Abstract
Coronaviruses cause several human diseases, including severe acute respiratory syndrome. The global coronavirus disease 2019 (COVID-19) pandemic has become a huge threat to humans. Intensive research on the pathogenic mechanisms used by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is urgently needed—notably to identify potential drug targets. Clinical studies of patients with COVID-19 have shown that gastrointestinal disorders appear to precede or follow the respiratory symptoms. Here, we review gastrointestinal disorders in patients with COVID-19, suggest hypothetical mechanisms leading to gut symptoms, and discuss the potential consequences of gastrointestinal disorders on the outcome of the disease. Lastly, we discuss the role of the gut microbiota during respiratory viral infections and suggest that targeting gut dysbiosis may help to control the pathogenesis of COVID-19.
Keywords: SARS-CoV-2, COVID-19, gastrointestinal symptoms, gut microbiota, microbial dysbiosis, disease outcomes
In this review, Trottein and Sokol present hypothetical mechanisms leading to gut symptoms in patients with COVID-19 and discuss their potential consequences on disease severity. They also discuss the role of the gut microbiota in disease and the potential interest of targeting it to improve COVID-19 pathogenesis.
Main Text
Coronaviruses are enveloped RNA viruses containing a large (25 to 32 kb), single-stranded, positive-sense RNA genome. These viruses circulate continuously in human populations and generally cause mild respiratory diseases, including the common cold. In contrast, the zoonotic severe acute respiratory syndrome coronavirus (SARS-CoV) and the Middle East respiratory syndrome coronavirus (MERS-CoV) cause severe respiratory diseases and are associated with a high mortality rate when they spread to humans (Fehr et al., 2017). No specific antiviral drugs or vaccines have been approved for combating infections by SARS and MERS. In late 2019, a new infectious respiratory disease (now termed coronavirus disease 2019 [COVID-19]) emerged in Wuhan, China (Wang et al., 2020a; Zhu et al., 2020). This disease spread rapidly across China and to other countries. The World Health Organization recently declared the COVID-19 outbreak to be a pandemic. SARS-CoV-2 is the etiological agent of COVID-19 (Zhu et al., 2020). A phylogenetic analysis has shown that SARS-CoV-2 is most closely related to SARS-CoV (nucleotide similarity: 89.1%). COVID-19 is characterized by acute pathological outcomes, including pneumonia and acute respiratory distress syndrome (ARDS), and has a substantial mortality rate (Chen et al., 2020). ARDS during a SARS-CoV-2 infection is also associated with dysregulation of cytokine production, barrier leakage, and organ dysfunction. COVID-19 is more severe in individuals with comorbidities (diabetes mellitus, cardiovascular diseases, and hypertension) and in the elderly (Guan et al., 2020; Wu and McGoogan, 2020).
SARS-CoV-2 Can Target the Gastrointestinal Tract in Humans
Both SARS-CoV and SARS-CoV-2 can infect host cells when the viral spike protein binds to its cell surface receptor angiotensin-converting enzyme II (ACE2). With the exception of human coronavirus (hCoV) NL63, the seven other hCoVs have other host cell receptors (also exopeptidases). ACE2 is a homolog of angiotensin-converting enzyme (ACE), the central enzyme in the renin-angiotensin-aldosterone system (RAAS), which is crucial for the physiology and pathology of all organs (Gheblawi et al., 2020). ACE2 maintains the homeostasis of RAAS as a negative regulator. Virus entry also requires cleavage of the spike protein (priming) by a type II transmembrane serine protease called TMPRSS2. This priming step is essential for fusion of the viral and cell membranes (Matsuyama et al., 2010; Hoffmann et al., 2020; Zhou et al., 2020a). Additional proteases (including furin) might be involved in SARS-CoV-2 priming (Coutard et al., 2020). SARS-CoV-2’s binding affinity for human ACE2 is 10 to 20 times greater than that of SARS-CoV; this disparity might result from differences in spike protein methylation and/or cleavage sites (Jin et al., 2020; Wang et al., 2020b). Hence, the expression of ACE2 is essential for SARS-CoV-2’s entry into host cells. As discussed later, changes in ACE2’s functional activity might critically affect the disease outcome. In humans, ACE2 and TMPRSS2 are strongly expressed in lung tissue in general and by epithelial cells in particular. This strong expression explains why the lung appears to be the most vulnerable target organ during COVID-19. A recent single-cell analysis revealed that more than 80% of ACE2-expressing pulmonary cells were type II alveolar cells (Zhao et al., 2020); hence, this cell type might have a crucial role in coronavirus invasion and replication. Bronchial transient secretory cells (with a phenotype between those of goblet cells and ciliated cells) also express high levels of ACE2 (Lukassen et al., 2020). Furthermore, epithelial cells in the upper respiratory tract (specifically nasal goblet/secretory cells and ciliated cells) express both ACE2 and TMPRSS2 (Sungnak et al., 2020). This combined expression in the upper airways may be correlated with enhanced transmission of the virus. ACE2 is also expressed in many extrapulmonary tissues, including the heart, liver, kidney, and intestine (Crackower et al., 2002; Hamming et al., 2004; Hashimoto et al., 2012). In the latter, high levels of ACE2 are found on the luminal surface of differentiated epithelial cells in the small intestine, whereas levels are lower in the crypt cells and in the colon (Hashimoto et al., 2012; Liang et al., 2020). Human proximal and distal enterocytes coexpress dipeptidyl peptidase-4 (the receptor for MERS-CoV) and alanine aminopeptidase (the receptor for hCoV-229E) (Liang et al., 2020). In the intestine, ACE2 acts as a coreceptor for nutrient uptake and especially amino acid absorption from food (Hashimoto et al., 2012). On the one hand, the gut might be a major entry site for SARS-CoV-2; this would suggest that consumption of contaminated food can propagate the virus in humans. On the other hand, the intestinal expression of ACE2 might have essential implications for fecal-oral transmission and thus the containment of viral spreading. As seen during previous coronavirus outbreaks, around half of all COVID-19 patients have detectable SARS-CoV-2 RNA in their stools—even when it is no longer found in the respiratory tract (Zhang et al., 2020; Xiao et al., 2020b; Wang et al., 2020c). The virus has also been detected in gastrointestinal histological samples and by endoscopy (Xiao et al., 2020a; Lin et al., 2020). Importantly, infectious viruses were detected in fecal samples of COVID-19 patients, suggesting that the digestive tract might be a site of viral replication and activity (Xiao et al., 2020b; Zhou et al., 2020c). Several studies using human small intestinal organoids have shown that SARS-CoV-2 replicates in enterocytes (Zhou et al., 2020c; Zang et al., 2020; Lamers et al., 2020). The presence of virus in the stools and its fecal persistence suggest that fecal-oral transmission is possible. A study (mouse system) reported that intragastric inoculation of SARS-CoV-2 causes productive infection and leads to pulmonary pathological changes (Sun et al., 2020). Collectively, the lung is not the only site targeted by SARS-CoV-2, and enteric infection occurs. Here, we review gastrointestinal disorders in patients with COVID-19, suggest hypothetical mechanisms leading to gut symptoms, and discuss the potential consequences of gastrointestinal disorders on the outcome of the disease.
SARS-CoV-2 Causes Gastrointestinal Disorders in Humans
Various research groups have analyzed the epidemiological and clinical characteristics of gastrointestinal disorders in patients with COVID-19. The percentage of patients with gastrointestinal disorders (as evidenced by vomiting, nausea, and/or diarrhea) varies depending on the study. In China, Jin and colleagues enrolled 651 patients on admission, i.e., before treatment with antiviral or antibiotic drugs, which might have biased the analysis (Jin et al., 2020). Interestingly, 11.4% of COVID-19 patients had at least one gastrointestinal tract symptom, the most common being diarrhea (in 5% to 8% of patients). In the study by Jin and colleagues, the intestinal disorders lasted a median of 4 days and appeared to precede the respiratory symptoms. Gastrointestinal symptoms were more frequent in severe/critical cases of COVID-19 (23%) than in mild COVID-19 (8%) (Jin et al., 2020). In other studies performed in China and Hong Kong, the proportion of COVID-19 patients (n total = 254, 59, 204, 58, 138, and 1,099, respectively) who developed gastrointestinal disorders was 26%, 25.4%, 18.6%, 11%, 13.7%, and 8.7%, respectively (Zhou et al., 2020b; Cheung et al., 2020; Pan et al., 2020; Lin et al., 2020; Wang et al., 2020c, Guan et al., 2020). Other studies (of 40 COVID-19 patients in Europe and 278 and 318 COVID-19 patients in the United States) reported that 55%, 35%, and 61% of the patients, respectively, displayed gastrointestinal signs and symptoms (Effenberger et al., 2020; Nobel et al., 2020; Redd et al., 2020). Importantly, meta-analyses of studies totaling 4,243, 6,686, and 10,890 patients found that the pooled prevalence of gastrointestinal symptoms was 17.6%, 15%, and 10%, respectively (Cheung et al., 2020; Mao et al., 2020; Sultan et al., 2020). SARS-CoV-2 infection triggers an inflammatory response in the gut, as evidenced by elevated fecal levels of calprotectin (a marker protein expressed mainly by neutrophils) (Effenberger et al., 2020). Lin and colleagues also observed that although the presence of SARS-CoV-2 in the stool does not correlate with gastrointestinal symptoms, the presence of SARS-CoV-2 in intestinal tissue is generally associated with severe gastrointestinal symptoms (Lin et al., 2020). The use of human small intestinal organoids will be instrumental in better analyzing the modalities of enterocyte infection and its impact on the inflammatory response. Furthermore, it will be interesting in the future to determine the consequences of pre-existing digestive diseases (i.e., inflammatory bowel disease) on gastrointestinal disorders caused by SARS-CoV-2 and on the severity of COVID-19.
Potential Mechanisms of Gastrointestinal Disorders in COVID-19
Gastrointestinal disorders in COVID-19 patients are likely to have several etiological factors. The recruitment of inflammatory cells (i.e., from the bone marrow) and/or the systemic and local production of inflammatory cytokines are likely to have a role. In this setting, local inflammation might weaken the epithelial barrier. Cellular damage induced directly by viral replication and spreading might also actively contribute to injury and inflammation of the gut epithelium. Gastrointestinal symptoms in COVID-19 patients might also result from dysfunction of ACE2. As mentioned earlier, ACE2 is a key regulatory enzyme in the RAAS, and the latter is known to influence immune functions and inflammation (Ranjbar et al., 2019). ACE2 is an essential regulator of intestinal homeostasis, and a lack of this enzyme accentuates the intestine’s susceptibility to inflammation (Hashimoto et al., 2012). The intestinal phenotype is (at least partly) due to ACE2’s non-catalytic function. A lack of ACE2 alters the expression of the neutral amino acid transporter B0AT1 on intestinal epithelial cells, which lowers the intake of tryptophan and thus reduces the production of nicotinamide and (potentially) other tryptophan-derived metabolites with a critical role in intestinal homeostasis (Hashimoto et al., 2012; Agus et al., 2018). Binding of the viral spike protein to ACE2 leads to reduced expression of the latter but does not change the expression of ACE (Kuba et al., 2005). During SARS-CoV infections, the expression of ACE2 is considerably reduced—at least in the lungs (Kuba et al., 2005). Given the key role of ACE2 in intestinal homeostasis, reduced expression and impaired function of this protein during a SARS-CoV infection might substantially contribute to local disorders and pathogenesis. It remains to be seen whether drugs able to modulate ACE2 expression, such as ACE inhibitors or angiotensin II receptor blockers (used in hypertension and diabetes), can modulate gastrointestinal disorders, including gut dysbiosis, in the context of SARS-CoV-2 infection. Along with inflammation and dysfunction of ACE2, other mechanisms might be implicated in gastrointestinal symptoms in COVID-19 patients. Because hypoxia is a major clinical symptom in COVID-19 patients (Cavezzi et al., 2020) and is known to be critical in intestinal homeostasis, including microbiota composition and function (Singhal and Shah., 2020), it is also likely that oxygen deprivation might be important in gastrointestinal disorders and disease severity. Recent evidence suggests that SARS-CoV-2 may affect the central nervous system (Paniz-Mondolfi et al., 2020; Helms et al., 2020). Regarding the importance of the gut-brain axis, one can speculate that this may play a role in gastrointestinal disorders during SARS-CoV-2 infection. It is possible that the enteric nervous system could be affected by SARS-CoV-2, either by direct viral infection or through the elicited immune response (e.g., inflammatory cytokines), amplifying diarrhea and possibly stimulating the vagus nerve to promote vomiting.
ACE2 Dysfunction Alters the Composition of the Gut Microbiota
It is well established that the gut microbiota has a critical role in intestinal homeostasis and that changes in its composition and function activity are involved in local inflammation (Blander et al., 2017; Lavelle and Sokol, 2020). A lack of ACE2 leads to substantial alteration in the composition of the gut microbiota in mice; this partly results from reduced production of the antimicrobial peptides that control the gut’s microbial community (Hashimoto et al., 2012). Moreover, disruption of the ACE/ACE2 axis during pulmonary hypertension (loss of ACE2) is associated with alteration of the gut microbiota in humans (Santisteban et al., 2016; Kim et al., 2020). One can speculate that the decrease in ACE2 availability during SARS-COV-2 infection is enough to alter the composition of the gut microbiota. Recent pilot studies suggest gut microbiota alterations during SARS-CoV-2 infection (Yu et al., 2020; Gu et al., 2020b; Zuo et al., 2020). In a cohort of 30 COVID-19 patients (fecal sample analysis using 16S rRNA gene sequencing), infection was associated with a significant decrease of bacterial diversity and abundance (Gu et al., 2020b). Significant changes in gut microbial communities were noticed, including lower relative abundance of beneficial (butyrate-producing) bacteria, such as several genera from the Ruminococcaceae and Lachnospiraceae families. However, significantly higher relative abundance occurred of opportunistic pathogens, including Streptococcus, Rothia, Veillonella, and Actinomyces. Of interest, the gut microbial signature of patients with COVID-19 was different from that of H1N1 patients (Gu et al., 2020b). Enrichment of opportunistic pathogens and depletion of beneficial commensals was also observed in a (longitudinal) study using deep shotgun metagenomics (15 COVID-19 patients) (Zuo et al., 2020). In particular, enrichment of opportunistic pathogens known to cause bacteremia, including Clostridium hathewayi, Actinomyces viscosus, and Bacteroides nordii, was observed. Lower relative abundance of beneficial commensals, including the anti-inflammatory bacterium Faecalibacterium prausnitzii, Alistipes onderdonkii, Roseburia, and Lachnospiraceae taxa, was also noticed. The authors found a correlation between rise or drop of these bacteria and disease severity. Of note, altered microbiota composition persisted after clearance of SARS-CoV-2 and resolution of respiratory symptoms, indicating that resiliency is long lasting (Zuo et al., 2020). Hence, SARS-CoV-2 infection modifies the composition of the gut microbiota in humans. It is possible that reduced levels of commensal bacteria with important physiological functions, such as butyrate producers, may promote the overgrowth of intestinal conditional pathogenic bacteria. Whether these changes increase intestinal mucosal permeability and endotoxin concentrations in the blood, ultimately triggering inflammation and cytokine release exacerbation, remains to be investigated (Figure 1 ). Further analyses are urgently needed in larger human cohorts to confirm and detail gut microbiota alterations in COVID-19 patients. Studies should prospectively include asymptomatic COVID-19-confirmed subjects and patients at disease onset, during disease course, and over the long term after discovery to delineate the role of microbiome changes in SARS-CoV-2 infection and post-infection recovery. It will be critical to consider patients treated or not with antibiotics (and other medications) and the comorbidity status of those patients. The consequences of SARS-CoV-2 infection on the gut microbiota should also be investigated in relevant animal models of COVID-19. Possible models include non-human primates, ferrets, human-ACE2-expressing mice, and hamsters (Callaway, 2020). The impact of SARS-CoV-2 on the gut microbiota’s metabolic output also remains to be defined. This aspect is of particular importance because in a mouse model, the altered gut microbiota associated with ACE2 deficiency (1) favors intestinal inflammation and (2) confers susceptibility to colitis when transferred to wild-type animals (Hashimoto et al., 2012). In parallel, it remains to be determined whether alterations in the microbiota influence the extraintestinal outcomes of COVID-19. Altogether, direct enterocyte infection, disruption of the enteric ACE2 axis (and interference with nutrient absorption), hypoxia, alterations of the enteric nervous system and local immune response, and changes in inflammatory cytokine levels might lead to adverse intestinal outcomes (including dysbiosis) in patients with COVID-19.
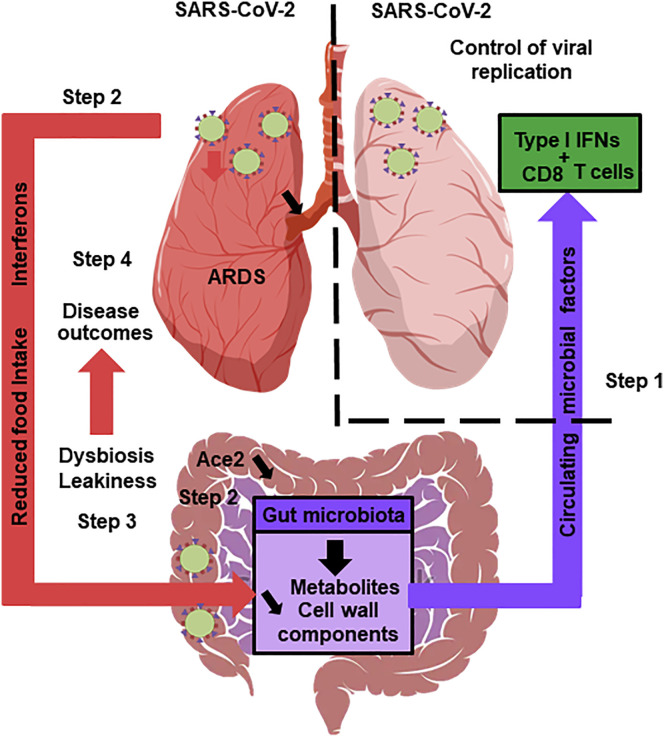
Figure 1.
The Gut Microbiota’s Hypothetical Role in SARS-CoV-2 Infection
A healthy gut microbiota boosts the lungs’ antiviral response, including interferon production and the effector function of CD8+ T cells (step 1). Soon after infection, SARS-CoV-2 replicates in the intestinal compartment and decreases the expression and activity of ACE2 (step 2). This leads to gut dysbiosis and gastrointestinal symptoms (step 3). Meanwhile, viral infection of the lungs triggers the systemic production of interferons and elicits weight loss, both of which trigger gut dysbiosis (step 3). Changes in the composition and functional activity of the gut microbiota and impairment of the gut’s barrier function contribute to disease outcomes, including ARDS, a systemic cytokine storm, and multiorgan dysfunction (step 4).
The Potential Role of the Gut Microbiota in COVID-19 Outcomes
Evidence suggests that the gut microbiota can remotely boost the host’s response to respiratory viral infections. Paradoxically, dysbiosis of the microbiota can also worsen the outcome of the disease. The gut microbiota’s role in hCoV infections has yet to be defined. In the setting of influenza infections, the results of experiments with antibiotic treatment (to deplete the residual microbiota) have demonstrated that the gut microbiota is critical for controlling viral replication (Ichinohe et al., 2011; Abt et al., 2012; Steed et al., 2017; Bradley et al., 2019). Bacterial cell wall components and bacterial metabolites (such as desaminotyrosine) favor the production of type I interferons and inflammasome-dependent cytokines by pulmonary cells. The gut microbiota also boosts CD8+ T cell effector function, a process that contributes to viral clearance (Ichinohe et al., 2011). Whereas altering the gut microbiota with antibiotics increases the severity of the infection, stimulating the microbiome with a high-fiber diet has the opposite effect (Trompette et al., 2018). The stimulatory mechanisms involve short-chain fatty acids (SCFAs, the end products of dietary fiber fermentation by commensal bacteria) that are absorbed by and act on immune cells to reduce the inflammatory component of infection. SCFAs also enhance the effector activity of CD8+ T cells by stimulating cellular metabolism (Trompette et al., 2018). Similar protective effects have been observed with the respiratory syncytial virus (Antunes et al., 2019). It remains to be seen whether the gut microbiota also participates in early control (through innate immunity) and late control (through adaptive immunity) of coronavirus replication, and this topic warrants investigation (Figure 1). It would also be useful to investigate the effect of SCFA-promoting high-fiber diets and/or probiotics on the outcome of SARS-CoV-2 infection.
Patients with respiratory infections generally have gut-dysfunction-related complications that worsen the clinical course. For example, this type of gut-lung crosstalk has been described during influenza infections in both humans and animal models. Severe influenza A virus infection is associated with intestinal disorders and gut microbiota alterations (Wang et al., 2014; Qin et al., 2015; Deriu et al., 2016; Groves et al., 2018; Yildiz et al., 2018; Sencio et al., 2020). These changes were attributed partly to infection-related reduction in food consumption (Sencio et al., 2020) and to interferon production (Wang et al., 2014; Deriu et al., 2016). We recently looked at whether microbiota conditioned by influenza A virus enhanced susceptibility to bacterial superinfection, a major cause of mortality during epidemics and pandemics (McCullers, 2014). In fecal transfer experiments, we showed that the dysbiotic microbiota remotely compromises the lung’s defenses against pneumococcal infection. In mechanistic terms, a reduction in SCFA production is associated with a decrease in the alveolar macrophages’ bactericidal activity (Sencio et al., 2020).
Whether gut disorders (including microbiota alterations) influence on outcomes in COVID-19 is still an open question. Evidence suggests that SARS-CoV-2 infection alters the gut-blood barrier and thus leads to systemic dissemination of bacteria, endotoxins, and microbial metabolites (Wang et al., 2020c; Huang et al., 2020; Guan et al., 2020). This might affect the host’s initial response to SARS-CoV-2 infection and might then contribute to multisystem dysfunction, septic shock, and the systemic inflammation storm seen in the second phase of SARS-CoV-2 infection, which typically result in death (Guan et al., 2020; Huang et al., 2020; Wang et al., 2020c). Clinical studies have demonstrated that the presence of a gastrointestinal disorder in COVID-19 patients is associated with a more aggressive clinical course, including ARDS, liver injury, a higher body temperature, and shock (Jin et al., 2020). Moreover, the risk factors for severity and mortality in COVID-19 (such as diabetes) (Singh et al., 2020) are known to be associated with disturbances of the intestinal microbiota and, in particular, low production of SCFAs. This is particularly the case for patients with metabolic syndrome (obesity, high blood pressure, and diabetes) who exhibit severity factors for viral infections (including respiratory infections) (Badawi et al., 2018; Honce and Schultz-Cherry, 2019). The role of intestinal dysbiosis in this setting needs to be investigated, e.g., by performing fecal transfer experiments in relevant animal models. We speculate that ACE2 disruption, changes in microbial profiles, and intestinal inflammation and leakiness during SARS-CoV infection strongly amplify systemic pathways in comorbid individuals and exacerbate underlying pathologies (e.g., diabetes and hypertension). SARS-CoV-2 infection may also deteriorate the course of human inflammatory bowel diseases (Effenberger et al., 2020). The mortality rate associated with SARS-CoV-2 infection is higher in older adults, who also tend to display chronic low-grade inflammation with underlying microbiota alterations and gut leakiness. In the setting of COVID-19, can we exploit the gut microbiota for the patient’s benefit? At present, no direct clinical evidence suggests that modulation of the gut microbiota has therapeutic value in patients with COVID-19. However, it is tempting to speculate that targeting gut microbiota might be a therapeutic option.
Concluding Remarks
The complex pathogenesis of SARS-CoV-2 infection is only starting to be deciphered. Along with the respiratory tract, the gastrointestinal tract is an entry and replication site for SARS-CoV-2. The gut constitutes one of the main extrapulmonary target organs with regard to symptoms and is a potential route for virus dissemination; its role therefore needs to be actively explored. In particular, several lines of evidence suggest that SARS-CoV-2 infection is associated with alteration of the gut microbiota. It remains to be determined whether the gut microbiota influences the gastrointestinal and pulmonary signs and symptoms of COVID-19 and overall mortality. Putative changes in the gut microbiota’s composition and functional activity might be biomarkers of disease severity. Lastly, and if the gut microbiota does prove to affect the disease’s severity and mortality rate, targeting the microbiota’s various components might be an attractive therapeutic strategy. In this setting, conventional approaches like antibiotic treatment targeting pathobionts (preferably with a narrow-spectrum drug), probiotics, and fecal transfer might be relevant. However, more specific interventions (e.g., live biotherapies or microbiota-derived metabolites) based on an in-depth understanding of the mechanisms involved are attractive.
Acknowledgments
We acknowledge support from our funding agencies Institut National de la Santé et de la Recherche Médicale, the Centre National de la Recherche Scientifique (CNRS), the University of Lille, the Pasteur Institute of Lille, the Région des Hauts-de-France, and l’Agence Nationale de la Recherche (AAP générique 2017, ANR-17-CE15-0020-01, ACROBAT) (to F.T.). F.T. received salary support from CNRS, and H.S. received salary support from Sorbonne Université and Saint Antoine Hospital. We thank Corinne Grangette and Isabelle Wolowczuk (Centre d’Infection et d’Immunité de Lille) for critical comments on the manuscript.
References
- Abt M.C., Osborne L.C., Monticelli L.A., Doering T.A., Alenghat T., Sonnenberg G.F., Paley M.A., Antenus M., Williams K.L., Erikson J. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agus A., Planchais J., Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe. 2018;23:716–724. doi: 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- Antunes K.H., Fachi J.L., de Paula R., da Silva E.F., Pral L.P., Dos Santos A.Á., Dias G.B.M., Vargas J.E., Puga R., Mayer F.Q. Microbiota-derived acetate protects against respiratory syncytial virus infection through a GPR43-type 1 interferon response. Nat. Commun. 2019;10:3273–3289. doi: 10.1038/s41467-019-11152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badawi A., Velummailum R., Ryoo S.G., Senthinathan A., Yaghoubi S., Vasileva D., Ostermeier E., Plishka M., Soosaipillai M., Arora P. Prevalence of chronic comorbidities in dengue fever and West Nile virus: A systematic review and meta-analysis. PLoS ONE. 2018;13:e0200200. doi: 10.1371/journal.pone.0200200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander J.M., Longman R.S., Iliev I.D., Sonnenberg G.F., Artis D. Regulation of inflammation by microbiota interactions with the host. Nat. Immunol. 2017;18:851–860. doi: 10.1038/ni.3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley K.C., Finsterbusch K., Schnepf D., Crotta S., Llorian M., Davidson S., Fuchs S.Y., Staeheli P., Wack A. Microbiota-driven tonic interferon signals in lung stromal cells protect from influenza virus infection. Cell Rep. 2019;28:245–256.e4. doi: 10.1016/j.celrep.2019.05.105. [DOI] [PubMed] [Google Scholar]
- Callaway E. Labs rush to study coronavirus in transgenic animals—some are in short supply. Nature. 2020;579:183. doi: 10.1038/d41586-020-00698-x. [DOI] [PubMed] [Google Scholar]
- Cavezzi A., Troiani E., Corrao S. COVID-19: hemoglobin, iron, and hypoxia beyond inflammation. A narrative review. Clin. Pract. 2020;10:1271–1277. doi: 10.4081/cp.2020.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K.S., Hung I.F.N., Chan P.P.Y., Lung K.C., Tso E., Liu R., Ng Y.Y., Chu M.Y., Chung T.W.H., Tam A.R. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.03.065. Published online April 3, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742–104746. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crackower M.A., Sarao R., Oudit G.Y., Yagil C., Kozieradzki I., Scanga S.E., Oliveira-dos-Santos A.J., da Costa J., Zhang L., Pei Y. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002;417:822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- Deriu E., Boxx G.M., He X., Pan C., Benavidez S.D., Cen L., Rozengurt N., Shi W., Cheng G. Influenza virus affects intestinal microbiota and secondary Salmonella infection in the gut through type I interferons. PLoS Pathog. 2016;12:e1005572. doi: 10.1371/journal.ppat.1005572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effenberger M., Grabherr F., Mayr L., Schwaerzler J., Nairz M., Seifert M., Hilbe R., Seiwald S., Scholl-Buergi S., Fritsche G. Faecal calprotectin indicates intestinal inflammation in COVID-19. Gut. 2020 doi: 10.1136/gutjnl-2020-321388. Published online April 20, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Channappanavar R., Perlman S. Middle East Respiratory Syndrome: Emergence of a Pathogenic Human Coronavirus. Annu. Rev. Med. 2017;68:387–399. doi: 10.1146/annurev-med-051215-031152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheblawi M., Wang K., Viveiros A., Nguyen Q., Zhong J.C., Turner A.J., Raizada M.K., Grant M.B., Oudit G.Y. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: Celebrating the 20th anniversary of the discovery of ACE2. Circ. Res. 2020;126:1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves H.T., Cuthbertson L., James P., Moffatt M.F., Cox M.J., Tregoning J.S. Respiratory Disease following Viral Lung Infection Alters the Murine Gut Microbiota. Front. Immunol. 2018;9:182–193. doi: 10.3389/fimmu.2018.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S., Chen Y., Wu Z., Chen Y., Gao H., Lv L., Guo F., Zhang X., Luo R., Huang C. Alterations of the gut microbiota in patients with COVID-19 or H1N1 Influenza. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa709. Published online June 4, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., China Medical Treatment Expert Group for Covid-19 Clinical characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M.L.C., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T., Perlot T., Rehman A., Trichereau J., Ishiguro H., Paolino M., Sigl V., Hanada T., Hanada R., Lipinski S. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms J., Kremer S., Merdji H., Clere-Jehl R., Schenck M., Kummerlen C., Collange O., Boulay C., Fafi-Kremer S., Ohana M. Neurologic features in Severe SARS-CoV-2 infection. N. Engl. J. Med. 2020;382:2268–2270. doi: 10.1056/NEJMc2008597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honce R., Schultz-Cherry S. Impact of obesity on Influenza A Virus pathogenesis, immune response, and evolution. Front. Immunol. 2019;10:1071–1084. doi: 10.3389/fimmu.2019.01071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T., Pang I.K., Kumamoto Y., Peaper D.R., Ho J.H., Murray T.S., Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. USA. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X., Lian J.S., Hu J.H., Gao J., Zheng L., Zhang Y.M., Hao S.R., Jia H.Y., Cai H., Zhang X.L. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut. 2020;69:1002–1009. doi: 10.1136/gutjnl-2020-320926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Rigatto K., Gazzana M.B., Knorst M.M., Richards E.M., Pepine C.J., Raizada M.K. Altered gut microbiome profile in patients with pulmonary arterial hypertension. Hypertension. 2020;75:1063–1071. doi: 10.1161/HYPERTENSIONAHA.119.14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., van Schayck J.P., Mykytyn A.Z., Duimel H.Q. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020 doi: 10.1126/science.abc1669. Published online May 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavelle A., Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020;17:223–237. doi: 10.1038/s41575-019-0258-z. [DOI] [PubMed] [Google Scholar]
- Liang W., Feng Z., Rao S., Xiao C., Xue X., Lin Z., Zhang Q., Qi W. Diarrhoea may be underestimated: a missing link in 2019 novel coronavirus. Gut. 2020;69:1141–1143. doi: 10.1136/gutjnl-2020-320832. [DOI] [PubMed] [Google Scholar]
- Lin L., Jiang X., Zhang Z., Huang S., Zhang Z., Fang Z., Gu Z., Gao L., Shi H., Mai L. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020;69:997–1001. doi: 10.1136/gutjnl-2020-321013. [DOI] [PubMed] [Google Scholar]
- Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T., Winter H., Meister M., Veith C., Boots A.W. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39:e105114. doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao R., Qiu Y., He J.S., Tan J.Y., Li X.H., Liang J., Shen J., Zhu L.R., Chen Y., Iacucci M. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID-19: a systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2020;5:667–678. doi: 10.1016/S2468-1253(20)30126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers J.A. The co-pathogenesis of influenza viruses with bacteria in the lung. Nat. Rev. Microbiol. 2014;12:252–262. doi: 10.1038/nrmicro3231. [DOI] [PubMed] [Google Scholar]
- Nobel Y.R., Phipps M., Zucker J., Lebwohl B., Wang T.C., Sobieszczyk M.E., Freedberg D.E. Gastrointestinal symptoms and coronavirus disease 2019: a case-control study from the United States. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.04.017. Published online April 12, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L., Mu M., Yang P., Sun Y., Wang R., Yan J., Li P., Hu B., Wang J., Hu C. Clinical characteristics of COVID-19 patients with digestive symptoms in Hubei, China: A descriptive, cross-Sectional, multicenter study. Am. J. Gastroenterol. 2020;115:766–773. doi: 10.14309/ajg.0000000000000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paniz-Mondolfi A., Bryce C., Grimes Z., Gordon R.E., Reidy J., Lednicky J., Sordillo E.M., Fowkes M. Central nervous system involvement by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) J. Med. Virol. 2020;92:699–702. doi: 10.1002/jmv.25915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin N., Zheng B., Yao J., Guo L., Zuo J., Wu L., Zhou J., Liu L., Guo J., Ni S. Influence of H7N9 virus infection and associated treatment on human gut microbiota. Sci. Rep. 2015;5:14771–14784. doi: 10.1038/srep14771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjbar R., Shafiee M., Hesari A., Ferns G.A., Ghasemi F., Avan A. The potential therapeutic use of renin-angiotensin system inhibitors in the treatment of inflammatory diseases. J. Cell. Physiol. 2019;234:2277–2295. doi: 10.1002/jcp.27205. [DOI] [PubMed] [Google Scholar]
- Redd W.D., Zhou J.C., Hathorn K.E., McCarty T.R., Bazarbashi A.N., Thompson C.C., Shen L., Chan W.W. Prevalence and characteristics of gastrointestinal symptoms in patients with SARS-CoV-2 infection in the United States: A multicenter cohort study. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.04.045. Published online April 22, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santisteban M.M., Kim S., Pepine C.J., Raizada M.K. Brain-gut-bone marrow axis: Implications for hypertension and related therapeutics. Circ. Res. 2016;118:1327–1336. doi: 10.1161/CIRCRESAHA.116.307709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sencio V., Barthelemy A., Tavares L.P., Machado M.G., Soulard D., Cuinat C., Queiroz-Junior C.M., Noordine M.L., Salomé-Desnoulez S., Deryuter L. Gut dysbiosis during influenza contributes to pulmonary pneumococcal superinfection through altered short-chain fatty acid production. Cell Rep. 2020;30:2934–2947.e6. doi: 10.1016/j.celrep.2020.02.013. [DOI] [PubMed] [Google Scholar]
- Singh A.K., Gupta R., Ghosh A., Misra A. Diabetes in COVID-19: prevalence, pathophysiology, prognosis and practical considerations. Diabetes Metab. Syndr. 2020;14:303–310. doi: 10.1016/j.dsx.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal R., Shah Y.M. Oxygen battle in the gut: hypoxia and hypoxia-inducible factors in metabolic and inflammatory responses in the intestine. J. Biol. Chem. 2020 doi: 10.1074/jbc.REV120.011188. Published online June 5, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steed A.L., Christophi G.P., Kaiko G.E., Sun L., Goodwin V.M., Jain U., Esaulova E., Artyomov M.N., Morales D.J., Holtzman M.J. The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science. 2017;357:498–502. doi: 10.1126/science.aam5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan S., Altayar O., Siddique S.M., Davitkov P., Feuerstein J.D., Lim J.K., Falck-Ytter Y., El-Serag H.B., American Gastroenterological Association AGA institute rapid review of the gastrointestinal and liver manifestations of COVID-19, meta-analysis of international data, and recommendations for the consultative management of patients with COVID-19. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.05.001. Published online May 11, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.-H., Chen Q., Gu H.-J., Yang G., Wang Y.-X., Huang X.-Y., Liu S.-S., Zhang N.-N., Li X.-F., Xiong R. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.05.020. Published online May 27, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., HCA Lung Biological Network SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompette A., Gollwitzer E.S., Pattaroni C., Lopez-Mejia I.C., Riva E., Pernot J., Ubags N., Fajas L., Nicod L.P., Marsland B.J. Dietary fiber confers protection against flu by shaping Ly6c− patrolling monocyte hematopoiesis and CD8+ T cell metabolism. Immunity. 2018;48:992–1005.e8. doi: 10.1016/j.immuni.2018.04.022. [DOI] [PubMed] [Google Scholar]
- Wang J., Li F., Wei H., Lian Z.-X., Sun R., Tian Z. Respiratory influenza virus infection induces intestinal immune injury via microbiota-mediated Th17 cell-dependent inflammation. J. Exp. Med. 2014;211:2397–2410. doi: 10.1084/jem.20140625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and important lessons from the Coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese center for disease control and prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. Published online February 24, 2020. [DOI] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833.e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Sun J., Xu Y., Li F., Huand X., Li H., Zhoa J., Huang J., Zhao J. Infectious SARS-CoV-2 in feces of patient with Severe COVID-19. Emerg. Infect. Dis. 2020;26 doi: 10.3201/eid2608.200681. Published online May 18, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz S., Mazel-Sanchez B., Kandasamy M., Manicassamy B., Schmolke M. Influenza A virus infection impacts systemic microbiota dynamics and causes quantitative enteric dysbiosis. Microbiome. 2018;6:9–25. doi: 10.1186/s40168-017-0386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Tong Y., Shen G., Fu A., Lai Y., Zhou X., Yan Y., Wang Y., Pan Y., Yu Z. Immunodepletion with hypoxemia: A potential high risk subtype of coronavirus disease 2019. medRxiv. 2020 doi: 10.1101/2020.03.03.20030650. [DOI] [Google Scholar]
- Zang R., Gomez Castro M.F., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5:eabc3582. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Du R.H., Li B., Zheng X.S., Yang X.L., Hu B., Wang Y.Y., Xiao G.F., Yan B., Shi Z.L., Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microbes Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Zhao Z., Wang Y., Zhou Y., Ma Y., Zuo W. Single-cell RNA expression profiling of ACE2, the putative receptor of Wuhan 2019-nCov. bioRxiv. 2020 doi: 10.1101/2020.01.26.919985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Zhao N., Shu Y., Han S., Chen B., Shu X. Effect of gastrointestinal symptoms in patients with COVID-19. Gastroenterology. 2020;158:2294–2297. doi: 10.1053/j.gastro.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li C., Liu X., Chiu M.C., Zhao X., Wang D., Wei Y., Lee A., Zhang A.J., Chu H. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 2020;2020:13. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., China Novel Coronavirus Investigating and Research Team A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo T., Zhang F., Lui G.C.Y., Yeoh Y.K., Li A.Y.L., Zhan H., Wan Y., Chung A., Cheung C.P., Chen N. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.05.048. Published online May 19, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]