Abstract
The objective was to identify the transcriptomic profile of in vitro-derived embryos with high competence to establish and maintain gestation. Embryos produced with X-sorted sperm were cultured from day 5 to day 7 in serum-free medium containing 10 ng/ml recombinant bovine colony-stimulating factor 2 (CSF2) or vehicle. The CSF2 was administered because this molecule can increase blastocyst competence for survival after embryo transfer. Blastocysts were harvested on day 7 of culture and manually bisected. One demi-embryo from a single blastocyst was transferred into a synchronized recipient and the other half was used for RNA-seq analysis. Using P < 0.01 and a fold change >2-fold or <0.5 fold as cutoffs, there were 617 differentially expressed genes (DEG) between embryos that survived to day 30 of gestation vs those that did not, 470 DEG between embryos that survived to day 60 and those that did not, 432 DEG between embryos that maintained pregnancy from day 30 to day 60 vs those where pregnancy failed after day 30, and 635 DEG regulated by CSF2. Pathways and ontologies in which DEG were overrepresented included many related to cellular responses to stress and cell survival. It was concluded that gene expression in the blastocyst is different between embryos that are competent to establish and maintain pregnancy vs those that are not. The relationship between expression of genes related to cell stress and subsequent embryonic survival probably reflects cellular perturbations caused by embryonic development taking place in the artificial environment associated with cell culture.
Keywords: transcriptome, pregnancy, blastocyst, CSF2, embryonic survival
Summary sentence: Embryos produced in vitro that are capable of maintaining gestation have a gene expression pattern that diverges from embryos that fail to establish pregnancy.
Introduction
Despite its successful development from the zygote stage, the continued survival of the blastocyst-stage embryo is problematic. Only ~50–75% of bovine embryos used for embryo transfer are competent to survive to term after transfer, with reduced competence for embryos produced in vitro [1]. Indeed, pregnancy rates for transfer of embryos produced in vitro are lower than for those produced in vivo by superovulation [2]. In part, reduced competence of embryos produced in vitro reflects an absence of maternally derived regulatory molecules such as CSF2 and di dickkopf WNT signaling pathway inhibitor 1 that can enhance post-transfer survival of embryos produced in vitro [3, 4].
It is likely that the ability of the blastocyst to sustain development to term depends upon its transcriptome, which encodes for proteins required by the embryo for cellular maintenance, execution of subsequent developmental events involving differentiation and lineage commitment and communication with the female reproductive tract. In vitro production of embryo is accompanied by changes in the transcriptome of the embryo [5–7]. Identification of gene expression profiles that confer a blastocyst with the capacity for sustained development could lead to methods for regulation of embryonic gene expression to improve survival of the embryo produced either in vivo or in vitro. In addition, identification of genes whose expression is associated with high embryo competence could lead to insights into identification of genetic loci important for genetic variation in fertility, using either a candidate gene approach [8, 9] or by combining genome-wide association studies with transcriptome data [10–12].
Studies performed by El-Sayed et al. [13] with bovine embryos produced in vitro and by Salilew-Wondim et al. [14] and Ghanem et al. [15] with embryos produced by superovulation have identified groups of up to 41–70 genes whose expression in the blastocyst differed between those that could establish and maintain pregnancy from those that either did not establish pregnancy or in which pregnancy was subsequently lost. The approach was to bisect blastocysts, transfer the larger portion of the embryo to a recipient female, and use the remaining portion for analysis of gene expression with a microarray platform containing either up to 2219 probes [13, 15] or 24 128 probes [14].
Here, we repeated these experiments with embryos produced in vitro in either the presence or absence of CSF2. To maximize identification of the number of differentially expressed genes (DEG), gene expression was evaluated using RNA-Seq. CSF2 was included as a treatment because, in addition to increasing blastocyst competence for embryo survival [3, 4], treatment of bovine embryos with CSF2 from days 5 to 7 of development (during the period of the morula-to-blastocyst transition) modifies gene expression of the morula and blastocyst-stage embryo [16–18]. In addition, CSF2 exerts long-term actions on the embryo that result in changes in trophoblast elongation at day 15 of pregnancy [19], fetal and placental gene expression at day 86 of gestation [20], and postnatal effects on calf growth [21]. The experiment was performed with female embryos to reduce variation due to embryo sex and because effects of CSF2 on the bovine embryo are different for female embryos than male embryos [18, 19]. It was hypothesized that the transcriptome of a blastocyst able to maintain pregnancy until day 30 and day 60 of gestation differs from a blastocyst that dies after transfer and that one action of CSF2 would be to alter expression of genes identified as differentially expressed between embryos that established pregnancy vs those that did not.
Methods
Animals
All animal procedures were approved by the University of Florida Institutional Animal Care and Use Committee and were conducted in accordance with SSR guidelines and standards. The experiment was conducted on a commercial farm located in Lee, Florida (30°25′10″N 83°17′59″W). Holstein virgin heifers (n = 16), 12 month of age, were used as oocyte donors, and another group of 12-month-old Holstein heifers (n = 45) was used as embryo transfer recipients. Donors and recipients were housed in free-stall barns with sand bedding and were fed ad libitum a total mixed ration formulated for growing Holstein heifers.
Ovum pickup
The 16 Holstein heifers used as oocyte donors were randomly subdivided into two groups of 8 each. Each heifer was subjected to two rounds of follicle superstimulation followed by oocyte collection by transvaginal ultrasound-guided follicular aspiration [oocyte pickup (OPU)]. For one group, the oocytes were used for CSF2 treatment in the first round and for control treatment in the second round. For the other group, oocytes were used for control in the first round and for CSF2 in the second round.
The procedure for the first round of follicle collection involved injection of 100 μg gonadorelin hydrochloride (Factrel, Zoetis, Kalamazoo, MI), i.m., on day 0, ablation of all follicles ≥5 mm by transvaginal ultrasound-guided follicle ablation and insertion of a controlled intravaginal progesterone releasing device (CIDR—Eazi-Breed CIDR, Zoetis, Kalamazoo, MI) on day 7, 6 injections i.m., of 30 mg follicle stimulating hormone (FSH; Folltropin-V, Vetoquinol, Fort Worth, TX), 12 h apart beginning at day 9 (48 h after follicle ablation), and CIDR removal and oocyte recovery about 30 h after the last FSH injection. The procedure for the second round of follicle collection was initiated 8 days after the first OPU by follicle ablation and CIDR insertion. Another six injections, i.m., of 30 mg FSH were administered 12 h apart beginning 48 h after follicle ablation. Removal of the CIDR and the second OPU occurred at 30 h after the last FSH injection.
For follicle ablation, an epidural block consisting of 5 ml of 2% (w/v) lidocaine was administered, ovarian follicles were visualized using an Aloka-SSD 500 ultrasound unit equipped with a 7.5 mHz microconvex probe (Aloka, Tokyo, Japan) enclosed in a plastic needle guide, and follicles ablated using an 18-gauge, 5.5 cm needle (Watanabe Tecnologia Aplicada, Campinas, Brazil). Cumulus oocyte complexes (COC) were harvested by OPU using the same procedure as for follicle ablation except that each follicle >5 mm was aspirated using an 18-gauge, 55 cm needle connected to a vacuum pump (Pioneer Medical Inc., Melrose, MA), which was regulated to achieve a flow rate of 15 ml/min. The follicular aspirate was collected into a single 50 ml conical tube containing 10 ml of oocyte washing medium (MOFA Global, Verona, WI) supplemented with IU/ml sodium heparin.
In vitro production of embryos
All procedures for in vitro maturation, fertilization, and embryo culture were performed while keeping oocytes and embryos from a single heifer in a single group and without mixing oocytes and embryos from other donors. The aspirate from each donor heifer was filtered through a 68 μm embryo filter (SPI, Canton, TX) into a 100 mm ×100 mm square petri dish with gridlines (Fisher Scientific, Hampton, NH). Contents of the dish were searched using a dissecting microscope and COCs recovered from each individual donor using a wiretrol pipette (Drummond, Broomall, PA).
Recovered oocytes were washed three times in oocyte washing medium (MOFA Global) and placed into 2 ml microcentrifuge tubes containing 1.5 ml oocyte maturation medium (BO-HEPES-IVM, IVF Bioscience, Falmouth, Cornwall, UK) that had been pre-warmed at 38.5 °C. The tubes containing the collected oocytes were placed into a portable incubator (Micro Q Technologies, Conshohocken, PA) set at 38.5 °C. After collection, COC were transported to the laboratory where they completed maturation for 22 h inside the portable incubator at 38.5 °C.
Following maturation, COCs from a single donor were washed once in HEPES-TALP [22] and placed into 60 μl drops of IVF-TALP [22] overlaid with mineral oil. X-sorted semen from a single Holstein bull was purified using Puresperm (Nidacon International, MoIndal, Sweden) [18] and added to each fertilization drop at a concentration of 2 × 106 spermatozoa/ml. Following addition of sperm, 3.5 μl of a solution of 0.5 mM penicillamine, 0.25 mM hypotaurine, and 25 μM epinephrine prepared as described by Ortega et al. [22] were added to each fertilization drop. Sperm and COCs were co-incubated for 18 h at 38.5 °C in 5% CO2 in humidified air. Afterwards, putative zygotes were denuded of cumulus cells by suspension in HEPES-TALP containing 1000 U/ml hyaluronidase and vortexing for 2 min. Presumptive zygotes from each donor heifer were then cultured in 25 μl drops of synthetic oviduct fluid-bovine embryo 2 (SOF-BE2; [22]) overlaid with mineral oil at 38.5 °C in a humidified atmosphere of 5% O2, 5% CO2, and 90% N2. Embryos derived from half of the donors were treated at day 5 after insemination with the replacement of 2.5 μl of the culture drop with SOF-BE2 supplemented with 100 ng/ml recombinant bovine CSF2 (Novartis, Basle, Switzerland) to produce a final concentration of 10 ng/ml of CSF2. Embryos derived from the other half of the donors (control) were treated by replacement of 2.5 μl of the medium in the culture drop with 2.5 μl of vehicle (SOF-BE2).
The percentage of oocytes that cleaved was assessed on day 3 after insemination and the percentage of oocytes that developed to the blastocyst stage was determined at day 7 after insemination, respectively. Blastocyst stage (blastocyst, expanded and hatching) embryos were harvested at day 7 and subjected to embryo bisection.
Embryo bisection
Blastocysts collected on day 7 of culture were transferred individually into 50 μl drops of embryo biopsy medium (BO-biopsy, IVF Bioscience) placed on the surface of an epoxy-printed well slide. While visualizing the embryo using an inverted microscope with ×20 magnification (Nikon, Diaphot, Tokyo, Japan), a microblade fixed to a micromanipulator was gently pressed against the middle of the embryo so that trophectoderm (TE) and inner cell mass (ICM) cells were evenly distributed on both sides (Figure 1). The microblade was slowly moved forward and backward until the embryo was completely bisected and the zona pellucida was removed. An example of the bisection process is depicted in Supplemental File 1, Movie S1. The microblade used was custom built according to the dimensions of BD Beaver microblade 30° produced by Beaver Visitec International (Waltham, MA). One portion of the bisected blastocyst (the smaller or less intact portion if bisection was not uniform) was directly placed into a 0.2 ml tube containing 50 μl of RNAlater (Invitrogen, Carlsbad, CA) and stored at −80 °C until RNA-seq analysis. The other demi-embryo was placed into a 25 μl drop of SOF-BE2 covered in mineral oil and maintained inside an incubator at 38.5 °C in a humidified atmosphere of 5% O2, 5% CO2, and 90% N2 for 30 min for re-establishment of the blastocoele after bisection (Supplemental File 2, Figure S1). The demi-embryo was then placed into a plate containing embryo transfer medium (BO-Transfer, IVF Bioscience), loaded into a 0.25 ml embryo transfer straw, and transported to the farm in a portable incubator at 38.5 °C (Micro Q Technologies).
Figure 1.
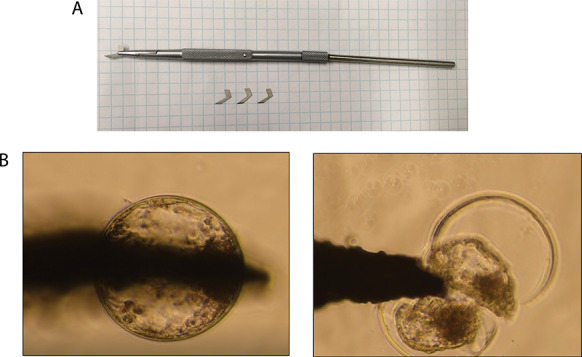
Embryo bisection. A. Custom built microblade fixed to a micromanipulator’s arm. The microblades were custom built with the dimensions of BD BeaverTM microblade 30° produced by Beaver Visitec International (Waltham, MA). B. Embryo bisection being performed with a microblade fixed to a micromanipulator using an inverted microscope with ×20 magnification (Nikon, Diaphot, Tokyo, Japan). The microblade was gently pressed against the middle of the embryo so that TE and ICM cells were evenly distributed on both sides.
Embryo transfer and pregnancy diagnosis
Recipient heifers were subjected to an ovulation synchronization protocol consisting of 100 μg gonadorelin hydrochloride (Factrel, Zoetis) i.m., and intravaginal insertion of a CIDR on day −9 (Day 0 = day of anticipated ovulation), 25 mg prostaglandin-F2α (Lutalyse, Zoetis), i.m., and CIDR removal on day −3, and 100 μg gonadorelin hydrochloride on day −1. At day 7 after anticipated ovulation (8 days after the last gonadorelin injection), each recipient was subjected to examination of the ovaries by transrectal ultrasonography (Aloka SSD 500, 5 MHz linear transducer) to confirm the presence of a corpus luteum. Recipients received an epidural block of 5 ml of 2% (w/v) lidocaine and then, randomly, a single demi-embryo was transferred into the ipsilateral uterine horn relative to the corpus luteum. Pregnancy diagnosis was carried out by transrectal ultrasonography on day 30 and day 60 of gestation.
Statistical analysis of data on embryonic development and pregnancy success
Data were analyzed using the GLIMMIX procedure of Statistical Analysis System (SAS Software 9.4; SAS institute Inc., Cary, NC). All dependent variables were considered to have a binomial distribution. For the percent of oocytes that cleaved, percent of oocytes that developed to the blastocyst stage at day 7 after insemination, percent of cows pregnant per embryo transfer at day 30 and day 60, and for pregnancy loss between day 30 and day 60, the model included the fixed effect of treatment and the random effect of replicate. The level of significance was considered as P < 0.05.
RNA extraction, cDNA library preparation, and RNA-Seq
Total RNA from each sample was isolated using the RNeasy Micro Kit (Qiagen, Germantown, MD) according to the manufacturer’s cell protocol with the following adjustments for samples maintained in RNAlater; 250 μl of 100% ethanol was used instead of the recommended 350 μl of 70% (v/v) ethanol and isolated RNA was eluted in 12 μl RNase-free water instead of the recommended 22 μl RNase-free water. Barcoded fragment libraries were constructed using the Bovine Custom Any Deplete Ovation SoLo RNA-Seq System from NuGEN (San Carlos, CA) following the manufacturer’s protocol. DNA integrity of the libraries was assessed using the High Sensitivity D1000 ScreenTape on the Agilent 4200 TapeStation (Santa Clara, CA) and DNA concentration was measured using the KAPA qPCR kit (Boston, MA) following the manufacturer’s protocol. Samples were uniquely barcoded using the eight base pair single index adaptors in the adaptor plate provided with the Nugen kit. A total of 20–25 samples were combined for each pool. Samples were sequenced with 1 × 75 bp single-end reads on an Illumina NEXT-seq (Illumina, Inc. San Diego, CA) at the Stanford Functional Genomics Facility (Stanford, CA) according to the manufacturer’s protocol. Sequence reads for each sample were mapped to the annotated bovine reference genome (UMD3.1.70) using CLC genomics workbench 12.0 (www.qiagenbioinformatics.com) to obtain read counts for each gene. Data have been deposited in the Gene Expression Omnibus of the National Center for Biotechnology Information and are accessible through GEO Series accession number GSE130954 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE130954).
Bioinformatics
The following procedures were performed with the limma package for the R software [23]. Multidimensional scaling (MDS) plots of distances between gene expression profiles were performed with the edgeR package using all expressed genes to identify outliers (Supplemental File 2, Figure S2). A total of five samples appeared as outliers in a multidimensional scaling plot (two CSF2 and one control embryos not pregnant at day 30, one control embryo pregnant at day 30 and day 60, and one control embryo pregnant at day 30 but lost by day 60). These five samples also had the highest proportion of zero counts (Supplemental File 3, Table S1). The five samples were excluded from the analyses as were three samples that did not express both TE [actin alpha 2, smooth muscle (ACTA2), GATA binding protein 2 and 3 (GATA2, and GATA3), caudal type homeobox 2 (CDX2)] and ICM [Nanog homoeobox (NANOG), DNA methyltransferase 3 alpha (DNMT3A), H2A.Z variant histone 1 (H2AFZ)] markers [24]. It was confirmed that all samples came from female embryos based on lack of expression of Y-linked genes [sex determining region Y (SRY), DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked (DDX3Y), eukaryotic translation initiation factor 1A, Y-linked (EIF1AY), ubiquitously transcribed tetratricopeptide repeat containing, Y-linked (UTY)] [25]. Genes with low expression counts (less than 1 count per million reads) in 6 or more of the 37 retained samples were filtered out before normalization. Thus, 7031 high-quality transcripts were retained for further analysis.
The normalization method applied was weighted trimmed mean of M-values [26]. Observation weights were used to apply a robust estimate of the negative binomial dispersion parameter for each gene. This method is employed to avoid outlier genes, which can lead to the detection of false positives. Next, the observation weights were used for estimating regression parameters. The method uses an iterative procedure where weights are calculated from residuals and estimates are made after re-weighting [27]. Finally, a quasi-likelihood negative binomial generalized log-linear model was fit to read counts for each transcript and conduct genewise statistical tests for the coefficient contrast [28]. The matrix of contrast was build based on the comparisons between the groups.
An embryo was classified based on the pregnancy outcome. Embryos where recipients were not pregnant at day 30 were classified as non-pregnant (NP), embryos where recipients were pregnant at both day 30 and day 60 were classified as pregnant–pregnant (PP), and embryos where recipients were pregnant at day 30 but not at day 60 were classified as pregnant-nonpregnant (PNP). Statistical analysis was performed by one-way ANOVA, applying an F-test to identify DEG between comparisons. Three series of comparisons were performed. The first series evaluated DEG based on pregnancy outcomes for data from all embryos (control and CSF2). Comparisons were based on pregnancy status at day 30 [PP + PNP (n = 14) vs NP (n = 23)], pregnancy status at day 60 [PP (n = 9) vs NP + PNP (n = 28)], and pregnancy loss between day 30 and day 60 [PP (n = 9) vs PNP (n = 5)]. A MDS scaling plot of DEG [false discovery rate (FDR) of P < 0.05 and a fold change >2 or <0.5] was constructed using the limma package of R to visualize differences between groups (Figure 2). The second series (one comparison) evaluated DEG due to treatment with CSF2 (n = 16) or control (n = 21) regardless of pregnancy outcome. The third series evaluate DEG based on pregnancy outcomes at day 30 for each treatment group (CSF2 or control) separately. For CSF2, the comparison was PP + PNP (n = 6) vs NP (n = 10), whereas, for control, the comparison was PP + PNP (n = 8) vs NP (n = 13). Data on pregnancy outcomes at day 60 were not evaluated within each treatment group because of the absence of pregnancy loss for the CSF2 group.
Figure 2.
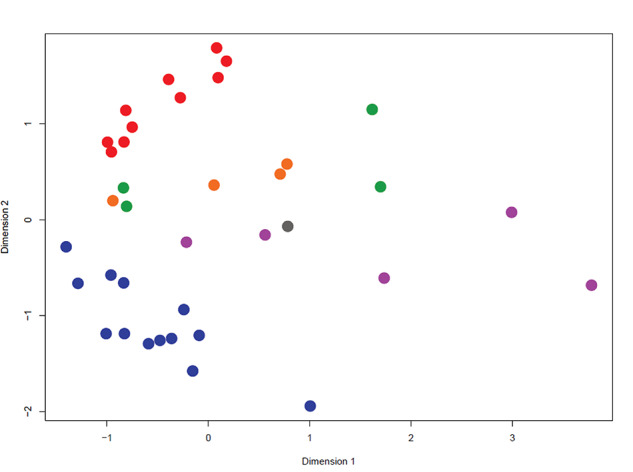
Multidimensional scaling plot based on differentially expressed genes (FDR of P < 0.05 and a fold change >2 or <0.5). Blue = NP-control, red = NP-CSF2, green = PP-control, purple = PP-CSF2, orange = PNP-control, and gray = PNP-CSF2. NP, not pregnant at day 30; PP, pregnant at day 60; PNP, pregnant and day 30 but not at day 60.
Differentially expressed genes were defined using two different criteria. The first was those with P < 0.01 and a fold change of either >2 or <0.5 for each pairwise comparison. The second, more stringent criterion was genes with an FDR-adjusted P < 0.05 and a fold change >2 or <0.5.
Functional analysis of DEG was conducted using the list of genes with a P-value <0.01 and a fold change of either >2 or <0.5. This list of genes was used rather than the more stringent list based on an FDR-adjusted P <0.05 to increase sensitivity of identifying relevant ontologies. Characteristics of DEG were determined using Ingenuity Pathway Analysis (IPA, Qiagen, Redwood City, CA, USA) for (1) clustering of DEG into molecular and cellular functions in which DEG were overexpressed, (2) molecular and cellular functions and canonical pathways predicted to be either activated or inhibited as a result of DEG, and (3) identification of putative transcription factors, ligand-dependent nuclear receptors, translational regulators, miRNA, and selected other regulators predicted to be activated or inhibited. Activation status was considered significant when the z-score was ≤−2.0 or ≥2.0. Disease pathways were excluded.
Results
Embryonic development and pregnancy outcomes
There was no effect of CSF2 on the percent of putative zygotes that cleaved after insemination (least-squares mean ± SEM; 87.2 ± 3.6% vs 85.7 ± 3.5% for control and CSF2, respectively) or that developed to the blastocyst stage at day 7 after insemination (31.4 ± 5.0% vs 28.6 ± 4.5% for control and CSF2, respectively). A total of 45 demi-embryos were transferred into recipients (25 control and 20 CSF2). Treatment with CSF2 did not significantly affect the percent of recipients pregnant on day 30 or 60 of gestation or on pregnancy loss between day 30 and day 60 (Table 1). Note, however, that only 1 of 6 pregnancies (16.6%) from the CSF2 group were lost between day 30 and day 60, whereas 5 of 10 pregnancies (50.0%) in the control group at day 30 were lost by day 60 (Table 1).
Table 1.
Effect of treatment with CSF2 (10 ng/ml) from day 5 to day 7 of culture on pregnancy outcomes after transfer of demi-embryos to recipients.
| CSF2 (ng/ml) | Pregnancy success, % (no pregnant/no transferred) | Pregnancy loss, % (lost/total) | |
|---|---|---|---|
| Day 30 | Day 60 | ||
| 0.0 (control) | 40% (10/25) | 20% (5/25) | 50% (5/10) |
| 10.0 | 30% (6/20) | 25% (5/20) | 17% (1/6) |
| P | 0.50 | 0.51 | 0.22 |
Differential expression of genes between embryos that maintained pregnancy to day 30 vs those that did not
The number of DEG for all comparisons is summarized in Table 2. There were 617 DEG using P < 0.01 and >2-fold or <0.5-fold difference as cutoffs. Of these, 341 were upregulated and 276 were downregulated in embryo that maintained pregnancy to day 30. Using a more stringent criteria, a FDR-adjusted P < 0.05 and a fold-change of >2-fold or <0.5, 370 DEG were identified in embryos that maintained pregnancy to day 30 (215 upregulated and 155 downregulated). The list of DEG is shown in Supplemental File 3, Table S2.
Table 2.
Numbers of differentially expressed genes for various comparisons.
| Comparison | Number of samples | No. of differentially expressed genes | |
|---|---|---|---|
| Fold change, >2 or <0.5; P < 0.01 | Fold change, >2 or <0.5; FDR, <0.05 | ||
| Pregnant day 30 vs nonpregnant day 30 | 14 vs 23 | 617 | 370 |
| Pregnant day 60 vs nonpregnant day 30 | 9 vs 23 | 470 | 255 |
| Pregnancy survival (pregnant day 30 and 60 vs pregnant at day 30 but non-pregnant at day 60) | 9 vs 5 | 432 | 0 |
| CSF2 vs control | 16 vs 21 | 635 | 420 |
| Control embryos, pregnant day 30 vs nonpregnant, day 30 | 8 vs 13 | 465 | 55 |
| CSF2 embryos, pregnant day 30 vs nonpregnant, day 30 | 6 vs 10 | 590 | 246 |
Molecular and cellular functions overrepresented in DEG were determined by IPA (kSupplemental File 3, Table S3). The top six functions based on P value were DNA replication, recombination and repair (53 genes), cellular assembly and organization (52 genes), cellular function and maintenance (62 genes), lipid metabolism (36 genes), small molecule biochemistry (57 genes), and cell death and survival (191 genes). The biological functions predicted to be increased in embryos competent to maintain pregnancy to day 30 were homologous recombination of cells (7 molecules), oxidation of long-chain fatty acids (7 genes), and oxidation of palmitic acid (5 genes) (Figure 3). Organismal death was predicted to be decreased (z = −1.9) in embryos that survived to day 30 (129 genes) (Supplemental File 2, Figure S3). Upstream regulators predicted to be activated were NFKB inhibitor alpha (NFKBIA) (9 genes), peroxisome proliferator activated receptor gamma (PPARG) (13 genes), the RAB, member RAS oncogene family like 6 (RABL6) (4 genes), and miR-30c-5p (8 genes), while the one upstream regulator predicted to be inhibited was lysine demethylase 5A (KDM5A) (8 genes) (Figure 4).
Figure 3.
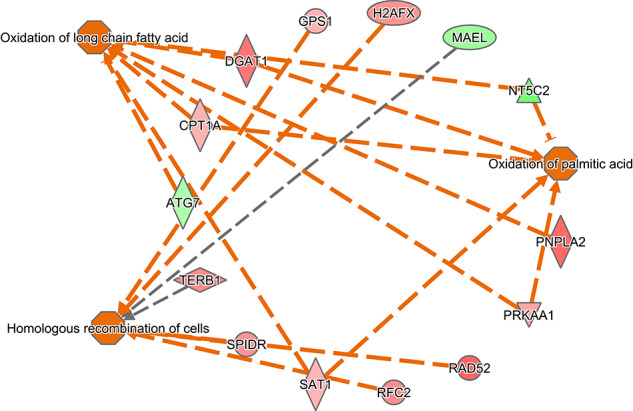
Functions predicted to be activated in embryos that survived to day 30. Genes in red were upregulated in embryos that survived to day 30 and genes in green were downregulated. Orange lines represent relationships that lead to activation and gray lines represent situations where the effect is not predicted.
Figure 4.
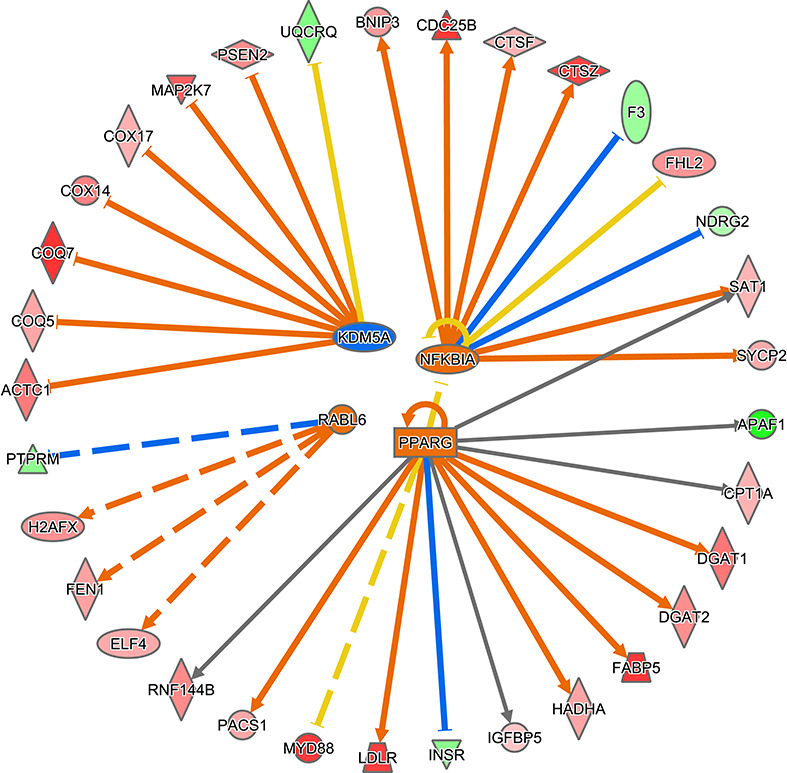
Predicted upstream regulators of DEG for embryo survival to day 30. NFKB1, PPARG, and mIR-30c-5p were predicted to be upregulated in embryos that survived to day 30 while KDM5A was predicted to be inhibited. Genes in red were upregulated in embryos that survived to day 30 and genes in green were downregulated. Orange lines represent relationships that lead to activation, blue lines represent relationships that lead to inhibition, yellow lines represent relationships inconsistent with the predicted state, and gray lines represent situations where the effect is not predicted.
Differential expression of genes between embryos that maintained pregnancy to day 60 vs those that did not
The list of DEG is shown in Supplemental File 3, Table S4. There were 470 DEG using P < 0.01 and >2-fold or <0.5-fold difference as cutoffs. Of these, 337 were upregulated in embryos that survived to day 60 and 133 were downregulated. Using a more stringent criteria, a FDR-adjusted P < 0.05, and a fold-change of >2-fold or <0.5, there were 255 DEG in embryos that survived to day 60 (206 upregulated and 49 downregulated). There were 349 DEG in common for maintenance of pregnancy to day 30 and day 60, using P < 0.01 and >2-fold or <0.5-fold difference as cutoffs, with 269 upregulated genes and 80 downregulated genes (i.e., 74% of the DEG for embryo survival to day 60 were also DEG for embryo survival to day 30).
Molecular and cellular functions overrepresented in DEG are summarized in Supplemental File 3, Table S5. The top six functions based on P value were DNA replication and repair (50 genes), lipid metabolism (33 genes), small molecule biochemistry (50 genes), cell morphology (43 genes), cellular assembly and organization (37 genes), and cell death and survival (152 genes). Based on DEG, several biological functions were predicted to be increased in embryos that maintained pregnancy to day 60 including cell survival (67 genes), removal of cells (8 genes), oxidation of long-chain fatty acids (5 genes), and repair of DNA (20 genes) (Supplemental File 2, Figure S4). Organismal death (102 genes) and quantity of insulin in blood (11 genes) were predicted to be decreased (Supplemental File 2, Figure S4). In addition, the canonical pathway for D-myo-inositol (1,4,5,6)-tetraphosphate biosynthesis (9 genes) was predicted to be activated and, as illustrated in Supplemental File 2, Figure S6, the WNT/Ca+ pathway was predicted to be decreased (4 genes).
Upstream regulators predicted to be activated in embryos that survived to day 60 were RB1 (17 molecules), NFKBIA (10 molecules), and peroxisome proliferator activated receptor alpha (PPARA) (11 molecules), while KDM5A (7 molecules), homeobox A10(HOXA10) (8 molecules), and miR-122-5p (5 molecules) were predicted to be inhibited (Supplemental File 2, Figure S7).
Differential expression of genes between embryos that maintained pregnancy from day 30 to day 60 vs those where pregnancy failed after day 30
The list of DEG is shown in Supplemental File 3, Table S6. There were 432 DEG using P < 0.01 and >2-fold or <0.5-fold difference as cutoffs. Of these, 315 were upregulated and 117 were downregulated in embryos that maintained pregnancy from day 30 to day 60 compared to those that failed to maintain pregnancy after day 30. Using an FDR-adjusted P < 0.05 and a fold-change of >2-fold or <0.5 as a cutoff, there were no DEG.
Molecular and cellular functions overrepresented in DEG are summarized in Supplemental File 3, Table S7. The top six functions based on P value were cell death and survival (131 genes), gene expression (89 genes), carbohydrate metabolism (13 genes), molecular transport (25 genes), small molecule biochemistry (30 genes), and lipid metabolism (19 molecules). Biological functions predicted to be increased in embryos that maintained pregnancy from day 30 to day 60 included those related to expression of RNA (86 genes), cell survival (59 genes), and organization of cytoplasm (58 genes), while organismal death (98 genes) was predicted to be inhibited (Supplemental File 2, Figure S8). In addition, the canonical pathway for apoptosis signaling (5 genes) was predicted to be activated (Supplemental File 2, Figure S9).
Upstream regulators predicted to be activated in embryos that survived from day 30 to day 60 were MYC proto-oncogene, bHLH transcription factor (MYC) (25 genes) and PPARA (9 genes), while the microRNA let-7 was predicted to be inhibited (11 genes) (Supplemental File 2, Figure S10).
Comparison of differentially expressed genes associated with embryo survival to day 30 and day 60 and for pregnancy maintenance between day 30 and day 60
There were 64 genes that were shared between DEG for maintenance of pregnancy until day 30 and pregnancy maintenance between day 30 and day 60, 140 genes that were shared between DEG for maintenance of pregnancy until day 60 and pregnancy maintenance between day 30 and day 60, and 61 genes shared between all three sets of DEG (embryos survival at day 30, day 60, and day 30 to day 60. The list of the genes in common for all three analyses is listed in Supplemental File 3, Table S8 and the molecular and cellular functions in which the genes are overrepresented in in Supplemental File 3, Table S9. Of the 61 genes, 7 were associated with lipid metabolism, 12 were associated with molecular transport, and 12 with small molecular biochemistry. There were no functions or canonical pathways predicted to change in activity because of DEG. There were no significant predicted upstream regulators.
Differentially expressed genes associated with inner cell mass, trophectoderm, or hypoblast
The list of DEG was screened to identify genes overexpressed in ICM [24]. The only gene identified as differentially expressed was interferon-tau 3g (IFNT), which was upregulated 56.4-fold in embryos that survived to day 30 (P = 0.0007), 103.9-fold in embryos that survived to day 60 (P = 0.0002), and 12.1 fold in embryos that survived from day 30 to day 60 (P = 022). Another gene reported to be important for formation of TE in bovine blastocysts, WW domain binding protein 1 (WBP1) [48], was upregulated 14.8-fold in embryos establishing pregnancy to day 30 (P = 0.012), 118.8-fold in embryos establishing pregnancy to day 60 (P = 0.0002), and 4417.4-fold in embryos that survived from day 30 to day 60 (P = 0.0052). Notch receptor 2 (NOTCH2), a gene implicated in TE formation in the mouse [29], was upregulated 64.7-fold in embryos establishing pregnancy to day 30 (P = 0.00005) and 41.2-fold in embryos establishing pregnancy to day 60 (P = 0.002). It was also downregulated 1.3-fold in embryos that survived from day 30 to day 60 (P = 0.58). TEA domain transcription factor 4 (TEAD4), which is involved in TE formation in the cow [30], was increased 1040.5-fold for embryos that survived from day 30 to day 60 as compared to those that did not.
Comparison of differentially expressed genes associated with embryo survival with similar results in the literature
The group of genes found to be related to competence of an embryo to survive to day 30 and day 60 of gestation was compared to other lists of genes that have been reported to be related to embryo survival to term [13–15]. Only three genes were in common with DEG in other papers. In particular, coenzyme Q7, hydroxylase (COQ7) [13], SEM1 26S proteasome complex subunit (SEM1) [15], and fatty acid binding protein 5 (FABP5) [14] were differentially expressed in the same direction between embryos that survived to day 30 and those that did not. Among the genes differently expressed in embryos that survived to day 60 in the current experiment, COQ7 and FABP5 were identified to be differently expressed in the same direction by El-Sayed et al. [13] and Salilew-Wondim et al. [14], respectively. ATPase H+ transporting V1 subunit F (ATP6V1F) and SEM1 were also identified to be differently expressed in the same direction by Ghanem et al. [15]. When comparing the genes differently regulated in embryos that survived from day 30 to day 60 vs died after day 30 with genes associated with maintenance of gestation to term [13–15], only WD repeat domain 47 (WDR47) was identified to be regulated in the same manner by Ghanem et al. [15].
Genes regulated by CSF2
The list of DEG is shown in Supplemental File S2, Table S10. There were 635 DEG using P < 0.01 and >2-fold or <0.5-fold difference as cutoffs, regardless of pregnancy outcome. Of these, 374 were upregulated and 261 were downregulated in CSF2-treated embryos. Using a more stringent criteria, a FDR-adjusted P <0.05 and a fold-change of >2-fold or <0.5, there were 420 DEG in CSF2-treateed embryos (262 upregulated and 158 downregulated).
Molecular and cellular functions overrepresented in DEG are summarized in Supplemental File 3, Table S11. The top functions were cell-to-cell signaling and interaction (17 genes), cell compromise (22 genes), post-translational modifications (51 genes), cell death and survival (190 genes), RNA post-transcriptional modification (5 genes), and carbohydrate metabolism (23 genes). As shown in Supplemental File 2, Figure S11, biological functions predicted to be increased based on changes in gene expression caused by CSF2 were an increase in accumulation of sphingolipid (5 genes) and cell death of epithelial cell lines (18 genes), while growth failure (35 genes) and aplasia or hypoplasia (32 genes) were predicted to be decreased.
The upstream regulators predicted to be activated were nuclear factor, erythroid 2 like 2 (NFE2L2) (11 molecules), PR/SET domain 1 (PRDM1) (4 genes), and thyroid hormone receptor alpha (THRA) (4 genes). Upstream regulators predicted to be inhibited were GLI family zinc finger 1 (GLI1) (5 genes), inhibitor of DNA binding 3, HLH protein (ID3) (5 genes), signal transducer and activator of transcription 6 (STAT6) (8 genes), and NK2 homeobox 3 (NKX2–3) (4 genes) (Supplemental File 2, Figure S12).
The group of genes regulated by CSF2 was compared with DEG found to be related to competence of an embryo to survive until day 30 of gestation. There were 54 upregulated and 28 downregulated DEG that overlapped between the genes regulated by CSF2 and genes associated with embryo survival to day 30.
The DEG for CSF2 were also screened for genes associated with ICM, TE, and hypoblast, as described above. As shown in Supplemental File 3, Table S10, embryos treated with CSF2 experienced a 44.2-fold increase in expression of muscleblind like splicing regulator 3 (CDX2) (P = 0.00163), a 63-fold upregulation of WBP1 (P = 0.00025), a 250.3-fold decrease in expression of MBNL3 (P = 0.0002), and a 76.8-fold increase in TEAD4 (P = 0.0023). In addition, Yes associated protein 1 (YAP1), which is involved in TE formation in cattle [31], was increased 9.8-fold by CSF2 (P = 0.0214), and large tumor suppressor kinase 2 (LATS2), which binds angiomotin (AMOT), was increased 271.3-fold (P < 0.0001).
Comparison of genes regulated by CSF2 in bovine blastocyst with the literature
The genes differently expressed in embryos treated with CSF2 in the current experiment were compared to genes previously reported to be regulated by CSF2 in ICM and TE of day 8 bovine blastocysts [17]. Only three genes were regulated by CSF2 in a similar manner in both experiments. DEAD-box helicase 28 (DDX28) (ICM) and FosB proto-oncogene, AP-1 transcription factor subunit (FOSB) (TE) were upregulated by CSF2 in both experiments and post-GPI attachment to proteins phospholipase 3 (PGAP3) (ICM) was downregulated in both experiments.
Differential expression of genes between embryos that maintained pregnancy to day 30 vs those that did not within control and CSF2 groups
The list of DEG associated with embryonic survival to day 30, as determined separately for control embryos and CSF2 embryos, is shown in Supplemental File 3, Tables S12 and S13, respectively. Within the control group, there were 465 DEG using P < 0.01 and >2-fold or <0.5-fold difference as cutoffs. Of these, 303 were upregulated and 162 were downregulated. Using a more stringent criteria, a FDR-adjusted P <0.05 and a fold-change of >2-fold or <0.5, there were 55 DEG in control embryos that maintained pregnancy (54 upregulated and 1 downregulated). Within the CSF2 group, there were 591 DEG using P < 0.01 and >2-fold or <0.5-fold difference as cutoffs. Of these, 324 were upregulated and 267 were downregulated in CSF2 embryos that maintained pregnancy. Using a more stringent criteria, a FDR-adjusted P <0.05, and a fold-change of >2-fold or <0.5, there were 246 DEG (153 upregulated and 93 downregulated).
The DEG for embryonic survival to day 30 for CSF2 embryos was compared to the DEG for embryo survival to day 30 in the control group. There were 27 upregulated and 10 downregulated genes that overlapped between the two sets of DEG. The list of these genes is in Supplemental File 3, Table S14 and the molecular and cellular functions in which the genes are overrepresented is summarized in Supplemental File 3, Table S15. Of the 27 genes, 13 were associated with cell morphology, cellular assembly and organization, cellular function, and maintenance and 17 with cell death and survival. There were no functions or canonical pathways predicted to change in activity because of the DEG. The one significant predicted upstream regulator was RB transcriptional corepressor 1 (RB1) (regulated five genes), which was predicted to be activated.
Discussion
The present results confirm earlier findings that there are significant differences in gene expression patterns between blastocysts that established pregnancy after transfer into recipients vs those that either did not establish pregnancy or which will experience embryonic death after day 30 of gestation [13–15]. Furthermore, CSF2, which can increase post-transfer survival of the blastocyst of the cow [3, 4], mouse [32], and human [33], also alters the blastocyst transcriptome. Thus, the transcriptome of the blastocyst is related to its subsequent ability to survive and develop to at least 60 days of gestation.
A summary of changes in gene expression associated with embryo survival and CSF2 treatment is shown in Figure 5. Changes in gene expression associated with pregnancy outcomes and CSF2 treatment were large. For example, after correcting for FDR there were 370 genes whose expression differed between embryos that established a pregnancy until day 30 vs those that did not. The identification of large numbers of DEG as compared to earlier experiments [13–15] probably reflects the use of RNA-Seq as compared to microarray [34] as well as the large number of samples analyzed.
Figure 5.
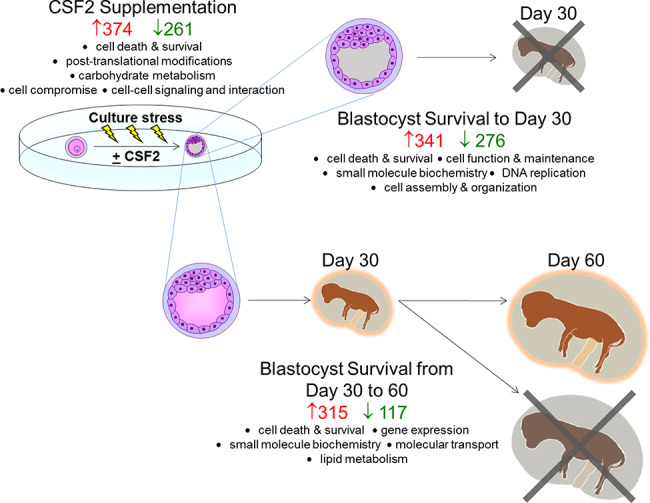
Summary of changes in gene expression in the blastocyst produced in vitro related to subsequent survival to day 30 and day 60 of pregnancy and treatment with CSF2. The number of upregulated and downregulated genes for each comparison is illustrated in red and green, respectively. The top molecular and cellular functions in which differentially expressed genes were overrepresented are listed. The predominance of terms related to cellular responses to stress (cell death and survival, cell compromise, DNA replication, recombination and repair, and cellular assembly and organization) is indicative of the importance of embryo adjustment to stress associated with in vitro production and embryo culture for pregnancy outcomes.
There were associations of embryonic survival with expression of a variety of genes and gene networks. One of the most notable type of biological functions related to embryonic survival were associated with responses to cellular stress. Thus, for example, one of the biological functions predicted to decrease in embryos that survived to day 30 was organismal death. Similarly, it was predicted that alterations in gene expression for embryos that survived to day 60 would cause increased cell survival and repair of DNA. Biological functions predicted to be activated in embryos that maintained pregnancy from day 30 to day 60 included cell survival, while organismal death and apoptosis signaling were predicted to be reduced in blastocysts which subsequently survived from day 30 to day 60 or gestation. Interestingly, one of the genes increased in embryos that survived to day 30 and day 60 was IFNT, a gene that is crucial for signaling the mother to prevent luteolysis [35]. Transcription of this gene is regulated by stress because heat shock increased expression of IFNT in bovine blastocysts [36].
The relationship between expression of cell stress genes and subsequent embryonic survival probably reflects cellular perturbations caused by embryonic development taking place in the artificial environment associated with cell culture. Indeed, the proportion of embryos that can develop to the blastocyst stage is greater in vivo than in vitro [37]. So too is pregnancy rate after transfer of embryos into recipient females [2]. In two studies [37, 38], blastocysts produced in vitro had higher expression of several genes involved in responses to reactive oxygen as compared to embryos that developed in vivo. Moreover, embryos with superior competence to develop to the blastocyst stage (as determined by time after fertilization when blastocyst formation occurs) had higher transcript abundance for antioxidant genes as well as the transcription factor NRF2 (i.e., NFE2L2) [36], which functions to regulate expression of antioxidant genes [40].
Actions of CSF on the embryo also involve modification of stress responses. Among the molecular and cellular functions containing an overrepresentation of DEG regulated by CSF2 was that for cell death and survival. In addition, one of the predicted upstream regulators for CSF2 was NFE2L2, which, as stated previously, is a transcription factor that binds to antioxidant response elements in promoter region of target genes [37]. Additional evidence that CSF2 regulates stress responses comes from the literature. Treatment with CSF2 can block apoptosis in the bovine embryo [41] and CSF2 also regulated expression of cell stress genes in the mouse [42]. It has been noted that CSF2 increases the percent of embryos becoming a blastocyst when the overall proportion of embryos becoming blastocysts is low but that CSF2 is actually inhibitory to blastocyst development when the overall amount of blastocyst development is high [43]. Perhaps, CSF2 increases competence of embryos to become blastocysts only when cell stress is high.
Additional evidence that there was a relationship between an embryo’s ability to develop normally in the artificial environment of the culture dish and subsequent competence to establish pregnancy was provided by the fact that many DEG associated with pregnancy outcomes at day 30 and day 60 were related to lipid metabolism. Moreover, PPARA or PPARG, which can regulate lipid and glucose metabolism [44, 45], was predicted to be activated in embryos that could survive to day 30 (PPARG), day 60 (PPARA), or from day 30 to day 60 (PPARA). Culture of bovine embryos causes alterations in lipid metabolism as shown by increased accumulation of cytoplasmic lipid droplets [46, 47] and alterations in expression of genes involved in lipid metabolism [37–39]. Thus, embryos that are more able to resist perturbations in lipid metabolism caused by culture may be more capable to establish pregnancy.
There were no consistent differences between embryos that established pregnancy or not in expression of genes characteristic of ICM or TE, suggesting that embryonic survival was not determined by the relative proportion of cells of the two lineages in the demi-embryo that was transferred following biopsy. However, NOTCH2 was overexpressed in embryos that survived to day 30 and day 60. Notch signaling has been implicated in formation of TE in the mouse [29]. Further evidence that characteristics of TE cells may be important for embryonic survival was the finding that expression of WBP1 was higher for embryos that survived to day 30, day 60, and from day 30 to day 60 than for embryos that failed to survive to those endpoints. Moreover, CSF2 increased expression of CDX2, LATS2, TEAD4, WBP1, and YAP1. WBP1 is a putative binding protein for YAP1 and is important for blastocyst formation and development of TE in cattle [22]. YAP1 interacts with TEAD4 to promote CDX2 expression and TE formation [30, 31]. LATS2 is an important component of Hippo signaling; inhibition of a downstream effector of LATS2, AMOT, decreased TE formation in cattle [31].
A significant proportion of pregnancies that become established and persist to days 28–30 of gestation are subsequently lost spontaneously. In the current experiment, 37.5% of pregnancies at day 30 were lost by day 60. This number might be high because of the use of demi-embryos, which likely have a higher mortality rate compared to intact blastocysts. For lactating cows bred by artificial insemination, it has been estimated that 12% of pregnancies at day 28 are lost by day 60 [48]. Causes for this loss are unknown. Although it is likely that events occurring early in development can program the embryo in a way that affects the likelihood of subsequent pregnancy loss. Evidence for this idea comes from experiments such as the present and other experiments [13–15] that gene expression at day 7 is associated with subsequent pregnancy loss between day 30 and day 60. Also, treatments applied early in development (e.g., exposure to CSF2; [3]) can modify subsequent pregnancy loss after initial diagnosis at day 28 of gestation. A total of 15% of the genes associated with embryo survival from day 30 to day 60 were also associated with survival to day 30 (64 of 432), indicating that, for the most part, genes and gene networks important for continued development after day 30 were distinct from those for establishment of pregnancy at day 30.
A large proportion of genes (74%; 349 of 470) that were associated with embryo survival to day 36 were also associated with survival to day 30. This is to be expected because of the biological association between embryonic survival at day 30 and at day 60 (an embryo cannot survive to day 60 unless it also survives to day 30). The fact that this proportion was not even larger probably reflects that some mechanisms for embryo survival to day 60 are different than for survival to day 30 (see preceding paragraph) and differences in statistical power between comparisons.
An important question is whether expression of genes important for embryonic survival in control embryos is the same or different than the genes important for embryonic survival in CSF2-treated embryos. Loss of power associated with analysis of the subsets of data consisting of only control or CSF2-treated embryos means that it is likely that not every gene that is similarly related to embryonic survival in both groups of embryos will be statistically significant. It is notable, though, that only a small number of genes (37) were in common between DEG associated with embryo survival to day 30 in control and CSF2 groups, suggesting that there are differences in the genes important for embryonic survival among the groups. One implication of this result is that the specific conditions to produce embryos in vitro are likely to change which genes are important for embryonic survival. Consistent with this idea was the finding that few genes related to embryonic survival found previously [13–15] were also associated with embryonic survival in the present experiment. Similarly, few of the genes whose expression in blastocysts was regulated by CSF2 in an earlier experiment [17] were similarly regulated in the present study. There are many possible reasons for lack of reproducibility including the system used to produce embryos, genetic differences in the embryos used in each experiment, and the fact that the current experiment was limited to female embryos. Perhaps, there are key genes that are critical for embryonic survival or actions of CSF2 but these are obscured by other changes in gene expression that reflect non-critical changes in cellular function.
The four genes that were associated with embryonic survival in this experiment and earlier ones [13–15] were ATP6V1F, COQ7, FABP5, and SEM1. Each of these genes may be involved in allowing embryonic survival to stress. ATP6V1F encodes a component of vacuolar ATPase, which is a multisubunit enzyme that mediates acidification of intracellular organelles such as lysosomes [49, 50]. COQ7 encodes a protein that is the central regulatory factor of CoQ complex [51]. CoQ is a lipid soluble molecule that carries electrons from complexes I and II to complex III of the mitochondrial respiratory chain [52]. COQ7 also has antioxidant properties and regulates permeability transition pores that can lead to mitochondrial swelling and cell death through apoptosis or necrosis [52]. Interestingly, COQ7 is stabilized by COQ9 [53], which contains a mutation important for fertility and ovarian function in cattle [8, 54, 55]. SEM1 encodes a multifunctional protein that plays a role in several processes including the BRCA2 complex required for homologous recombination [56], and the 26S proteasome complex that recognizes and degrades ubiquitylated proteins [56]. Finally, FABP5 encodes a fatty acid binding protein, which can participate in transcriptional regulation through activation of peroxisome proliferator-activated receptors [57, 58]. Known gene targets for these receptors are involved in cellular glucose and lipid homeostasis [59–61], differentiation [62, 63], and resistance to apoptosis [63, 64].
Only DDX28 and FOSB (both upregulated) and PGAP3 (downregulated) were regulated by CSF2 in the same direction in the current experiment and an earlier study [17]. DDX28 is an RNA helicase that participates in formation of mitochondrial RNA granules and mitoribosomes [65]. FOSB is a component of the AP-1 transcription factor complex, which participates in a wide variety of cell functions including trophoblast invasiveness in the human [66]. PGAP3 is a phospholipase involved in remodeling of glycosylphosphatidylinositol membrane-protein anchors [67].
Among the transcriptional factors activated in embryos that maintained gestation to day 30 or 60 were NFKBIA and RB1. NFKBIA encodes a protein, IkBα that binds to NFκB dimers to inhibit translocation to the nucleus [68]. Once in the cytoplasm NFκB is degraded and cell proliferation is inhibited [69] and differentiation promoted [70]. Similarly, RB1 is an important protein involved in cell cycle arrest and induction of differentiation [71]. Perhaps, the actions of these transcription factors promote differentiation of the blastocyst. Among the transcription factors inhibited in embryos that maintained gestation until day 60 was HOXA10, which has also been described to play an important role in the regulation of cell differentiation [72, 73].
Another transcriptional regulator predicted to be activated in embryos that maintained gestation from day 30 to day 60 was MYC. MYC has been described to be an important gene for embryonic stem cell self-renewal and pluripotency [74]. As mentioned before, PPARA, which was predicted to be activated in embryos that survived to day 60 and from day 30 to day 60, is involved not only in cellular glucose and lipid homeostasis [44, 45] but also in differentiation [63, 64] and resistance to apoptosis [62, 63]. Finally, let-7, which encodes a microRNA involved in repression of cell proliferation [75], was also activated in embryos that were able to maintain gestation from day 30 to day 60. The regulation of the expression of these transcription factors and microRNA in embryos able to maintain gestation reinforces the importance of biological functions related to cell proliferation, differentiation, and repression of apoptosis for embryonic competence to develop to term.
Actions of CSF2 on gene expression also involve predicted changes in transcriptional regulators. Besides NFE2L2, which has been mentioned above, another upstream regulator predicted to be activated was PRDM1 (also known as BLIMP1). PRDM1 is critical for germ cell specification [76] and differentiation of trophoblast stem cells [77, 78]. Two transcriptional regulators predicted to be inhibited by CSF2, GLI1 and ID3, are mediators of Hedgehog signaling. In the mouse, sonic hedgehog is expressed in the TE of the blastocyst; knockdown leads to reduced yolk sac vascularization and formation of the placental labyrinth [79]. These findings, as well as the observation that CSF2 upregulates several genes involved in TE differentiation (CDX2, LATS2, TEAD4, WBP1, and YAP1), point to the TE as a possible target for actions of CSF2 to increase competence of embryos to establish pregnancy.
It should be noted that all the embryos analyzed for gene expression were females. The decision to limit analysis to female embryos was based on the fact that there are differences in gene expression between male and female blastocysts [80], and more importantly, CSF2 affects female embryos differently than male embryos [18, 19]. Taken together, results suggest that how an individual embryo can respond to alterations in cellular function caused by the stress of culture is an important determinant of its subsequent ability to develop in utero to day 30 and day 60 of pregnancy. Given that the embryo produced in vivo does not experience the same environment as for the in vitro produced embryo, it remains to be seen whether different gene networks are important for long-term survival of embryo produced in vivo.
An interesting question for further research is whether some of the variation among embryos for competence to survive after transfer is genetic in origin. If so, the use of assisted reproductive technologies to produce embryos could change the frequency of specific alleles in the offspring in a way that affects postnatal function. Differences in postnatal performance between offspring produced by in vitro fertilization vs those that developed completely in vivo have been noted for mice [81, 82] and cattle [83]. While epigenetic dysfunction is probably the major cause for alterations in postnatal phenotype, a change in gene frequencies should be considered also.
Conflict of interest
The authors have declared that no conflict of interest exists.
Supplementary Material
Acknowledgments
The authors thank the owners and staff of Full Circle Dairy LCC (Lee, Florida) for the use of cows and facilities and, in particular, Eric Diepersloot for help in implementing the study. The authors also thank the University of Bonn and, in particular, Franca Rings for training on embryo bisection. Thanks are also extended to Zoetis, Inc. for performing RNA Seq. We extend thanks to Odinei Marques for help in conducting the field experiment. This research was supported by Zoetis Inc. and NIH R01 HD088352.
References
- 1. McMillan WH. Statistical models predicting embryo survival to term in cattle after embryo transfer. Theriogenology 1998; 50:1053–1070. [DOI] [PubMed] [Google Scholar]
- 2. Ferraz PA, Burnley C, Karanja J, Viera-Neto A, Santos JE, Chebel RC, Galvão KN. Factors affecting the success of a large embryo transfer program in Holstein cattle in a commercial herd in the southeast region of the United States. Theriogenology 2016; 86:1834–1841. [DOI] [PubMed] [Google Scholar]
- 3. Loureiro B, Bonilla L, Block J, Fear JM, Bonilla AQ, Hansen PJ. Colony-stimulating factor 2 (CSF-2) improves development and post-transfer survival of bovine embryos produced in vitro. Endocrinology 2009; 150:5046–5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Denicol AC, Block J, Kelley DE, Pohler KG, Dobbs KB, Mortensen CJ, Ortega MS, Hansen PJ. The WNT signaling antagonist Dickkopf-1 directs lineage commitment and promotes survival of the preimplantation embryo. FASEB J 2014; 28:3975–3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smith SL, Everts RE, Tian XC, Du F, Sung LY, Rodriguez-Zas SL, Jeong BS, Renard JP, Lewin HA, Yang X. Global gene expression profiles reveal significant nuclear reprogramming by the blastocyst stage after cloning. Proc Natl Acad Sci 2005; 102:17582–17587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smith SL, Everts RE, Sung LY, Du F, Page RL, Henderson B, Rodriguez-Zas SL, Nedambale TL, Renard JP, Lewin HA, Yang X, Tian XC. Gene expression profiling of single bovine embryos uncovers significant effects of in vitro maturation, fertilization and culture. Mol Reprod Dev 2009; 76:38–47. [DOI] [PubMed] [Google Scholar]
- 7. Corcoran D, Rizos D, Fair T, Evans AC, Lonergan P. Temporal expression of transcripts related to embryo quality in bovine embryos cultured from the two-cell to blastocyst stage in vitro or in vivo. Mol Reprod Dev 2007; 74:972–977. [DOI] [PubMed] [Google Scholar]
- 8. Cochran SD, Cole JB, Null DJ, Hansen PJ. Discovery of single nucleotide polymorphisms in candidate genes associated with fertility and production traits in Holstein cattle. BMC Genet 2013; 14:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ortega MS, Denicol AC, Cole JB, Null DJ, Taylor JF, Schnabel RD, Hansen PJ. Association of single nucleotide polymorphisms in candidate genes previously related to genetic variation in fertility with phenotypic measurements of reproductive function in Holstein cows. J Dairy Sci 2017; 100:3725–3734. [DOI] [PubMed] [Google Scholar]
- 10. Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, Eyler AE, Denny JC, GTEx Consortium, Nicolae DL, Cox NJ, Im HK. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet 2015; 47:1091–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BW, Jansen R, de Geus EJ, Boomsma DI, Wright FA, Sullivan PF, Nikkola E et al. . Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet 2016; 48:245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heinig M. Using gene expression to annotate cardiovascular GWAS loci. Front Cardiovasc Med 2018; 5:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. El-Sayed A, Hoelker M, Rings F, Salilew D, Jennen D, Tholen E, Sirard MA, Schellander K, Tesfaye D. Large-scale transcriptional analysis of bovine embryo biopsies in relation to pregnancy success after transfer to recipients. Physiol Genomics 2006; 28:84–96. [DOI] [PubMed] [Google Scholar]
- 14. Salilew-Wondim D, Hölker M, Rings F, Ghanem N, Ulas-Cinar M, Peippo J, Tholen E, Looft C, Schellander K, Tesfaye D. Bovine pretransfer endometrium and embryo transcriptome fingerprints as predictors of pregnancy success after embryo transfer. Physiol Genomics 2010; 42:201–218. [DOI] [PubMed] [Google Scholar]
- 15. Ghanem N, Salilew-Wondim D, Gad A, Tesfaye D, Phatsara C, Tholen E, Phatsara C, Tholen E, Looft C, Schellander K, Hoelker M. Bovine blastocysts with developmental competence to term share similar expression of developmentally important genes although derived from different culture environments. Reproduction 2011; 142:551–564. [DOI] [PubMed] [Google Scholar]
- 16. Loureiro B, Oliveira LJ, Favoreto MG, Hansen PJ. Colony-stimulating factor 2 inhibits induction of apoptosis in the bovine preimplantation embryo. Am J Reprod Immunol 2011; 65:578–588. [DOI] [PubMed] [Google Scholar]
- 17. Ozawa M, Sakatani M, Dobbs KB, Kannampuzha-Francis J, Hansen PJ. Regulation of gene expression in the bovine blastocyst by colony stimulating factor 2. BMC Res Notes 2016; 9:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Siqueira LGB, Hansen PJ. Sex differences in response of the bovine embryo to colony-stimulating factor 2. Reproduction 2016; 152:645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dobbs KB, Gagné D, Fournier E, Dufort I, Robert C, Block J, Sirard MA, Bonilla L, Ealy AD, Loureiro B, Hansen PJ. Sexual dimorphism in developmental programming of the bovine preimplantation embryo caused by colony-stimulating factor 2. Biol Reprod 2014; 91. [DOI] [PubMed] [Google Scholar]
- 20. Siqueira LG, Tribulo P, Chen Z, Denicol AC, Ortega MS, Negrón-Pérez VM, Kannampuzha-Francis J, Pohler KG, Rivera RM, Hansen PJ. Colony-stimulating factor 2 acts from days 5 to 7 of development to modify programming of the bovine conceptus at day 86 of gestation. Biol Reprod 2017;96(4):743–757. [DOI] [PubMed] [Google Scholar]
- 21. Kannampuzha-Francis J, Denicol AC, Loureiro B, Kaniyamattam K, Ortega MS, Hansen PJ. Exposure to colony stimulating factor 2 during preimplantation development increases postnatal growth in cattle. Mol Reprod Dev 2015; 82:892–897. [DOI] [PubMed] [Google Scholar]
- 22. Ortega MS, Kurian JJ, McKenna R, Hansen PJ. Characteristics of candidate genes associated with embryonic development in the cow: Evidence for a role for WBP1 in development to the blastocyst stage. PLoS One 2017; 12:e0178041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Negrón-Pérez VM, Zhang Y, Hansen PJ. Single-cell gene expression of the bovine blastocyst. Reproduction 2017; 154:627–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chang T-C, Yang Y, Retzel EF, Liu W-S. Male-specific region of the bovine Y chromosome is gene rich with a high transcriptomic activity in testis development. Proc Natl Acad Sci USA 2013; 110:12373–12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 2010; 11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou X, Lindsay H, Robinson MD. Robustly detecting differential expression in RNA sequencing data using observation weights. Nucleic Acids Res 2014; 42:e91–e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lund SP, Nettleton D, McCarthy DJ, Smyth GK. Detecting differential expression in RNA-sequence data using quasi-likelihood with shrunken dispersion estimates. Stat Appl Genet Mol Biol 2012; 11. [DOI] [PubMed] [Google Scholar]
- 29. Rayon T, Menchero S, Nieto A, Xenopoulos P, Crespo M, Cockburn K, Cañon S, Sasaki H, Hadjantonakis AK, Pompa JL, Rossant J, Manzanares M. Notch and hippo converge on CDX2 to specify the trophectoderm lineage in the mouse blastocyst. Dev Cell 2014; 30:410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Akizawa H, Kobayashi K, Bai H, Takahashi M, Kagawa S, Nagatomo H, Kawahara M. Reciprocal regulation of TEAD4 and CCN2 for the trophectoderm development of the bovine blastocyst. Reproduction 2018; 155:563–571. [DOI] [PubMed] [Google Scholar]
- 31. Negrón-Pérez VM, Hansen PJ. Role of yes-associated protein 1, angiomotin, and mitogen-activated kinase kinase 1/2 in development of the bovine blastocyst. Biol Reprod 2018; 98:170–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sjöblom C, Roberts CT, Wikland M, Robertson SA. Granulocyte-macrophage colony-stimulating factor alleviates adverse consequences of embryo culture on fetal growth trajectory and placental morphogenesis. Endocrinology 2005; 146:2142–2153. [DOI] [PubMed] [Google Scholar]
- 33. Ziebe S, Loft A, Povlsen BB, Erb K, Agerholm I, Aasted M, Gabrielsen A, Hnida C, Zobel DP, Munding B, Bendz SH, Robertson SA. A randomized clinical trial to evaluate the effect of granulocyte-macrophage colony-stimulating factor (GM-CSF) in embryo culture medium for in vitro fertilization. Fertil Steril 2013; 99:1600, e2–1609. [DOI] [PubMed] [Google Scholar]
- 34. Zhao S, Fung-Leung W-P, Bittner A, Ngo K, Liu X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One 2014; 9:e78644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Forde N, Lonergan P. Interferon-tau and fertility in ruminants. Reproduction 2017; 154:33–43. [DOI] [PubMed] [Google Scholar]
- 36. Hickman CF, Clinton M, Ainslie A, Ashworth CJ, Rooke JA. Heat shock induces interferon-TAU gene expression by in vitro-produced bovine blastocysts. Am J Reprod Immunol 2013; 70:177–181. [DOI] [PubMed] [Google Scholar]
- 37. Gad A, Hoelker M, Besenfelder U, Havlicek V, Cinar U, Rings F, Held E, Dufort I, Sirard MA, Schellander K, Tesfaye D. Molecular mechanisms and pathways involved in bovine embryonic genome activation and their regulation by alternative in vivo and in vitro culture conditions. Biol Reprod 2012; 87:1–13. [DOI] [PubMed] [Google Scholar]
- 38. Heras S, De Coninck DIM, Van Poucke M, Goossens K, Bogado Pascottini O, Van Nieuwerburgh F, Deforce D, De Sutter P, Leroy JL, Gutierrez-Adan A, Peelman L, Van Soom A. Suboptimal culture conditions induce more deviations in gene expression in male than female bovine blastocysts. BMC Genomics 2016; 17:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Amin A, Gad A, Salilew-Wondim D, Prastowo S, Held E, Hoelker M, Rings F, Tholen E, Neuhoff C, Looft C, Schellander K, Tesfaye D. Bovine embryo survival under oxidative-stress conditions is associated with activity of the NRF2-mediated oxidative-stress-response pathway. Mol Reprod Dev 2014; 81:497–513. http://www.ncbi.nlm.nih.gov/pubmed/25057524. [DOI] [PubMed] [Google Scholar]
- 40. Lacher SE, Lee JS, Wang X, Campbell MR, Bell DA, Slattery M. Beyond antioxidant genes in the ancient Nrf2 regulatory network. Free Radic Biol Med. 2015; 88:452–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Loureiro B, Block J, Favoreto MG, Carambula S, Pennington KA, Ealy AD, Hansen PJ. Consequences of conceptus exposure to colony-stimulating factor 2 on survival, elongation, interferon-τ secretion, and gene expression. Reproduction 2011; 141:617–624. [DOI] [PubMed] [Google Scholar]
- 42. Chin PY, Macpherson AM, Thompson JG, Lane M, Robertson SA. Stress response genes are suppressed in mouse preimplantation embryos by granulocyte-macrophage colony-stimulating factor (GM-CSF). Hum Reprod 2009; 24:2997–3009. [DOI] [PubMed] [Google Scholar]
- 43. Dobbs KB, Khan FA, Sakatani M, Moss JI, Ozawa M, Ealy AD, Hansen PJ. Regulation of pluripotency of inner cell mass and growth and differentiation of trophectoderm of the bovine embryo by colony stimulating factor 2. Biol Reprod 2013; 89:141. [DOI] [PubMed] [Google Scholar]
- 44. Semple RK, Chatterjee VKK, O’Rahilly S. PPAR and human metabolic disease. J Clin Invest 2006; 116:581–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kersten S, Stienstra R. The role and regulation of the peroxisome proliferator activated receptor alpha in human liver. Biochimie 2017; 136:75–84. [DOI] [PubMed] [Google Scholar]
- 46. Abe H, Otoi T, Tachikawa S, Yamashita S, Satoh T, Hoshi H. Fine structure of bovine morulae and blastocysts in vivo and in vitro. Anat Embryol (Berl) 1999; 199:519–527. http://www.ncbi.nlm.nih.gov/pubmed/10350132. [DOI] [PubMed] [Google Scholar]
- 47. Rizos D, Ward F, Duffy P, Boland MP, Lonergan P. Consequences of bovine oocyte maturation, fertilization or early embryo development in vitro versus in vivo: Implications for blastocyst yield and blastocyst quality. Mol Reprod Dev 2002; 61:234–248. [DOI] [PubMed] [Google Scholar]
- 48. Wiltbank MC, Baez GM, Garcia-Guerra A, Toledo MZ, Monteiro PLJ, Melo LF, Ochoa JC, Santos JE, Sartori R. Pivotal periods for pregnancy loss during the first trimester of gestation in lactating dairy cows. Theriogenology 2016; 86:239–253. [DOI] [PubMed] [Google Scholar]
- 49. Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol 2007; 8:917–929. [DOI] [PubMed] [Google Scholar]
- 50. Wagner CA, Finberg KE, Breton S, Marshansky V, Brown D, Geibel JP. Renal vacuolar H+-ATPase. Physiol Rev 2004; 84:1263–1314. [DOI] [PubMed] [Google Scholar]
- 51. Cascajo MV, Abdelmohsen K, Noh JH, Fernández-Ayala DJM, Willers IM, Brea G, López-Lluch G, Valenzuela-Villatoro M, Cuezva JM, Gorospe M, Siendones E, Navas P. RNA-binding proteins regulate cell respiration and coenzyme Q biosynthesis by post-transcriptional regulation of COQ7. RNA Biol 2016; 13:622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Turunen M, Olsson J, Dallner G. Metabolism and function of coenzyme Q. Biochim Biophys Acta 2004; 1660:171–199. [DOI] [PubMed] [Google Scholar]
- 53. Lohman DC, Forouhar F, Beebe ET, Stefely MS, Minogue CE, Ulbrich A, Stefely JA, Sukumar S, Luna-Sánchez M, Jochem A, Lew S, Seetharaman J et al. . Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis. Proc Natl Acad Sci USA 2014; 111:E4697–E4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ortega MS, Wohlgemuth S, Tribulo P, Siqueira LGB, Null DJ, Cole JB, Sirard MA, Bonilla L, Ealy AD, Loureiro B, Hansen PJ. A single nucleotide polymorphism in COQ9 affects mitochondrial and ovarian function and fertility in Holstein cows. Biol Reprod 2017; 96:652–663. [DOI] [PubMed] [Google Scholar]
- 55. Zolini AM, Ortiz WG, Estrada-Cortes E, Ortega MS, Dikmen S, Sosa F, Giordano JO, Hansen PJ. Interactions of human chorionic gonadotropin with genotype and parity on fertility responses of lactating dairy cows. J Dairy Sci 2019; 102:846–856. [DOI] [PubMed] [Google Scholar]
- 56. Kragelund BB, Schenstrøm SM, Rebula CA, Panse VG, Hartmann-Petersen R. DSS1/Sem1, a multifunctional and intrinsically disordered protein. Trends Biochem Sci 2016; 41:446–459. [DOI] [PubMed] [Google Scholar]
- 57. Bao Z, Malki MI, Forootan SS, Adamson J, Forootan FS, Chen D, Foster CS, Rudland PS, Ke Y. A novel cutaneous fatty acid-binding protein-related signaling pathway leading to malignant progression in prostate cancer cells. Genes Cancer 2013; 4:297–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Armstrong EH, Goswami D, Griffin PR, Noy N, Ortlund EA. Structural basis for ligand regulation of the fatty acid-binding protein 5, peroxisome proliferator-activated receptor β/δ (FABP5-PPAR β/δ) Signaling Pathway 2014;289:14941–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Barak Y, Liao D, He W, Ong ES, Nelson MC, Olefsky JM, Boland R, Evans RM. Effects of peroxisome proliferator-activated receptor on placentation, adiposity, and colorectal cancer. Proc Natl Acad Sci 2002; 99:303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Akiyama TE, Lambert G, Nicol CJ, Matsusue K, Peters JM, Brewer HB, Gonzalez FJ. Peroxisome proliferator-activated receptor β/δ regulates very low density lipoprotein production and catabolism in mice on a Western diet. J Biol Chem 2004; 279:20874–20881. [DOI] [PubMed] [Google Scholar]
- 61. Cohen SM, Ogawa S, Shulman RG. 13C NMR studies of gluconeogenesis in rat liver cells: Utilization of labeled glycerol by cells from euthyroid and hyperthyroid rats. Proc Natl Acad Sci 1979; 76:1603–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nadra K, Anghel SI, Joye E, Tan NS, Basu-Modak S, Trono D, Wahli W, Desvergne B. Differentiation of trophoblast giant cells and their metabolic functions are dependent on peroxisome proliferator-activated receptor β/δ. Mol Cell Biol 2006; 26:3266–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tan NS, Michalik L, Noy N, Yasmin R, Pacot C, Heim M, Flühmann B, Desvergne B, Wahli W. Critical roles of PPARβ/δ in keratinocyte response to inflammation. Genes Dev 2001; 15:3263–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Di-Poï N, Tan NS, Michalik L, Wahli W, Desvergne B. Antiapoptotic role of PPARβ in keratinocytes via transcriptional control of the Akt1 signaling pathway. Mol Cell 2002; 10:721–733. http://www.ncbi.nlm.nih.gov/pubmed/12419217. [DOI] [PubMed] [Google Scholar]
- 65. Tu YT, Barrientos A. The human mitochondrial DEAD-box protein DDX28 resides in RNA granules and functions in mitoribosome assembly. Cell Rep 2015; 10:854–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Renaud SJ, Kubota K, Rumi MA, Soares MJ. The FOS transcription factor family differentially controls trophoblast migration and invasion. J Biol Chem 2014; 289:5025–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Maeda Y, Tashima Y, Houjou T, Fujita M, Yoko-o T, Jigami Y, Taguchi R, Kinoshita T. Fatty acid remodeling of GPI-anchored proteins is required for their raft association. Mol Biol Cell 2007;18(4):1497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Malek S, Huxford T, Ghosh G. IκBα functions through direct contacts with the nuclear localization signals and the DNA binding sequences of NF-κB. J Biol Chem 1998; 273:25427–25435. [DOI] [PubMed] [Google Scholar]
- 69. Zhang W, Cheng P, Hu W, Yin W, Guo F, Chen A, Huang H. Inhibition of microRNA-384-5p alleviates osteoarthritis through its effects on inhibiting apoptosis of cartilage cells via the NF-κB signaling pathway by targeting SOX9. Cancer Gene Ther 2018; 25:326–338. [DOI] [PubMed] [Google Scholar]
- 70. Ruijtenberg S, Heuvel S. Coordinating cell proliferation and differentiation: antagonism between cell cycle regulators and cell type-specific gene expression. Cell Cycle 2016; 15:196–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Beshiri ML, Holmes KB, Richter WF, Hess S, Islam ABM, Yan Q, Plante L, Litovchick L, Gévry N, Lopez-Bigas N, Kaelin WG Jr, Benevolenskaya EV. Coordinated repression of cell cycle genes by KDM5A and E2F4 during differentiation. Proc Natl Acad Sci 2012; 109:18499–18504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yamamoto M, Kuroiwa A. HOXA-11 and HOXA-13 are involved in repression of MyoD during limb muscle development. Dev Growth Differ 2003; 45:485–498. [DOI] [PubMed] [Google Scholar]
- 73. Gross S, Krause Y, Wuelling M, Vortkamp A. HOXA11 and HOXD11 regulate chondrocyte differentiation upstream of RUNX2 and SHOX2 in mice. PLoS One 2012; 7:e43553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Varlakhanova NV, Cotterman RF, deVries WN, Morgan J, Donahue LR, Murray S, Knowles BB, Knoepfler PS. myc maintains embryonic stem cell pluripotency and self-renewal. Differentiation 2010; 80:9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, Chin L, Brown D et al. . The let-7 MicroRNA represses cell proliferation pathways in human cells. Cancer Res 2007; 67:7713–7722. [DOI] [PubMed] [Google Scholar]
- 76. Ohinata Y, Payer B, O'Carroll D, Ancelin K, Ono Y, Sano M, Barton SC, Obukhanych T, Nussenzweig M, Tarakhovsky A, Saitou M, Surani MA. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 2005; 436:207–213. [DOI] [PubMed] [Google Scholar]
- 77. Mould A, Morgan MA, Li L, Bikoff EK, Robertson EJ. Blimp1/Prdm1 governs terminal differentiation of endovascular trophoblast giant cells and defines multipotent progenitors in the developing placenta. Genes Dev 2012; 26:2063–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Nelson AC, Mould AW, Bikoff EK, Robertson EJ. Mapping the chromatin landscape and Blimp1 transcriptional targets that regulate trophoblast differentiation. Sci Rep 2017; 7:6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Pan YB, Gong Y, Ruan HF, Pan LY, Wu XK, Tang C, Wang CJ, Zhu HB, Zhang ZM, Tang LF, Zou CC, Wang HB et al. . Sonic hedgehog through Gli2 and Gli3 is required for the proper development of placental labyrinth. Cell Death Dis 2015; 6:e1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bermejo-Alvarez P, Rizos D, Rath D, Lonergan P, Gutierrez-Adan A. Sex determines the expression level of one third of the actively expressed genes in bovine blastocysts. Proc Natl Acad Sci USA 2010; 107:3394–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Calle A, Miranda A, Fernandez-Gonzalez R, Pericuesta E, Laguna R, Gutierrez-Adan A. Male mice produced by in vitro culture have reduced fertility and transmit organomegaly and glucose intolerance to their male offspring. Biol Reprod 2012;87:34. [DOI] [PubMed] [Google Scholar]
- 82. Donjacour A, Liu X, Lin W, Simbulan R, Rinaudo PF. In vitro fertilization affects growth and glucose metabolism in a sex-specific manner in an outbred mouse model. Biol Reprod 2014;90:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Siqueira LGB, Dikmen S, Ortega MS, Hansen PJ. Postnatal phenotype of dairy cows is altered by in vitro embryo production using reverse X-sorted semen. J Dairy Sci 2017; 100:5899–5908. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


