Abstract
Purpose
Little is known about how many insured patients receive pharmacogenetic testing. We describe trends of single-gene pharmacogenetic testing in a US managed care population, and demographic and clinical characteristics of patients who received a test.
Methods
We leveraged a random sample of nearly 11 million patients from a data set of paid medical and pharmacy claims to identify patients with at least one claim indicating receipt of at least one of these single-gene pharmacogenetic tests: CYP2C19, CYP2D6, CYP2C9, VKORC1, UGT1A1, and HLA class 1 typing.
Results
From 1 January 2013 to 30 September 2017, 5712 patients received at least one pharmacogenetic test (55% female; mean age = 43 years). The median number of tests per patient was 3 (mean = 2.7, max = 12); 54% were processed through Managed Medicare/Medicaid, while 45% were processed through commercial insurance. The total number of pharmacogenetic tests received more than doubled from 2013 (n = 1955) to 2015 (n = 4192), then decreased slightly in 2016 (n = 3946). The most common test was CYP2C19 (n = 4719), and “long-term (current) use of other medications” was the most common diagnosis.
Conclusion
Pharmacogenetic testing through patients’ insurance was low, but more than doubled from 2013 to 2016. This study highlights the need to better understand utilization patterns and insurance coverage for pharmacogenetic tests.
Keywords: pharmacogenetic, pharmacogenomic, testing, insurance, managed care
INTRODUCTION
Personalized medicine leverages a person’s unique characteristics, such as their environment, lifestyle, and genetic makeup, to individually tailor strategies for the prevention and treatment of disease.1 Pharmacogenetics is a component of personalized medicine that studies the impact of genetic variation on interindividual variability in drug disposition, response, and adverse effects.2 Many actionable drug–gene pairs exist across a variety of therapeutic areas. For example, genetically mediated CYP2C19 poor metabolism is associated with decreased response to clopidogrel, an antiplatelet medication;3 CYP2C19 and CYP2D6 genetic variants are associated with altered metabolism of certain selective serotonin reuptake inhibitors and tricyclic antidepressants used in the treatment of mental health disorders;4,5 and HLA-B genetic variants are associated with increased risk of cutaneous and noncutaneous hypersensitivity reactions to certain drugs (e.g., abacavir, carbamazepine, oxcarbazepine, phenytoin, and allopurinol).6–9 However, uptake of pharmacogenetic testing in the clinical setting has been slow.10 Barriers to widespread adoption include lack of evidence supporting the clinical utility of testing, lack of physician and patient knowledge, and limited insurance coverage and reimbursement of testing.11–15
The who, what, and when of pharmacogenetic testing are largely determined by insurance coverage of the tests, which is low and varies across tests and health plans.16–18 Coverage determination is informed by the availability of evidence supporting clinical utility.13,16,17 Data from 2012 indicate that coverage of pharmacogenetic tests across a handful of commercial plans was only 30% of the tests reviewed, and coverage was inconsistent from insurer to insurer.17 The most commonly covered pharmacogenetic tests among commercial insurers and Medicare at that time included CYP2C19 for clopidogrel, CYP2D6 for tetrabenazine, HLA-B*15:02 for carbamazepine (only for persons of Asian ancestry), and HLA-B*57:01 for abacavir.17,19
Another factor complicating the insurance coverage landscape is that documentation of pharmacogenetic testing in claims and medical records has been inconsistent and has changed over time. The Current Procedural Terminology (CPT) coding system is used to code medical services and procedures, including pathology and laboratory tests that encompass pharmacogenetic testing.20 Documentation of pharmacogenetic testing is only as good as the codes available. Prior to 2012, specific CPT codes were not available for pharmacogenetic tests and they were typically assigned a molecular pathology procedure code.21 In 2012, the American Medical Association (AMA) assigned unique CPT codes to pharmacogenetic tests known to detect genetic variants associated with specific drugs; these specific codes were implemented in 2013 and most recently updated in 2018.21,22
Implementing pharmacogenetic testing in the clinical setting has the potential to improve patient care by decreasing failed treatment attempts due to medication ineffectiveness and/or adverse effects and by increasing effectiveness of improperly dosed medications. Insurance coverage plays an integral role in the clinical expansion of testing, yet little is known about how many insured patients receive pharmacogenetic testing, what types of tests they receive, and for what clinical reasons. To the best of our knowledge, no other studies have comprehensively examined coverage of single-gene pharmacogenetic tests among a large, insured population that includes different types of insurance. The purpose of this study is to describe the landscape of pharmacogenetic testing in a managed care population. Specifically, we describe the trends of single-gene pharmacogenetic testing between 2013 and 2017, the demographic and clinical characteristics of patients who received a pharmacogenetic test through their insurance, and the coverage and billed amounts of the pharmacogenetic tests in this cohort. We consider only single-gene pharmacogenetic tests because CPT codes had not been implemented for multiple-gene pharmacogenetic panel tests during the period for which data for the managed care population were available.
MATERIALS AND METHODS
Data source
This descriptive retrospective cross-sectional study leveraged IQVIA® PharMetrics. This data source includes health plan information for over 75 million unique patients and is nationally representative in terms of age and gender distributions of patients in the US commercially insured population.23 The paid medical and pharmacy claims are from 75 US health plans. Claim-level variables include service date, place of service, CPT procedure codes, International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM) and ICD-10 diagnosis codes, and prescriptions filled (date, medication identification code, and days supplied). The University of Colorado Skaggs School of Pharmacy and Pharmaceutical Sciences holds a license for a random sample of nearly 11 million patients from IQVIA® PharMetrics. This study was approved as exempt by the Colorado Multiple Institutional Review Board.
Study population
We identified patients of all ages from the random sample of IQVIA® PharMetrics who were enrolled in a health plan at some point from 1 January 2013 through the end of the available data (30 September 2017). We then identified patients with at least one procedure code indicating a pharmacogenetic test processed through their health plan, with a service date from 1 January 2013 through 30 September 2017. We included single-gene pharmacogenetic tests identified using specific CPT procedure codes: CYP2C19 (CPT code 81225), CYP2D6 (CPT code 81226), CYP2C9 (CPT code 81227), VKORC1 (CPT code 81355), HLA class 1 typing (CPT code 81381), and UGT1A1 (CPT code 81350). A list of the tests and corresponding clinically actionable medications is provided in Table 1. We excluded patients not enrolled in a health plan during the month of the test. UGT1A1 tests were excluded if the patient ever had a claim indicating disorders of porphyrin and bilirubin metabolism. HLA class 1 typing tests were excluded if the patient ever had any diagnosis or procedure codes indicating a solid organ or allogeneic bone marrow transplant. Patients could have received more than one type of pharmacogenetic test during the study period, and could have received the same test more than once on different dates.
Table 1.
Description of single-gene pharmacogenetic tests.
| CPT code | Single-gene pharmacogenetic test | Examples of clinically actionable medicationsa |
|---|---|---|
| 81225 | CYP2C19 (cytochrome P450, family 2, subfamily C, polypeptide 19) (e.g., drug metabolism), gene analysis, common variants (e.g., *2, *3, *4, *8, *17) | Clopidogrel, voriconazole, selective serotonin reuptake inhibitors (i.e., citalopram, escitalopram), tertiary amine tricyclic antidepressants (e.g., amitriptyline) |
| 81226 | CYP2D6 (cytochrome P450, family 2, subfamily D, polypeptide 6)(e.g., drug metabolism), gene analysis, common variants (e.g., *2, *3, *4, *5, *6, *9, *10, *17, *19, *29, *35, *41, *1XN, *2XN, *4XN) | Secondary and tertiary amine tricyclic antidepressants (e.g., amitriptyline, nortriptyline), selective serotonin reuptake inhibitors (e.g., paroxetine, fluvoxamine); opioids (e.g., codeine, oxycodone, tramadol), atomoxetine, ondansetron, pitolisant, tamoxifen |
| 81227 | CYP2C9 (cytochrome P450, family 2, subfamily C, polypeptide 9) (e.g., drug metabolism), gene analysis, common variants (e.g.,*2, *3,*5, *6) | Phenytoin, siponimid, warfarin |
| 81355 | VKORC1 (vitamin K epoxide reductase complex, subunit 1) (e.g., warfarin metabolism), gene analysis, common variant(s) (e.g., −1639G>A, c.173+1000C>T) | Warfarin |
| 81381 | HLA class I typing, high resolution (i.e., alleles or allele groups); 1 allele or allele group (e.g., B*57:01P), each | Abacavir, allopurinol, carbamazepine, oxcarbazepine, phenytoin |
| 81350 | UGT1A1 (UDP glucuronosyltransferase 1 family, polypeptide A1) (e.g., irinotecan metabolism), gene analysis, common variants (e.g., *28, *36, *37) | Atazanavir, irinotecan |
CPT Current Procedural Terminology.
aClinically actionable, as defined by Clinical Pharmacogenetics Implementation Consortium (CPIC) Level A designation.
Measures
Measures of interest included diagnoses received on the same service date as the pharmacogenetic test (up to 12 diagnoses are provided on each claim), coverage amount of the test, and demographic characteristics of patients receiving the tests. We identified the most common diagnoses for each type of pharmacogenetic test using ICD-9-CM and ICD-10 diagnostic codes recorded on the same day as the test. Coverage amount of the test was measured using the allowed cost, defined as the contracted or accepted reimbursable amount for covered medical services or supplies that the health plan agrees to pay to service providers. If the test was represented in the data by more than one claim on the same date, the allowed costs were added across all claims for that specific test on that date to calculate the total allowed cost for that test for that patient. Available patient demographic characteristics include year of birth, gender, region of residence (East, South, Midwest, and West), and insurance type. Insurance types included Medicare Risk and Medicaid administered through private carriers (i.e., managed Medicare/Medicaid), and commercial insurance.
Statistical analysis
We used descriptive statistics to characterize demographic and clinical characteristics of patients receiving pharmacogenetic tests. Specifically, counts and percentages described categorical variables, while means, medians, and ranges described continuous variables (including coverage and billed amounts). Associations between patient characteristics and types of pharmacogenetic tests received were not explored because patients could receive more than one type of pharmacogenetic test and therefore groups would not be mutually exclusive. We did, however, compare demographic characteristics between patients with commercial insurance and patients with managed Medicare/Medicaid using chi-square tests for categorical variables and a t-test for age in years (a normally distributed, continuous variable). Two-sided tests and an alpha cutoff of 0.05 were used to determine statistical significance. We examined the trend of testing by plotting the annual count of each type of test from 2013 through 2017, and examined trends in costs by plotting annual mean coverage amounts and annual mean billed amounts from 2013 through 2017. We used SAS version 9.4 for all data management and statistical calculations.24
RESULTS
From the nearly 11 million patients included in the IQVIA® PharMetrics random sample, 4,655,570 were enrolled in a health plan at some point from 1 January 2013 through the end of the available data (30 September 2017). From this 4.66 million, we identified 5712 patients (0.12%) who received a total of 15,382 single-gene pharmacogenetic tests of interest from January 2013 to September 2017. The most common pharmacogenetic test was CYP2C19, followed by CYP2D6 and CYP2C9 (Table 2). The median number of tests received per patient was 3 (mean = 2.7, range 1–12). Just over half of the patients (55%) were female, and the average age was 43 years. For just over half (54%), the pharmacogenetic test claim was processed through managed Medicare/Medicaid; for 45%, the claim was processed through commercial insurance; insurance type was indicated as other or unknown for the remaining 1%. Patients with commercial insurance were less likely to be female compared with patients with managed Medicare/Medicaid (53% vs. 57% respectively, p < 0.05); they were also older (mean age 43 years vs. 42 years, p < 0.05). There was a significant association between type of insurance and region of residence (p < 0.05): 61% of patients with commercial insurance lived in the East or South regions of the United States (compared with 38% of patients with managed Medicare/Medicaid), while 62% of patients with managed Medicare/Medicaid lived in the Midwest or West regions of the United States (compared with 39% of patients with commercial insurance).
Table 2.
Most common diagnoses reported for each single-gene pharmacogenetic test of interesta.
| Diagnosis reported on pharmacogenetic test claim | CYP2C19 (N = 4719) N (%) | CYP2D6 (N = 3775) N (%) | CYP2C9 (N = 3289) N (%) | HLA class 1 (N = 1821) N (%) | VKORC1 (N = 1587) N (%) | UGT1A1 (N = 191) N (%) |
|---|---|---|---|---|---|---|
| Long-term (current) use of medications | 966 (20.5%) | 883 (23.4%) | 705 (21.4%) | 110 (6.0%) | 534 (33.7%) | 48 (25.1%) |
| Depression | 976 (20.1%) | 940 (24.9%) | 826 (25.1%) | 507 (27.8%) | 165 (10.4%) | 25 (13.1%) |
| Anxiety | 622 (13.2%) | 592 (15.7%) | 523 (15.9%) | 299 (16.4%) | 117 (7.4%) | 32 (16.8%) |
| Lipid disorders | 664 (14.1%) | 221 (5.8%) | 228 (6.9%) | 73 (4.0%) | 204 (12.8%) | 15 (7.8%) |
| Hypertension | 563 (11.9%) | 310 (8.2%) | 310 (9.4%) | 43 (2.4%) | 293 (18.5%) | 29 (15.2%) |
| Pain/low back pain | 436 (9.2%) | 440 (11.7%) | 389 (11.8%) | 41 (2.2%) | 238 (15.0%) | 58 (30.4%) |
| ADHD | 515 (10.9%) | 519 (13.8%) | 460 (14.0%) | 301 (16.5%) | 53 (3.3%) | 25 (13.1%) |
| HIV | 0 | 0 | 0 | 544 (29.9%) | 0 | 0 |
ADHD attention deficit–hyperactivity disorder.
aTop three diagnoses for each test are indicated in italics.
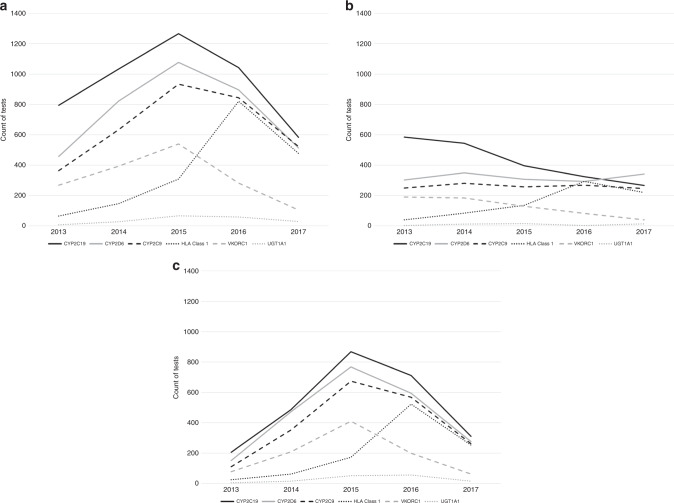
Figure 1a graphically displays time trends for annual counts of each pharmacogenetic test from 2013 to 2017 (2017 includes data only through 30 September). The number of patients receiving at least one of these tests nearly doubled from 2013 (n = 893) to 2015 (n = 1567), then decreased slightly in 2016 (n = 1372) and again in 2017 (n = 733). With the exception of HLA class 1, annual counts of each test increased steadily from 2013 to 2015, then decreased between 2015 and 2017. Counts of HLA class 1 testing, on the other hand, continued to increase from 2015 to 2016, then decreased in 2017. Figure 1b, c display time trends for patients with commercial insurance and managed Medicare/Medicaid, respectively. These stratified results indicate that while the count of tests increased over time for patients under a managed Medicare/Medicaid plan, they generally decreased over time for patients covered by commercial insurance.
Fig. 1. Annual counts of each type of single-gene pharmacogenetic test from 2013 to 2017.
Note: 2017 only includes data through 30 September 2017. (a) Annual counts from 2013 to 2017 for all tests (i.e., commercial insurance, managed Medicare/ Medicaid, and other/unknown insurance). (b) Annual counts from 2013 to 2017 for all tests covered by commercial insurance. (c) Annual counts from 2013 to 2017 for all tests covered by managed Medicare/Medicaid.
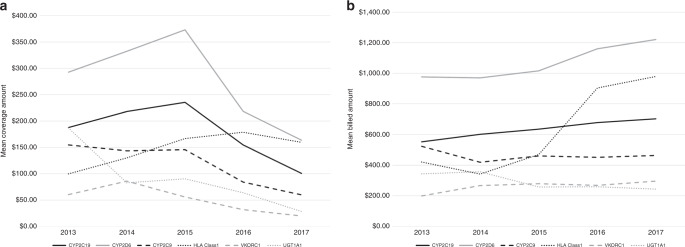
Coverage amount (i.e., mean allowed cost) for most of the pharmacogenetic tests of interest decreased from 2015 to 2017 (Fig. 2a). The single-gene test with the highest coverage amount was CYP2D6, with the total coverage amount for one test ranging from $0 to $6780 (mean from 2013 to 2017 = $288.60). The test with the lowest coverage amount was VKORC1, ranging from $0 to $830 (mean from 2013 to 2017 = $57.79). These temporal trends for the coverage amount for the single-gene tests coincide with the trends of the annual count of each type of test, which also decrease after 2015 (Fig. 1a). In general, coverage amounts were higher under commercial plans compared with managed Medicare/Medicaid (data not presented in figure). For CYP2D6, the test with the highest coverage amount, the total coverage amount for one test ranged from $0 to $6780 (mean from 2013 to 2017 = $317.43) for commercial insurance and ranged from $0 to $2432 (mean from 2013 to 2017 = $270.73) for managed Medicare/Medicaid. The test with the lowest coverage amount overall, VKORC1, ranged from $0 to $748.71 (mean from 2013 to 2017 = $70.68) for commercial insurance and ranged from $0 to $830 (mean from 2013 to 2017 = $49.42) for managed Medicare/Medicaid. While mean coverage amounts for most single-gene pharmacogenetic tests decreased from 2013 to 2017 (Fig. 2a), mean billed amounts tended to increase (Fig. 2b). The most drastic increase was seen for HLA class 1—the mean billed amount doubled from 2013 ($421) to 2017 ($980).
Fig. 2. Mean coverage amount (i.e., allowed cost) and billed amount for each single-gene pharmacogenetic test of interest from 2013 to 2017, for all tests (i.e., commercial insurance, managed Medicare/Medicaid, and other/unknown insurance).
Note: 2017 only includes data through 30 September 2017. (a) Mean allowed cost from 2013 to 2017. Allowed cost is defined as the contracted or accepted reimbursable amount for covered medical services or supplies that the health plan agrees to pay to service providers. (b) Mean billed amount from 2013 to 2017. Billed amount is defined as the amount billed for services provided by the servicing provider or facility.
The most commonly documented diagnosis for CYP2C19, CYP2D6, CYP2C9, UGT1A1, and VKORC1 was “long-term (current) use of other medications”, with the most commonly documented diagnosis for HLA class 1 typing being HIV (Table 2). Depression or anxiety was the second most common diagnosis for all tests except VKORC1. Pain, including specifically coded low back pain, was a commonly documented diagnosis for VKORC1 and UGT1A1. Such a diagnosis is likely indicative of a comorbidity.
DISCUSSION
We leveraged a large sample of paid medical and pharmacy claims for patients in managed care plans across the United States to identify patients receiving CYP2C19, CYP2D6, CYP2C9, VKORC1, UGT1A1, and HLA class 1 typing pharmacogenetic tests from 2013 through 2017. To the best of our knowledge, this is the first study examining coverage of single-gene pharmacogenetic tests among a large insured population that includes different types of insurance, including managed Medicare/Medicaid and commercial insurance. Overall, the number of single-gene pharmacogenetic tests was low; however, there was an uptick in testing from 2013 to 2015. CYP2C19, CYP2D6, and CYP2C9 were the most commonly received tests, respectively. All pharmacogenetic tests identified from this data source were covered at least partially through the patients’ insurance.
A unique aspect of our study was the broad catchment, including commercial insurance and managed Medicare/Medicaid. While patients with managed Medicare/Medicaid represent over half of this cohort receiving pharmacogenetic tests, patients with managed Medicare/Medicaid comprise less than 25% of the 75 million patients represented in IQVIA® PharMetrics. We observed an increase in single-gene pharmacogenetic testing from 2013 to 2015, which subsequently started to decline in 2015, coinciding with a decline in coverage amounts. It is unknown if this indicates a decrease in costs of testing, or a decrease in coverage of testing. The timing of the decline we observed likely coincides with a change in Medicare policy in October 2015 that limited coverage for CYP2C19 and CYP2D6 testing to defined indications, and deemed CYP2C9 and VKORC1 testing for all medications as not reasonable or necessary.25 The decrease also likely coincides with gaining popularity of multiple-gene panel tests and direct-to-consumer genetic testing (which were not captured in this study).26–29
Our findings are consistent with a recent study by Lynch and colleagues of Medicare beneficiaries, which showed that CYP pharmacogenetic tests were the most common tests received by this group in 2013.30 Likewise, “long-term (current) use of other medication” was the most common diagnosis for CYP pharmacogenetic tests among both Medicare cohorts.30 In our entire cohort, we identified patients with paid claims for these single-gene pharmacogenetic tests, indicating some level of insurance coverage. Weitzel and colleagues reported an 85% reimbursement rate for outpatient claims for CYP2C19 genotyping for clopidogrel in the setting of percutaneous coronary intervention (PCI) during the first month of their pharmacogenomic program in 2013.14
While few studies have systematically assessed insurance coverage patterns of pharmacogenetic tests for inherited (germline) variants, more work has been conducted in the field of oncology and noninherited (somatic) variants. Lu and colleagues reported variability in insurance coverage policies and prior authorization requirements for single- and multiple-gene pharmacogenetic tests for somatic variants influencing cancer treatment.19 However, insurance coverage of precision medicine-related tests in oncology is gaining traction. For example, in 2018 the Centers for Medicare and Medicaid Services (CMS) issued a National Coverage Determination for next-generation sequencing tests that detect genetic variants in solid tumors of patients with advanced cancer, thus providing information for cancer diagnosis and treatment.31–33 Along the same lines, the field is seeing an increase in coverage for exome sequencing. For example, Douglas and colleagues reported a shift from no coverage among the five largest private payers in 2015 to over 50% coverage by the 15 largest payers in 2017 for the use of exome sequencing in pediatric patients with neurodevelopmental disorders.34
The landscape of germline pharmacogenetic testing is beginning to mirror some of the advances observed in the somatic and exome testing space, moving from single-gene tests to more comprehensive multiple-gene panels. However, it is only recently that germline pharmacogenetic multiple-gene panel tests have been included in coverage policies. On 1 October 2019, United Healthcare issued a coverage decision indicating “the use of multi-gene panels to guide therapy decisions is proven medically necessary for antidepressants and antipsychotics medication” when certain criteria are met, including a diagnosis of major depressive disorder or anxiety and at least one failed prior medication.35
While these recent developments are promising, the reimbursement landscape for pharmacogenetic tests, particularly multiple-gene tests, still remains uncertain, as payers grapple with questions about clinical utility and application, economic utility, and impact on coverage policies.36–39 Lack of evidence is often cited as a major barrier to pharmacogenetic test use and insurance coverage. Phillips and colleagues suggested some potential solutions for remedying this evidence gap: (1) document what evidence is lacking, (2) standardize testing procedures and interpretation, (3) provide incentives for closing evidence gaps, and (4) develop creative approaches to obtaining evidence.40 While their framework was focused on the example of trastuzumab and HER2 testing, most of the suggested solutions can be applied to germline pharmacogenetic testing as well.
There are several limitations of our study that deserve to be addressed. First, we leveraged insurance claims data to conduct this landscape study, thus we cannot make generalizations about the receipt of pharmacogenetic tests outside of US managed care systems. Patients who used direct-to-consumer genetic tests or who paid entirely out of pocket are not represented in this data source. We also did not capture patients who received one of these tests through their insurance but had it coded using a general CPT code (e.g., 81479, “Unlisted molecular pathology procedure”). Second, as of the time these data were extracted from IQVIA® PharMetrics, CPT codes had not been implemented for multiple-gene panel tests so only single-gene pharmacogenetic tests were captured and described. Lastly, we cannot determine whether the testing was conducted in a reactive manner (as a pharmacogenetically relevant medication was prescribed or to explain the occurrence of medication-related side effects) or a preemptive manner (before a pharmacogenetically relevant medication was needed, typically using a multiple-gene panel).
Regardless of these limitations, this is the first study known to take a broad look at receipt of single-gene pharmacogenetic tests covered at least partially through insurance using a large population of patients. Our findings provide insight to temporal trends of single-gene testing and reasons for testing. This study also highlights the need for widespread implementation of procedure and billing codes to capture multiple-gene panel tests. Secondary analysis of existing administrative data sets such as insurance claims and electronic medical records provides a platform for robust and powerful evaluations of the clinical utility of pharmacogenetic testing. Such studies will continue to build the evidence necessary to expand insurance coverage and increase the judicious clinical use of pharmacogenetic tests in this era of personalized and precision medicine.
Acknowledgements
This work was supported, in part, by National Institutes of Health (NIH) grant K08HL125725 (D.P.K.). Contents are the authors’ sole responsibility and do not necessarily represent official NIH views. The statements, findings, conclusions, views, and opinions contained and expressed in this publication are based in part on data obtained under license from IQVIA®. Source: IQVIA® PharMetrics, 1 January 2007–30 September 2017, IQVIA®. The statements, findings, conclusions, views, and opinions contained and expressed herein are not necessarily those of IQVIA® or any of its affiliated or subsidiary entities.
Disclosure
The authors declare no conflicts of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.US National Library of Medicine. Genetics Home Reference. What is precision medicine? https://ghr.nlm.nih.gov/primer/precisionmedicine/definition. Accessed 25 November 2019.
- 2.Relling M, Evans W. Pharmacogenomics in the clinic. Nature. 2015;526:343–350. doi: 10.1038/nature15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scott SA, Sangkuhl K, Stein CM, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther. 2013;94:317–323. doi: 10.1038/clpt.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hicks JK, Bishop JR, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin Pharmacol Ther. 2015;98:127–134. doi: 10.1002/cpt.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hicks JK, Sangkuhl K, Swen JJ, et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther. 2017;102:37–44. doi: 10.1002/cpt.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caudle KE, Rettie AE, Whirl-Carrillo M, et al. Clinical pharmacogenetics implementation consortium guidelines for CYP2C9 and HLA-B genotypes and phenytoin dosing. Clin Pharmacol Ther. 2014;96:542–548. doi: 10.1038/clpt.2014.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin MA, Hoffman JM, Freimuth RR, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for HLA-B genotype and abacavir dosing: 2014 update. Clin Pharmacol Ther. 2014;95:499–500. doi: 10.1038/clpt.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phillips EJ, Sukasem C, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium guideline for HLA genotype and use of carbamazepine and oxcarbazepine: 2017 update. Clin Pharmacol Ther. 2018;103:574–581. doi: 10.1002/cpt.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saito Y, Stamp LK, Caudle KE, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for human leukocyte antigen B (HLA-B) genotype and allopurinol dosing: 2015 update. Clin Pharmacol Ther. 2016;99:36–37. doi: 10.1002/cpt.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunnenberger HM, Crews KR, Hoffman JM, et al. Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Annu Rev Pharmacol Toxicol. 2015;55:89–106. doi: 10.1146/annurev-pharmtox-010814-124835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frueh F. Real-world clinical effectiveness, regulatory transparency and payer coverage: three ingredients for translating pharmacogenomics into clinical practice. Pharmacogenomics. 2010;11:657–660. doi: 10.2217/pgs.10.46. [DOI] [PubMed] [Google Scholar]
- 12.Scott SA. Personalizing medicine with clinical pharmacogenetics. Genet Med. 2011;13:987–995. doi: 10.1097/GIM.0b013e318238b38c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trosman JR, Van Bebber SL, Phillips KA. Health technology assessment and private payers’ coverage of personalized medicine. J Oncol Pract. 2011;7((Suppl 3)):18s–24s. doi: 10.1200/JOP.2011.000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weitzel KW, Elsey AR, Langaee TY, et al. Clinical pharmacogenetics implementation: approaches, successes, and challenges. Am J Med Genet C Semin Med Genet. 2014;166C:56–67. doi: 10.1002/ajmg.c.31390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zineh I, Lesko LJ. Pharmacogenetics in medicine: barriers, critical factors and a framework for dialogue. Pers Med. 2009;6:359–361. doi: 10.2217/pme.09.27. [DOI] [PubMed] [Google Scholar]
- 16.Cohen J, Wilson A, Manzolillo K. Clinical and economic challenges facing pharmacogenomics. Pharmacogenomics J. 2012;13:378–388. doi: 10.1038/tpj.2011.63. [DOI] [PubMed] [Google Scholar]
- 17.Hresko A, Haga S. Insurance coverage policies for personalized medicine. J Pers Med. 2012;2:201–216. doi: 10.3390/jpm2040201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park SK, Thigpen J, Lee IJ. Coverage of pharmacogenetic tests by private health insurance companies. J Am Pharm Assoc (2003). 2019 Dec 13; 10.1016/j.japh.2019.10.003 [Epub ahead of print]. [DOI] [PubMed]
- 19.Lu CY, Loomer S, Ceccarelli R, et al. Insurance coverage policies for pharmacogenomic and multi-gene testing for cancer. J Pers Med. 2018;8:E19.. doi: 10.3390/jpm8020019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Human Genome Research Institute. Coverage and reimbursement of genetic tests. 2018. https://www.genome.gov/19016729/coverage-and-reimbursement-of-genetic-tests. Accessed 9 April 2019.
- 21.Hefti E, Blanco J. Documenting pharmacogenomic testing with Current Procedure Terminology (CPT) codes, a review of past and present practices. J AHIMA. 2016;87:56–59. [PMC free article] [PubMed] [Google Scholar]
- 22.American Medical Association. CPT 2018 Professional Edition. Chicago, IL; 2018.
- 23.IQVIA® PharMetrics Users Guide & Data Dictionary. Plymouth Meeting, PA: IQVIA®; 2017.
- 24.The SAS System for Windows Version 9.4. Cary, NC. SAS Institute. 2016.
- 25.Centers for Medicare and Medicaid Services. Local coverage determination (LCD): CYP2C19, CYP2D6, CYP2C9, and VKORC1 genetic testing (L35698). 2015. https://www.cms.gov/medicare-coverage-database/details/lcd-details.aspx?LCDId=35698&ver=21&articleId=55941&Cntrctr=All&UpdatePeriod=779&bc=AQAACAAAQAAA&. Accessed 25 November 2019.
- 26.Allyse MA, Robinson DH, Ferber MJ, Sharp RR. Direct-to-consumer testing 2.0: emerging models of direct-to-consumer genetic testing. Mayo Clin Proc. 2018;93:113–120. doi: 10.1016/j.mayocp.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 27.Bousman CA, Zierhut H, Muller DJ. Navigating the labyrinth of pharmacogenetic testing: a guide to test selection. Clin Pharmacol Ther. 2019;106:309–312. doi: 10.1002/cpt.1432. [DOI] [PubMed] [Google Scholar]
- 28.Phillips KA, Deverka PA, Hooker GW, Douglas MP. Genetic test availability and spending: where are we now? Where are we going? Health Aff (Millwood) 2018;37:710–716. doi: 10.1377/hlthaff.2017.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vo TT, Bell GC, Owusu Obeng A, Hicks JK, Dunnenberger HM. Pharmacogenomics implementation: considerations for selecting a reference laboratory. Pharmacotherapy. 2017;37:1014–1022. doi: 10.1002/phar.1985. [DOI] [PubMed] [Google Scholar]
- 30.Lynch JA, Berse B, Dotson WD, Khoury MJ, Coomer N, Kautter J. Utilization of genetic tests: analysis of gene-specific billing in Medicare claims data. Genet Med. 2017;19:890–899. doi: 10.1038/gim.2016.209. [DOI] [PubMed] [Google Scholar]
- 31.Centers for Medicare and Medicaid Services. Decision memo for next generation sequencing (NGS) for Medicare beneficiaries with advanced cancer (CAG-00450N). https://www.cms.gov/medicare-coverage-database/details/nca-decision-memo.aspx?NCAId=290&CoverageSelection=National&KeyWord=next+generation+sequencing&KeyWordLookUp=Title&KeyWordSearchType=And&FriendlyError=MissingCALID&bc=gAAAACAAQAAA&. Accessed 25 November 2019.
- 32.Turna Ray. CMS to revisit national coverage determination on NGS tumor testing. https://www.genomeweb.com/molecular-diagnostics/cms-revisit-national-coverage-determination-ngs-tumor-testing#.XOa9E_lKjIU. Accessed 25 November 2019.
- 33.Phillips KA. Evolving payer coverage policies on genomic sequencing tests: beginning of the end or end of the beginning? JAMA. 2018;319:2379–2380. doi: 10.1001/jama.2018.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Douglas MP, Parker SL, Trosman JR, Slavotinek AM, Phillips KA. Private payer coverage policies for exome sequencing (ES) in pediatric patients: trends over time and analysis of evidence cited. Genet Med. 2019;21:152–160. doi: 10.1038/s41436-018-0043-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.UnitedHealthcare pharmacogenomic testing policy number: 2019T0587E https://www.genomeweb.com/sites/default/files/uhc_pharmacogenetic-testing-10012019.pdf. Accessed 25 November 2019.
- 36.Keeling NJ, Rosenthal MM, West-Strum D, Patel AS, Haidar CE, Hoffman JM. Preemptive pharmacogenetic testing: exploring the knowledge and perspectives of US payers. Genet Med. 2019;21:1224–1232. doi: 10.1038/gim.2017.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chambers JD, Saret CJ, Anderson JE, Deverka PA, Douglas MP, Phillips KA. Examining evidence in US payer coverage policies for multi-gene panels and sequencing tests. Int J Technol Assess Health Care. 2017;33:534–540. doi: 10.1017/S0266462317000903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ginsburg GS, Phillips KA. Precision medicine: from science to value. Health Aff (Millwood) 2018;37:694–701. doi: 10.1377/hlthaff.2017.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phillips KA, Deverka PA, Trosman JR, et al. Payer coverage policies for multigene tests. Nature Biotechnol. 2017;35:614–617. doi: 10.1038/nbt.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phillips KA. Closing the evidence gap in the use of emerging testing technologies in clinical practice. JAMA. 2008;300:2542–2544. doi: 10.1001/jama.2008.754. [DOI] [PMC free article] [PubMed] [Google Scholar]