Abstract
In general, beef cattle long-distance transportation from cow-calf operations to feedlots or from feedlots to abattoirs is a common situation in the beef industry. The aim of this study was to determine the effect of rumen-protected methionine (RPM) supplementation on a proposed gene network for muscle fatigue, creatine synthesis (CKM), and reactive oxygen species (ROS) metabolism after a transportation simulation in a test track. Angus × Simmental heifers (n = 18) were stratified by body weight (408 ± 64 kg; BW) and randomly assigned to dietary treatments: 1) control diet (CTRL) or 2) control diet + 8 gr/hd/day of top-dressed rumen-protected methionine (RPM). After an adaptation period to Calan gates, animals received the mentioned dietary treatment consisting of Bermuda hay ad libitum and a soy hulls and corn gluten feed based supplement. After 45 days of supplementation, animals were loaded onto a trailer and transported for 22 hours (long-term transportation). Longissimus muscle biopsies, BW and blood samples were obtained on day 0 (Baseline), 43 (Pre-transport; PRET), and 46 (Post-transport; POST). Heifers’ average daily gain did not differ between baseline and PRET. Control heifer’s shrink was 10% of BW while RPM heifers shrink was 8%. Serum cortisol decreased, and glucose and creatine kinase levels increased after transportation, but no differences were observed between treatments. Messenger RNA was extracted from skeletal muscle tissue and gene expression analysis was performed by RT-qPCR. Results showed that AHCY and DNMT3A (DNA methylation), SSPN (Sarcoglycan complex), and SOD2 (Oxidative Stress-ROS) were upregulated in CTRL between baseline and PRET and, decreased between pre and POST while they remained constant for RPM. Furthermore, CKM was not affected by treatments. In conclusion, RPM supplementation may affect ROS production and enhance DNA hypermethylation, after a long-term transportation.
Introduction
Long-distance road transportation (> 400 km) is a stressful event that most cattle experience at least once in their lifetime [1]. It is important to consider that road transportation includes more than just the shipping itself; it should also considerthe assembly and loading of animals at their place of origin, confinement on a moving or stationary vehicle, unloading, and penning at their final destination [2]. Several research studies have been conducted in the last few decades, in an effort to achieve a better understanding of the characteristics of road transportation and its effects on cattle [3–6]. Analysis of these effects will improve animal welfare and help reduce the economic losses associated with transportation [7].
Cattle have to potentially stand for long periods of time during transportation, thus muscles are constantly in tension [8]. Cattle transportation in the United States usually takes many hours, since the distance between cow-calf operations or stocker operations and feedlots is considerable [9]. One of the negative effects of long-term transportation is muscle fatigue, which can be defined as the reversible decrease of force generation by a muscle after its intense and repetitive use. In other words, muscle fatigue manifests as an inability to continue a motor task at the required intensity, eventually leading to exhaustion [10].
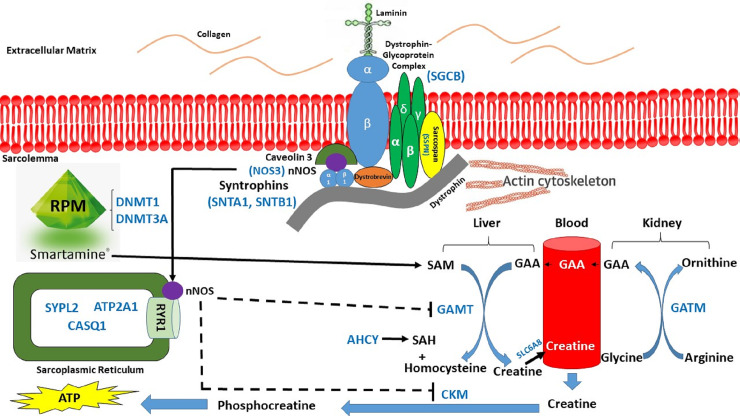
Our hypothesis was that rumen-protected methionine (RPM) supplementation during a preconditioning period of 45 days prior to transportation could provide additional methyl groups required for the activation of the creatine precursor called S-Adenosyl methionine (SAM), which will generate additional energy (ATP) required to mitigate muscle fatigue [11]. The proposed muscle fatigue gene network is as follows (Fig 1): RPM provided by Smartamine® will incorporate additional methyl groups required for the synthesis of creatine in the liver by guanidinoacetate N-methyltransferase (GAMT). Glycine amidinotransferase, mitochondrial (GATM) is involved in creatine biosynthesis, whereby it catalyzes the transfer of a guanido group from L-arginine to glycine, resulting in guanidinoacetic acid (GAA), the immediate precursor of creatine. In skeletal muscle, the dystroglycan complex works as a transmembrane linkage between the extracellular matrix and the cytoskeleton. α-dystroglycan is extracellular and binds to laminin in the basement membrane, while β-dystroglycan is a transmembrane protein and binds to dystrophin. Dystrophin attaches to intracellular actin cables. In this way, the dystroglycan complex, which links the extracellular matrix to the intracellular actin cables, is thought to provide structural integrity in muscle tissues. The neuronal form of nitric oxide (nNOS) inhibits GAMT and the enzyme creatine kinase present in muscle (CKM), responsible for the conversion of creatine to phosphocreatine which releases the energy (ATP) required to cope against muscle fatigue (Fig 1).
Fig 1. The proposed effect of administration of RPM on alleviation of muscle fatigue in beef cattle stressed due to long-duration transportation.
The objective of this study was to analyze the effect of administration of RPM on a proposed muscle fatigue gene network during pre- and post- exposure to a long-duration transportation simulation. Furthermore, we aimed to determine if RPM induces changes in expression of genes related to oxidative stress and DNA methylation.
Materials and methods
All the procedures for this study were conducted in accordance with a protocol approved by the Institutional Animal Care and Use Committee of Auburn University (IACUC Protocol #2017‐3129). Eighteen Angus × Simmental heifers were divided in two groups of 9 each, with the same average body weight (BW; 408 ± 64 kg) and age (375 ± 44 days old) per treatment. Animals belonged to the E. V. Smith Research Center Beef Unit located in Shorter, AL (36075), and they were relocated to the Beef Cattle Evaluation Center, Auburn University, Auburn, AL (36849) for the duration of the study. After a successful 21-day adaptation period to a Calan Gate System (Northwood, NH), both groups received two pounds per day of a diet consisting of soyhulls (50%) and corn gluten feed (50%) with bermudagrass hay ad libitum. Additionally, during the following 45 days, nine heifers received supplement containing 8 gr/hd/day as a fixed rate of top-dressed RPM (Smartamine M, Adisseo Inc., Antony, France), whereas the remaining nine heifers received supplement without the addition of RPM (CTRL).
After 45 days on treatment, both groups were loaded onto a 32’ × 7’ steel gooseneck trailer (Circle W trailers, McKenzie, AL 36456) at a stocking density of 1.15 m2/hd. Subsequently, animals were transported via pickup truck and trailer from the Beef Cattle Evaluation Center located at 405 Shug Jordan Parkway, Auburn, AL 36832 to the National Center for Asphalt Technology (NCAT) Test Track located at 1600 Lee Road 151, Opelika, AL 36804 (~18 miles apart). The transportation simulation took place on a 1.7-mile oval test track at NCAT, where animals remained loaded and transported for 22 hours, at a constant speed of 45 mph. A team of three truck drivers followed a modified version of the NCAT driver safety schedule which consisted of stops once per hour (5 minutes per stop) to rest, and every three hours for fuel (~10 minutes per fuel stop). Every time the truck stopped, visual evaluations of the animals were performed to check their well-being (i.e. panting, respiration rate, proportion of heifers resting or standing).
Animals were weighed at the beginning of the adaptation period to Calan Gate System (Day– 21), at the beginning of the study (Day 0), 2 days before transportation (Day 43, pre-transportation, PRET), and immediately after transportation (Day 46, post-transportation, POST).
The first skeletal muscle biopsy was performed at the beginning of the administration of RPM (Day 0 or baseline—BASE). The second biopsy was performed 2 days prior to transportation simulation (PRET), and a final biopsy was performed right after unloading the trailer following transportation simulation (POST). These biopsies were taken in order to assess muscle fatigue-related gene expression changes due to RPM supplementation throughout the preconditioning period, during and after transportation. Animals’ IDs were recorded so that the same animals were biopsied at each time point to provide repeated measures sampling.
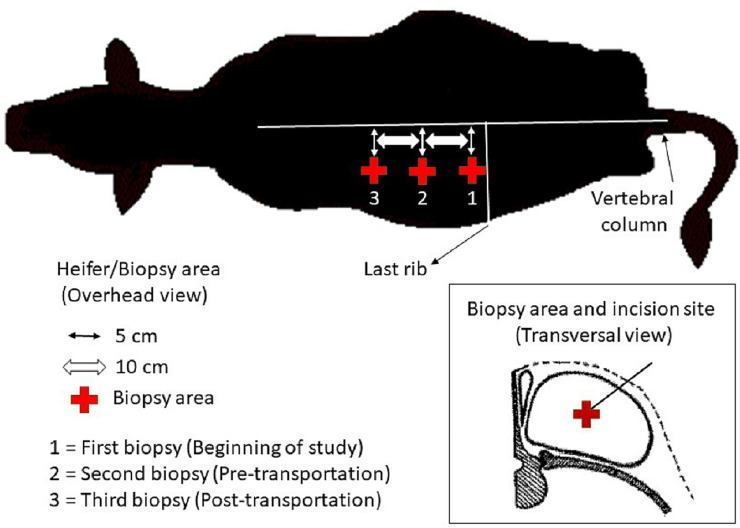
Skeletal muscle biopsies of Longissimus dorsi muscle were performed for epigenetic regulation (DNA methylation) and gene expression analysis using real-time quantitative PCR (RT-qPCR). Each muscle sample was obtained from the left side and moving up the side toward the head (longitudinally) with each sequential biopsy. Anesthesia injections of 5 mL of Lidocaine HCl 2% (VetOne®, Boise, ID) were placed before perform each biopsy incision. A muscle biopsy core (500–600 mg) was removed using a biopsy needle (12-gauge × 16 cm) inserted in a biopsy gun (Bard Magnum MG#1522, Tempe, AZ). The first biopsy area coincided with the Longissimus dorsi muscle at the level of the last rib. The second biopsy was performed 10 cm towards the head from the first biopsy area, and the final biopsy was 10 cm towards the head from the second biopsy area. The incision sites for all biopsies were located 5 cm down the vertebral column with the aim to take samples from the center of the Longissimus dorsi muscle (Fig 2). Ten mL blood samples were collected via jugular venipuncture at BASE, PRET and POST to determine hormonal and metabolic changes. Cortisol concentrations were measured at the Auburn University Endocrine Diagnostics Laboratory with the IMMULITE 2000 (Siemens Healthcare Diagnostics, Deerfield, IL, USA), which uses a solid-phase competitive enzyme-amplified chemiluminescent immunoassay. Creatine kinase and glucose analysis were performed at Auburn University Clinical Pathology Laboratory using methods previously described [12, 13].
Fig 2. Overhead and transversal view of skeletal muscle biopsy area on a beef heifer.
The protocols used for RNA extraction, primer design and testing, cDNA synthesis and RT-qPCR analysis were previously described [14]. RNA integrity number average was 7 ± 0.5. The following genes were selected (S1 Table): as internal controls: Mitochondrial Ribosome-Associated GTPase 1 (MTG1), Ribosomal Protein S15a (RPS15A) and, Ubiquitously Expressed Prefoldin Like Chaperone (UXT). Furthermore, for the dystrophin-glycoprotein complex, we selected Sarcoglycan Beta (SGCB), Syntrophin alpha 1 (SNTA1), Syntrophin beta 1 (SNTB1) and Sarcospan (SSPN). For genes present in the sarcoplasmic reticulum, we selected ATPase Sarcoplasmic/Endoplasmic Reticulum Ca++ Transporting 1 (ATP2A1), Calsequestrin (CASQ1) and Synaptophysin Like 2/MG29 (SYPL2). For the creatine synthesis pathway, we considered Creatine Kinase M-type (CKM), Guanidinoacetate N-methyltransferase (GAMT), Glycine amidinotransferase (GATM) and, Solute carrier family 6 member 8 (SLC6A8). For genes related to oxidative stress, we selected the following genes: Superoxide dismutase 1 (SOD1), Superoxide dismutase 2 (SOD2), Nitric Oxide synthase 3 (NOS3), Nuclear factor kappa B subunit 1 (NFKB1), NAD(P)H quinone dehydrogenase 1 (NQO1) and, PPARg coactivator alpha (PGC1a). Finally, for DNA methylation we selected DNA Methyltransferase 1 (DNMT1), DNA Methyltransferase 3 alpha (DNMT3A) and Adenosylhomocysteinase (AHCY). Further information is available in supporting tables (S2–S5 Tables).
Statistical analysis
Real time quantitative PCR data were analyzed using the MIXED procedure of SAS (SAS 9.4 Institute, Cary, NC, USA). Before statistical analysis, RT-qPCR data was normalized using quantities values of expression of each gene under study, divided by the geometric mean of 3 housekeeping genes (i.e., UXT, MTG1 and RPS15A). Then, normalized data was transformed to fold-change relative to day 0 (i.e. BASE). To estimate standard errors at day 0 and prevent biases in statistical analysis, normalized RT-qPCR data were transformed to obtain a perfect mean of 1.0 at day 0, leaving the proportional difference between the biological replicate. The same proportional change was calculated at all other time points to obtain a fold-change relative to day 0. Fixed effects in the statistical model for each variable analyzed (i.e. genes, blood metabolite) included treatment, time on experiment and treatment × time on experiment interactions when appropriate (e.g. mRNA expression over time). Gene expression data analysis included a repeated-measures statement with an autoregressive covariate structure. Animal performance and blood metabolite levels were also analyzed using the MIXED procedure of SAS, and treatment was the fixed effect in the statistical model. The random effect in all models was heifer within treatment. Average daily gain data was analyzed using PROC GLM procedure of SAS.
The statistical model used was: Yijl = μ + Ci + Tj + Sl + (C × T)ij + εijl; where, Yijl is the background-adjusted normalized fold change or blood data value; μ is the overall mean; Ci is the fixed effect of time (3 levels); Tj is the fixed effect of treatment (2 levels); Sl is the random effect of heifer nested within treatment; C × T is the interactions of time by treatment and εijl is the random error (0, σe2) associated with Yijl.
Results
Animal performance
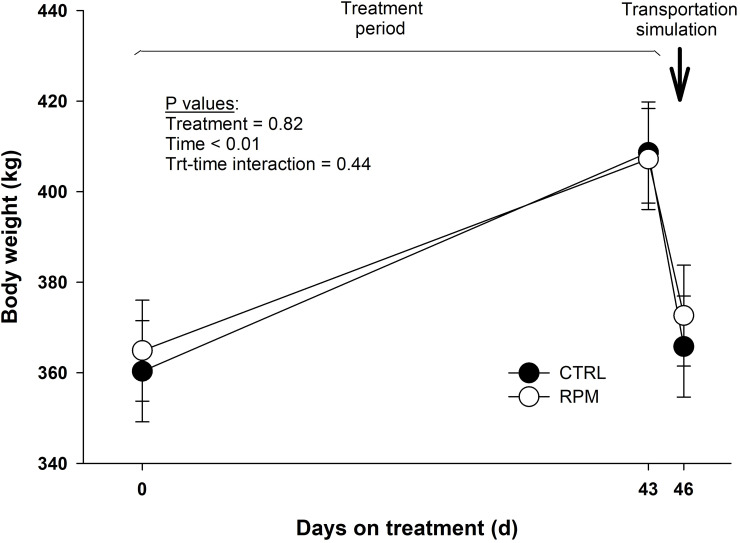
Body weight did not present a treatment × time interaction (P = 0.44), although it showed a decrease after transportation in CTRL and RPM heifers (P < 0.01) (Fig 3). Shrink loss was calculated by difference in body weight before and after transportation. Control heifers lost 43 kg (10% shrink), while RPM heifers lost 35 kg (8% shrink). The P values for shrink loss in kilograms and by percent were not significant (P = 0.29 and P = 0.28, respectively). Furthermore, average daily gain (ADG) did not differ between treatments (P = 0.41) during the preconditioning period (i.e., between BASE and PRET). Heifers in RPM group had an ADG of 0.98 kg/day, whereas heifers in CTRL an ADG of 1.12 kg/day(S1 Fig).
Fig 3. Body weight of beef heifers that received rumen-protected methionine (RPM) and heifers control (CTRL) that did not received RPM.
Day 0 (BASE), day 43 (PRET) and day 46 (POST). Statistical significant differences where declared at P < 0.05 and tendencies at P > 0.05 and < 0.1.
Serum cortisol, glucose, and creatine kinase
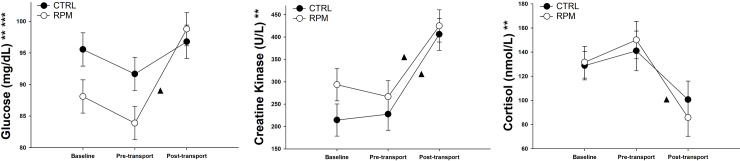
Glucose levels showed a treatment × time interaction (P < 0.05) and a time effect (P < 0.05). There was not a significant difference (P = 0.14) in glucose concentration in both treatments between BASE and PRET Heifers in RPM group had a significant increase (P < 0.05) in glucose levels between PRET (83.9 mg/dL) and POST (98.8 mg/dL); and heifers in CTRL did not show a significant difference between PRET (91.7 mg/dL), and POST (96.8 mg/dL) (Fig 4).
Fig 4. Serum glucose, creatine kinase and cortisol levels of beef heifers that received rumen-protected methionine (RPM) for 45 days before transportation and heifers control (CTRL) that did not received RPM.
Statistically significant differences were declared at P < 0.05 and tendencies at P > 0.05 and < 0.1.
Circulating creatine kinase levels did not show a treatment × time interaction (P = 0.16). However, creatine kinase concentration showed a time effect (P < 0.05). Both groups showed a significant increase between PRET and POST (Fig 4). Finally, serum cortisol concentrations did not present a treatment × time interaction (P = 0.68). However, there was an important reduction in cortisol levels after transportation in CTRL and RPM heifers (Fig 4).
Gene expression
Dystrophin-glycoprotein complex
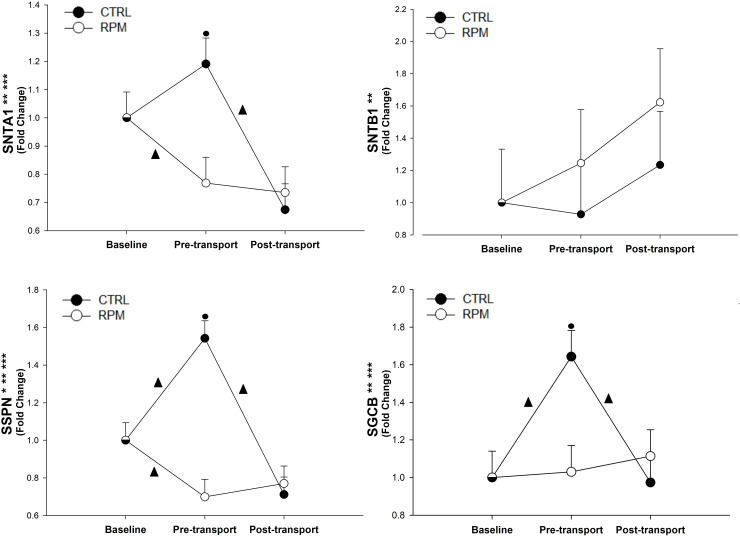
There was a treatment × time interaction (P < 0.05) and time effect (P < 0.05) for SGCB, SNTA1 and SSPN (Fig 5). The mRNA expression of SGCB in CTRL group was upregulated between BASE and PRET (P < 0.05), and then it was decreased between PRET and POST (P < 0.05). In RPM treatment, SNTA1 mRNA expression was downregulated between BASE and PRET (P < 0.05) and downregulated in CTRL between PRET and POST (P < 0.05). Expression of SSPN had a treatment × time interaction (P < 0.01), treatment effect (P < 0.05), and time effect (P < 0.01). Its expression was upregulated in CTRL between BASE and PRET (P < 0.05), and then downregulated between PRET and POST (P < 0.05). However, SSPN expression decreased in RPM between BASE and PRET (P < 0.05). The mRNA expression of SGCB, SSPN, and STNA1 in CTRL were greater than RPM (P < 0.05) at PRET. Finally, SNTB1 did not showed a significant treatment × time interaction (P = 0.34) (Fig 5).
Fig 5. Expression of genes related to the dystrophin-glycoprotein complex in Longissimus dorsi muscle of beef heifers that received rumen-protected methionine (RPM) for 45 days before transportation and heifers control (CTRL) that did not received RPM.
Statistically significant differences were declared at P < 0.05 and tendencies at P > 0.05 and < 0.1. Treatment × time interaction (***), time effect (**) and treatment effect (*). Tendencies are denoted if symbols (*, ** or ***) are underlined. Symbols (▲) on lines denote significant differences (P < 0.05) between two time points for the same treatment, symbols (●) denote significant differences (P < 0.05) between treatment at the same time point. Because the data presented in this chart are fold-change values, the baseline reference value is 1 for RPM and CTRL, therefore, there is overlap between symbols with error bars at baseline.
Sarcoplasmic reticulum
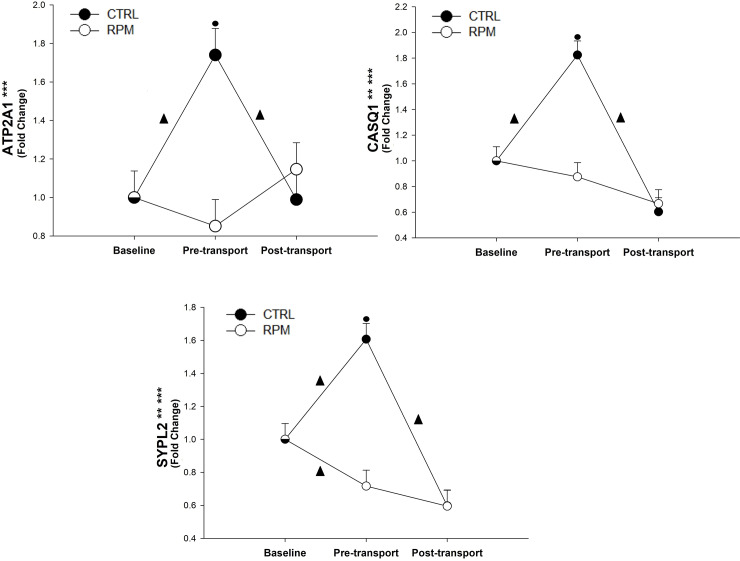
There was a treatment × time interaction (P < 0.01) and a time effect (P < 0.01) for both CASQ1 and SYPL2 (Fig 6). Furthermore, ATP2A1 showed only a treatment × time interaction (P < 0.01). In all cases, ATP2A1, CASQ1 and SYPL2 mRNA expression for CTRL was significantly greater (P < 0.05) than RPM at PRET. Furthermore, ATP2A1, CASQ1 and SYPL2 expressions for CTRL increased (P < 0.05) between BASE and PRET; however, their level of expression decreased (P < 0.05) between PRET and POST. In addition, SYPL2 had a significant downregulation (P < 0.05) between BASE and PRET in RPM heifers (Fig 6).
Fig 6. Expression of genes located at the sarcoplasmic reticulum in Longissimus dorsi muscle of beef heifers that received rumen-protected methionine (RPM) for 45 days before transportation and heifers control (CTRL) that did not received RPM.
Statistically significant differences were declared at P < 0.05 and tendencies at P > 0.05 and < 0.1. Treatment × time interaction (***), time effect (**) and treatment effect (*). Tendencies are denoted if symbols (*, ** or ***) are underlined. Symbols (▲) on lines denote significant differences (P < 0.05) between two time points for the same treatment, symbols (●) denote significant differences (P < 0.05) between treatment at the same time point. Because the data presented in this chart are fold-change values, the baseline reference value is 1 for RPM and CTRL, therefore, there is overlap between symbols with error bars at baseline.
Creatine synthesis
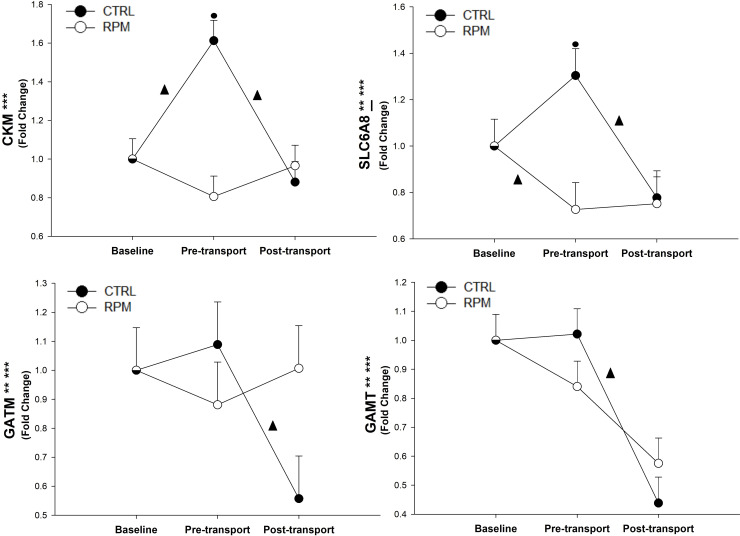
Expression of CKM showed a significant treatment × time interaction (P < 0.01). The mRNA expression of GAMT and GATM showed a treatment × time interaction (P < 0.05) and a time effect (P < 0.01). The mRNA expression of SLC6A8 showed a treatment × time interaction (P < 0.01) and a tendency for a time effect (P < 0.08). In addition, CKM and SLC6A8 mRNA expression on CTRL was greater (P < 0.05) than RPM at PRET. Finally, CKM expression was increased (P < 0.05) between BASE and PRET in CTRL, but its expression was decreased (P < 0.05) between PRET and POST. In CTRL animals, GATM and GAMT expressions were downregulated (P < 0.05) between PRET and POST. In RPM heifers, SLC6A8 expression was decreased (P < 0.05) and increased in CTRL heifers (P < 0.05) between BASE and PRET, while SLC6A8 decreased between PRET and POST for CTRL but did not change for RPM. Finally, SLC6A8 had a tendency for a time effect (P = 0.07) (Fig 7).
Fig 7. Expression of genes related to the creatine synthesis pathway in Longissimus dorsi muscle of beef heifers that received rumen-protected methionine (RPM) for 45 days before transportation and heifers control (CTRL) that did not received RPM.
Statistically significant differences were declared at P < 0.05 and tendencies at P > 0.05 and < 0.1. Treatment × time interaction (***), time effect (**) and treatment effect (*). Tendencies are denoted if symbols (*, ** or ***) are underlined. Symbols (▲) on lines denote significant differences (P < 0.05) between two time points for the same treatment, symbols (●) denote significant differences (P < 0.05) between treatment at the same time point. Because the data presented in this chart are fold-change values, the baseline reference value is 1 for RPM and CTRL, therefore, there is overlap between symbols with error bars at baseline.
DNA methylation
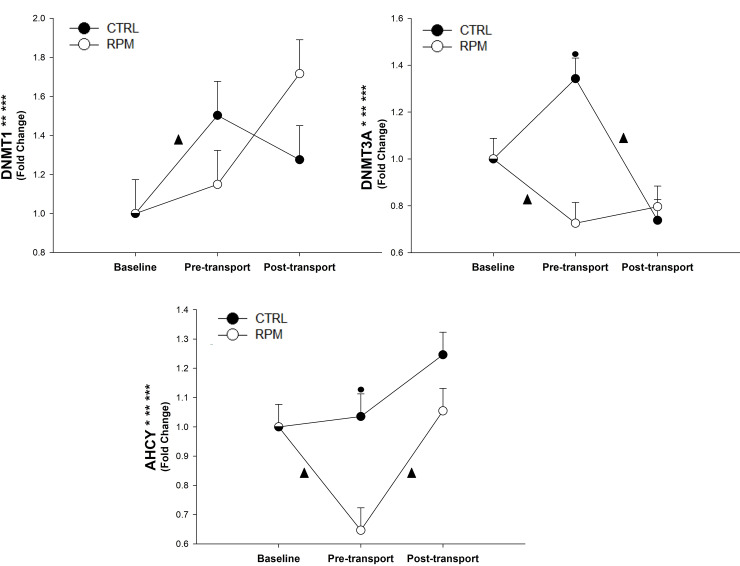
There was a treatment × time interaction (P = 0.05), and a time effect (P < 0.01) for DNMT1 mRNA expression. Furthermore, DNMT3A showed a treatment × time interaction (P < 0.01), a treatment effect (P = 0.05), and a time effect (P < 0.01). Furthermore, there was an increase of DNMT1 mRNA expression in RPM between PRET and POST. For DNMT3A, CTRL had a greater expression (P < 0.05) at PRET than RPM. Finally, DNMT3A mRNA expression decreased (P < 0.05) in RPM between BASE and PRET. Similarly, DNMT3A expression was downregulated (P < 0.05) in CTRL between PRET and POST.
There was a significant treatment × time interaction (P < 0.01), treatment effect (P < 0.05), and time effect (P < 0.01) for AHCY mRNA expression. In addition, AHCY mRNA expression in CTRL was greater (P < 0.05) than RPM at PRET. There was a significant downregulation of AHCY (P < 0.05) in RPM between BASE and PRET; however, it was significantly upregulated (P < 0.05) between PRET and POST (Fig 8).
Fig 8. Expression of genes related to DNA methylation in Longissimus dorsi muscle of beef heifers that received rumen-protected methionine (RPM) for 45 days before transportation and heifers control (CTRL) that did not received RPM.
Statistically significant differences were declared at P < 0.05 and tendencies at P > 0.05 and < 0.1. Treatment × time interaction (***), time effect (**) and treatment effect (*). Tendencies are denoted if symbols (*, ** or ***) are underlined. Symbols (▲) on lines denote significant differences (P < 0.05) between two time points for the same treatment, symbols (●) denote significant differences (P < 0.05) between treatment at the same time point. Because the data presented in this chart are fold-change values, the baseline reference value is 1 for RPM and CTRL, therefore, there is overlap between symbols with error bars at baseline.
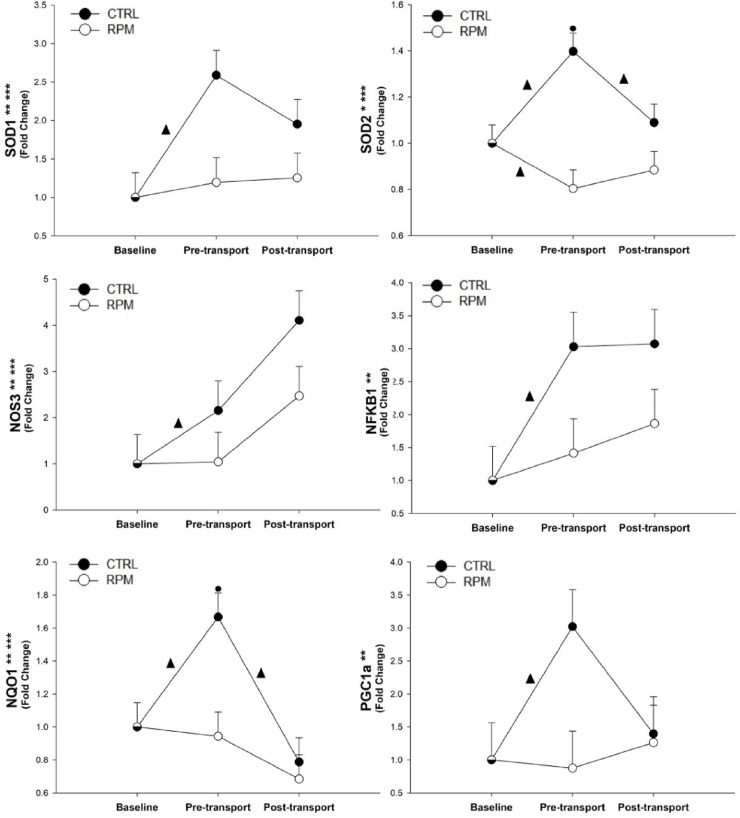
Oxidative stress
There was a significant treatment × time interaction (P < 0.05), and a time effect (P < 0.01), for SOD1, NQO1, and NOS3 mRNA expression. Furthermore, SOD2 showed a significant treatment × time interaction (P < 0.01), and treatment effect (P < 0.05). There was not a significant treatment × time interaction (P > 0.05) for PGC1α and NFKB1. At PRET, CTRL had a greater expression (P < 0.05) for SOD2, and NQO1 expressions in comparison to RPM. There was a significant increase (P < 0.05) in SOD1, NOS3, NFKB1, and PGC1α mRNA expressions in CTRL between BASE and PRET. In CTRL heifers, SOD2 mRNA expression presented upregulation (P < 0.05) between BASE and PRET; however, its expression was downregulated between PRET and POST. In the case of SOD2 expression in RPM, a significant decrease (P < 0.05) in expression was observed between BASE and PRET, which remained steady between PRET and POST. Finally, NQO1 mRNA expression increased significantly (P < 0.05) in CTRL between BASE and PRET; however, a significant decrease (P < 0.05) was observed between PRET and POST, while it remained steady for RPM group (Fig 9).
Fig 9. Expression of genes related to oxidative stress in Longissimus dorsi muscle of beef heifers that received rumen-protected methionine (RPM) for 45 days before transportation and heifers control (CTRL) that did not received RPM.
Statistically significant differences were declared at P < 0.05 and tendencies at P > 0.05 and < 0.1. Treatment × time interaction (***), time effect (**) and treatment effect (*). Tendencies are denoted if symbols (*, ** or ***) are underlined. Symbols (▲) on lines denote significant differences (P < 0.05) between two time point for the same treatment, symbols (●) denote significant differences (P < 0.05) between treatment at the same time point. Because the data presented in this chart are fold-change values, the baseline reference value is 1 for RPM and CTRL, therefore, there is overlap between symbols with error bars at baseline.
Discussion
Animal performance
With respect to animal performance, CTRL group showed no difference in ADG compared with RPM group (1.12 kg/d vs. 0.98 kg/d, respectively). There are no previous studies that report the performance of beef heifers under RPM supplementation; however, our results support Chen et al. (2011), who evaluated RPM supplementation (Smartamine®) in dairy cows, and reported no difference in ADG with a control group [15]. In addition, Torrentera et al. (2017) tested the performance of Holstein steers (127 ± 4.9 kg) under the inclusion of 0.032%, 0.064%, 0.096%, or 0.128% of RPM. They obtained superior results with RPM supplementation when Smartamine® was included at a level of 0.096% of dietary dry matter (DM) [16]. In our study, the RPM supplementation was 0.07% of DM; inclusion up to 0.09% might possibly have results that are more notable.
The most important parameter to assess, from the beef production standpoint, is the body weight loss or shrink associated with transportation. Numerous factors affect shrink during livestock transportation. Transportation duration and distance travelled are especially critical. In our study, animals were loaded on the trailer for 22 hours; however, because of the stops to rest, the net transportation time was 20 hours. Interestingly, Lambooy & Hulsegge (1988) studied the effect of 24 hours transportation in pregnant heifers, and reported body weight losses of 8% on average [17]. This result is similar to the shrink loss we observed for RPM heifers.
Environmental conditions during the transportation simulation day was retrieved from AWIS (Agricultural Weather Information Service, Inc.) at the Auburn Mesonet weather station (Auburn_CR10_92–18, Lee County) which is located 5.2 miles from NCAT. The average temperature was 5°C, relative humidity was 48.5%, and no rain was recorded during the 22 hours of transportation nor on the previous or subsequent days. According to Gonzalez et al. (2012), 20–25 hours transportation with an ambient temperature of ~10°C will produce a ~8% of BW losses on feeder cattle, and ~10% of BW losses on cull cattle [18]. Thus, the environmental conditions during our trial were not detrimental for body weight losses. However, RPM supplementation may affect shrink during warmer seasons in the Southeast.
Serum cortisol, glucose, and creatine kinase
Transportation is a stressful event that causes physiological and endocrine changes in animals’ metabolism. One indicator of these changes is cortisol, a glucocorticoid hormone released from the adrenal gland in response to stress [19]. Several studies have shown an increase in cortisol levels due to transportation in cattle, especially in the first hours of the trip [19, 20]. The release of blood cortisol increases when the transportation starts, reaching its highest point at ~ 4 hours, and then it starts to decline until reaching similar levels to the beginning of transportation. This level is maintained for the next 24 hours post-transportation [21]. However, these studies assessed the effect of shorter transportation time (i.e., 9 hours maximum), in comparison to a longer transportation period as in this study. Nevertheless, evidence of the reduction of blood cortisol level due to a long transportation could be associated to a familiarization of the animal with the environment inside the truck [22]. In total, our truck was stopped for 160 minutes during the 22-hour trip, either to rest or to refuel, and visual evaluations of the animals’ status took place during these stops. The small amount of time that the truck was stopped, could be beneficial for both, driver and animals. Frequent rest stops with loading and unloading may be more stressful than continuously being in the vehicle, consequently, this practice would likely be detrimental for the well-being of the animals [8].
Blood glucose level is used as a metabolic indicator of energy status in cattle. Our results suggest that glucose levels in blood during the preconditioning period are comparable to previous studies that reported 80.53 mg/dL of glucose in growing steers consuming a high forage diet [23]. Tarrant et al (1992) assessed the metabolic changes on Friesian steers after a 24-hour transportation. They reported PRET glucose levels of 89.2 mg/dL and POST glucose levels of 106.30 mg/dL, both of which are comparable to our results [24]. In addition, Earley et al (2010) reported blood glucose levels of growing Charolais bulls of 77.5 mg/dL at PRET, and 104.5 mg/dL immediately after a 24-hour transportation was complete [25]. However, the increase in glucose in RPM during transportation could be related to greater glycogenolysis, associated with the increase in catecholamine’s and glucocorticoids which were released after long-duration transportation stress [26].
Finally, creatine kinase (CK) in blood is commonly used in studies related to muscle fatigue for assessing the activity of the muscle [27, 28]. Even though, there are some differences in the serum CK levels in male and females [29], there is not a clear pattern. Previous studies reported increased CK activity due to transportation in cattle [24]. Tarrant et al (1992) reported 234 U/L of increase in CK after 24-hour transportation with a stocking density (1.13 m2/hd) similar to the one used in our study (1.15 m2/hd). However, we reported a lower increase in circulating CK during transportation for RPM and CTRL (P = 0.16), which may be related to the BW of the animals. Tarrant et al (1992) used Friesian bulls with average BW of 618 kg (Min, 537 kg; Max 900 kg) at pre-transportation; and in our study, heifers had an average BW of 408 kg (Min, 351 kg; Max 472 kg) at PRET. The lower BW of heifers may cause a decrease in CK units per liter of blood compared to steers with greater BW [24]. In addition, Earley et al (2010) reported an increase in circulating CK levels after 24-hours of transportation in Charolais bulls with BW of 367 ± 35 kg. In that study, before transportation, average CK level was 162.3 U/L, which is lower than in our study at PRET (CTRL, 227.4 U/L; RPM, 266.7 U/L). However, circulating CK after 24-hours transportation was 496 U/L, which is similar to heifers’ POST CK levels measured in this study (CTRL, 406 U/L; RPM, 424.8 U/L) [25]. In conclusion, beyond the rise in glucose and CK levels after transportation, no significant effect was observed due to the addition of RPM in heifers’ diet pre-transportation. Counter to our hypothesis, there was not a greater CK concentration, which could mitigate the effect of muscle fatigue due to greater amounts of creatine phosphate synthetized from RPM administration.
Gene expression
Dystrophin-glycoprotein complex
Dystrophin-glycoprotein complex (DGC) is a key element for the integrity of the sarcolemma, composed primarily of dystrophin and sarcoglycan complexes. The function of DGC is associated with the anchoring of sarcolemma to the actin cytoskeleton [30]. The DGC is formed by the dystroglycan complex, composed by dystrophin, the syntrophins, α- and β-dystroglycan; and sarcoglycan complex (SG), which is formed by four different transmembrane glycoproteins called α-, ß-, γ-, and δ -sarcoglycan [31]. Dystrophin, a cytoskeletal actin-binding protein, is one of the major proteins designed for the integrity of skeletal muscle fibers [32]. The dystroglycan complex is responsible for binding the basal lamina through α-dystroglycan and laminin [33] and attaches dystrophin at its C-terminal domain via β-dystroglycan [31]. At the same time, dystrophin N-terminal binds actin generating a link between the cytoskeleton and the extracellular matrix [34].
Sarcospan (SSPN), a transmembrane protein, is a DGC member [35] with a role in stabilizing the sarcolemma. Additionally, SSPN may serve as a chaperone in the DGC assembly and may help enhance localization of other adhesion complex proteins [36]. To the best of our knowledge, there is no literature reporting the effects of fatigue on SSPN expression. Therefore, it would be possible that CTRL group had more stabilization in the sarcolemma by SSPN upregulation between BASE and PRET. However, in a previous study, SSPN deficient mice present normal creatine kinase levels, sarcolemma integrity, and force generation aptitude [36].
The gene expression regulation of Syntrophin isoforms is an important factor for the sarcolemma integrity and binding signaling proteins. If one isoform is missed or deleted, another can replace it with no functional effects observed [37]. The absence α-Syntrophin 1 (SNTA1) gene in mutant mice did not show reduced force recovery in a fatigue stimulation trial [38]. Mice lacking α- and ß-syntrophins (SNTA1 and SNTB1, respectively) produce a decrease in skeletal muscle function [37]. It is possible to infer that sarcolemma integrity might not be affected by RPM supplementation nor fatigue due to transportation, since both SNTA1 and SNTB1 expression did not differ between treatments. Similarly, ß-sarcoglycan (SGCB), a gene that encodes for a transmembrane protein in the sarcoglycan complex, showed a similar pattern of expression in comparison to SNTA1 and SSPN, indicating its possible correlation with the whole Dystrophin-Glycoprotein complex. Because SSPN was the only gene that showed a treatment effect x time interaction in the Dystrophin-Glycoprotein complex, it would not be accurate to declare that the sarcolemma maintains its stability in CTRL group during transportation due to the unique upregulation of SSPN at PRET. Consequently, administration of RPM may not alter skeletal muscle membrane integrity in growing heifers. Further analysis of more genes related to Dystrophin-Glycoprotein complex and their phenotypic effects will help to fully understand the mechanism behind changes on skeletal muscle membrane’s integrity under this stressful condition.
Sarcoplasmic reticulum
The regulation of Ca2+ ions in muscle cells has been widely studied in different species; however, little is known about the effect of fatigue on beef cattle. In a previous study, a downregulation of CASQ1 protein expression in mice after an endurance trial [39] was reported. These results are consistent with our study for the CTRL group, because there was a downregulation of CASQ1 between PRET and POST. Similarly, reduction in activity of ATP2A1 in fatigued skeletal muscle was reported in rats [40] and humans [41].Even though, we did not measure Ca2+ levels in skeletal muscle, the downregulation of CASQ1, ATP2A1, and SYPL2 in CTRL during transportation may be related to Ca2+ depletion after a repetitive use of muscle [42]. These responses were not observed in heifers that received RPM between BASE and PRET. It has been shown that, after a methylation process of sarcolemma throughout SAM as a methyl donor agent, the Na+-Ca+ exchange is inhibited in the cardiac muscle. In skeletal muscle, phospholipid methyltransferase activity is highly localized in sarcoplasmic reticulum and present to a lesser extent in sarcolemma. Methylation of phosphatidylethanolamine modulates sarcoplasmic reticulum phospholipid composition, which in turn alters the energetic efficiency of SERCA (Ca2+ uptake / ATP hydrolysis) ion pump, decreasing the skeletal muscle metabolic rate [43–45]. Our results can be explained by a possible inhibition of Ca2+ uptake under an excess of methylation in the sarcoplasmic reticulum of RPM.
The administration of RPM downregulated only one of the genes that we selected to study the sarcoplasmic reticulum complex, SYPL2. It is possible that the methyl groups provided by RPM inhibited SYPL2 expression. Nagaraj et al (2000) reported that wild-type mice and mice lacking MG29, the protein that SYPL2 encodes, exhibited lower force generation after fatigue in the diaphragm, a skeletal mixed-fiber muscle like the Longissimus Dorsi [46]. In addition, mice lacking SYPL2 did not show decreased force generation after fatigue compared with wild-type mice, indicating the effect of MG29 can be compensated in a muscle fatigue scenario for that species. In RPM, ATP2A1 and CASQ1 remained unchanged among time points, suggesting that the greater availability of methyl groups does not modify Ca2+ regulation in skeletal muscle or inhibits the expression of genes related to sarcoplasmic reticulum. Skeletal muscle fatigue impairs Ca2+ uptake as previously described [47]. Down-regulation in the expression of all the genes analyzed related to sarcoplasmic reticulum between PRET and POST in CTRL may suggest that Ca2+ metabolism could be impaired under fatigue conditions. In contrast, RPM did not show a significant difference between PRET and POST. Thus, the stabilization of the expression of genes related to sarcoplasmic reticulum could potentially explain a better Ca2+ uptake by the skeletal muscle [47, 48].
Creatine synthesis pathway
To the best of our knowledge, there are no published nutrigenomics studies of muscle fatigue during transportation stress in cattle, and because of this, we utilized information from other speciesCreatine biosynthesis occurs mainly in the kidney, pancreas and liver, and involves two steps [49]. First, glycine amidinotransferase (GATM or AGAT) catalyzes the transamidination of guanidine group from arginine to glycine, producing ornithine and guanidinoacetate (GAA) [50]. Second, guanidinoacetate methyl transferase (GAMT) catalyzes the addition of a methyl group from S-adenosyl methionine (SAM) to GAA, producing creatine and S-adenosyl homocysteine (SAH) [51]. Once creatine is formed, it reaches different tissues via its transporter called solute carrier family 6 member 8 (SLC6A8) [52].
In our study, the difference in GATM expression can be explained by the maintenance of the expression in RPM at POST, in comparison to CTRL, which was downregulated. In mice, GATM upregulation in skeletal muscle does not compensate deficiency in SLC6A8 [53]. Thus, it is possible that the GATM expression in CTRL does not make up for decreased expression of CKM and SLC6A8 at POST.
In addition, GAMT deficiency has a negative effect in force maintenance in both, high-intensity electrical stimulation, and lower electrical stimulation frequency in mice [54]. The downregulation of GAMT expression in CTRL between PRET and POST would explain a decrease in creatine synthesis due to muscle fatigue since CKM expression was downregulated in CTRL for the same period of time.
In skeletal muscle of Thoroughbreds horses, no difference in CKM expression was found under moderate and high intensity exercise on a treadmill [55]. Samples were taken before the exercise, immediately after exercise, and 4 hours later. It has been shown that muscle force generation vary depending on the activity of the muscle. For example, muscle generates larger forces at moments of high speeds (i.e. running) compared with lower speed of muscle use (i.e. walking) [56]. Consequently, the velocity of muscle contraction has an important influence in energy use. For example, energy expenditure during an active movement (i.e. walking) is greater compared with less motion activities (i.e. standing) [57]. In our study, animals were not doing an intense exercise but they were certainly contracting theirs muscles in order to maintain equilibrium inside the truck for a long period of time, it is possible to compare these results with ours. However, even though RPM treatment remained stable among time points for CKM expression, it is not possible to explain the upregulation in CTRL heifers between BASE and PRET.
Interestingly, SLC6A8 shows the same pattern as CKM expression for both treatments among all time points. Since SLC6A8 encodes for the major creatine transporter, the decrease of creatine levels produces an inhibition of SLC6A8 expression, similarly to results reported in previous studies with creatine transporter deficiency in mice [52].
Oxidative stress
The generation of free radical elements such as Nitric Oxide (NO-) and superoxide (O2-) is a result of normal cellular metabolism [58]. Superoxide suffers reductions generating reactive oxygen species (ROS) [i.e., O2-, hydrogen peroxide (H2O2) and, hydroxyl radicals (OH-)] during resting conditions. Nitric oxide can be inactivated by coupling with O2-, producing the oxidant proxynitrite (ONOO-) as a result [59]. Nitric oxide creates reactive nitrogen species (RNS) like peroxynitrous acid (HNO3-), ONOO- and other nitrogen-derived oxidants [60]. These products are released in low concentration during resting, and they are necessary for normal cellular function [59]. For example, these compounds are involved in muscle regeneration, acting with other elements like growth factors and chemokines. Additionally, ROS promote mitochondriogenesis during exercise, which generates higher ATP levels for cell utilization [61]. However, skeletal muscle generates oxidants at high rates during muscle contraction, and they either became blockers of myogenic differentiation, or cause injuries by oxidative damage [62]. A single bout of exhaustive exercise leads to strong increases in ROS, which cannot be buffered by endogenous antioxidants, particularly in untrained individuals [63]. Therefore, high levels of ROS results in contractile dysfunction and fatigue [64]. In contrast, in our study, a prolonged period of water deprivation and utilization of energy reserves, which occur during long-term transportation of livestock, ultimately result in dehydration and a switch to a heightened gluconeogenic state while sparing muscle protein degradation [65]. The generated state of energy deprivation produces an increase in oxidative stress [66].
Exercise and fatigue increase muscle ONOO- and O2- producing an excess of oxidants in the cellular milieu. Nevertheless, cellular defense mechanisms against oxidative damage can effectively degrade these products [67]. Superoxide dismutases (SODs) are oxidoreductases generated by cells as a protection against oxidants. They are capable of spontaneous dismutation of O2- into H2O2. Consequently, they also help reducing the production of ONOO-.
Mammals present three isoforms of SOD, and they are differentiated by their composition and site of action. The cytosolic SOD1 (Cu/ZnSOD) is the main intracellular dismutase, which has enzymatic activity dependent on the presence of copper and zinc [68]. The mitochondrial SOD2 (MnSOD) has enzymatic activity dependent on the presence of manganese. Manganese plays a catalytic role of the dismutation of O2- to H2O2, similar to SOD1 [59]. The major site of O2- generation is the mitochondrial respiratory chain; consequently, SOD2 is a very important regulator of ROS equilibrium in the cell [69]. Finally, an extracellular dismutase called SOD3 (ecSOD) catalyzes ROS in the extracellular matrix. It is mostly expressed in internal organs and blood vessels. Skeletal muscle does not present high expression of SOD3, thus, it was not subject to analysis in our study [59, 70].
In our study, expression of SOD1 and SOD2 was significantly activated in CTRL heifers during the pre-conditioning period compared to RPM heifers. The lack of response in RPM heifers could be interpreted as a better controlled oxidant—antioxidant balance within skeletal muscle. We base our hypothesis on the observation that regular exercise appears to gradually increase the level of adaptation by the repeated activation of antioxidant genes and proteins [71]. We believe that after a continuous contraction of muscles during the 22 hours on the trailer, RPM heifers responded better than CTRL heifers in the adaption to a stressful situation and, they were able to reach homeostasis in their ROS production by the end of the study. Thus, activation of SOD1 and SOD2 was not in demand.
As reported in our study, BW losses of 8% in RPM, and 10% in CTRL were caused by the transportation, and up to 50% of that BW loss may be tissue partitioning [18]. During the 45-day preconditioning period with RPM supplementation, most ROS-related genes had an upregulation pattern in CTRL heifers. Methionine is a precursor of S-adenosylmethionine and hydrogen sulfide, which alleviate oxidant stress and protect the tissue from the damage [72]. Therefore, RPM supplementation might prevent an oxidative stress response in the RPM group before and after transportation contrary to CTRL, which showed an upregulation in ROS-related genes during the same period of time. The upregulation in CTRL compared with RPM before transportation may be related to a lower antioxidant capacity due to a minor methionine residues content in skeletal muscle cells. Methionine residues are known as ROS scavengers [73]. The greater expression of superoxide agents in CTRL as compared to RPM may be related to the oxidative stress imbalance due to tissue partitioning in heifers’ adipose and skeletal muscle tissues, due to the long-duration transportation without access to water and feed [74, 75].Furthermore, the occurrence of oxidative stress due to starvation has been reported in different species [76–78]. Fasting rats had a higher hepatic oxidative stress imbalance due to different processes of tissue degradation such as lipid peroxidation, protein misfolding, and glycolysis [79]. In addition, ROS generation increased in skeletal muscle after 24 hours of fasting in mice [80].
Metabolic stress inherent to endurance exercise has been demonstrated to stimulate mitochondrial biogenesis [81]. Mitochondrial synthesis is stimulated by the PGC-1α—NRF1—TFAM pathway [63]. Therefore, PGC-1α is the first stimulator of mitochondrial biogenesis [82], and its expression increases with exercise training [83]. PGC-1α is a key player in the adaptation of muscle cells to exercise [63]. Oxidative stress increases expression of PGC-1α [84], and PGC-1α controls cellular antioxidant homeostasis by stimulating SOD2, catalase, glutathione peroxidase 1 (GPx1), and uncoupling protein (UCP) [85]. In our study, neither administration of RPM nor transportation stress affected PGC-1α expression, although the peak in PGC-1α expression at PRET in CTRL heifers leads us to suggest that RPM could be producing an inhibition in the expression of this gene. Furthermore, the presence of additional methionine could be producing a more balanced environment in the RPM heifer’s skeletal muscle, where the activation of the oxidative stress-related gene network was not required. In contrast to our results, PGC-1α is upregulated in liver of fasting mice [86]. However, PGC-1α stabilization in RPM during transportation could mean that tissue partitioning caused by fasting metabolism was not related to the expression of this gen. Furthermore, the effect of mice lacking PGC-1α on substrate utilization from fed to fasted state was not relevant [87]. This could explain the lack of significant differences in PGC-1α.
The generation of nitric oxide (NO) in endothelial cells is produced by nitric oxide synthetase Type III or eNOS, which is encoded by NOS3. The presence of O2- can stimulate the development of fatigue. Thus, the inactivation of O2- by NO may be protective for the muscle cell [88]. Our results showed that the addition of RPM did not modify the expression of NOS3, suggesting that O2- remained unchanged among time points and an upregulation of the gene expression might not have been required. There could be a possible relationship between mithocondrogenesis, for PGC-1α and NOS3. Nisoli et al (2014) supports our results, showing that eNOS -/- muscle cells have lower mitochondriogenesis activity, and lower expression of PGC-1α [89].
Finally, NQO1 is a member of an antioxidant defense system. An acute bout of exercise at sufficient intensity activates this antioxidant enzyme, among others [90]. In contrast, in patients with chronic fatigue syndrome, NQO1 was downregulated [91]. Similar to other antioxidant genes assessed in this study, NQO1 expression remained unchanged among time points in RPM heifers. The higher concentration of available methyl groups provided by the addition of RPM could decrease ROS production. Consequently, the activation of NQO1 would not be necessary for the muscle cells after the transportation simulation was performed.
DNA methylation
DNA methylation is the major regulator of epigenetics in the genome of mammalians by the covalent addition of a methyl group (-CH3) to cytosine in combination of CpG dinucleotide called “CpG” island [92]. Enzymes capable of adding methyl groups to the hemimethylated and unmethylated DNA are called DNA methyltransferase (DNMT), like DNMT1 and DNMT3A, respectively [93]. Thus, DNMT1 and DNMT3A gene expression in skeletal muscle would potentially explain epigenetic changes due to muscle fatigue. DNMT3A presents a catalytic activity of de novo DNA methyltransferases while, DNMT1 works maintaining methylation of the CpG sites [94]. Furthermore, a cooperation between DNMT1 and DNMT3B was observed in humans; when disrupted, DNA methylation was decrease by 95% in colon rectal cancer cell [95].
The addition of RPM to the diet increases methionine availability [96]. Consequently, it can be transferred to S-adenosylmethionine (SAM), which is a molecule considered as the universal methyl donor [97]. Similar to our results, Osorio et al. (2014) did not obtain a significant difference in hepatic DNMT1 expression in periparturient dairy cows under Smartamine® supplementation. Furthermore, in contrast to our results, hepatic DNMT3A showed a greater expression after 11 days under Smartamine® supplementation during the pre-partum period [98]. However, the difference was mitigated after calving, and no difference was obtained from 11 to 31 days of supplementation. In addition, there was a similar pattern of expression of DNMT1 and DNMT3A in CTRL across time points, similar to results from Rhee et al (2002). Although, this pattern was not observed in RPM [95].
The AHCY gene provides instructions for producing the enzyme S-adenosylhomocysteine hydrolase, an important inhibitor of the majority of the methytransferases, affecting DNA methylation, RNA, and protein methylation in humans [99]. Specifically, S-adenosylhomocysteine hydrolase controls the chemical reaction that converts S-adenosylhomocysteine to adenosine and homocysteine [100]. This chemical reaction also plays an important role in regulating the addition of methyl groups to other compounds (i.e., methylation) [101]. Therefore, the principal role of AHCY in metabolism is to hydrolyse and efficiently remove S-adenosylhomocysteine, the by-product of transmethylation reactions and one of the most potent methyltransferase inhibitors [102]. S-adenosylhomocysteine hydrolase (AHCY) is the only mammalian enzyme able to hydrolyze S-adenosyl-l-homocysteine, and it binds to DNMT1 during DNA replication [101]. Hypermethylation of the genome can occur due to DNMT1 overexpression caused by AHCY regulation in vitro [101]. Furthermore, in isolated guinea pig heart muscles, adrenaline secretion inhibits S-adenosylhomocysteine hydrolase activity by a calcium mediated mechanism [103].
Stress can also directly influence the transcriptional regulation at the epigenetic level. In a previous study, prenatal transportation stress in Brahman bull calves was assessed at the methylome level. Results showed at least 10% more methylation in stressed calves as compared to controls [104]. In our study, RPM produce the inhibition of AHCY during preconditioning. However, under the stress of transportation, AHCY had activation in RPM heifers, coinciding with DNMT1 upregulation. Furthermore, in a previous study, the transcriptional repression of DNMT1 by glucocorticoid exposure is considered as a proxy for stress response [105]. Hypermethylation plays a role in controlling the physiological response to stressors by regulating the release of glucocorticoids in response to challenges [106]. Our results indicate that preconditioning with RPM supplementation might produce an activation of the process of DNA methylation during transportation stress. The animal’s benefit due to this metabolic response (i.e., hypermethylation) to long-term transportation stress still needs to be elucidated.
Conclusion
There was not a significant reduction in shrink with RPM; however, the utilization of more animals in further studies, could make this difference significant. In addition, serum glucose, creatine kinase and cortisol levels were not affected by the administration of RPM, suggesting that the catalytic processes generated due to high stress level could not be diminished by a greater methionine availability.
Contrary to our hypothesis, rumen-protected methionine during a preconditioning period of 45 days does not have an effect in CKM neither genes related to creatine synthesis in beef cattle transported for a period of 22 hours. Expression of genes related to muscle integrity and fatigue, creatine synthesis, sarcoplasmic reticulum and Dystrophin-glycoprotein complex were not affected by the administration of RPM in beef heifer’s skeletal muscle. However, our results indicate that administration of 8 gr/hd/ day of RPM could promote a better response to transportation stress in heifer’s skeletal muscle due to a possibly more stable redox balance after long-duration transportation. Perhaps, evaluating the Semitendinosus muscle, which might suffer more tension during transportation, would make these results more clear. However, we wanted to focus on the ribeye area (i.e., Longissimus dorsi) where the most expensive beef cuts are located. In conclusion, preconditioning growing Angus × Simmental heifers with RPM pre-transportation may have an effect on oxidative stress (SOD2), DNA methylation (DNMT3A) and hypermethylation (AHCY).
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Best hits using BLASTN (http://www.ncbi.nlm.nih.gov) are shown.
(DOCX)
(DOCX)
Acknowledgments
The authors would like to thank to Buzz Powell, Assistant Director & Test Track Manager, Michael Heitzman, Assistant Director, Jennifer Dukes, Pavement Test Track, drivers and employees from the National Center of Asphalt Technology (NCAT) for their invaluable contributions to this research study. Thanks also to Daniel Luchini from Adisseo who collaborated donating the rumen-protected methionine (Smartamine®) for this research project. Finally, the authors would like to thank Robert Britton and George Richburg for their extraordinary collaboration during the course of this study.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was funded by the USDA National Institute of Food and Agriculture, Hatch program - Project No. ALA013-1-17015. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Fike K, Spire MF. Transportation of cattle. Vet Clin North Am Food Anim Pract. 2006. July;22(2):305–20. 10.1016/j.cvfa.2006.03.012 [DOI] [PubMed] [Google Scholar]
- 2.Tarrant PV. Transportation of cattle by road. Applied Animal Behaviour Science 1990;28:153–70. [Google Scholar]
- 3.Earley B, Murray M, Prendiville DJ, Pintado B, Borque C, Canali E. The effect of transport by road and sea on physiology, immunity and behaviour of beef cattle. Res Vet Sci. 2012. June;92(3):531–41. 10.1016/j.rvsc.2011.04.002 [DOI] [PubMed] [Google Scholar]
- 4.Hagenmaier JA, Reinhardt CD, Bartle SJ, Henningson JN, Ritter MJ, Calvo-Lorenzo MS, et al. Effect of handling intensity at the time of transport for slaughter on physiological response and carcass characteristics in beef cattle fed ractopamine hydrochloride. J Anim Sci. 2017. May;95(5):1963–76. 10.2527/jas.2016.0821 [DOI] [PubMed] [Google Scholar]
- 5.Holman DB, Timsit E, Amat S, Abbott DW, Buret AG, Alexander TW. The nasopharyngeal microbiota of beef cattle before and after transport to a feedlot. BMC Microbiol. 2017. March 22;17(1):70 10.1186/s12866-017-0978-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng L, He C, Zhou Y, Xu L, Xiong H. Ground transport stress affects bacteria in the rumen of beef cattle: A real-time PCR analysis. Anim Sci J. 2017. May;88(5):790–7. 10.1111/asj.12615 [DOI] [PubMed] [Google Scholar]
- 7.Losada-Espinosa N, Villarroel M, Maria GA, Miranda-de la Lama GC. Pre-slaughter cattle welfare indicators for use in commercial abattoirs with voluntary monitoring systems: A systematic review. Meat Sci. 2018. April;138:34–48. 10.1016/j.meatsci.2017.12.004 [DOI] [PubMed] [Google Scholar]
- 8.Grandin T. Assessment of stress during handling and transport. J Anim Sci. 1997. January;75(1):249–57. 10.2527/1997.751249x [DOI] [PubMed] [Google Scholar]
- 9.Grandin T. Chapter 9: Cattle Transport. In: Grandin T, editor. Livestock Handling and Transport. 3rd ed Fort Collins, Colorado, U.S.A: CABI International; 2007. [Google Scholar]
- 10.Finsterer J, Mahjoub SZ. Fatigue in healthy and diseased individuals. Am J Hosp Palliat Care. 2014. August;31(5):562–75. 10.1177/1049909113494748 [DOI] [PubMed] [Google Scholar]
- 11.Ulrey CL, Liu L, Andrews LG, Tollefsbol TO. The impact of metabolism on DNA methylation. Hum Mol Genet. 2005. April 15;14 Spec No 1:R139–47. [DOI] [PubMed] [Google Scholar]
- 12.Oliver IT. A spectrophotometric method for the determination of creatine phosphokinase and myokinase. Biochem J. 1955. September;61(1):116–22. 10.1042/bj0610116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosalki SB. An improved procedure for serum creatine phosphokinase determination. J Lab Clin Med. 1967. April;69(4):696–705. [PubMed] [Google Scholar]
- 14.Novak TE, Rodriguez-Zas SL, Southey BR, Starkey JD, Stockler RM, Alfaro GF, et al. Jersey steer ruminal papillae histology and nutrigenomics with diet changes. J Anim Physiol Anim Nutr (Berl). 2019. November;103(6):1694–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen ZH, Broderick GA, Luchini ND, Sloan BK, Devillard E. Effect of feeding different sources of rumen-protected methionine on milk production and N-utilization in lactating dairy cows. J Dairy Sci. 2011. April;94(4):1978–88. 10.3168/jds.2010-3578 [DOI] [PubMed] [Google Scholar]
- 16.Torrentera N, Carrasco R, Salinas-Chavira J, Plascencia A, Zinn RA. Influence of methionine supplementation of growing diets enriched with lysine on feedlot performance and characteristics of digestion in Holstein steer calves. Asian-Australas J Anim Sci. 2017. January;30(1):42–50. 10.5713/ajas.16.0181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambooy E, Hulsegge B. Long-Distance Transport of Pregnant Heifers by Truck. Applied Animal Behaviour Science. 1988. August;20(3–4):249–58. [Google Scholar]
- 18.Gonzalez LA, Schwartzkopf-Genswein KS, Bryan M, Silasi R, Brown F. Factors affecting body weight loss during commercial long haul transport of cattle in North America. J Anim Sci. 2012. October;90(10):3630–9. 10.2527/jas.2011-4786 [DOI] [PubMed] [Google Scholar]
- 19.Ishizaki H, Kariya Y. Road transportation stress promptly increases bovine peripheral blood absolute NK cell counts and cortisol levels. J Vet Med Sci. 2010. June;72(6):747–53. 10.1292/jvms.09-0441 [DOI] [PubMed] [Google Scholar]
- 20.Buckham Sporer KR, Burton JL, Earley B, Crowe MA. Transportation stress in young bulls alters expression of neutrophil genes important for the regulation of apoptosis, tissue remodeling, margination, and anti-bacterial function. Vet Immunol Immunopathol. 2007. July 15;118(1–2):19–29. 10.1016/j.vetimm.2007.04.002 [DOI] [PubMed] [Google Scholar]
- 21.Sporer KR, Xiao L, Tempelman RJ, Burton JL, Earley B, Crowe MA. Transportation stress alters the circulating steroid environment and neutrophil gene expression in beef bulls. Vet Immunol Immunopathol. 2008. February 15;121(3–4):300–20. 10.1016/j.vetimm.2007.10.010 [DOI] [PubMed] [Google Scholar]
- 22.Van Engen NK, Stock ML, Engelken T, Vann RC, Wulf LW, Karriker LA, et al. Impact of oral meloxicam on circulating physiological biomarkers of stress and inflammation in beef steers after long-distance transportation. J Anim Sci. 2014. February;92(2):498–510. 10.2527/jas.2013-6857 [DOI] [PubMed] [Google Scholar]
- 23.Vasconcelos JT, Sawyer JE, Tedeschi LO, McCollum FT, Greene LW. Effects of different growing diets on performance, carcass characteristics, insulin sensitivity, and accretion of intramuscular and subcutaneous adipose tissue of feedlot cattle. J Anim Sci. 2009. April;87(4):1540–7. 10.2527/jas.2008-0934 [DOI] [PubMed] [Google Scholar]
- 24.Tarrant PV, Kenny FJ, Harrington D, Murphy M. Long-Distance Transportation of Steers to Slaughter—Effect of Stocking Density on Physiology, Behavior and Carcass Quality. Livest Prod Sci. 1992. February;30(3):223–38. [Google Scholar]
- 25.Earley B, Murray M, Prendiville DJ. Effect of road transport for up to 24 hours followed by twenty-four hour recovery on live weight and physiological responses of bulls. BMC Vet Res. 2010. July 20;6:38 10.1186/1746-6148-6-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Damtew A, Erega Y., Ebrahim H., Tsegaye S. and Msigie D. The Effect of long Distance Transportation Stress on Cattle: a Review. Biomed J Sci & Tech Res. 2018;3(3):3304–8. 10.1016/j.bj.2019.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edwards RH. Muscle fatigue and pain. Acta Med Scand Suppl. 1986;711:179–88. 10.1111/j.0954-6820.1986.tb08948.x [DOI] [PubMed] [Google Scholar]
- 28.Newham DJ, Jones DA, Edwards RH. Large delayed plasma creatine kinase changes after stepping exercise. Muscle Nerve. 1983. June;6(5):380–5. 10.1002/mus.880060507 [DOI] [PubMed] [Google Scholar]
- 29.Mpakama T, Chulayo AY, Muchenje V. Bruising in Slaughter Cattle and Its Relationship with Creatine Kinase Levels and Beef Quality as Affected by Animal Related Factors. Asian Austral J Anim. 2014. May;27(5):717–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michele DE, Campbell KP. Dystrophin-glycoprotein complex: post-translational processing and dystroglycan function. J Biol Chem. 2003. May 2;278(18):15457–60. 10.1074/jbc.R200031200 [DOI] [PubMed] [Google Scholar]
- 31.Gumerson JD, Michele DE. The Dystrophin-Glycoprotein Complex in the Prevention of Muscle Damage. J Biomed Biotechnol. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tidball JG, Wehling-Henricks M. The role of free radicals in the pathophysiology of muscular dystrophy. J Appl Physiol (1985). 2007. April;102(4):1677–86. [DOI] [PubMed] [Google Scholar]
- 33.Han R, Kanagawa M, Yoshida-Moriguchi T, Rader EP, Ng RA, Michele DE, et al. Basal lamina strengthens cell membrane integrity via the laminin G domain-binding motif of alpha-dystroglycan. Proc Natl Acad Sci U S A. 2009. August 4;106(31):12573–9. 10.1073/pnas.0906545106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stone MR, O'Neill A, Catino D, Bloch RJ. Specific interaction of the actin-binding domain of dystrophin with intermediate filaments containing keratin 19. Mol Biol Cell. 2005. September;16(9):4280–93. 10.1091/mbc.e05-02-0112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crosbie RH, Heighway J, Venzke DP, Lee JC, Campbell KP. Sarcospan, the 25-kDa transmembrane component of the dystrophin-glycoprotein complex. J Biol Chem. 1997. December 12;272(50):31221–4. 10.1074/jbc.272.50.31221 [DOI] [PubMed] [Google Scholar]
- 36.Lebakken CS, Venzke DP, Hrstka RF, Consolino CM, Faulkner JA, Williamson RA, et al. Sarcospan-deficient mice maintain normal muscle function. Mol Cell Biol. 2000. March;20(5):1669–77. 10.1128/mcb.20.5.1669-1677.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim MJ, Whitehead NP, Bible KL, Adams ME, Froehner SC. Mice lacking alpha-, beta 1-and beta 2-syntrophins exhibit diminished function and reduced dystrophin expression in both cardiac and skeletal muscle. Hum Mol Genet. 2019. February 1;28(3):386–95. 10.1093/hmg/ddy341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yokota T, Miyagoe-Suzuki Y, Ikemoto T, Matsuda R, Takeda S. alpha1-Syntrophin-deficient mice exhibit impaired muscle force recovery after osmotic shock. Muscle Nerve. 2014. May;49(5):728–35. 10.1002/mus.23990 [DOI] [PubMed] [Google Scholar]
- 39.Kinnunen S, Manttari S. Specific effects of endurance and sprint training on protein expression of calsequestrin and SERCA in mouse skeletal muscle. J Muscle Res Cell Motil. 2012. June;33(2):123–30. 10.1007/s10974-012-9290-0 [DOI] [PubMed] [Google Scholar]
- 40.Byrd SK, Bode AK, Klug GA. Effects of exercise of varying duration on sarcoplasmic reticulum function. J Appl Physiol (1985). 1989. March;66(3):1383–9. [DOI] [PubMed] [Google Scholar]
- 41.Li JL, Wang XN, Fraser SF, Carey MF, Wrigley TV, McKenna MJ. Effects of fatigue and training on sarcoplasmic reticulum Ca(2+) regulation in human skeletal muscle. J Appl Physiol (1985). 2002. March;92(3):912–22. [DOI] [PubMed] [Google Scholar]
- 42.Westerblad H, Lee JA, Lannergren J, Allen DG. Cellular mechanisms of fatigue in skeletal muscle. Am J Physiol. 1991. August;261(2 Pt 1):C195–209. 10.1152/ajpcell.1991.261.2.C195 [DOI] [PubMed] [Google Scholar]
- 43.Vemuri R, Philipson KD. Protein methylation inhibits Na+-Ca2+ exchange activity in cardiac sarcolemmal vesicles. Biochim Biophys Acta. 1988. April 22;939(3):503–8. 10.1016/0005-2736(88)90097-1 [DOI] [PubMed] [Google Scholar]
- 44.Panagia V, Elimban V, Ganguly PK, Dhalla NS. Decreased Ca2+-binding and Ca2+-ATPase activities in heart sarcolemma upon phospholipid methylation. Mol Cell Biochem. 1987;78(1):65–71. 10.1007/BF00224425 [DOI] [PubMed] [Google Scholar]
- 45.Verkerke ARP, Ferrara PJ, Lin CT, Johnson JM, Ryan TE, Maschek JA, et al. Phospholipid methylation regulates muscle metabolic rate through Ca(2+) transport efficiency. Nat Metab. 2019. September;1(9):876–85. 10.1038/s42255-019-0111-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagaraj RY, Nosek CM, Brotto MA, Nishi M, Takeshima H, Nosek TM, et al. Increased susceptibility to fatigue of slow- and fast-twitch muscles from mice lacking the MG29 gene. Physiol Genomics. 2000. November 9;4(1):43–9. 10.1152/physiolgenomics.2000.4.1.43 [DOI] [PubMed] [Google Scholar]
- 47.Tupling AR. The sarcoplasmic reticulum in muscle fatigue and disease: role of the sarco(endo)plasmic reticulum Ca2+-ATPase. Can J Appl Physiol. 2004. June;29(3):308–29. 10.1139/h04-021 [DOI] [PubMed] [Google Scholar]
- 48.Allen DG, Lamb GD, Westerblad H. Skeletal muscle fatigue: cellular mechanisms. Physiol Rev. 2008. January;88(1):287–332. 10.1152/physrev.00015.2007 [DOI] [PubMed] [Google Scholar]
- 49.Demant TW, Rhodes EC. Effects of creatine supplementation on exercise performance. Sports Med. 1999. July;28(1):49–60. 10.2165/00007256-199928010-00005 [DOI] [PubMed] [Google Scholar]
- 50.Choe CU, Nabuurs C, Stockebrand MC, Neu A, Nunes P, Morellini F, et al. L-arginine:glycine amidinotransferase deficiency protects from metabolic syndrome. Hum Mol Genet. 2013. January 1;22(1):110–23. 10.1093/hmg/dds407 [DOI] [PubMed] [Google Scholar]
- 51.Iqbal F, Hoeger H, Lubec G, Bodamer O. Biochemical and behavioral phenotype of AGAT and GAMT deficient mice following long-term Creatine monohydrate supplementation. Metab Brain Dis. 2017. December;32(6):1951–61. 10.1007/s11011-017-0092-3 [DOI] [PubMed] [Google Scholar]
- 52.Russell AP, Ghobrial L, Wright CR, Lamon S, Brown EL, Kon M, et al. Creatine transporter (SLC6A8) knockout mice display an increased capacity for in vitro creatine biosynthesis in skeletal muscle. Front Physiol. 2014;5:314 10.3389/fphys.2014.00314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stockebrand M, Sasani A, Das D, Hornig S, Hermans-Borgmeyer I, Lake HA, et al. A Mouse Model of Creatine Transporter Deficiency Reveals Impaired Motor Function and Muscle Energy Metabolism. Front Physiol. 2018;9:773 10.3389/fphys.2018.00773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kan HE, Buse-Pot TE, Peco R, Isbrandt D, Heerschap A, de Haan A. Lower force and impaired performance during high-intensity electrical stimulation in skeletal muscle of GAMT-deficient knockout mice. Am J Physiol Cell Physiol. 2005. July;289(1):C113–9. 10.1152/ajpcell.00040.2005 [DOI] [PubMed] [Google Scholar]
- 55.Gu J, MacHugh DE, McGivney BA, Park SD, Katz LM, Hill EW. Association of sequence variants in CKM (creatine kinase, muscle) and COX4I2 (cytochrome c oxidase, subunit 4, isoform 2) genes with racing performance in Thoroughbred horses. Equine Vet J Suppl. 2010. November(38):569–75. 10.1111/j.2042-3306.2010.00181.x [DOI] [PubMed] [Google Scholar]
- 56.Arnold EM, Hamner S. R., Seth A., Millard M., Delp S. L. How muscle fiber lengths and velocities affect muscle force generation as humans walk and run at different speeds. Journal of Experimental Biology 2013;216:2150–60. 10.1242/jeb.075697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Creasy SA, Rogers RJ, Byard TD, Kowalsky RJ, Jakicic JM. Energy Expenditure During Acute Periods of Sitting, Standing, and Walking. J Phys Act Health. 2016. June;13(6):573–8. 10.1123/jpah.2015-0419 [DOI] [PubMed] [Google Scholar]
- 58.Jackson MJ, Pye D, Palomero J. The production of reactive oxygen and nitrogen species by skeletal muscle. J Appl Physiol (1985). 2007. April;102(4):1664–70. [DOI] [PubMed] [Google Scholar]
- 59.Fukai T, Ushio-Fukai M. Superoxide dismutases: role in redox signaling, vascular function, and diseases. Antioxid Redox Signal. 2011. September 15;15(6):1583–606. 10.1089/ars.2011.3999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ferreira LF, Reid MB. Muscle-derived ROS and thiol regulation in muscle fatigue. J Appl Physiol (1985). 2008. March;104(3):853–60. [DOI] [PubMed] [Google Scholar]
- 61.Wright DC, Han DH, Garcia-Roves PM, Geiger PC, Jones TE, Holloszy JO. Exercise-induced mitochondrial biogenesis begins before the increase in muscle PGC-1 alpha expression. J Biol Chem. 2007. January 5;282(1):194–9. 10.1074/jbc.M606116200 [DOI] [PubMed] [Google Scholar]
- 62.Barbieri E, Sestili P. Reactive oxygen species in skeletal muscle signaling. J Signal Transduct. 2012;2012:982794 10.1155/2012/982794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Steinbacher P, Eckl P. Impact of oxidative stress on exercising skeletal muscle. Biomolecules. 2015. April 10;5(2):356–77. 10.3390/biom5020356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reid MB, Moylan JS. Beyond atrophy: redox mechanisms of muscle dysfunction in chronic inflammatory disease. J Physiol. 2011. May 1;589(Pt 9):2171–9. 10.1113/jphysiol.2010.203356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parker AJ, Dobson GP, Fitzpatrick LA. Physiological and metabolic effects of prophylactic treatment with the osmolytes glycerol and betaine on Bos indicus steers during long duration transportation. J Anim Sci. 2007. November;85(11):2916–23. 10.2527/jas.2006-193 [DOI] [PubMed] [Google Scholar]
- 66.Mracek T, Pecina P, Vojtiskova A, Kalous M, Sebesta O, Houstek J. Two components in pathogenic mechanism of mitochondrial ATPase deficiency: energy deprivation and ROS production. Exp Gerontol. 2006. July;41(7):683–7. 10.1016/j.exger.2006.02.009 [DOI] [PubMed] [Google Scholar]
- 67.Nikolaidis MG, Jamurtas AZ, Paschalis V, Fatouros IG, Koutedakis Y, Kouretas D. The effect of muscle-damaging exercise on blood and skeletal muscle oxidative stress: magnitude and time-course considerations. Sports Med. 2008;38(7):579–606. 10.2165/00007256-200838070-00005 [DOI] [PubMed] [Google Scholar]
- 68.Wu CY, Steffen J, Eide DJ. Cytosolic superoxide dismutase (SOD1) is critical for tolerating the oxidative stress of zinc deficiency in yeast. Plos One. 2009. September 16;4(9):e7061 10.1371/journal.pone.0007061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu Y, Rosen DG, Zhou Y, Feng L, Yang G, Liu J, et al. Mitochondrial manganese-superoxide dismutase expression in ovarian cancer: role in cell proliferation and response to oxidative stress. J Biol Chem. 2005. November 25;280(47):39485–92. 10.1074/jbc.M503296200 [DOI] [PubMed] [Google Scholar]
- 70.Folz RJ, Crapo JD. Extracellular superoxide dismutase (SOD3): tissue-specific expression, genomic characterization, and computer-assisted sequence analysis of the human EC SOD gene. Genomics. 1994. July 1;22(1):162–71. 10.1006/geno.1994.1357 [DOI] [PubMed] [Google Scholar]
- 71.Radak Z, Zhao Z, Koltai E, Ohno H, Atalay M. Oxygen consumption and usage during physical exercise: the balance between oxidative stress and ROS-dependent adaptive signaling. Antioxid Redox Signal. 2013. April 1;18(10):1208–46. 10.1089/ars.2011.4498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bin P, Huang R, Zhou X. Oxidation Resistance of the Sulfur Amino Acids: Methionine and Cysteine. Biomed Res Int. 2017;2017:9584932 10.1155/2017/9584932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Celi P, Gabai G. Oxidant/Antioxidant Balance in Animal Nutrition and Health: The Role of Protein Oxidation. Front Vet Sci. 2015;2:48 10.3389/fvets.2015.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Z, Liang M, Li H, Cai L, He H, Wu Q, et al. l-Methionine activates Nrf2-ARE pathway to induce endogenous antioxidant activity for depressing ROS-derived oxidative stress in growing rats. J Sci Food Agric. 2019. August 15;99(10):4849–62. 10.1002/jsfa.9757 [DOI] [PubMed] [Google Scholar]
- 75.Grisenti TG. Lactational Performance and Energy Partitioning of Dairy Cows Supplemented with N-Acetyl-L-Methionine During Mid to Late Lactation. Logan, Utah: Utah State University; 2017. [Google Scholar]
- 76.Morales AE, Perez-Jimenez A, Hidalgo MC, Abellan E, Cardenete G. Oxidative stress and antioxidant defenses after prolonged starvation in Dentex dentex liver. Comp Biochem Physiol C Toxicol Pharmacol. 2004. October;139(1–3):153–61. 10.1016/j.cca.2004.10.008 [DOI] [PubMed] [Google Scholar]
- 77.Schraag S, Mandach U, Schweer H, Beinder E. Metabolic changes, hypothalamo-pituitary-adrenal axis and oxidative stress after short-term starvation in healthy pregnant women. J Perinat Med. 2007;35(4):289–94. 10.1515/JPM.2007.076 [DOI] [PubMed] [Google Scholar]
- 78.Sorensen M, Sanz A, Gomez J, Pamplona R, Portero-Otin M, Gredilla R, et al. Effects of fasting on oxidative stress in rat liver mitochondria. Free Radic Res. 2006. April;40(4):339–47. 10.1080/10715760500250182 [DOI] [PubMed] [Google Scholar]
- 79.Wasselin T, Zahn S, Maho YL, Dorsselaer AV, Raclot T, Bertile F. Exacerbated oxidative stress in the fasting liver according to fuel partitioning. Proteomics. 2014. August;14(16):1905–21. 10.1002/pmic.201400051 [DOI] [PubMed] [Google Scholar]
- 80.Qi ZT, He Q, Ji L, Ding SZ. Antioxidant Supplement Inhibits Skeletal Muscle Constitutive Autophagy rather than Fasting-Induced Autophagy in Mice. Oxid Med Cell Longev. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Di Donato DM, West DW, Churchward-Venne TA, Breen L, Baker SK, Phillips SM. Influence of aerobic exercise intensity on myofibrillar and mitochondrial protein synthesis in young men during early and late postexercise recovery. Am J Physiol Endocrinol Metab. 2014. May 1;306(9):E1025–32. 10.1152/ajpendo.00487.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vina J, Gomez-Cabrera MC, Borras C, Froio T, Sanchis-Gomar F, Martinez-Bello VE, et al. Mitochondrial biogenesis in exercise and in ageing. Adv Drug Deliv Rev. 2009. November 30;61(14):1369–74. 10.1016/j.addr.2009.06.006 [DOI] [PubMed] [Google Scholar]
- 83.Mathai AS, Bonen A, Benton CR, Robinson DL, Graham TE. Rapid exercise-induced changes in PGC-1alpha mRNA and protein in human skeletal muscle. J Appl Physiol (1985). 2008. October;105(4):1098–105. [DOI] [PubMed] [Google Scholar]
- 84.Kang C, O'Moore KM, Dickman JR, Ji LL. Exercise activation of muscle peroxisome proliferator-activated receptor-gamma coactivator-1 alpha signaling is redox sensitive. Free Radical Bio Med. 2009. November 15;47(10):1394–400. [DOI] [PubMed] [Google Scholar]
- 85.Kang C, Li Ji L. Role of PGC-1alpha signaling in skeletal muscle health and disease. Ann N Y Acad Sci. 2012. October;1271:110–7. 10.1111/j.1749-6632.2012.06738.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yoon JC, Puigserver P, Chen GX, Donovan J, Wu ZD, Rhee J, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001. September 13;413(6852):131–8. 10.1038/35093050 [DOI] [PubMed] [Google Scholar]
- 87.Gudiksen A, Pilegaard H. PGC-1alpha and fasting-induced PDH regulation in mouse skeletal muscle. Physiol Rep. 2017. April;5(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kobzik L, Reid MB, Bredt DS, Stamler JS. Nitric oxide in skeletal muscle. Nature. 1994. December 8;372(6506):546–8. 10.1038/372546a0 [DOI] [PubMed] [Google Scholar]
- 89.Nisoli E, Falcone S, Tonello C, Cozzi V, Palomba L, Fiorani M, et al. Mitochondrial biogenesis by NO yields functionally active mitochondria in mammals. P Natl Acad Sci USA. 2004. November 23;101(47):16507–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ji LL. Oxidative stress during exercise: implication of antioxidant nutrients. Free Radic Biol Med. 1995. June;18(6):1079–86. 10.1016/0891-5849(94)00212-3 [DOI] [PubMed] [Google Scholar]
- 91.Pietrangelo T, Mancinelli R, Toniolo L, Montanari G, Vecchiet J, Fano G, et al. Transcription profile analysis of vastus lateralis muscle from patients with chronic fatigue syndrome. Int J Immunopathol Pharmacol. 2009. Jul-Sep;22(3):795–807. 10.1177/039463200902200326 [DOI] [PubMed] [Google Scholar]
- 92.Robertson KD, Jones PA. DNA methylation: past, present and future directions. Carcinogenesis. 2000. March;21(3):461–7. 10.1093/carcin/21.3.461 [DOI] [PubMed] [Google Scholar]
- 93.Fatemi M, Hermann A, Gowher H, Jeltsch A. Dnmt3a and Dnmt1 functionally cooperate during de novo methylation of DNA. Eur J Biochem. 2002. October;269(20):4981–4. 10.1046/j.1432-1033.2002.03198.x [DOI] [PubMed] [Google Scholar]
- 94.Tajima S, Suetake I, Takeshita K, Nakagawa A, Kimura H. Domain Structure of the Dnmt1, Dnmt3a, and Dnmt3b DNA Methyltransferases. Adv Exp Med Biol. 2016;945:63–86. 10.1007/978-3-319-43624-1_4 [DOI] [PubMed] [Google Scholar]
- 95.Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002. April 4;416(6880):552–6. 10.1038/416552a [DOI] [PubMed] [Google Scholar]
- 96.Graulet B, Richard C, Robert JC. Methionine availability in plasma of dairy cows supplemented with methionine hydroxy analog isopropyl ester. J Dairy Sci. 2005. October;88(10):3640–9. 10.3168/jds.S0022-0302(05)73049-6 [DOI] [PubMed] [Google Scholar]
- 97.Martinov MV, Vitvitsky VM, Banerjee R, Ataullakhanov FI. The logic of the hepatic methionine metabolic cycle. Biochim Biophys Acta. 2010. January;1804(1):89–96. 10.1016/j.bbapap.2009.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Osorio JS, Ji P, Drackley JK, Luchini D, Loor JJ. Smartamine M and MetaSmart supplementation during the peripartal period alter hepatic expression of gene networks in 1-carbon metabolism, inflammation, oxidative stress, and the growth hormone-insulin-like growth factor 1 axis pathways. J Dairy Sci. 2014. December;97(12):7451–64. 10.3168/jds.2014-8680 [DOI] [PubMed] [Google Scholar]
- 99.James SJ, Melnyk S, Pogribna M, Pogribny IP, Caudill MA. Elevation in S-adenosylhomocysteine and DNA hypomethylation: potential epigenetic mechanism for homocysteine-related pathology. J Nutr. 2002. August;132(8 Suppl):2361S–6S. 10.1093/jn/132.8.2361S [DOI] [PubMed] [Google Scholar]
- 100.Turner MA, Yang XD, Yin D, Kuczera K, Borchardt RT, Howell PL. Structure and function of S-adenosylhomocysteine hydrolase. Cell Biochem Biophys. 2000;33(2):101–25. 10.1385/CBB:33:2:101 [DOI] [PubMed] [Google Scholar]
- 101.Ponnaluri VKC, Esteve PO, Ruse CI, Pradhan S. S-adenosylhomocysteine Hydrolase Participates in DNA Methylation Inheritance. J Mol Biol. 2018. July 6;430(14):2051–65. 10.1016/j.jmb.2018.05.014 [DOI] [PubMed] [Google Scholar]
- 102.Motzek A, Knezevic J, Switzeny OJ, Cooper A, Baric I, Beluzic R, et al. Abnormal Hypermethylation at Imprinting Control Regions in Patients with S-Adenosylhomocysteine Hydrolase (AHCY) Deficiency. Plos One. 2016. March 14;11(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Suarez J, HernandezMunoz R, Sanchez L, deSanchez VC. Adrenaline inhibits S-adenosylhomocysteine hydrolase activity by a calcium mediated mechanism in isolated guinea pig heart muscles. Faseb J. 1996. March 8;10(3):2277-. [Google Scholar]
- 104.Littlejohn BP, Price DM, Neuendorff DA, Carroll JA, Vann RC, Riggs PK, et al. Prenatal transportation stress alters genome-wide DNA methylation in suckling Brahman bull calves. J Anim Sci. 2018. December 3;96(12):5075–99. 10.1093/jas/sky350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Klengel T, Binder EB. Epigenetics of Stress-Related Psychiatric Disorders and Gene x Environment Interactions. Neuron. 2015. June 17;86(6):1343–57. 10.1016/j.neuron.2015.05.036 [DOI] [PubMed] [Google Scholar]
- 106.Taff CC, Campagna L, Vitousek MN. Genome-wide variation in DNA methylation is associated with stress resilience and plumage brightness in a wild bird. Mol Ecol. 2019. August;28(16):3722–37. 10.1111/mec.15186 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Best hits using BLASTN (http://www.ncbi.nlm.nih.gov) are shown.
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.