Abstract
To perceive fluctuations in light quality, quantity, and timing, higher plants have evolved diverse photoreceptors including UVR8 (a UV-B photoreceptor), cryptochromes, phototropins, and phytochromes (Phys). In contrast to plants, prokaryotic oxygen-evolving photosynthetic organisms, cyanobacteria, rely mostly on bilin-based photoreceptors, namely, cyanobacterial phytochromes (Cphs) and cyanobacteriochromes (CBCRs), which exhibit structural and functional differences compared with plant Phys. CBCRs comprise varying numbers of light sensing domains with diverse color-tuning mechanisms and signal transmission pathways, allowing cyanobacteria to respond to UV-A, visible, and far-red lights. Recent genomic surveys of filamentous cyanobacteria revealed novel CBCRs with broader chromophore-binding specificity and photocycle protochromicity. Furthermore, a novel Cph lineage has been identified that absorbs blue-violet/yellow-orange light. In this minireview, we briefly discuss the diversity in color sensing and signal transmission mechanisms of Cphs and CBCRs, along with their potential utility in the field of optogenetics.
Keywords: color sensing, cyanobacteria, cyanobacterial phytochromes, cyanobacteriochromes, signal transmission
INTRODUCTION
Most organisms have evolved a number of receptors that can sense environmental fluctuations, which is a critical advantage for survival. Bilin-based photoreceptors are one of the best studied light sensing photoswitches in plants, algae, cyanobacteria, and other bacterial species. Unlike higher plants that employ blue light photoreceptors, cryptochromes, phototropins, and FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 (FKF1) and ZEITLUPE (ZTL) as additional players in various photobiological responses, cyanobacteria mostly utilize bilin phytochromes (Cphs) and cyanobacteriochromes (CBCRs) in various photobiological responses (Hwang et al., 2019; Rockwell and Lagarias, 2020).
Cphs and canonical plant phytochromes (Phys) are modular in structure, with an N-terminal photosensory core module (PCM) and a C-terminal output regulatory module. PAS (Period/Arnt/Single-minded), GAF (cGMP phosphodiesterase/Adenylyl cyclase/FhlA), and PHY (phytochrome-specific) domains form the PCM. While the GAF domain is necessary for forming the bilin adduct, PAS and PHY domains are involved in bilin lyase activity and the stabilization of 15E lit photostate, respectively (Rockwell and Lagarias, 2017). In addition to Cphs, another group of bilin-based photoreceptors known as CBCRs are widespread among cyanobacteria. In contrast to canonical Cphs, CBCRs sense a wide range of light wavelengths including near-ultraviolet (UV), visible, and far-red (FR) spectra, and this sensitivity is enabled only by the GAF domain since CBCRs lack the PAS and PHY domains (Fushimi and Narikawa, 2019). Such absorption of spectral difference is called color (spectral) tuning, which is attributable to varied effective π conjugation length of the bilin chromophore that originates from either the chromophore-binding plasticity of the apoprotein or chromophore-binding geometry of the pocket and ionic environments. The mechanisms underlying such color tunings involve bilin chromophore species and cysteine (Cys) residues in the GAF domain, which are further regulated by different types of residues surrounding the bilin-binding pocket that affect the tilted geometry and protonation status of bilins. Recently, phylogenetic analysis, transient absorption spectroscopy, crystallography, and molecular genetics revealed novel CBCRs and Cphs that exhibit broader chromophore-binding specificity and protochromicity of the lit state. For instance, in a newly established tandem cysteine cyanobacterial phytochrome (TCCP) lineage, a second Cys present within the GAF domain is responsible for blue-violet/yellow-orange photoconversion (Rockwell et al., 2011; Song et al., 2020). In this review, we summarize the biochemical properties and photobiological functions of some Cphs and CBCRs, and based on their properties, we propose future perspectives for the application of Cphs and CBCRs in various fields including optogenetics.
CYANOBACTERIAL PHOTOBIOLOGICAL RESPONSES
Cyanobacteria represent the first organisms on primitive Earth that utilized solar radiation and evolved oxygen to acquire chemical energy (ATP) and the reducing power (NADPH) necessary for cellular processes. The ability of cyanobacteria to harness solar energy exceeds their photosynthetic capability. Cyanobacteria exploit the quality, quantity, and photoperiodicity of solar radiation to perform various photobiological responses including cell growth, chromatic acclimation, phototactic movements, hormogonia development, circadian rhythms, biofilm formation, and UV-absorbing compound production (Table 1 and references therein). These responses are directly or indirectly related to optimal photosynthesis but avoid either limited or excess light environments.
Table 1.
Domain structure and biological functions of cyanobacterial photoreceptors
| Photoreceptora | Signal transmissionb | Response | Organism (reference) | |
|---|---|---|---|---|
AtPhy
|
Single-Cys/GAF (PΦB) | Phosphorelay | Growth and development | Arabidopsis thaliana (Franklin and Quail, 2010) |
BphP
|
Single-Cys/PAS (BV) | Phosphorelay | LH4 synthesis | R. palustri (Evans et al., 2005) |
Cph1
|
Single-Cys (PCB) | Phosphorelay | Growth | Synechocystis (Fiedler et al., 2007) |
ToTCCP
|
Dual-Cys (PCB) | n.a. | n.a. | Tolypothrix PCC 7910 (Song et al., 2020) |
Cph2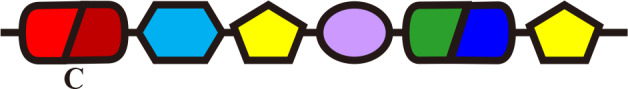
|
Single-Cys (PCB) | 2nd messenger | Growth, phototaxis | Synechocystis (Fiedler et al., 2007; Wilde et al., 2002) |
RcaE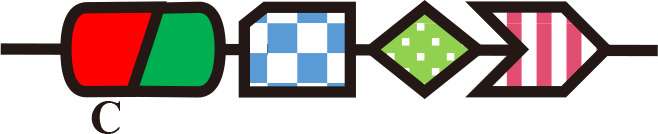
|
Single-Cys (PCB) | Phosphorelay | Chromatic acclimation | F. diplosiphon (Hirose et al., 2013) |
SyCcaS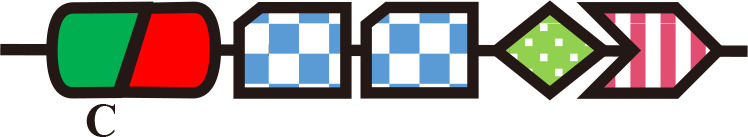
|
Single-Cys (PCB) | Phosphorelay | Chromatic acclimation | Synechocystis (Hirose et al., 2013) |
Slr1393-RGS
|
Single-Cys (PCB) | Phosphorelay | n.a. | Synechocystis (Chen et al., 2012) |
TePixJ
|
Dual-Cys (PVB) | Phosphorelay | Phototaxis | T. elongatus BP-1 (Ishizuka et al., 2007; 2011) |
UirS
|
Dual-Cys (PVB) | Phosphorelay | Phototaxis | Synechocystis (Song et al., 2011) |
SesA
|
Dual-Cys (PVB) | 2nd messenger | Cell aggregation | T. elongatus (Enomoto and Ikeuchi, 2020; Enomoto et al., 2014) |
UGS1
|
Dual-Cys (PCB) | n.a. | n.a. | Microcoleus IPPAS B353 (Cho et al., 2015) |
UGS2
|
Dual-Cys (PCB) | n.a. | n.a. | Microcoleus IPPAS B353 (Cho et al., 2015) |
Anacy_4718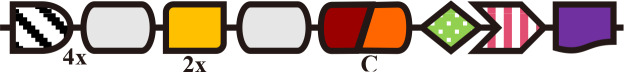
|
Single-Cys (PCB, BV) | n.a. | n.a. | A. cylindrica PCC 7122 (Rockwell et al., 2016) |
| n.a. | n.a. | UV protectant synthesis | Fischerella PCC 9339 (Yang et al., 2018a) | |
PΦB, phytochromobilin; BV, biliverdin; PCB, phycocyanobilin; PVB, phycoviolobilin; , GAF;
, GAF; , histidine (His) kinases, adenylyl cyclases, methyl-accepting proteins, and phosphatases (HAMP);
, histidine (His) kinases, adenylyl cyclases, methyl-accepting proteins, and phosphatases (HAMP); , phytochrome-specific (PHY);
, phytochrome-specific (PHY); , Period/Arnt/Single-minded (PAS)/the C-terminal end of PAS;
, Period/Arnt/Single-minded (PAS)/the C-terminal end of PAS; , His kinase;
, His kinase; , diguanylate phosphodiesterase;
, diguanylate phosphodiesterase; , ATPase domain of His kinase;
, ATPase domain of His kinase; , methyl-accepting domain;
, methyl-accepting domain; , response regulator;
, response regulator; , cystathionine β-synthase;
, cystathionine β-synthase; , receiver domain;
, receiver domain; , diguanylate cyclase;
, diguanylate cyclase; , PAS; n.a., not available.
, PAS; n.a., not available.
Photocycles are represented as color-coded 15Z/15E states; gray color indicates the absence of photocycle.
Experimentally verified signal transmission pathways such as phosphorelay and 2nd messenger are shown.
Growth of the unicellular cyanobacterium Synechocystis PCC 6803 (hereafter referred to as Synechocystis) in red (R) and FR light is controlled by Cph1 and Cph2 in an antagonistic manner. Disruption of Cph1 decreased the growth of Synechocystis under FR light, while disruption of Cph2 decreased its growth under R light (Fiedler et al., 2007). Mutation of Cph2 altered growth rate and biofilm formation in response to trophic condition changes, implying that Cph2 is involved in the regulation of the primary energy metabolism (Schwarzkopf et al., 2014). Similar to other photosynthetic organisms, Synechocystis exhibits differential growth under different light wavelengths, leading to an unbalanced excitation of photosystems, thus hampering the photosynthetic utilization of solar radiation in an unbalanced and inefficient manner. Under such conditions, bilin composition of cyanobacterial light-harvesting antenna phycobilisomes is adjusted, allowing cyanobacteria to balance the proportion of light absorption between two photosystems. This type of response is known as chromatic acclimation (CA). To date, six types of CA have been reported (Sanfilippo et al., 2019; Wiltbank and Kehoe, 2019). Various components of CA include RcaE and DpxA in Fremyella diplosiphon, CcaS in Synechocystis and Nostoc punctiforme, and the R/FR knotless Cph, RfpA in Leptolyngbya sp. JSC-1; all of these four proteins harbor a histidine (His) kinase domain, which acts as the sensor kinase in a signal transduction cascade.
Cyanobacteria lack flagella but exhibit phototaxis in response to variation in light quantity and quality. However, Synechocystis exhibits type IV pili-dependent twitching motility (Bhaya, 2004), while the filamentous cyanobacterium N. punctiforme exhibits gliding motility (Khayatan et al., 2015) to relocate to another location for optimal photosynthesis. In Synechocystis, positive (Yoshihara and Ikeuchi, 2004) and negative (Song et al., 2011) phototaxes are mediated by two different CBCRs, namely, PixJ and UirS, respectively. By contrast, in Synechococcus elongatus, a single 5-GAF domain photoreceptor, PixJse, senses the direction of illumination by wavelengths that induce both positive and negative phototactic movements (Yang et al., 2018b). Orthologs of bacterial methyl-accepting chemotaxis proteins such as SyPixJ (Yoshihara et al., 2004), TePixJ (Ishizuka et al., 2006), and AnPixJ (Narikawa et al., 2008) are mostly involved in such taxis.
Like other microorganisms, cyanobacteria form biofilms, where cells are mostly attached to and grow on a surface and produce extracellular polymers. In Synechocystis, these surfaces include exopolysaccharides, the S-layer and pili (Allen et al., 2019). In filamentous Leptolyngbya and Scytonema, biofilm formation involves hormogonia, whose sticky ends are adhered to a surface (Maldener et al., 2014). Formation of biofilm in Thermosynechococcus is mediated by the bacterial second messenger, cyclic diguanosine monophosphate (c-di-GMP) (Agostoni et al., 2016). Three CBCRs, SesA/B/C, in the blue/green light (ON/OFF)-c-di-GMP switch control sessility and motility in planktonic communities (Enomoto et al., 2014; 2015), where blue/green light penetration is restricted to upper layers (Enomoto and Ikeuchi, 2020).
In addition, cyanobacteria sense diurnal photoperiod to adjust photosynthetic and respiratory activities. In S. elongatus PCC 7942, the circadian clock regulates genes at dusk and dawn in a promoter-dependent manner. Regulation of promoters is period-specific, which in turn leads to diurnal regulation of energy metabolism, cellular division, and chromosome architecture modification. KaiA, the first component of the S. elongatus oscillator (KaiABC), and CikA (circadian input kinase A) PsR domains act as environmental sensors, detecting the redox state of the quinone pool during the transition from day to night (Cohen and Golden, 2015).
Most heterocystous or non-heterocystous filamentous cyanobacteria form hormogonia, i.e., motile short filaments formed during asexual reproduction from vegetative cells or trichomes (Marsac, 1994). Hormogonium differentiation in the cyanobacterium Calothrix sp. PCC 7601 is stimulated by R light and inhibited by green light (Damerval et al., 2007). In N. punctiforme, a methyl-accepting chemotaxis protein (MCP)-like photoreceptor, PtxD, is involved in the phototaxis of hormogonia (Campbell et al., 2015).
Photoinhibitory light environments trigger the production of sunscreen pigments in some cyanobacteria. For instance, in response to high light or UV irradiance, mycosporine-like amino acids and scytonemins (Rastogi et al., 2014) are produced and accumulated in the outer cell wall spaces in halophilic cyanobacteria such as Euhalothece sp. and Microcoleus sp., several Nodularia species, and Scytonema hofmanii (Sinha and Häder, 2008; Yang et al., 2020). Sensing and signaling components of these photobiological responses are likely mediated by bilin photoreceptors, i.e., Cphs and CBCRs, although further investigation is needed.
DIVERSE COLOR SENSING ABILITY OF Cphs AND CBCRs
Since the first cyanobacterial genome of Synechocystis was reported, the discovery of diverse photoreceptors and their color-tuning mechanisms has largely depended on the identification of a vast number of cyanobacterial proteins containing PAS-GAF-PHY domains (Yeh et al., 1997). Whole-genome sequencing of cyanobacterial species including Microcoleus IPAS B373 (Cho et al., 2015), Euhalothece Z-M001 (Yang et al., 2020), and Tolypothrix PCC7910 (Song et al., 2020) revealed the absence of the HY2 gene, which encodes phytochromobilin (PФB) synthase; therefore, PФB, a bilin chromophore covalently bound to canonical Phys in higher plants, is absent in these cyanobacteria. Instead, these cyanobacteria harbor the pcyA gene, which encodes a phycocyanobilin (PCB):ferredoxin oxidoreductase that catalyzes four-electron reduction of biliverdin IXα (BV) to PCB, a major cofactor of Cphs and CBCRs (Fujita et al., 2015; Fushimi and Narikawa, 2019). The number of Cphs and CBCRs varies among cyanobacteria, with three in Euhalothece, eight in Synechocystis, nine in Microcoleus IPAS B353, 12 in the chlorophyll d-containing cyanobacterium Acaryochloris marina, 18 in N. punctiforme, and 36 in Tolypothrix PCC 7910. The total number of bilin photoreceptors in a cyanobacterium is roughly proportional to its genome size (Cho et al., 2015). Additionally, in cyanobacteria, CBCRs are more abundant than Cphs, and the abundance ratio of blue CBCRs to red CBCRs is likely related to light conditions in the natural habitat. For instance, Microcoleus IPAS B353 is enriched in near-UV and violet CBCRs but lacks red/green and green/red CBCRs, suggesting that the enrichment of short wavelength-absorbing CBCRs is critical for acclimation of cyanobacteria to high light environments in their natural habitat (Cho et al., 2015).
The PCB-bound holo-Cphs make spectral difference with PФB- or BV-bound Cphs because of differences in the number of π electrons; PCB has two- or four-electrons less than PФB and BV. Consequently, the spectra of Cph1- and knotless Cph2-bound PCB chromophores show a slight blue shift compared with PФB-bound PhyB and BV-bound BphP (Rockwell and Lagarias, 2010). However, unlike canonical Phys or bacterial-type Phys BphP, the newly identified TCCPs shift their light sensing maximum into the violet spectral region (Rockwell et al., 2011; Song et al., 2020). The color sensing diversity of TCCPs seems to be related to the protonated 15E bilin chromophores with different ionization state of the non-canonical ‘second’ Cys sulfhydryl group (Fig. 1; Song et al., 2020). This expanded range of light absorption by Cphs is also observed in some eukaryotic algal species including Cyanophora paradoxa (Song et al., 2020).
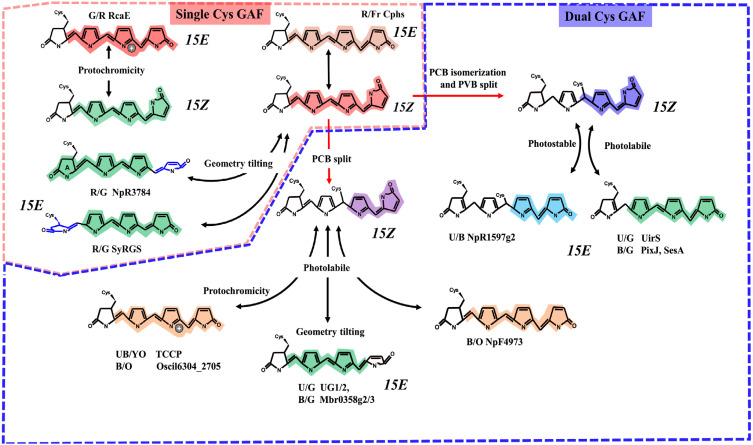
Fig. 1. Color-tuning mechanisms adopted by cyanobacterial phytochromes (Cphs) and cyanobacteriochromes (CBCRs).
Cphs and CBCRs exhibit reversible photoisomerization at C15-C16 (R/FR photocycle). Single-cysteine (Cys) red/green or green/red CBCRs bind to phycocyanobilin (PCB) in the dark state, exhibiting red and green absorption maxima. Upon red or green illumination, green or red light-absorbing forms are formed by lowering effective π-conjugation length via protochromicity of bilin (RcaE) or geometry tilting by the A-ring (SyRGS) or D-ring (NpR3784). A second Cys residue located at the DXCF or CXXR/K motif forms a second thioether linkage at C10, yielding a blue light-absorbing dark state. Some dual-Cys CBCRs isomerize PCB to phycoviolobilin (PVB), forming violet or blue light-absorbing dark state. These adducts are either photolabile or photostable upon photoisomerization, yielding green or blue-absorbing lit states. Such variation is further enhanced by bilin protochromicity or tilted geometry, yielding violet, teal, orange and red -absorbing photoproducts. In dual-Cys cyanobacterial phytochromes, tandem cysteine cyanobacterial phytochrome (TCCP)-specific Cys is responsible for the bilin split, and lit state tuning is related to the protonated lit form with different ionization state of the Cys thiol group. Black double arrows and red forward arrow represent reversible photoisomerization around the C15=C16 double bond of a bilin and second thioether linkage formation with or without bilin isomerization, respectively.
Unlike canonical Phys and Cphs, CBCRs only require the GAF domain(s) to bind to a bilin and then to exhibit photocycle encompassing near-UV to FR light (Ikeuchi and Ishizuka, 2008; Table 1, Fig. 1). Like Cphs, CBCRs are categorized into two subgroups, based on the number of Cys residues in the GAF domain that form covalent adducts to apoproteins (Fushimi and Narikawa, 2019; Rockwell and Lagarias, 2017; Rockwell et al., 2015). Singly- or doubly-bound Cys residues of GAF domains form thioether linkages with the chromophore because of their auto-lyase activity. The canonical Cys residue in the CH motif of the GAF domain binds to the C31 (PCB and PVB) or C32 (BV) atom of bilin (Fushimi and Narikawa, 2019), forming a red/green photocycle, unlike the canonical R/FR photocycle, mostly because of the conserved phenylalanine (Phe) and aspartic acid (Asp) residues that partially deconjugate the D-ring (Rockwell et al., 2014) or A-ring in Slr1393G3 (SyRGS) CBCR (Buhrke et al., 2020), respectively, by trapping bilin in a twisted geometry (Lim et al., 2018; Xu et al., 2020). By contrast, protochromicity of bilin chromophores determine green/red (Hirose et al., 2013) or blue/orange (Sato et al., 2019) photocycles. For instance, the green light-absorbing dark state contains deprotonated chromophore, which is protonated in the R light-absorbing lit state via the involvement of protochromic triad residues (Hirose et al., 2013).
Some CBCRs contain additional Cys residues located in the highly conserved DXCF motif or the weakly conserved CXXR/K motif in the insertion loop (insert-Cys) that forms the second thioether linkage via the C10 atom of their dark states (Cho et al., 2017; Rockwell et al., 2011; 2014). In many dual-Cys CBCRs, this second thioether linkage is unstable and light-labile, forming additional reversible thioether adducts. Accordingly, these CBCRs can sense blue or violet light in the dark state and teal, green, yellow, or orange light in the lit state. In the case of teal-DXCF CBCRs, tilting of the D-ring by Phe residues leads to teal-absorbing photoproducts (Rockwell et al., 2014). Similarly, insert-Cys CBCRs adopt both tuning mechanisms, namely, photolabile second thioether linage and tilted geometry (Cho et al., 2017). By contrast, blue/orange CBCR Oscil6304_2705 combines protochromic and two-Cys photocycles in separate time scales (Sato et al., 2019). Unlike CBCRs above mentioned, the second thioether linkage is formed upon illumination of the dark state, yielding red/blue photocycle (Narikawa et al., 2014).
Like Cphs, cyanobacterial CBCRs mainly bind to PCB. However, some CBCRs also bind to BV (Fushimi et al., 2016; Narikawa et al., 2015), phycoviolobilin (PVB) (Ishizuka et al., 2007; Song et al., 2011; Cho et al., 2015), and PΦB (Rockwell et al., 2016). PVB adduct is formed by the isomerization of a covalent PCB adduct, shortening the π-conjugated system in the dark (Ishizuka et al., 2011; Rockwell et al., 2012). This conversion of PCB into PVB is DXCF-CBCRs subfamily specific (Rockwell et al., 2012) although residue(s) involved have not been characterized yet. In several cases, some CBCRs and Phys covalently bind to porphyrin compounds, which are considered as contaminants when expressed in a heterologous system (Fischer et al., 2005; Rockwell et al., 2016; Wagner et al., 2008). Despite the characterization of such tuning mechanisms, some tuning mechanisms remain unknown among the newly discovered CBCRs such as Fr/O (Rockwell et al., 2016).
Signal transmission diversity
Despite the development of molecular genetic tools in a few model species such as F. diplosiphon, Synechocystis, S. elongatus PCC 7942, and Thermosynechococcus vulcanus, the physiological functions of most CBCRs remain elusive (Wendt and Pakrasi, 2019). Nonetheless, several cyanobacterial photoreceptors have been studied in detail, indicating that signal transmission is accompanied mostly by phosphotransfer or c-di-GMP-initiated downstream pathways (Table 1). Phosphorelay is observed in several signal transmission pathways involved in the autophosphorylation of His residues by His kinases, followed by phosphotransfer to cognate response regulators. For instance, the CBCR UirS is an atypical membrane-bound His kinase, and its cognate AraC family response regulator, UirR, functions as a UV-A-sensing two-component signaling system in Synechocystis (Song et al., 2011). The DNA-binding transcriptional regulator, UirR, targets the adjacent PatA family response regulator, LsiR, which functions as a signal output regulator to reverse the twitching orientation of Synechocystis cells from positive to negative under unidirectional UV-A illumination. Activation of UirR by UirS is mediated by phosphorelay upon the UV-A-dependent activation of UirS (Song et al., 2011). In addition, phototactic motility in Synechocystis, controlled by the blue/green MCP domain-containing SyPixJ (previously known as TaxD; Yoshihara and Ikeuchi, 2004), is subjected to a light-dependent phosphorelay cascade. CA signal transmission is also regulated by phosphorelay between sensors and response regulators. Upon green light illumination, SyCcaS is autophosphorylated, and the phosphate is transferred to its cognate response regulator CcaR (Castillo-Hair et al., 2019; Hirose et al., 2008; 2010).
SesA CBCR induces cell aggregation in T. vulcanus via c-di-GMP signaling (Enomoto et al., 2014; 2015). Blue light-activated SesA activates the GGDEF domain, leading to the generation of c-di-GMP (Enomoto et al., 2014), although the activation mechanism remains elusive. Similarly, the GAF domain of Cph2 activates c-di-GMP signaling involved in blue light-dependent inhibition of positive phototaxis (Wilde et al., 2002). The N-terminal domain of SesA contains two tandem repeats of the cystathionine beta-synthase (CBS) domain that binds to nucleotides such as ATP, ADP, and AMP, which possibly regulate SesA activity at the chromophorylation step. The CBB domain-containing Slr2111 allosterically regulates chromophorylation of the red/green sensor (RGS) Slr1393. ATP-bound CBS-Slr2111 dissociates from RGS, allowing the chromophorylation of Slr1393. By contrast, AMP-bound Slr2111 maintains a strong interaction with Slr1393, hindering chromophorylation of Slr1393 (He et al., 2018). Thus, CBCRs with or without CBS domains seem to be post-translationally activated by cellular energy charge (ATP/[ADP + AMP] ratio), although this hypothesis needs further verification.
PERSPECTIVES
Color and signal transmission diversity of Cphs and CBCRs arise from the presence of light sensing and signal transmitting domain(s) in various combinations. Such a variety of signal input and output domains provides advantages in regulating in situ cellular functions such as photosynthesis, growth, development, and circadian rhythm. Despite the well-characterized color-tuning and signal-relaying mechanisms, the role of multiple GAFs in a single CBCR is largely unknown. Genomes of cyanobacteria such as N. punctiforme and Tolypothrix PCC 7910 encode several multiple-GAF CBCRs. This either represents redundancy for signal addition and amplification, or plays a regulatory role, for instance, in controlling the direction of movement (Yang et al., 2018b). Comparative X-ray crystallography or cryogenic electron microscopy (cryo-EM) image analysis of the whole structure of wild-type and mutant (domain-less) CBCRs in the dark, intermediate, and lit states, and development of genetic transformation toolboxes for naturally untransformed cyanobacterial strains, could help elucidate the unknown biological roles of previously characterized cyanobacterial photoreceptors. Additionally, CBCRs stand out as ideal sources for optogenetic toolboxes (Castillo-Hair et al., 2019; Fushimi et al., 2019; Ramakrishnan and Tabor, 2016), considering the pros and cons of Phys (Rockwell et al., 2016). The broad range of light inputs of CBCRs and signal transmitting pathways seem advantageous in the field of fluorescence probes (Chernov et al., 2017; Oliinyk et al., 2019; Ong et al., 2018) and light-controlled subcellular functions (Milias-Argeitis et al., 2016; Tandar et al., 2019) including protein–protein interactions, gene expression and c-di-GMP/-AMP-dependent signaling cascades (Blain-Hartung et al., 2018; Klausen et al., 2019). The recently characterized porphyrin-binding and pH-sensitive CBCRs are particularly appealing for application in porphyrin sequestration and as fluorescence reporters for diagnosing abnormal cells with varied intra- and inter-cellular pH values.
ACKNOWLEDGMENTS
This work was supported by grants from the Next-Generation BioGreen 21 Program, Rural Development Administration (PJ013118), the KIST Open Research Program (2E30642-20-152), and the Collaborative Genome Program funded by the Ministry of Oceans and Fisheries (20180430), Korea.
Footnotes
AUTHOR CONTRIBUTIONS
Y.V., H.W.Y., and Y.I.P. wrote the manuscript. Y.I.P. secured fundings.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Agostoni M., Waters C.M., Montgomery B.L. Regulation of biofilm formation and cellular buoyancy through modulating intracellular cyclic di-GMP levels in engineered cyanobacteria. Biotechnol. Bioeng. 2016;113:311–319. doi: 10.1002/bit.25712. [DOI] [PubMed] [Google Scholar]
- Allen R., Rittmann B.E., Curtiss R. Axenic biofilm formation and aggregation by Synechocystis sp. strain PCC 6803 are induced by changes in nutrient concentration and require cell surface structures. Appl. Environ. Microbiol. 2019;85:1–33. doi: 10.1128/AEM.02192-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaya D. Light matters: phototaxis and signal transduction in unicellular cyanobacteria. Mol. Microbiol. 2004;53:745–754. doi: 10.1111/j.1365-2958.2004.04160.x. [DOI] [PubMed] [Google Scholar]
- Blain-Hartung M., Rockwell N.C., Moreno M.V., Martin S.S., Gan F., Bryant D.A., Lagarias J.C. Cyanobacteriochrome-based photoswitchable adenylyl cyclases (cPACs) for broad spectrum light regulation of cAMP levels in cells. J. Biol. Chem. 2018;293:8473–8483. doi: 10.1074/jbc.RA118.002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhrke D., Battocchio G., Wilkening S., Blain-Hartung M., Baumann T., Schmitt F.J., Friedrich T., Mroginski M.A., Hildebrandt P. Red, orange, green: light- and temperature-dependent color tuning in a cyanobacteriochrome. Biochemistry. 2020;59:509–519. doi: 10.1021/acs.biochem.9b00931. [DOI] [PubMed] [Google Scholar]
- Campbell E.L., Hagen K.D., Chen R., Risser D.D., Ferreira D.P., Meeks J.C. Genetic analysis reveals the identity of the photoreceptor for phototaxis in hormogonium filaments of Nostoc punctiforme. J. Bacteriol. 2015;197:782–791. doi: 10.1128/JB.02374-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Hair S.M., Baerman E.A., Fujita M., Igoshin O.A., Tabor J.J. Optogenetic control of Bacillus subtilis gene expression. Nat. Commun. 2019;10:3099. doi: 10.1038/s41467-019-10906-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhang J., Luo J., Tu J.M., Zeng X.L., Xie J., Zhou M., Zhao J.Q., Scheer H., Zhao K.H. Photophysical diversity of two novel cyanobacteriochromes with phycocyanobilin chromophores: photochemistry and dark reversion kinetics. FEBS J. 2012;279:40–54. doi: 10.1111/j.1742-4658.2011.08397.x. [DOI] [PubMed] [Google Scholar]
- Chernov K.G., Redchuk T.A., Omelina E.S., Verkhusha V.V. Near-infrared fluorescent proteins, biosensors, and optogenetic tools engineered from phytochromes. Chem. Rev. 2017;117:6423–6446. doi: 10.1021/acs.chemrev.6b00700. [DOI] [PubMed] [Google Scholar]
- Cho S.M., Jeoung S.C., Song J.Y., Kupriyanova E.V., Pronina N.A., Lee B.W., Jo S.W., Park B.S., Choi S.B., Song J.J., et al. Genomic survey and biochemical analysis of recombinant candidate cyanobacteriochromes reveals enrichment for near UV/violet sensors in the halotolerant and alkaliphilic cyanobacterium Microcoleus IPPAS B353. J. Biol. Chem. 2015;290:28502–28514. doi: 10.1074/jbc.M115.669150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.M., Jeoung S.C., Song J.Y., Song J.J., Park Y.I. Hydrophobic residues near the bilin chromophore-binding pocket modulate spectral tuning of insert-Cys subfamily cyanobacteriochromes. Sci. Rep. 2017;7:1–12. doi: 10.1038/srep40576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S.E., Golden S.S. Circadian rhythms in cyanobacteria. Microbiol. Mol. Biol. Rev. 2015;79:373–385. doi: 10.1128/MMBR.00036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damerval T., Guglielmi G., Houmard J., de Marsac N.T. Hormogonium differentiation in the cyanobacterium Calothrix: a photoregulated developmental process. Plant Cell. 2007;3:191–201. doi: 10.2307/3869288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto G., Ikeuchi M. Blue-/green-light-responsive cyanobacteriochromes are cell shade sensors in red-light replete niches. iScience. 2020;23:100936. doi: 10.1016/j.isci.2020.100936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto G., Ni-Ni-Win, Narikawa R., Ikeuchi M. Three cyanobacteriochromes work together to form a light color-sensitive input system for c-di-GMP signaling of cell aggregation. Proc. Natl. Acad. Sci. U. S. A. 2015;112:8082–8087. doi: 10.1073/pnas.1504228112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto G., Nomura R., Shimada T., Win N.N., Narikawa R., Ikeuchi M. Cyanobacteriochrome SesA is a diguanylate cyclase that induces cell aggregation in Thermosynechococcus. J. Biol. Chem. 2014;289:24801–24809. doi: 10.1074/jbc.M114.583674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans K., Fordham-Skelton A.P., Mistry H., Reynolds C.D., Lawless A.M., Papiz M.Z. A bacteriophytochrome regulates the synthesis of LH4 complexes in Rhodopseudomonas palustris. Photosynth. Res. 2005;85:169–180. doi: 10.1007/s11120-005-1369-7. [DOI] [PubMed] [Google Scholar]
- Fischer A.J., Rockwell N.C., Jang A.Y., Ernst L.A., Alan S., Duan Y., Lei H., Lagarias J.C. Multiple roles of a conserved GAF domain tyrosine residue in cyanobacterial and plant phytochromes. Biochemistry. 2005;44:15203–15215. doi: 10.1021/bi051633z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler B., Broc D., Schubert H., Rediger A., Börner T., Wilde A. Involvement of cyanobacterial phytochromes in growth under different light qualitities and quantities. Photochem. Photobiol. 2007;79:551–555. doi: 10.1111/j.1751-1097.2004.tb01275.x. [DOI] [PubMed] [Google Scholar]
- Franklin K.A., Quail P.H. Phytochrome functions in Arabidopsis development. J. Exp. Bot. 2010;61:11–24. doi: 10.1093/jxb/erp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Tsujimoto R., Aoki R. Evolutionary aspects and regulation of tetrapyrrole biosynthesis in cyanobacteria under aerobic and anaerobic environments. Life. 2015;5:1172–1203. doi: 10.3390/life5021172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushimi K., Miyazaki T., Kuwasaki Y., Nakajima T., Yamamoto T., Suzuki K., Ueda Y., Miyake K., Takeda Y., Choi J.H., et al. Rational conversion of chromophore selectivity of cyanobacteriochromes to accept mammalian intrinsic biliverdin. Proc. Natl. Acad. Sci. U. S. A. 2019;116:8301–8309. doi: 10.1073/pnas.1818836116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushimi K., Nakajima T., Aono Y., Yamamoto T., Win N.N., Ikeuchi M., Sato M., Narikawa R. Photoconversion and fluorescence properties of a red/green-type cyanobacteriochrome AM1_C0023g2 that binds not only phycocyanobilin but also biliverdin. Front. Microbiol. 2016;7:588. doi: 10.3389/fmicb.2016.00588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushimi K., Narikawa R. Cyanobacteriochromes: photoreceptors covering the entire UV-to-visible spectrum. Curr. Opin. Struct. Biol. 2019;57:39–46. doi: 10.1016/j.sbi.2019.01.018. [DOI] [PubMed] [Google Scholar]
- He Q., Tang Q.Y., Sun Y.F., Zhou M., Gärtner W., Zhao K.H. Chromophorylation of cyanobacteriochrome Slr1393 from Synechocystis sp. PCC 6803 is regulated by protein Slr2111 through allosteric interaction. J. Biol. Chem. 2018;293:17705–17715. doi: 10.1074/jbc.RA118.003830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y., Narikawa R., Katayama M., Ikeuchi M. Cyanobacteriochrome CcaS regulates phycoerythrin accumulation in Nostoc punctiforme, a group II chromatic adapter. Proc. Natl. Acad. Sci. U. S. A. 2010;107:8854–8859. doi: 10.1073/pnas.1000177107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y., Rockwell N.C., Nishiyama K., Narikawa R., Ukaji Y., Inomata K., Lagarias J.C., Ikeuchi M. Green/red cyanobacteriochromes regulate complementary chromatic acclimation via a protochromic photocycle. Proc. Natl. Acad. Sci. U. S. A. 2013;110:4974–4979. doi: 10.1073/pnas.1302909110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y., Shimada T., Narikawa R., Katayama M., Ikeuchi M. Cyanobacteriochrome CcaS is the green light receptor that induces the expression of phycobilisome linker protein. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9528–9533. doi: 10.1073/pnas.0801826105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang D.Y., Park S., Lee S., Lee S.S., Imaizumi T., Song Y.H. GIGANTEA regulates the timing stabilization of CONSTANS by altering the interaction between FKF1 and ZEITLUPE. Mol. Cells. 2019;42:693–701. doi: 10.14348/molcells.2019.0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M., Ishizuka T. Cyanobacteriochromes: a new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochem. Photobiol. Sci. 2008;7:1159–1167. doi: 10.1039/b802660m. [DOI] [PubMed] [Google Scholar]
- Ishizuka T., Kamiya A., Suzuki H., Narikawa R., Noguchi T., Kohchi T., Inomata K., Ikeuchi M. The cyanobacteriochrome, TePixJ, isomerizes its own chromophore by converting phycocyanobilin to phycoviolobilin. Biochemistry. 2011;50:953–961. doi: 10.1021/bi101626t. [DOI] [PubMed] [Google Scholar]
- Ishizuka T., Narikawa R., Kohchi T., Katayama M., Ikeuchi M. Cyanobacteriochrome TePixJ of Thermosynechococcus elongatus harbors phycoviolobilin as a chromophore. Plant Cell Physiol. 2007;48:1385–1390. doi: 10.1093/pcp/pcm106. [DOI] [PubMed] [Google Scholar]
- Ishizuka T., Shimada T., Okajima K., Yoshihara S., Ochiai Y., Katayama M., Ikeuchi M. Characterization of cyanobacteriochrome TePixJ from a thermophilic cyanobacterium Thermosynechococcus elongatus strain BP-1. Plant Cell Physiol. 2006;47:1251–1261. doi: 10.1093/pcp/pcj095. [DOI] [PubMed] [Google Scholar]
- Khayatan B., Meeks J.C., Risser D.D. Evidence that a modified type IV pilus-like system powers gliding motility and polysaccharide secretion in filamentous cyanobacteria. Mol. Microbiol. 2015;98:1021–1036. doi: 10.1111/mmi.13205. [DOI] [PubMed] [Google Scholar]
- Klausen C., Kaiser F., Stüven B., Hansen J.N., Wachten D. Elucidating cyclic AMP signaling in subcellular domains with optogenetic tools and fluorescent biosensors. Biochem. Soc. Trans. 2019;47:1733–1747. doi: 10.1042/BST20190246. [DOI] [PubMed] [Google Scholar]
- Lim S., Yu Q., Gottlieb S.M., Chang C.W., Rockwell N.C., Martin S.S., Madsen D., Lagarias J.C., Ames J.B. Correlating structural and photochemical heterogeneity in cyanobacteriochrome NpR6012g4. Proc. Natl. Acad. Sci. U. S. A. 2018;115:4387–4392. doi: 10.1073/pnas.1720682115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldener I., Summers M.L., Sukenik A. Cellular differentiation in filamentous cyanobacteria. In: E. Flores and A. Herrero., editor. The Cell Biology of Cyanobacteria. Academic Press; London, United Kingdom: 2014. pp. 263–291. [Google Scholar]
- Marsac N.T. Differentiation of hormogonia and relationships with other biological processes. In: D.A. Bryant., editor. The Molecular Biology of Cyanobacteria. Kluwer Academic; Dordrecht, Netherlands: 1994. pp. 825–842. [DOI] [Google Scholar]
- Milias-Argeitis A., Rullan M., Aoki S.K., Buchmann P., Khammash M. Automated optogenetic feedback control for precise and robust regulation of gene expression and cell growth. Nat. Commun. 2016;7:1–11. doi: 10.1038/ncomms12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narikawa R., Enomoto G., Ni Ni W., Fushimi K., Ikeuchi M. A new type of dual-cys cyanobacteriochrome GAF domain found in cyanobacterium Acaryochloris marina, which has an unusual red/blue reversible photoconversion cycle. Biochemistry. 2014;53:5051–5059. doi: 10.1021/bi500376b. [DOI] [PubMed] [Google Scholar]
- Narikawa R., Fukushima Y., Ishizuka T., Itoh S., Ikeuchi M. A novel photoactive GAF domain of cyanobacteriochrome AnPixJ that shows reversible green/red photoconversion. J. Mol. Biol. 2008;380:844–855. doi: 10.1016/j.jmb.2008.05.035. [DOI] [PubMed] [Google Scholar]
- Narikawa R., Nakajima T., Aono Y., Fushimi K., Enomoto G., Itoh S., Sato M., Ikeuchi M. A biliverdin-binding cyanobacteriochrome from the chlorophyll d-bearing cyanobacterium Acaryochloris marina. Sci. Rep. 2015;5:1–10. doi: 10.1038/srep07950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliinyk O.S., Shemetov A.A., Pletnev S., Shcherbakova D.M., Verkhusha V.V. Smallest near-infrared fluorescent protein evolved from cyanobacteriochrome as versatile tag for spectral multiplexing. Nat. Commun. 2019;10:279. doi: 10.1038/s41467-018-08050-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong N.T., Olson E.J., Tabor J.J. Engineering an E. coli near-infrared light sensor. ACS Synth. Biol. 2018;7:240–248. doi: 10.1021/acssynbio.7b00289. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan P., Tabor J.J. Repurposing Synechocystis PCC6803 UirS-UirR as a UV-violet/green photoreversible transcriptional regulatory tool in E. coli. ACS Synth. Biol. 2016;5:733–740. doi: 10.1021/acssynbio.6b00068. [DOI] [PubMed] [Google Scholar]
- Rastogi R.P., Sinha R.P., Moh S.H., Lee T.K., Kottuparambil S., Kim Y.J., Rhee J.S., Choi E.M., Brown M.T., Häder D.P., et al. Ultraviolet radiation and cyanobacteria. J. Photochem. Photobiol. 2014;141:154–169. doi: 10.1016/j.jphotobiol.2014.09.020. [DOI] [PubMed] [Google Scholar]
- Rockwell N.C., Lagarias J.C. A brief history of phytochromes. Chemphyschem. 2010;11:1172–1180. doi: 10.1002/cphc.200900894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell N.C., Lagarias J.C. Phytochrome diversification in cyanobacteria and eukaryotic algae. Curr. Opin. Plant Biol. 2017;37:87–93. doi: 10.1016/j.pbi.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell N.C., Lagarias J.C. Phytochrome evolution in 3D: deletion, duplication, and diversification. New Phytol. 2020;225:2283–2300. doi: 10.1111/nph.16240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell N.C., Martin S.S., Feoktistova K., Lagarias J.C. Diverse two-cysteine photocycles in phytochromes and cyanobacteriochromes. Proc. Natl. Acad. Sci. U. S. A. 2011;108:11854–11859. doi: 10.1073/pnas.1107844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell N.C., Martin S.S., Gulevich A.G., Lagarias J.C. Phycoviolobilin formation and spectral tuning in the DXCF cyanobacteriochrome subfamily. Biochemistry. 2012;51:1449–1463. doi: 10.1021/bi201783j. [DOI] [PubMed] [Google Scholar]
- Rockwell N.C., Martin S.S., Gulevich A.G., Lagarias J.C. Conserved phenylalanine residues are required for blue-shifting of cyanobacteriochrome photoproducts. Biochemistry. 2014;53:3118–3130. doi: 10.1021/bi500037a. [DOI] [PubMed] [Google Scholar]
- Rockwell N.C., Martin S.S., Lagarias J.C. Identification of DXCF cyanobacteriochrome lineages with predictable photocycles. Photochem. Photobiol. Sci. 2015;14:929–941. doi: 10.1039/C4PP00486H. [DOI] [PubMed] [Google Scholar]
- Rockwell N.C., Martin S.S., Lagarias J.C. Identification of cyanobacteriochromes detecting far-red light. Biochemistry. 2016;55:3907–3919. doi: 10.1021/acs.biochem.6b00299. [DOI] [PubMed] [Google Scholar]
- Sanfilippo J.E., Garczarek L., Partensky F., Kehoe D.M. Chromatic acclimation in cyanobacteria: a diverse and widespread process for optimizing photosynthesis. Ann. Rev. Microbiol. 2019;73:407–433. doi: 10.1146/annurev-micro-020518-115738. [DOI] [PubMed] [Google Scholar]
- Sato T., Kikukawa T., Miyoshi R., Kajimoto K., Yonekawa C., Fujisawa T., Unno M., Eki T., Hirose Y. Protochromic absorption changes in the two-cysteine photocycle of a blue/orange cyanobacteriochrome. J. Biol. Chem. 2019;294:18909–18922. doi: 10.1074/jbc.RA119.010384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzkopf M., Yoo Y.C., Huckelhoven R., Park Y.M., Proels R.K. Cyanobacterial phytochrome2 regulates heterotrophic metabolism and has a function in the heat and high-light stress response. Plant Physiol. 2014;164:2157–2166. doi: 10.1104/pp.113.233270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R.P., Häder D.P. UV-protectants in cyanobacteria. Plant Sci. 2008;174:278–289. doi: 10.1016/j.plantsci.2007.12.004. [DOI] [Google Scholar]
- Song J.Y., Cho H.S., Cho J.I., Jeon J.S., Lagarias J.C., Park Y.I. Near-UV cyanobacteriochrome signaling system elicits negative phototaxis in the cyanobacterium Synechocystis sp. Proc. Natl. Acad. Sci. U. S. A. 2011;108:10780–10785. doi: 10.1073/pnas.1104242108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.Y., Lee H.Y., Yang H.W., Song J.J., Lagarias J.C., Park Y.I. Spectral and photochemical diversity of tandem cysteine cyanobacterial phytochromes. J. Biol. Chem. 2020;295:6754–6766. doi: 10.1074/jbc.RA120.012950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandar S.T., Senoo S., Toya Y., Shimizu H. Optogenetic switch for controlling the central metabolic flux of Escherichia coli. Metab. Eng. 2019;55:68–75. doi: 10.1016/j.ymben.2019.06.002. [DOI] [PubMed] [Google Scholar]
- Wagner J.R., Zhang J., von Stetten D., Günther M., Murgida D.H., Mroginski M.A., Walker J.M., Forest K.T., Hildebrandt P., Vierstra R.D. Mutational analysis of Deinococcus radiodurans bacteriophytochrome reveals key amino acids necessary for the photochromicity and proton exchange cycle of phytochromes. J. Biol. Chem. 2008;283:12212–12226. doi: 10.1074/jbc.M709355200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt K.E., Pakrasi H.B. Genomics approaches to deciphering natural transformation in cyanobacteria. Front. Microbiol. 2019;10:1259. doi: 10.3389/fmicb.2019.01259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde A., Fiedler B., Borner T. The cyanobacterial phytochrome Cph2 inhibits phototaxis towards blue light. Mol. Microbiol. 2002;44:981–988. doi: 10.1046/j.1365-2958.2002.02923.x. [DOI] [PubMed] [Google Scholar]
- Wiltbank L.B., Kehoe D.M. Diverse light responses of cyanobacteria mediated by phytochrome superfamily photoreceptors. Nat. Rev. Microbiol. 2019;17:37–50. doi: 10.1038/s41579-018-0110-4. [DOI] [PubMed] [Google Scholar]
- Xu X., Port A., Wiebeler C., Zhao K.H., Schapiro I., Gärtner W. Structural elements regulating the photochromicity in a cyanobacteriochrome. Proc. Natl. Acad. Sci. U. S. A. 2020;117:2432–2440. doi: 10.1073/pnas.1910208117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Cozad M.A., Holland D.A., Zhang Y., Luesch H., Ding Y. Photosynthetic production of sunscreen shinorine using an engineered cyanobacterium. ACS Synth. Biol. 2018a;7:664–671. doi: 10.1021/acssynbio.7b00397. [DOI] [PubMed] [Google Scholar]
- Yang H.W., Song J.Y., Cho S.M., Kwon H.C., Pan C.H., Park Y.I. Genomic survey of salt acclimation-related genes in the halophilic cyanobacterium Euhalothece sp. Z-M001. Sci. Rep. 2020;10:676. doi: 10.1038/s41598-020-57546-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Lam V., Adomako M., Simkovsky R., Jakob A., Rockwell N.C., Cohen S.E., Taton A., Wang J., Lagarias J.C., et al. Phototaxis in a wild isolate of the cyanobacterium Synechococcus elongatus. Proc. Natl. Acad. Sci. U. S. A. 2018b;115:E12378–E12387. doi: 10.1073/pnas.1812871115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K.C., Wu S.H., Murphy J.T., Lagarias J.C. A cyanobacterial phytochrome two-component light sensory system. Science. 1997;277:1505–1508. doi: 10.1126/science.277.5331.1505. [DOI] [PubMed] [Google Scholar]
- Yoshihara S., Ikeuchi M. Phototactic motility in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Photochem. Photobiol. Sci. 2004;3:512–518. doi: 10.1039/b402320j. [DOI] [PubMed] [Google Scholar]
- Yoshihara S., Katayama M., Geng X., Ikeuchi M. Cyanobacterial phytochrome-like PixJ1 holoprotein dhows novel reversible photoconversion between blue- and green-absorbing forms. Plant Cell Physiol. 2004;45:1729–1737. doi: 10.1093/pcp/pch214. [DOI] [PubMed] [Google Scholar]