsenic cycling was an active feature of Neoarchean (2.8–2.5 billion years ago (Gya)) environments17. This report came amidst a vigorous debate concerning the evolution of arsenic oxyanion utilization in respiration. Arsenite can serve as an electron donor to stimulate chemolithoauthotrophic growth during aerobic18 and anaerobic19,20 respiration. Arsenite can also be exploited as an electron donor to fuel photolithoautotrophic growth during anoxygenic photosynthesis21,22. The ability to use arsenate as a terminal electron acceptor during anaerobic respiration is a trait found among phylogenetically diverse Bacteria and Archaea23. Arsenate respiration is catalyzed by the respiratory arsenate reductase (Arr). An anaerobic arsenite oxidase (Arx) mediates the oxidation of arsenite during anaerobic respiration. A different enzyme, aerobic arsenite oxidase (Aio), catalyzes the oxidation of arsenite during aerobic respiration. Both Aio and Arx can function in photosynthetic organisms to exploit arsenite as an electron donor during anoxygenic photosynthesis.
The catalytic subunits of Arr and Arx, ArrA and ArxA, appear to be closely related members of the DMSOR family20, though both seem to be only distantly related to the catalytic subunit of Aio, AioA18. Phylogenies constructed using the neighbor-joining method have been used previously to support the idea that AioA was the primordial arsenite oxidase catalytic subunit and present in the last universal common ancestor (LUCA) of the two prokaryotic domains1,24-26, given that bacterial and archaeal homologs have formed separate monophyletic clades in these tree topologies. Presumably, chemolithoautotrophic arsenite oxidation via AioA would have been coupled to the reduction of the nitrogen oxyanions nitrate and nitrite and nitric oxide27, whilst the resulting arsenate would have been rapidly reduced via abiotic geochemical mechanisms. Other researchers, using geochemical and physiological arguments, have demonstrated that it is ArxA, not AioA, that couples chemolithoautotrophic arsenite oxidation to nitrogen oxyanion reduction in modern environments21,23, suggesting that the ArxA/ArrA lineage sustained an active arsenic biogeochemical cycle consisting of arsenite oxidation and respiratory arsenate reduction throughout the Archean Eon (~ 4.0–2.5 Gya).
This is fundamentally an evolutionary question and requires the most sophisticated tools of phylogenetic analyses in order to address it. Yet, such analyses are lacking for the DMSOR family, despite the diverse substrates utilized by DMSORs and their central role in core biogeochemical cycles. Thus, evolutionary relationships among members of this family remain poorly resolved. The only previous comprehensive phylogenetic analysis of the family employed the neighbor-joining method and lacked a root that would allow for a relative ordination of when these enzymes, and their associated biochemical functions, diversified from the broader family through geologic time13. Similarly, phylogenetic arguments regarding the deep antiquity of AioA employed neighbor joining phylogenies and featured unrooted topologies. More recent phylogenetic analyses of DMSOR family enzymes have exploited maximum likelihood methods, but focused on a small subset of enzymes28,29 and either lacked a root altogether or used enzymes within the family to root the tree.
The neighbor-joining method has long been superseded by likelihood-driven methods (e.g., Bayesian30 and maximum likelihood31 analyses) that offer more robust statistical evaluation of nodes within tree topologies as well as superior phylogenetic resolution. Moreover, recent advances in our knowledge of the probable physiology of LUCA32 has provided an opportunity to ordinate DMSOR phylogenies with a root that is equally as ancient as this family. To this end, the aldehyde:ferredoxin oxidoreductase family is an outstanding candidate. The aldehyde:ferredoxin family was also likely a part of LUCA’s physiology32 and harbors a tungstopterin co-factor14 similar to the DMSOR family. Finally, a wealth of genomic data has been generated since the previous comprehensive phylogenetic analysis of this family (e.g.,33,34), which enables much more extensive taxon sampling. The improved methodologies of likelihood-based phylogenetic methods, coupled with a wider array of genomes to mine for sequence variation, offers an ability to rigorously separate genuine vertical inheritance from LUCA from ancient lateral gene transfer events.
Taking advantage of all of these developments, we constructed maximum likelihood phylogenies consisting of 1,568 members of this family using the aldehyde:ferredoxin family as a root. We focused on a number of lineages beyond ArxA/ArrA and AioA. Phylogenetic analyses have consistently found that ArxA/ArrA is closely related to a lineage comprised of the catalytic subunits of the respiratory polysulfide reductase of Wolinella succinogenes (PsrA)35, thiosulfate reductase of Salmonella enterica serovar Typhymurium (PhsA)36, and selenite reductase of Bacillus selenitireducens (SrrA)37, as well as a lineage comprised of the catalytic subunits of the respiratory tetrathionate reductase of S. eneterica serovar Typhymurium (TtrA)38, the selenate reductase of B. selenatarsenatis39, and the arsenate reductase of the archaeon Pyrobaculum aerophilum40, whilst AioA is more closely related to formate dehydrogenase N catalytic subunits (FdhG)1,24-26. Rigorous phylogenetic analyses of these DMSOR members and closely related lineages has revealed several novel insights into the evolution of DMSORs. Our tree topologies robustly support a hydrogenotrophic methanogenic origin for this family, with formylmethanofuran dehydrogenase subunit B (FwdB/FmdB) constituting the most deeply branching lineage. We show that the 21st amino acid selenocysteine (Sec) is likely the ancestral ligand for the Mo/W atom of the Mo/W-bisPGD cofactor. Finally, our analysis marks a substantial advance in our knowledge of the evolution of arsenic respiration. We demonstrate that the ArxA/ArrA lineage is more deeply rooted in the tree topology than AioA, consistent with an Archean origin for chemo- and photolithoautotrophic arsenite oxidation in anoxic environments. Consistent with this, we show that AioA diversified from the assimilatory nitrate reductase lineage, rather than constituting a primordial enzyme present in LUCA.
Results and discussion
Systematics and topology of DMSOR members
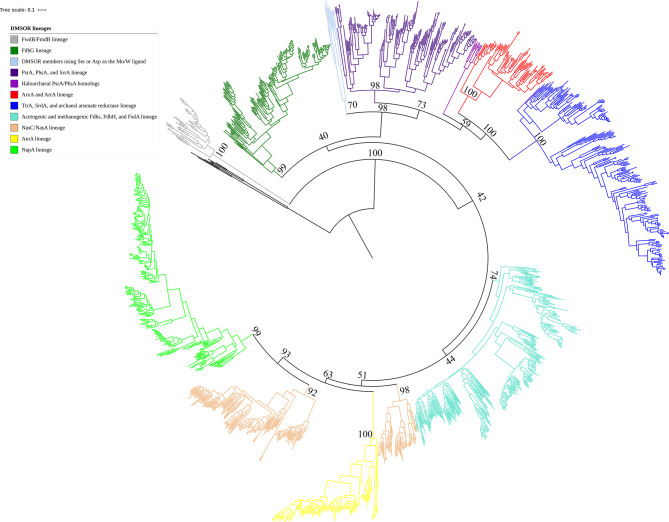
We sampled a total of fifteen physiologically distinct enzymes from the DMSOR family (Table 1) to reconstruct a maximum likelihood analysis using RAxML41. We produced phylogenies using untrimmed (Figs. 1, 2) and trimmed (Fig. S1) protein sequences. The phylogenies were rooted at the midpoint, which independently positioned the aldehyde:ferredoxin family as the root. Both untrimmed and trimmed phylogenies robustly supported positioning the formylmethanofuran dehydrogenase subunit B (FwdB for homologs containing W and FmdB for homologs containing Mo) within the tree topology as the most ancestral DMSOR lineage with a bootstrap support of 100. FwdB/FmdB is a member of a staggeringly complex multi-subunit formylmethanofuran dehydrogenase that was observed either as a dimer of six heterohexameric subunits or a tetramer of six heterohexameric subunits in a recently reported crystal structure42. While formylmethanofuran dehydrogenase catalyzes the reduction of CO2 and methanofuran to formylmethanofuran, the first step in hydrogenotrophic methanogenesis, structural data revealed that the FwdB/FmdB subunit specifically reduces CO2 to formate using ferredoxin as a physiological electron donor42. The crystal structures additionally confirmed that FwdB/FmdB harbors an N terminal [4Fe-4S] cluster in addition to the Mo/W-bisPGD cofactor, with the Mo/W atom coordinated by either a Sec or Cys residue. The subunit is composed of three domains analogous to the first three domains of formate dehydrogenases and formate dehydrogenase catalytic subunits. An additional subunit in the complex, FwdD/FmdD bears structural similarity to the fourth domain of these DMSOR members.
Table 1.
Enzyme lineages included in the phylogenetic analysis, their function, cellular localization, and Mo/W ligand.
| Mo/W-bisPGD catalytic subunit | DMSOR family lineage | Substrate | Function | Localization | Mo/W ligand |
|---|---|---|---|---|---|
| Polysulfide reductase (PsrA) | PsrA/PhsA/SrrA | Sn2− | Reduces polysulfide in anaerobic respiration | Periplasm | Cys |
| Thiosulfate reductase (PhsA) | PsrA/PhsA/SrrA | S2O32− | Reduces thiosulfate in anaerobic respiration | Periplasm | Cys |
| Selenite reductase (SrrA) | PsrA/PhsA/SrrA | SeO32− | Reduces selenite in anaerobic respiration | Periplasm | Cys |
| Arsenite oxidase (ArxA) | ArxA/ArrA | AsO33− | Oxidizes arsenite as an electron donor in anaerobic respiration or anoxygenic photosynthesis | Periplasm | Cys |
| Arsenate reductase (ArrA) | ArxA/ArrA | AsO43− | Reduces arsenate in anaerobic respiration | Periplasm | Cys |
| Tetrathionate reductase (TtrA) | TtrA/SrdA/archaeal arsenate reductase | S4O62− | Reduces tetrathionate in anaerobic respiration | Periplasm | Cys |
| Selenate reductase (SrdA) | TtrA/SrdA/archaeal arsenate reductase | SeO42− | Reduces selenate in anaerobic respiration | Periplasm | Cys |
| Archaeal arsenate reductase | TtrA/SrdA/archaeal arsenate reductase | AsO43− | Reduces arsenate in anaerobic respiration in some archaea | Periplasm | Cys |
| Formylmethano-furan dehydrogenase (FwdB/FmdB) | ? | CO2 | Reduces CO2 to formate in hydrogen-otrophic methano-genesis | Cytoplasm | Sec/Cys |
| Formate dehydrogenase N (FdhG) | FdhG | HCOO−1 | Oxidizes formate as an electron donor in anaerobic respiration | Periplasm | Sec/Cys |
| NAD-dependent formate dehydrogenase | ? | CO2 | Reduces CO2 to formate during acetogenesis | Cytoplasm | Sec/Cys |
| F420-dependent formate dehydrogenase | ? | HCOO−1 | Oxidizes formate to CO2 during hydrogeno-trophic methano-genesis | Cytoplasm | Sec/Cys |
| Formate hydrogen lyase (FdhH) | ? | HCOO−1 | Oxidizes excess formate to carbon dioxide during fermentative growth | Cytoplasm | Sec/Cys |
| Arsenite oxidase (AioA) | AioA | AsO33- | Oxidizes arsenite as an electron donor in aerobic respiration and anoxygenic photosynthesis | Periplasm | No ligand |
| Assimilatory nitrate reductase (NasC/NasA) | NasC/NasA | NO3− | Reduces nitrate to nitrite for assimilation into macro-molecules | Cytoplasm | Cys |
| Periplasmic nitrate reductase (NapA) | NapA | NO3− | Reduces nitrate to nitrite, can fulfill various physiological functions, including respiration, redox homeostasis, and assimilation | Periplasm | Cys |
Question marks denote that particular DMSOR representatives have not previously been incorporated into phylogenetic analyses, thus their position within the family has not yet been resolved.
Figure 1.
Maximum likelihood phylogeny of 1,568 DMSOR family protein sequences. All sequences came from cultured organisms with sequenced genomes. The lineage associated with each clade is indicated in the figure. The scale bar refers to the number of amino acid substitutions per site. The bootstrap support for crucial nodes in our phylogeny is provided in text.
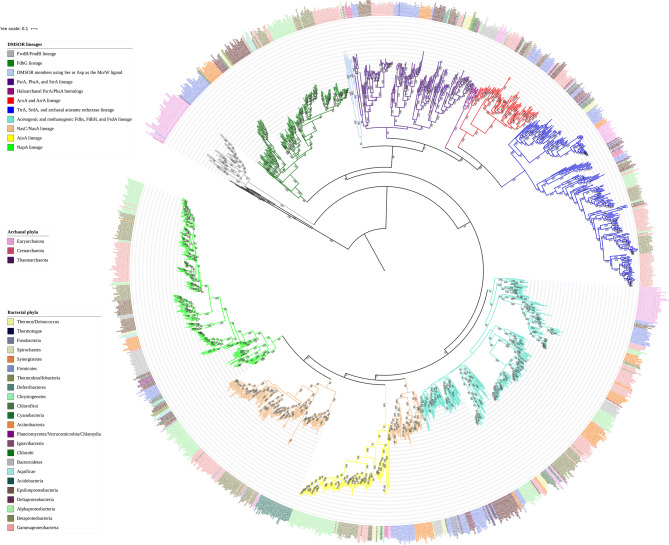
Figure 2.
This is the same phylogeny provided in Fig. 1 with a shorthand code at each branch in the tree indicating the organism from whose genome the protein homolog was found, along with the predicted lineage of the putative DMSOR based off sequence homology to and similar operon organization with the query sequence. The key to each organism’s code is found in the Supplemental Information. The name used for each protein lineage is consistent with the rest of the text. The phylum level affiliation of each organism is indicated by the color of the text of each code name as described in the figure legend. The scale bar refers to the number of amino acid substitutions per site. Bootstrap support for all nodes is denoted in text at each respective node.
The robust placement of FwdB/FmdB as a basal lineage to formate dehydrogenases suggests that the fusion of the FwdB/FmdB and FwdD/FmdD subunits was essential for the diversification of these lineages from the DMSOR family. Formate dehydrogenases have traditionally been classified into three groups. The first group comprises a collection of periplasmic formate dehydrogenases that are canonically expressed during respiratory growth on nitrate and function to oxidize formate to CO2. These include the FdhG catalytic subunits of FdhN and another formate dehydrogenase (FdhO) expressed in Escherichia coli during respiratory growth on nitrate in low O2 concentrations43. The FdhN complex has been studied extensively (e.g.,44,45) but the evolutionary relationship between the FdhG catalytic subunits of FdhO and FdhN is unclear. The similar operon organization of the FdhN and FdhO complexes and high sequence identity (~ 75%) of the FdhG catalytic subunits14 suggest that these complexes are closely related. Consistent with this traditional taxonomy, we found that putative FdhG homologs did indeed form a coherent monophyletic clade with a robust bootstrap support of 99 in the untrimmed phylogeny and 98 in the trimmed phylogeny.
The remaining formate dehydrogenases are classified into two distinct groups, with one group comprising formate hydrogen lyases (FdhH) that functions as a member of a multi-subunit complex to reduce excess formate to CO2 and H2 during fermentative growth43. FdhH from E. coli has been extensively studied46,47. The other group comprises a disparate collection of NAD+-dependent formate dehydrogenases (FdsA) found in aerobic bacteria that function to oxidize excess formate to CO2, and, like FdhH, does not appear to be linked to energy conservation48,49. FdsA of Cupriavidus necator (formerly Ralstonia eutropha), however, can catalyze the reverse reaction (CO2 reduction to formate) in vitro50. Curiously, this taxonomy neglects the formate dehydrogenases that function in acetogenesis and hydrogenotrophic methanogenesis. The acetogenic formate dehydrogenases function, like the FwdB/FmdB subunit, to reduce CO2 to formate in the first step of acetogenesis, whereas the methanogenic formate dehydrogenases function to oxidize formate to CO2 to provide a source of CO2 for hydrogenotrophic methanogenesis in the presence of formate51. Our phylogeny demonstrates that classifying formate dehydrogenases by their biochemical and physiological function does not accurately reflect their evolutionary history. Acetogenic and methanogenic formate dehydrogenases, FdhH, and FdsA homologs all clustered together in a monophyletic clade with bootstrap support of 74 in the untrimmed phylogeny and 79 in the trimmed phylogeny. Thus, these various physiologically diverse formate dehydrogenases constitute members of a single evolutionary lineage.
The only substantive difference between the untrimmed and trimmed phylogenies concerns the exact positioning of the FdhG subunit within the DMSOR family. The untrimmed phylogeny positions the FdhG lineage within a clade of DMSOR members that include the respiratory dimethyl sulfoxide reductase (DmsA), respiratory nitrate reductase (NarG), PsrA/PhsA/SrrA, ArxA/ArrA, and TtrA/SrdA/archaeal arsenate reductase lineages that interact with the membrane quinone pool during anaerobic respiration using the canonical subunits identified by Rothery et al.13. The other formate dehydrogenase lineage clusters within a clade comprising assimilatory nitrate reductase catalytic subunits (NasA/NasC), AioA, and NapA. None of the members of this clade are associated with the canonical four [4Fe-4S] cluster electron transfer or membrane anchor subunits. While the most basal members of this clade, the acetogenic and methanogenic formate dehydrogenases, participate in energy conservation, the FdhH and FdsA homologs no longer retain this function, and the NasA/NasC subunits function exclusively in nitrogen assimilation52. While NapA can function as a respiratory nitrate reductase, Nap can also reduce nitrate for assimilation and redox homeostasis16.
This topology is consistent with the evolutionary principle of parsimony, in that it suggests that the association of DMSOR members with characteristic electron transfer and membrane anchor subunits arose once early in the evolution of DSMORs and co-evolved with these representatives through multiple diversification events. However, the trimmed phylogeny offers a topology wherein the FdhG lineage clusters with the various formate dehydrogenases, NasC/NasA, AioA, and NapA. The trimmed phylogeny also positions the FdhG lineage basal to the other formate dehydrogenases, which would suggest that components of the anaerobic electron transport chain evolved before enzyme mediated catalysis for each step of the pathways for acetogenesis and methanogenesis had fully evolved. Bootstrap support for both scenarios is weak, and thus both topologies are provided for consideration.
Both untrimmed and trimmed phylogenies position the PsrA/PhsA/SrrA, ArxA/ArrA, and TtrA/SrdA lineages as a monophyletic clade sister to DMSOR members utilizing a Ser and Asp residue to coordinate the Mo/W atom of the Mo/W-bisPGD cofactor. These members include NarG, DmsA, respiratory chlorate reductase catalytic subunit (ClrA), and the respiratory selenate reductase of Thauera selenatis5,14. The clade comprising the PsrA/PhsA/SrrA, ArxA/ArrA, and TtrA/SrdA/archaeal arsenate reductase lineages all utilize a Cys residue to coordinate the Mo/W atom, and also share oxidoreductase activity toward a diverse array of sulfur intermediates and arsenic and selenium oxyanions. Bootstrap support for clustering these various DMSOR members as a coherent monophyletic clade separate from the DMSOR members using Ser and Asp as Mo/W ligands is 98 in the untrimmed phylogeny and 97 in the trimmed phylogeny. The strong bootstrap support for a substantial, deeply branched clade of DMSOR members that specifically exploit chalcophiles as electron donors and terminal electron acceptors in anaerobic respiration is a powerful demonstration of the strong selective force that chalcophile utilization has exerted on the diversification of this family.
The NasC/NasA, AioA, and NapA catalytic subunits cluster with the various formate dehydrogenases, and both tree topologies suggest that this formate dehydrogenase lineage is ancestral to the NasC/NasA lineage, though bootstrap support for this is weak. However, the diversification of the various formate dehydrogenases from the family clearly predated the diversification of the archaeal and bacterial domains from LUCA, as the bacterial formate dehydrogenases form a single monophyletic clade from the substantial number of archaeal representatives (Fig. 2). This is certainly not the case for NasC/NasA. There are only two archaeal homologs present in the NasC/NasA lineage, and both were clearly inherited via lateral gene transfer, given their derived position within the lineage. The NasC/NasA and NapA catalytic subunits coordinate the Mo/W atom using a Cys residue, while the AioA subunit is the only known member of this family that doesn’t use an amino acid ligand for cofactor coordination5,14. The assimilatory and periplasmic nitrate reductase sequences were incorporated into our analysis when it became apparent that we could not precisely root AioA sequences within the DSMOR family without them.
Our phylogenetic analysis revealed that these sequences were required because the AioA lineage likely diversified from NasC/NasA, as neither the untrimmed nor trimmed phylogenies could position AioA as a separate lineage from NasC/NasA with robust bootstrap support. This is in striking contrast to the NapA lineage, which clearly formed a monophyletic clade separate from NasC/NasA with a bootstrap support of 93 in the untrimmed and trimmed phylogenies. This close evolutionary relationship between NasC/NasA and AioA is quite surprising, given that such a relationship has never previously been postulated. Nevertheless, this is consistent with the physiological function of these DMSORs, which are utilized predominantly in oxic and suboxic environments. Further, this would argue for a later appearance of the NasC/NasA/AioA lineage as ammonium was the abundant form of nitrogen in the Archean53, obviating the need for an assimilatory nitrate reductase. A number of physiological studies have established that assimilatory nitrate reductase is expressed in both bacteria and archaea exclusively under oxic and suboxic conditions54-58, and in cyanobacteria assimilatory nitrate reduction occurs concomitantly with the evolution of O2 during oxygenic photosynthesis59. Likewise, AioA functions predominantly to oxidize arsenite during aerobic respiration, even in arsenite oxidizing representatives occupying the deepest branches of the lineage (Fig. 3), including representatives from the Haloarchaea60 and the Aquificales61. In fact, the only demonstration of AioA catalyzing anaerobic arsenite oxidation comes from Chloroflexus aurantiacus, which can exploit arsenite as a source of reducing equivalents during anoxygenic photosynthesis22. The potential for phototrophic arsenite oxidation exists also in certain strains of Chlorobium limnicola and Chlorobium phaeobacteroides as indicated by the presence of Aio homologs in their genomes. Many strains of Chloroflexus and Chlorobium species, however, lack Aio, indicating this ability is highly variable and a trait acquired through horizontal gene transfer16.
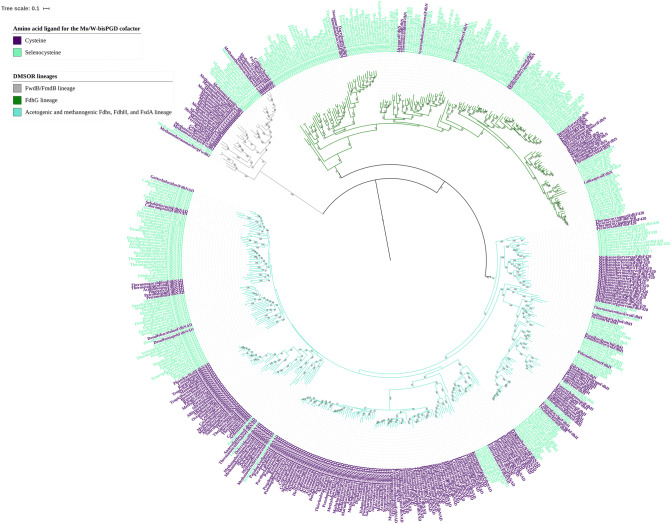
Figure 3.
Sub-pruned portion of the maximum likelihood phylogeny showing the FwdB/FmdB, FdhG, and various formate dehydrogenase lineages. The clade for each lineage is indicated by the color of the branches. The text labels for each protein representative in the family is color coded by whether the Mo or W atom is coordinated by a Cys or a Sec residue. The scale bar refers to the number of amino acid substitutions per site. Bootstrap support for all nodes ≥ 60 is denoted in text at the respective node.
Relative ordination of DMSOR diversifications
Our phylogenies cannot assess when, in the evolutionary record, lineages of DMSOR family enzymes diversified absent geochemical evidence for these metabolisms in dated lithologies of the fossil record. Nonetheless, the phylogeny can be used to determine whether lineages could have plausibly diverged from the DMSOR family prior to the divergence of LUCA into the Bacteria and Archaea domains. This can be assessed by determining whether homologs from the Bacteria form coherent monophyletic clades distinct from archaeal homologs within lineages of these enzymes. The two formate dehydrogenase lineages clearly fit this criterion (Fig. 2), particularly the lineage of physiologically diverse formate dehydrogenases which has a substantial number of representatives from the Archaea. It is also possible that the PsrA/PhsA/SrrA lineage diversified from the DMSOR family prior to LUCA, given that archaeal PsrA/PhsA/SrrA representatives form a coherent monophyletic clade. However, these representatives cluster with a monophyletic group of bacterial PsrA/PhsA/SrrA homologs. All of these archaeal homologs come from hyperthermophiles, and the most deeply branched bacterial homolog comes from the thermophilic bacterium Thermosyntropha lipolytica, which was isolated in a syntrophic coculture with a methanogenic archaeon62. We contend that it is likely that the PsrA/PhsA/SrrA lineage diversified from the family before the diversification of LUCA into the bacterial and archaeal domains of life. Subsequently, a lateral gene transfer event occurred between the domains, explaining the extensive vertical inheritance through many phyla observed in bacterial representatives in this clade.
The positioning of the FwdB/FmdB subunit basal to the two formate dehydrogenase lineages necessitates that this lineage diversified from the family before the diversification of the two prokaryotic domains, despite the absence of bacterial FwdB/FmdB representatives. The deep antiquity of the FwdB/FmdB and acetogenic and methanogenic formate dehydrogenase lineages is fully consistent with the hypothesis that hydrogenotrophic methanogenesis and acetogenesis represent the remnants of the earliest form of energy conservation, and that the Wood–Ljungdahl pathway represents the ancestral mechanism for carbon fixation4,32,63. Our phylogeny indicates that LUCA was additionally capable of energy conservation using formate as an electron donor and sulfur intermediates such as polysulfide as a terminal electron acceptor in a membrane-embedded electron transport chain.
It is curious to note that members of the three oldest DMSOR lineages have been shown to utilize Sec to coordinate the Mo/W atom of the Mo/W-bisPGD cofactor. This includes a number of representatives from the FwdB/FmdB lineage64-66, acetogenic NAD+-dependent formate dehydrogenases67,68, methanogenic F420-dependent formate dehydrogenases69,70, FdhH from E. coli46, and a number of FdhG catalytic subunits44,45,71. Indeed, formate dehydrogenases represent the most common selenoproteins in the genomes of Sec utilizing bacteria and archaea72-74. It has recently been established that the ability to synthesize and incorporate Sec into oxidoreductases was likely a feature of the metabolism of LUCA32,75. However, our phylogenetic analyses are the first to investigate the evolutionary history of selenoproteins (the oxidoreductases that incorporate a Sec residue) themselves. It was therefore of interest to determine if Sec use within these lineages constitutes the ancestral state. We mapped the distribution of Sec residues amongst the FwdB/FmdB, formate dehydrogenase, and FdhG lineages (Fig. 3). We found Sec utilizing homologs in each of the three lineages in deeply branched positions within the tree topology. Thus, Sec use either evolved independently in these three lineages early in their diversification or, as is more likely, Sec utilization represents the ancestral state for the earliest DMSOR representatives with the loss of Sec occurring independently at multiple points throughout all three lineages. It should be said that an ancestral sequence reconstruction analysis could statistically assess the likelihood that Sec represents the ancestral state for Mo/W coordination, but bioinformatic tools that recognize and analyze Sec residues have yet to be developed.
The ArxA/ArrA, TtrA/SrdA/archaeal arsenate reductase, NasC/NasA, AioA, and NapA lineages all unambiguously diversified from the DMSOR family after the bacterial and archaeal domains had diverged. The association of the NasC/NasA and AioA lineages with aerobic metabolism indicates that these lineages did not diversify from the family until a reservoir of O2 had been established on Earth surface environments. Additionally, representatives from both the AioA and the TtrA/SrdA/archaeal arsenate reductase lineages occupy some of the most recently derived branches in the tree topology. Our results starkly contradict previous findings that TtrA/SrdA/archaeal arsenate reductase13 and AioA1,24-26 represent lineages that had diversified from the family before the two prokaryotic domains had been established.
In the case of the TtrA/SrdA/archaeal arsenate reductase lineage, we demonstrate conclusively that the presence of archaeal homologs constitutes multiple lateral gene transfer events between Bacteria and Archaea. For the AioA lineage, the most ancestral homolog in the tree topology is from B. oryziterrae, the only member of the Firmicutes we found whose genome contained an AioA homolog, with a bootstrap support of 100. The second most ancestral AioA representatives comes from the haloarchaea, not the hyperthermophilic archaea, with a bootstrap support of 81. This is in contrast to the phylogenies of genuinely primordial enzymes, in which homologs from thermophilic clostridia and hyperthermophilic members of Archaea constitute the basal lineages of enzyme representatives for each domain, consistent with the likely hydrothermal origin of LUCA32. This is further buttressed by the basal position of a number of NasC/NasA representatives with respect to AioA, given that the diversification of NasC/NasA is unambiguously younger than the diversification of life into two domains.
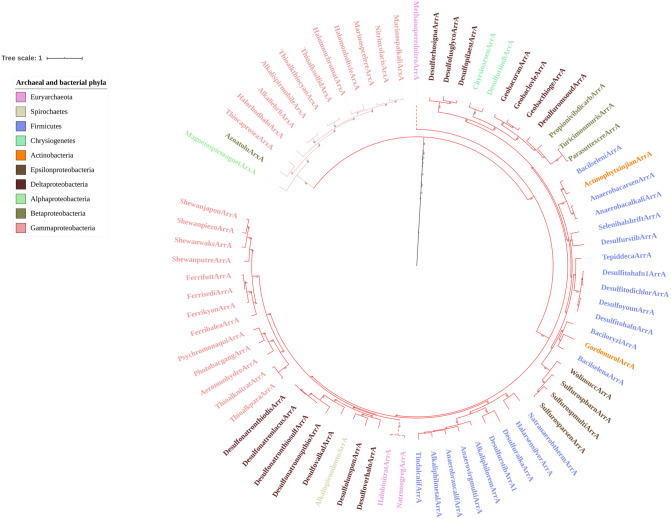
The topology of the ArxA/ArrA lineage (Fig. 4) provides substantial evidence that this lineage diversified from the DMSOR family before the diversification of the AioA lineage. It has previously been proposed that ArxA homologs constitute a distinct monophyletic clade within the broader ArxA/ArrA lineage20. Our phylogeny decisively demonstrates that not only is this robustly supported, with a bootstrap support of 100, but that the ArxA clade is ancestral to the ArrA clade. This further undermines the hypothesis that AioA represents a primordial DMSOR member in LUCA that mediated an Archean arsenic biogeochemical cycle consisting exclusively of anaerobic arsenite oxidation. It has consistently been proposed that respiratory arsenate reduction was the initial selective pressure for the diversification of the ArxA/ArrA lineage as the Earth’s atmosphere began to accumulate O2 over the course of the Neoarchean and GOE1,25,26. Thus, ArrA should predate the diversification of the ArxA clade if anaerobic arsenite oxidation was initially mediated by AioA. The basal position of the ArxA clade instead argues that autotrophic arsenite oxidation was not a feature of LUCA, but rather evolved sometime later in the Archean. Furthermore, the greater ancestry of the ArxA/ArrA lineage with respect to AioA indicates that respiratory arsenate reduction was also widespread on the Archean Earth.
Figure 4.
Sub-pruned portion of the maximum likelihood phylogeny showing the ArxA/ArrA lineage. The putative ArxA homologs are identified both by the text label and a lighter shade of red for the branches comprising the clade. The phylum level affiliation of each organism is indicated by the color of the text of each code name as described in the figure legend. The scale bar refers to the number of amino acid substitutions per site. Bootstrap support for all nodes ≥ 60 is denoted in text at the respective node.
Finally, it should be noted that NasC/NasA was the only lineage in our analysis that did not form a monophyletic clade, and it could be supposed that one clade comprises NasC/NasA, whereas the other clade constitutes members misidentified as NasC/NasA. However, both clades contain biochemically and genetically characterized assimilatory nitrate reductases that we used as queries to collect sequences. This includes NasC from B. subtilis76 from the basal clade and NarB/NasA from Synechcoccus elongatus PCC 794277 and Azotobacter vinelandii78 from the younger clade. Another potential explanation for the failure of NasC/NasA sequences to form a monophyletic clade could come from the observation that assimilatory nitrate reductases are found to have two different molecular weights, and thus some members of the lineage have sequence insertions in the primary sequence. The assimilatory nitrate reductase catalytic subunits of B. subtilis and S. elongatus PCC 7,942, for instance, are approximately 710 amino acids long and weigh approximately 70 kDa76,79. Proteobacterial and haloarchaeal assimilatory nitrate reductase catalytic subunits are approximately 950 amino acids long and weigh approximately 95 kDa55,78,80,81. A similar phenomenon has been reported for the NapA lineage, with the Desulfovibrio desulfuricans NapA homolog having a molecular weight of 70 kDa and the Rhodobacter sphaeroides NapA homolog having a molecular weight of 90 kDa16. These differences in the primary sequence of the assimilatory and periplasmic nitrate reductase catalytic subunits, however, are not associated with sequence insertions at specific regions of the primary sequence, but vary considerably between different taxa, and none of these sequence insertions occur at regions of the catalytic subunits essential for catalysis (e.g., the N-terminal [4Fe–4S] cluster, the Mo/W-bisPGD binding sites, or substate binding funnel). Thus, differences in the sequence length of NasA/NasC and NapA homologs are unlikely to explain the presence of two distinct monophyletic clades of NasA/NasC representatives. Regardless, the cyanobacterial NarB/NasA homologs cluster with the proteobacterial NasA homologs, not the B. subtilis NasC homologs, and the monophyly of NapA homologs was robustly supported. Finally, our phylogenetic analysis is fully consistent with a previous study of the evolution of the NapA lineage16, underscoring that NapA originated from NasC/NasA and that the most ancient NapA representatives come from the Deltaproteobacteria, Bacteroidetes, and Firmicutes phyla.
Implications for future evolutionary study of ancient molybdo- and tungsto-enzymes
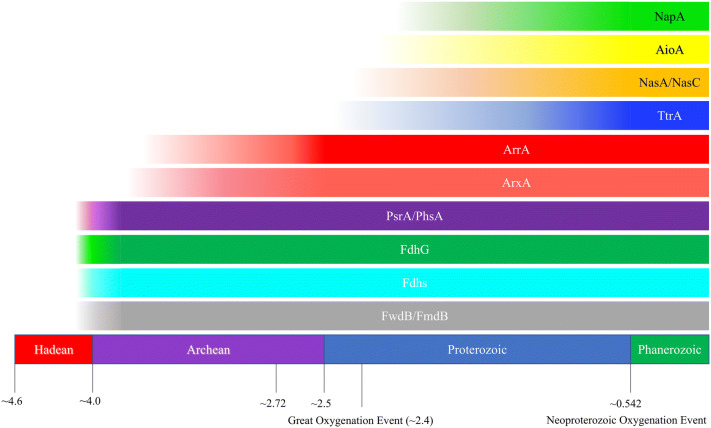
Our work has presented the first comprehensive phylogenetic analysis of members of the DMSOR family using maximum likelihood methods and appropriate outgroups to produce rooted phylogenies. This analysis has provided the first robust relative ordination of when specific lineages of DMSORs diversified from the family through geologic time (Fig. 5). We demonstrate that a sufficient phylogenetic signal exists to identify the oldest lineage of DMSOR members (FwdB/FmdB), and to provide a relative ordination for when lineages diversified from the family. Achieving this requires a substantial amount of sequence data for each lineage, as well as the most statistically rigorous phylogenetic tools available. We hope this work establishes conclusively that neighbor-joining methods are inadequate for studying this ~ 3.7 Gya assemblage of enzymes, and that appropriate roots are available for establishing a relative ordination of their diversification. Such analyses are essential for understanding the evolution of global biogeochemical cycles. We illustrate, for example, that Sec has been essential to the origin and evolution of crucial reactions in the global carbon biogeochemical cycle, including acetogenesis, methanogenesis, and carbon fixation. This underscores the deep antiquity of the global selenium biogeochemical cycle and emphasizes the importance of selenium to nascent life in catalyzing central reactions in energy conservation and autotrophic growth. We also show that while the arsenic biogeochemical cycle did not originate from LUCA, the Archean arsenic biogeochemical cycle was far more complex than originally conceived, with arsenite oxidation and arsenate reduction likely sustaining microbial communities. While our focus here was primarily on enzymes using chalcophilc elements as substrates, future analyses of other DMSOR members may further illuminate how organisms and biogeochemical cycles have co-evolved as the Earth’s oxidation state has increased over time.
Figure 5.
Inferred time of divergence of lineages from the DMSOR family based on our phylogenetic analysis as well as consideration of the biogeochemistry of DMSOR substrates and the physiological role (e.g., function in aerobic respiration and O2-dependence) of DMSOR lineages positioned over the geologic time-scale.53 The arsenic-containing Tumbiana stromatolites are found at ~ 2.72 Ga.17.
Methods
Sequence selection
Protein representatives from DMSOR family lineages where the function of the enzyme was previously elucidated by biochemical, genetic, or structural methods were selected to be BLAST queries to construct a comprehensive library of sequences. For the families we analyzed, these included PsrA from W. succinogenes35, PhsA from S. enterica serovar Typhimurium36, SrrA from B. selenitireducens37, ArrA from Chrysiogenes arsenatis82 and B. selenitireducens83, ArxA from A. ehrlichii19, TtrA from S. enterica serovar Typhimurium38, SrdA from B. selenatarsenatis39, the arsenate reductase of P. aerophilum40, AioA from A. faecalis84 and Rhizobium sp. NT-2685, FmdB from Methanosarcina barkeri86, FmdB and FwdB from Methanothermobacter wolfei87 FdhG from E. coli44, D. gigas45, and D. desulfuricans71, FdhH from E. coli46, NAD-dependent formate dehydrogenases from Moorella thermoacetica88 and Peptoclostridium acidaminophilum68, and the F420-dependent formate dehydrogenases of Methanococcus maripaludis70 and M. vannielli89. These queries were blasted against the NCBI database using the DELTA-BLAST tool from the National Center for Biotechnology Information (NCBI) web server90.
Candidates were selected if the sequence came from an organism that had been isolated in a pure culture or defined co-culture, aligned over at least 95% of the query with an amino acid identify of at least 30%, if the sequence length was consistent with the sequence length of the query, and if the primary sequence contained motifs considered characteristic of the enzyme family (e.g., a twin-arginine translocation motif, a [4Fe–4S] or [3Fe–4S] cluster binding motif, and a Mo/W-bisPGD binding motif). Candidates were additionally screened using the Integrated Microbial Genomics (IMG) platform91 to view the genomic context of the putative homolog. Sequences were retained only if the primary sequences between the NCBI database and IMG database were conserved, and if the operon contained other subunits consistent with the operon structure described in model organisms previously (e.g., a four [4Fe–4S] cluster containing protein, a [2Fe–2S] Rieske protein, a membrane anchor).
Additional well-characterized representatives from other DMSOR family lineages were included in the analysis to provide a fuller context of where the analyzed lineages fit in the larger family. These proteins included NarG from E. coli92, P. aerophilium93, and Thermus thermophilus94, DmsA from E. coli9, chlorate reductase (ClrA) from Ideonella dechloratans95, SerA from T. selenatis96, dimethyl sulfide dehydrogenase (DdhA) from Rhodovulum sulfidophilum97, TorA from E. coli98 and Shewanella massilia99, and BisC from E. coli100. Well-characterized members of the aldehyde:ferredoxin reductase family were included as outgroups. These proteins included the aldehyde ferredoxin oxidoreductases (AORs) of M. thermoacetica101 and Pyrococcus furiosus102, the formaldehyde ferredoxin oxidoreductases (FORs) of Pyr. furiosus103 and Thermococcus litoralis104, and the glyceraldehyde-3-phosphate ferredoxin oxidoreductases (GAPORs) of Pyr. furiosus105, M. maripaludis106, and P. aerophilum107.
Verification of selenocysteine residues in DMSOR members
Several DMSOR members, including the formyl methanofuran dehydrogenase B subunit108, formate dehydrogenase G subunit44,45, and formate dehydrogenase H46, have been shown to coordinate the Mo/W atom of the Mo/W-bisPGD co-factor with a Sec residue. This residue is located at a specific position within the primary sequence of these proteins and shares the active site of the subunits with other conserved amino acid residues (such as a neighboring His residue). These well-characterized representatives were used as query sequences in DELTA-BLAST searches for putative FmdB/FwdB, FdhG, and FdhH homologs. A putative member of these lineages was determined to be a Sec-containing homolog if the Sec residue aligned with the Sec-containing query sequence in multiple sequence alignments. Furthermore, neighboring conserved residues mentioned in X-ray crystallographic studies also had to align with query sequences before we confidently identified a putative DMSOR member as a selenoprotein. No homologs were found over the course of DELTA-BLAST searches that had more than one Sec residue in the primary sequence, or with a Sec residue that did not align with Sec residue of the active site of the well-characterized query sequences.
Phylogenetic analysis
A total of 1,568 sequences were included for the phylogenetic analysis. The sequences were aligned using the online platform of MAFFT for large-scale sequence alignments109 using the G-INS-1 method. Untrimmed alignments were analyzed directly. Trimmed alignments were prepared using the trimal tool110. The alignments were trimmed such that all columns with gaps in more than 20% of DMSOR sequences, or with a similarity score below 0.001 were omitted, with the caveat that 60% of the columns be conserved for the analysis. The amino acid selection model that best fit our data was chosen using the ModelFinder program111, which compared 170 amino acid selection models. The ModelFinder program found that the LG4M amino acid substitution model112, which is comprised of four separate gamma distributed amino acid substitution matrices to better capture heterogeneities in mutation rates at different sites in protein alignments, was the best model for both the untrimmed and trimmed alignments. Maximum likelihood phylogenies were constructed using RAxML Version 841 using the LG4M model to find the best scoring tree. The phylogenetic analysis consisted of a rapid bootstrap search followed by a search for the most likely tree topology. The autoMRE criterion was used to determine when bootstraps had converged sufficiently to use to find the most likely topology, rather than specifying a certain number of bootstraps. The model selection and maximum likelihood analyses were performed using the CIPRES gateway portal113. All phylogenies were visualized using Interactive Tree of Life (iTOL) platform114.
Supplementary information
Acknowledgements
We thank the Bayer School for Natural and Environmental Sciences for support of M.W., N.J.K., and A.M.A.Z.
Author contributions
M.W. was responsible for sequence retrieval, verification, alignments, phylogenetic analyses, and tree construction with additional assistance from A.M.A.Z. and N.J.K. J.J. provided the expertise in the phylogenetic analyses and robustness of the data, while P.B. and R.S.O. contributed their expertise in molybdoenzymes and biogeochemistry respectively. M.W. and J.F.S. were primarily responsible for writing and editing the manuscript, with all authors contributing to the final editing.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-67892-9.
References
- 1.Schoepp-Cothenet B, et al. The ineluctable requirement for the trans-iron elements molybdenum and/or tungsten in the origin of life. Sci. Rep. 2012;2:263. doi: 10.1038/srep00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nitschke W, Russell MJ. Beating the acetyl coenzyme A-pathway to the origin of life. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2013;368:20120258. doi: 10.1098/rstb.2012.0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schoepp-Cothenet B, et al. On the universal core of bioenergetics. Biochim. Biophys. Acta. 2013;1827:79–93. doi: 10.1016/j.bbabio.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Sousa FL, et al. Early bioenergetic evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20130088. doi: 10.1098/rstb.2013.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grimaldi S, Schoepp-Cothenet B, Ceccaldi P, Guigliarelli B, Magalon A. The prokaryotic Mo/W-bisPGD enzymes family: A catalytic workhorse in bioenergetic. Biochim. Biophys. Acta. 2013;1827:1048–1085. doi: 10.1016/j.bbabio.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 6.Abin CA, Hollibaugh JT. Transcriptional response of the obligate anaerobe Desulfuribacillus stibiiarsenatis MLFW-2T to growth on antimonate and other terminal electron acceptors. Environ. Microbiol. 2019;21:618–630. doi: 10.1111/1462-2920.14503. [DOI] [PubMed] [Google Scholar]
- 7.Shi L-D, et al. Multi-omics reveal various potential antimonate reductases from phylogenetically diverse microorganisms. Appl. Microbiol. Biotechnol. 2019;103:9119–9129. doi: 10.1007/s00253-019-10111-x. [DOI] [PubMed] [Google Scholar]
- 8.Bilous PT, Cole ST, Anderson WF, Weiner JH. Nucleotide sequence of the dmsABC operon encoding the anaerobic dimethylsulphoxide reductase of Escherichia coli. Mol. Microbiol. 1988;2:785–795. doi: 10.1111/j.1365-2958.1988.tb00090.x. [DOI] [PubMed] [Google Scholar]
- 9.Weiner JH, MacIsaac DP, Bishop RE, Bilous PT. Purification and properties of Escherichia coli dimethyl sulfoxide reductase, an iron-sulfur molybdoenzyme with broad substrate specificity. J. Bacteriol. 1988;170:1505–1510. doi: 10.1128/jb.170.4.1505-1510.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cammack R, Weiner JH. Electron paramagnetic resonance spectroscopic characterization of dimethyl sulfoxide reductase of Escherichia coli. Biochemistry. 1990;29:8410–8416. doi: 10.1021/bi00488a030. [DOI] [PubMed] [Google Scholar]
- 11.Schindelin H, Kisker C, Hilton J, Rajagopalan KV, Rees DC. Crystal structure of DMSO reductase: Redox-linked changes in molybdopterin coordination. Science. 1996;272:1615–1621. doi: 10.1126/science.272.5268.1615. [DOI] [PubMed] [Google Scholar]
- 12.Schneider F, et al. Crystal structure of dimethyl sulfoxide reductase from Rhodobacter capsulatus at 1.88 Å resolution. J. Mol. Biol. 1996;263:53–69. doi: 10.1006/jmbi.1996.0555. [DOI] [PubMed] [Google Scholar]
- 13.Rothery RA, Workun GJ, Weiner JH. The prokaryotic complex iron–sulfur molybdoenzyme family. Biochim. Biophys. Acta. 2008;1778:1897–1929. doi: 10.1016/j.bbamem.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Hille R, Hall J, Basu P. The mononuclear molybdenum enzymes. Chem. Rev. 2014;114:3963–4038. doi: 10.1021/cr400443z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McEwan AG, Ridge JP, McDevitt CA, Hugenholtz P. The DMSO reductase family of microbial molybdenum enzymes: Molecular properties and role in the dissimilatory reduction of toxic elements. Geomicrobiol. J. 2002;19:3–21. [Google Scholar]
- 16.Sparacino-Watkins C, Stolz JF, Basu P. Nitrate and periplasmic nitrate reductases. Chem. Soc. Rev. 2014;43:676–706. doi: 10.1039/c3cs60249d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sforna MC, et al. Evidence for arsenic metabolism and cycling by microorganisms 2.7 billion years ago. Nat. Geosci. 2014;7:811–815. [Google Scholar]
- 18.Stolz JF, Basu P, Santini JM, Oremland RS. Arsenic and selenium in microbial metabolism. Annu. Rev. Microbiol. 2006;60:107–130. doi: 10.1146/annurev.micro.60.080805.142053. [DOI] [PubMed] [Google Scholar]
- 19.Zargar K, Hoeft S, Oremland R, Saltikov CW. Identification of a novel arsenite oxidase gene, arxA, in the haloalkaliphilic, arsenite-oxidizing bacterium Alkalilimnicola ehrlichii strain MLHE-1. J. Bacteriol. 2010;192:3755–3762. doi: 10.1128/JB.00244-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zargar K, et al. ArxA, a new clade of arsenite oxidase within the DMSO reductase family of molybdenum oxidoreductases. Environ. Microbiol. 2012;14:1635–1645. doi: 10.1111/j.1462-2920.2012.02722.x. [DOI] [PubMed] [Google Scholar]
- 21.Kulp TR, et al. Arsenic(III) fuels anoxygenic photosynthesis in hot spring biofilms from Mono Lake California. Science. 2008;321:967–970. doi: 10.1126/science.1160799. [DOI] [PubMed] [Google Scholar]
- 22.Stolz JF. Gaia and her microbiome. FEMS Microbiol. Ecol. 2017;93:flw247. doi: 10.1093/femsec/fiw247. [DOI] [PubMed] [Google Scholar]
- 23.Oremland RS, Saltikov CW, Wolfe-Simon F, Stolz JF. Arsenic in the evolution of Earth and extraterrestrial ecosystems. Geomicrobiol. J. 2009;26:522–536. [Google Scholar]
- 24.Lebrun E, et al. Arsenite oxidase, an ancient bioenergetic enzyme. Mol. Biol. Evol. 2003;20:686–693. doi: 10.1093/molbev/msg071. [DOI] [PubMed] [Google Scholar]
- 25.Duval S, Ducluzeau A-L, Nitschke W, Schoepp-Cothenet B. Enzyme phylogenies as markers for the oxidation state of the environment: The case of respiratory arsenate reductase and related enzymes. BMC Evol. Biol. 2008;8:206. doi: 10.1186/1471-2148-8-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Lis R, Nitschke W, Duval S, Schoepp-Cothenet B. Arsenics as bioenergetic substrates. Biochim. Biophys. Acta. 2013;1827:176–188. doi: 10.1016/j.bbabio.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 27.Ducluzeau A-L, et al. Was nitric oxide the first deep electron sink? Trends Biochem. Sci. 2009;34:9–15. doi: 10.1016/j.tibs.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 28.Harel A, Häggblom MM, Falkowski PG, Yee N. Evolution of prokaryotic respiratory molybdoenzymes and the frequency of their genomic co-occurrence. FEMS Microbiol. Ecol. 2016;92:187. doi: 10.1093/femsec/fiw187. [DOI] [PubMed] [Google Scholar]
- 29.Edwardson CF, Hollibaugh JT. Metatranscriptomic analysis of prokaryotic communities active in sulfur and arsenic cycling in Mono Lake, California, USA. ISME J. 2017;11:2195–2208. doi: 10.1038/ismej.2017.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–2314. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- 31.Huelsenbeck JP, Crandall KA. Phylogeny estimation and hypothesis testing using maximum likelihood. Annu. Rev. Ecol. Syst. 1997;28:437–466. [Google Scholar]
- 32.Weiss MC, et al. The physiology and habitat of the last universal common ancestor. Nat. Microbiol. 2016;1:16116. doi: 10.1038/nmicrobiol.2016.116. [DOI] [PubMed] [Google Scholar]
- 33.Wu D, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukherjee S, et al. 1,003 reference genomes of bacterial and archaeal isolates expand coverage of the tree of life. Nat. Biotechnol. 2017;35:676–683. doi: 10.1038/nbt.3886. [DOI] [PubMed] [Google Scholar]
- 35.Krafft T, et al. Cloning and nucleotide sequence of the psrA gene of Wolinella succinogenes polysulphide reductase. Eur. J. Biochem. 1992;206:503–510. doi: 10.1111/j.1432-1033.1992.tb16953.x. [DOI] [PubMed] [Google Scholar]
- 36.Heinzinger NK, Fujimoto SY, Clark MA, Moreno MS, Barrett EL. Sequence analysis of the phs operon in Salmonella typhimurium and the contribution of thiosulfate reduction to anaerobic energy metabolism. J. Bacteriol. 1995;177:2813–2820. doi: 10.1128/jb.177.10.2813-2820.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wells M, et al. Respiratory selenite reductase from Bacillus selenitireducens strain MLS10. J. Bacteriol. 2019;201:e00614–e618. doi: 10.1128/JB.00614-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hensel M, Hinsley AP, Nikolaus T, Sawers G, Berks BC. The genetic basis of tetrathionate respiration in Salmonella typhimurium. Mol. Microbiol. 1999;32:275–287. doi: 10.1046/j.1365-2958.1999.01345.x. [DOI] [PubMed] [Google Scholar]
- 39.Kuroda M, et al. Molecular cloning and characterization of the srdBCA operon, encoding the respiratory selenate reductase complex, from the selenate-reducing bacterium Bacillus selenatarsenatis SF-1. J. Bacteriol. 2011;193:2141–2148. doi: 10.1128/JB.01197-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cozen AE, et al. Transcriptional map of respiratory versatility in the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. J. Bacteriol. 2009;191:782–794. doi: 10.1128/JB.00965-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagner T, Ermler U, Shima S. The methanogenic CO2 reducing-and-fixing enzyme is bifunctional and contains 46 [4Fe–4S] clusters. Science. 2016;354:114–117. doi: 10.1126/science.aaf9284. [DOI] [PubMed] [Google Scholar]
- 43.Sawers G. The hydrogenases and formate dehydrogenases of Escherichia coli. Antonie Van Leeuwenhoek. 1994;66:57–88. doi: 10.1007/BF00871633. [DOI] [PubMed] [Google Scholar]
- 44.Jormakka M, Törnroth S, Byrne B, Iwata S. Molecular basis of proton motive force generation: Structure of formate dehydrogenase-N. Science. 2002;295:1863–1868. doi: 10.1126/science.1068186. [DOI] [PubMed] [Google Scholar]
- 45.Raaijmakers H, et al. Gene sequence and the 1.8 Å crystal structure of the tungsten-containing formate dehydrogenase from Desulfovibrio gigas. Structure. 2002;10:1261–1272. doi: 10.1016/s0969-2126(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 46.Boyington JC, Gladyshev VN, Khangulov SV, Stadtman TC, Sun PD. Crystal structure of formate dehydrogenase H: Catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster. Science. 1997;275:1305–1308. doi: 10.1126/science.275.5304.1305. [DOI] [PubMed] [Google Scholar]
- 47.Khangulov SV, Gladyshev VN, Dismukes GC, Stadtman TC. Selenium-containing formate dehydrogenase H from Escherichia coli: A molybdopterin enzyme that catalyzes formate oxidation without oxygen transfer. Biochemistry. 1998;37:3518–3528. doi: 10.1021/bi972177k. [DOI] [PubMed] [Google Scholar]
- 48.Oh JI, Bowien B. Structural analysis of the fds operon encoding the NAD+-linked formate dehydrogenase of Ralstonia eutropha. J. Biol. Chem. 1998;273:26349–26360. doi: 10.1074/jbc.273.41.26349. [DOI] [PubMed] [Google Scholar]
- 49.Niks D, Duvvuru J, Escalona M, Hille R. Spectroscopic and kinetic properties of the molybdenum-containing, NAD+-dependent formate dehydrogenase from Ralstonia eutropha. J. Biol. Chem. 2016;291:1162–1174. doi: 10.1074/jbc.M115.688457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu X, Niks D, Mulchandani A, Hille R. Efficient reduction of CO2 by the molybdenum-containing formate dehydrogenase from Cupriavidus necator (Ralstonia eutropha) J. Biol. Chem. 2017;292:16872–16879. doi: 10.1074/jbc.M117.785576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stock T, Rother M. Selenoproteins in Archaea and Gram-positive bacteria. Biochim. Biophys. Acta. 2009;1790:1520–1532. doi: 10.1016/j.bbagen.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 52.Luque-Almagro VM, et al. Bacterial nitrate assimilation: Gene distribution and regulation. Biochem. Soc. Trans. 2011;39:1838–1843. doi: 10.1042/BST20110688. [DOI] [PubMed] [Google Scholar]
- 53.Catling DC, Zahnle KJ. The Archean atmosphere. Sci. Adv. 2020;6:eaax1420. doi: 10.1126/sciadv.aax1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Glaser P, Danchin A, Kunst F, Zuber P, Nakano MM. Identification and isolation of a gene required for nitrate assimilation and anaerobic growth of Bacillus subtilis. J. Bacteriol. 1995;177:1112–1115. doi: 10.1128/jb.177.4.1112-1115.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martínez-Espinosa RM, Marhuenda-Egea FC, Bonete MJ. Assimilatory nitrate reductase from the haloarchaeon Haloferax mediterranei: Purification and characterisation. FEMS Microbiol. Lett. 2001;204:381–385. doi: 10.1016/s0378-1097(01)00431-1. [DOI] [PubMed] [Google Scholar]
- 56.Kilic V, Kilic GA, Kutlu HM, Martínez-Espinosa RM. Nitrate reduction in Haloferax alexandrinus: The case of assimilatory nitrate reductase. Extremophiles. 2017;21:551–561. doi: 10.1007/s00792-017-0924-4. [DOI] [PubMed] [Google Scholar]
- 57.Ruiz B, et al. The nitrate assimilatory pathway in Sinorhizobium meliloti: Contribution to NO production. Front. Microbiol. 2019;10:1526. doi: 10.3389/fmicb.2019.01526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hidalgo-García A, et al. Rhizobium etli produces nitrous oxide by coupling the assimilatory and denitrification pathways. Front. Microbiol. 2019;10:980. doi: 10.3389/fmicb.2019.00980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flores E, Frías JE, Rubio LM, Herrero A. Photosynthetic nitrate assimilation in cyanobacteria. Photosyn. Res. 2005;83:117–133. doi: 10.1007/s11120-004-5830-9. [DOI] [PubMed] [Google Scholar]
- 60.Ordoñez OF, Rasuk MC, Soria MN, Contreras M, Farías ME. Haloarchaea from the Andean Puna: Biological role in the energy metabolism of arsenic. Microb. Ecol. 2018;76:695–705. doi: 10.1007/s00248-018-1159-3. [DOI] [PubMed] [Google Scholar]
- 61.Härtig C, et al. Chemolithotrophic growth of the aerobic hyperthermophilic bacterium Thermocrinis ruber OC 14/7/2 on monothioarsenate and arsenite. FEMS Microbiol. Ecol. 2014;90:747–760. doi: 10.1111/1574-6941.12431. [DOI] [PubMed] [Google Scholar]
- 62.Svetlitshnyi V, Rainey F, Wiegel J. Thermosyntropha lipolytica gen. nov., sp. nov., a lipolytic, anaerobic, alkalitolerant, thermophilic bacterium utilizing short- and long-chain fatty acids in syntrophic coculture with a methanogenic archaeum. Int. J. Syst. Bacteriol. 1996;46:1131–1137. doi: 10.1099/00207713-46-4-1131. [DOI] [PubMed] [Google Scholar]
- 63.Martin W, Baross J, Kelley D, Russell MJ. Hydrothermal vents and the origin of life. Nat. Rev. Microbiol. 2008;6:805–814. doi: 10.1038/nrmicro1991. [DOI] [PubMed] [Google Scholar]
- 64.Bult CJ, et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 65.Slesarev AI, et al. The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens. Proc. Natl. Acad. Sci. USA. 2002;99:4644–4649. doi: 10.1073/pnas.032671499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hendrickson EL, et al. Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J. Bacteriol. 2004;186:6956–6969. doi: 10.1128/JB.186.20.6956-6969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Andreesen JR, Ljungdahl LG. Nicotinamide adenine dinucleotide phosphate-dependent formate dehydrogenase from Clostridium thermoaceticum: Purification and properties. J. Bacteriol. 1974;120:6–14. doi: 10.1128/jb.120.1.6-14.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Graentzdoerffer A, Rauh D, Pich A, Andreesen JR. Molecular and biochemical characterization of two tungsten- and selenium-containing formate dehydrogenases from Eubacterium acidaminophilum that are associated with components of an iron-only hydrogenase. Arch. Microbiol. 2003;179:116–130. doi: 10.1007/s00203-002-0508-1. [DOI] [PubMed] [Google Scholar]
- 69.Jones JB, Dilworth GL, Stadtman TC. Occurrence of selenocysteine in the selenium-dependent formate dehydrogenase of Methanococcus vannielii. Arch. Biochem. Biophys. 1979;195:255–260. doi: 10.1016/0003-9861(79)90351-5. [DOI] [PubMed] [Google Scholar]
- 70.Wood GE, Haydock AK, Leigh JA. Function and regulation of the formate dehydrogenase genes of the methanogenic archaeon Methanococcus maripaludis. J. Bacteriol. 2003;185:2548–2554. doi: 10.1128/JB.185.8.2548-2554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Costa C, Teixeira M, LeGall J, Moura JJG, Moura I. Formate dehydrogenase from Desulfovibrio desulfuricans ATCC 27774: Isolation and spectroscopic characterization of the active sites (heme, iron-sulfur centers and molybdenum) JBIC. 1997;2:198–208. [Google Scholar]
- 72.Zhang Y, Romero H, Salinas G, Gladyshev VN. Dynamic evolution of selenocysteine utilization in bacteria: A balance between selenoprotein loss and evolution of selenocysteine from redox active cysteine residues. Genome Biol. 2006;7:R94. doi: 10.1186/gb-2006-7-10-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rother M, Krzycki JA. Selenocysteine, pyrrolysine, and the unique energy metabolism of methanogenic Archaea. Archaea. 2010;2010:453642. doi: 10.1155/2010/453642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peng T, Lin J, Xu Y-Z, Zhang Y. Comparative genomics reveals new evolutionary and ecological patterns of selenium utilization in bacteria. ISME J. 2016;10:2048–2059. doi: 10.1038/ismej.2015.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mariotti M, et al. Evolution of selenophosphate synthetases: Emergence and relocation of function through independent duplications and recurrent subfunctionalization. Genome Res. 2015;25:1256–1267. doi: 10.1101/gr.190538.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ogawa K, et al. The nasB operon and nasA gene are required for nitrate/nitrite assimilation in Bacillus subtilis. J. Bacteriol. 1995;177:1409–1413. doi: 10.1128/jb.177.5.1409-1413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Suzuki I, Sugiyama T, Omata T. Primary structure and transcriptional regulation of the gene for nitrite reductase from the cyanobacterium Synechococcus PCC 7942. Plant Cell Physiol. 1993;34:1311–1320. [Google Scholar]
- 78.Gangeswaran R, Lowe DJ, Eady RR. Purification and characterization of the assimilatory nitrate reductase of Azotobacter vinelandii. Biochem. J. 1993;289(Pt 2):335–342. doi: 10.1042/bj2890335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rubio LM, Flores E, Herrero A. Purification, cofactor analysis, and site-directed mutagenesis of Synechococcus ferredoxin-nitrate reductase. Photosyn. Res. 2002;72:13–26. doi: 10.1023/A:1016078700839. [DOI] [PubMed] [Google Scholar]
- 80.Lin JT, Goldman BS, Stewart V. Structures of genes nasA and nasB, encoding assimilatory nitrate and nitrite reductases in Klebsiella pneumoniae M5al. J. Bacteriol. 1993;175:2370–2378. doi: 10.1128/jb.175.8.2370-2378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Blasco R, Castillo F, Martínez-Luque M. The assimilatory nitrate reductase from the phototrophic bacterium, Rhodobacter capsulatus E1F1, is a flavoprotein. FEBS Lett. 1997;414:45–49. doi: 10.1016/s0014-5793(97)00968-x. [DOI] [PubMed] [Google Scholar]
- 82.Krafft T, Macy JM. Purification and characterization of the respiratory arsenate reductase of Chrysiogenes arsenatis. Eur. J. Biochem. 1998;255:647–653. doi: 10.1046/j.1432-1327.1998.2550647.x. [DOI] [PubMed] [Google Scholar]
- 83.Afkar E, et al. The respiratory arsenate reductase from Bacillus selenitireducens strain MLS10. FEMS Microbiol. Lett. 2003;226:107–112. doi: 10.1016/S0378-1097(03)00609-8. [DOI] [PubMed] [Google Scholar]
- 84.Ellis PJ, Conrads T, Hille R, Kuhn P. Crystal structure of the 100 kDa arsenite oxidase from Alcaligenes faecalis in two crystal forms at 1.64 Å and 2.03 Å. Structure. 2001;9:125–132. doi: 10.1016/s0969-2126(01)00566-4. [DOI] [PubMed] [Google Scholar]
- 85.Warelow TP, Pushie MJ, Cotelesage JJH, Santini JM, George GN. The active site structure and catalytic mechanism of arsenite oxidase. Sci. Rep. 2017;7:1757. doi: 10.1038/s41598-017-01840-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Karrasch M, Börner G, Thauer RK. The molybdenum cofactor of formylmethanofuran dehydrogenase from Methanosarcina barkeri is a molybdopterin guanine dinucleotide. FEBS Lett. 1990;274:48–52. doi: 10.1016/0014-5793(90)81326-j. [DOI] [PubMed] [Google Scholar]
- 87.Schmitz RA, Albracht SP, Thauer RK. A molybdenum and a tungsten isoenzyme of formylmethanofuran dehydrogenase in the thermophilic archaeon Methanobacterium wolfei. Eur. J. Biochem. 1992;209:1013–1018. doi: 10.1111/j.1432-1033.1992.tb17376.x. [DOI] [PubMed] [Google Scholar]
- 88.Yamamoto I, Saiki T, Liu SM, Ljungdahl LG. Purification and properties of NADP-dependent formate dehydrogenase from Clostridium thermoaceticum, a tungsten-selenium-iron protein. J. Biol. Chem. 1983;258:1826–1832. [PubMed] [Google Scholar]
- 89.Jones JB, Stadtman TC. Selenium-dependent and selenium-independent formate dehydrogenases of Methanococcus vannielii. Separation of the two forms and characterization of the purified selenium-independent form. J. Biol. Chem. 1981;256:656–663. [PubMed] [Google Scholar]
- 90.Boratyn GM, et al. Domain enhanced lookup time accelerated BLAST. Biol. Direct. 2012;7:12. doi: 10.1186/1745-6150-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Markowitz VM, et al. The integrated microbial genomes (IMG) system. Nucl. Acids Res. 2006;34:D344–D348. doi: 10.1093/nar/gkj024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bertero MG, et al. Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat. Struct. Biol. 2003;10:681–687. doi: 10.1038/nsb969. [DOI] [PubMed] [Google Scholar]
- 93.Afshar S, Johnson E, de Vries S, Schröder I. Properties of a thermostable nitrate reductase from the hyperthermophilic archaeon Pyrobaculum aerophilum. J. Bacteriol. 2001;183:5491–5495. doi: 10.1128/JB.183.19.5491-5495.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ramírez-Arcos S, Fernández-Herrero LA, Berenguer J. A thermophilic nitrate reductase is responsible for the strain specific anaerobic growth of Thermus thermophilus HB8. Biochim. Biophys. Acta. 1998;1396:215–227. doi: 10.1016/s0167-4781(97)00183-8. [DOI] [PubMed] [Google Scholar]
- 95.Thorell HD, Stenklo K, Karlsson J, Nilsson T. A gene cluster for chlorate metabolism in Ideonella dechloratans. Appl. Environ. Microbiol. 2003;69:5585–5592. doi: 10.1128/AEM.69.9.5585-5592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schröder I, Rech S, Krafft T, Macy JM. Purification and characterization of the selenate reductase from Thauera selenatis. J. Biol. Chem. 1997;272:23765–23768. doi: 10.1074/jbc.272.38.23765. [DOI] [PubMed] [Google Scholar]
- 97.McDevitt CA, Hugenholtz P, Hanson GR, McEwan AG. Molecular analysis of dimethyl sulphide dehydrogenase from Rhodovulum sulfidophilum: Its place in the dimethyl sulphoxide reductase family of microbial molybdopterin-containing enzymes. Mol. Microbiol. 2002;44:1575–1587. doi: 10.1046/j.1365-2958.2002.02978.x. [DOI] [PubMed] [Google Scholar]
- 98.Méjean V, et al. TMAO anaerobic respiration in Escherichia coli: Involvement of the tor operon. Mol. Microbiol. 1994;11:1169–1179. doi: 10.1111/j.1365-2958.1994.tb00393.x. [DOI] [PubMed] [Google Scholar]
- 99.Czjzek M, et al. Crystal structure of oxidized trimethylamine N-oxide reductase from Shewanella massilia at 2.5 Å resolution. J. Mol. Biol. 1998;284:435–447. doi: 10.1006/jmbi.1998.2156. [DOI] [PubMed] [Google Scholar]
- 100.Pierson DE, Campbell A. Cloning and nucleotide sequence of bisC, the structural gene for biotin sulfoxide reductase in Escherichia coli. J. Bacteriol. 1990;172:2194–2198. doi: 10.1128/jb.172.4.2194-2198.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.White H, Strobl G, Feicht R, Simon H. Carboxylic acid reductase: A new tungsten enzyme catalyses the reduction of non-activated carboxylic acids to aldehydes. Eur. J. Biochem. 1989;184:89–96. doi: 10.1111/j.1432-1033.1989.tb14993.x. [DOI] [PubMed] [Google Scholar]
- 102.Mukund S, Adams MW. The novel tungsten-iron-sulfur protein of the hyperthermophilic archaebacterium, Pyrococcus furiosus, is an aldehyde ferredoxin oxidoreductase. Evidence for its participation in a unique glycolytic pathway. J. Biol. Chem. 1991;266:14208–14216. [PubMed] [Google Scholar]
- 103.Hu Y, Faham S, Roy R, Adams MWW, Rees DC. Formaldehyde ferredoxin oxidoreductase from Pyrococcus furiosus: The 1.85 Å resolution crystal structure and its mechanistic implications. J. Mol. Biol. 1999;286:899–914. doi: 10.1006/jmbi.1998.2488. [DOI] [PubMed] [Google Scholar]
- 104.Mukund S, Adams MW. Characterization of a novel tungsten-containing formaldehyde ferredoxin oxidoreductase from the hyperthermophilic archaeon, Thermococcus litoralis. A role for tungsten in peptide catabolism. J. Biol. Chem. 1993;268:13592–13600. [PubMed] [Google Scholar]
- 105.Mukund S, Adams MW. Glyceraldehyde-3-phosphate ferredoxin oxidoreductase, a novel tungsten-containing enzyme with a potential glycolytic role in the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem. 1995;270:8389–8392. doi: 10.1074/jbc.270.15.8389. [DOI] [PubMed] [Google Scholar]
- 106.Park M-O, Mizutani T, Jones PR. Glyceraldehyde-3-phosphate ferredoxin oxidoreductase from Methanococcus maripaludis. J. Bacteriol. 2007;189:7281–7289. doi: 10.1128/JB.00828-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Reher M, Gebhard S, Schönheit P. Glyceraldehyde-3-phosphate ferredoxin oxidoreductase (GAPOR) and nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN), key enzymes of the respective modified Embden–Meyerhof pathways in the hyperthermophilic crenarchaeota Pyrobaculum aerophilum and Aeropyrum pernix. FEMS Microbiol. Lett. 2007;273:196–205. doi: 10.1111/j.1574-6968.2007.00787.x. [DOI] [PubMed] [Google Scholar]
- 108.Vorholt JA, Vaupel M, Thauer RK. A selenium-dependent and a selenium-independent formylmethanofuran dehydrogenase and their transcriptional regulation in the hyperthermophilic Methanopyrus kandleri. Mol. Microbiol. 1997;23:1033–1042. doi: 10.1046/j.1365-2958.1997.2931653.x. [DOI] [PubMed] [Google Scholar]
- 109.Nakamura T, Yamada KD, Tomii K, Katoh K. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics. 2018;34:2490–2492. doi: 10.1093/bioinformatics/bty121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Le SQ, Dang CC, Gascuel O. Modeling protein evolution with several amino acid replacement matrices depending on site rates. Mol. Biol. Evol. 2012;29:2921–2936. doi: 10.1093/molbev/mss112. [DOI] [PubMed] [Google Scholar]
- 113.Miller, M. A., Pfeiffer, W. & Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In 2010 Gateway Computing Environments Workshop (GCE) 1–8 (2010). 10.1109/GCE.2010.5676129.
- 114.Letunic I, Bork P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.