Abstract
Coronavirus disease 2019 (COVID-19) has become a worldwide threat to humans, and neutralizing antibodies have therapeutic potential. We have purified more than 1,000 memory B cells specific to SARS-CoV-2 S1 or its RBD (receptor binding domain) and obtain 729 paired heavy- and light-chain fragments. Among these, 178 antibodies test positive for antigen binding, and the majority of the top 17 binders with EC50 below 1 nM are RBD binders. Furthermore, we identify 11 neutralizing antibodies, eight of which show IC50 within 10 nM, and the best one, 414-1, with IC50 of 1.75 nM. Through epitope mapping, we find three main epitopes in RBD recognized by these antibodies, and epitope-B antibody 553-15 could substantially enhance the neutralizing abilities of most of the other antibodies. We also find that 515-5 could cross neutralize the SARS-CoV pseudovirus. Altogether, our study provides 11 potent human neutralizing antibodies for COVID-19 as therapeutic candidates.
Keywords: COVID-19, SARS-CoV-2, neutralizing antibodies, epitope, cross-neutralizing antibody, human antibodies, coronavirus, RBD, spike protein, infection
Graphical Abstract
Wan et al. identify 11 potent neutralizing antibodies against COVID-19 from 11 convalescent patients. These human neutralizing antibodies target three separate epitopes on the receptor binding domain (RBD) of the SARS-CoV-2 spike protein, and epitope-B antibody (553-15) can enhance the neutralizing abilities of most other antibodies.
Introduction
Over the last two decades in the 21st century, the outbreaks of several viral infectious diseases affected millions of people (Bauch and Oraby, 2013; Callaway et al., 2020; Chang et al., 2016; Huang et al., 2020; Peiris et al., 2003; Wu et al., 2020b; Zhou et al., 2020). Among these, three coronaviruses, SARS-CoV, MERS-CoV (Middle East respiratory syndrome coronavirus), and SARS-CoV-2 (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, 2020), have received significant attention especially due to the current outbreak of COVID-19 caused by SARS-CoV-2, and high mortality rates of the infected individuals. Most patients died due to severe pneumonia and multi-organ failure (Callaway et al., 2020; Chen et al., 2020b). Despite the existence of rare exceptions such as asymptomatic carriers, it is generally believed that if the infected individuals could not develop effective adaptive immune responses for viral clearance to prevent sustained infection, there are high chances for transformation into severe acute respiratory infection. Supporting this idea, treatment with convalescent plasma to COVID-19 patients showed significant clinical improvement and decreased viral load within days (Chen et al., 2020a). However, the sources of convalescent plasma are limited, and could not be amplified; therefore, effective and scalable treatments are still urgently needed.
Owing to recent rapid development of single-cell cloning technology, the process of antibody identification has been greatly shortened, from years to months. Therefore, human and humanized neutralizing antibodies represent great hopes for a prompt development of therapeutics in treating infectious diseases. In support of this, cocktail treatment of three mixed antibodies recognizing different epitopes, with one of them able to robustly neutralize, was successfully used in the curation of a British Ebola patient (Cao et al., 2018; Davey et al., 2016). Regarding coronaviruses, neutralizing antibodies against MERS were tested as effective in animals (Stalin Raj et al., 2018), although SARS-CoV neutralizing antibodies did not meet human requirements due to lack of patients after the development. Interestingly one of them, S309, was shown to be able to cross react with, and neutralize, SARS-CoV-2 (Pinto et al., 2020). However, the RBD (receptor binding domain) regions (which are key targets for viral neutralization, also see below) only share 74% amino acid sequence identity between the two SARS viruses (Tian et al., 2020), raising concerns about the effectiveness of SARS-CoV neutralizing antibodies against SARS-CoV-2.
The spike proteins of coronaviruses play an essential role in viral entry into human target cells. The S1 region, especially the RBD, primes the viral particle to human cell surface through the interaction with the receptor protein angiotensin I converting enzyme 2 (ACE2; Tai et al., 2020), which then triggers an infusion process primarily mediated by the S2 region (Walls et al., 2020). The primary amino acid sequences of the spike proteins of SARS-CoV and SARS-CoV-2 share 76% amino acid sequence identity throughout the full coding regions, with 79.59% similarity and 74% identity in RBD domains (Chan et al., 2020; Ou et al., 2020; Tian et al., 2020). Structure analyses revealed high 3D similarity between the spike proteins of the two viruses, and both trimerize and interact with ACE2 through the RBD domains (Wrapp et al., 2020). Importantly, the interaction between the SARS-CoV-2 RBD and ACE2 was assessed at around 1.2 nM (Hoffmann et al., 2020; Walls et al., 2020), which is 4-fold stronger than the SARS-CoV RBD. While this enhanced affinity may explain a much stronger spreading ability of SARS-CoV-2, it also suggests that finding potent neutralizing antibodies targeting SARS-CoV-2 RBD could be more challenging.
While this manuscript was under preparation, identification of multiple human neutralizing monoclonal antibodies had been reported (Cao et al., 2020; Chen et al., 2020c; Chi et al., 2020; Wang et al., 2020; Wu et al., 2020d). While these groups and ours all employed similar approaches and obtained authentic viral neutralizing antibodies, the performances of these antibodies varied in different assays, e.g., the correlation between binding affinities, pseudoviral and authentic viral neutralizing abilities. Nevertheless, more human antibodies either directly neutralizing SARS-CoV-2 or opsonizing free viral particles for rapid immune clearances are still needed.
Here, we report the identification of 178 S1 and RBD binding human monoclonal antibodies from the memory B cells of 11 recently recovered patients. A total of 17 antibodies showed binding affinities (EC50) lower than 1 nM. We further identified eight antibodies showing robust authentic viral neutralizing activities, and the best one, 414-1, showed neutralizing IC50 at 1.75 nM. Through epitope mapping, we found the eight antibodies bound three different RBD epitopes, and epitope-B antibody 553-15 could substantially enhance neutralizing abilities of most other neutralizing antibodies.
Results
Serological Responses and Single B Cell Isolation
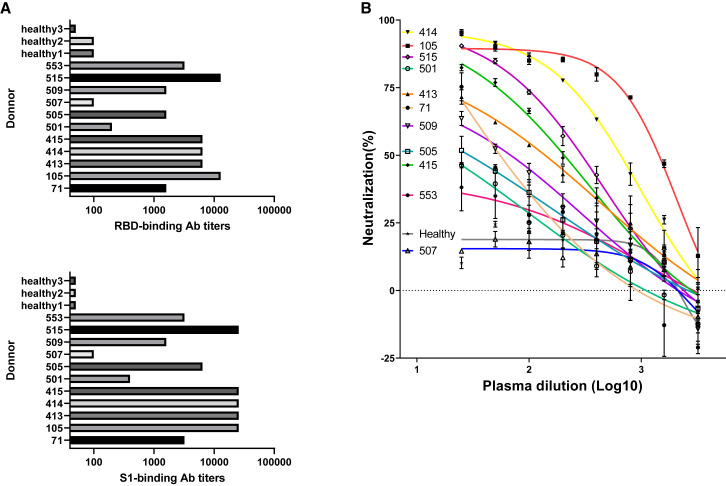
We screened sera samples from 11 patients recently recovered from COVID-19, and found all individuals showed certain levels of serological responses, with #507 and #501 being the weakest, to SARS-CoV-2 spike RBD and S1 proteins (Figure 1 A). We also found that 10 sera, except for 507, showed neutralization abilities against SARS-CoV-2 pseudoviral infection of HEK293T cells stably expressing human ACE2 (Figure 1B). Such observations, i.e., the sera from different individuals displayed a wide range of antibody responses, were consistent with a recent report (Wu et al., 2020a). Of note, the #509 blood sample was obtained on the second day after hospitalization (Table S2); the sera already showed weak S1 antigen response and pseudoviral neutralizing activities.
Figure 1.
Serological Responses of 11 Convalescent COVID19 Patients
(A and B) Spike protein binding (A) and pseudoviral neutralizing tests (B) of donor plasma. RBD and S1 were coated at 1 μg/mL for binding ELISA. Plasma samples of heathy donors were used as controls. The mean values and standard deviations of two technical replicates are shown in a pseudo-typed viral neutralization assay.
The RBD domain in the S1 region of SARS-CoV-2 spike protein is the critical region mediating viral entry through host receptor ACE2. Using recombinant RBD and S1 antigens, we then isolated RBD and S1-bound memory B cells for antibody identification using the PBMCs (peripheral blood mononuclear cells) from the 11 individuals by fluorescence activated cell sorting (Figure S1A).
Sequences encoding immunoglobulin heavy (IGH) and light (IGL) chains were amplified from single B cell complementary DNA samples after reverse transcription and then cloned through homologous recombination into mammalian expressing vectors (Robbiani et al., 2017). Overall, 729 naturally paired IGH and IGL clones were obtained, and the numbers of clones derived from each individual were listed in Table S1.
Identification and Characterization of Human Monoclonal Antibodies to SARS-CoV-2
In order to screen for SARS-CoV-2 spike antigen specific monoclonal antibodies, we used two primary assays based on ELISA (enzyme linked immunosorbent assay) and FCA (flow cytometry assay), respectively. Among the 729 candidate antibodies expressed in HEK293E cells, 178 were positive for RBD or S1 binding (Figure 2 A). All the positive clones were then sequenced. Notably, almost all (98.6%) of the sequences obtained were unique ones (Figure 2B), similar to what were previously reported in Ebola (Fan et al., 2020; Zhang et al., 2016) and yellow fever (Calvert et al., 2016) studies.
Figure 2.
The Identification and Characterization of Spike Protein-Specific Monoclonal Antibodies
(A) Characteristics of antibodies binding with RBD and S-ECD. RBD and S-ECD dual binders, red dots; binders for S-ECD only, purple; binders for RBD only, green; and weaker binders, black. Authentic neutralizing antibodies are represented by bigger dots.
(B) Maximum-likelihood phylogenetic tree analysis of the heavy chains of all sequenced monoclonal antibodies. Different colors indicate antibodies identified from individual patients.
(C) Summary of the performance of the top 29 monoclonal antibodies in the indicated assays.
Based on the ranking of ELISA values and FCA positivities, we focused on 29 antibodies for further characterization. We first measured the precise values of EC50 by ELISA, and identified 17 strong binders for S-ECD (extracellular domain) and RBD with EC50 below 1 nM, and the most potent one showing EC50 at 0.057 nM (8.55 ng/mL; Figures 2C and S2A). Of note, among these 29 antibodies, four antibodies (413-3, 414-4, 105-28, and 105-41) were FCA-negative but bound recombinant RBD relatively well in ELISA (Figure 2C). On the other hand, we also identified another four antibodies (505-8, 414-3-1, 505-17, and 515-5) that showed strong FCA positivity but with barely detectable ELISA signals (Figure 2C). These findings indicated certain conformational differences might exist between the recombinant and membrane-bound S antigens.
We also noticed that the majority of the 17 strong binders were RBD binders, except for 413-2, 553-13, and 553-18 (Figure 2C). And we further confirmed that 553-13 and 553-18 were NTD (N-terminal domain of spike protein) binders (Figure S2C). These results indicated that the RBD represents the primary immunogenic region of the S protein.
Identification of Neutralizing Antibodies by Pseudotyped and Authentic Viral Infection Assays
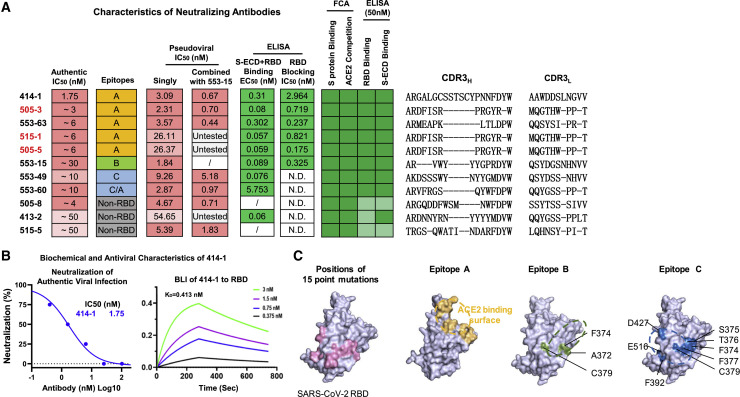
To identify neutralizing antibodies, we first employed pseudoviral infection assays using HEK293T-ACE2 cells. From all the antibodies tested, we found a total of 16 pseudoviral neutralizing antibodies (Figure 2C, and Figure 3 A, column 3). Among these, 11 could neutralize authentic virus entry into Vero-E6 cells, and eight of them showed potent IC50 within 10 nM (Figure 3A, column 1). We next characterized the best one, 414-1, which was able to effectively block authentic viral entry at IC50 of 1.75 nM (Figure 3B, left). We also tested 414-1 expressing in Chinese hamster ovary (CHO) cells, and found it could achieve ~300 mg/L without any optimization, suggesting great potential in therapeutic development. Furthermore, the RBD binding affinity of 414-1 was also tested by BLI (bio-layer interferometry assay), and showed comparable KD of 0.413 nM (Figure 3B, right).
Figure 3.
Identification and Epitope Mapping of Neutralizing Antibodies
(A) Summary of the indicated characteristics of all authentic viral neutralizing antibodies. Column 1, IC50 of authentic SARS-CoV-2 neutralization; column 2, epitope-mapping results (note that although 553-60 showed an identical pattern to 553-49 in the mutagenesis assay, it partially completed with 414-1, and therefore was classified as epitope C/A); column 3, IC50 of pseudoviral neutralizing results; column 4, cocktail IC50 of pseudoviral neutralizing results of the indicated antibodies together with 553-15 (note that 505-3 was used as representative for 515-1 and 505-5); columns 5 and 6, binding ELISA and blocking ELISA results; columns 7 and 8, FCA results of the binding and ACE2 competition to freshly expressed membrane-bound S protein (color shading in the heatmaps represents magnitude of intensities—light green, weak; darker green, strong); and columns 9 and 10, results of ELISA binding to RBD or S-ECD. Red highlights the antibodies with similar CDR3 sequences.
(B) Left, neutralization results of 414-1 against authentic virus (SARS-CoV-2-SH01) using Vero-E6 (n = 2). Right, BLI examination of 414-1 and RBD binding affinity.
(C) Left, positions of the 15 designed single amino acid replacements in the non-ACE2 binding surface of RBD, purple; middle-left, ACE2 binding surface, yellow; middle-right, amino acid replacements that affected epitope-B antibody 553-15 binding to RBD, green; right, amino acid replacements that affected the RBD bindings of 553-49 and 553-60, blue. Proposed epitopes A, B, and C were circled.
Since complementarity-determining region 3 (CDR3) is the most critical region for antibody diversity, we then aligned the CDR3 sequences of the heavy (CDR3H) and light (CDR3L) chains of the 11 authentic viral neutralizing antibodies and found nine unique ones (Figure 3A, right). Interestingly, 505-3, 505-5, and 515-1 shared identical CDR3H and CDR3L sequences, with only a few amino acid variations outside the CDR regions. These three antibodies indeed behaved very similarly in all assays except for pseudoviral neutralization assay (Figure 3A, column 3) tests. Of note, they were obtained from two individuals (#505 and #515).
Epitope Mapping of the Neutralizing Antibodies
To understand the neutralizing mechanism of the 11 antibodies, we performed epitope-mapping experiments for the eight RBD binders (note that 505-8, 413-2, and 515-5 were non-RBD binders; Figures 2C and 3A). We first utilized RBD-ACE2 blocking ELISA, and found that six of the RBD binders could effectively compete ACE2 binding to RBD with IC50 below 3 nM (Figure 3A, column 6), indicating their binding epitopes overlapping with the ACE2 binding surface (Figure 3C, middle left).
We then carried out BLI competition and mutagenesis assays for further analyses. Due to the identical CDR3 sequence and the high similarities among 505-3, 515-1, and 505-5 mentioned above, we only chose 505-3 as the representative in these assays. Based on BLI competition results, the eight RBD binders could be classified into three groups, namely epitopes A, B, and C (Figure 3A, column 2). We then utilized 15 point mutants of recombinant RBD (Wu et al., 2020c), each having one amino acid residue replaced in the non-ACE2 binding surface (Figure 3C, left) to locate these epitopes. All epitope-A antibodies were largely unaffected by these mutations (Figure S3C, left), indicating that epitope A is limited within ACE2 binding surface (Figure 3C, middle left), considering that they complete ACE2 binding in blocking ELISA, as mentioned above. Epitope-B antibody, 553-15, was sensitive to the F374A, A372T, and C379A mutations (Figure S3C, middle) and also complete ACE2 in blocking ELISA (Figure 3A, column 6); therefore, we speculated that epitope B should include these residues and partially overlap with the ACE2 binding surface (Figure 3C, middle right). Finally, residues critical for epitope-C antibody binding were shown in Figure 3C (right panel). Since they did not complete ACE2 in blocking ELISA (Figure 3A, column 6), we proposed epitope C at the indicated area of RBD (Figure 3C, right).
Notable, despite that the epitope C and non-RBD binders could not block ACE2 binding for RBD (Figure 3A, column 6), they all recognized freshly expressed membrane-bound S protein and completed ACE2 binding in FCA quite well (Figure 3A, columns 7 and 8), indicating they might execute their neutralizing activities through a membrane-dependent mechanism.
Cocktail Design for Neutralizing Antibodies
Since eight of the 11 authentic viral neutralizing antibodies were mapped to three different epitopes in RBD with the other three being non-RBD binders, these antibodies bound at least four different regions in S protein. We then wanted to examine whether certain cocktail combinations could significantly enhance the neutralizing abilities of these antibodies. After systematic investigation using pseudoviral assay, we found that epitope-B antibody, 553-15, could significantly enhance the neutralizing abilities of epitope-A antibodies (414-1, 505-3, and 553-63) by around 5 folds (Figure 3A, columns 3 and 4). Interestingly, 553-15 also potentiated 553-60 and 505-8 in the same assay.
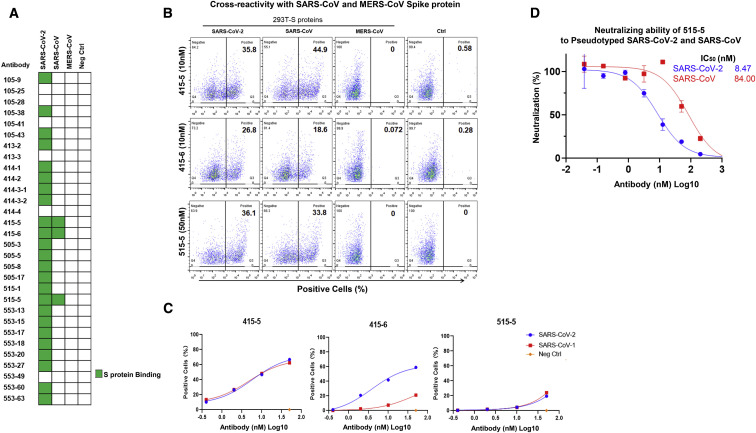
Cross Reactivity with SARS-CoV Spike Protein
The spike proteins of SARS-CoV-2 share 76% and 35% of amino acid identities with SARS-CoV and MERS-CoV, respectively. Therefore, we wondered whether our antibodies could cross react with the S proteins of these two other coronaviruses. In order to do so, we overexpressed the S proteins of SARS-CoV-2, SARS-CoV, and MERS-CoV in HEK293T, and tested the cross reactivities by flow cytometry analyses. From this exercise, we found three antibodies, 415-5, 415-6, and 515-5, cross recognizing SARS-CoV S, but not MERS-CoV S (Figures 4A and 4B). 415-5 and 515-5 shared similar S protein affinities between SARS-CoV-2 and SARS-CoV, but 415-6 had much lower affinity toward SARS-CoV S compared to SARS-CoV-2 S (Figure 4C). Furthermore, 515-5 showed relative weaker but detectable neutralizing activity of pseudotyped SARS-CoV with IC50 at 84 nM (Figure 4D), indicating a potential for further development against the homologous SARS family virus.
Figure 4.
Cross Reactivity and Neutralization with SARS-CoV
Cross-reactivity analysis of the indicated antibodies against S proteins of SARS-CoV and MERS-CoV. Flow cytometry analyses were performed using HEK293T cells expressing the S proteins of SARS-CoV-2, SARS-CoV, and MERS-CoV; 50 nM of the indicated antibodies were used. Non-transfected HEK293T were used as controls.
(A) Heatmap summary of the results.
(B) Flow cytometry analyses of the three cross-reactive antibodies.
(C) Binding curves of the three cross-reactive antibodies.
(D) Cross neutralization of 515-5 to pseudoviruses of SARS-CoV-2 and SARS-CoV. Data were obtained from a representative experiment containing two replicates. Data are represented as mean ± SD, and n = 2.
Discussion
Within the last few weeks, several groups have published a panel of full human neutralizing antibodies against COVID19 (Brouwer et al., 2020; Cao et al., 2020; Chen et al., 2020c; Chi et al., 2020; Wang et al., 2020; Wu et al., 2020d). Here, we have added 11 more to the reservoir, with eight of them showed neutralizing IC50 within 10 nM.
In our study, in order to provide good therapeutic candidates of monoclonal antibodies, we have implemented multiple approaches during the triage processes before authentic viral testing. Our triage approaches included: (1) RBD and S binding abilities by ELISA; (2) the ability to recognize membrane-bound S protein by FCA; (3) ACE2 competition abilities using both ELISA and FCA; and (4) the pseudotyped and authentic viral neutralization assay. The best three antibodies, 414-1, 505-3, and 553-63, showed consistent performances in all of these assays (Figure S5). The other five antibodies with IC50 within 10 nM for authentic viral neutralization also showed relatively consistent performances in most of the assays (Figure S5). In general, seven of the top eight neutralizing antibodies showed robust binding in ELISA (Figure 3A, columns 1 and 5). The only one exception is 505-8; however, it functioned well in membrane-bound S binding and competition assays detected by FCA (Figure 3A, columns 7 and 8; also see below), indicating that binding abilities are the fundamental quality for neutralizing activities in our study. And we observed a general increase of neutralizing IC50 values compared to binding affinity EC50 values. However, compared with other studies mentioned above, we found that the leading antibodies reported in the studies by Cao et al. (2020), Chi et al. (2020), and Wu et al. (2020d) showed great neutralizing abilities, with IC50 ranging from 0.1 to 1 nM, and EC50 of 0.8 to 70 nM. Similarly, a 100-fold enhancement of neutralizing activity compared to the affinity was also reported by Chi et al. (2020) for their leading antibodies. The reason for such a discrepancy is still unknown at this point, but conformation differences between recombinant S and RBD protein and their natural forms may be one possibility; another possibility may be that assay conditions could vary among different groups.
Among the top 29 antibodies (Figure 2C), 24 were obtained from B cells isolated using the S1 protein. We found the majority, 18 of 24, are RBD binders; these findings indicated that the RBD region is the primary epitope for immune responses. Similar observations were also reported elsewhere for SARS-CoV-2 (Cao et al., 2020; Wu et al., 2020d) as well as SARS-CoV (Berry et al., 2010; Yuan et al., 2017). We also found another four antibodies, 505-8, 414-3-1, 505-17, and 515-5, showed barely any ELISA signals toward both RBD and S, but could robustly bind freshly expressed S protein in A549 membrane (Figure 2C, columns 4 and 6). These included two neutralizing antibodies 505-8 (mentioned above) and 515-5, indicating that the recombinant RBD or S protein may differ from the membrane-bound S in terms of 3D conformation. We would have missed these antibodies if we had used ELISA, alone, for antibody triage. Therefore, future antibody study should consider multiple approaches for the initial identification and for quality control.
Through epitope mapping, we could largely locate the epitopes of the eight RBD binding neutralizing antibodies. Based on all analyses, we propose that epitope A locates at the top of RBD where direct ACE2 interaction occurs (Figure 3C). Epitope-A antibodies also represented the largest group of neutralizing antibodies in our study. Interestingly, we found that all recently published human SARS-CoV-2 neutralizing antibodies with structure information bound the same or similar areas on RBD (Figure S3D). The findings of epitopes B and C, and the non-RBD binders in neutralizing SARS-CoV-2, are of importance, particularly for epitope-B antibody 553-15, which could substantially potentiate several other antibodies for their neutralizing abilities. Future structure analysis will be needed for detailed understanding of how the antibody binding of these different epitopes result in neutralization of SARS-CoV-2 infection.
It’s encouraging to see the increasing numbers of human neutralizing antibodies against COVID-19. The combination of different clones of potent neutralizing human monoclonal antibodies recognizing different vulnerable sites of SARS-CoV2 can prevent the occurrence of mutant escapes when administered clinically. Therefore, more candidate antibody sequences could greatly facilitate the therapeutic development in curing COVID-19 patients.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-Mouse IgG Secondary Antibody HRP conjugated | SAB | Cat# L3032; RRID:AB_895481 |
| Goat anti-Rabbit IgG Secondary Antibody HRP conjugated | SAB | Cat# L3012; RRID:AB_895483 |
| HRP-Conjugated 6∗His, His-Tag Antibody | Proteintech | Cat# HRP-66005; RRID:AB_2857904 |
| F(ab’)2-Goat anti-Mouse IgG (H+L) Secondary Antibody, HRP | Invitrogen | Cat# A24512; RRID:AB_2535981 |
| F(ab’)2-Goat anti-Human IgG (H+L) Secondary Antibody, HRP | Invitrogen | Cat# A24464; RRID:AB_2535933 |
| Goat F(ab’)2 Anti-Human IgG - Fc (PE), pre-adsorbed | Abcam | Cat# ab98596; RRID:AB_10673825 |
| Bacterial and Virus Strains | ||
| SARS-CoV-2 strain nCoV-SH01 | Fudan University | N/A |
| SARS-CoV-2 pseudotyped virus | This paper | N/A |
| Top10 Competent Cells | SMART-Lifesciences | Cat# SLB010 |
| Biological Samples | ||
| PBMCs from COVID-19 convalescent patients | Shanghai Public Health Clinical Center | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| S Protein RBD-SD1 (Mammalian,C-mFc) | Novoprotein Scientific Inc. | Cat# DRA38 |
| ACE-2 (Mammalian,C-6His) | Novoprotein Scientific Inc. | Cat# C419 |
| SARS-CoV-2 Spike protein (ECD, His & Flag Tag) | Genscript | Cat# Z03481 |
| Mutant RBD proteins | Ying Tianlei Lab | N/A |
| Certified Fetal Bovine Serum (FBS) | BI | Cat# 04-001-1A |
| Polyethylenimine (PEI) | Sigma | Cat# 764604 |
| DPBS, no calcium, no magnesium | GIBCO | Cat# 14190144 |
| TMB Substrate Solution | SMART-Lifesciences | Cat# SLR006 |
| rProtein A Beads 4FF | SMART-Lifesciences | Cat# SA012C |
| 0.4% (w/v) Trypan blue stain | ThermoFisher | Cat# 15250061 |
| EDTA 0.5M, pH8.0, RNase-free | ThermoFisher | Cat# AM9260G |
| Critical Commercial Assays | ||
| Bright-Glo™ Luciferase Assay System | Promega | Cat# E2620 |
| Smart Assembly Cloning kit | SMART-Lifesciences | Cat# SC004250 |
| Experimental Models: Cell Lines | ||
| HEK293E | Xu Yanhui Lab | N/A |
| Vero-E6 cells | ATCC | Cat# CRL-1586 |
| Modified CHO cells | ZHEJIANG HISUN PHARMACEUTICAL Co., LTD. | N/A |
| A549 | ATCC | Cat# CCL-185 |
| Recombinant DNA | ||
| SARS-CoV-2 (YP_009724390.1) | Genscript | N/A |
| SARS-CoV (NP_828851.1) | Genscript | N/A |
| MERS (AFS88936.1) | Genscript | N/A |
| pCDNA3.4 modified | This paper | N/A |
| Software and Algorithms | ||
| Pymol | Pymol | N/A |
| GraphPad Prism 8 | GraphPad Prism | N/A |
| RAxML | Kozlov et al., 2019 | https://doi.org/10.1093/bioinformatics/btz305 |
Resource Availability
Lead Contact
Further information and requests for resources and reagents may be directed to and will be fulfilled by the Lead Contact, Fei Lan (fei_lan@fudan.edu.cn)
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
This study did not generate any unique datasets or code.
Experimental Model and Subject Details
Cell lines and Viruses
Vero-E6, A549-Spike (A549 expressing SARS-CoV-2 Spike protein), and A549-ACE2 (A549 expressing human ACE2) cell lines were supplied by Shanghai Public Health Clinical Center, Fudan University. Pseudovirus of SAR2-CoV-2 was generated by Shanghai Public Health Clinical Center, and Fudan University and SARS-CoV-2-SH01 was from BSL-3 of Fudan University. The experiments involving authentic COVID-19 virus were performed in Fudan University biosafety level 3 (BSL-3) facility. The overall study was reviewed and approved by the SHAPHC Ethics Committee (approval no. 2020-Y008-01).
Method Details
B cell sorting and single cell RT-PCR
Samples of peripheral blood for serum or PBMCs isolation were obtained from Shanghai Public Health Clinical Center, per 5 mL blood. PBMCs were purified using the gradient centrifugation method with Ficoll and cryopreserved in 90% heat-inactivated fetal bovine serum (FBS) supplemented with 10% dimethylsulfoxide (DMSO), storage in liquid nitrogen.
The fluorescently labeled S1 bait was previously prepared by incubating 5 μg of His tag-S1 protein with Anti His tag antibody-PE (Phycoerythrin) for at least 1 hr at 4°C in the dark, RBD bait performed as before. PBMCs were stained using 7AAD, anti-human CD19 (Allophycocyanin, APC), IgM [PE-Cy7], IgG (fluorescein isothiocyanate, FITC), PE labeled Antigen. Single antigen specific memory B cells were sorted on BD FACS Aria II into 96-well PCR plates (Axygen) containing 10 μl per well of lysis buffer (10 mM Dulbecco’s phosphate buffered saline, DPBS, 4 U Mouse RNase Inhibitor, NEB). The final sorted cells were live cells, CD19+ IgM- IgG+ and antigen+. Plates were immediately frozen on dry ice and stored at −80°C or processed for cDNA synthesis.
Single cell RT-PCR reactions were performed following a published protocol using random N5 for initial priming of the first chain synthesis (Ho et al., 2016). Reverse transcription and subsequent PCR amplification of heavy and light chain variable genes were performed using SuperScript III (Life Technologies). First and second PCR reactions were performed in 50 μL volume with 5 μL of reaction product using PCR mixture (SMART-Lifesciences). We used the primer sets published by Smith et al. (2009). PCR products were then purified using DNA FragSelect XP Magnetic Beads (SMART-Lifesciences) and cloned into human IgG1, lambda or kappa expression plasmids for antibody expression by seamless cloning method (see below). After transformation, individual colonies were picked for sequencing and characterization. Sequences were analyzed using IMGT/ V-QUEST (http://www.imgt.org/IMGT_vquest) and IgBlast (IgBLAST, http://www.ncbi.nlm.nih.gov/igblast).
Expression and purification of human monoclonal antibodies
The antibody VH/VL and constant region genes were then amplified and cloned into expression vector pcDNA3.4 using SMART Assembly Cloning Kit (SMART-Lifesciences), subsequently antibodies plasmids were amplified in competent cells (SMART-Lifesciences). HEK293E cells were transfected using polyethylenimine (PEI, Sigma), after 4-5 days of cell culture, antibodies purification was processed from supernatants. Antibodies were purified by Protein A magnetic beads (SMART-Lifesciences) for 30 min at room temperature, then eluted by 100 mM glycine pH 3.0 and quickly neutralized by TrisCl pH 7.4.
Enzyme linked immunosorbent assay (ELISA)
96-well plates (Falcon and MATRIX) were coated overnight at 4°C with 0.5 μg/mL SARS-CoV-2 RBD-mFC (Novoprotein Scientific Inc.), and 0.6 μg/mL SARS-CoV-2 S-ECD (GenScript). After washes with PBST (SMART-Lifesciences), the plates were blocked using 3% non-fat milk in PBST for 1 h at 37°C. Series dilutions of antibodies in PBST were added to each well and incubated at 37°C for 1 h. Then the antibodies were removed and washed 3 times by PBST (phosphate buffer solution with tween), HRP-conjugated anti-human IgG Fab antibody (Sigma) was added at the dilution of 1:10,000 in PBST containing 3% BSA (Sangon Biotech) and incubated at 37°C for 0.5 h. After washing with PBST three times, tetramethylbenzidine (TMB) solution (SMART-Lifesciences) was added to the microplate and incubated at room temperature for 5-10 min, followed by adding 1M HCl to terminate the reaction. The OD450 absorbance was detected by Synergy HT Microplate Reader (Bio-Tek). The curves and EC50 were analyzed by GraphPad Prism 8.0.
Bio-layer interferometry assay (BLI)
Measuring Kinetics of antibodies interact with S protein RBD using by Bio-layer Interferometry. RBD diluted in a 2-fold series by assay buffer (20mM Morpholinepropanesulfonic acid (MOPs), 50 nM KCl, pH 7.4).
Flow cytometry assays (FCA)
For Figure 4, flow cytometry analyses were performed to detect the binding abilities of antibodies to Spike protein in HEK293T cells freshly expressing of SARS-CoV-2, SARS-CoV and MERS-CoV. Briefly, 10 thousand cells in 100 μl were incubated with indicated antibodies for 30 min at room temperature, after twice washes then PE-labeled goat anti-human IgG-Fc antibody was added (1:5,000; Abcam) for 30 min, followed by flow cytometry analyses.
Flow cytometry analysis was also performed to detect the ACE2 competition (Figures 3A and S3). Then 10 thousand S expressing cells in 100 μl were first incubated with free ACE2 at 50nM for 30 min at room temperature, then different concentrations of antibodies were added for 30 min, followed by incubation with PE-labeled goat anti-human IgG-Fc antibody (1:5000; Abcam) for 30 min and analyzed by flow cytometry.
For Figure S4, to test S protein expression on cell membrane, recombinant ACE2-Cter-6xHis labeled by rabbit anti-His-PE antibody in 1.2:1 (n:n) ratio was added to 10 thousand cells expressing S protein. Detection was done similarly as above.
Virus neutralization assay (pseudotyped and authentic)
Coding fragments of SARS-CoV-2 (YP_009724390.1),SARS-CoV (NP_828851.1) and MERS (AFS88936.1) Spike proteins were synthesized (GenScript) and cloned into pcDNA3.1. SARS-CoV-2, SARS-CoV and MERS pseudotyped viruses were produced as previously described (Qiu et al., 2013). Briefly, pseudovirus were generated by co-transfection of 293T cells with pNL4-3.Luc.R-E- backbone and the SARS-CoV-2 spike protein expression plasmid and the supernatants were harvested after 48 hr, and followed by centrifuge at 2,000 rpm for 5 min and stored in −80°C.
The neutralization assay was performed as the following steps. Pseudovirus was diluted in complete DMEM mixed with or without an equal volume (50 μl) of diluted serum or antibody and then incubated at 37°C for 1 h. The mixtures were then transferred to 96-well plate seeded with 20,000 293T-ACE2 cells for 12h and incubated at 37°C for additional 48 hr. Assays were developed with bright glo luciferase assay system (Promega), and the relative light units (RLU) were read on a luminometer (Promega GloMax 96). The titers of neutralizing antibodies and activities of plasma were calculated as 50% inhibitory dose (ID50) compared to virus control.
Cocktail neutralization assay was performed with 2 antibodies by 1:1 (n:n), and calculated IC50 by total antibodies concentrate ion.
All experiments related to authentic virus were done in BSL-3. Monoclonal antibodies were incubated with 200 PFU SARS-CoV-2 SH01 at 37°C for 1 hour before added into Vero-E6 cell culture (96-well plate, 4 × 104 cells per well), and the cells were continued for 48 hr before microscopic analyses for CPE (cytopathic effect).
Quantification and Statistical Analyses
The descriptive statistics mean ± SEM or mean ± SD were determined for continuous variables as noted. EC50 and IC50 values in this study were determined after log10 transformation of antibody concentration using a 4-parameters nonlinear fit analysis (Prism V8.0, GraphPad). Technical and biological replicates are indicated in the figure legends.
Acknowledgments
We appreciate Novoprotein Scientific Inc. for gifting SARS-CoV-2 Spike, ACE2 and related recombinant proteins. Prof. Lu Lu and Prof. Zhigang Lu from Fudan University provided the 293T-ACE2 cell line and helpful suggestions. We thank Prof. Lilin Ye from the Third Military Medical University for helpful suggestions and manuscript writing. We thank Yiwei Tang from Fudan University for help in drawing the maximum-likelihood phylogenetic tree. The HEK293E cell line was a gift from Prof. Yanhui Xu, Fudan University. This work was supported by the National Major Science and Technology Projects of China (2018ZX10301403); the National Key Research and Development Program of China (2016YFA0101800 and 2018YFA0108700); the National Natural Science Foundation of China (31925010); the Shanghai Municipal Science and Technology Major Project (2017SHZDZX01); the Zhejiang University Special Scientific Research Fund for COVID-19 Prevention and Control (2020XGZX023); the Shanghai Pujiang Program (19PJ1409100); Natural Science Foundation of China (81761128007); and Shanghai Committee of Science and Technology (18DZ2293000).
Author Contributions
F.L., J.X., Y.L., and Y.X. conceived the project; J.W., S.X., and L.D. performed most of experiments with assistance from Y.W.; C.G. performed the authentic virus neutralization; Y.W. performed the BLI assay; B.R. performed binding surface site analysis; C.L. provided point mutation RBD proteins; S.W., K.C., C.H., D.Z., S.Y., C.Q., C.Z., L.N., Z.G., J.J., X.Z., X.W., T.Y., and H.W. helped to improve the experiments; all authors contributed to data analyses; and F.L., J.W., S.X., L.D., and Y.L. wrote the manuscript.
Declaration of Interests
Fei Lan is a shareholder and a scientific advisor of Active Motif China. Yanan Lu, Fei Lan, Jianqing Xu, Longfei Ding, Jinkai Wan, Shenghui Xing, and Yongheng Wang are listed as inventors on a patent application related to this work.
Published: July 3, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107918.
Supplemental Information
References
- Bauch C.T., Oraby T. Assessing the pandemic potential of MERS-CoV. Lancet. 2013;382:662–664. doi: 10.1016/S0140-6736(13)61504-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J.D., Hay K., Rini J.M., Yu M., Wang L., Plummer F.A., Corbett C.R., Andonov A. Neutralizing epitopes of the SARS-CoV S-protein cluster independent of repertoire, antigen structure or mAb technology. MAbs. 2010;2:53–66. doi: 10.4161/mabs.2.1.10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. bioRxiv. 2020 doi: 10.1126/science.abc5902. 2020.2005.2012.088716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E., Cyranoski D., Mallapaty S., Stoye E., Tollefson J. The coronavirus pandemic in five powerful charts. Nature. 2020;579:482–483. doi: 10.1038/d41586-020-00758-2. [DOI] [PubMed] [Google Scholar]
- Calvert A.E., Dixon K.L., Piper J., Bennett S.L., Thibodeaux B.A., Barrett A.D.T., Roehrig J.T., Blair C.D. A humanized monoclonal antibody neutralizes yellow fever virus strain 17D-204 in vitro but does not protect a mouse model from disease. Antiviral Res. 2016;131:92–99. doi: 10.1016/j.antiviral.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao P., Bai H., Wang X., Che J. Role of the Ebola membrane in the protection conferred by the three-mAb cocktail MIL77. Sci. Rep. 2018;8:17628. doi: 10.1038/s41598-018-35964-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020 doi: 10.1016/j.cell.2020.05.025. Published online May 18, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.-W., Kok K.-H., Zhu Z., Chu H., To K.K.-W., Yuan S., Yuen K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C., Ortiz K., Ansari A., Gershwin M.E. The Zika outbreak of the 21st century. J. Autoimmun. 2016;68:1–13. doi: 10.1016/j.jaut.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Xiong J., Bao L., Shi Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect. Dis. 2020;20:398–400. doi: 10.1016/S1473-3099(20)30141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Li R., Pan Z., Qian C., Yang Y., You R., Zhao J., Liu P., Gao L., Li Z. Human monoclonal antibodies block the binding of SARS-CoV-2 spike protein to angiotensin converting enzyme 2 receptor. Cell. Mol. Immunol. 2020;17:647–649. doi: 10.1038/s41423-020-0426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y. A potent neutralizing human antibody reveals the N-terminal domain of the Spike protein of SARS-CoV-2 as a site of vulnerability. bioRxiv. 2020 doi: 10.1126/science.abc6952. 2020.2005.2008.083964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey R.T., Jr., Dodd L., Proschan M.A., Neaton J., Neuhaus Nordwall J., Koopmeiners J.S., Beigel J., Tierney J., Lane H.C., Fauci A.S., PREVAIL II Writing Group. Multi-National PREVAIL II Study Team A randomized, controlled trial of ZMapp for Ebola virus infection. N. Engl. J. Med. 2016;375:1448–1456. doi: 10.1056/NEJMoa1604330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan P., Chi X., Liu G., Zhang G., Chen Z., Liu Y., Fang T., Li J., Banadyga L., He S. Potent neutralizing monoclonal antibodies against Ebola virus isolated from vaccinated donors. MAbs. 2020;12:1742457. doi: 10.1080/19420862.2020.1742457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho I.Y., Bunker J.J., Erickson S.A., Neu K.E., Huang M., Cortese M., Pulendran B., Wilson P.C. Refined protocol for generating monoclonal antibodies from single human and murine B cells. J. Immunol. Methods. 2016;438:67–70. doi: 10.1016/j.jim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov A.M., Darriba D., Flouri T., Morel B., Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35:4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Chu C.M., Cheng V.C., Chan K.S., Hung I.F., Poon L.L., Law K.I., Tang B.S., Hon T.Y., Chan C.S., HKU/UCH SARS Study Group Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020 doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Qiu C., Huang Y., Zhang A., Tian D., Wan Y., Zhang X., Zhang W., Zhang Z., Yuan Z., Hu Y. Safe pseudovirus-based assay for neutralization antibodies against influenza A(H7N9) virus. Emerg. Infect. Dis. 2013;19:1685–1687. doi: 10.3201/eid1910.130728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Bozzacco L., Keeffe J.R., Khouri R., Olsen P.C., Gazumyan A., Schaefer-Babajew D., Avila-Rios S., Nogueira L., Patel R. Recurrent potent human neutralizing antibodies to Zika virus in Brazil and Mexico. Cell. 2017;169:597–609 e511. doi: 10.1016/j.cell.2017.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K., Garman L., Wrammert J., Zheng N.Y., Capra J.D., Ahmed R., Wilson P.C. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat. Protoc. 2009;4:372–384. doi: 10.1038/nprot.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalin Raj V., Okba N.M.A., Gutierrez-Alvarez J., Drabek D., van Dieren B., Widagdo W., Lamers M.M., Widjaja I., Fernandez-Delgado R., Sola I. Chimeric camel/human heavy-chain antibodies protect against MERS-CoV infection. Sci. Adv. 2018;4:eaas9667. doi: 10.1126/sciadv.aas9667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W., He L., Zhang X., Pu J., Voronin D., Jiang S., Zhou Y., Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292 e286. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Wang A., Liu M., Wang Q., Chen J., Xia S., Ling Y., Zhang Y., Xun J., Lu L. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv. 2020 2020.2003.2030.20047365. [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Li C., Xia S., Tian X., Kong Y., Wang Z., Gu C., Zhang R., Tu C., Xie Y. Identification of human single-domain antibodies against SARS-CoV-2. Cell Host Microbe. 2020;27:891–898 e895. doi: 10.1016/j.chom.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., Lu G., Wu Y., Yan J., Shi Y. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Gui M., Niu X.F., He S.H., Wang R.K., Feng Y.P., Kroeker A., Zuo Y.N., Wang H., Wang Y. Potent neutralizing monoclonal antibodies against Ebola virus infection. Sci Rep-Uk. 2016;6:25856. doi: 10.1038/srep25856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code.