Abstract
Background
Aberrant MET receptor tyrosine kinase (RTK) activation leads to invasive tumor growth in different types of cancer. Overexpression of MET and its ligand hepatocyte growth factor (HGF) occurs more frequently in glioblastoma (GBM) than in low-grade gliomas. Although we have shown previously that HGF-autocrine activation predicts sensitivity to MET tyrosine kinase inhibitors (TKIs) in GBM, whether it initiates tumorigenesis remains elusive.
Methods
Using a well-established Sleeping Beauty (SB) transposon strategy, we injected human HGF and MET cDNA together with a short hairpin siRNA against Trp53 (SB-hHgf.Met.ShP53) into the lateral ventricle of neonatal mice to induce spontaneous glioma initiation and characterized the tumors with H&E and immunohistochemistry analysis. Glioma sphere cells also were isolated for measuring the sensitivity to specific MET TKIs.
Results
Mixed injection of SB-hHgf.Met.ShP53 plasmids induced de novo glioma formation with invasive tumor growth accompanied by HGF and MET overexpression. While glioma stem cells (GSCs) are considered as the tumor-initiating cells in GBM, both SB-hHgf.Met.ShP53 tumor sections and glioma spheres harvested from these tumors expressed GSC markers nestin, GFAP, and Sox 2. Moreover, specific MET TKIs significantly inhibited tumor spheres’ proliferation and MET/MAPK/AKT signaling.
Conclusions
Overexpression of the HGF/MET axis along with p53 attenuation may transform neural stem cells into GSCs, resulting in GBM formation in mice. These tumors are primarily driven by the MET RTK pathway activation and are sensitive to MET TKIs. The SB-hHgf.Met.ShP53 spontaneous mouse glioma model provides a useful tool for studying GBM tumor biology and MET-targeting therapeutics.
Keywords: genetically engineered mouse model, glioblastoma, glioma initiating cells, HGF/MET signaling, targeted therapy
Key Points.
We demonstrate that HGF/MET overexpression along with Trp53 inhibition induces spontaneous glioma in mice.
We established an HGF/MET-driven glioma mouse model for studying GBM biology.
We show that HGF/MET-driven GBMs are sensitive to MET inhibitors.
Importance of the Study.
Elevated HGF/MET axis provokes RAS/RTK/PI3K pathway activation, the leading signaling alteration in primary GBM. MET overexpression is a marker of the “mesenchymal” subtype and also is required for GSC maintenance and its invasive repopulation. While HGF or MET transgenic mice were shown to initiate liver or breast cancers, a glioma phenotype has not been reported, raising the question of whether the HGF/MET axis may drive glioma initiation. Here, we show that overexpression of HGF and MET, along with Trp53 inhibition, induces spontaneous glioma in mice. Moreover, the GSCs derived from these tumors are sensitive to MET and MEK inhibitors. These results not only provide the evidence that HGF/MET elevation may transform NSCs into GSCs, resulting in GBM initiation and progression, but also suggest that HGF-autocrine GBMs are primarily driven by the MET RTK pathway that may be inhibited by MET or MEK inhibitors.
Glioblastoma (GBM), WHO grade IV glioma, is the most common primary brain tumor. It has a bleak prognosis because tumor cells invade into the normal tissue of the central nervous system (CNS) without a clear margin for surgical resection, resulting in early recurrence.1 Overall, GBMs account for 15% of all primary brain tumors with median survival time as short as 9–12 months, and the 5-year survival rate is only about 5%.2,3 Using in silico analysis, the Cancer Genome Atlas (TCGA) has established comprehensive genomic profiles that classified GBM into 4 molecular subtypes. While the “classical” signature represents a more proliferative phenotype, a “mesenchymal” signature is indicative of a more invasive phenotype; both are associated with poor prognosis and short survival time.4,5 Theoretically, within the heterogeneous GBM, a subset of poorly differentiated yet highly self-renewing glioma cells, termed glioma stem cells (GSCs), have invasive and tumorigenic properties and are responsible for therapeutic resistance.6,7 To date, GSCs have been isolated from primary GBMs or patient-derived xenograft (PDX) models to facilitate our understanding of GBM invasiveness and its vulnerability to chemo- and radiation therapy.
Aberrant MET pathway activation is one of the major mechanisms leading to RAS/RTK/PI3K pathway alteration, which occurred in 88% of primary GBM.8 Overexpression of MET and its ligand hepatocyte growth factor (HGF) occurs more frequently in primary GBM than in low-grade glioma and is associated with poor prognosis.9,10 Genomic characterization further shows that MET overexpression is a marker of the “mesenchymal” phenotype, the most invasive type of GBM with the shortest survival time.4,11 In addition to primary GBM, recent studies have reported significant enrichment of MET-exon-14-skipping (METex14), PTPRZ1-MET (ZM) fusion, and MET amplification in secondary GBM, among which the co-occurrence of METex14 and ZM reached 14% of cases and is associated with poor prognosis.12,13 Although secondary GBM has a much better prognosis than primary GBM and constitutes only 5% of all GBMs, overexpressing the METex14–ZM fusion transcript in U87MG cells induced a more invasive phenotype, supporting MET as a therapeutic target in secondary GBM.13 Notably, MET activation is essential in maintaining GSC’s stem-like phenotype.14,15 Blocking the MET pathway depletes GSCs in PDX models,16 supporting the rationale of targeting MET in GBM.1,17,18 Since MET inhibitors are being evaluated in clinical trials in different types of cancer, including GBM, understanding the role of the HGF/MET axis in gliomagenesis and progression may accelerate the development of MET-targeted therapy.
Genetically engineered mouse models are widely used to study human diseases. Using the Sleeping Beauty (SB) transposon system, Wiesner et al.19 were able to show that injecting mixed oncogene-coding plasmids such as NRAS, EGFR vIII, and AKT may induce de novo glioma growth in mice with phenotypes resembling human GBM pathology. The embedded luciferase reporter in the SB constructs further allows the use of real-time bioluminescence imaging (BLI) to monitor tumor growth, providing a useful tool for studying MET oncogenic activation mediated glioma initiation and progression. Using TCGA dataset analysis, we previously showed that HGF and MET expression levels correlate in primary GBM tumor specimens and that overexpression of HGF and MET occurred in approximately 30% of GBM patients.20 Using GBM PDX models, we further profiled an HGF genomic signature that may facilitate the identification of GBMs that are suitable for treatment with MET inhibitors.21 However, whether HGF-autocrine activation directly leads to glioma formation has not been tested. In this study, we show that the overexpression of HGF and MET using SB transposon technology induces glioma formation in the murine brain. We also show that isolated GSCs from these tumors propagate HGF-autocrine activation and are sensitive to MET inhibitors.
Materials and Methods
Plasmid Vectors
Plasmids pT2/C-Luc/PGK-SB100 (the SB transposase expression plasmid coding luciferase as the reporter gene), pT/CAGGS-NRASV12 (a construct coding a mutant NRAS), and pT2/shP53/GFP4 (a short hairpin siRNA against Trp53 constructed with green fluorescent protein) were created as previously described.22 To construct human HGF and MET coding plasmids, the full-length human HGF or MET cDNAs were PCR amplified from pMOG or MG-pRS24 plasmids (provided by Dr George Vande Woude, Van Andel Research Institute) with the primers engineered with SnaBI and XbaI compatible enzyme sites (FspI and AvrII). The hHGF and hMET cDNAs were further cloned into the pENTR1 Gateway vector using SnaBI and XbaI and then moved to the PT3.5-CAGG-DEST plasmid by an LR Clonase reaction (Invitrogen) as described previously.23 The SB constructs are summarized in Figure 1A. The primers are as follows: hMET forward—NNNTGCGCAGCCACCATGAAG GCCCCCGC; hMET reverse—NNNCCTAGGCTATGATGTCTCCCAGAAGG; hHGF forward—NNNTGCGCAGCCACCATGTGGGTGACCAAAC; hHGF reverse—NNNCCTAGG CTATGAC TGTGGTACC.
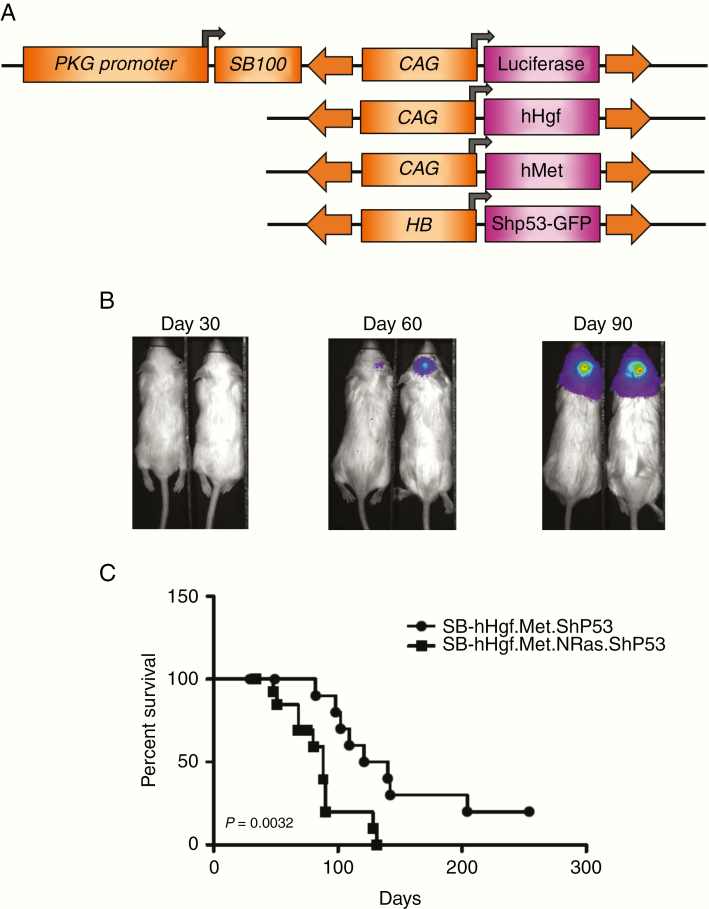
Figure 1.
Sleeping Beauty (SB) transposon-mediated HGF/MET transfection and real-time imaging in mice. (A) The SB expression vector uses the phosphoglycerate kinase (PGK) promoter to regulate SB transposase expression. The same plasmid backbone also harbors the firefly luciferase (FLuc) as a reporter regulated by the CAG promoter. Neonatal mice were injected with plasmids coding hHGF, hMET, and Shp53-GFP along with the backbone vector to initiate tumor growth in the brain. (B) Real-time, noninvasive bioluminescent imaging of tumor growth with SB-hHgf.Met.Shp53 mouse model. (C) Survival of SB-hHgf.Met.Shp53 and SB-hHgf.Met.NRas.Shp53 mice (survival time = 130.5 vs 88 days, P = .0032).
Animal Models, Plasmid Injections, and Bioluminescence Imaging
FVB/N strain mice were purchased from the Charles River Corporation and breeding pairs were monitored each day at the Vivarium of the Van Andel Research Institute (VARI). Given previous observations that neonatal mice are tolerant to human neoantigen delivered within 24 h of birth and that glioma penetrance in mice is reduced in mice older than 3 days, only neonatal mice with age less than 2 days were used for the studies.22 Mixed plasmid injection is described in Supplementary Methods. Athymic/nude mice were used for testing glioma initiation and tumor growth inhibition using isolated glioma sphere cells, which is also described in Supplementary Methods. All studies involving animals were approved by the VARI and East Tennessee State University (ETSU) Institutional Animal Care and Use Committees.
Immunohistochemistry Staining of GBM Orthotopic Tumors
At the time of necropsy, mouse brains with tumors were dissected, fixed with 10% neutral buffered formalin (Sigma-Aldrich), and embedded into paraffin blocks for H&E and immunohistochemistry (IHC) staining using standard clinical techniques in the ETSU pathology laboratory (Supplementary Methods).
Cell Lines and Drugs
The SB-026 and SB-033 neurospheres were generated from mouse 26 and mouse 33, respectively, which received SB-hHgf.Met.ShP53 plasmid injections. To establish SB-026 and SB-033 spheres, tumor tissues were excised and isolated using accutase (Invitrogen) and grown in the DMEM/F12 serum-free medium supplemented with B27 (Invitrogen), EGF (20 ng/mL, R&D), bFGF (20 ng/mL, R&D), and 1% penicillin and streptomycin (Invitrogen). To eliminate the possible clonal selection effect by EGF or bFGF, both growth factors were withdrawn from the culture medium after sphere cells started to passage. M114 cells (NIH3T3 cells stably transfected with mouse Met and mouse Hgf)24 were grown in DMEM (Invitrogen) supplemented with 10% FBS. V-4084 and SGX523 are specific MET tyrosine kinase inhibitors (TKIs). V-4084 was kindly provided by Vertex Pharmaceuticals. SGX523, EGFR inhibitor erlotinib, and MEK inhibitor PD0325901 were purchased from Selleck. All compounds were dissolved in DMSO at 0.01 M and aliquots were stored at −80°C until use. To treat cells in vitro, stock solutions were serially diluted using culture medium at the indicated concentrations.
Immunofluorescent Staining
SB-026 and SB-033 cells were seeded at 5000 cells per well in chamber slides (Lab-Tek) for neurosphere formation. Antibodies and staining are described in Supplementary Methods.
RT-PCR and Western Blot Analysis
Total RNA extraction, PCR primers and analysis, Western blotting, and antibodies are all described in Supplementary Methods.
Cell Proliferation Assay
Cells were seeded into a 96-well plate at 5 × 103 cells per well and grown for 48 h at 37°C followed by treatment with V-4084 or SGX523 at the indicated concentrations. Triplicate wells were used for each concentration. After an additional 5 days, cell proliferation was determined using the CellTiter 96 aqueous (MTS) assay (Promega). After MTS reagent was added into each well and incubated for another 3 h at 37ºC, the number of viable cells was measured by the absorbance (OD = 490) using a microplate reader (BioTek).
Statistical Analysis
Survival time was graphed using Prism 5 software (GraphPad Software) and analyzed by log-rank test (P < .05). The therapeutic efficacy of small molecule inhibitors was analyzed using the Student’s t-test (P < .05) as previously described.25
Results
Overexpression of HGF and MET Induces Spontaneous Glioma Growth in Mice
Using the SB transposon system, Wiesner et al.22 have previously shown that mixed injection of oncogene-coding plasmids into the lateral ventricle of neonatal mice induced spontaneous glioma formation. For this study, a modified SB vector (pT2/C-Luc/PGK-SB100) with enhanced transposase expression was used to test whether overexpression of HGF and MET induces glioma formation in mice. Given that p53 inhibition alone or in combination with EGFRvIII did not induce gliomas in mice,22 we used pT2/ShP53/GFP4, a plasmid with a small hairpin RNA against p53 that is linked to co-expression of green fluorescence protein (GFP), to enhance the oncogenic activity of HGF/MET axis (Figure 1A).5,8 This SB vector coinjected with an empty transposon and pT2/ShP53/GFP4 did not induce glioma in mice (Supplementary Table 1). To perform the injection, plasmids pT2/C-Luc/PGK-SB100, pT3.5/ CAGGS-huHgf, pT3.5/GAGGS-huMet, and pT2/ShP53/GFP4 were mixed at 1:1:1:1 ratio (µg/µg) and intracerebrally injected into FVB/N neonatal mice (n = 14). All mice were subjected to BLI at 24 h post-injection as an indication for transient plasmid uptake and once every month thereafter for tumor initiation and growth (Figure 1B). After excluding 4 mice that died of post-surgical hydrocephalus or significant weight loss, 8 out of 10 remaining mice (80%) developed lethal glioma growth with median survival time at 130.5 days (SB-hHgf.Met.ShP53, Figure 1C). Given that RTK/RAS/PI3K signaling accounts for the top pathway alteration in GBM, and that MET activates RAS as part of its downstream signaling,8 we tested whether overexpression of NRAS along with HGF/MET axis may accelerate glioma tumor growth. With 2 independent injections, a total of 11 mice were analyzed, and all grew tumors with a median survival time that is significantly shorter than HGF/MET/ShP53 mice (Figure 1C, SB-hHgf.Met.NRas.ShP53 vs SB-hHgf.Met.ShP53, 88 vs 130.5 days, P = .0032).
SB-hHgf.Met.ShP53 Mice Grow Invasive Glioma Expressing GSC Markers
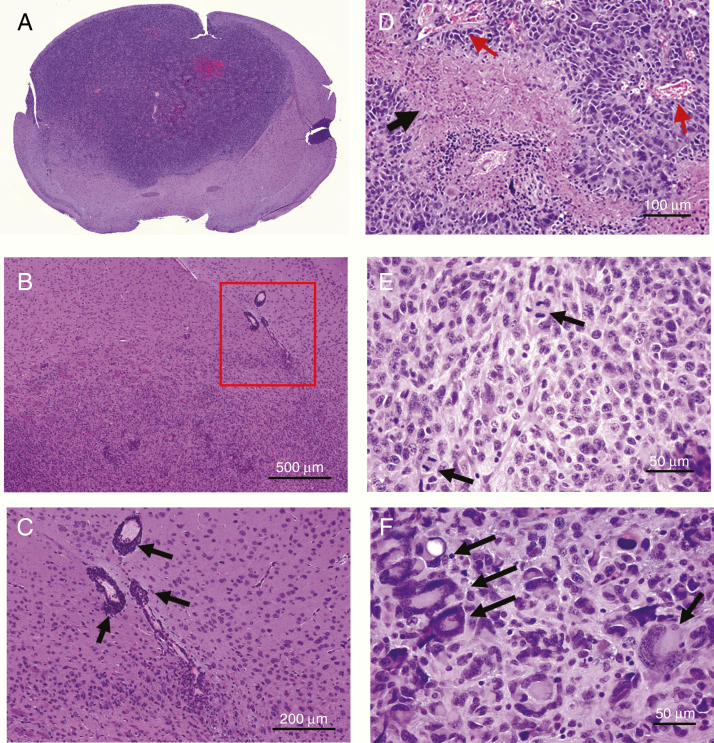
To characterize the SB-hHgf.Met.ShP53 tumors, we first sectioned the paraffin-embedded tumor blocks for histology analysis using H&E staining. We show tumor cells infiltrating into normal parenchyma without a clear border, demonstrating an invasive phenotype (Figure 2A–C). A high density of tumor cells accumulating around the blood vessels were also seen, showing a peri-vascular tumor growth (Figure 2B, squared and Figure 2C, arrowed). We also observed geographic necrosis in tumor sections (Figure 2D, arrowed) with nearby neoangiogenesis (Figure 2D, arrowed in red). Actual microvascular proliferation (endothelial), however, was not impressive. While microvascular proliferation is commonly present in GBM, the cellular anaplasia, mitotic activity, and intrinsic tumor necrosis are sufficient for the diagnosis of GBM.26 Additional GBM hallmarks also illustrated include mitoses (Figure 2E) and tumor giant cells (Figure 2F).
Figure 2.
Characterization of SB-huHgf.Met.Shp53 mouse model. (A–C) Representative images demonstrating the invasive nature. Low and high magnifications to show the infiltrative tumor borders with growth in the perivascular space (A and B). (C) Higher magnification of the area shown in the square in B. Arrows indicate the tumor cells around the blood vessels. (D–F) Representative images showing GBM hallmarks such as geographic necrosis (arrow) and neovasculature formation at the edge of necrosis (arrows in red); (E) mitoses are relatively numerous (arrows); and (F) giant tumor cells and giant tumor nuclei are also common (arrows).
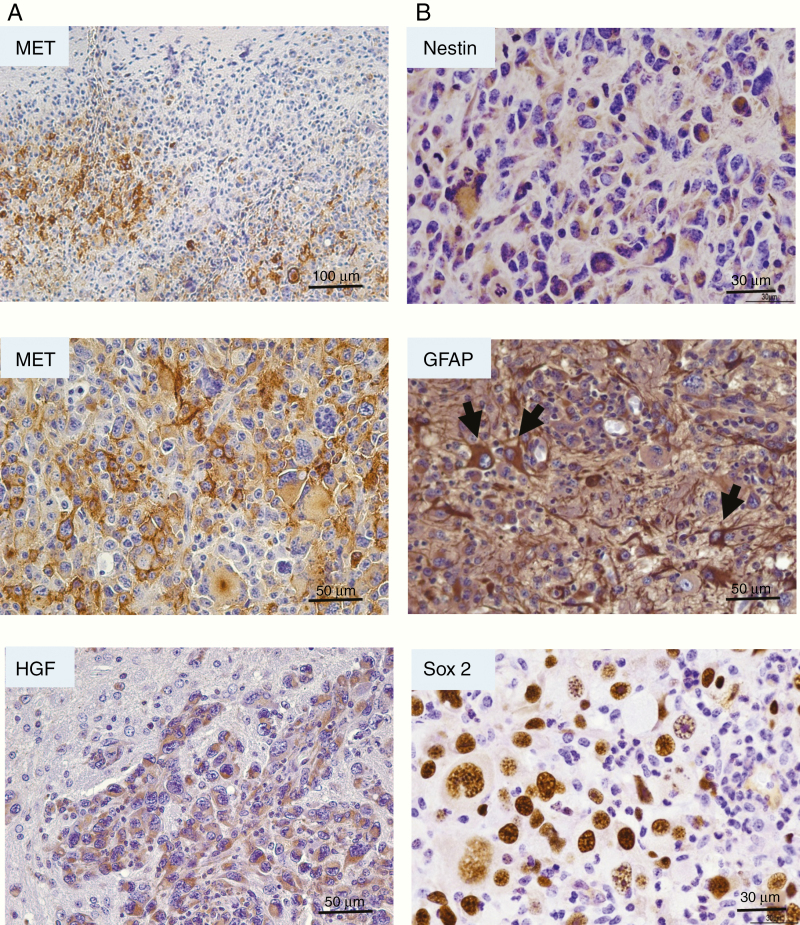
We then performed IHC staining to analyze the HGF and MET expressions in these tumors. We show that SB-hHgf.Met.Shp53 tumors overexpress MET in tumor cells as compared with the surrounding normal brain cells, demonstrating an infiltrative border (Figure 3A). At the higher magnification, we show a high expression of MET localized at the tumor cell membrane, however, with overall expression varying from low to high within the tumors (Figure 3A), suggesting a heterogeneous nature of these tumors. HGF was also overexpressed in tumor cells relative to the normal tissues, showing a cytoplasmic location (Figure 3A).
Figure 3.
IHC analysis of HGF/MET expression and NSC markers in SB-hHgf.Met.Shp53 glioma mouse model. (A) Representative images of MET and HGF expression in SB-hHgf.Met.ShP53 tumors as determined by IHC staining. (B) Representative images of NSC marker expression including Nestin, GFAP (arrows point to cells with obvious similarity to reactive-type astrocytes, but likely represent neoplastic cells) and Sox2 in tumor sections as determined by IHC staining.
Since MET is reported to play an essential role in maintaining GSC properties,14,15 we further stained SB-hHgf.Met.ShP53 tumors with glial fibrillary acid protein (GFAP), nestin, and Sox II, the neural stem cell (NSC) or progenitor cell markers indicating self-renewal and pluripotency (Figure 3B). We show that all 3 markers were expressed in the tumor sections. These results suggest that overexpression of HGF/MET may transform NSCs and progenitor cells into GSCs, which leads to GBM formation.27
Characterization of GSChgf/met Cells
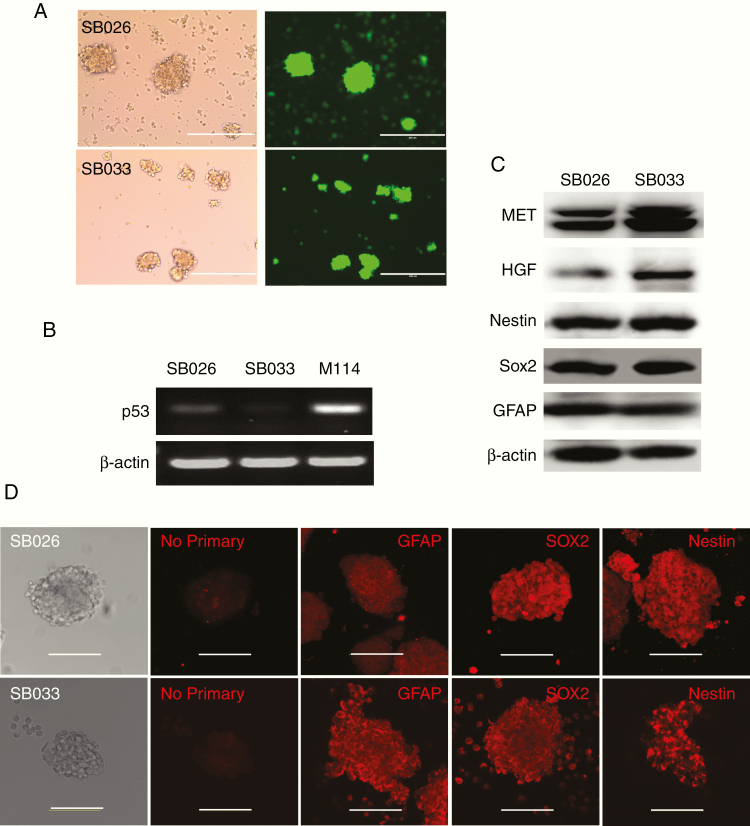
With IHC staining, the SB-hHgf.Met.Shp53 tumors showed tumor-specific expression of nestin and Sox 2, but a heterogeneous expression of GFAP, a marker for astrocytes, which was more evident in some apparent reactive astrocytes than tumor cells (Figure 3B GFAP, arrowed). To confirm the GFAP expression in tumor cells, 2 neurosphere cell lines (SB-026 and SB-033) were generated from SB-hHgf.Met.ShP53 tumors and characterized for expression markers. Both SB-026 and SB-033 spheres expressed strong GFP (Figure 4A), indicating sufficient uptake of the ShP53/GFP plasmid. Since we did not generate wild-type p53 glioma spheres from this study, the levels of p53 suppression in SB-026 and SB-033 were tested by RT-PCR, showing a significant reduction as compared with M114 cells,24 the normal mouse fibroblast cells overexpressing mouse Hgf/Met (Figure 4B). Both Western blot (Figure 4C) and confocal analysis (Figure 4D) confirmed the expression of GFAP, nestin, and Sox 2 in SB026 and SB033 spheres, ruling out reactive astrocytes as the only source of GFAP expression in SB-huHgf.Met.Shp53 tumors. Importantly, on intracranial injection, both SB-026 and SB-033 cells initiate glioma growth in nude mice. SB-026 sphere-derived intracranial tumors propagate GBM histological hallmarks including infiltrative border, perivascular tumor growth, and necrotic center. Mitoses and giant cells are also shown (Supplementary Figure 1). These results indicate that HGF-autocrine activation of MET RTK is the driving force that transforms NSCs into GSCs to initiate glioma growth in these mice.
Figure 4.
Characterization of neurospheres derived from SB-huHgf.Met.Shp53 tumors. (A) Representative image showing SB-026 and SB-033 forms tumor spheres that are GFP positive. Scale bar refers to 400 µm. (B) p53 mRNA levels in SB-026, SB-033, and M114 cells as determined by RT-PCR. (C) HGF, MET, GFAP, Nestin, and SOX2 expression levels in SB-026 and SB-033 as determined by Western blot. (D) GFAP, Nestin, and SOX2 expression levels in SB-026 and SB-033 as shown by confocal microscopy. Scale bar refers to 100 µm. A “No primary” control was included to indicate the background staining due to secondary antibody alone.
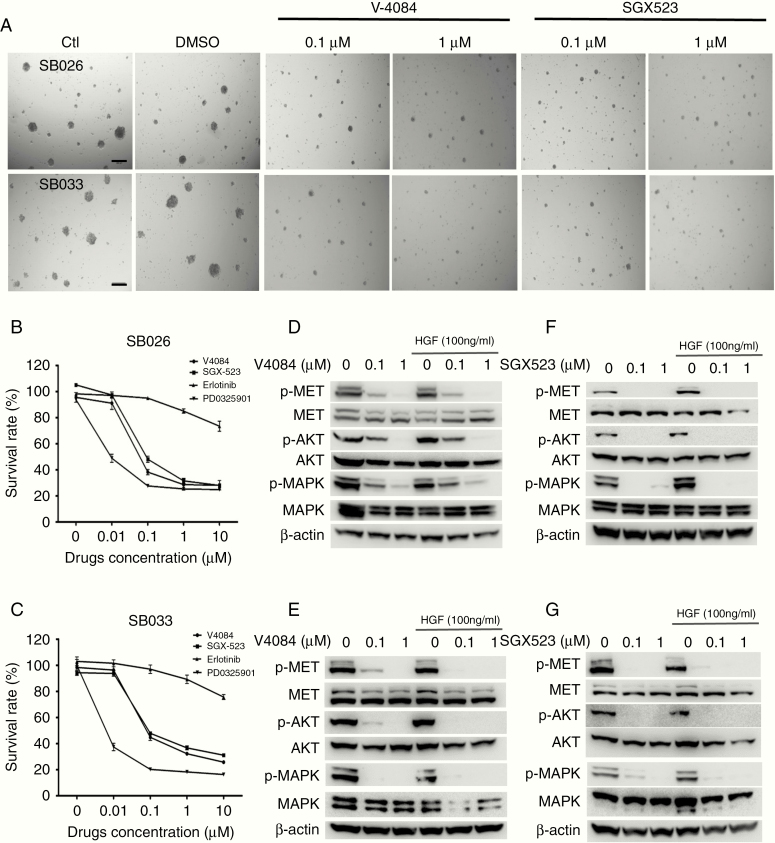
SB-026 and SB-033 Are Sensitive to Specific MET TKIs
Using GBM and HCC models, we previously reported that tumor cells overexpressing both HGF and MET often propagate an HGF-autocrine activation of MET RTK, indicating sensitivity to MET TKIs.20,21,25 To test whether this applies to SB-026 and SB-033, we treated these cells with V-4084 and SGX523 for 5 days and monitored spheroid formation. We show that both V-4084 and SGX523 at 0.1–1 µM significantly reduced the size of SB-026 and SB-033 spheroids (Figure 5A). The 5-day survival rate of SB-026 and SB-033 after V-4084 and SGX523 treatment was quantified using MTS assay, with erlotinib (an EGFR inhibitor) and PD-325901 (a MEK inhibitor targeting MAPK downstream from MET) tested for comparison (Figure 5B and C). We show that V-4084 and SGX523 at 0.1 µM and higher concentration significantly inhibited SB-026 (V-4084 vs SGX523: 61.7% vs 51.9%, P < .001) and SB-033 (V-4084 vs SGX523: 55.2% vs 55.1%, P < .001) proliferation. In contrast, erlotinib (1 µM) only marginally inhibited SB-026 and SB-033 (15% and 11%, respectively, P < .05), suggesting a nonspecific targeting. Notably, PD-0325901 inhibited both SB-026 and SB-033 at 0.01 µM (51.1% and 62.6%, respectively, P < .001), showing a higher efficacy than MET TKIs. At the protein expression level, both SB-026 and SB-033 expressed HGF, MET, and p-MET, demonstrating an HGF-autocrine activation of MET. Both V-4084 (Figure 5D and E) and SGX523 (Figure 5F and G) treatment significantly inhibit MET/MAPK/AKT signaling regardless of additional HGF stimulation. We further tested whether V-4084 inhibited SB-033 tumor growth in vivo. While an initial single dose of V-4080 at 30 mg/kg for 8 days showed marginal inhibition, increasing dosing to 60 mg/kg for an additional 3 days significantly inhibited SB-033 tumor growth in mice (Supplementary Figure 2, 1595 vs 647 mm3, P < .05). While the non-orthotopic model is limited by the lack of the appropriate microenvironment for tumor growth and therefore may not reproduce the therapeutic efficacy or pharmaceutical kinetics precisely, mechanistically, these results indicate that the HGF/MET axis is the driving force in SB-026 and SB-023 tumor growth and that targeting MET/MAPK signaling will be effective for treating HGF-autocrine GBMs.
Figure 5.
MET inhibitors suppress HGF-autocrine neurosphere formation. (A) Representative images of SB-026 and SB-033 tumor spheres formation under V-4084 and SGX523 treatment. SB-026 and SB-033 cells were cultured in a 96-well plate for various treatments as indicated. Triplicates were used for each treatment. Images were taken at 5 days after treatment under a light microscope. Scale bar = 200 µm. (B and C) Quantification of SB-026 and SB-033 spheroid formation assay as described in A. After treating SB-026 (B) and SB-033 (C) cells with V-4084 or SGX523 at indicated concentrations, cell proliferation at day 5 was measured using the MTS assay as described in the Methods section. Survival rate (%) = Absorbance (OD = 490) of treated cells/Absorbance (OD = 490) of control untreated cells × 100%. Data represent the average of 2 independent assays. A short bar refers to the standard deviation. (D and E) Specific MET inhibitor V-4084 at indicated concentrations inhibits MET, MAPK/AKT pathways in SB-026 and SB-033 spheres as shown by Western blot. (F and G) Specific MET inhibitor SGX523 at indicated concentrations inhibits MET, MAPK/AKT pathways in SB-026 and SB-033 spheres as shown by Western blot.
Discussion
Aberrant HGF/MET signaling activation is known to promote cancer initiation and malignant progression.28,29 Genetically engineered mouse models have been widely used to test the tumor initiation role of HGF/MET in different cancer types. Previously, overexpression of HGF alone resulted in significant liver enlargement and triggers hepatocellular adenomas and/or carcinomas in mice.30,31 Knocking in mutant Met in mice induced diverse mammary carcinomas mimicking human basal breast cancer.32 Overexpression of Met alone or in combination with beta-catenin is sufficient for developing hepatocellular carcinoma.33,34 However, none of these mouse models showed brain tumor phenotypes. Although HGF/MET expression in human primary brain tumors is found to increase with the grade of malignancy35 and is a prognostic marker of short survival time in GBM,10 whether the HGF/MET axis directly triggers brain tumor formation has not been tested. In this study, we show that overexpression of HGF/MET, along with p53 suppression, induced de novo glioma formation in mice. These tumors infiltrate the brain and the perivascular spaces, are mitotically active, and show intrinsic, geographic necrosis and neoangiogenesis, pathological hallmarks that recapitulate human GBM histology. These results suggest that expression of HGF or MET is not sufficient for initiating brain tumors, while a co-expression of both ligand and receptor especially preconditioned with loss of function of tumor-suppressor genes is likely required. Although the SB-based gene transfer has the potential to cause insertional mutagenesis, the tumors developed in this model did not likely result from randomized mutations for 2 reasons: (1) the negative control plasmids did not induce any glioma in mice (Supplementary Table 1) and (2) the SB033 tumors are sensitive to V-4084, a MET-specific inhibitor, demonstrating a level of MET dependency (Supplementary Figure 1).
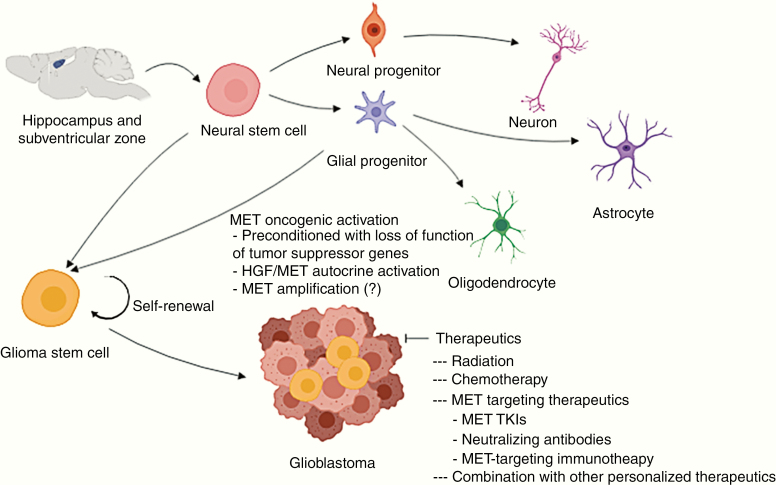
During normal CNS development, NSCs are primarily located in the hippocampus and subventricular zone. These are undifferentiated precursor cells defined by their capacity for self-renewal and multipotency, which may further amplify into multi-lineage progenitor cells and differentiate into neurons, astrocytes, and oligodendrocytes to populate the nervous system. However, GSCs within a brain tumor may arise from normal NSCs or neural progenitor cells that harbor genetic alterations such as mutations and oncogene overexpression.1,36,37 Previous studies have shown that loss of Pten in combination with p53 converted neural and glioma stem cells into glial tumors.38,39 While p53 suppression, alone or in combination with EGFR vIII, is insufficient to induce glioma formation in mice, injecting plasmids encoding NRAS, EGFRVIII, and AKT along with p53 inhibition into the neonatal mice subventricular zone induces de novo glioma formation.22 In this study, we delivered the SB transposon coded plasmids into the lateral cerebral ventricle, a method with a demonstrated capacity of transfecting cells that are enriched in the subventricular zone where NSCs reside.22 Although the exact “cell of origin” of the glioma initiation in this model remains elusive, we observed NSC markers nestin, GFAP, and Sox 2 expression27 in both SB-hHgf.Met.ShP53 tumors and SB-026, SB-033 spheres. More importantly, both cell lines initiated tumor growth in mice. Therefore, we propose that intracerebroventricular injection of SB-hHgf.Met.ShP53 plasmids into neonatal mice may initiate HGF-autocrine activation in NSCs and progenitor cells, transform these cells into GSCs, and ultimately, initiate glioma growth and progression in these mice (Figure 6). MET activation is essential in maintaining GSC’s stem-like phenotype.14,15 The fact that MET TKIs V-4084 and SGX523 and MEK inhibitor PD-0325901 significantly inhibited SB-026 and SB-033 proliferation in vitro and tumor growth in vivo support the application of targeting MET/MAPK signaling pathway for treating gliomas with HGF/MET overexpression. Because MET-targeted inhibitors are all designed to block the ATP binding in the MET RTK domain rather than blocking MET binding to HGF, our results only demonstrate a MET dependency in this model. However, whether the tumors are driven by an HGF-autocrine activation remains undetermined. Therefore, it would be necessary to further test the tumors for sensitivity to anti-HGF or anti-MET neutralizing antibodies to further distinguish the HGF dependency.25
Figure 6.
Proposed mechanism of MET RTK alteration induced gliomagenesis and therapeutics. During normal neurogenesis, neural stem cells are primarily located in the hippocampus and subventricular zone where they proliferate into multi-lineage progenitor cells and differentiate into mature neural cells. When preconditioned with loss of function of tumor-suppressor genes, such as TP53 mutation, MET oncogenic activation such as HGF-autocrine activation may transform NSCs or progenitor cells into GSCs, which ultimately lead to glioma initiation. While these tumors are primarily driven by MET RTK signaling and are sensitive to MET-targeting therapy, the extensive clinical literature indicating the accumulation of many other molecular abnormalities in clinically evident infiltrating gliomas (especially glioblastoma) likely requires consideration of combining MET-targeting therapy with other targeted therapies to overcome resistance and recurrence.
While we show in this study that overexpression of HGF/MET induces gliomagenesis, it is not clear whether HGF or MET alone may be sufficient. Specifically, MET amplification occurs in approximately 4% of GBM patients.8 Although early studies have not reported that HGF or MET transgenic mouse models develop glioma as a phenotype,30,31,34 it is possible that the dose of HGF or MET transgenes in previous models was not high enough to initiate tumors in the brain. Since the SB transposon system allows for the quantification of the concentration of plasmid DNA for injection, it is worthwhile to apply injections to deliver a higher dose of human HGF or MET, either alone or in combination with deletions of glioma suppressor genes, such as TP53, PTEN, or NF1.5 Such studies may elucidate how ligand-independent MET activation may regulate GBM initiation and progression (Figure 6). With the SB transposon system as a tool, another important application is to study how HGF-autocrine activation may crosstalk with other tumor-driver genes or suppressor genes in determining the therapeutic response to anticancer treatments. We have shown that introducing NRAS into the HGF/MET/ShP53 axis may accelerate glioma growth and significantly shorten the survival time of the animals (Figure 1C). Thus, future studies may test how other GBM core pathway components, such as p53 and RB signaling pathways, may regulate HGF/MET axis in gliomagenesis and progression.8 Recently, CRISPER/cas9 has become a powerful technology for gene editing. Combining the use of CRISPER/cas9 and SB approaches has been shown to be an effective approach in screening for tumor-driver genes and therapeutic targets for functional validation.40 Future studies may use CRISPR/cas9 to improve this SB-hHgf.Met.ShP53 mouse model in multi-target gene delivery as a better system for studying brain tumor biology and therapeutic strategies.
In conclusion, using a well-established SB transposon strategy, we demonstrate that mixed injection of plasmid DNAs coding human HGF and MET, along with microRNA-mediated siRNA against p53, results in de novo glioma formation in mice. These tumors show invasive phenotypes that resemble human GBM pathology. Moreover, the neurospheres isolated from SB-hHgf.Met.Shp53 tumors expressed human HGF and MET along with NSC markers including nestin, GFAP, and Sox 2 and are sensitive to specific MET TKIs. These results suggest that HGF-autocrine activation may transform NSCs into GSCs that ultimately lead to GBM initiation. Successful targeted therapy relies on clinically relevant preclinical models. MET inhibitors are being evaluated in clinical trials against multiple cancer types including brain tumors.28,41 Although MET-targeted therapy in GBM patients has not been successful, a recent clinical trial has shown that a combination of MET and VEGFR inhibitors (onartuzumab plus bevacizumab vs placebo plus bevacizumab) significantly improved both progression-free survival and overall survival in the mesenchymal subtype of recurrent GBM patients with high HGF expression.42 This result supports our previous findings that HGF is a predictive marker for MET-targeted therapy and that developing biomarker analysis for patient stratification is necessary in future clinical trials targeting MET.20,21 While our previous studies have used immune-compromised mice bearing human GBM cell lines or PDXs, the SB-hHgf.Met.ShP53 mouse glioma model provides an immune-competent microenvironment for studying GBM tumor biology and MET-targeting therapeutics and will be of value for studying cancer immunotherapy against malignant glioma.
Supplementary Material
Acknowledgments
We thank Dr George Vande Woude for providing pMOG and MG-pRS24 plasmids, Rolf B. Fritz for assisting confocal analysis, and Dr Phillip R. Musich for reviewing the manuscript. We also thank Vertex Pharmaceuticals for providing V-4084.
Conflict of interest statement. D.A.L. is the co-founder and co-owner of several biotechnology companies including NeoClone Biotechnologies, Inc., Discovery Genomics, Inc. (recently acquired by Immusoft, Inc.), B-MoGen Biotechnologies, Inc. (recently acquired by the Bio-Techne corporation), and Luminary Therapeutics, Inc. He consults for Genentech, Inc., which is funding some of his research. D.A.L. holds equity in and serves as the Chief Scientific Officer of Surrogen, a subsidiary of Recombinetics, a genome-editing company. The business of all these companies is unrelated to the contents of this manuscript. All other authors declare no conflict of interest.
Funding
This work was supported by the ETSU start-up fund, East Tennessee State University Research Development Committee Major Grant (to Q.X.), and American Cancer Society Professor award, National Institutes of Health U54CA210190 (to D.A.L.).
Authorship Statement
Y.Q., A.M., J.K., B.T., A.Q., C.L., B.S., L.K., and Q.X. performed the majority of the experiments and analysis. B.T. constructed the Sleeping Beauty plasmids. J.P. performed pathological staining and J.B.S. reviewed slides. K.T. provided material. D.A.L. and Q.X. performed study design and data interpretation. Q.X. wrote the paper with feedback from Y.J., K.T., J.B.S., and D.A.L. All authors read and approved the manuscript.
References
- 1. Xie Q, Mittal S, Berens ME. Targeting adaptive glioblastoma: an overview of proliferation and invasion. Neuro Oncol. 2014;16(12):1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Johnson DR, Omuro AMP, Ravelo A, et al. Overall survival in patients with glioblastoma before and after bevacizumab approval. Curr Med Res Opin. 2018;34(5):813–820. [DOI] [PubMed] [Google Scholar]
- 3. Ostrom QT, Gittleman H, Fulop J, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro Oncol. 2015;17(Suppl 4):iv1–iv62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. [DOI] [PubMed] [Google Scholar]
- 5. Verhaak RG, Hoadley KA, Purdom E, et al. ; Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu G, Yuan X, Zeng Z, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. [DOI] [PubMed] [Google Scholar]
- 8. Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hoelzinger DB, Demuth T, Berens ME. Autocrine factors that sustain glioma invasion and paracrine biology in the brain microenvironment. J Natl Cancer Inst. 2007;99(21):1583–1593. [DOI] [PubMed] [Google Scholar]
- 10. Petterson SA, Dahlrot RH, Hermansen SK, et al. High levels of c-Met is associated with poor prognosis in glioblastoma. J Neurooncol. 2015;122(3):517–527. [DOI] [PubMed] [Google Scholar]
- 11. De Bacco F, Casanova E, Medico E, et al. The MET oncogene is a functional marker of a glioblastoma stem cell subtype. Cancer Res. 2012;72(17):4537–4550. [DOI] [PubMed] [Google Scholar]
- 12. Bao ZS, Chen HM, Yang MY, et al. RNA-seq of 272 gliomas revealed a novel, recurrent PTPRZ1-MET fusion transcript in secondary glioblastomas. Genome Res. 2014;24(11):1765–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hu H, Mu Q, Bao Z, et al. Mutational landscape of secondary glioblastoma guides MET-targeted trial in brain tumor. Cell. 2018;175(6):1665–1678.e1618. [DOI] [PubMed] [Google Scholar]
- 14. Li Y, Li A, Glas M, et al. c-Met signaling induces a reprogramming network and supports the glioblastoma stem-like phenotype. Proc Natl Acad Sci U S A. 2011;108(24):9951–9956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Joo KM, Jin J, Kim E, et al. MET signaling regulates glioblastoma stem cells. Cancer Res. 2012;72(15):3828–3838. [DOI] [PubMed] [Google Scholar]
- 16. Rath P, Lal B, Ajala O, et al. In vivo c-met pathway inhibition depletes human glioma xenografts of tumor-propagating stem-like cells. Transl Oncol. 2013;6(2):104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boccaccio C, Comoglio PM. The MET oncogene in glioblastoma stem cells: implications as a diagnostic marker and a therapeutic target. Cancer Res. 2013;73(11):3193–3199. [DOI] [PubMed] [Google Scholar]
- 18. Prados MD, Byron SA, Tran NL, et al. Toward precision medicine in glioblastoma: the promise and the challenges. Neuro Oncol. 2015;17(8):1051–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wiesner S, Decker S, Larson J, et al. De novo induction of genetically engineered brain tumors in mice using plasmid DNA. Cancer Res. 2009;69(2):431–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xie Q, Bradley R, Kang L, et al. Hepatocyte growth factor (HGF) autocrine activation predicts sensitivity to MET inhibition in glioblastoma. Proc Natl Acad Sci U S A. 2012;109(2):570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johnson J, Ascierto ML, Mittal S, et al. Genomic profiling of a hepatocyte growth factor-dependent signature for MET-targeted therapy in glioblastoma. J Transl Med. 2015;13:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wiesner SM, Decker SA, Larson JD, et al. De novo induction of genetically engineered brain tumors in mice using plasmid DNA. Cancer Res. 2009;69(2):431–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moriarity BS, Rahrmann EP, Beckmann DA, et al. Simple and efficient methods for enrichment and isolation of endonuclease modified cells. PLoS One. 2014;9(5):e96114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shinomiya N, Gao CF, Xie Q, et al. RNA interference reveals that ligand-independent met activity is required for tumor cell signaling and survival. Cancer Res. 2004;64(21):7962–7970. [DOI] [PubMed] [Google Scholar]
- 25. Kou J, Musich PR, Staal B, et al. Differential responses of MET activations to MET kinase inhibitor and neutralizing antibody. J Transl Med. 2018;16(1):253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 27. Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. [DOI] [PubMed] [Google Scholar]
- 28. Thewke D, Kou J, Fulmer M, Xie Q. The HGF/MET signaling and therapeutics in cancer. In: Shinomiya N, Kataoka H, Xie Q, eds. Regulation of Signal Transduction in Human Cell Research. Singapore: Springer Nature; 2018:155–182. [Google Scholar]
- 29. Shinomiya N, Xie Q, Vande Woude G. Met activation and carcinogenesis. In: Shinomiya N, Kataoka H, Xie Q, eds. Regulation of Signal Transduction in Human Cell Research. Singapore: Springer Nature; 2018:129–154. [Google Scholar]
- 30. Sakata H, Takayama H, Sharp R, Rubin JS, Merlino G, LaRochelle WJ. Hepatocyte growth factor/scatter factor overexpression induces growth, abnormal development, and tumor formation in transgenic mouse livers. Cell Growth Differ. 1996;7(11):1513–1523. [PubMed] [Google Scholar]
- 31. Xie Q, Su Y, Dykema K, et al. Overexpression of HGF promotes HBV-induced hepatocellular carcinoma progression and is an effective indicator for met-targeting therapy. Genes Cancer. 2013;4(7–8):247–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Graveel CR, DeGroot JD, Su Y, et al. Met induces diverse mammary carcinomas in mice and is associated with human basal breast cancer. Proc Natl Acad Sci U S A. 2009;106(31):12909–12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tward AD, Jones KD, Yant S, et al. Distinct pathways of genomic progression to benign and malignant tumors of the liver. Proc Natl Acad Sci U S A. 2007;104(37):14771–14776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang R, Ferrell LD, Faouzi S, Maher JJ, Bishop JM. Activation of the Met receptor by cell attachment induces and sustains hepatocellular carcinomas in transgenic mice. J Cell Biol. 2001;153(5):1023–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Koochekpour S, Jeffers M, Rulong S, et al. Met and hepatocyte growth factor/scatter factor expression in human gliomas. Cancer Res. 1997;57(23):5391–5398. [PubMed] [Google Scholar]
- 36. Tang Y, Yu P, Cheng L. Current progress in the derivation and therapeutic application of neural stem cells. Cell Death Dis. 2017;8(10):e3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang P, Lathia JD, Flavahan WA, Rich JN, Mattson MP. Squelching glioblastoma stem cells by targeting REST for proteasomal degradation. Trends Neurosci. 2009;32(11):559–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zheng H, Ying H, Yan H, et al. Pten and p53 converge on c-Myc to control differentiation, self-renewal, and transformation of normal and neoplastic stem cells in glioblastoma. Cold Spring Harb Symp Quant Biol. 2008;73:427–437. [DOI] [PubMed] [Google Scholar]
- 39. Zheng H, Ying H, Yan H, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455(7216):1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gao M, Liu D. CRISPR/Cas9-based Pten knock-out and Sleeping Beauty transposon-mediated Nras knock-in induces hepatocellular carcinoma and hepatic lipid accumulation in mice. Cancer Biol Ther. 2017;18(7):505–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wen PY, Schiff D, Cloughesy TF, et al. A phase II study evaluating the efficacy and safety of AMG 102 (rilotumumab) in patients with recurrent glioblastoma. Neuro Oncol. 2011;13(4):437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cloughesy T, Finocchiaro G, Belda-Iniesta C, et al. Randomized, double-blind, placebo-controlled, multicenter phase ii study of onartuzumab plus bevacizumab versus placebo plus bevacizumab in patients with recurrent glioblastoma: efficacy, safety, and hepatocyte growth factor and O6-methylguanine-DNA methyltransferase biomarker analyses. J Clin Oncol. 2017;35(3):343–351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.