Abstract
Background
Non-typhoidal Salmonella (NTS) serotypes Typhimurium and Enteritidis are a major cause of bloodstream infections in children in sub-Saharan Africa but their reservoir is unknown. We compared pairs of NTS blood and stool isolates (with the same NTS serotype recovered in the same patient) for genetic similarity.
Methods
Between November 2013 and April 2017, hospital-admitted children (29 days to 14 years) with culture-confirmed NTS bloodstream infections were enrolled in a cross-sectional study at Kisantu Hospital, DR Congo. Stool cultures for Salmonella were performed on a subset of enrolled children, as well as on a control group of non-febrile hospital-admitted children. Pairs of blood and stool NTS isolates were assessed for genetic similarity by multiple-locus variable-number of tandem repeats (MLVA) and genomics analysis.
Results
A total of 299 children with NTS grown from blood cultures (Typhimurium 68.6%, Enteritidis 30.4%, other NTS 1.0%) had a stool sample processed; in 105 (35.1%) of them NTS was detected (Typhimurium 70.5%, Enteritidis 25.7%, other NTS 3.8%). A total of 87/105 (82.9%) pairs of blood and stool NTS isolates were observed (representing 29.1% of the 299 children). Among 1598 controls, the proportion of NTS stool excretion was 2.1% (p < 0.0001). MLVA types among paired isolates were identical in 82/87 (94.3%) pairs (27.4% of the 299 children; 61/66 (92.4%) in Typhimurium and 21/21 (100%) in Enteritidis pairs). Genomics analysis confirmed high genetic similarity within 41/43 (95.3%) pairs, showing a median SNP difference of 1 (range 0–77) and 1 (range 0–4) for Typhimurium and Enteritidis pairs respectively. Typhimurium and Enteritidis isolates belonged to sequence types ST313 lineage II and ST11 respectively.
Conclusion
Nearly 30% of children with NTS bloodstream infection showed stool excretion of an NTS isolate with high genetic similarity, adding to the evidence of humans as a potential reservoir for NTS.
Author summary
Non-typhoidal Salmonella (NTS) is an important cause of bloodstream infections in sub-Saharan Africa, but little is known about its reservoir and transmission. We assessed the possibility of a human reservoir by culturing stool samples from children with NTS found in their blood. Next, we compared the obtained blood and stool NTS isolates for genetic similarity. We found that a high proportion of children with NTS in their blood also had NTS in their stool (35.1%) when compared to a control group of children without suspicion of bloodstream infection (2.1%). We observed that Salmonella Typhimurium and Salmonella Enteritidis were the main serotypes isolated from blood and stool, and that their proportions were similar within the different populations. We found that 94.3% of the paired NTS blood and stool isolates (or 27.4% of children with NTS in their blood and a stool culture done) were genetically similar to each other. This observation adds to the evidence of a potential human reservoir for NTS.
Introduction
Non-typhoidal Salmonella (NTS) is a leading cause of bloodstream infections (BSI) with an estimated global burden of invasive NTS infections of 535,000 cases and a case fatality rate of 14.5% in 2017. The burden is highest in sub-Saharan Africa (sSA) and in children under five years of age [1]. Salmonella enterica subspecies enterica serotypes Typhimurium and Enteritidis are the most common causes of NTS BSI [2–5]. The pathogenicity of NTS differs between high-income countries and sSA with a higher risk of invasive disease and associated mortality in the latter [6]; this can be partly explained by the emergence of distinct and invasive clades of Typhimurium and Enteritidis serotypes across sSA [7–10]. Unlike Salmonella Typhi for which the role of human carriers is pivotal in the transmission [3,11], little is known about the reservoirs and transmission of NTS in sSA, but environmental, zoonotic and human sources have been hypothesized [3,12,13]. The role of NTS excretion in the stool has not yet been elucidated and it is unknown whether the NTS strains in the blood of BSI patients are also excreted in stool [3,13].
The objectives of this study were to determine the proportion of NTS stool excretion among hospital-admitted children with NTS BSI and non-febrile hospitalized children as a control group, and to assess the genetic similarity of paired blood and stool NTS isolates (i.e. identical NTS serotypes obtained from the same patient in the course of the hospital admission).
Methods
Study site and study period
The study took place from November 2013 to April 2017 at the hospital of Saint-Luc in Kisantu (HSLK), Kongo-Central province, Democratic Republic of the Congo (DR Congo). HSLK is the major sampling site of the microbiological surveillance network organized by the Institut National de Recherche Biomédicale (INRB), Kinshasa, DR Congo, and the Institute of Tropical Medicine, Antwerp, Belgium [4,14]. Over a 10-year period (2007–2017), NTS ranked first as a cause of BSI in children accounting for 63.8% of culture-confirmed BSI [14,15].
Study design
A cross-sectional study in which hospital-admitted patients aged 29 days to 14 years old with culture-confirmed NTS BSI were enrolled (further referred to as “NTS BSI group”). Enrollment depended on the availability of regular clinical staff during office hours. As soon as possible after enrollment, a stool sample was collected for culture. Stool cultures were also sampled in a control group selected by convenience and consisting of non-febrile hospital-admitted children, as they were presumed to be most closely related to the NTS BSI group in terms of demographics and residency or referring health center. A single stool culture was sampled per patient–repeat cultures were removed from analysis and only the first NTS isolate was considered.
Blood and stool cultures
Blood cultures were collected and processed as previously described [4,15]. About 1 g of stool was suspended in 10 ml selenite broth (Oxoid, Basingstoke, UK and BD DifcoTM, Becton, Dickinson and Company, Franklin Lakes, NJ, U.S.), and incubated at 36°C for 18–24 hours. Next, 10 μl of selenite broth was inoculated on two plates of Salmonella-Shigella (SS) agar (Oxoid) which were incubated at 36°C for 18–24 hours. In case of absence of growth on the SS plate, the plate was re-incubated for another 24 hours. Two colonies suspected of Salmonella per SS plate were inoculated on a Kligler Iron Agar (KIA) tube (Oxoid) and incubated at 36°C for 18–24 hours. KIA tubes suggestive of Salmonella species were further identified by their biochemical characteristics [2,16]. Salmonella spp. isolates from blood and stool were stored in tubes with Tryptone Soy Agar (Oxoid) and shipped to INRB for serotyping and ITM for confirmation and molecular testing. Salmonella spp. isolates were serotyped using commercial antisera (Sifin, Berlin, Germany; and VisionTM, Pro-Lab Diagnostics, Richmond Hill, Canada) according to the Kaufman-White scheme [17].
Multiple-locus variable-number of tandem repeats analysis (MLVA)
MLVA typing was performed at Sciensano (Brussels, Belgium) on Salmonella Typhimurium and Salmonella Enteritidis isolates from blood and stool. MLVA typing was performed using a 5-loci MLVA scheme for both serotypes (STTR9-STTR5-STTR6-STTR10-STTR3 for Salmonella Typhimurium and SENTR7-SENTR5-SENTR6-SENTR4-SE-3 for Salmonella Enteritidis) [18,19]. MLVA types were reported as a string of five numbers representing the number of repeats at the corresponding locus or “NA” in case a PCR product was not obtained for that locus.
Whole genome sequencing (WGS) and maximum likelihood phylogenetic tree analysis
A subset of NTS pairs recovered during the first months of the study (see further) were subjected to whole-genome sequencing (listed in S1 Table). Total DNA was extracted using the Gentra Puregene extraction kit (Qiagen, Hilden, Germany), a sequencing library was prepared using the TruSeq library prep kit (Illumina, San Diego, CA, USA) and sequencing was done on the Illumina 1500 HiSeq platform at the University of Antwerp sequencing facility (Belgium). Salmonella Typhimurium and Salmonella Enteritidis sequences were respectively mapped to the Salmonella Typhimurium D23580 (Accession Number = NC_016854) and Salmonella Enteritidis P125109 (Accession Number = AM933172) reference sequences, using SMALT v0.7.4. Variation detection was performed using samtools mpileup v0.1.19 (-d 1000 -DSugBf) and bcftools v0.1.19 [20]. Per serovar, a pseudo-genome was constructed by substituting the base call at each site in the BCF file into the reference genome. Uncertain sites were substituted with an N. Insertions with respect to the reference genome were ignored and deletions with respect to the reference genome were filled with Ns in the pseudo-genome. Plasmids and recombinant regions (prophages and CRISPR sequences) were detected using Phaster and Gubbins v1.4.10 and were removed from the core genome alignment [21–23]. Single nucleotide polymorphism (SNP) sites were extracted using snp-sites and used to construct a maximum likelihood phylogeny with RAxML v8.2.8 with substitution model GTRCAT [24,25]. Support for nodes on the trees was assessed using 1000 bootstrap replicates. Trees were rooted on Salmonella Typhi 10040_15 (Accession Number = ERS1574281) [26]. The number of SNPs on the branches was calculated using Sankoff Parsimony [27–29]. Phylogenetic trees were visualized using iTOL [30].
Definitions
Paired NTS isolates (NTS pairs) were defined as corresponding blood and stool NTS isolates with identical serotype recovered from the same patient. Co-presence was defined as corresponding blood and stool NTS isolates with different serotypes recovered from the same patient. Identical MLVA types for the Typhimurium serotype were defined as isolates with variation in none or one of the rapidly changing loci (STTR5, STTR6 and STTR10) and no variation in the stable loci (STTR3 or STTR9) [31]; identical MLVA types for the Enteritidis serotype were defined as isolates with variation in none or one of the five loci [32]. For WGS, genetic similarity was based on the relatedness of the paired isolates in the phylogenetic tree.
Data analysis
Data were encoded into an Excel database (Microsoft, Redmond, Washington). Data were characterized by proportions, percentages, ratios, medians, 25–75% interquartile ranges (IQR) and ranges. Differences between proportions were tested for significance using the χ2 test or the McNemar’s test in case of correlated proportions; medians were compared using the Mann-Whitney U test. A p-value of < 0.05 was considered significant.
Ethical issues
Ethical approval was granted by the Institutional Review Board of the Institute of Tropical Medicine in Antwerp (Belgium), the Ethics Committees of the University of Antwerp (Belgium) and the School of Public Health of the University of Kinshasa (DR Congo). Oral informed consent was requested from the guardian of the participants; additional oral assent was requested from participants of 12 years and older. The reasons for oral rather than written consent included the absence of potential risk and individual benefit of stool collection and analysis (non-invasive, no relevance for patient care) as well as the potential impact on the already stressful situation of the participant and his/her caretakers and the need for timely antibiotic treatment. The use of oral consent was approved by the above mentioned ethical committees. A logbook at the pediatric ward was used to document the oral consent.
Results
Among 1052 children with NTS grown from blood cultures, 299 (28.4%) had a stool sample processed (Table 1). Their median age (available for 298/299 (99.7%) children) was 1.5 (IQR 0.9–2.6) years; 78.2% (233/298) were < 36 months and 92.3% (275/298) were < 5 years old. The control group consisted of 1598 children; their median age (available for 100% of children) was 1.8 (IQR 0.9–3.8) years. The median age between both groups did not differ significantly (p = 0.02) and the male-to-female ratio was identical (1:0.9). Overall, the most common diagnoses (n = 4349) of children admitted for non-febrile diseases during the study period were sickle cell anemia (31.6%), malnutrition (23.4%) and amoebiasis (6.4%). Patient inclusion was consistent over the study period, varying between 19.8% (2014) and 36.8% (2013) respectively (S1 Table).
Table 1. Proportions and serotype distributions of non-typhoidal Salmonella blood and stool isolates among children with culture-confirmed bloodstream infection versus a control group of non-febrile hospital-admitted children.
| NTS blood isolates | NTS stool isolates | |||||
|---|---|---|---|---|---|---|
| NTS serotype | All children with NTS BSI (n = 1052) |
Enrolled children with NTS BSI and stool culture done (NTS BSI group) (n = 299) |
p-value | Enrolled children with NTS BSI and NTS stool excretion (n = 105) |
Control group (n = 1598) with NTS stool excretion (n = 34) |
p-value |
| Typhimurium | 645 (61.3%) | 205 (68.6%) | 0.001 | 74 (70.5%) | 25 (73.5%) | 0.98 |
| Enteritidis | 402 (38.2%) | 91 (30.4%) | 27 (25.7%) | 9 (26.5%) | ||
| Other NTS | 5* (0.5%) | 3$ (1.0%) | 4± (3.8%) | 0 (0.0%) | ||
| NTS stool excretion | NA | NA | 105/299 (35.1%) (95% CI: 29.8–40.9%) |
34/1598 (2.1%) (95% CI: 1.5–3.0%) |
<0.0001 | |
*1 Salmonella Aberdeen, 1 Salmonella Abony, 1 Salmonella Chandans, 1 Salmonella Stanleyville, 1 Salmonella Urbana
$1 Salmonella Aberdeen, 1 Salmonella Stanleyville and 1 Salmonella non-typable isolate
±2 Salmonella Virchow and 2 Salmonella non-typable isolates. Abbreviations: BSI = bloodstream infection, CI = confidence interval, NA = not applicable, NTS = non-typhoidal Salmonella.
Table 1 lists the proportions and serotype distributions of the NTS stool isolates. NTS stool excretion in the NTS BSI group was 35.1% versus 2.1% in the control group (p < 0.0001). NTS serotypes included mainly Typhimurium and Enteritidis, with the former being the most frequent. Serotype distributions were similar among blood cultures and stool cultures from both NTS BSI and the control group; however, a small but significant difference (p = 0.001) was found between the total NTS BSI group and the children with NTS BSI and a stool culture done. The proportions of stool cultures grown did not differ significantly between the dry and the rainy season (36.8% (21/57) versus 34.7% (84/242) respectively, p = 0.84) (S1 Table).
Table 2 lists the serotype distributions of the NTS blood and stool isolates from 299 children with a NTS BSI, of whom 105 (35.1%) had NTS isolated from both blood and stool. In 87 of these children (87/105, 82.9%; 87/299, 29.1%), pairs of blood and stool NTS were recovered; representing 66/74 (89.2%) and 21/27 (77.8%) of the Typhimurium and Enteritidis stool culture isolates respectively. In the remaining 18/105 (17.1%) children, serotypes of NTS blood and stool isolates were different ("co-presence"). The median delay between blood and stool sampling within the NTS pairs and NTS co-presence was 2 days and 1.5 day respectively (IQR of 1–3 days). For 9.2% (8/87) of the NTS pairs, stool sampling was done before or on the same day as the blood sampling (1 and 7 pairs respectively). Of note, two NTS pairs had a delay between blood and stool sampling of more than 14 days (16 days and 43 days); the corresponding patients had extended hospital stays because of suspicion of tuberculosis and treatment for malnutrition. (S2 Table)
Table 2. Serotype distribution of non-typhoidal Salmonella blood and stool isolates from 299 children with non-typhoidal Salmonella bloodstream infection.
| Stool cultures | |||||
|---|---|---|---|---|---|
| Blood cultures | Typhimurium | Enteritidis | Other NTS | No growth of Salmonella | Total |
| Typhimurium | 66* | 6‡ | 1‡ ± | 132 | 205 |
| Enteritidis | 8‡ | 21* | 3‡ † | 59 | 91 |
| Other NTS | 0 | 0 | 0 | 3$ | 3 |
| Total | 74 | 27 | 4 | 194 | 299 |
The grey shaded cells represent all non-typhoidal Salmonella isolates cultured from stool in children with NTS BSI (= 105/299, 35.1%).
*Paired NTS isolates
‡Co-presence of non-identical NTS serotypes
±1 Salmonella non-typable isolate
†2 Salmonella Virchow and 1 Salmonella non-typable isolate
$1 Salmonella Aberdeen, 1 Salmonella Stanleyville and 1 Salmonella non-typable isolate. Abbreviations: BSI = bloodstream infection, NTS = non-typhoidal Salmonella.
MLVA types were identical in 61/66 (92.4%) Typhimurium and 21/21 (100%) Enteritidis pairs; for both serotypes combined, this proportion was 94.3% (82/87) (Tables 3 and 4), representing 27.4% of 299 children with NTS BSI in whom stool cultures were done. The two Typhimurium pairs with a delay between blood and stool sampling of 16 days and 43 days had identical MLVA types (S2 Table). MLVA types of the stool NTS recovered from the control group were similar to those of the NTS pairs (S3 Table).
Table 3. Overview of MLVA types of Salmonella Typhimurium paired isolates recovered from stool versus blood.
| Typhimurium MLVA types from blood | |||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Typhimurium MLVA types from stool | 2-4-12-7-0210 | 2-5-10-7-0210 | 2-5-11-7-0210 | 2-5-13-8-0210 | 2-5-14-8-0210 | 2-5-15-8-0210 | 2-5-16-8-0210 | 2-6-7-9-0210 | 2-6-9-9-0210 | 2-6-18-9-0210 | 2-6-NA-9-0210 | 2-7-14-6-0210 | 2-7-15-6-0210 | 2-7-9-9-0210 | 2-8-10-8-0210 | 2-8-11-8-0210 | 2-8-11-NA-0210 | 2-8-12-8-0210 | 2-8-12-9-0210 | 2-8-14-8-0210 | 2-8-14-9-0210 | 2-9-11-7-0210 | 2-9-11-8-0210 | 2-9-12-7-0210 | 2-NA-9-7-0210 | 3-7-13-8-0210 | 3-7-14-8-0210 | 3-NA-10-7-0210 | 3-NA-12-7-0210 | 3-NA-7-7-0210 | Total |
| 2-4-12-7-0210 | 0 | 1* | 1 | ||||||||||||||||||||||||||||
| 2-5-10-7-0210 | 3 | 3 | |||||||||||||||||||||||||||||
| 2-5-11-7-0210 | 1 | 1 | |||||||||||||||||||||||||||||
| 2-5-13-8-0210 | 2 | 2 | |||||||||||||||||||||||||||||
| 2-5-14-8-0210 | 1 | 1 | |||||||||||||||||||||||||||||
| 2-5-15-8-0210 | 5 | 5 | |||||||||||||||||||||||||||||
| 2-5-16-8-0210 | 1 | 1 | |||||||||||||||||||||||||||||
| 2-6-7-9-0210 | 2 | 2 | |||||||||||||||||||||||||||||
| 2-6-9-9-0210 | 23 | 1+ | 1+ | 25 | |||||||||||||||||||||||||||
| 2-6-18-9-0210 | 0 | 0 | |||||||||||||||||||||||||||||
| 2-6-NA-9-0210 | 0 | 0 | |||||||||||||||||||||||||||||
| 2-7-14-6-0210 | 0 | 0 | |||||||||||||||||||||||||||||
| 2-7-15-6-0210 | 1+ | 1 | 2 | ||||||||||||||||||||||||||||
| 2-7-9-9-0210 | 1 | 1 | |||||||||||||||||||||||||||||
| 2-8-10-8-0210 | 1* | 0 | 1 | ||||||||||||||||||||||||||||
| 2-8-11-8-0210 | 0 | 1+ | 1 | ||||||||||||||||||||||||||||
| 2-8-11-NA-0210 | 1* | 0 | 1 | ||||||||||||||||||||||||||||
| 2-8-12-8-0210 | 2 | 2 | |||||||||||||||||||||||||||||
| 2-8-12-9-0210 | 1 | 1 | |||||||||||||||||||||||||||||
| 2-8-14-8-0210 | 1 | 1 | |||||||||||||||||||||||||||||
| 2-8-14-9-0210 | 1 | 1 | |||||||||||||||||||||||||||||
| 2-9-11-7-0210 | 1 | 1 | |||||||||||||||||||||||||||||
| 2-9-11-8-0210 | 0 | 0 | |||||||||||||||||||||||||||||
| 2-9-12-7-0210 | 2 | 2 | |||||||||||||||||||||||||||||
| 2-NA-9-7-0210 | 5 | 5 | |||||||||||||||||||||||||||||
| 3-7-13-8-0210 | 0 | 1+ | 1 | ||||||||||||||||||||||||||||
| 3-7-14-8-0210 | 0 | 0 | |||||||||||||||||||||||||||||
| 3-NA-10-7-0210 | 1* | 1 | 2 | ||||||||||||||||||||||||||||
| 3-NA-12-7-0210 | 1* | 1 | 2 | ||||||||||||||||||||||||||||
| 3-NA-7-7-0210 | 1 | 1 | |||||||||||||||||||||||||||||
| Total | 0 | 3 | 1 | 2 | 1 | 7 | 2 | 3 | 23 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 3 | 1 | 1 | 1 | 1 | 1 | 2 | 5 | 0 | 1 | 1 | 1 | 1 | 66 |
The grey shaded cells represent concordance of MLVA serotype of paired blood and stool samples. Data represent numbers.
+NTS pairs with MLVA types with variation in 1 of the rapidly changing loci (STTR5, STTR6 and STTR10) and no variation in the stable loci (STTR3 or STTR9): these are considered as identical isolates
*Typhimurium pairs with MLVA types with variation in > 1 of the rapidly changing loci (STTR5, STTR6 and STTR10) or variation in the stable loci (STTR3 or STTR9): these are considered as different isolates. Abbreviations: MLVA = multiple-locus variable-number of tandem repeats analysis, STTR = Salmonella Typhimurium tandem repeat.
Table 4. Overview of MLVA types of Salmonella Enteritidis paired isolates recovered from stool versus blood.
| Enteritidis MLVA types from blood | |||||||
|---|---|---|---|---|---|---|---|
| Enteritidis MLVA types from stool | 2-12-3-3-NA | 2-13-3-3-NA | 2-14-3-3-NA | 2-15-3-3-NA | 2-17-3-3-NA | 2-18-3-3-NA | Total |
| 2-12-3-3-NA | 2 | 2 | |||||
| 2-13-3-3-NA | 4 | 4 | |||||
| 2-14-3-3-NA | 1 | 1 | |||||
| 2-15-3-3-NA | 1+ | 7 | 8 | ||||
| 2-17-3-3-NA | 3 | 3 | |||||
| 2-18-3-3-NA | 3 | 3 | |||||
| Total | 3 | 4 | 1 | 7 | 3 | 3 | 21 |
The grey shaded cells represent concordance of MLVA serotype of paired blood and stool samples. Data represent numbers.
+Enteritidis pair with MLVA types with variation in one of the five loci: these are considered as identical isolates. Abbreviations: MLVA = multiple-locus variable-number of tandem repeats analysis.
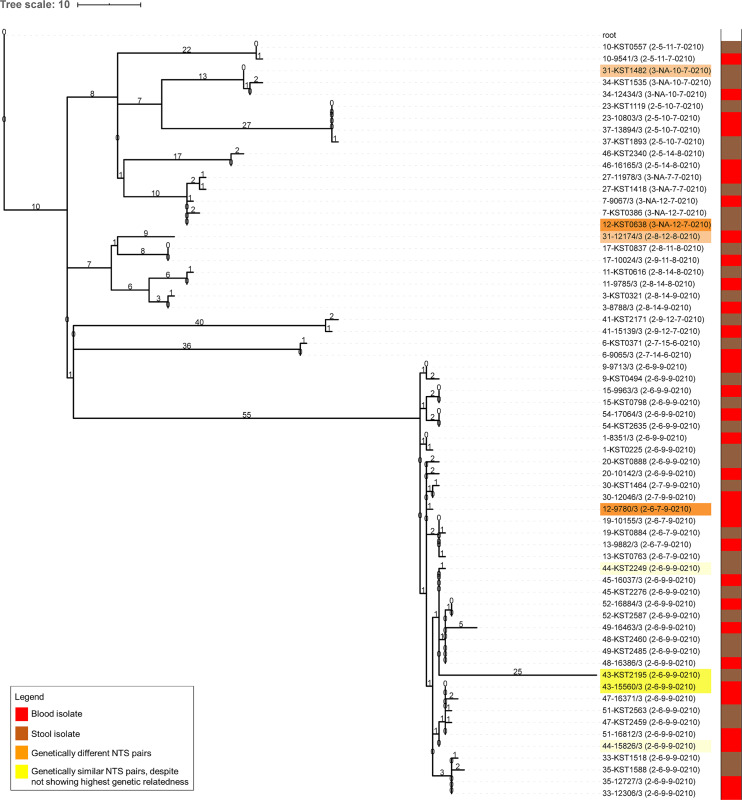
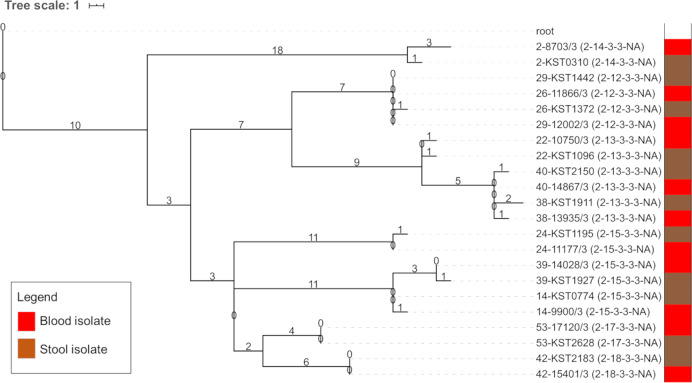
A genomics analysis was done on a subset of 43 NTS pairs consisting of 32/66 Typhimurium and 11/21 Enteritidis pairs. All analyzed Typhimurium isolates belonged to the ST313 sequence type lineage II and II.1 [8,33], while all Enteritidis isolates belonged to ST11 (S2 Table). A phylogenetic analysis (Figs 1 and 2) showed high genetic similarity for 30/32 (93.8%) Typhimurium and all Enteritidis pairs (11/11, 100%). The median number of SNP difference among NTS pairs was 1 (range 0–77) for Typhimurium pairs and 1 (range 0–4) for Enteritidis pairs. The Typhimurium pair with a delay between blood and stool sampling of 43 days had a difference of 1 SNP.
Fig 1. Phylogenetic tree of Salmonella Typhimurium pairs.
Maximum likelihood phylogenetic tree of Salmonella Typhimurium paired isolates, mapped on reference strain Salmonella Typhimurium D23580 (Accession Number = NC_016854) and rooted on Salmonella Typhi 10040_15 (Accession Number = ERS1574281). The tree scale represents the number of SNPs, which are also annotated on the branches. The isolate names have a prefix representing the pair number followed by the study number of each isolate (S2 Table) and a suffix between brackets indicating the MVLA type. Isolates highlighted in orange are pairs showing low genetic similarity and were considered genetically different (orange highlight, pairs N° 12 and 31); isolates highlighted in yellow are pairs that despite not showing highest genetic relatedness were considered genetically similar based on the MLVA type, the SNP difference and the bootstrap values of the branch nodes (yellow highlight, pairs N° 43 and 44). The bar on the right indicates the specimen of isolation (blood isolate (red) versus stool isolate (brown)).
Fig 2. Phylogenetic tree of Salmonella Enteritidis pairs.
Maximum likelihood phylogenetic tree of Salmonella Enteritidis paired isolates, mapped on reference strain Salmonella Enteritidis P125109 (Accession Number = AM933172) and rooted on Salmonella Typhi 10040_15 (Accession Number = ERS1574281). The tree scale represents the number of SNPs, which are also annotated on the branches. The isolate names have a prefix representing the pair number followed by the study number of each isolate (S2 Table) and a suffix between brackets indicating the MVLA type. The bar on the right indicates the specimen of isolation (blood isolate (red) versus stool isolate (brown)).
Agreement between MLVA and WGS was nearly perfect, with 39 Typhimurium and Enteritidis pairs showing identical MLVA types and high genetic similarity in the phylogenetic tree and two Typhimurium pairs showing different MLVA types and low genetic similarity. The latter pairs showed a difference of 45 and 77 SNPs and were considered genetically different (Fig 1, pairs highlighted in orange). In contrast, two Typhimurium pairs showed identical MLVA types but were not the most closely related isolates (Fig 1, pairs highlighted in yellow), with a difference of 3 and 28 SNPs respectively. For the latter pair, the stool isolate (sampled 6 days after blood) accumulated 25 of the 28 SNPs independently. As the genomic differences were small and the branch bifurcations lacked a full bootstrap support for these pairs (around 60%; S1 Fig); we considered both pairs genetically similar.
Discussion
The present study demonstrated that nearly 30% of children with NTS BSI had stool excretion of a NTS isolate with high genetic similarity to the corresponding blood culture isolate. In contrast, NTS stool excretion was observed in only 2.1% of children admitted to the hospital without suspicion of BSI. Among the NTS blood and stool culture pairs, Salmonella Typhimurium and Salmonella Enteritidis belonged to ST313 lineage II and ST11 respectively.
Unlike for Salmonella Typhi the role of carriers in the transmission of NTS is unclear. NTS stool excretion has mainly been studied among patients recovering from NTS gastroenteritis in high-income countries [34,35]: median duration of stool excretion in children ≤ 5 years old is 7 weeks, with 2.6% excreting beyond a 1 year period [35]. In older children, durations are shorter (median 3–4 weeks). Furthermore, the duration of stool excretion is shorter for Typhimurium and antibiotic treatment may prolong excretion [34–36]. Even less is known about stool excretion of invasive NTS isolates in sSA. Most studies surveyed asymptomatic community members or food handlers [37,38]. Cross-sectional community-based studies showed 1.0% and 2.4% of NTS stool excretion among healthy persons in Senegal and Guinea-Bissau respectively [37]. Likewise, a recent community study in rural Kongo-Central in DR Congo showed 3.4% of NTS stool excretion (two consecutive stool samples) [39], which was in line with the proportion in the present control group (2.1%, single stool sample).
The high rate of NTS pairs (29.1% but probably higher given the single stool sampling under antibiotic coverage) provides incremental evidence for a human reservoir of invasive NTS. It complements studies showing NTS stool excretion among household members of index patients with NTS BSI [12,40]. In addition, MLVA types of stool isolates from the control group were similar to those of the NTS pairs from the NTS BSI group, which is in line with previous findings from the same area in DR Congo [39]. However, depending on the concentration of NTS excreted in the stool (which is currently not known), growth of NTS in contaminated food or water may be needed to reach the infectious dose as is the case in high-resource settings [13]. Further, the current evidence on transmission of NTS does not exclude animals as a reservoir of NTS [41].
The finding of NTS pairs may also shed a light on the pathophysiology of NTS infections. The stool excretion of NTS can be interpreted as intestinal infection from the onset of symptoms but might also represent intestinal colonization with bacterial translocation from the gut and subsequent systemic dissemination. This "typhoid fever-like" scenario would be in line with the clinical presentation of invasive NTS infections in sSA, i.e. febrile systemic fever with non-specific clinical features, without gastroenteritis [13] but with increased gut permeability driven by Plasmodium falciparum sequestration and altered gut microbiota associated with malnutrition [42,43].
The cross-sectional study design had limitations. Children and their parents were addressed for enrollment only after blood culture growth. As a consequence, there was a delay between blood and stool sampling and stool sampling was done under antibiotic treatment. In addition, the study was set up in a capacity building program and relied on regular clinical staff for participant recruitment during working hours which meant that only a few patients per day were included. Likewise, the control group was selected by convenience. We do not believe that this had a significant impact on the results, as the control group was large (1598 children), thereby ensuring a wide variety of clinical presentations. Due to the limited diagnostic platforms available, it was not possible to make a reliable clinical diagnosis for the individual control patients, however, the most common diagnoses of children admitted for non-febrile diseases during the study period were known (see results). Collecting stool samples in small and often severely ill children proved to be difficult. Only a single stool sample was collected while it is known that sampling on consecutive days increases yields [44]. Lastly, the date of onset of symptoms was not registered, nor was the occurrence of gastrointestinal symptoms. As to its strengths, blood cultures were systematically sampled based on harmonized indications and the hospital staff got the trust of the patient's families facilitating enrollment [14,15].
Future research is needed to put the present findings into perspective. A prospective study with blood and stool cultures directly after onset of symptoms and with consecutive sampling during convalescence could clarify onset and duration of NTS stool excretion. Assessing the concentration of NTS in stool samples could give an idea about its infectivity (e.g. 106 to 107 NTS organisms per gram of feces in case of NTS gastroenteritis) [34]. A difficult point will be the assessment of the infectious dose, as human challenge experiments typically address adult patients whereas children have the highest risk for NTS BSI [42,45]. As to the aforementioned studies, WGS should cover the diversity of infecting organisms within a particular specimen and time-point, i.e. by assessing multiple isolates per sample [46,47].
In conclusion, the present study, conducted in an endemic area for NTS showed that nearly 30% of children with NTS bloodstream infection had stool excretion of an NTS isolate with high genetic similarity. This observation adds to the evidence of humans as a potential reservoir for NTS.
Supporting information
Checklist of items that should be included in reports of observational studies.
(DOC)
The rainy season at the Kisantu sampling site comprises the months October to May, the dry season comprises the months June to September. Abbreviations: NTS = non-typhoidal Salmonella.
(DOCX)
Abbreviations: MLVA = multiple-locus variable-number of tandem repeats analysis, MLST = multi-locus sequence type, ND = no data, NA = not applicable, SNP = single nucleotide polymorphism, WGS = whole genome sequencing.
(DOCX)
Abbreviations: MLVA = multiple-locus variable-number of tandem repeats analysis, NA = not applicable, NTS = non-typhoidal Salmonella.
(DOCX)
Maximum likelihood phylogenetic tree of Salmonella Typhimurium paired isolates, mapped on reference strain Salmonella Typhimurium D23580 (Accession Number = NC_016854) and rooted on Salmonella Typhi 10040_15 (Accession Number = ERS1574281). The bootstrap values of the branch nodes are indicated on the branches. The branch lengths represent the nucleotide substitution rate in the core SNP alignment of 58148 SNPs. The isolate names have a prefix representing the pair number followed by the study number of each isolate (S2 Table) and a suffix between brackets indicating the MVLA type. The bar on the right indicates the specimen of isolation (blood isolate (red) versus stool isolate (brown)).
(TIF)
Acknowledgments
We would like to thank all clinical and laboratory staff of the Hospital of Saint-Luc in Kisantu, DR Congo, for their contribution and dedication to the microbiological surveillance network and this study. We would like to thank Edmonde Bonebe and Immaculée Kahindo for the processing of bacterial isolates at the Institut National de Recherche Biomédicale (Kinshasa, DR Congo). We would like to thank Marleen Verlinden and Tessa de Block of the Institute of Tropical Medicine (Antwerp, Belgium) for the processing of the bacterial isolates for confirmation and genome sequencing. We are grateful to the Pathogen Informatics Unit and Simon Harris from The Wellcome Sanger Institute (Cambridge, UK) for informatics support.
Data Availability
The sequence data that support the findings of this study have been deposited in ENA (EMBL), under study ERP120852, with accession numbers as listed in S2 Table. The source data underlying Table 1, Table 2 and S1 Table are available on Figshare (doi 10.6084/m9.figshare.11611680). The source data underlying Tables 3A and 3B are provided in S2 Table.
Funding Statement
This work was funded by the Belgian Directorate of Development Cooperation (DGD) through Project 2.01 of the Third Framework Agreement between the Belgian DGD and the Institute of Tropical Medicine, Belgium, and by the Baillet-Latour fund. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Stanaway JD, Parisi A, Sarkar K, Blacker BF, Reiner RC, Hay SI, et al. The global burden of non-typhoidal Salmonella invasive disease: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect Dis. 2019;3099: 1–13. 10.1016/s1473-3099(19)30418-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lunguya O, Lejon V, Phoba MF, Bertrand S, Vanhoof R, Glupczynski Y, et al. Antimicrobial Resistance in Invasive Non-typhoid Salmonella from the Democratic Republic of the Congo: Emergence of Decreased Fluoroquinolone Susceptibility and Extended-spectrum Beta Lactamases. PLoS Negl Trop Dis. 2013;7: 2–10. 10.1371/journal.pntd.0002103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crump JA, Sjölund-Karlsson M, Gordon MA, Parry CM. Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin Microbiol Rev. 2015;28: 901–937. 10.1128/CMR.00002-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalonji LM, Post A, Phoba MF, Falay D, Ngbonda D, Muyembe JJ, et al. Invasive Salmonella infections at multiple surveillance sites in the Democratic Republic of the Congo, 2011–2014. Clin Infect Dis. 2015;61: S346–S353. 10.1093/cid/civ713 [DOI] [PubMed] [Google Scholar]
- 5.Uche IV, MacLennan CA, Saul A. A Systematic Review of the Incidence, Risk Factors and Case Fatality Rates of Invasive Nontyphoidal Salmonella (iNTS) Disease in Africa (1966 to 2014). PLoS Negl Trop Dis. 2017;11 10.1371/journal.pntd.0005118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crump JA, Heyderman RS. A perspective on invasive Salmonella disease in Africa. Clin Infect Dis. 2015;61: S235–S240. 10.1093/cid/civ709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kingsley RA, Msefula CL, Thomson NR, Kariuki S, Holt KE, Gordon MA, et al. Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 2009;19: 2279–2287. 10.1101/gr.091017.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okoro CK, Kingsley RA, Connor TR, Harris SR, Parry CM, Al-Mashhadani MN, et al. Intracontinental spread of human invasive Salmonella Typhimurium pathovariants in sub-Saharan Africa. Nat Genet. 2012;44: 1215–1221. 10.1038/ng.2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feasey NA, Hadfield J, Keddy KH, Dallman TJ, Jacobs J, Deng X, et al. Distinct Salmonella Enteritidis lineages associated with enterocolitis in high-income settings and invasive disease in low-income settings. Nat Genet. 2016;48: 1211–1217. 10.1038/ng.3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okoro CK, Barquist L, Connor TR, Harris SR, Clare S, Stevens MP, et al. Signatures of Adaptation in Human Invasive Salmonella Typhimurium ST313 Populations from Sub-Saharan Africa. PLoS Negl Trop Dis. 2015;9 10.1371/journal.pntd.0003611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gal-Mor O, Boyle EC, Grassl GA. Same species, different diseases: How and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Front Microbiol. 2014;5: 1–10. 10.3389/fmicb.2014.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kariuki S, Revathi G, Kariuki N, Kiiru J, Mwituria J, Muyodi J, et al. Invasive multidrug-resistant non-typhoidal Salmonella infections in Africa: Zoonotic or anthroponotic transmission? J Med Microbiol. 2006;55: 585–591. 10.1099/jmm.0.46375-0 [DOI] [PubMed] [Google Scholar]
- 13.Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA. Invasive non-typhoidal salmonella disease: An emerging and neglected tropical disease in Africa. Lancet. 2012;379: 2489–2499. 10.1016/S0140-6736(11)61752-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tack B, Phoba MF, Van Puyvelde S, Kalonji LM, Hardy L, Barbé B, et al. Salmonella Typhi from blood cultures in the democratic republic of the congo: A 10-year surveillance. Clin Infect Dis. 2019;68: S130–S137. 10.1093/cid/ciy1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tack B, Phoba M, Barbé B, Kalonji L, Hardy L, Van Puyvelde S, et al. Increase in non-typhoidal Salmonella bloodstream infections in the Democratic Republic of the Congo (2015–2017): emergence of O5-negative Salmonella Typhimurium and alarming rates of resistance. PLoS Negl Trop Dis. 2020;14: e0008121 10.1371/journal.pntd.0008121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lunguya O, Lejon V, Phoba MF, Bertrand S, Vanhoof R, Verhaegen J, et al. Salmonella Typhi in the Democratic Republic of the Congo: fluoroquinolone decreased susceptibility on the rise. PLoS Negl Trop Dis. 2012;6: 3–8. 10.1371/journal.pntd.0001921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimont P, Weill F-X. Antigenic formulae of the Salmonella servovars. WHO Collab Cent Ref Res Salmonella. 2007; 1–167. Available: http://www.pasteur.fr/ip/portal/action/WebdriveActionEvent/oid/01s-000036-089%5Cnpapers2://publication/uuid/CA3447A0-61BF-4D62-9181-C9BA78AF0312 [Google Scholar]
- 18.Wuyts V, Mattheus W, De Laminne De Bex G, Wildemauwe C, Roosens NHC, Marchal K, et al. MLVA as a tool for public health surveillance of human Salmonella Typhimurium: Prospective study in Belgium and evaluation of MLVA loci stability. PLoS One. 2013;8 10.1371/journal.pone.0084055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bertrand S, De Lamine De Bex G, Wildemauwe C, Lunguya O, Phoba MF, Ley B, et al. Multi locus variable-number tandem repeat (MLVA) typing tools improved the surveillance of Salmonella Enteritidis: A 6 years retrospective study. PLoS One. 2015;10: 1–13. 10.1371/journal.pone.0117950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44: W16–W21. 10.1093/nar/gkw387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: A Fast Phage Search Tool. Nucleic Acids Res. 2011;39: 347–352. 10.1093/nar/gkq749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43: e15 10.1093/nar/gku1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Page AJ, Taylor B, Delaney AJ, Soares J, Seemann T, Keane JA, et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb genomics. 2016;2: e000056 10.1099/mgen.0.000056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phoba MF, Barbé B, Lunguya O, Masendu L, Lulengwa D, Dougan G, et al. Salmonella enterica serovar Typhi Producing CTX-M-15 Extended Spectrum β-Lactamase in the Democratic Republic of the Congo. Clin Infect Dis. 2017;65: 1229–1231. 10.1093/cid/cix342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fitch WM. Toward Defining the Course of Evolution: Minimum Change for a Specific Tree Topology. Syst Zool. 1971;20: 406–416. 10.2307/2412116 [DOI] [Google Scholar]
- 28.Sankoff D. Minimal mutation trees of sequences. SIAM J Appl Math. 1975;28: 35–42. 10.1137/0128004 [DOI] [Google Scholar]
- 29.Sankoff D, Rousseau P. Locating the vertices of a steiner tree in an arbitrary metric space. Math Program. 1975;9: 240–246. 10.1007/BF01681346 [DOI] [Google Scholar]
- 30.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44: W242–W245. 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dimovski K, Cao H, Wijburg OLC, Strugnell RA, Mantena RK, Whipp M, et al. Analysis of Salmonella enterica serovar Typhimurium variable-number tandem-repeat data for public health investigation based on measured mutation rates and whole-genome sequence comparisons. J Bacteriol. 2014;196: 3036–3044. 10.1128/JB.01820-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hopkins KL, Peters TM, de Pinna E, Wain J. Standardisation of multilocus variable-number tandemrepeat analysis (MLVA) for subtyping of Salmonella enterica serovar Enteritidis. Eurosurveillance. 2011;16. [PubMed] [Google Scholar]
- 33.Van Puyvelde S, Pickard D, Vandelannoote K, Heinz E, Barbé B, de Block T, et al. An African Salmonella Typhimurium ST313 sublineage with extensive drug-resistance and signatures of host adaptation. Nat Commun. 2019;10: 1–12. 10.1038/s41467-018-07882-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gal-Mor O. Persistent infection and long-term carriage of typhoidal and nontyphoidal Salmonellae. Clin Microbiol Rev. 2019;32: 1–31. 10.1128/CMR.00088-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haeusler GM, Curtis N. Non-typhoidal Salmonella in children: microbiology, epidemiology and treatment. Adv Exp Med Biol. 2013;764: 13–26. 10.1007/978-1-4614-4726-9_2 [DOI] [PubMed] [Google Scholar]
- 36.Marzel A, Desai PT, Goren A, Schorr YI, Nissan I, Porwollik S, et al. Persistent infections by nontyphoidal Salmonella in humans: Epidemiology and genetics. Clin Infect Dis. 2016;62: 879–886. 10.1093/cid/civ1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Im J, Nichols C, Bjerregaard-Andersen M, Sow AG, Løfberg S, Tall A, et al. Prevalence of Salmonella Excretion in Stool: A Community Survey in 2 Sites, Guinea-Bissau and Senegal. Clin Infect Dis. 2016;62: s50–s55. 10.1093/cid/civ789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bradbury RS, Barbé B, Jacobs J, Jallow AT, Camara KC, Colley M, et al. Enteric pathogens of food sellers in rural Gambia with incidental finding of Myxobolus species (Protozoa: Myxozoa). Trans R Soc Trop Med Hyg. 2015;109: 334–339. 10.1093/trstmh/trv020 [DOI] [PubMed] [Google Scholar]
- 39.Mbuyi-Kalonji L, Barbé B, Nkoji G, Madinga J, Roucher C, Linsuke S, et al. Non-typhoidal Salmonella intestinal carriage in a Schistosoma mansoni endemic community in a rural area of the Democratic Republic of Congo. PLoS Negl Trop Dis. 2020;14: e0007875 10.1371/journal.pntd.0007875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Post AS, Diallo SN, Guiraud I, Lompo P, Tahita MC, Maltha J, et al. Supporting evidence for a human reservoir of invasive non-Typhoidal Salmonella from household samples in Burkina Faso. Akullian A, editor. PLoS Negl Trop Dis. 2019;13: e0007782 10.1371/journal.pntd.0007782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parsons BN, Humphrey S, Salisbury AM, Mikoleit J, Hinton JCD, Gordon MA, et al. Invasive Non-Typhoidal Salmonella Typhimurium ST313 Are Not Host-Restricted and Have an Invasive Phenotype in Experimentally Infected Chickens. PLoS Negl Trop Dis. 2013;7: 1–8. 10.1371/journal.pntd.0002487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takem EN, Roca A, Cunnington A. The association between malaria and nontyphoid Salmonella bacteraemia in children in sub-Saharan Africa: A literature review. Malar J. 2014;13 10.1186/1475-2875-13-400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lim SH, Methé BA, Knoll BM, Morris A, Obaro SK. Invasive non-typhoidal Salmonella in sickle cell disease in Africa: Is increased gut permeability the missing link? J Transl Med. 2018;16 10.1186/s12967-018-1622-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buchwald DS, Blaser MJ. A review of human salmonellosis: II. Duration of excretion following infection with nontyphi Salmonella. Rev Infect Dis. 1984;6: 345–56. 10.1093/clinids/6.3.345 [DOI] [PubMed] [Google Scholar]
- 45.Gibani MM, Jin C, Darton TC, Pollard AJ. Control of invasive Salmonella disease in Africa: Is there a role for human challenge models? Clin Infect Dis. 2015;61: S266–S271. 10.1093/cid/civ673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mather AE, Vaughan TG, French NP. Molecular approaches to understanding transmission and source attribution in nontyphoidal Salmonella and their application in Africa. Clin Infect Dis. 2015;61: S259–S265. 10.1093/cid/civ727 [DOI] [PubMed] [Google Scholar]
- 47.Octavia S, Wang Q, Tanaka MM, Sintchenko V, Lan R. Genomic variability of serial human isolates of Salmonella enterica serovar Typhimurium associated with prolonged carriage. J Clin Microbiol. 2015;53: 3507–3514. 10.1128/JCM.01733-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Checklist of items that should be included in reports of observational studies.
(DOC)
The rainy season at the Kisantu sampling site comprises the months October to May, the dry season comprises the months June to September. Abbreviations: NTS = non-typhoidal Salmonella.
(DOCX)
Abbreviations: MLVA = multiple-locus variable-number of tandem repeats analysis, MLST = multi-locus sequence type, ND = no data, NA = not applicable, SNP = single nucleotide polymorphism, WGS = whole genome sequencing.
(DOCX)
Abbreviations: MLVA = multiple-locus variable-number of tandem repeats analysis, NA = not applicable, NTS = non-typhoidal Salmonella.
(DOCX)
Maximum likelihood phylogenetic tree of Salmonella Typhimurium paired isolates, mapped on reference strain Salmonella Typhimurium D23580 (Accession Number = NC_016854) and rooted on Salmonella Typhi 10040_15 (Accession Number = ERS1574281). The bootstrap values of the branch nodes are indicated on the branches. The branch lengths represent the nucleotide substitution rate in the core SNP alignment of 58148 SNPs. The isolate names have a prefix representing the pair number followed by the study number of each isolate (S2 Table) and a suffix between brackets indicating the MVLA type. The bar on the right indicates the specimen of isolation (blood isolate (red) versus stool isolate (brown)).
(TIF)
Data Availability Statement
The sequence data that support the findings of this study have been deposited in ENA (EMBL), under study ERP120852, with accession numbers as listed in S2 Table. The source data underlying Table 1, Table 2 and S1 Table are available on Figshare (doi 10.6084/m9.figshare.11611680). The source data underlying Tables 3A and 3B are provided in S2 Table.