Abstract
Powdery mildew is an important foliar disease of barley (Hordeum vulgare L.) caused by the biotrophic fungus Blumeria graminis f. sp. hordei (Bgh). The understanding of the resistance mechanism is essential for future resistance breeding. In particular, the identification of race-nonspecific resistance genes is important because of their regarded durability and broad-spectrum activity. We assessed the severity of powdery mildew infection on detached seedling leaves of 267 barley accessions using two poly-virulent isolates and performed a genome-wide association study exploiting 201 of these accessions. Two-hundred and fourteen markers, located on six barley chromosomes are associated with potential race-nonspecific Bgh resistance or susceptibility. Initial steps for the functional validation of four promising candidates were performed based on phenotype and transcription data. Specific candidate alleles were analyzed via transient gene silencing as well as transient overexpression. Microarray data of the four selected candidates indicate differential regulation of the transcription in response to Bgh infection. Based on our results, all four candidate genes seem to be involved in the responses to powdery mildew attack. In particular, the transient overexpression of specific alleles of two candidate genes, a potential arabinogalactan protein and the barley homolog of Arabidopsis thaliana’s Light-Response Bric-a-Brac/-Tramtrack/-Broad Complex/-POxvirus and Zinc finger (AtLRB1) or AtLRB2, were top candidates of novel powdery mildew susceptibility genes.
Introduction
Crop protection is mostly provided by the application of chemical agents, which can be harmful to the environment while a more sustainable disease control is resistance breeding [1,2]. Nonetheless, this often requires extensive field-testing of plant material, due to the influence of the environment on plant diseases. Considering this, characterizing plant resistance mechanisms provides important hints for selection and thus assists breeding [1,3,4].
One relevant foliar disease of barley (Hordeum vulgare L. subsp. vulgare) and wheat (Triticum aestivum L.) is powdery mildew (PM), which is caused by the biotrophic fungi Blumeria graminis f. sp. hordei (Bgh) and Blumeria graminis f. sp. tritici (Bgt), respectively [5,6]. Both fungi grow usually only on the corresponding host [7] while infection with either Bgt or Bgh can lead to severe yield losses of wheat and barley [8,9], which are the second and fourth most produced grains in the world, respectively [10]. Moreover, barley is a model organism to study plant-pathogen interactions in cereals. For instance, important resistance loci like Mla, MlLa, and Mlg [11,12], as well as susceptibility factors like Mlo [13–15] have been identified in barley and some of them are even relevant for non-cereal species [13,16,17]. Resistance can be classified into: a) race-specific, which is active against a particular pathogen species (race/isolate); and b) race-nonspecific [18,19]. The resistance provided by race-nonspecific resistance genes is mostly independent of the pathogen race/isolate, the time of its onset, and the plant developmental stage or the environment [18–20]. The use of monogenic resistance genes provides usually strong race-specific protection but results often in an early breakdown of the resistance caused by the co-evolution of host and pathogen. These ‘boom-and-bust’ cycles occur particularly if the same resistant cultivar is grown on a large area across some years [4]. One exception to this rule is the race-nonspecific PM resistance conferred by loss-of-function mutations in the Mlo gene, which was durable during several decades [13–15]. This gene represents a crucial susceptibility factor necessary for fungal development and virulence [13]. The underlying mechanism of the recessive mlo resistance is related to basal defense reactions, which are of special interest for plant breeding due to their regarded durability and broad-spectrum activity [13,17,21]. Nonetheless, despite the promising potential of race-nonspecific resistance, its use is quite limited in breeding. This is presumably due to a lack of general understanding of race-nonspecific mechanisms, which are usually of polygenic nature and thus more complicated to handle in breeding [21].
Domestication and high selection intensities have reduced genetic diversity in modern material [22,23] and landraces, as well as wild crop ancestors, may contain valuable genetic variation for agronomical traits awaiting to be mined [24–26]. Genome-wide association (GWA) studies proved to be versatile in the identification of candidate genes or loci in genetically diverse material [27,28]. Particularly, GWA studies have been successfully used to identify PM resistance genes/loci [29–31]. Provided a highly diverse population, the mapping resolution of GWAS depends to a great extent on marker density [32]. In this regard, exome capture delivers highly dense data in a cost-efficient manner and focuses on variations in protein-encoding regions [33–35]. For most breeding applications a significantly associated marker could be enough [36], but for the understanding of a resistance mechanism or its regulation, it is necessary to identify the corresponding causal gene and its function in vivo. Despite its importance, the functional validation of candidates identified by GWA studies is often not provided [37,38]. In particular, in the barley-Bgh pathosystem, transient assays are an established tool for the initial validation of identified candidates [39]. Analyses of race-specific PM loci revealed strong allelic effects indicating the importance of the exploitation of this diversity [40,41]. In contrast, knowledge of allele specificities of race-nonspecific resistance genes is quite limited [31] and in particular, the functional validation of alleles in different genetic backgrounds is usually missing.
We started our approach by evaluating the potential race-nonspecific seedling resistance or susceptibility of 267 barley genotypes based on detached leaves against two poly-virulent Bgh isolates. Further, we selected 201 genotypes to create a collection, which represents a broad proportion of the genetic diversity existent in barley and evaluated its suitability to conduct a GWA study. We fitted a mixed linear model for 424,567 single nucleotide polymorphisms (SNPs) to identify potentially important factors associated with race-nonspecific, quantitative plant responses against PM. In particular, we began to validate and analyze specifically allelic effects to gain first insights into the functionality of four promising candidate genes underlying two association peaks on chromosome 5H that co-localize with the seedling PM resistance locus Rbgq15 [11].
Material and methods
Plant material
Based on an initial characterization of the resistance against the PM isolate D35/3 (JKI-242) of 459 genotypes of the barley Whealbi (EU FP7 n°FP7- 613556) collection, a representative panel of 267 genotypes was chosen (hereafter, phenotyping-panel), spanning the complete range of PM susceptibility. Passport data of the phenotyping-panel (S1 Table) were obtained from the official germplasm description for the barley Whealbi collection (https://wheat-urgi.versailles.inra.fr/Projects/Achieved-projects/Whealbi) and were complemented with data from Bustos-Korts et al. [42]. The susceptible cultivar Morex and the moderately susceptible cultivar Roland were included as positive controls for infection efficiency during the experiments.
Leaf segments of JB Flavour and Roland were used for the weekly maintenance of the Blumeria graminis (DC.) E. O. Speer f. sp. hordei (Bgh) isolates D35/3 (JKI-242) and RiIII (JKI-75), respectively. The plants were grown for 7d under long-day conditions (16h) at 20°C in plant growth chambers. A mixture of Maythorpe:Ingrid:Golden-Promise (2:2:1) cultivars grown for 6 d in a glasshouse chamber under long-day conditions at 22°C/20°C, was used for the maintenance of the isolate CH4.8 (JKI-247). Infected plants and inoculated material were incubated at 20°C in plant growth chambers at long-day conditions.
Powdery mildew resistance screening
The barley genotypes of the phenotyping-panel were grown in 9x6 multi-pot trays filled with compost soil from the IPK nursery, without fertilization. They were grown for 12d in a glasshouse chamber under long-day conditions at 20°C/16°C (day/night temperature). The PM isolates D35/3 and RiIII were selected regarding their poly-virulent nature to a smaller set of 33 barley genotypes known for their differential responses against PM (S2 Table). A detached leaf assay was conducted for the resistance scoring, in which second leaves of seedlings were cut and placed on standard 4-well multi-titter plates filled with 1% Phytoagar solution supplemented with benzimidazole (20 mg/l). In this essay, leaves were inoculated with fresh PM spores (8-10 conidia/mm2) and disease symptoms were estimated (scored by eye) 7d after inoculation [31,43]. To reveal the quantitative characteristic of the PM infection, the infected leaf area was scored as the estimated percentage of leaf area covered by macroscopically visible disease symptoms. In this sense, a decreased percentage of infected leaf area is interpreted as enhanced quantitative resistance while higher percentage values correspond to increased quantitative susceptibility. We introduce here the term “experiment” to describe all those individual plants that were grown and inoculated simultaneously. In each experiment and for each isolate, five different plants (technical replicates) were inoculated per tested genotype while each genotype of the phenotyping-panel was assessed in at least three different experiments (experimental replicates) per isolate.
Exome capture data
The exomes of a total of 403 genotypes of the barley Whealbi collection were genotyped by a hybridization-based sequence capture platform [42]. SNP data were then subtracted for the overlapping accessions between the Whealbi collection and our phenotyping-panel. The final set of 424,567 SNPs was generated by removing SNPs with a minor allele frequency <5% leading to an average SNP density of 6.9 SNP/kb.
Processing of phenotypic data
To determine the association between the two isolates, the arithmetic means per genotype per isolate were calculated from the values normalized with the control cultivar Morex after outlier test correction (ROUT Q = 1%, performed with GraphPad [44] Prism version 7.01 for Windows). The significance of Pearson correlation coefficients between the disease scores of D35/3 and RiIII was assessed by a two-tailed t-test. After removing 21 completely resistant genotypes coming from the phenotyping panel (S1 Table, see Results for further details), 201 genotypes having SNP and phenotypic data were retained for further analyses. Here onwards these 201 genotypes are referred to as the GWA-panel.
Linear mixed models [45,46] are useful for estimating genotype performance from non-orthogonal phenotypic data [47,48]. We used a two-step mixed model approach to obtain the best linear unbiased estimations (BLUE) of genotypes from the GWA-panel. In the first step, the following linear mixed model was separately fitted to the data of each experiment:
| (1) |
where y(i)jk is the non-normalized detached leaf assay value for the jth genotype evaluated within the kth plate in the ith experiment, μ corresponds to the common mean of the material, gj is the effect of jth genotype, pk denotes the effect of the kth plate, and e(i)jk is the residual variation. The common mean and genotypic effects were assumed as fixed factors, while the rest of the factors were considered random. In a second step, the genotype BLUEs from each experiment were analyzed together using the model:
| (2) |
where yij is the BLUE of fungal infection for the jth genotype evaluated in the ith experiment, μ denotes the common mean across experiments, gj corresponds to the effect of the jth genotype across experiments, ξi indicates the effect of the ith experiment and eij is the residual variation. Common mean and genotypic effects were considered fixed, while the rest of the factors were assumed as random. Furthermore, genotypic variances (σg2) were estimated for each experiment and across experiments using Eqs (1) and (2) but considering genotypes as random. In parallel, error variances () for each experiment and across experiments were computed as the squared mean standard error of the difference between two genotypes according to Eqs (1) and (2), respectively. Variance estimates were subsequently used to compute the repeatability of each experiment and isolate as well as the heritability for each isolate according to Piepho and Möhring [49]:
| (3) |
Based on the calculated BLUEs, an artificial maximal infection (Max) was generated in silico for each genotype of the GWA-panel by selecting the higher infection value of the two isolates.
Population structure and linkage disequilibrium
Pairwise Rogers’ distances (RDs) [50] were estimated among genotypes using SNP data of the GWA-panel and were subjected to hierarchical cluster analysis for studying the population structure. Further, the Pearson correlation coefficients of the RD and the phenotypic distance matrices from the genotypes of the GWA-panel were assessed by a Mantel test implemented in R [51] Version 3.5.1.
The pairwise linkage disequilibrium (LD) as r2 of the significant SNPs of the selected candidate genes was determined by TASSEL5 [52] using 50 SNPs as sliding window size and excluding heterozygous SNPs.
Genome-wide association study
A standard linear mixed model [53] was fitted at each SNP position on the plant responses of D35/3 and RiIII as well as their maximal infection (Max):
| (4) |
where y is the vector containing the BLUEs across experiments of the genotypes of the GWA-panel, β denotes a vector of fixed effects containing the common population mean, the effects of the subpopulations portrayed by the RD and the allele substitution effect of the tested marker, g corresponds to a vector containing the random genetic background effects of the genotypes in the GWA-panel, X and Z represent design matrices that relate y to β and g, respectively, while e is a vector of random errors. Within the X matrix, those columns corresponding to subpopulations are the memberships of genotypes to them. Vectors g and e were assumed as g~N(0,σg2K), e~N(0,σe2I), where K is a marker-derived kinship matrix calculated as 2*(1-RD) [54], I is an identity matrix, while σg2 and σe2 are the corresponding variances. The genome-wide multiple-test Type I error was controlled by using the effective number of markers (Meff_G) necessary to explain 99.8% of the chromosome-wise calculated LD matrices as proposed by Gao et al. [55]. By dividing the nominal Type I error rate (αe) of 0.05 by Meff_G = 1,414 a significance threshold of -log10(P) = 4.45 was obtained. Mixed model Eqs (1), (2) and (4) were fitted using ASReml-R [56] in R environment [57] Version 2.15.
At least 2kb of the flanking genomic regions surrounding the significant SNPs of the Max trait from the barley reference genome [34] were extracted via the IPK Galaxy Server and used for a ‘BLASTN’ analysis with the IPK BLAST Server [58] and the NCBI server. Details about the used databases are provided in the S3 Table.
Characterization and functional validation of the selected candidate genes
Significant SNPs for the Max trait located within annotated gene models [34] were curated by comparison of the gene models with homologous sequences identified in the related species Aegilops tauschii subsp. tauschii and Brachypodium distachyon. Combinations of the selected SNPs within the curated gene model defined the corresponding alleles. Therefore, the term allele in this context corresponds to an SNP haplotype. Means of the infected leaf area of all genotypes carrying the respective alleles were calculated and the significance of the difference between the corresponding alleles was assessed via a two-tailed t-test. Linear models were fitted in R environment [51] Version 3.5.1 to determine the proportion of the variation that is explained by the candidate genes.
The closest protein homologs of the candidate genes were identified via BLAST analyses. In case of the related species A. tauschii and B. distachyon, the protein sequences of the NCBI BLAST hits were selected, and the homologs of Arabidopsis thaliana and rice (Oryza sativa ssp. japonica cv. Nipponbare) were identified via ‘BLASTP’ against the TAIR server [59] and the rice genome annotation project server [60], respectively. For candidate gene CG-4, the protein sequences described by Kimura et al. [61] were chosen. The off-targets for RNAi predicted by the si-Fi21 tool [62] were assumed as candidate homologs of barley. The obtained sequences per candidate were aligned with the online tool ‘Clustal Omega’ [63] (S1A-S4A Figs in S1 File) and domains were predicted with the ‘Pfam 32.0 sequence search’ [64] and the ‘NLS mapper’ [65] for candidate CG_3 using default settings. The nucleotide sequences of the defined candidate alleles were generated based on the annotated SNPs and the obtained sequencing results of the cloned alleles. These sequences were translated into putative proteins with the ‘ExPASy -Translate tool’ [66]. The generated protein sequences of CG_4 were analyzed with the ‘SignalP 5.0 tool’ [67].
The expression of candidate genes was analyzed in Microarray data published by Delventhal et al. [68] based on the SCRI_Hv35_44k_v1 chip (Agilent_20599). The corresponding U35 sequence identifier and probe-IDs of the candidates were identified via BLAST against the HarvEST barley database (S3 Table). The significance of the difference in gene expression was assessed from three biological replicates via unpaired two-tailed t-test between inoculated and non-inoculated samples per time point.
Particle bombardment
For the Transient-Induced Gene Silencing (TIGS), hairpin constructs were created from cDNA fragments of 190-500bp length amplified with the Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific, Waltham, USA). For this purpose, total RNA was extracted from 7d old leaves of the cultivar Morex using the RNeasy® Plant Mini Kit (Qiagen, Hilden, Germany) and subsequently treated with DNase I (Thermo Fisher Scientific). Afterwards, cDNA was generated using the RevertAid M-MuLV RT Kit and Oligo(dT)18 primer (Thermo Fisher Scientific) following the manufacturer’s instructions. The candidate primers (S4 Table) were designed based on the prediction of the si-Fi21 tool [62] for the candidate regions with the most efficient small interfering RNAs (siRNAs) without predicted off-targets. The corresponding primer positions were indicated in the gene models in S1B-S4B Figs in S1 File. Cloning and particle bombardment was performed similarly to the method described by Douchkov et al. [69]. After 3 d, the leaf segments of cultivar Morex and WB-052 (HOR2573) were inoculated with the PM isolate CH4.8 (spore density: 450–530 conidia/mm2; S2 Table). To determine the susceptibility index (SI), the proportion of the GUS-stained cells with growing haustorium to the total number of transformed (GUS-stained) cells was calculated. Then, the relative SI (proportion of the SI of the test gene to the SI of the empty vector pIPKTA30N) was calculated. The significance of the difference was assessed from five independent experiments via one-sample two-tailed t-test from log2-transformed values against the hypothetical value ‘0’.
For the overexpression constructs, the full-length candidate gene sequence was amplified from genomic DNA extracted with the DNeasy® Plant Mini Kit (Qiagen) according to the manufacturer´s instructions. The amplification was performed with the Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific). Primer details were provided in the S4 Table and S1B-S4B Figs in S1 File. The different alleles were amplified either from the susceptible genotype Morex or the resistant WB-052 and the corresponding fragments were gel-purified with the GeneJET PCR purification Kit (Thermo Fisher Scientific). The blunt-end cloning into pIPKTA9 [70] was performed similarly as described for the TIGS constructs but using SmaI (Thermo Fisher Scientific) instead of SwaI. The correct orientation of the insert was determined by PCRs and sequencing. The particle bombardment and the microscopical analysis were performed similarly to the TIGS assay, but the PM inoculation was performed after 4h (spore density: 150–250 conidia/mm2) and pIPKTA9 served as empty vector control. Each construct was tested in four biological replicates. All work reported in this study was performed following the appropriate biosecurity measures according to the German legislation.
Results
The diverse barley material showed high variation for powdery mildew seedling resistance
The phenotyping-panel includes spring and winter as well as two- and six-rowed barley types (S1 Table). Furthermore, this panel is constituted by modern elite (25%) and traditional material like landraces and old cultivars (70%), as well as by wild barley accessions (5%). To cover key world production regions of barley, the 267 genotypes of the phenotyping-panel were selected mainly from Europe (23%), the Middle-East (22%), Africa (20%), and Asia (17%). In the detached leaf assays, the phenotyping-panel spanned the complete range of susceptibility (Fig 1and S1 Table) against the two poly-virulent PM isolates, which were virulent against 22 of the most common (European) resistance specificities (S2 Table).
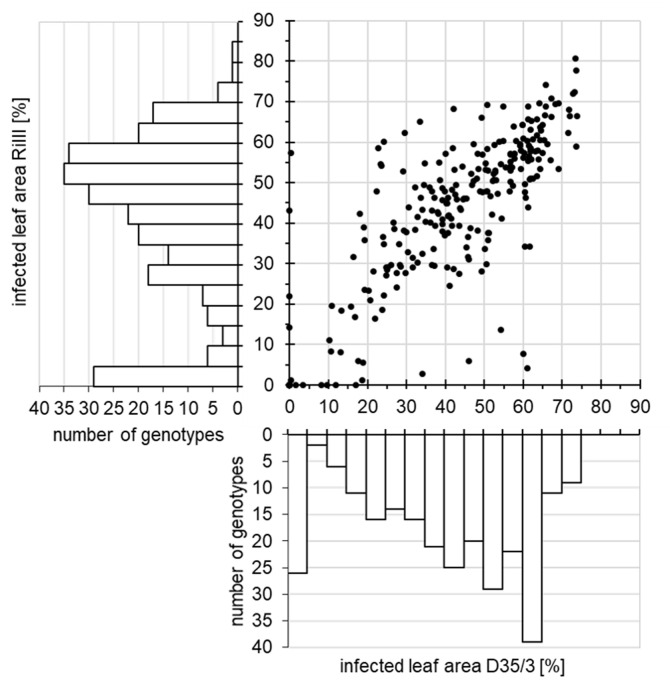
Fig 1. Biplot of the phenotypic distribution of the 267 Hordeum vulgare accessions included in the phenotyping-panel.
Phenotypic values correspond to the arithmetic mean (n ≥3) of the normalized infected leaf area [in %] for the Blumeria graminis f. sp. hordei isolates D35/3 vs. RiIII and the corresponding absolute frequencies of genotypes per interval and isolate.
Genotype responses against both isolates were strongly correlated (r = 0.81, p-value <0.0001). The majority of the material of the phenotyping-panel was susceptible to at least one isolate but also 21 completely resistant genotypes against both isolates (arithmetic means of the normalized infected leaf area ≤ 1.5%) were identified (Fig 1and S1 Table). These genotypes were excluded from further analysis because their phenotype resembled the classical qualitative resistance and the focus of the study was put on the genotypes with a quantitative resistance phenotype. In addition, forty-five genotypes of the phenotyping-panel lack exome capture data [42], leading to the final GWA-panel with 201 genotyped barley genotypes having data on quantitative PM resistance. The high heritability (D35/3: 0.88 and RiIII: 0.87) and reproducibility of the different experiments (D35/3: 0.65-0.95 and RiIII: 0.70-0.93) indicate that the observed phenotypic variation is mostly genetically determined.
Population structure does not affect the powdery mildew resistance
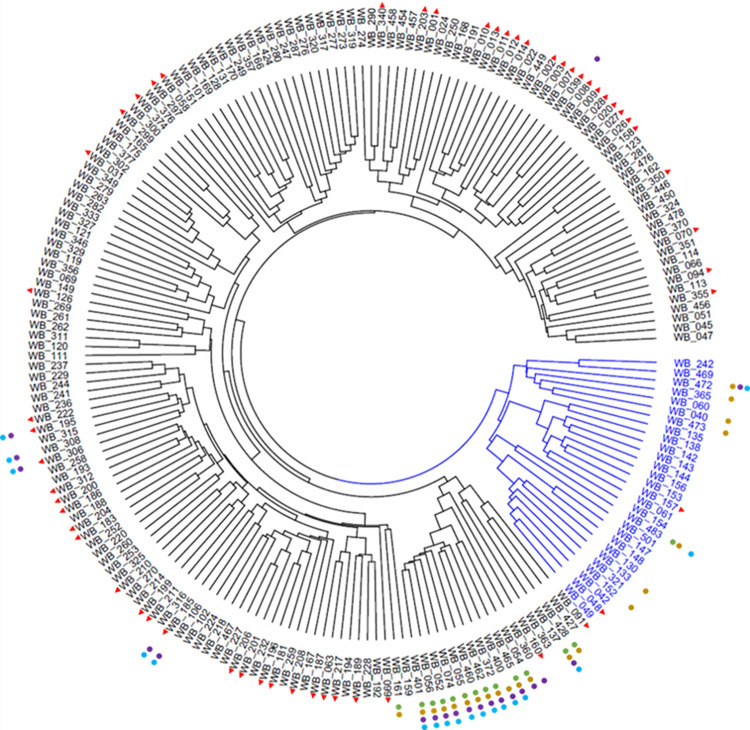
The RDs indicate the presence of two major clusters within the GWA-panel, where 28 genotypes were more closely related to each other than to the rest of the collection forming a small subpopulation (Fig 2).
Fig 2. Hierarchical cluster analysis of the 201 Hordeum vulgare accessions of the GWA-panel based on the Rogers‘ distance.
The identified subpopulation of 28 genotypes is highlighted in blue, modern material is labeled with red arrow-heads and carriers of the resistant alleles are indicated with colored dots: green–CG_1; ochre–CG_2; violet–CG_3; blue–CG_4.
Most of these landraces and wild barleys originate in South East Asia (75%) or the Middle-East (18%). Nevertheless, the subpopulation does not affect PM resistance because the arithmetic means of the infected leaf area of the two clusters (28 genotypes: 46.0 ± 19.9% and 173 genotypes: 49.9 ± 15.3%) are statistically not different from the one of the GWA-panel (49.4 ± 16.0%). Four further sub-clusters were mainly determined by the modern material or by traditional cultivars and landraces of North Africa and mixed origins (Fig 2and S1 Table). Regarding the low association of RDs and phenotypic distances (rMax = 0.12, p-value <0.001; rRiIII = 0.10, p-value <0.002 and rD35/3 = 0.08, p-value <0.001), only marginal effects of the population structure on the association study are expected. Therefore, the sub-clusters were not explicitly modeled during the GWA study.
Identification of candidate genes based on a genome-wide association study
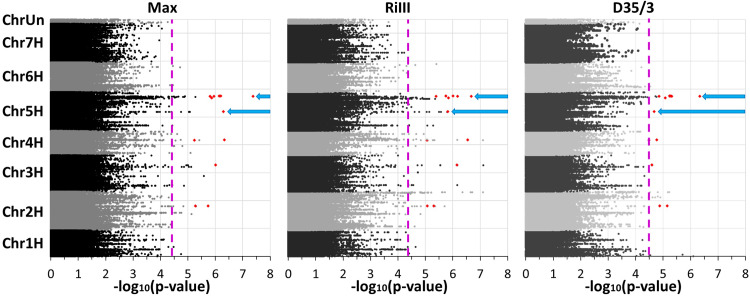
In addition to the single isolate resistance scores, an artificial maximal infection (Max) was created in silico, based on the highest infection value from both isolate sets. This Max trait was generated under the assumption that effects on the resistance caused by the different isolates were minimized and that the observed effects represent thus race-nonspecific responses. The linear mixed model in Eq (4) was fitted for the BLUEs of the three traits (RiIII, D35/3, and Max) at each of the 424,567 selected SNP positions. Based on the significance threshold of -log10 (p-value) = 4.45, a total of 214 SNPs located in 15 loci and distributed over six chromosomes, were significantly associated with PM resistance (Fig 3and S5 Table). Among them, 17 SNPs were in particular significantly associated with all three traits.
Fig 3. Manhattan plots of the–log10-transformed p-values for Max trait, RiIII, and D35/3 severities.
The significance threshold of -log10 (p-value) = 4.45 is depicted as a pink dotted line and the blue arrows represent the peaks from which the four candidate genes have been selected. The 17 SNPs, which were significantly associated with all three traits, are indicated as red diamonds.
Chromosome 7H was the only chromosome without significant associations. Among these associations, five peaks on chromosomes 2H to 5H were simultaneously significant for the three traits while two peaks co-localize with the seedling PM resistance locus Rbgq15 on chromosome 5H [11]. The involvement of this genomic region in the barley-Bgh interaction was further supported by recent results of Hunt et al. and Gupta et al. [71,72]. To further assess this promising region on chromosome 5H (S5 Fig in S1 File), the reference genome [34] was used to define candidate genes, whose SNPs were found to be associated with the Max trait. Within the first and the second peak, three and twelve candidate genes were identified respectively. The until now identified race-nonspecific resistance genes display various functions [1] and thus, the annotated candidate gene function was not considered as sufficient criterium to select promising candidate genes. Instead, the identified candidate gene models (S1A-S4A Figs in S1 File) were verified by comparison to similar sequences of (predicted) genes in related species, full-length cDNA and EST sequences to minimize the risk of the selection of pseudogenes (S6 Table). Further, the presence of transcripts in public databases, as a proof that the gene is expressed, and the possibility to design specific primers that amplify these genes from different genetic backgrounds formed also part of the selection criteria. As a result, four candidate genes, denoted as CG_1 to CG_4, respectively, were considered for further analyses (Table 1and S6 Table).
Table 1. Annotated information of the selected candidate genes mapping on chromosome 5H along with their corresponding significantly associated Single Nucleotide Polymorphisms (SNPs).
| -log10(P) | |||||||
|---|---|---|---|---|---|---|---|
| Candidate | Gene-IDa | Startb | Endb | SNPc | Max | RiIII | D35/3 |
| CG_1 | HORVU5Hr1 | 554454779 | 554457103 | SNP554456783chr5H | 5.07 | 5.79 | 3.46 |
| G078000 | |||||||
| CG_2 | HORVU5Hr1 | 554737443 | 554738375 | SNP554738172chr5H | 6.30 | 5.82 | 4.68 |
| G078330 | |||||||
| CG_3 | HORVU5Hr1 | 649208466 | 649212390 | SNP649211044chr5H | 4.83 | 3.89 | 4.36 |
| G116860 | |||||||
| CG_4 | HORVU5Hr1 | 650973514 | 650980482 | SNP650973939chr5H | 5.96 | 5.39 | 5.32 |
| G117650 | SNP650976700chr5H | 7.38 | 6.68 | 6.34 | |||
a Annotation in Morex [34].
b Physical start and end gene position in the reference genome in base pairs.
c Name of the significant SNP.
Altered transcript levels after powdery mildew attack
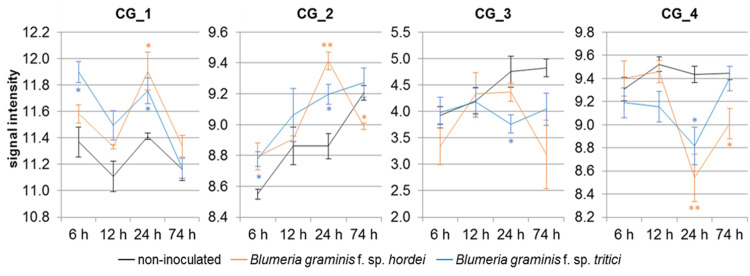
The transcript levels of all selected candidate genes are altered in PM attacked epidermis cells compared to the non-inoculated control (Fig 4) based on a transcript-profiling Microarray [68].
Fig 4. Relative expression of the four Candidate Genes (CG) in powdery mildew attacked epidermis cells.
The quantile-normalized signal intensities represent the relative expression of each CG based on Microarray data [68] for non-inoculated, Blumeria graminis f. sp. hordei (isolate CH4.8) or Blumeria graminis f. sp. tritici (isolate FAL 92315) inoculated epidermal peels of the resistant barley cultivar Vada for the indicated time points post-inoculation. The data embody arithmetic means (n = 3) plus standard error of the mean. * p-value < 0.05; ** p-value < 0.01.
In the case of CG_1, the detected pattern indicates that the expression is controlled by the circadian clock, while the inoculation with adapted PM fungus Bgh as well as the non-adapted fungus Bgt led to increased transcript levels. Moreover, elevated transcript levels were observed for CG_2 after inoculation with the non-adapted fungus. Nevertheless, the attack of the adapted fungus led to a transcript peak after 24h, which then declines significantly. The general expression of CG_3 is lower compared to the other candidate genes and the transcript level decreases after Bgt inoculation. Furthermore, the transcript of CG_4 is reduced 24h after PM attack. This effect seems to last longer after inoculation with Bgh than with Bgt.
Defined alleles vary in the powdery mildew resistance and are in linkage disequilibrium
Regarding the significantly associated SNPs, each selected candidate gene was present in two specific alleles, which differ significantly (p-value <0.0001) in their PM resistance (Table 2).
Table 2. Candidate alleles, their powdery mildew infection values as arithmetic means with standard error based on the infected leaf area from the Max BLUEs and their presence (n: Number of genotypes carrying them) in the GWA-panel.
| Allele | CG_1 | CG_2 | CG_3 | CG_4 |
|---|---|---|---|---|
| Resistant | 22.89 ± 3.28% | 24.76 ± 2.48% | 28.38 ± 3.48% | 24.24 ± 3.09% |
| (n = 14) | (n = 21) | (n = 20) | (n = 18) | |
| Susceptible | 51.14 ± 0.97% | 51.91 ± 0.94% | 51.50 ± 0.98% | 51.51 ± 0.95% |
| (n = 185) | (n = 179) | (n = 179) | (n = 181) |
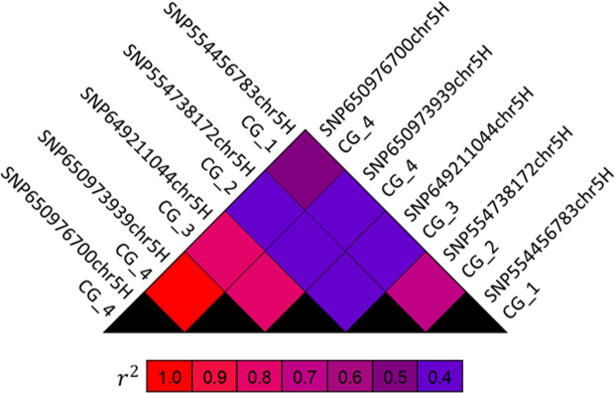
Additionally, the significant SNPs of the candidates were in LD (r2 > 0.4), with the highest r2 observed between the two SNPs of CG_4 (Fig 5).
Fig 5. Heatmap-plot of the linkage disequilibrium levels as r2 values for the Single-Nucleotide Polymorphisms (SNPs) of the selected Candidate Genes (CG).
The corresponding p-values for each r2 value were < 0.0001 based on the two-sided Fisher’s exact test.
Interestingly, the carriers of the resistant alleles, present in 7-10% of the GWA-panel (Table 2), cluster mainly together in a sub-cluster that is defined by Ethiopian landraces (Fig 2).
To estimate the effect of the different candidates, a simple linear regression model was fitted on the GWA-panel data for each candidate separately. In addition, a multiple linear regression model including all candidates at once was also fitted. Based on the simple regression models, the candidate genes explain the following proportion of the phenotypic variation: CG_1: 23.4%; CG_2: 31.0%; CG_3: 16.7%; and CG_4: 27.3%. Moreover, the multiple regression model considering all candidates explains only 36.0% of the phenotypic variation.
Sequence alterations between the selected candidate alleles
For each candidate gene, we defined two alleles; a potential resistant and a susceptible allele (Table 2). Since the susceptible alleles originate from Morex, all changes in resistant alleles were according to its sequence (S1A-S4A Figs in S1 File). The majority of the annotated and detected SNPs were located in non-coding regions. In the case of CG_1, three synonymous SNPs and one SNP that restores a conserved amino acid (Ile64Val) were detected (S1A Fig in S1 File). The significant SNP is located at the beginning of the 3’UTR (untranslated region). The protein sequences of both alleles from CG_2 were identical because only one synonymous SNP was determined and the significant SNP is located in the first intron (S2 Fig in S1 File). In the resistant allele of CG_3, three synonymous, and eight amino acid exchanges altering the protein sequence were identified (S3A Fig in S1 File). Most of these exchanged amino acids were conservative replacements in both, conserved and non-conserved regions. The only exception was a non-conservative amino acid exchange of the conserved Proline370 to Leucine (S3A Fig in S1 File). The significant SNP is located in the fourth intron. In the case of CG_4, one of the significant SNPs is located in the eleventh intron, while the other one causes an amino acid exchange of the conserved Alanine373 to Threonine (S4 Fig in S1 File). A new in-frame start codon, introduced by an SNP in the 5’UTR of CG_4 is potentially adding 51 additional amino acids to the N-terminus of the annotated protein (S4A Fig in S1 File). Protein sequence analysis by the SignalP server (http://www.cbs.dtu.dk/services/SignalP/) predicts the absence of a signal peptide in both alleles. No further sequence polymorphism was found in the available sequence information, particularly since none of the alleles of this candidate could be recovered as full-length E. coli clone.
Initial functional validation of the candidates
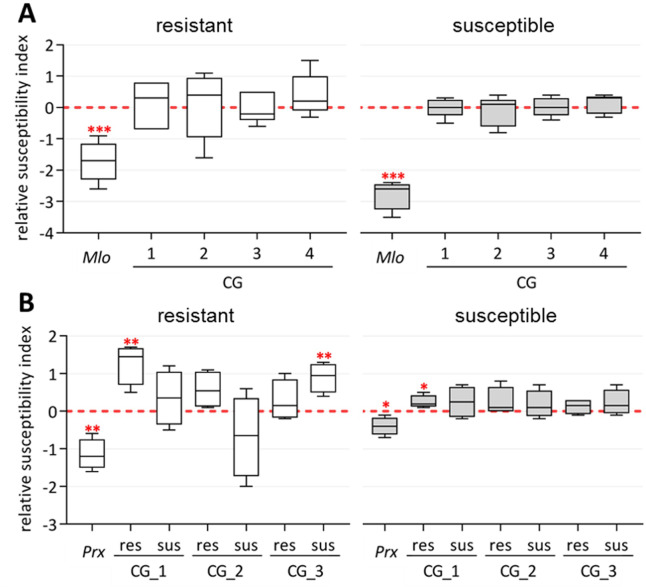
To test the different alleles in vivo, leaf segments of a race-specific resistant genotype (WB-052), which carries all resistant alleles, and a susceptible genotype (Morex) carrying the susceptible alleles were bombarded with different constructs. For the TIGS approach, hairpin RNAi constructs targeting the candidate genes were generated and tested [69], however, without a statistically significant effect on the resistance of either genotype (Fig 6A).
Fig 6. Box plots of the log2-transformed relative susceptibility indices of transiently transformed Hordeum vulgare epidermis cells.
The error bars represent the minimal and maximal values of the susceptibility indices relative to the empty vector control for the transient silencing (n = 5) (A) of the indicated candidates (CG) and for the transient overexpression (n = 4) (B) in a resistant (WB-052, hollow boxes) and a susceptible barley genotype (Morex, gray-shaded boxes) after inoculation with the Blumeria graminis f. sp. hordei isolate CH4.8, whereas the putative resistant (res) and the putative susceptible (sus) alleles were tested in the transient overexpression. The corresponding positive controls for the transient silencing: the barley Mlo gene (pIPKTA36) and for the transient overexpression: a wheat Class III peroxidase (Prx, pJP01) are depicted [43,69]. * p-value < 0.05; ** p-value < 0.01; *** p-value < 0.001.
As mentioned before, obtaining E. coli clones expressing the full-length coding sequence for GC_4 failed in all attempts. In this regard, only the full-length alleles of CG_1, CG_2, and CG_3 were cloned using genomic DNA to capture also the SNPs located in non-coding regions for the overexpression constructs (S1-S4 Figs in S1 File). The overexpression of the resistant allele of CG_1 enhances significantly the susceptibility in terms of an increased relative susceptibility index for both, resistant and susceptible background (Fig 6B). Moreover, the overexpression of any of the two alleles of CG_2 did not significantly affect the pathogen resistance in any of the two tested backgrounds. Last but not least, the overexpression of the susceptible allele of CG_3 enhances the susceptibility only in the resistant background.
Discussion
Powdery mildew resistance under controlled conditions
Detached leaf assays have been applied for the characterization of race-specific [73–75], as well as for race-nonspecific PM resistance genes [31]. PM isolates used in our study were poly-virulent and displayed overlapping virulence spectra (S2 Table), which is reflected in the high correlation between genotype responses against both isolates (Fig 1). Only five of the 22 inefficient resistance specificities were specific for one of the isolates. Moreover, cross-talk of defense-related and wound responsive genes has been observed in the past [76] thus, leaves displaying stress or wound responses were excluded from our analyses. The PM response assessed by us on detached seedling leaves under controlled conditions presented high repeatability (0.65–0.93) as well as heritability estimates (0.87–0.88), which evidence the high quality of the data subsequently used for the GWA study.
Identification of candidate genes in a highly diverse genotype panel
Despite the 28 genotypes with shared geographical origins forming a subpopulation (Fig 2and S1 Table), only a marginal impact of the population structure is expected because the mean infection of this group did not differ significantly from the rest of the GWA-panel and the correlation between genetic and phenotypic distances is in general low. In contrast, genotypes carrying detected resistant alleles display as a group a significantly reduced mean PM infection (Table 2).
In previous studies, genotype panels were restricted to specific geographical regions [30,77], the cultivation status [29,78], or were examined in a candidate gene-based approach [31], thus limiting the genetic diversity being studied. We were able to identify a total of 214 significantly associated SNPs (S5 Table) across the three traits (Max, RiIII, and D35/3). Furthermore, our approach demonstrates that it is possible to use a highly diverse barley panel to identify candidate genes associated with PM susceptibility.
For functional validation, we focused on two promising association peaks on chromosome 5H (Fig 3) co-localizing with the seedling PM resistance locus Rbgq15 [11]. These last authors proposed the lipoxygenase2 as the causal gene. Nevertheless, none of the SNPs in the identified corresponding candidate lipoxygenase genes (HORVU5Hr1G093700, HORVU5Hr1G093770, and HORVU5Hr1G117110) were significantly associated in our study (S5 Fig in S1 File). Furthermore, recent results of Hunt et al. [71] and Gupta et al. [72] identified the same genomic region either as a target region of small RNAs involved in the barley-Bgh interaction or as QTL involved in adult plant resistance, respectively. In addition, the potential race-nonspecific resistance gene HvLSD1b (LESION SIMULATING DISEASE RESISTANCE 1), i.e. HvLOL3 (LSD one-like 3; HORVU5Hr1G093280) [31], and Ror2 (Required for mlo-specified resistance; HORVU5Hr1G103590) are located in this genomic region [79]. These data strengthen the importance of the here-identified candidate region of chromosome 5H. Nevertheless, it should be mentioned that the association of SNPs is influenced by several factors like for instance, genetic heterogeneity, and that the most significant markers do not always correspond with causal variants [36,80]. Additionally, it has to be considered that the used exome capture data represent only 1.23% of the barley genome [33]. Besides, the platform design, as well as the candidate gene identification, relied on the annotated genes in the PM susceptible reference genotype Morex, which presumably left additional (candidate) genes carried by the identified PM resistant varieties unexplored [2]. At this point, genotyping with higher coverage as currently done in the BARN project (http://www.eracaps.org/joint-calls/era-caps-funded-projects/) might allow further evaluation of this particular question.
Selected candidate genes involved in the barley responses to powdery mildew
We identified a total of 214 significantly associated SNPs (S5 Table), but for the functional validation, only four candidate genes were selected (Table 1) from two promising peaks mapping on chromosome 5H (Fig 3). Linkage-mapping studies have shown the influence of loci mapping on the long arm of chromosome 5H on nonhost [7] and adult-plant [72] resistance against PM. Particularly, Gupta et al. [72] reported associated markers that span a 7.6 Mbp region on chromosome 5H and are located between 619.7 and 627.3 Mbp on the reference sequence of Morex. In this respect, our four candidate genes map close (Table 1) but are most likely not related to those previously reported QTL (S5 Fig in S1 File). Moreover, the two promising association peaks of our study co-localize with a QTL potentially involved in the barley-Bgh interaction [11,71]. A very recent transcriptome analysis [81] comparing two barley genotypes with contrasting responses to PM proposed a list of candidate genes controlling the response to PM infection. Interestingly, none of the four candidate genes of our study can be found among their proposed genes, which further highlights the novelty of our findings. Further analysis of significantly associated markers of the present study and the corresponding candidate genes can provide new valuable insights into the barley-Bgh plant pathosystem.
Each of the four selected candidate genes explained 17-31% of the phenotypic variation, while this magnitude amounts to 36% for all candidates together. The obtained effect sizes are similar to the reported 6.2–24.8% for Rbgq15 [11], further supporting the hypothesis that the here presented peaks correspond to this QTL. Only a small increase of the effect size was observed between the different models, which could be related to the LD (Fig 5) or a lack of complete additivity between the significant candidate SNPs.
Analyses of race-specific resistance genes indicate specific allelic effects [40,41]. In the present study, two distinct alleles could be annotated for each selected candidate (S1-S4 Figs in S1 File), which display a clear effect on the PM resistance (Table 2). Also, the transient assays confirm the allelic effects of the candidates (Fig 6B). Nevertheless, it has to be considered that the carriers of the potential resistant alleles cluster together (Fig 2). In particular, the resistance of WB-052 (HOR2573) might be independent of the here presented candidate genes since it was described as a donor of the MlLa-H resistance locus on chromosome 2H [82]. On the other hand, it cannot be ruled out that the observed resistance of WB-052 is caused by the combined effects of the MlLa-H resistance specificity and our identified candidate genes.
Because of the moderate LD between the significant candidate SNPs (Fig 5), it is possible that the observed PM resistance (Table 2) could be related to other (hidden) resistance specificities. Nonetheless, the transcript levels of all four candidate genes were altered in epidermal cells attacked by the adapted (Bgh) as well as the non-adapted (Bgt) PM fungus (Fig 4). These last results further support the involvement of the candidates in the PM responses (Table 3).
Table 3. Evidence for the candidates to be involved in Powdery Mildew (PM) responses based on the gene function, literature, the observed effect, the expressional regulation after PM attack, and the results of the transient assays.
| Candidate | Annotateda | Proposedb | Literature | Effectc | Regulationd | TIGSe | OXf |
|---|---|---|---|---|---|---|---|
| CG_1 | Unknown | AGP | Liang et al. [83] | NEGATIVE | UP | NO | YES |
| Wang et al. [84] | |||||||
| CG_2 | DPMT subunit | DPMS3 | Jadid et al. [85] | NEGATIVE | UP/DOWN | NO | NO |
| Häweker et al. [86] | |||||||
| CG_3 | BTB/POZ/Kelch-associated protein | LRB1/2 | Qu et al. [87] | NEGATIVE | DOWN | NO | YES |
| Christians et al. [88] | |||||||
| Xie et al. [89] | |||||||
| CG_4 | FEN 1-B | FEN 1-B | NO | NEGATIVE | DOWN | NO | NA |
a Annotated function: DPMT—dolichol-phosphate-mannosyl-transferase; BTB—Bric-a-Brac/-Tramtrack/-Broad Complex and POZ—POxvirus and Zinc-finger; FEN—flap endonuclease.
b Proposed function based on potential Arabidopsis homologs: AGP–arabinogalactan protein; DPMS3 –dolichol-phosphate-mannose-synthase 3; LRB—light-response BTB.
c Sign of the average difference in infected leaf area between resistant and susceptible alleles.
d Transcript regulation in PM attacked barley epidermis peels.
e Effect of transient silencing.
f Effect of transient overexpression; NA–not analyzed.
To gain further insights into the candidate gene functions, potential homologs of related grasses and model organisms were identified (S1A-S4A Figs in S1 File). For CG_1 no function is annotated (Table 3) and according to the homologs of Aegilops tauschii subsp. tauschii and Brachypodium distachyon, the candidate belongs to the conserved family of ‘UPF0664 stress-induced protein C29B12.11c’ proteins. Regarding the rice and the Arabidopsis homologs, the candidate (Table 3) might be a plasma membrane-bound arabinogalactan protein (AGP) [90,91]. In most homologs, a PH-GRAM-WBP2 (Pleckstrin-Homology-Glucosyltransferases, Rab-like GTPase activators and Myotubularins_WW-binding-protein 2) domain (domain family: cd13214) was annotated. Considering the high sequence identity to our candidate, this domain is conserved (S1A Fig in S1 File). In Magnaporthe oryzae-inoculated rice, a plasma membrane-bound AGP epitope was specifically upregulated in epidermis cells [83]. Similarly, the transcript level of CG_1 is elevated after PM inoculation (Fig 4). Results from Wang et al. [84] further support the hypothesis that CG_1 might function as a circadian clock-regulated defense gene (Fig 4). The overexpression of the potential resistant allele leads to enhanced susceptibility in both analyzed backgrounds (Fig 6B). This dominant-negative effect on the PM resistance in presence of an active race-specific resistance might be related to the inhibited regulation by the circadian clock, which seems to be naturally maintained even after PM attack (Fig 4).
CG_2 is annotated as ‘dolichol-phosphate-mannosyl-transferase subunit’ (Table 3) similarly to the annotations of the A. tauschii and the Arabidopsis homologs (S2A Fig in S1 File). In most identified homologs, a dolichol-phosphate-mannosyl-transferase subunit 3 domain is predicted, supporting the hypothesis that the candidate acts as a homolog of the Arabidopsis ‘dolichol-phosphate-mannose-synthase 3’ (DPMS3). DPMS3 is required for the production of isoprenyl-linked glycans [85]. N-glycans are essential for development and immune responses [86,92] and the extensive modification of the plant glycan structure after the fungal attack [93] further supports a candidate function in the response to PM attack. The significant transcript declines after 24h (Fig 4) and indicates that the adapted fungus could be specifically able to circumvent innate immune responses. In this respect, it might explain the negative outcome of the transient overexpression (Fig 6B). It is possible that Bgh could silence the (candidate) transcript similar to a mechanism described in Botrytis cinerea, which produces small RNAs perturbing the host immune signaling [94].
The candidate CG_3 and its homologs are annotated as ‘BTB/POZ/Kelch-associated proteins’ (Table 3). The ‘Bric-a-Brac/-Tramtrack/-Broad Complex (BTB)/-POxvirus and Zinc-finger (POZ)’ motif is a common protein-protein interaction motif [87]. In Arabidopsis, two protein homologs with high sequence identity (S3A Fig in S1 File), firstly described as POZ/BTB Containing-Protein1 and 2 (POB1/2) and later as Light-Response BTB1 and 2 (LRB1/2) [88], were identified. Similar high sequence identity was observed between CG_3 and its potential barley paralog and the two rice orthologs (S3A Fig in S1 File). Interestingly, this paralog seems not to be associated with PM resistance. Whether this is a specific candidate effect or caused by genetic heterogeneity [36] needs further investigation. With exception of the nuclear localization signal (NLS), the annotated BTB and BACK (BTB And C-terminal-Kelch) domains were conserved (S3A Fig in S1 File), thus supporting the hypothesis that CG_3 is an ortholog of AtLRB1/2. The LRBs constitute Cullin3-based E3 ubiquitin ligases involved in flowering, light-signaling [95], and jasmonate-mediated defense reactions [87]. Particularly, PhyB is essential for light-mediated defense responses [96], while altered resistance responses were described for various phyB mutants and pathogens [89,97–99]. The role of CG_3 as a regulator of PhyB in barley needs further investigation. In contrast to the ubiquitous transcription of the AtLRBs [88], only low transcript levels were detected in barley and were further reduced after PM attack (Fig 4). The enhanced susceptibility observed in the transient overexpression of the potentially susceptible candidate allele (Fig 6B) supports this hypothesis; although this effect is only observed in the resistant background (Fig 6B). A higher expression of the natural allele may have no additional effect on Morex’s susceptibility. On the other hand, potential dominance effects might mask the effects of the potential resistant candidate alleles, in particular regarding the still active natural allele in the corresponding genotype and the race-specific resistance of WB-052.
CG_4 is annotated as ‘flap endonuclease (FEN) 1-B’ (Table 3) and the corresponding barley homolog as FEN1-A (S4A Fig in S1 File). A similar protein pair was described in rice. Additional FEN1-B homologs were identified in related grass species (S4A Fig), but in Arabidopsis FEN1 is a single-copy gene [100]. Particularly, the XPG (Xeroderma Pigmentosum Complementation Group G) N-terminal domain and the XPG Internal-region seem to be conserved (S4A Fig). In Arabidopsis, FEN1 is essential for genome stability, development, and transcriptional gene silencing [101,102]. Nevertheless, FEN1-B proteins seem to fulfill distinct functions besides their regulatory role in cell proliferation [61]. The OsFEN1 transcripts are expressed in proliferating tissue and OsFEN1-B additionally in mature leaves [61]. Similarly, FEN1-B (CG_4) transcripts are detected in mature leaf epidermal cells (Fig 4) and are significantly reduced after PM attack. This result supports the candidate involvement in PM responses. Nonetheless, the candidate validation via transient overexpression is pending because the full-length alleles could not be cloned, whereas the transient gene silencing did not lead to significant results for either candidate gene (Fig 6A), although the positive control (Mlo) indicate the success of the assay. In this context, it has to be considered that this gene has a strong effect on the PM resistance [69].
Altogether, the here presented results give first insights into the functionality of the selected candidate genes and their involvement in race-nonspecific resistance, because: (a) the annotated functions indicate that all four candidates participate in basic (regulatory) processes (Table 3); (b) the candidate transcript levels were altered after PM attack of the adapted as well as the non-adapted fungus (Fig 4) and (c) the enhanced susceptibility caused by the overexpression of CG_1 (an AGP) and CG_3, the barley homolog of AtLRB1/2 (Fig 6B), occurs even after the inoculation with a PM isolate, which was not used for the phenotyping and (d) in the presence of a race-specific resistance background. Furthermore, the results of this work further confirm the complexity of the biological/genetic mechanisms underlying race-nonspecific resistance/susceptibility against fungal plant diseases. At this stage and although promising, these results should be further validated in future research by using other functional techniques such as stable transformants or mutants, before they can have any practical application in barley breeding.
Supporting information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
Acknowledgments
We would like to acknowledge the technical support of Jeyaraman Rajaraman and Daniela Nowara to the present study. We thank Kerstin Flath from the Institute for Plant Protection in Field Crops and Grassland, Julius Kühn-Institute (JKI) Kleinmachnow for the kind gift of Bgh isolates and corresponding virulence spectra. Patrick Schweizer passed away before the submission of the final version of this manuscript. Albert W. Schulthess accepts responsibility for the integrity and validity of the data collected and analyzed.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
MP was supported by the WHEALBI project (EU FP7 n°FP7- 613556). AWS and DD were supported by the Federal Ministry of Education and Research of Germany (Grant FKZ031B0184A). FL was supported by funds of the China Scholarship Council (CSC; Grant no. 201606760065). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Niks RE, Qi X, Marcel TC. Quantitative resistance to biotrophic filamentous plant pathogens: concepts, misconceptions, and mechanisms. Annu Rev Phytopathol. 2015;53:445–470. 10.1146/annurev-phyto-080614-115928 [DOI] [PubMed] [Google Scholar]
- 2.Sánchez-Martín J, Keller B. Contribution of recent technological advances to future resistance breeding. Theor Appl Genet. 2019;132:713–732. 10.1007/s00122-019-03297-1 [DOI] [PubMed] [Google Scholar]
- 3.Dangl JL, Horvath DM, Staskawicz BJ. Pivoting the plant immune system from dissection to deployment. Science. 2013;341:746–751. 10.1126/science.1236011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown JKM. Durable resistance of crops to disease: a Darwinian perspective. Annu Rev Phytopathol. 2015;53:513–539. 10.1146/annurev-phyto-102313-045914 [DOI] [PubMed] [Google Scholar]
- 5.Wyand RA, Brown JKM. Genetic and forma specialis diversity in Blumeria graminis of cereals and its implications for host-pathogen co-evolution. Mol Plant Pathol. 2003;4:187–198. 10.1046/j.1364-3703.2003.00167.x [DOI] [PubMed] [Google Scholar]
- 6.Glawe DA. The powdery mildews: A review of the world’s most familiar (yet poorly known) plant pathogens. Annu Rev Phytopathol. 2008;46:27–51. 10.1146/annurev.phyto.46.081407.104740 [DOI] [PubMed] [Google Scholar]
- 7.Romero CCT, Vermeulen JP, Vels A, Himmelbach A, Mascher M, et al. Mapping resistance to powdery mildew in barley reveals a large-effect nonhost resistance QTL. Theor Appl Genet. 2018;131:1031–1045. 10.1007/s00122-018-3055-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu J, Zhou Y, Shang Y, Hua W, Wang J, Jia Q, et al. Genetic evidence of local adaption and long distance migration in Blumeria graminis f. sp. hordei populations from China. J Gen Plant Pathol. 2016;82:69–81. 10.1007/s10327-016-0643-1 [DOI] [Google Scholar]
- 9.Draz IS, Esmail SM, Abou-Zeid MAEH, Essa TAEM. Powdery mildew susceptibility of spring wheat cultivars as a major constraint on grain yield. Ann Agric Sci. 2019;64:39–45. 10.1016/j.aoas.2019.05.007 [DOI] [Google Scholar]
- 10.FAOSTAT-Statistics-Division. Food and Agriculture Organization of the United Nations. FAOSTAT Database, Rome, Italy. 2019. Available from: http://www.fao.org/faostat/en/#data/QC (Accessed on 11 September 2019).
- 11.Aghnoum R, Marcel TC, Johrde A, Pecchioni N, Schweizer P, Niks RE. Basal host resistance of barley to powdery mildew: Connecting quantitative trait loci and candidate genes. Mol Plant-Microbe Interact. 2010;23:91–102. 10.1094/MPMI-23-1-0091 [DOI] [PubMed] [Google Scholar]
- 12.Jørgensen JH, Wolfe M. Genetics of powdery mildew resistance in barley. CRC Crit Rev Plant Sci. 1994;13:97–119. 10.1080/07352689409701910 [DOI] [Google Scholar]
- 13.Kusch S, Panstruga R. mlo-Based Resistance: An apparently universal “weapon” to defeat powdery mildew disease. Mol Plant-Microbe Interact. 2017;30:179–189. 10.1094/MPMI-12-16-0255-CR [DOI] [PubMed] [Google Scholar]
- 14.Büschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, et al. The barley Mlo gene: A novel control element of plant pathogen resistance. Cell. 1997;88:695–705. 10.1016/s0092-8674(00)81912-1 [DOI] [PubMed] [Google Scholar]
- 15.Jørgensen JH. Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica. 1992;63:141–152. 10.1007/BF00023919 [DOI] [Google Scholar]
- 16.Eissa HF, Hassanien SE, Ramadan AM, El-Shamy MM, Saleh OM, Shokry AM, et al. Developing transgenic wheat to encounter rusts and powdery mildew by overexpressing barley chi26 gene for fungal resistance. Plant Methods. 2017;13:41 10.1186/s13007-017-0191-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niks RE, Marcel TC. Nonhost and basal resistance: How to explain specificity? New Phytol. 2009;182:817–828. 10.1111/j.1469-8137.2009.02849.x [DOI] [PubMed] [Google Scholar]
- 18.Chen X. Review article: High-temperature adult-plant resistance, key for sustainable control of stripe rust. Am J Plant Sci. 2013;04:608–627. 10.4236/ajps.2013.43080 [DOI] [Google Scholar]
- 19.Parlevliet JE. Resistance of the non-race-specific type In: Roelfs AP, Bushnell WR, editors. The Cereal Rusts, Volume II: Diseases, Distribution, Epidemiology, and Control. Academic Press; 1985. p. 501–525. 10.1016/B978-0-12-148402-6.50024-9 [DOI] [Google Scholar]
- 20.Li Z, Lan C, He Z, Singh RP, Rosewarne GM, Chen X, et al. Overview and application of QTL for adult plant resistance to leaf rust and powdery mildew in wheat. Crop Sci. 2014;54:1907–1925. 10.2135/cropsci2014.02.0162 [DOI] [Google Scholar]
- 21.Kou Y, Wang S. Broad-spectrum and durability: understanding of quantitative disease resistance. Curr Opin Plant Biol. 2010;13:181–185. 10.1016/j.pbi.2009.12.010 [DOI] [PubMed] [Google Scholar]
- 22.Kilian B, Özkan H, Kohl J, Von Haeseler A, Barale F, Deusch O, et al. Haplotype structure at seven barley genes: Relevance to gene pool bottlenecks, phylogeny of ear type and site of barley domestication. Mol Genet Genomics. 2006;276:230–241. 10.1007/s00438-006-0136-6 [DOI] [PubMed] [Google Scholar]
- 23.Pourkheirandish M, Komatsuda T. The importance of barley genetics and domestication in a global perspective. Ann Bot. 2007;100:999–1008. 10.1093/aob/mcm139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Mittal N, Leamy LJ, Barazani O, Song BH. Back into the wild—Apply untapped genetic diversity of wild relatives for crop improvement. Evol Appl. 2017;10:5–24. 10.1111/eva.12434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmid K, Kilian B, Russell J. Barley domestication, adaptation and population genomics In: Stein N, Muehlbauer GJ, editors. The Barley Genome, Compendium of Plant Genomes. Springer International Publishing; 2018. p. 317–336. 10.1007/978-3-319-92528-8_17 [DOI] [Google Scholar]
- 26.McCouch S. Agriculture: Feeding the future. Nature. 2013;499:23–24. 10.1038/499023a [DOI] [PubMed] [Google Scholar]
- 27.Zhu C, Gore M, Buckler ES, Yu J. Status and prospects of association mapping in plants. Plant Genome J. 2008;1:5–20. 10.3835/plantgenome2008.02.0089 [DOI] [Google Scholar]
- 28.Brachi B, Morris GP, Borevitz JO. Genome-wide association studies in plants: The missing heritability is in the field. Genome Biol. 2011;12:232 10.1186/gb-2011-12-10-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ames N, Dreiseitl A, Steffenson BJ, Muehlbauer GJ. Mining wild barley for powdery mildew resistance. Plant Pathol. 2015;64:1396–1406. 10.1111/ppa.12384 [DOI] [Google Scholar]
- 30.Bengtsson T, Åhman I, Manninen O, Reitan L, Christerson T, Jensen JD, et al. A novel QTL for powdery mildew resistance in nordic spring barley (Hordeum vulgare L. ssp. vulgare) revealed by genome-wide association study. Front Plant Sci. 2017;8:1954 10.3389/fpls.2017.01954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spies A, Korzun V, Bayles R, Rajaraman J, Himmelbach A, Hedley PE, et al. Allele mining in barley genetic resources reveals genes of race-non-specific powdery mildew resistance. Front Plant Sci. 2012;2:113 10.3389/fpls.2011.00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flint-Garcia SA, Thornsberry JM, Buckler ES. Structure of linkage disequilibrium in plants. Annu Rev Plant Biol. 2003;54:357–374. 10.1146/annurev.arplant.54.031902.134907 [DOI] [PubMed] [Google Scholar]
- 33.Mascher M, Richmond TA, Gerhardt DJ, Himmelbach A, Clissold L, Sampath D, et al. Barley whole exome capture: A tool for genomic research in the genus Hordeum and beyond. Plant J. 2013;76:494–505. 10.1111/tpj.12294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T, et al. A chromosome conformation capture ordered sequence of the barley genome. Nature. 2017;544:427–433. 10.1038/nature22043 [DOI] [PubMed] [Google Scholar]
- 35.Warr A, Robert C, Hume D, Archibald A, Deeb N, Watson M. Exome sequencing: Current and future perspectives. G3 Genes, Genomes, Genet. 2015;5:1543–1550. 10.1534/g3.115.018564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Platt A, Vilhjálmsson BJ, Nordborg M. Conditions under which genome-wide association studies will be positively misleading. Genetics. 2010;186:1045–1052. 10.1534/genetics.110.121665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bartoli C, Roux F. Genome-wide association studies in plant pathosystems: Toward an ecological genomics approach. Front Plant Sci. 2017;8:763 10.3389/fpls.2017.00763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burghardt LT, Young ND, Tiffin P. A Guide to genome-wide association mapping in plants. Curr Protoc Plant Biol. 2017;2:22–38. 10.1002/cppb.20041 [DOI] [PubMed] [Google Scholar]
- 39.Panstruga R. A golden shot: How ballistic single cell transformation boosts the molecular analysis of cereal-mildew interactions. Mol Plant Pathol. 2004;5:141–148. 10.1111/j.1364-3703.2004.00208.x [DOI] [PubMed] [Google Scholar]
- 40.Seeholzer S, Tsuchimatsu T, Jordan T, Bieri S, Pajonk S, Yang W, et al. Diversity at the Mla powdery mildew resistance locus from cultivated barley reveals sites of positive selection. Mol Plant-Microbe Interact. 2010;23:497–509. 10.1094/MPMI-23-4-0497 [DOI] [PubMed] [Google Scholar]
- 41.Bourras S, McNally KE, Ben-David R, Parlange F, Roffler S, Praz CR, et al. Multiple avirulence loci and allele-specific effector recognition control the Pm3 race-specific resistance of wheat to powdery mildewopen. Plant Cell. 2015;27:2991–3012. 10.1105/tpc.15.00171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bustos-Korts D, Dawson IK, Russell J, Tondelli A, Guerra D, Ferrandi C, et al. Exome sequences and multi-environment field trials elucidate the genetic basis of adaptation in barley. Plant J. 2019; 99:1172–1191. 10.1111/tpj.14414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altpeter F, Varshney A, Abderhalden O, Douchkov D, Sautter C, Kumlehn J, et al. Stable expression of a defense-related gene in wheat epidermis under transcriptional control of a novel promoter confers pathogen resistance. Plant Mol Biol. 2005;57:271–283. 10.1007/s11103-004-7564-7 [DOI] [PubMed] [Google Scholar]
- 44.GraphPad. Prism 7.01 for Windows, GraphPad Software, La Jolla California USA. La Jolla California USA. La Jolla California USA; 2017. Available from: www.graphpad.com.
- 45.Henderson CR. Best linear unbiased estimation and prediction under a selection model. Biometrics. 1975;31:423–447. 10.2307/2529430 [DOI] [PubMed] [Google Scholar]
- 46.Piepho HP, Büchse A, Emrich K. A Hitchhiker’s Guide to mixed models for randomized experiments. J Agron Crop Sci. 2003;189:310–322. 10.1046/j.1439-037X.2003.00049.x [DOI] [Google Scholar]
- 47.González MY, Philipp N, Schulthess AW, Weise S, Zhao Y, Börner A, et al. Unlocking historical phenotypic data from an ex situ collection to enhance the informed utilization of genetic resources of barley (Hordeum sp.). Theor Appl Genet. 2018;131:2009–2019. 10.1007/s00122-018-3129-z [DOI] [PubMed] [Google Scholar]
- 48.Philipp N, Weise S, Oppermann M, Börner A, Graner A, Keilwagen J, et al. Leveraging the use of historical data gathered during seed regeneration of an ex situ genebank collection of wheat. Front Plant Sci. 2018;9:609 10.3389/fpls.2018.00609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Piepho HP, Möhring J. Computing heritability and selection response from unbalanced plant breeding trials. Genetics. 2007;177:1881–1888. 10.1534/genetics.107.074229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rogers JS. Measures of genetic similarity and genetic distance In: Wheeler MR, editor. Studies in genetics, VII. Publ. 7213. Austin, USA: University of Texas Publication; 1972. p. 145–153. [Google Scholar]
- 51.R Development Core Team. R: a language and environment for statistical computing. Version 3.5.1. Vienna, Austria: R Foundation for Statistical Computing; 2017. Available from: https://www.r-project.org/ [Google Scholar]
- 52.Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. 10.1093/bioinformatics/btm308 [DOI] [PubMed] [Google Scholar]
- 53.Yu J, Pressoir G, Briggs WH, Bi IV, Yamasaki M, Doebley JF, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 2006;38:203–208. 10.1038/ng1702 [DOI] [PubMed] [Google Scholar]
- 54.Melchinger AE, Messmer MM, Lee M, Woodman WL, Lamkey KR. Diversity and Relationships among U.S. Maize Inbreds Revealed by Restriction Fragment Length Polymorphisms. Crop Sci. 1991;31:669 10.2135/cropsci1991.0011183X003100030025x [DOI] [Google Scholar]
- 55.Gao X, Starmer J, Martin ER. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol. 2008;32:361–369. 10.1002/gepi.20310 [DOI] [PubMed] [Google Scholar]
- 56.Butler D, Cullis BR, Gilmour AR, Gogel BJ. ASReml-R reference manual, release 3.0 Brisbane: Queensland Department of Primary Industries; 2009. Available from: https://asreml.kb.vsni.co.uk/wp-content/uploads/sites/3/2018/02/ASReml-3-User-Guide.pdf [Google Scholar]
- 57.R Development Core Team. R: a language and environment for statistical computing. Version 2.15. Vienna, Austria: R Foundation for Statistical Computing; 2013. Available from: https://www.r-project.org/ [Google Scholar]
- 58.Deng W, Nickle DC, Learn GH, Maust B, Mullins JI. ViroBLAST: A stand-alone BLAST web server for flexible queries of multiple databases and user’s datasets. Bioinformatics. 2007;23:2334–2336. 10.1093/bioinformatics/btm331 [DOI] [PubMed] [Google Scholar]
- 59.Huala E, Dickerman AW, Garcia-Hernandez M, Weems D, Reiser L, LaFond F, et al. The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 2001;29:102–105. 10.1093/nar/29.1.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 2007;35:883–887. 10.1093/nar/gkl976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kimura S, Furukawa T, Kasai N, Mori Y, Kitamoto HK, Sugawara F, et al. Functional characterization of two flap endonuclease-1 homologues in rice. Gene. 2003;314:63–71. 10.1016/s0378-1119(03)00694-2 [DOI] [PubMed] [Google Scholar]
- 62.Lück S, Kreszies T, Strickert M, Schweizer P, Kuhlmann M, Douchkov D. siRNA-Finder (si-Fi) software for RNAi-target design and off-target prediction. Front Plant Sci. 2019;10:1023 10.3389/fpls.2019.01023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47:427–432. 10.1093/nar/gky995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci U S A. 2009;106:10171–10176. 10.1073/pnas.0900604106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. 10.1093/nar/gkg563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 2019;37:420–423. 10.1038/s41587-019-0036-z [DOI] [PubMed] [Google Scholar]
- 68.Delventhal R, Rajaraman J, Stefanato FL, Rehman S, Aghnoum R, McGrann GRD, et al. A comparative analysis of nonhost resistance across the two Triticeae crop species wheat and barley. BMC Plant Biol. 2017;17:232 10.1186/s12870-017-1178-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Douchkov D, Nowara D, Zierold U, Schweizer P. A High-Throughput Gene-Silencing System for the Functional Assessment of Defense-Related Genes in Barley Epidermal Cells. Mol Plant-Microbe Interact. 2005;18:755–761. 10.1094/MPMI-18-0755 [DOI] [PubMed] [Google Scholar]
- 70.Zimmermann G, Bäumlein H, Mock HP, Himmelbach A, Schweizer P. The multigene family encoding germin-like proteins of barley. Regulation and function in basal host resistance. Plant Physiol. 2006;142:181–192. 10.1104/pp.106.083824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hunt M, Banerjee S, Surana P, Liu M, Fuerst G, Mathioni S, et al. Correction to: small RNA discovery in the interaction between barley and the powdery mildew pathogen. BMC Genomics. 2019;20:697 10.1186/s12864-019-6012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gupta S, Vassos E, Sznajder B, Fox R, Khoo KHP, Loughman R, et al. A locus on barley chromosome 5H affects adult plant resistance to powdery mildew. Mol Breed. 2018;38:103 10.1007/s11032-018-0858-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dreiseitl A. Heterogeneity of powdery mildew resistance revealed in accessions of the ICARDA wild barley collection. Front Plant Sci. 2017;8:202 10.3389/fpls.2017.00202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Silvar C, Flath K, Kopahnke D, Gracia MP, Lasa JM, Casas AM, et al. Analysis of powdery mildew resistance in the Spanish barley core collection. Plant Breed. 2011;130:195–202. 10.1111/j.1439-0523.2010.01843.x [DOI] [Google Scholar]
- 75.Šurlan-Momirović G, Flath K, Silvar C, Branković G, Kopahnke D, Knežević D, et al. Exploring the Serbian GenBank barley (Hordeum vulgare L. subsp. vulgare) collection for powdery mildew resistance. Genet Resour Crop Evol. 2016;63:275–287. 10.1007/s10722-015-0246-2 [DOI] [Google Scholar]
- 76.van Loon LC, Rep M, Pieterse CMJ. Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol. 2006;44:135–162. 10.1146/annurev.phyto.44.070505.143425 [DOI] [PubMed] [Google Scholar]
- 77.Gupta S, D’Antuono M, Bradley J, Li C, Loughman R. Identification and expression of adult plant resistance in barley to powdery mildew (Blumeria graminis f. sp. hordei) in Australia. Euphytica. 2015;203:595–605. 10.1007/s10681-014-1280-4 [DOI] [Google Scholar]
- 78.Rajaraman J, Douchkov D, Lück S, Hensel G, Nowara D, Pogoda M, et al. Evolutionarily conserved partial gene duplication in the Triticeae tribe of grasses confers pathogen resistance. Genome Biol. 2018;19:116 10.1186/s13059-018-1472-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, et al. SNARE-protein-mediated disease resistance at the plant cell wall. Nature. 2003;425:973–7. 10.1038/nature02076 [DOI] [PubMed] [Google Scholar]
- 80.Korte A, Farlow A. The advantages and limitations of trait analysis with GWAS: A review. Plant Methods. 2013;9:29 Plant Methods. 10.1186/1746-4811-9-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li Y, Guo G, Zhou L, Chen Y, Zong Y, Huang J, et al. Transcriptome Analysis Identifies Candidate Genes and Functional Pathways Controlling the Response of Two Contrasting Barley Varieties to Powdery Mildew Infection. Int J Mol Sci. 2019;21:151 10.3390/ijms21010151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hoseinzadeh P, Zhou R, Mascher M, Himmelbach A, Niks RE, Schweizer P, et al. High resolution genetic and physical mapping of a major powdery mildew resistance locus in barley. Front Plant Sci. 2019;10:146 10.3389/fpls.2019.00146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liang Y, Zhao J, Wang C, Jing Y, Chengyun L, Baldwin TC. Infection with blast fungus (Magnaporthe orzyae) leads to increased expression of an arabinogalactan-protein epitope in both susceptible and resistant rice cultivars. Physiol Mol Plant Pathol. 2018;102:136–143. 10.1016/j.pmpp.2018.01.002 [DOI] [Google Scholar]
- 84.Wang W, Barnaby JY, Tada Y, Li H, Tör M, Caldelari D, et al. Timing of plant immune responses by a central circadian regulator. Nature. 2011;470:110–115. 10.1038/nature09766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jadid N, Mialoundama A, Heintz D, Ayoub D, Erhardt M, Mutterer J, et al. DOLICHOL PHOSPHATE MANNOSE SYNTHASE1 mediates the biogenesis of isoprenyl-linked glycans and influences development, stress response, and ammonium hypersensitivity in Arabidopsis. Plant Cell. 2011;23:1985–2005. 10.1105/tpc.111.083634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Häweker H, Rips S, Koiwa H, Salomon S, Saijo Y, Chinchilla D, et al. Pattern recognition receptors require N-glycosylation to mediate plant immunity. J Biol Chem. 2010;285:4629–4636. 10.1074/jbc.M109.063073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qu N, Gan W, Bi D, Xia S, Zhang Y, Li X. Two BTB proteins function redundantly as negative regulators of defense against pathogens in Arabidopsis. Botany. 2010;88:953–960. 10.1139/B10-067 [DOI] [Google Scholar]
- 88.Christians MJ, Gingerich DJ, Hua Z, Lauer TD, Vierstra RD. The light-response BTB1 and BTB2 proteins assemble nuclear ubiquitin ligases that modify phytochrome B and D signaling in Arabidopsis. Plant Physiol. 2012;160:118–134. 10.1104/pp.112.199109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xie XZ, Xue YJ, Zhou JJ, Zhang B, Chang H, Takano M. Phytochromes regulate SA and JA signaling pathways in rice and are required for developmentally controlled resistance to Magnaporthe grisea. Mol Plant. 2011;4:688–696. 10.1093/mp/ssr005 [DOI] [PubMed] [Google Scholar]
- 90.Marmagne A, Ferro M, Meinnel T, Bruley C, Kuhn L, Garin J, et al. A high content in lipid-modified peripheral proteins and integral receptor kinases features in the Arabidopsis plasma membrane proteome. Mol Cell Proteomics. 2007;6:1980–1996. 10.1074/mcp.M700099-MCP200 [DOI] [PubMed] [Google Scholar]
- 91.Marmagne A, Roue MA, Ferro M, Rolland N, Alcon C, Joyard J, et al. Identification of new intrinsic proteins in Arabidopsis plasma membrane proteome. Mol Cell Proteomics. 2004;3:675–691. 10.1074/mcp.M400001-MCP200 [DOI] [PubMed] [Google Scholar]
- 92.Saijo Y, Tintor N, Lu X, Rauf P, Pajerowska-Mukhtar K, Häweker H, et al. Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 2009;28:3439–3449. 10.1038/emboj.2009.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chaliha C, Rugen MD, Field RA, Kalita E. Glycans as modulators of plant defense against filamentous pathogens. Front Plant Sci. 2018;9:928 10.3389/fpls.2018.00928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hua C, Zhao JH, Guo HS. Trans-kingdom RNA silencing in plant–fungal pathogen interactions. Mol Plant. 2018;11:235–244. 10.1016/j.molp.2017.12.001 [DOI] [PubMed] [Google Scholar]
- 95.Shu K, Yang W. E3 ubiquitin ligases: ubiquitous actors in plant development and abiotic stress responses. Plant Cell Physiol. 2017;58:1461–1476. 10.1093/pcp/pcx071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ballaré CL. Light regulation of plant defense. Annu Rev Plant Biol. 2014;65:335–363. 10.1146/annurev-arplant-050213-040145 [DOI] [PubMed] [Google Scholar]
- 97.Cerrudo I, Caliri-Ortiz ME, Keller MM, Degano ME, Demkura P V., Ballaré CL. Exploring growth-defence trade-offs in Arabidopsis: phytochrome B inactivation requires JAZ10 to suppress plant immunity but not to trigger shade-avoidance responses. Plant Cell Environ. 2017;40:635–644. 10.1111/pce.12877 [DOI] [PubMed] [Google Scholar]
- 98.Griebel T, Zeier J. Light regulation and daytime dependency of inducible plant defenses in Arabidopsis: phytochrome signaling controls systemic acquired resistance rather than local defense. Plant Physiol. 2008;147:790–801. 10.1104/pp.108.119503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kazan K, Manners JM. The interplay between light and jasmonate signalling during defence and development. J Exp Bot. 2011;62:4087–4100. 10.1093/jxb/err142 [DOI] [PubMed] [Google Scholar]
- 100.Shultz RW, Tatineni VM, Hanley-Bowdoin L, Thompson WF. Genome-wide analysis of the core DNA replication machinery in the higher plants Arabidopsis and rice. Plant Physiol. 2007;144:1697–1714. 10.1104/pp.107.101105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang J, Xie S, Zhu JK, Gong Z. Requirement for flap endonuclease 1 (FEN1) to maintain genomic stability and transcriptional gene silencing in Arabidopsis. Plant J. 2016;87:629–640. 10.1111/tpj.13224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang Y, Wen C, Liu S, Zheng L, Shen B, Tao Y. Shade avoidance 6 encodes an Arabidopsis flap endonuclease required for maintenance of genome integrity and development. Nucleic Acids Res. 2016;44:1271–1284. 10.1093/nar/gkv1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.