Abstract
Triple-negative breast cancer (TNBC) is an aggressive subtype of breast cancer that lacks expression of the three most common receptors present on other subtypes, leaving it unsusceptible to current targeted or hormonal therapies. In this study, we introduce an alternative treatment strategy for TNBC that exploits its overexpression of Notch1 receptors and its underexpression of the tumor suppressive microRNA (miRNA) miR-34a. Studies have shown that introducing mimics of miR-34a to TNBC cells effectively inhibits cancer growth, but miR-34a cannot be administered in the clinic without a carrier. To enable delivery of miR-34a to TNBC cells, we encapsulated miR-34a mimics in poly(lactic-co-glycolic acid) nanoparticles (NPs) that were functionalized with Notch1 antibodies to produce N1-34a-NPs. In addition to binding Notch1 receptors overexpressed on the surface of TNBC cells, the antibodies in this formulation enable suppression of Notch signaling through signal cascade interference. Herein, we present the results of in vitro experiments that demonstrate N1-34a-NPs can regulate Notch signaling and downstream miR-34a targets in TNBC cells to induce senescence and reduce cell proliferation and migration. These studies demonstrate that NP-mediated co-delivery of miR-34a and Notch1 antibodies is a promising alternative treatment strategy for TNBC, warranting further optimization and in vivo investigation in future studies.
Keywords: gene regulation, miRNA replacement therapy, multivalency, Notch signaling, cancer nanomedicine, signal cascade interference, poly(lactic-co-glycolic acid), nanoparticles, oncology, drug delivery
Graphical Abstract
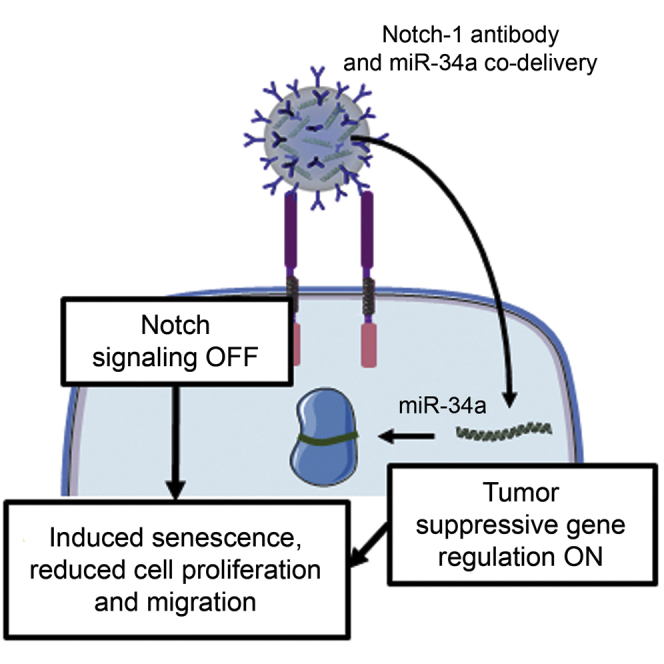
Triple-negative breast cancer is an aggressive disease with poor clinical outcomes. This study demonstrates that nanoparticles that co-deliver Notch1 antibodies and miR-34a to triple-negative breast cancer cells can reduce cell proliferation and migration and activate senescence, indicating that simultaneous regulation of Notch signaling and miR-34a is a promising treatment strategy.
Introduction
Triple-negative breast cancer (TNBC) is an aggressive subtype of breast cancer that accounts for 15%–25% of all breast cancer cases.1,2 Its characteristic lack of expression of common receptors present on other subtypes of breast cancer (the estrogen, progesterone, and human epidermal growth factor 2 receptors) leaves TNBC unsusceptible to current targeted or hormonal therapies, leading to high mortality and recurrence rates.1, 2, 3 In this study, we introduce a promising alternative treatment strategy for TNBC that exploits its overexpression of Notch1 receptors4, 5, 6 to enable combination antibody-mediated signal cascade interference and microRNA (miRNA) replacement therapy.7
miRNA replacement therapy involves the delivery of exogenous miRNA mimics to diseased cells to regulate gene expression. Upon cell entry, miRNA mimics bind specific messenger RNA (mRNA) molecules with either perfect or imperfect complementarity to induce mRNA degradation or translational repression, respectively.8 In TNBC, the tumor suppressor miRNA miR-34a is frequently downregulated or deleted, leading to enhanced cell survival, proliferation, and migration.9,10 Delivering miR-34a mimics to these cells can effectively regulate downstream expression of a broad network of genes, resulting in phenotypic changes such as reduced proliferation and migration and onset of apoptosis or senescence.10, 11, 12 Unfortunately, naked miRNA mimics cannot be used clinically, as they experience significant barriers to delivery.8 For effective miRNA replacement therapy, miRNA mimics must avoid early clearance and degradation in the bloodstream, cross blood vessel walls, travel through the extracellular matrix of the tumor microenvironment, enter the target cell, escape endosomes, and load into the RNA-induced silencing complex (RISC) to enable gene regulation.8 In this work, we encapsulate miR-34a mimics in poly(lactic-co-glycolic acid) (PLGA) nanoparticles (NPs) to overcome these delivery challenges.
Several NP formulations have been developed to deliver miR-34a mimics to TNBC.13, 14, 15, 16 Our group has previously created two layer-by-layer (LbL) NP systems that utilize either gold nanoshell13 or PLGA14 cores for TNBC therapy via miR-34a replacement. LbL NP formulations for miR-34a delivery to TNBC cells have also been developed using nanodiamond, protamine, and folic acid constructs.15 Finally, hyaluronic acid-chitosan NPs co-encapsulating miR-34a and doxorubicin (DOX) have been shown to suppress the expression of Bcl-2 to enhance the therapeutic efficacy of DOX.16 Taken together, these studies demonstrate the potential for enabling delivery of miR-34a mimics in combination with other therapeutic modalities as an alternative treatment strategy for TNBC. Intriguingly, several of these studies14,16 and others17, 18, 19 have shown that miR-34a delivery can regulate Notch signaling, showing there is crosstalk between the pathways and suggesting that combining miR-34a delivery with Notch inhibition may be a potent therapeutic approach. Herein, we present PLGA NPs loaded with miR-34a mimics and functionalized with Notch1 antibodies as a system that can simultaneously regulate Notch1 signaling and enable tumor suppressive gene regulation via miR-34a delivery.
In this work, we coated miR-34a-loaded NPs with Notch1 antibodies to produce N1-34a-NPs that not only deliver miR-34a to TNBC cells, but also inhibit the Notch signaling pathway, which is aberrantly expressed in TNBC.5,6 The overexpression of Notch1 receptors on the surface of TNBC cells serves as a resource for NP attachment, which may facilitate NP retention in the tumor microenvironment.20 In this study, we show that after binding TNBC cells, N1-34a-NPs can prevent Notch signaling activation by locking Notch1 receptors in a non-responsive state and blocking receptor-ligand interactions. Critically, our group and others have previously demonstrated that antibody-NP conjugates are much more effective than freely delivered antibodies because antibody-NP conjugates exhibit multivalent binding that yields enhanced signal cascade interference.21,22 Thus, by functionalizing our NP formulation with Notch1 antibodies, we can suppress Notch signaling while also delivering miR-34a to the desired cells. We present the results of in vitro studies that confirm N1-34a-NPs can effectively regulate both Notch signaling targets and miR-34a downstream targets in TNBC cells to induce senescence and reduce cell proliferation and migration. These exciting observations provide evidence warranting further investigation of this system as an effective treatment for TNBC.
Results
Characterization of Antibody and miRNA Loading in N1-34a-NPs and Analysis of NP Interaction with TNBC Cells
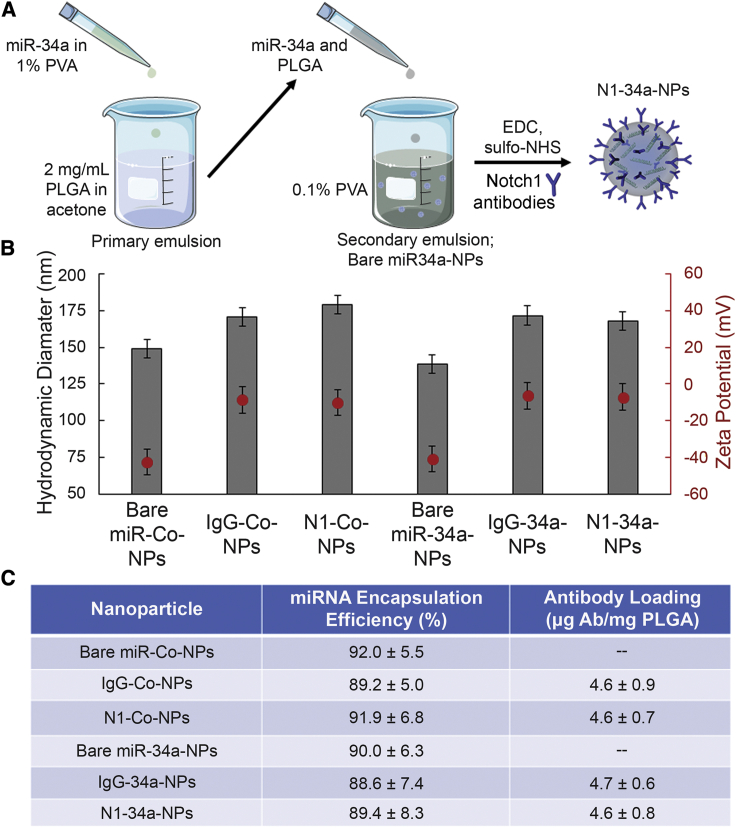
N1-34a-NPs and control NPs carrying either non-silencing miRNA (miR-Co), nonspecific immunoglobulin G (IgG) antibodies, or both were synthesized as depicted in Figure 1A and characterized using dynamic light scattering (DLS), zeta potential, OliGreen, and enzyme-linked immunosorbent assay (ELISA) measurements, which demonstrated that IgG or Notch1 antibodies were successfully conjugated to the surface of miR-Co- or miR-34a-loaded NPs. Prior to antibody conjugation, miRNA-loaded NPs had a hydrodynamic diameter and zeta potential of approximately 140 nm and −40 mV, respectively, with 0.37 nmol miR-Co and 0.36 nmol miR-34a encapsulated per 2 mg of PLGA (Figures 1B and 1C). After IgG or Notch1 antibody conjugation, the hydrodynamic diameter of NPs increased by approximately 20 nm and the zeta potential increased to around −8 mV, which indicates successful antibody attachment (Figure 1B). Antibody attachment was confirmed by ELISA measurements, which showed that the NPs contained approximately 4.6 μg of antibodies per mg of PLGA. OliGreen assays indicated that approximately 0.01 nmol of miRNA was lost from the NPs during the antibody functionalization procedure, leaving around 0.35 nmol of miRNA encapsulated per 2 mg of PLGA (Figure 1C). Altogether, these data demonstrate that miRNA can be successfully loaded in antibody-functionalized PLGA NPs.
Figure 1.
Synthesis and Characterization of N1-34a-NPs
(A) Scheme depicting N1-34a-NP synthesis. (B) Hydrodynamic diameter and zeta potential of bare miR-Co-NPs, bare miR-34a-NPs, IgG-Co-NPs, IgG-34a-NPs, N1-Co-NPs, and N1-34a-NPs. Error bars indicate standard error. (C) Encapsulation efficiency of miRNA and loading of antibodies on the different NP formulations used in this study. Data represent means and standard deviations.
Following NP synthesis and characterization, we evaluated the interaction of Notch1 and IgG antibody-conjugated NPs with MDA-MB-231 TNBC cells and MCF-10A healthy mammary epithelial cells using flow cytometry. For these studies, the NPs were loaded with Cy5-tagged miR-Co in order to facilitate fluorescence analysis of NP delivery to cells. TNBC cells were first treated with IgG-Cy5-NPs or N1-Cy5-NPs at doses of 50 nM miRNA for 1, 4, 8, 12, 16, or 24 h, and then the Cy5 signal in the cells was analyzed by flow cytometry. These data demonstrate that N1-34a-NPs exhibit a time-dependent interaction with MDA-MB-231 TNBC cells, but the cells show no preference for Notch1-functionalized NPs compared to non-specific IgG-functionalized NPs (Figure S1A). This prompted us to further probe the interaction of N1-Cy5-NPs with MDA-MB-231 TNBC cells and non-cancerous MCF-10A cells at the therapeutic dose used in subsequent studies (200 nM). These data indicate that N1-Cy5-NPs exhibit specific, enhanced interaction with MDA-MB-231 TNBC cells compared to MCF-10A cells (Figure S1B).
N1-34a-NPs Regulate Notch Signaling and Downstream miR-34a Targets
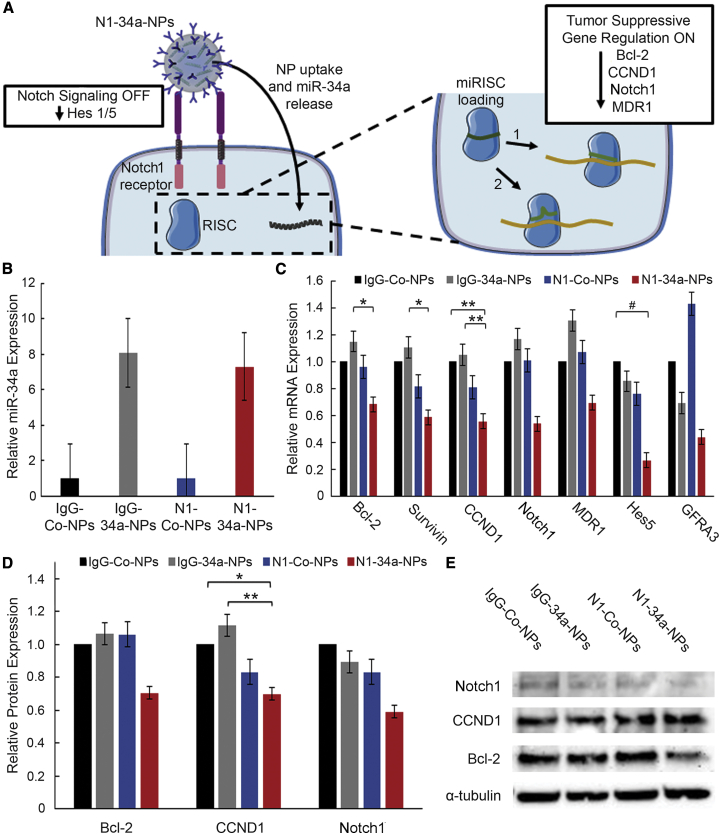
To determine the influence of N1-34a-NPs on downstream Notch and miR-34a signaling (Figure 2A), we evaluated the expression of miR-34a and its downstream targets (Bcl-2, survivin, CCND1, Notch1, MDR1, GFRA3) as well as Notch signaling targets (Hes1, Hes5) in TNBC cells treated with IgG-Co-NPs, IgG-34a-NPs, N1-Co-NPs, and N1-34a-NPs at 200 nM miRNA using real-time quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) and western blotting. The real-time qRT-PCR data demonstrate that miR-34a expression is elevated approximately 8-fold after treatment with IgG-34a-NPs and N1-34a-NPs (Figure 2B). However, IgG-34a-NPs, along with control IgG-Co-NPs and N1-Co-NPs, have minimal impact on downstream mRNA expression. In contrast, N1-34a-NPs substantially suppressed Bcl-2 by 32%, survivin by 41%, CCND1 by 45%, Notch1 by 46%, MDR1 by 31%, Hes5 by 73%, and GFRA3 by 56% (Figure 2C). Western blot analysis revealed similar results, as N1-34a-NPs reduced Bcl-2 expression by 29%, CCND1 by 30%, Notch1 by 41%, and Hes1 by 40% (Figures 2D and 2E; Figure S2). Taken together, these real-time qRT-PCR and western blot data indicate that N1-34a-NPs can increase miR-34a levels in TNBC cells and regulate both miR-34a downstream targets and the Notch signaling pathway.
Figure 2.
Nanoparticle Interaction with TNBC Cells and Evaluation of Gene Regulation
(A) Scheme of posited NP interaction with MDA-MB-231 TNBC cells. Upon cellular binding (see also Figure S1), N1-34a-NPs inhibit downstream Notch signaling through antibody-mediated signal cascade interference and also deliver miR-34a, which reduces the expression of several genes by guiding the RNA-induced silencing complex (miRISC) to targeted mRNA sequences with perfect (1) or imperfect (2) complementarity, resulting in mRNA degradation or translational repression. (B) Real-time qRT-PCR analysis of relative miR-34a levels after treatment with IgG-Co-NPs, IgG-34a-NPs, N1-Co-NPs, or N1-34a-NPs (n = 3). U6 was used as a control, and relative expression is normalized to that of cells treated with IgG-Co-NPs. Error bars indicate standard error. (C) Real-time qRT-PCR analysis of relative Bcl-2, survivin, CCND1, Notch1, MDR1, GFRA3, and Hes5 mRNA expression after treatment with IgG-Co-NPs, IgG-34a-NPs, N1-Co-NPs, or N1-34a-NPs (n = 3). GUSB was used as a control, and relative mRNA expression is normalized to that of cells treated with IgG-Co-NPs. Error bars indicate standard error. #p < 0.1, ∗p < 0.05, ∗∗p < 0.01. (D) Quasi-quantitative analysis of western blotting for normalized Bcl-2, CCND1, and Notch1 protein expression (n = 3). α-Tubulin was used as a control, and expression was normalized to expression in cells treated with IgG-Co-NPs. Error bars indicate standard error. ∗p < 0.05, ∗∗p < 0.01. (E) Representative western blot bands for Bcl-2, CCND1, Notch1, and α-tubulin protein levels. Bands are from a single blot that was stripped and probed for multiple targets.
N1-34a-NPs Induce Senescence and Reduce TNBC Cell Proliferation and Migration In Vitro
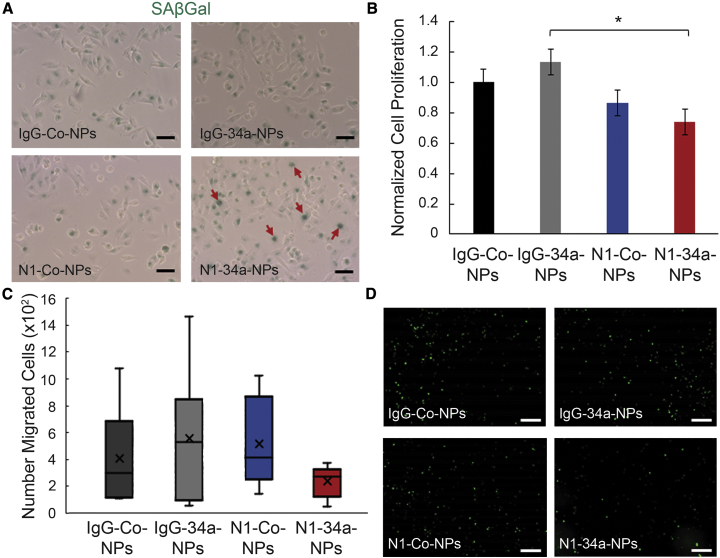
We further investigated the impact of IgG-Co-NPs, IgG-34a-NPs, N1-Co-NPs, and N1-34a-NPs on TNBC cell viability and function using a senescence-associated β-galactosidase (SAβGal) assay, an 5-ethynyl-2′-deoxyuridine (EdU) proliferation assay, and a transwell invasion assay. We selected these assays because miR-34a and Notch signaling have been highly implicated in the regulation of cell senescence,9,23, 24, 25 proliferation,23,25, 26, 27 and migration.9,23,26,27 The SAβGal assay showed that N1-34a-NPs begin inducing β-galactosidase activity in MDA-MB-231 TNBC cells after a 4-h treatment period, indicating an onset of senescence (Figure 3A). We also evaluated the effect of these NPs on cell proliferation after a 48-h treatment period using an EdU assay. The results demonstrate that N1-Co-NPs can reduce cell proliferation by 13%, which we attribute to the Notch inhibition mediated through the Notch1 antibodies on the NPs. Excitingly, N1-34a-NPs have an enhanced effect on cell proliferation, as they reduce EdU-positive cells by 26%, which is double the inhibition observed for NPs carrying only Notch1 antibodies (Figure 3B). Finally, we investigated the effect of N1-34a-NPs on cell migration after 48 h through a transwell invasion assay. This study showed that N1-34a-NPs could reduce TNBC cell migration by 34% relative to controls (Figures 3C and 3D; Figure S3). Altogether, these data indicate that N1-34a-NPs can functionally impair TNBC cells by activating senescence and inhibiting cell proliferation and migration.
Figure 3.
Effect of N1-34a-NPs on TNBC Cell Viability, Proliferation, and Migration
(A) Representative images of SAβGal assay of MDA-MB-231 cells treated with IgG-Co-NPs, IgG-34a-NPs, N1-Co-NPs, or N1-34a-NPs. Green color indicates β-galactosidase activity. Red arrows indicate cells with disperse staining. Scale bars= 50 μm. (B) Percent proliferation of MDA-MB-231 cells treated with IgG-Co-NPs, IgG-34a-NPs, N1-Co-NPs, or N1-34a-NPs (n = 4). Error bars indicate standard error. ∗p < 0.05. (C) Normalized number of GFP-expressing MDA-MB-231 cells migrated through the transwell insert after treatment with IgG-Co-NPs, IgG-34a-NPs, N1-Co-NPs, or N1-34a-NPs (n = 3). (D) Representative fluorescence images of GFP-expressing MDA-MB-231 cells migrated through the transwell insert after treatment with IgG-Co-NPs, IgG-34a-NPs, N1-Co-NPs, or N1-34a-NPs. See also Figure S2. Scale bars = 500 μm
Discussion
In this work, we used PLGA NPs as carriers of both miR-34a mimics and Notch1 antibodies to effectively treat TNBC by a combination of miRNA replacement therapy and antibody-mediated signal cascade interference. We showed by real-time qRT-PCR and western blotting that these N1-34a-NPs could deliver miR-34a to TNBC cells and inhibit the expression of several targets of miR-34a and Notch signaling, including Bcl-2, survivin, CCND1, Notch1, GFRA3, and MDR1 in vitro. This gene regulation induced senescence and reduced cell proliferation and migration, which is in agreement with the known roles of miR-34a and Notch signaling in cancer cell biology.9,23, 24, 25, 26, 27 With further optimization and in vivo evaluation, these N1-34a-NPs may offer a promising strategy to treat TNBC.
Future studies that continue this work should investigate the optimal loading of miRNA and antibody components within the NP formulation. First, by increasing the quantity of miRNA in each NP, this treatment strategy may achieve similar therapeutic effects with fewer NPs administered. This alteration mitigates the risk of experiencing adverse effects from the PLGA content and may improve the formulation’s performance in vivo by reducing the risk of release of tumor necrosis factor (TNF)-α.28
In addition to altering the loading of miRNA in these NPs, the density of the antibodies on the surface of the particles should be optimized in future work. As synthesized here, N1-34a-NPs have a diameter of approximately 170 nm and an antibody loading of 4.6 μg per mg of PLGA. These NPs preferentially bind MDA-MB-231 cells versus MCF-10A cells (Figure S1B), but they show no notable advantage in binding MDA-MB-231 cells compared to non-specific IgG-functionalized NPs. Notably, in previous work, we have created Notch1 antibody-functionalized NPs that are approximately 70 nm in diameter and contain 9.1 μg of antibodies per mg of PLGA; these NPs exhibit sustained preferential and specific binding of TNBC cells, which translates to enhanced tumor accumulation and retention compared to IgG-functionalized NPs in vivo.29 We posit that the difference in particle size and antibody loading density between N1-34a-NPs and our previous NP formulation is responsible for the disparity in obtaining preferential and specific binding to MDA-MB-231 TNBC cells. This would be in agreement with prior studies that show cellular binding of NPs is significantly impacted by the size and shape of the NP30, 31, 32, 33 and by the density of the targeting moiety presented on the surface of the NP.32,33 Future studies are needed to define the optimal size of the particles and density of Notch1 antibodies on these NPs to maximize TNBC cell-specific binding and potentially increase downstream Notch signaling inhibition.
While N1-34a-NPs did not display enhanced binding to TNBC cells in this work relative to IgG-functionalized NPs, we did find that Notch1 antibody functionalization was critical for eliciting significant therapeutic effect. The Notch signaling targets Hes5 and Hes1 were suppressed only with Notch1 antibody-coated NPs, which confirms that the amount of NP binding was sufficient to elicit signal cascade interference. Likewise, even though N1-34a-NPs and IgG-34a-NPs had similar levels of interaction with TNBC cells and showed similar increases in miR-34a levels, only the Notch1-functionalized NPs had a notable effect on downstream miR-34a targets, indicating that the inclusion of Notch1 antibodies improves the efficacy of this formulation. It is known that coating NPs with receptor-specific moieties such as Notch1 antibodies can alter the mechanism of endocytosis and subsequently influence the intracellular fate of the NP and any cargo it is carrying.34, 35, 36 This altered trafficking can affect the potency of the cargo, which is particularly important for the delivery of RNA molecules such as miR-34a that require cytosolic delivery to engage the RNA interference machinery and elicit gene regulation.37 NPs typically enter cells through clathrin- or caveolae-dependent uptake mechanisms that lead to lysosomal accumulation, although in some cases caveolae have been shown to fuse with caveosomes to avoid lysosomal trafficking.38, 39, 40 Future studies should elucidate the intracellular fate of N1-34a-NPs in order to provide insight to their mechanism of action and therapeutic potential.
Finally, future studies should also evaluate N1-34a-NPs in the in vivo setting. Notch signaling is primarily implicated in early stages of tumor development and in cancer stem cells.41,42 Additionally, Sethi and Kang42 have demonstrated that Notch signaling facilitates bone metastasis in later stages of tumor progression. However, its role in the primary tumor at these later stages has not been extensively explored. Reduced miR-34a expression has been associated with approximately 50% of TNBC cases and is correlated with poor prognosis.43,44 Future studies should examine the expression of the Notch1 receptor in tumors of various sizes and miR-34a expression in tumors of various origins and determine the therapeutic efficacy of N1-34a-NPs when these expression levels are different than those explored in this work.
In conclusion, this study demonstrates that miR-34a-loaded, Notch1 antibody-functionalized PLGA NPs can effectively regulate Notch signaling and downstream miR-34a targets in TNBC cells in vitro to induce senescence and reduce cell proliferation and migration. These N1-34a-NPs are a promising alternative treatment for aggressive cancers such as TNBC that display lower levels of miR-34a and are driven by overexpression of Notch signaling. With additional optimization, development, and implementation, these NPs may substantially improve patient outcomes.
Materials and Methods
Synthesis of NPs
miRNA-loaded PLGA NPs were synthesized using a double emulsion method.45 Briefly, PLGA (Lactel, 50:50 carboxylic acid terminated, 0.55–0.75 dL/g) was dissolved in acetone (VWR International) at 2 mg/mL. miR-Co and miR-34a (Dharmacon, prepared as duplexes and stored in duplex buffer at 20 μM; miRNA sequences are listed in Table S1) were mixed separately with a 1% poly(vinyl) alcohol (PVA) solution and then added dropwise to the PLGA in acetone at a concentration of 0.2 nmol/mg PLGA and stirred for 5 min to allow for homogenization. This mixture was subsequently added dropwise to a 0.1% PVA solution in a 1:3 volume ratio while stirring. This secondary emulsion continued to stir for 2 h, letting the acetone evaporate. The NPs were then purified using centrifugal filtration (Millipore, molecular weight cutoff [MWCO] of 50 kDa, 4,200 × g, 15 min) to remove unencapsulated miRNA and excess solvent. Rabbit anti-human IgG or Notch1 antibodies were conjugated to the surface of the bare NPs using 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) chemistry.46,47 Briefly, after centrifugal filtration, miRNA-loaded NPs were suspended in 4 mM EDC and 4 mM N-hydroxysulfosuccinimide sodium salt (sulfo-NHS) and incubated on a rocker at 4°C. IgG or Notch1 antibodies were then added to the solution for further incubation at 4°C. To remove free antibodies from solution after conjugation, the NPs were purified using trans-flow filtration (Spectrum, MWCO of 300 kDa). NPs were freshly prepared and used immediately for experiments.
Characterization of Antibody and miRNA Loading in NPs
Purified NPs were characterized by DLS and zeta potential measurements on a Litesizer 500 instrument (Anton Paar) before and after antibody conjugation, and the reported intensity-based hydrodynamic diameter is the average of three measurements. miRNA encapsulation was quantified using an OliGreen assay. During NP synthesis, all filtrate containing unencapsulated miRNA was concentrated and analyzed alongside a standard curve of known miRNA concentration.
Antibody loading on the NPs was quantified using a solution-based ELISA modified from a previously published protocol.48 IgG-Co-NPs, IgG-34a-NPs, N1-Co-NPs, N1-34a-NPs, bare miR-Co-NPs, or bare miR-34a-NPs were incubated with 10 μg/mL horseradish peroxidase (HRP)-conjugated anti-rabbit IgG antibodies for 1 h at room temperature. Unbound secondary antibodies were removed through centrifugation and the samples were suspended in 3% bovine serum albumin in phosphate-buffered saline (PBS). The samples were then developed in 3,3′,5,5′-tetramethylbenzidine (TMB) solution (TMB core; Bio-Rad) for 15 s before the reaction was stopped with 2 mM sulfuric acid. The absorbance was then measured at 450 nm on a Synergy H1 plate reader and compared to a standard curve of known HRP-IgG concentration to calculate the quantity of IgG or Notch1 antibodies conjugated per mg of PLGA.
Cell Culture
MDA-MB-231 TNBC cells (ATCC) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, VWR International) supplemented with 10% fetal bovine serum (FBS; Gemini Bio Products) and 1% penicillin-streptomycin (pen-strep; VWR International). MCF-10A cells (ATCC) were cultured in DMEM supplemented with 1% pen-strep, 5% FBS, 50 μg/mL bovine pituitary extract (Sigma), 0.5 μg/mL hydrocortisone (Sigma), 20 ng/mL human epidermal growth factor (STEMCELL Technologies), 10 μg/mL insulin (Thermo Fisher Scientific), and 100 ng/mL cholera toxin (Sigma). The cultures were maintained at 37°C in a 5% CO2 humidified environment. When cells reached 80%–90% confluency in T75 cell culture flasks, they were passaged or plated by detaching the cells from the flask using trypsin-EDTA (Thermo Fisher Scientific) and then counting cells with a hemocytometer.
Cellular Binding and Uptake of Cy5-Tagged miRNA-Loaded NPs
To analyze cellular binding and uptake of antibody-functionalized particles, PLGA NPs were loaded with miR-Co tagged with Cy5 (Dharmacon; excitation 647 nm/emission 665 nm) and functionalized as described above to create IgG-Cy5-NPs and N1-Cy5-NPs. For flow cytometric analysis of cellular binding and uptake, cells were plated at 3 × 104 cells per well (MDA-MB-231 cells) or 2 × 104 cells per well (MCF-10A cells) in a 24-well plate and incubated overnight. Cells were then treated with IgG-Cy5-NPs or N1-Cy5-NPs at 50 or 200 nM miRNA or were left untreated and incubated for 0, 1, 4, 8, 12, 16, or 24 h prior to rinsing with PBS. The cells were then lifted off the plate with trypsin-EDTA and resuspended in PBS. All cell suspensions were analyzed using an ACEA NovoCyte 2060 flow cytometer with the allophycocyanin (APC) (excitation, 640 nm; emission, 675/30 nm) channel. Density plots showing forward and side scatter data were used to create a primary gate for cells, excluding debris, prior to analyzing Cy5 content. Flow cytometric analysis was performed in triplicate.
Evaluating Changes in Gene Expression Induced by NPs
To analyze the effects of IgG-Co-NPs, IgG-34a-NPs, N1-Co-NPs, and N1-34a-NPs on gene expression in TNBC cells by real-time qRT-PCR, cells were seeded at 1.5 × 105 cells per well in a six-well plate and incubated overnight. Cells were then treated with NPs at 200 nM miRNA for 48 h. At the conclusion of the treatment period, cells were rinsed with PBS and mRNA was extracted using a Bioline Isolate II RNA mini kit. Real-time qRT-PCR for Bcl-2, MDR1, survivin, CCND1, Notch1, Hes5, and GFRA3 was then performed using SensiFAST SYBR one-step master mix on a LightCycler 96 (Roche), and gene expression was normalized to that of GUSB. Real-time qRT-PCR for miR-34a was performed using a TaqMan miRNA assay (Thermo Fisher Scientific), TaqMan miRNA reverse transcription kit (Thermo Fisher Scientific), and TaqMan universal PCR master mix (Thermo Fisher Scientific) on a peqSTAR thermal cycler (VWR International) and a LightCycler 96 (Roche). miR-34a expression was normalized to that of U6. These experiments were performed in triplicate and analyzed using a one-way ANOVA with a Tukey’s post hoc test. Primer sequences are listed in Table S2.
Investigating Changes in Protein Expression Induced by NPs
To further evaluate the effects of N1-34a-NPs on Bcl-2, CCND1, Notch1, and Hes1 by western blotting, MDA-MB-231 cells were plated at 1.5 × 105 cells per well in a six-well plate and incubated overnight. Cells were then treated as described for real-time qRT-PCR. After 48 h of treatment with NPs, the cells were rinsed with PBS and lysed in radioimmunoprecipitation assay (RIPA) buffer supplemented with Halt protease inhibitor in a 1:50 volume ratio. After removing membrane debris through centrifugation, the extracted protein was quantified using a DC (detergent compatible) protein assay (Bio-Rad), and 10 μg of protein was separated on 4%–12% Bis-Tris gels at 135 V for 60 min. Then, the protein was transferred to a 0.2-μm nitrocellulose membrane for 10 min using a power blotter system (Invitrogen). The membrane was subsequently blocked for 60 min in 5% milk in Tris-buffered saline with 0.1% Tween 20 (TBS-T) and then incubated with rabbit anti-human Bcl-2 (ProteinTech; 1:1,000), CCND1 (Cell Signaling Technology; 1:250), Notch1 (Cell Signaling Technology; 1:500), and Hes1 (Cell Signaling Technology; 1:500) antibodies in 5% milk in TBS-T overnight at 4°C. Mouse anti-human α-tubulin antibody (Cell Signaling Technology; 1:20,000) was used as the normalization control. After incubation in primary antibodies, membranes were washed three times in TBS-T and incubated with HRP-anti-rabbit or mouse IgG antibody (VWR International; 1:25,000) in 5% milk in TBS-T for 1 h at room temperature. Membranes were then washed twice in TBS-T and twice in TBS (without Tween 20), and protein bands were visualized using a Pierce enhanced chemiluminescence (ECL) detection solution (Thermo Scientific). Band densities were quantified in ImageJ, and Bcl-2, CCND1, Notch1, and Hes1 densities were normalized to that of α-tubulin prior to further normalizing treatment groups to the IgG-Co-NP group. The data shown represent the average band density across three trials and were analyzed using a one-way ANOVA with a Tukey’s post hoc test.
Evaluating the Effect of NPs on Cellular Senescence
To determine the effect of IgG-Co-NPs, IgG-34a-NPs, N1-Co-NPs, and N1-34a-NPs on cellular viability through a SAβGal assay, cells were seeded at 4 × 105 cells per well in a six-well plate and incubated overnight. Cells were then treated with NPs for 4 h and senescence was evaluated using a SAβGal kit (Cell Signaling Technology) per the manufacturer’s instructions. Stained cells were imaged on a Zeiss AxioObserver Z1 microscope equipped with a color camera.
Analyzing the Effect of NPs on Cell Proliferation
To analyze the effect of IgG-Co-NPs, IgG-34a-NPs, N1-Co-NPs, and N1-34a-NPs on cellular proliferation via an EdU assay, cells were seeded at 1.0 × 105 cells per well in a 12-well plate and incubated overnight. Cells were then treated with NPs at 200 nM miRNA for 48 h. At 16 h prior to the end of the treatment period, the cells were spiked with EdU at 10 μM. Cells were then prepared per the manufacturer’s instructions modified for a single-cell suspension. Briefly, cells were fixed in 4% formaldehyde and permeabilized with 0.5% Triton X-100. They were subsequently incubated in the Click-iT reaction cocktail, washed in PBS, and examined on an ACEA NovoCyte 2060 flow cytometer with the fluorescein isothiocyanate (FITC) (excitation, 488 nm; emission, 530/30 nm) channel. Density plots showing forward and side scatter data were used to create a primary gate for cells, excluding debris, prior to analyzing EdU-labeling azide content. Flow cytometric analysis was performed in quadruplicate and statistical analysis was performed using a one-way ANOVA with a Tukey’s post hoc test.
Investigating the Effect of NPs on Cell Migration
To evaluate the impact of IgG-Co-NPs, IgG-34a-NPs, N1-Co-NPs, and N1-34a-NPs on cellular migration using a transwell invasion assay, MDA-MB-231 GFP-expressing cells were plated at 1.5 × 104 cells per well in a 96-well plate and incubated overnight. Cells were then treated with NPs at 200 nM for 48 h, at which time the cells were washed with PBS and lifted off the plate using 0.25% trypsin-EDTA. The cells were pelleted (0.3 relative centrifugal force [RCF], 5 min) and resuspended in DMEM (without supplements) at 25,000 cells per mL. Cells were then seeded in transwell inserts (Corning Life Sciences) at 5,000 cells per 200 μL and 700 μL of DMEM (with FBS and pen-strep supplements) was added beneath the insert in a 24-well black-walled, glass-bottom plate (Cellvis). The transwell samples were imaged on a Zeiss AxioObserver Z1 microscope after 48 h. ImageJ software was subsequently used to analyze the number of cells migrated within the entire transwell, and the data presented represent the average of three trials normalized to the IgG-Co-NP group. Statistical analysis was performed using a one-way ANOVA with a Tukey’s post hoc test.
Author Contributions
Conceptualization, D.M.V. and E.S.D.; Methodology, D.M.V.; Formal Analysis, D.M.V.; Investigation, D.M.V.; Writing – Original Draft, D.M.V.; Writing – Review & Editing, D.M.V. and E.S.D.; Visualization, D.M.V.; Funding Acquisition, E.S.D.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award no. R35GM119659.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.06.003.
Supplemental Information
References
- 1.Bianchini G., Balko J.M., Mayer I.A., Sanders M.E., Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 2016;13:674–690. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turner N., Moretti E., Siclari O., Migliaccio I., Santarpia L., D’Incalci M., Piccolo S., Veronesi A., Zambelli A., Del Sal G., Di Leo A. Targeting triple negative breast cancer: is p53 the answer? Cancer Treat. Rev. 2013;39:541–550. doi: 10.1016/j.ctrv.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Griffiths C.L., Olin J.L. Triple negative breast cancer: a brief review of its characteristics and treatment options. J. Pharm. Pract. 2012;25:319–323. doi: 10.1177/0897190012442062. [DOI] [PubMed] [Google Scholar]
- 4.Mungamuri S.K., Yang X., Thor A.D., Somasundaram K. Survival signaling by Notch1: mammalian target of rapamycin (mTOR)-dependent inhibition of p53. Cancer Res. 2006;66:4715–4724. doi: 10.1158/0008-5472.CAN-05-3830. [DOI] [PubMed] [Google Scholar]
- 5.Speiser J., Foreman K., Drinka E., Godellas C., Perez C., Salhadar A., Erşahin Ç., Rajan P. Notch-1 and Notch-4 biomarker expression in triple-negative breast cancer. Int. J. Surg. Pathol. 2012;20:139–145. doi: 10.1177/1066896911427035. [DOI] [PubMed] [Google Scholar]
- 6.Speiser J.J., Erşahin C., Osipo C. The functional role of Notch signaling in triple-negative breast cancer. Vitam. Horm. 2013;93:277–306. doi: 10.1016/B978-0-12-416673-8.00013-7. [DOI] [PubMed] [Google Scholar]
- 7.Mollaei H., Safaralizadeh R., Rostami Z. MicroRNA replacement therapy in cancer. J. Cell. Physiol. 2019;234:12369–12384. doi: 10.1002/jcp.28058. [DOI] [PubMed] [Google Scholar]
- 8.Kapadia C.H., Luo B., Dang M.N., Irvin-Choy N., Valcourt D.M., Day E.S. Polymer nanocarriers for microRNA delivery. J. Appl. Polym. Sci. 2020;137:48651. doi: 10.1002/app.48651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 10.Li L., Yuan L., Luo J., Gao J., Guo J., Xie X. miR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clin. Exp. Med. 2013;13:109–117. doi: 10.1007/s10238-012-0186-5. [DOI] [PubMed] [Google Scholar]
- 11.Imani S., Wu R.-C., Fu J. MicroRNA-34 family in breast cancer: from research to therapeutic potential. J. Cancer. 2018;9:3765–3775. doi: 10.7150/jca.25576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adams B.D., Wali V.B., Cheng C.J., Inukai S., Booth C.J., Agarwal S., Rimm D.L., Győrffy B., Santarpia L., Pusztai L. miR-34a silences c-SRC to attenuate tumor growth in triple-negative breast cancer. Cancer Res. 2016;76:927–939. doi: 10.1158/0008-5472.CAN-15-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goyal R., Kapadia C.H., Melamed J.R., Riley R.S., Day E.S. Layer-by-layer assembled gold nanoshells for the intracellular delivery of miR-34a. Cell. Mol. Bioeng. 2018;11:383–396. doi: 10.1007/s12195-018-0535-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapadia C.H., Ioele S.A., Day E.S. Layer-by-layer assembled PLGA nanoparticles carrying miR-34a cargo inhibit the proliferation and cell cycle progression of triple-negative breast cancer cells. J. Biomed. Mater. Res. A. 2020;108:601–613. doi: 10.1002/jbm.a.36840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia Y., Deng X., Cao M., Liu S., Zhang X., Xiao X., Shen S., Hu Q., Sheng W. Nanodiamond-based layer-by-layer nanohybrids mediate targeted delivery of miR-34a for triple negative breast cancer therapy. RSC Advances. 2018;8:13789–13797. doi: 10.1039/c8ra00907d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng X., Cao M., Zhang J., Hu K., Yin Z., Zhou Z., Xiao X., Yang Y., Sheng W., Wu Y., Zeng Y. Hyaluronic acid-chitosan nanoparticles for co-delivery of miR-34a and doxorubicin in therapy against triple negative breast cancer. Biomaterials. 2014;35:4333–4344. doi: 10.1016/j.biomaterials.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Bu P., Chen K.-Y., Chen J.H., Wang L., Walters J., Shin Y.J., Goerger J.P., Sun J., Witherspoon M., Rakhilin N. A microRNA miR-34a-regulated bimodal switch targets Notch in colon cancer stem cells. Cell Stem Cell. 2013;12:602–615. doi: 10.1016/j.stem.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang Y., Tang Y., Cheng Y.S. miR-34a inhibits pancreatic cancer progression through Snail1-mediated epithelial-mesenchymal transition and the Notch signaling pathway. Sci. Rep. 2017;7:38232. doi: 10.1038/srep38232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park E.Y., Chang E., Lee E.J., Lee H.-W., Kang H.-G., Chun K.-H., Woo Y.M., Kong H.K., Ko J.Y., Suzuki H. Targeting of miR34a-NOTCH1 axis reduced breast cancer stemness and chemoresistance. Cancer Res. 2014;74:7573–7582. doi: 10.1158/0008-5472.CAN-14-1140. [DOI] [PubMed] [Google Scholar]
- 20.Valcourt D.M., Harris J., Riley R.S., Dang M., Wang J., Day E.S. Advances in targeted nanotherapeutics: from bioconjugation to biomimicry. Nano Res. 2018;11:4999–5016. doi: 10.1007/s12274-018-2083-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riley R.S., Day E.S. Frizzled7 antibody-functionalized nanoshells enable multivalent binding for Wnt signaling inhibition in triple negative breast cancer cells. Small. 2017;13 doi: 10.1002/smll.201700544. 1700544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott A.M., Wolchok J.D., Old L.J. Antibody therapy of cancer. Nat. Rev. Cancer. 2012;12:278–287. doi: 10.1038/nrc3236. [DOI] [PubMed] [Google Scholar]
- 23.Zhang L., Liao Y., Tang L. MicroRNA-34 family: a potential tumor suppressor and therapeutic candidate in cancer. J. Exp. Clin. Cancer Res. 2019;38:53. doi: 10.1186/s13046-019-1059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tazawa H., Tsuchiya N., Izumiya M., Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc. Natl. Acad. Sci. USA. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slabáková E., Culig Z., Remšík J., Souček K. Alternative mechanisms of miR-34a regulation in cancer. Cell Death Dis. 2017;8:e3100. doi: 10.1038/cddis.2017.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venkatesh V., Nataraj R., Thangaraj G.S., Karthikeyan M., Gnanasekaran A., Kaginelli S.B., Kuppanna G., Kallappa C.G., Basalingappa K.M. Targeting Notch signalling pathway of cancer stem cells. Stem Cell Investig. 2018;5 doi: 10.21037/sci.2018.02.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo H., Lu Y., Wang J., Liu X., Keller E.T., Liu Q., Zhou Q., Zhang J. Targeting the Notch signaling pathway in cancer therapeutics. Thorac. Cancer. 2014;5:473–486. doi: 10.1111/1759-7714.12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiong S., George S., Yu H., Damoiseaux R., France B., Ng K.W., Loo J.S. Size influences the cytotoxicity of poly (lactic-co-glycolic acid) (PLGA) and titanium dioxide (TiO2) nanoparticles. Arch. Toxicol. 2013;87:1075–1086. doi: 10.1007/s00204-012-0938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valcourt D.M., Dang M.N., Scully M.A., Day E.S. Nanoparticle-mediated co-delivery of Notch-1 antibodies and ABT-737 as a potent treatment strategy for triple-negative breast cancer. ACS Nano. 2020;14:3378–3388. doi: 10.1021/acsnano.9b09263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barua S., Yoo J.-W., Kolhar P., Wakankar A., Gokarn Y.R., Mitragotri S. Particle shape enhances specificity of antibody-displaying nanoparticles. Proc. Natl. Acad. Sci. USA. 2013;110:3270–3275. doi: 10.1073/pnas.1216893110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang W., Kim B.Y.S., Rutka J.T., Chan W.C.W. Nanoparticle-mediated cellular response is size-dependent. Nat. Nanotechnol. 2008;3:145–150. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- 32.Lee H., Odom T.W. Controlling ligand density on nanoparticles as a means to enhance biological activity. Nanomedicine (Lond.) 2015;10:177–180. doi: 10.2217/nnm.14.204. [DOI] [PubMed] [Google Scholar]
- 33.Elias D.R., Poloukhtine A., Popik V., Tsourkas A. Effect of ligand density, receptor density, and nanoparticle size on cell targeting. Nanomedicine (Lond.) 2013;9:194–201. doi: 10.1016/j.nano.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valcourt D.M., Dang M.N., Wang J., Day E.S. Nanoparticles for manipulation of the developmental Wnt, Hedgehog, and Notch signaling pathways in cancer. Ann. Biomed. Eng. 2019 doi: 10.1007/s10439-019-02399-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kou L., Sun J., Zhai Y., He Z. The endocytosis and intracellular fate of nanomedicines: implication for rational design. Asian J. Pharm. Sci. 2013;8:1–10. [Google Scholar]
- 36.Yameen B., Choi W.I., Vilos C., Swami A., Shi J., Farokhzad O.C. Insight into nanoparticle cellular uptake and intracellular targeting. J. Control. Release. 2014;190:485–499. doi: 10.1016/j.jconrel.2014.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dominska M., Dykxhoorn D.M. Breaking down the barriers: siRNA delivery and endosome escape. J. Cell Sci. 2010;123:1183–1189. doi: 10.1242/jcs.066399. [DOI] [PubMed] [Google Scholar]
- 38.Rejman J., Bragonzi A., Conese M. Role of clathrin- and caveolae-mediated endocytosis in gene transfer mediated by lipo- and polyplexes. Mol. Ther. 2005;12:468–474. doi: 10.1016/j.ymthe.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 39.Pelkmans L., Kartenbeck J., Helenius A. Caveolar endocytosis of simian virus 40 reveals a new two-step vesicular-transport pathway to the ER. Nat. Cell Biol. 2001;3:473–483. doi: 10.1038/35074539. [DOI] [PubMed] [Google Scholar]
- 40.Munsell E.V., Ross N.L., Sullivan M.O. Journey to the center of the cell: current nanocarrier design strategies targeting biopharmaceuticals to the cytoplasm and nucleus. Curr. Pharm. Des. 2016;22:1227–1244. doi: 10.2174/1381612822666151216151420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mollen E.W.J., Ient J., Tjan-Heijnen V.C.G., Boersma L.J., Miele L., Smidt M.L., Vooijs M.A.G.G. Moving breast cancer therapy up a Notch. Front. Oncol. 2018;8:518. doi: 10.3389/fonc.2018.00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sethi N., Kang Y. Notch signalling in cancer progression and bone metastasis. Br. J. Cancer. 2011;105:1805–1810. doi: 10.1038/bjc.2011.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeng Z., Chen X., Zhu D., Luo Z., Yang M. Low expression of circulating microRNA-34c is associated with poor prognosis in triple-negative breast cancer. Yonsei Med. J. 2017;58:697–702. doi: 10.3349/ymj.2017.58.4.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lodygin D., Tarasov V., Epanchintsev A., Berking C., Knyazeva T., Körner H., Knyazev P., Diebold J., Hermeking H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 45.Zhou J., Patel T.R., Fu M., Bertram J.P., Saltzman W.M. Octa-functional PLGA nanoparticles for targeted and efficient siRNA delivery to tumors. Biomaterials. 2012;33:583–591. doi: 10.1016/j.biomaterials.2011.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Srinivasan S., Manchanda R., Lei T., Nagesetti A., Fernandez-Fernandez A., McGoron A.J. Targeted nanoparticles for simultaneous delivery of chemotherapeutic and hyperthermia agents—an in vitro study. J. Photochem. Photobiol. B. 2014;136:81–90. doi: 10.1016/j.jphotobiol.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 47.Kocbek P., Obermajer N., Cegnar M., Kos J., Kristl J. Targeting cancer cells using PLGA nanoparticles surface modified with monoclonal antibody. J. Control. Release. 2007;120:18–26. doi: 10.1016/j.jconrel.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 48.Riley R.S., Melamed J.R., Day E.S. Enzyme-linked immunosorbent assay to quantify targeting molecules on nanoparticles. Methods Mol. Biol. 2018;1831:145–157. doi: 10.1007/978-1-4939-8661-3_11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.