Abstract
Glioblastoma multiforme (GBM) is a fatal malignant tumor that is characterized by diffusive growth of tumor cells into the surrounding brain parenchyma. However, the diffusive nature of GBM and its relationship with the tumor microenvironment (TME) is still unknown. Here, we investigated the interactions of GBM with the surrounding microenvironment in orthotopic xenograft animal models using two human glioma cell lines, U87 and LN229. The GBM cells in our model showed different features on the aspects of cell growth rate during their development, dispersive nature of glioma tumor cells along blood vessels, and invasion into the brain parenchyma. Our results indicated that these differences in the two models are in part due to differences in the expression of CXCR4 and STAT3, both of which play an important role in tumor progression. In addition, the GBM shows considerable accumulation of resident microglia and peripheral macrophages, but polarizes differently into tumor-supporting cells. These results suggest that the intrinsic factors of GBM and their interaction with the TME determine the diffusive nature and probably the responsiveness to non-cancer cells in the TME.
Keywords: glioblastoma, invasion, LN229, tumor microenvironment, U87
INTRODUCTION
Glioblastoma multiforme (GBM) is the most common and deadliest malignant primary brain tumor and represents about 15% of all primary brain tumors (Thakkar et al., 2014). The mortality rate of patients with GBM is high due to several reasons including genetic heterogeneity of tumor cells, blood-brain barrier to therapeutic agents, and diffusive growth of tumor cells into the surrounding brain parenchyma, the latter of which causes curative surgery to be inefficient. Therefore, a better understand of the microenvironment in promoting the growth and recurrence of these tumors is necessary.
Invasive tumor growth of GBM is mediated by the interaction of tumor cells with the surrounding tumor microenvironment (TME) within the brain (Quail and Joyce, 2013). Neoplastic tumor cells in TME develop the innate heterogeneity of GBM. In addition, genetically distinct immune cell populations exist within the brain TME, including tissue-resident microglia and peripheral macrophages/monocytes (Hambardzumyan et al., 2016). These immune cells constitute about 40% of the tumor mass and play a key role in glioma invasion through several mechanisms (Graeber et al., 2002).
The U87 cell line has an epithelial morphology and was obtained from a male patient of uncertain age in 1966 and deposited to the American Type Culture Collection (ATCC). In the past couple of decades, tremendous preclinical studies aiming to develop GBM therapy have utilized brain orthotopic models that contain U87 cells as human glioma cells. However, U87 cells can represent diffusive human glioma cells because of their rapid growth and demarcated, circumscript tumor margins, which may be a concern in some studies (Allen et al., 2016; Zeng et al., 2018).
The LN229 cell line was isolated from human patients with GBM (Van Meir et al., 1990). These cells have an epithelial morphology, grow with diffusive margins, and has been regarded to represent human GBM more closely than U87 cells.
In this study, we investigate the diffuse nature of GBM and its microenvironment in cellular and animal levels using two human glioma cell lines: U87 and LN229. We described the differences in growth rate, the mediators promoting the migration, the infiltrative nature of glioma cells along blood vessels, and the interaction with surrounding microglia and macrophages. Our data suggest that either the human LN229 or U87 cell line in mouse xenograft models of glioma will be useful for future research to develop drugs through molecular regulation of tumor cell infiltration.
MATERIALS AND METHODS
Study approval
All experimental protocols were approved by the Institutional Review Board of the Ajou University Hospital (No. AJIRB-GEN-GEN-10-175) and all animal protocols were approved by the Institutional Animal Care and Use Committee of the Ajou University School of Medicine (No. 2017-0052).
Cell culture and transduction
The LN229 and U87MG human glioma cell line were obtain from ATCC (USA). U87MG and LN229 were maintained in Dulbecco’s modified Eagle medium (DMEM; Welgene, Korea) supplemented with 5% fetal bovine serum (FBS; GE Healthcare Life Sciences, Australia), 100 U/ml penicillin, and 100 mg/ml streptomycin (Invitrogen, USA) in a 37°C incubator. The lentiviral vector LL3.7 encoding green fluorescence protein (GFP) (Addgene, USA) was used to transduce U87 or LN229 cell lines in the presence of 4 μg/ml polybrene. Two days later, transduced cells were sorted by fluorescence-activated cell sorting (FACS; BD Biosciences, USA). The resulting U87/GFP and LN229/GFP cells were expanded in growth medium (GM).
In vivo xenograft mouse model
Briefly, adult BALB/C nude mice (~20 g; Nara Biotech, Korea) were anesthetized and 1 × 105 cells each of U87/GFP or LN229/GFP cells in 3 μl of phosphate-buffered saline (PBS) were sequentially transplanted into the striatum (AP, +0.05 cm; ML, –0.18 cm; DV, –0.3 cm) using a stereotaxic device (Stoelting, USA). To assess the tumor volumes, the animals were subject to magnetic resonance imaging (MRI) analysis or sacrificed for H&E staining at the indicated time.
Measurement of tumor volume
MRI was performed with a 9.4-T animal MRI scanner (Biospec 94/30; Bruker BioSpin MRI GmbH, Germany). T2-weighted images were acquired with the following parameters: repetition time = 3,000 ms, echo time = 32 ms, average = 2, acquisition matrix = 256 × 256; 10 slices with 1-mm slice thickness, and flip angle = 90°. A total of 10 slices were scanned to cover the whole brain. Reconstruction of MRI images was performed with the Digital Imaging and Communications in Medicine software (DICOM Viewer R3.0 SP3; Philips Healthcare, Netherlands). Immediately after the MRI experiments, 2-mm-thick coronal brain slices were prepared using a mouse brain mold (CellPoint Scientific, USA) and processed for H&E staining or immunohistochemical analysis. The tumor volume was calculated using as the following formulation: V = (L × W × W)/2, where V is the tumor volume, W is the tumor width, L is the tumor length.
Tissue preparation
At the indicated time, animals were anesthetized with ketamine (100 mg/kg)/xylazine (10 mg/kg) (Yuhan, Korea), perfused transcardially with ice-cold saline, and then fixed with 10% neutral buffered formalin (NBF; BBC Biochemical, USA). The brains were post-fixed in 10% NBF, and dehydrated by serially soaking in 70%, 80%, 90%, 95%, 100% ethanol, and xylene, and eventually embedded in paraffin using a Leica TP1020 (Leica Microsystems, Germany).
Immunohistochemistry
Paraffin sections were prepared with a thickness of 5 μm and deparaffinized by reversing the dehydration process. The sections were placed in boiled citrate buffer (pH 6.0) for 10 min and incubated with 0.3% H2O2 for 30 min to inhibit endogenous peroxidase activity. After blocking nonspecific interactions via an incubation with 10% of the host serum at 24°C for 1 h, the samples were incubated at 4°C overnight with primary antibodies against STAT3 (1:300; Cell Signaling, USA), pSTAT3 (1:200; Cell Signaling), Iba1 (1:3,000; Wako, USA), IL-1β (1:200; Abcam, USA), and CD206 (1:1,000; Abcam). After washing in PBS-T (0.1% Tween X-100), the sections were treated with biotinylated goat anti-rabbit IgG (Vector Lab, USA) or biotinylated horse anti-mouse IgG (Vector Lab) in the blocking buffer. The reaction was amplified for 30 min with an avidin-biotin complex, using an ABC kit (Vector Lab) and visualized with 3,3’-diaminobenzidine (DAB; Sigma-Aldrich, USA). After dehydration through a graded series of alcohol, the sections were mounted in Shandon synthetic mountant (Thermo Scientific, UK) and cover-slipped.
For immunofluorescence staining, sections were incubated overnight at 4°C with antibodies against CD31 (1:200; Abcam), GFP (1:500; Abcam), and STAT3 (1:300; Cell Signaling). After washing 3 times for 5 min each in TBS containing 0.1% Tween-20, the immunoreactivity was visualized using Alexa Fluor 488- or 568-conjugated goat anti-mouse or rabbit IgG. After washing, the slides were air-dried and mounted with aqueous mounting media (SouthernBiotech, USA).
Sections without primary antibodies were processed in parallel as a negative control to monitor the nonspecific peroxidase staining of secondary antibodies in the damaged tissue. The brain sections scanned were under high power magnification (40×) on the Axio Scan.Z1 (Carl Zeiss, Germany) and the digital images were acquired.
The number of immunuoreactive cells per mm2 were manually counted in each field of interest by an investigator blinded to the groups. The number of GFP-positive cells around the blood vessels were counted in divided regions from the adjacent area to the remote areas. The number of Iba1-, IL-1β-, and CD206-positive cells were counted in the intra-tumoral and peri-tumoral areas of the striatum. The results are presented as mean counts per mm2 ± SE.
In vitro cell migration and invasion assay
A scratch wound assay with LN229/GFP and U87/GFP cultures was performed to determine the migration potential of the cells. GBM cells were plated onto 24-well plates at a density of 5 × 104 cells/cm2 and were allowed to grow for a 24-h period in serum-free media to reach a confluent monolayer. The serum-free media was removed and the confluent cell sheet was wounded by scratching the culture well surface with a 1 ml pipette tip. Fluorescence microscopic imaging was used to record a frame every 12 h for 24 h with an AxioCam single-channel camera and AxioVision software (Carl Zeiss). The GFP intensity was also measured in the wounded area at indicated times. The results from three independent experiments are presented as mean ± SE.
The migration/invasion assays were performed in a 24-well transwell cell culture chamber with an 8-μm pore polycarbonate membrane insert (Corning Costar, USA). GBM cells were plated at a density of 5 × 104 cells/cm2 with or without coated Matrigel (Corning Costar) in the upper compartment of the chamber. The lower compartment was loaded with bEND.3 cells at a density of 5 × 104 cells/cm2 or conditioned medium (CM) of bEND.3 cells, and the GBM cells were then incubated for 24 h at 37°C. The insert was washed with PBS and fixed with 10% NBF (BBC Biochemical). The cells at the upper surface of the membrane in the insert were removed with a cotton swab, and the fluorescence images of the invaded cells in the bottom surface were acquired with a fluorescence IX71 microscope (Olympus, Japan). The GFP levels remaining on the bottom surface of the entire membrane was presented as mean ± SE from three independent experiments.
In vitro experiment to mimic the interaction of GBM cells with TME
GBM cells were co-cultured with BV2 cells in a 6-well transwell cell culture chamber with a 0.4-μm pore polycarbonate membrane insert (Corning, USA). GBM cells were plated at a density of 2 × 105 cells/cm2 in the upper compartment of the chamber. The lower compartment was loaded with BV2 cells at a density of 5 × 104 cells/cm2. After co-culturing for 24 h, total RNA was extracted from BV2 cells, and the amount of NOS2 mRNA and ARG1 mRNA were quantified by quantitative real-time polymerase chain reaction (qRT-PCR).
RT-PCR analysis
Total RNA was extracted through Hybrid-R (GeneAll, Korea). One microgram of RNA template was reverse-transcribed using a SuperScript III First-Strand Synthesis System (Invitrogen). The qRT-PCR was performed using 100 ng of cDNA solution in a 20 μl reaction mixture containing 10 μl of power SYBR green PCR Master mix (Applied Biosystems, USA), and 1 μl of the forward primer, and 1 μl of the reverse primer. Relative mRNA expression was assessed using the comparative ΔΔCt method. GAPDH was used as an internal standard. The qRT-PCR primers are shown in Table 1.
Table 1.
Primers for qRT-PCR
| Gene | Forward (5′-3′) | Reverse (5′-3′) | Accession No. |
|---|---|---|---|
| CXCR4 | ACTACACCGAGGAAATGGGCT | CCCACAATGCCAGTTAAGAAGA | NM_001008540.2 |
| CXCL12 | CGCCAGAGCCAACGTCAAG | CTTGTTTAAAGCTTTCTCCAGGTAC | NM_013655.4 |
| hGAPDH | GTCTCCTCTGACTTCAACAGCG | ACCACCCTGTTGCTGTAGCCAA | NM_002046.7 |
| NOS2 | GTTCTCAGCCCAACAATACAAGA | GTGGACGGGTCGATGTCAC | NM_010927.4 |
| ARG1 | CTCCAAGCCAAAGTCCTTAGAG | AGGAGCTGTCATTAGGGACATC | NM_007482.3 |
| mGAPDH | AACTTTGGCATTGTGGAAGG | GGATGCAGGGATGATGTTCT | NM_001289726.1 |
Western blot
GBM cells were homogenized in a buffer consisting of 10 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.1% SDS, and protease inhibitors. The homogenates were centrifuged at 14,000 rpm for 15 min at 4°C. The supernatant was collected, and the total protein contents were determined using the BCA protein assay kit (Pierce, USA). Equal amounts of total protein were separated by SDS-PAGE and transferred to PVDF membranes (Bio-Rad, USA). The membranes were blocked in 5% nonfat milk for 1 h at room temperature followed by an overnight incubation at 4°C with primary antibodies for anti-STAT3 (1:1,000; Cell Signaling) and anti-pSTAT3 (1:500; Cell Signaling). Actin was used as a loading control for protein quantification. Membranes were then washed and incubated with a horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature. Protein bands were visualized using the ChemiDoc image system (Bio-Rad). Quantitative results are presented as the ratio of the band intensity of the protein of interest to the band intensity of actin.
Statistical analysis
Statistical analyses were carried out using Sigmaplot (Systat Software, USA). Data were analyzed by Student’s t-test or one-way ANOVA. Significant differences were further evaluated using Tukey’s honest significant difference post-hoc test. A P value < 0.05 was considered statistically significant. All data are presented as mean ± SE.
RESULTS
Comparison of characteristics between LN229/GFP cells and U87/GFP cells
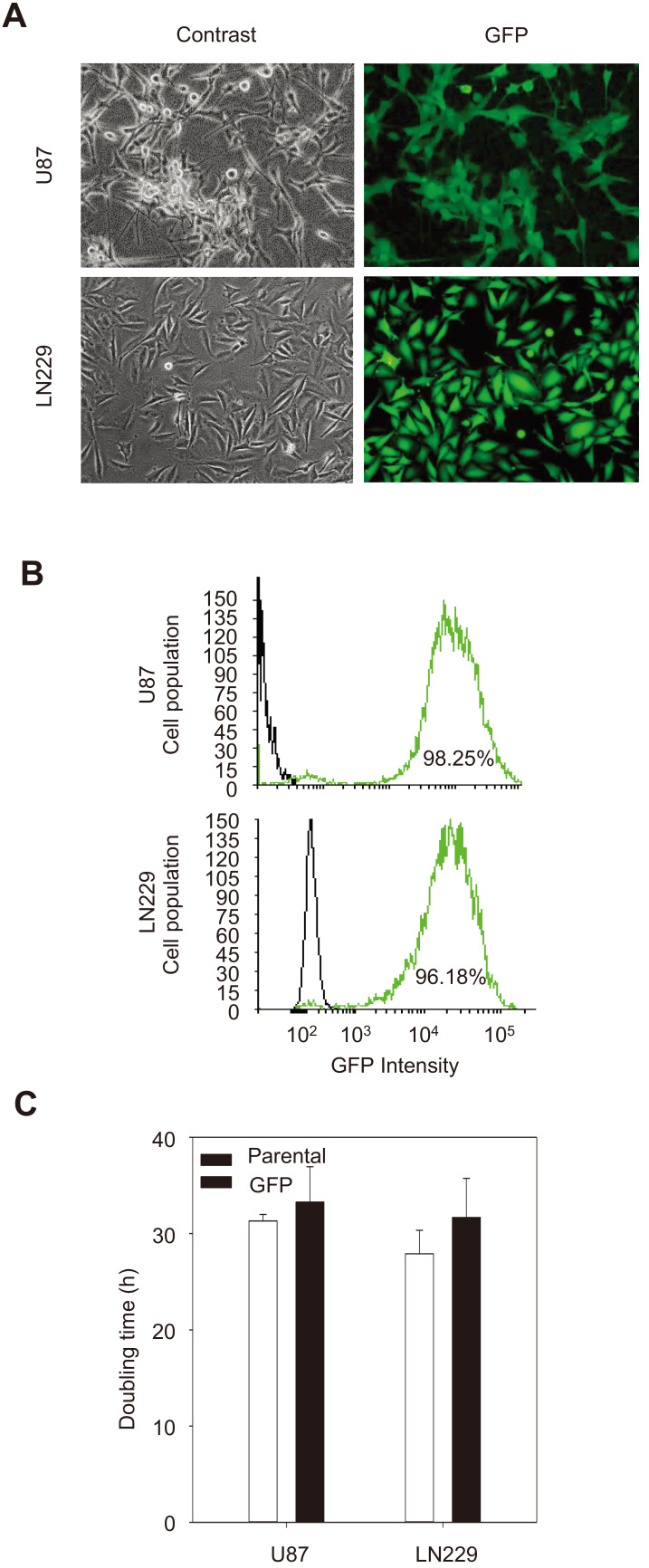
To better monitor the migration/invasion in vivo, the LN229 and U87 cells were labeled with GFP using a lentiviral vector pLL3.7 encoding GFP (Fig. 1A). Transduced cells were sorted using FACS to obtain LN229/GFP and U87/GFP cells, which were then expanded in GM. Both U87 and LN229 cells were labeled with GFP to a similar level (98% vs 96%) (Fig. 1B). Importantly the doubling time of both GBM cell lines was not altered by transduction and FACS sorting (Fig. 1C).
Fig. 1. Validation of GFP-expressing GBM cells.
(A) Phase contrast and fluorescence images of U87 and LN229 cells grown in monolayers. Note that the cell morphology is more uniform in LN229 cells (magnification, ×200). (B) Fluorescence-activated cell sorting analysis showing the U87/GFP and LN229/GFP cells had a > 95%. (C) Comparison of doubling times of U87 and LN229 cells before and after transduction with a GFP-expressing lentiviral vector.
Characterization of tumor growth in orthotopic xenograft mouse models
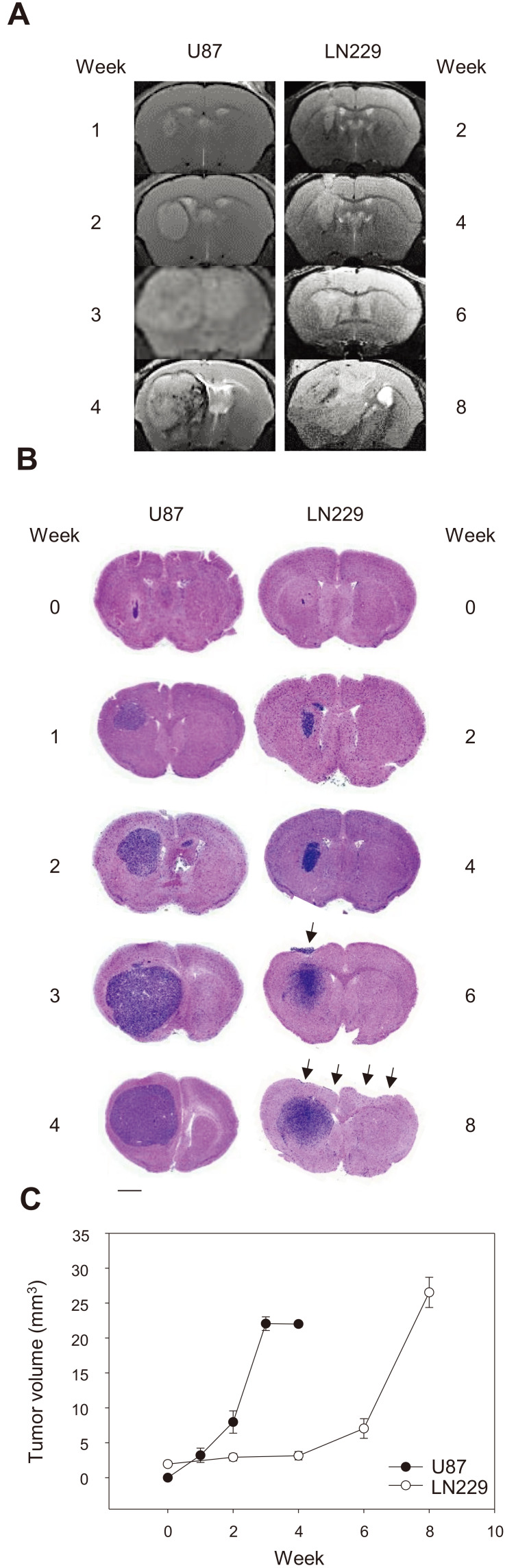
Next, we compared the tumor growth of U87/GFP and LN229/GFP cells in orthotopic GBM models. An MRI analysis of the GBM models revealed that U87/GFP cells grew rapidly and the growth peaked at 3 weeks. By comparison, LN229/GFP cells grew at a slower rate than U87/GFP cells and reached a peak at 8 weeks (Figs. 2A and 2B). We verified the MRI results with classical histological analysis. Simple H&E staining revealed the densely stained basophilic tumor mass in both GBM models. As shown by MRI, U87/GFP cells grew more compactly with demarcated margins. By comparison, LN229/GFP cells grew in a disperse manner from the main tumor and sometimes out of the meninges, which was easily lost during preparation of paraffin samples (arrows, Figs. 2B and 3B).
Fig. 2. Tumor growth in two GBM cell lines in orthotopic xenograft mouse models.
(A) T2-weighted MRI scans of mouse brains at the indicated time points after tumor transplantation. (B) H&E staining of brain sections showing the tumor xenografts of U87/GFP and LN229/GFP cells. Note the ectopic location of LN229/GFP cells (arrows). Scale bar = 1 mm. (C) Quantification of histological images at the indicated time showing the tumor volume as mean ± SE (3 representative sections per animal and 3 animals per group).
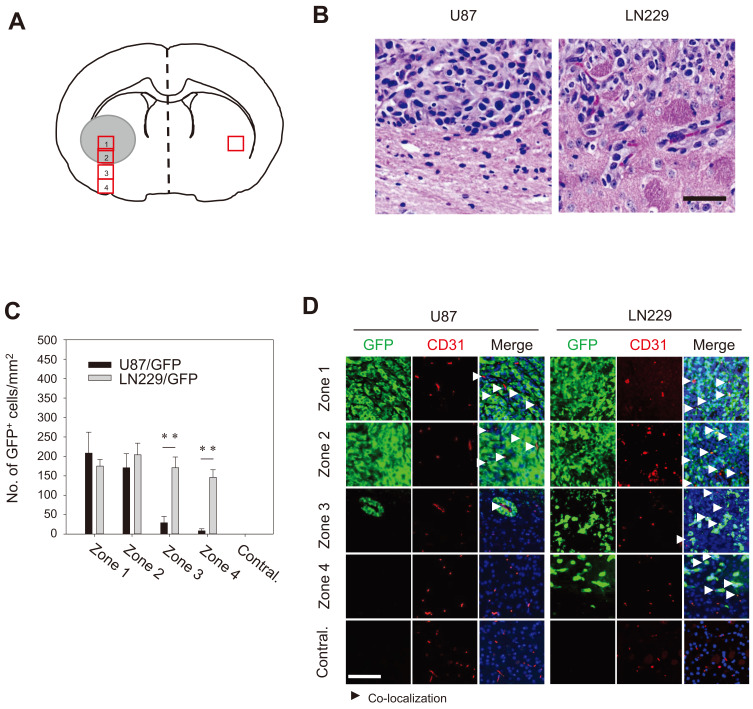
Fig. 3. Distinct distribution of GBM around the tumor center.
(A) A schematic presentation for region of interest showing zones 1 to 4 (red rectangles) were serially arranged in a 200-µm distance from the tumor center. Contralateral (Contral.) area corresponding to zone 2 in the contralateral hemisphere. (B) Bright field images of H&E-stained brain sections. Note the demarcating margin between zones 2 and 3 in U87/GFP–bearing tumors (2 weeks). The margin between zones 2 and 3 was diffuse for LN229/GFP cells (6 weeks). Scale bar = 50 µm. (C) Quantification of the number of GFP-positive cells in the proximity of blood vessel cells. The data are presented as mean ± SE per mm2 obtained from three animals per group. **P < 0.01. (D) The brain sections were immunostained for the expression of GFP and CD31 (an endothelial cell marker) in the indicated area. Arrowheads indicate co-localization/proximity of GFP- and CD31-positive cells. Note that dispersion of LN229/GFP cells along blood vessels in zones 3 and 4. Scale bar = 100 µm.
Tumor cell migration along blood vessels
Perivascular growth is the most common characteristics of GBM (Burden-Gulley et al., 2011; Cheng et al., 2013), which is achieved by migration and distribution of GBM cells to/near endothelial cells. The brain regions were divided into 1 to 4 zones from the tumor inoculation site to the ventral surface of the brain intratumoral area (zone 1), tumor-associated boundary (zone 2), and remote areas (zone 3 and zone 4) (Fig. 3A). H&E staining revealed the demarcating boundaries between zones 2 and 3 for U87/GFP cells but diffused boundaries for LN229/GFP cells. To determine whether the demarcating margin of U87/GFP cells and the diffused nature of LN229/GFP cells were due to the intrinsic tropism toward/along blood vessels, the brain sections from U87/GFP and LN229/GFP xenografts were stained with an antibody specific for the endothelial cell marker CD-31. U87/GFP cells were found together with blood vessels within zones 1 and 2 but scarcely found in zones 3 and 4. It was evident that U87/GFP cells were found closely associated with CD-31-positive vasculature in zone 3. By comparison, LN229/GFP cells were found not only in zone 1 and 2 but also abundantly in zones 3 and 4 (Figs. 3B and 3C). LN229/GFP cells were found in association with blood vessels in all zones (arrowheads in Fig. 3D). The results indicated that LN229 cells grow at a lower rate than U87 but infiltrate diffusively to disperse in a wide range.
Comparison of GBM cells migration and invasion activity
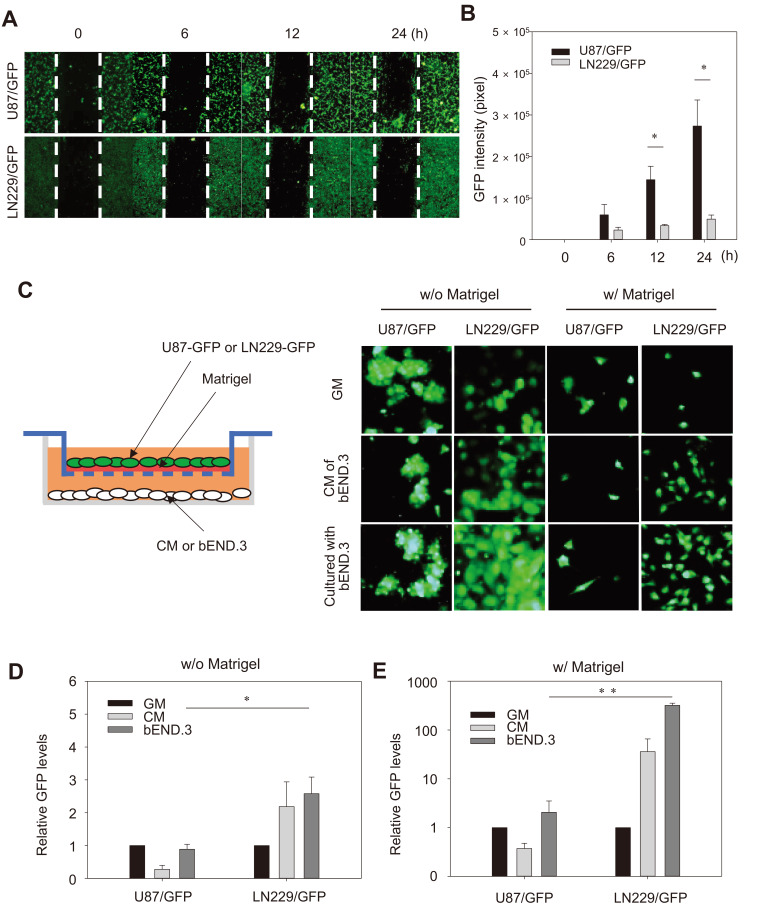
To test the intrinsic migration activity of GBM cells, a scratch assay using a pipette tip was performed to investigate whether the dispersal difference between the two GBM cells along the blood vessel was due to differences in intrinsic migratory activity. For U87/GFP cells, the GFP intensity in the scratched area increased significantly in a time-dependent manner, whereas it remained minimally for LN229/GFP cells (Cheng et al., 2013 Figs. 4A and 4B). This result indicates that U87 cells had higher intrinsic migratory activity than LN229 cells.
Fig. 4. In vitro migration and invasion assays.
(A) Fluorescence microscopic images of wound closure of U87/GFP and LN229/GFP cells cultured to a monolayer at the indicated time after scratching with a pipette. Scale bar = 400 µm. (B) Quantification of GFP intensity in the scratched area at the indicated time. Results from three independent experiments are presented as mean ± SE. *P < 0.05 compared to LN229/GFP at indicated time. (C) Schematic illustration of the transwell invasion assays. Representative fluorescence images of U87/GFP and LN229/GFP cells with (w/) or without (w/o) Matrigel in the upper chamber and bEND.3 cells or CM in the lower chamber. (D and E) The intensity of GFP levels measured at the bottom surface of membrane of the upper chamber. The data are presented as mean ± SE from three independent experiments. *P < 0.05, **P < 0.01.
To assess the invasiveness of GBM cells, a transwell invasion asssay was performed with or without Matrigel as an extracellular matrix (ECM) to better mimic in vivo conditions. The GFP-labeled GBM cells were plated in the upper chamber with a brain endothelial cell line while, bEND.3 cells or the CM of bEND.3 cells were added in the lower chamber (Fig. 4C). In the absence of Matrigel, co-culture with bEND.3 cells or its CM in the lower chamber enhanced migration of LN229/GFP cells compared to U87/GFP cells. The enhanced activity was specific to bEND.3 cells since the GM was less effective. The GFP levels decreased significantly by addition of Matrigel in the upper chamber for both GFP-labeled GBM cells (Figs. 4C and 4D). Importantly, co-culture with bEND.3 cells or its CM markedly increased the LN229/GFP cells, whereas they did not affect migration of U87/GFP (Figs. 4C and 4E). The results suggest that although intrinsic migratory activity was higher in U87 cells than LN229 cells in the absence of any stimuli, the invasion activity through ECM particularly toward the endothelial cells was much higher in LN229. The result also implies the presence of chemoattractants in the CM that interact with LN229 cells and promote its migration.
CXCL12 from bEND.3 cells promote migration of GBM cells
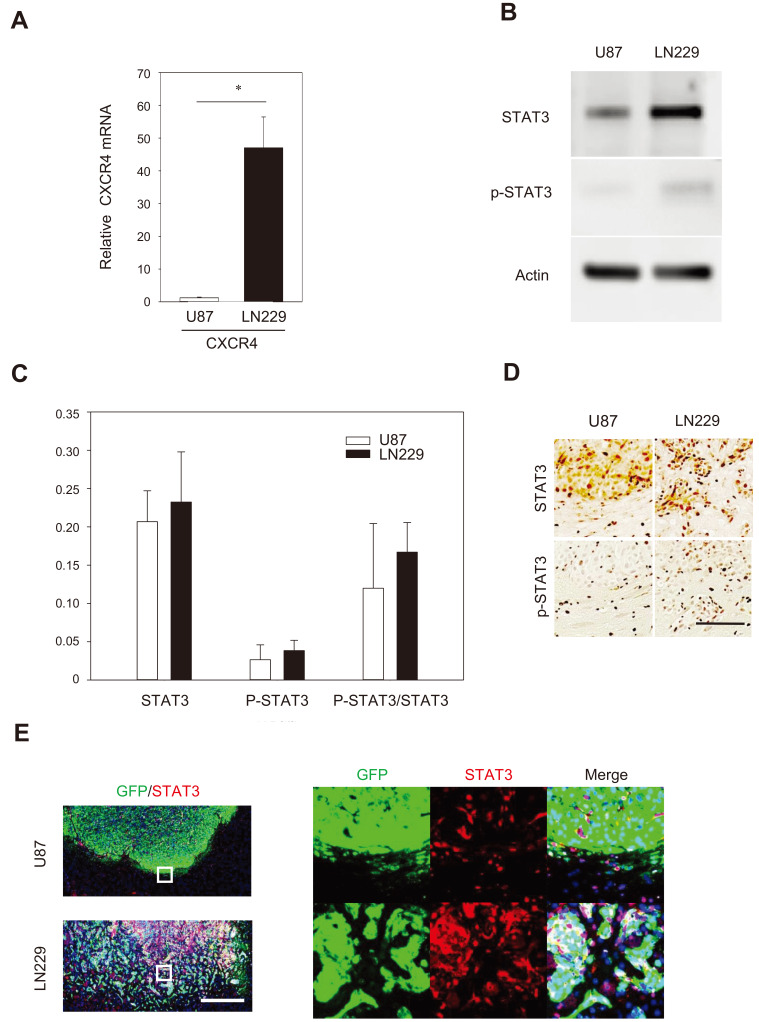
Recently, it was reported that endothelial cells promote migration of human GBM tumor cells by direct interaction through the chemokine CXCL12 (also known as stromal cell-derived factor 1), and its receptor CXCR4 (Rao et al., 2012; Yadav et al., 2016). CXCL12 was expressed to a level comparable to a housekeeping GAPDH in bEND.3 cells since the Ct value of CXCL12 was 2.5-fold higher than that of GAPDH in a RT-PCR analysis.
To assess whether the CXCL12-CXCR4 signaling differ in the two GBM cell lines, we investigated the expression levels of CXCR4 mRNA in LN229 and U87 cells. CXCR4 mRNA expression was higher in LN229 cells compared to U87 cells (Fig. 5A). It has been well established that the JAK2/STAT3 pathway initiates transcription of multiple cancer-associated genes promoting cell proliferation, migration, and invasion of neoplastic cells (Luwor et al., 2013).
Fig. 5. Comparison of expression of genes and proteins related to the interaction between GBM cells and brain-endothelial cells.
(A) High expression of CXCR4 in LN229/GFP cells. The data is presented as mean ± SE from three independent experiments. *P < 0.05. (B) Representative western blot analysis showing the endogenous expression of STAT3 and p-STAT3 in U87 and LN229 cells. (C) Densitometry analysis of STAT3 and p-STAT3 immunoblots showing no significant difference between GBM cell lines. (D) The STAT3 and p-STAT3 immunoreactivity in the zones 2 and 3 of GBM-bearing tumors. Scale bar = 100 µm. (E) Double immunostaining for the presence of STAT3 (red) and GFP (green) showing co-localization (or proximity) of GFP and STAT3 immunoreactivity in the LN229/GFP-bearing tumors. Scale bar = 500 µm.
STAT3 can be activated by CXCL12-CXCR4 (Ahr et al., 2005; Vila-Coro et al., 1999), we assessed STAT3 expression in two GBM cell lines by western blot analysis. Without activation in vitro, the expression levels of STAT3 and pSTAT3 were slightly higher in LN229 cells than U87 cell; however, the difference was insignificant (Figs. 5B and 5C). To assess STAT3 expression in vivo in GBM models, the brain tumor sections bearing U87/GFP and LN229/GFP cells were analyzed using anti-STAT3 and anti-pSTAT3 antibodies. The immunohistochemistry analysis revealed that level of both STAT3 and pSTAT3 were higher in the tumor bearing LN229/GFP cells than the tumor bearing U87/GFP cells (Fig. 5D). Double immunohistochemistry analysis indicated that high STAT3 immunoreactivity was colocalized with GFP signals in LN229/GFP-bearing tumors (Fig. 5E). By comparison, the STAT3 expression was weak in most U87/GFP-bearing tumors (Fig. 5E). The results suggest that LN229 cells respond to interact with the brain microenvironment in response to CXCL12.
Functional analysis of microglia responses to the GBM cells
In the TME, GBM shows considerable accumulation of resident microglia and peripheral macrophages, and their polarization into tumor-supporting cells (Dello et al., 2017; Lisi et al., 2017). Recent evidence suggests that exposure to glioma-derived chemokines induce constitutive STAT3 activation in microglia, with consequent suppression of antitumor mechanisms or even tolerance to tumor antigens (Hussain et al., 2006; Kostianovsky et al., 2008; Zhang et al., 2009). Conversely STAT3 has been shown to be constitutively active in a large percentage of GBM tissue and inactivation of STAT3 in GBM cell line causes microglia suppression (Luwor et al., 2013).
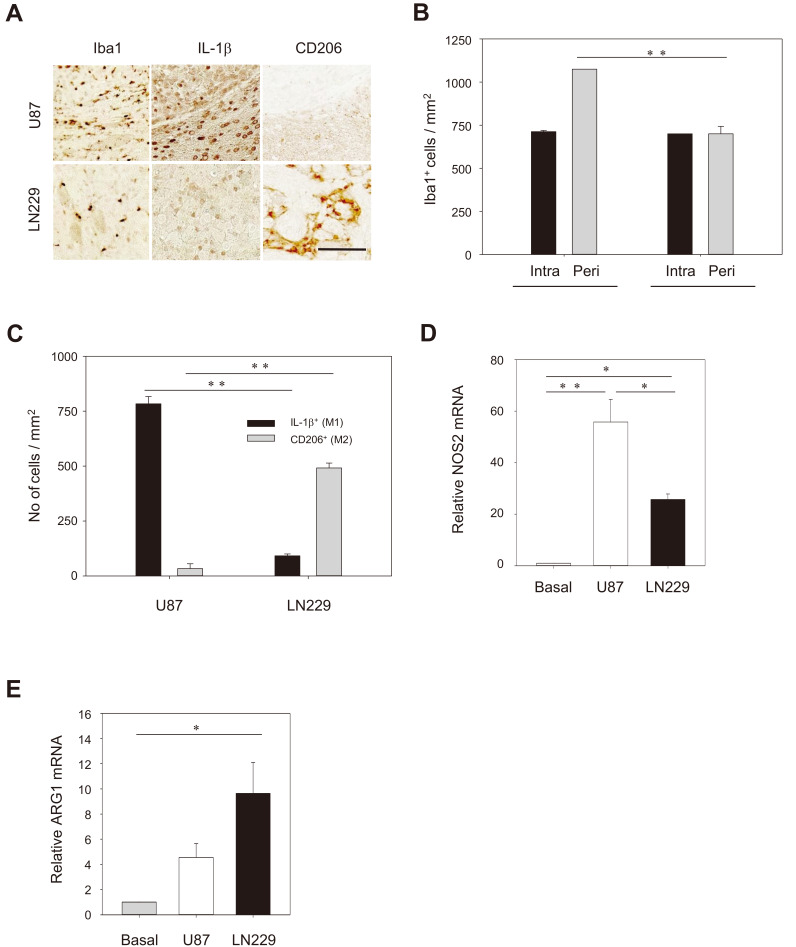
To assess whether different levels of STAT3 in U87 and LN229 cells in the brain tumors influences microglia in the TME, we evaluated M1- and M2-type microglia. Iba1-positive microglia was found in the intra-tumoral area (zone 2) for both U87 and LN229-bearing tumors whereas more microglia were found in the peri-tumoral region (boundary between zones 2 and 3) for LN229 than U87-bearing tumors (Vila-Coro et al., 1999 Figs. 6A and 6B). Interestingly, higher IL-1β–positive immunoreactivity was observed in the peritumoral region of U87-bearing tumors whereas more CD206-positive immunoreactivity was high in LN229-bearing tumors (Fig. 6A). Quantitative analysis indicated that the number of IL-1β–positive M1 types was higher in U87-bearing tumors whereas the number of CD206-positive M2 type was higher in LN229-bearing tumors (Fig. 6C). To determine the interaction of GBM cells and microglia, we co-cultured each of GBM cells in a transwell chamber with the BV2 microglial cell line, and performed a RT-PCR analysis. The mRNA level of NOS2, a M1-type marker, was dramatically increased in BV2 cells by 58- and 25-fold following co-culturing with U87 and LN229, respectively (Fig. 6D). By comparison, the mRNA level of ARG1, a M2-type marker, was significantly upregulated in BV2 cells profoundly after co-culturing with LN229 (10-fold) but to a lesser degree with U87 cells (4.2-fold) (Fig. 6E). These results suggest that LN229 cells, unlike U87 cells, may produce a polarization factor that induces conversion of microglia and macrophage to M2 type in the invasion process.
Fig. 6. GBM-induced polarization of microglia/macrophages.
(A) Immunostaining with anti-Iba1 showing the higher level of microglia/macrophages in the peri-tumoral area (a boundary between zones 2 and 3) with U87/GFP cells compared to the LN229/GFP cells. Note the difference for the IL-1β (a M1 type marker), and CD206 (a M2 type marker) in the peri-tumoral region. Scale bar = 100 µm. (B and C) Quantification of the number of cells that were immunoreactive to antibodies specific for Iba1 (B) and IL-1β and CD206 (C). The data are presented as mean ± SE per mm2 obtained from three animals per group. **P < 0.01. (D and E) RT-PCR analysis showing differential gene expression of NOS2 and ARG1 in BV2 cells following co-culture with U87/GFP or LN229/GFP cells. The data present mean ± SE from three independent experiments. *P < 0.05, **P < 0.01.
DISCUSSION
In this study, two types of GBM cell lines (U87 and LN229) that grow distinctively in the brain were utilized to delineate the cellular interactions of neoplastic cancers with surrounding TME using orthotopic xenograft mouse models.
The U87MG cell line that can be obtained from ATCC was derived from a male patient of unknown age. During the past few decades, tremendous preclinical studies aiming to develop GBM therapy have utilized brain orthotopic models that contain U87 cells as human glioma cells. U87 glioma cells grow with demarcated circumscript tumor margins. Therefore, there have been several concerns with respect to U87 cells because it can represent diffusive human glioma cells.
LN229 cell line was isolated from patients with GBM (Van Meir et al., 1990). This cell line has an epithelial morphology and exhibit mutations in p53, p16, and p14ARF. LN229 cells grow with diffusive margins and represent human GBM more closely than U87 cells. The orthotopic xenograft mouse model with U87 and LN229 cells may provide useful information to study the diffusive nature of glioma cells in addition to neoplastic characteristics. Understanding the functions of the myeloid lineage cells in the xenograft is necessary to precisely predict patient outcomes in the drug development (Masuda et al., 2018). Most studies were performed with one of the GBM cell lines independently of the other GBM cell lines. Here, we compared two GBM cell lines side by side with respect to the growth rates to form the tumor mass, diffusive nature of tumor margin, migration along the blood vessels, and interaction with resident microglia.
U87 cells grow rapidly to form the tumor mass compared to LN229 cells in the brain of an immune compromised nude mice (Fig. 2). U87 cells are compactly packed in the tumor mass with clear, demarcating tumor margins so that they are scarcely found in the remote area (zones 3 and 4) from the tumor center (Fig. 2). Our findings of the compact nature of U87 cells are consistent with other finding (Yu et al., 2018), and also suggest that U87 cells may not reflect clinical characteristics of GBM, Therefore, cautions should be exercised to interpret the data obtained from in vivo study with U87 (Delgado-López et al., 2020; Urbańska et al., 2014) although U87 still provides attractive in vitro GBM tumor models for drug screening.
By comparison, LN229 cells migrate significantly along blood vessels and are abundantly found in a disperse manner in zones 3 and 4 (Fig. 2), suggesting that they may respond to chemoattractants produced by brain endothelial cells. A number of migration-promoting molecules have been identified in dispersive GBM cells and brain endothelial cells in vivo (Armento et al., 2017; Bougnaud et al., 2016). In our in vitro assays to test intrinsic migration activity, U87 cells showed higher activity than LN229 cells. However, LN229 cells exhibited higher invasion activity than U87 cells in the presence of ECM and bEND.3 (Fig. 4). The bEND.3 cells are a brain-derived endothelial cell line that is known to express CXCL12, which verified by RT-PCR assays (Mueller et al., 2014). CXCR4 is the prominent receptor for CXCL12 and is also abundantly found in LN229 cells (Fig. 5A). The results suggest that LN229 cells migrate along the blood vessels in response to CXCL12 secreted by brain endothelial cells. Previous studies have shown that ionizing radiation increases CXCL12 expression via epigenetic modulation of its promoter and thereby increases the aggressiveness in the hepatocellular carcinoma model (Ahn et al., 2019). It would be of interest to delineate the role of the CXCL12-CXCR4 axis in recurrent GBM since X-ray irradiation is a part of a standard of care of GBM (Kazda et al., 2018).
In the TME of GBM, resident microglia are activated and peripheral macrophages are infiltrated from the systemic blood (Dello et al., 2017; Lisi et al., 2017). CXCL12 is also known to function as a chemokine and promote microglia/macrophages trafficking in gliomas (Kioi et al., 2010). In the TME, microglia/macrophages and tumor cells orchestrate to establish an immunosuppressive microenvironment, in which microglia/macrophages acquire alternatively activated (M2) phenotypes. The M2 type-cells further contributes to the local immunosuppression and thereby support tumor growth and invasion (Gjorgjevski et al., 2019; Walentynowicz et al., 2018). Our results showed that the number of Iba-1 positive microglia/macrophages in the intra-tumoral area was similar between two glioma cells but was significantly different in the peri-tumoral area (Fig. 6). Accordingly, the STAT3 expression was higher in the peri-tumoral area of LN229-bearing tumors (Fig. 5). STAT3 in GBM cells can induce microglia suppression (Luwor et al., 2013). Indeed, the M2 polarization of microglia/macrophage was much higher at the peri-tumoral area with LN229 cells compared to the U87 cells (Fig. 6). These results suggest that LN229 cells, unlike U87 cells, may promote the polarization of microglia and macrophage to the M2 type during invasion process. The M2-polarization by LN229 was verified by reduced expression of IL-1β and increased expression of Arg-1 in co-culture assays with BV2, a microglial cell line (Fig. 6).
In summary, we investigated the distinct nature of tumor growth as well as, diffusive nature, migration, invasion, and polarization of microglia/macrophages at the cellular and animal levels using two distinct GBM cell lines. The studies including our results provide a better understanding of the interactions of the GBM with the microenvironment and can eventually lead to the development of therapeutics by that regulate the dispersive nature and tumor suppressive environment of the GBM.
ACKNOWLEDGMENTS
This study was supported by the Ministry of Trade, Industry & Energy (MOTIE, Korea) under Biomedical Equipment-Industrial Technology Innovation Program (No. 10067378 to S.S.K.); and the Ministry of Food and Drug Safety in 2020 (18172MFDS182-5).
Footnotes
AUTHOR CONTRIBUTIONS
J.H. performed experiments. J.H., J.S.Y., D.Y.C., K.G.C., J.L., and S.S.K. analyzed the data. S.S.K. and H.S.K. supervised the research. J.H., J.S.Y., and H.S.K. wrote the manuscript. All authors read and approved the manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Ahn H.J., Hwang S.Y., Nguyen N.H., Lee I.J., Lee E.J., Seong J., Lee J.S. Radiation-induced CXCL12 upregulation via histone modification at the promoter in the tumor microenvironment of hepatocellular carcinoma. Mol. Cells. 2019;42:530–545. doi: 10.14348/molcells.2019.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahr B., Denizot M., Robert-Hebmann V., Brelot A., Biard-Piechaczyk M. Identification of the cytoplasmic domains of CXCR4 involved in Jak2 and STAT3 phosphorylation. J. Biol. Chem. 2005;280:6692–6670. doi: 10.1074/jbc.M408481200. [DOI] [PubMed] [Google Scholar]
- Allen M., Bjerke M., Edlund H., Nelander S., Westermark B. Origin of the U87MG glioma cell line: good news and bad news. Sci. Transl. Med. 2016;8:354re3. doi: 10.1126/scitranslmed.aaf6853. [DOI] [PubMed] [Google Scholar]
- Armento A., Ehlers J., Schötterl S., Naumann U. Molecular mechanisms of glioma cell motility. In: S.D. Vleeschouwer., editor. Glioblastoma. Codon Publications; Brisbane, Australia: 2017. pp. 73–94. [DOI] [PubMed] [Google Scholar]
- Bougnaud S., Golebiewska A., Oudin A., Keunen O., Harter P.N., Mäder L., Azuaje F., Fritah S., Stieber D., Kaoma T., et al. Molecular crosstalk between tumour and brain parenchyma instructs histopathological features in glioblastoma. Oncotarget. 2016;7:31955–31971. doi: 10.18632/oncotarget.7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burden-Gulley S.M., Qutaish M.Q., Sullivant K.E., Lu H., Wang J., Craig S.E.L., Basilion J.P., Wilson D.L., Brady-Kalnay S.M. Novel cryo-imaging of the glioma tumor microenvironment reveals migration and dispersal pathways in vivid three-dimensional detail. Cancer Res. 2011;71:5932–5940. doi: 10.1158/0008-5472.CAN-11-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Huang Z., Zhou W., Wu Q., Donnola S., Liu J.K., Fang X., Sloan A.E., Mao Y., Lathia J.D., et al. Glioblastoma stem cells generate vascular pericytes to support vessel function and tumor growth. Cell. 2013;153:139–152. doi: 10.1016/j.cell.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-López P.D., Saiz-López P., Gargini R., Sola-Vendrell E., Tejada S. A comprehensive overview on the molecular biology of human glioma: what the clinician needs to know. Clin. Transl. Oncol. 2020 Mar 28; doi: 10.1007/s12094-020-02340-8. [Epub]. https://doi.org/10.1007/s12094-020-02340-8. [DOI] [PubMed] [Google Scholar]
- Dello R.C., Lisi L., Tentori L., Navarra P., Graziani G., Combs C.K. Exploiting microglial functions for the treatment of glioblastoma. Curr. Cancer Drug Targets. 2017;17:267–281. doi: 10.2174/1568009616666160813191240. [DOI] [PubMed] [Google Scholar]
- Gjorgjevski M., Hannen R., Carl B., Li Y., Landmann E., Buchholz M., Bartsch J.W., Nimsky C. Molecular profiling of the tumor microenvironment in glioblastoma patients: correlation of microglia/macrophage polarization state with metalloprotease expression profiles and survival. Biosci. Rep. 2019;39:BSR20182361. doi: 10.1042/BSR20182361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber M.B., Scheithauer B., Kreutzberg G. Microglia in brain tumors. Glia. 2002;40:252–259. doi: 10.1002/glia.10147. [DOI] [PubMed] [Google Scholar]
- Hambardzumyan D., Gutmann D.H., Kettenmann H. The role of microglia and macrophages in glioma maintenance and progression. Nat. Neurosci. 2016;19:20–27. doi: 10.1038/nn.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S.F., Yang D., Suki D., Aldape K., Grimm E., Heimberger A.B. The role of human glioma-infiltrating microglia/macrophages in mediating antitumor immune responses. Neuro Oncol. 2006;8:261–279. doi: 10.1215/15228517-2006-008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazda T., Dziacky A., Burkon P., Pospisil P., Slavik M., Rehak Z., Jancalek R., Slampa P., Slaby O., Lakomy R. Radiotherapy of glioblastoma 15 years after the landmark Stupp's trial: more controversies than standards? Radiol. Oncol. 2018;52:121–128. doi: 10.2478/raon-2018-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kioi M., Vogel H., Schultz G., Hoffman R.M., Harsh G.R., Brown J.M. Inhibition of vasculogenesis, but not angiogenesis, prevents the recurrence of glioblastoma after irradiation in mice. J. Clin. Invest. 2010;120:694–705. doi: 10.1172/JCI40283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostianovsky A.M., Maier L.M., Anderson R.C., Bruce J.N., Anderson D.E. Astrocytic regulation of human monocytic/microglial activation. J. Immunol. 2008;181:5425–5432. doi: 10.4049/jimmunol.181.8.5425. [DOI] [PubMed] [Google Scholar]
- Lisi L., Ciotti G.M., Braun D., Kalinin S., Currò D., Dello R.C., Coli A., Mangiola A., Anile C., Feinstein D.L., et al. Expression of iNOS, CD163 and ARG-1 taken as M1 and M2 markers of microglial polarization in human glioblastoma and the surrounding normal parenchyma. Neurosci. Lett. 2017;645:106–112. doi: 10.1016/j.neulet.2017.02.076. [DOI] [PubMed] [Google Scholar]
- Luwor R.B., Stylli S.S., Kaye A.H. The role of Stat3 in glioblastoma multiforme. J. Clin. Neurosci. 2013;20:907–911. doi: 10.1016/j.jocn.2013.03.006. [DOI] [PubMed] [Google Scholar]
- Masuda J., Shigehiro T., Matsumoto T., Satoh A., Mizutani A., Umemura C., Saito S., Kijihira M., Takayama E., Seno A., et al. Cytokine expression and macrophage localization in xenograft and allograft tumor models stimulated with lipopolysaccharide. Int. J. Mol. Sci. 2018;19:E1261. doi: 10.3390/ijms19041261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller A.M., Yoon B.H., Sadiq S.A. Inhibition of hyaluronan synthesis protects against central nervous system (CNS) autoimmunity and increases CXCL12 expression in the inflamed CNS. J. Biol. Chem. 2014;289:22888–22899. doi: 10.1074/jbc.M114.559583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail D.F., Joyce J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S., Sengupta R., Choe E.J., Woerner B.M., Jackson E., Sun T., Leonard J., Piwnica-Worms J., Rubin J.B. CXCL12 mediates trophic interactions between endothelial and tumor cells in glioblastoma. PLoS One. 2012;7:e33005. doi: 10.1371/journal.pone.0033005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakkar J.P., Dolecek T.A., Horbinski C., Ostrom Q.T., Lightner D.D., Barnholtz-Sloan J.S., Villano J.L. Epidemiologic and molecular prognostic review of glioblastoma. Cancer Epidemiol. Biomarkers Prev. 2014;23:1985–1996. doi: 10.1158/1055-9965.EPI-14-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbańska K., Sokołowska J., Szmidt M., Sysa P. Glioblastoma multiforme: an overview. Contemp. Oncol. (Pozn). 2014;18:307–312. doi: 10.5114/wo.2014.40559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meir E., Diserens A.C., Ilamou M.F., Tribolet N. Human glioblastoma cells release interleukin 6 in vivo and in vitro. Cancer Res. 1990;50:6683–6688. [PubMed] [Google Scholar]
- Vila-Coro A.J., Rodríguez-Frade J.M., Martín D.A.A., Moreno-Ortíz M.C., Mellado M. The chemokine SDF-1alpha triggers CXCR4 receptor dimerization and activates the JAK/STAT pathway. FASEB J. 1999;13:1699–1710. doi: 10.1096/fasebj.13.13.1699. [DOI] [PubMed] [Google Scholar]
- Walentynowicz K.A., Ochocka N., Pasierbinska M., Wojnicki K., Stepniak K., Mieczkowski J., Ciechomska I.A., Kaminska B. In search for reliable markers of glioma-induced polarization of microglia. Front. Immunol. 2018;9:1329. doi: 10.3389/fimmu.2018.01329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V.N., Zamler D., Baker G.J., Kadiyala P., Erdreich-Epstein A., DeCarvalho A.C., Mikkelsen T., Castro M.G., Lowenstein P.R. CXCR4 increases in-vivo glioma perivascular invasion, and reduces radiation induced apoptosis: a genetic knockdown study. Oncotarget. 2016;7:83701–83719. doi: 10.18632/oncotarget.13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K.K., Taylor J.T., Pathmanaban O.N., Youshani A.S., Beyit D., Dutko-Gwozdz J., Benson R., Griffiths G., Peers I., Cueppens P., et al. High content screening of patient-derived cell lines highlights the potential of non-standard chemotherapeutic agents for the treatment of glioblastoma. PLoS One. 2018;13:e0193694. doi: 10.1371/journal.pone.0193694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Wang X., Wang J., Yi R., Long H., Zhou M., Luo Q., Zhai Z., Song Y., Qi S. The tumorgenicity of glioblastoma cell line U87MG decreased during serial in vitro passage. Cell. Mol. Neurobiol. 2018;38:1245–1252. doi: 10.1007/s10571-018-0592-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Alizadeh D., Van H.M., Kortylewski M., Yu H., Badie B. Stat3 inhibition activates tumor macrophages and abrogates glioma growth in mice. Glia. 2009;57:1458–1467. doi: 10.1002/glia.20863. [DOI] [PubMed] [Google Scholar]