Abstract
Staphylococcus aureus is a major cause of hospital- and community-acquired infections placing a significant burden on the healthcare system. With the widespread of multidrug-resistant bacteria and the lack of effective antibacterial drugs, fosfomycin has gradually attracted attention as an “old drug.” Thus, investigating the resistance mechanisms and epidemiology of fosfomycin-resistant S. aureus is an urgent requirement. In order to investigate the mechanisms of resistance, 11 fosfomycin-resistant S. aureus isolates were analyzed by PCR and sequencing. The genes, including fosA, fosB, fosC, fosD, fosX, and tet38, as well as mutations in murA, glpT, and uhpT were identified. Quantitative real-time PCR (qRT-PCR) was conducted to evaluate the expression of the target enzyme gene murA and the efflux pump gene tet38 under the selection pressure of fosfomycin. Furthermore, multilocus sequence typing (MLST) identified a novel sequence type (ST 5708) of S. aureus strains. However, none of the resistant strains carried fosA, fosB, fosC, fosD, and fosX genes in the current study, and 12 distinct mutations were detected in the uhpT (3), glpT (4), and murA (5) genes. qRT-PCR revealed an elevated expression of the tet38 gene when exposed to increasing concentration of fosfomycin among 8 fosfomycin-resistant S. aureus strains and reference strain ATCC 29213. MLST analysis categorized the 11 strains into 9 STs. Thus, the mutations in the uhpT, glpT, and murA genes might be the primary mechanisms underlying fosfomycin resistance, and the overexpression of efflux pump gene tet38 may play a major role in the fosfomycin resistance in these isolates.
Keywords: fosfomycin, Staphylococcus aureus, resistance mechanism, mutation, tet38
Introduction
Staphylococcus aureus is a kind of facultative anaerobe pathogenic Gram-positive coccus with strong resistance and tolerance to harsh environments (Wang et al., 2020). At present, S. aureus has become a significant pathogen of nosocomial infections, such as deep-seated skin and soft tissue infections (SSTI), endocarditis, and other life-threatening severe infections (Mehraj et al., 2016). In recent years, with the widespread use of antibiotics, the emergence of multidrug-resistant (MDR) S. aureus has become a major concern (Gatadi et al., 2019). In addition, the lack of effective clinical treatments against MDR S. aureus has rekindled the interest of clinicians in fosfomycin. It is an antimicrobial agent that was discovered in Streptomyces sp. It exhibits broad-spectrum activity against both Gram-positive and Gram-negative bacteria by inhibiting the peptidoglycan synthesis pathway, which is essential for the synthesis of the cell walls (Shorr et al., 2017). However, the number of fosfomycin-resistant S. aureus strains is increasing rapidly (Etienne et al., 1991).
Several mechanisms of fosfomycin resistance have been proposed, including the plasmid-encoded fosfomycin-modifying enzymes (FosA, FosB, FosC, FosD, and FosX) and the acquisition of chromosomal mutations (Nakaminami et al., 2008; Liu et al., 2017; Silver, 2017). Mutations in the MurA target enzyme and transporters (GlpT and UhpT) have been shown to be responsible for fosfomycin resistance (Michalopoulos et al., 2011). Additionally, the overexpression of target enzymes, MurA and Tet38 efflux pump, also contributes to fosfomycin resistance in S. aureus (Truong-Bolduc et al., 2018). Notably, there are no reports yet suggesting that fosfomycin can stimulate the expression of the efflux pump gene and mediate drug resistance. However, a few studies on S. aureus have described the drug sensitivity and resistance mechanism of fosfomycin in S. aureus, although they are not fully understood.
In the present study, we focus on the mutations of the target enzyme MurA, which catalyzes the initial step in the biosynthesis of peptidoglycan and transporters (GlpT and UhpT), as well as the overexpression of murA and tet38 efflux pump in 11 fosfomycin-resistant S. aureus. In addition, a strong correlation was established between fosfomycin resistance and efflux pump gene tet38 overexpression that has not been reported previously. The results of quantitative real-time PCR (qRT-PCR) indicated that the Tet38 efflux pump plays a vital role in fosfomycin resistance by pumping out the drug.
Materials and Methods
Bacterial Strains
In 2018, a total of 200 S. aureus isolates were obtained from the First Affiliated Hospital of Wenzhou Medical University, a comprehensive teaching hospital in China. The bacteria were identified by matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS; BioMérieux, Lyons, France). S. aureus ATCC 29213 (American Type Tissue Culture Collection, Manassas, VA, United States) was used as an endogenous control strain in antimicrobial susceptibility testing experiments. The study and consent procedure were approved by the Ethics Committee of the hospital.
Antimicrobial Susceptibility Testing
The minimum inhibitory concentration (MIC) of fosfomycin for each clinical strain was determined using the agar dilution method, wherein the media were supplemented with glucose-6-phosphate (25 mg/L), according to the recommendations of the Clinical and Laboratory Standards Institute [CLSI], 2018 (Ushanov et al., 2020). The data were interpreted according to the European Committee on Antimicrobial Susceptibility Testing criteria (available at http://www.eucast.org/clinical_breakpoints/) (susceptible, ≤32 mg/L; resistant, ≥64 mg/L), and the fosfomycin-resistant isolates were selected for further investigation. In addition, the MICs of fosfomycin-resistant S. aureus to other classes of antibiotics, including oxacillin, erythromycin, ciprofloxacin, levofloxacin, gentamicin, rifampicin, linezolid, vancomycin, and teicoplanin, were detected using the broth microdilution method.
Detection of Fosfomycin-Resistant Genes
The DNA of fosfomycin-resistant and fosfomycin-susceptible S. aureus isolates was extracted using a Biospin Bacterial Genomic DNA Extraction Kit (Bioflux, Tokyo, Japan) and was utilized as the template for PCR amplification of the fosA, fosB, fosC, fosD, fosX, glpT, uhpT, murA, and tet38 genes; the primers are listed in Table 1. The PCR products were sequenced by Beijing Genomics Institute Technology Co., Ltd. (Shanghai, China), and the sequences were aligned by BLAST on the NCBI platform1. The PCR products of uhpT, glpT, and murA were sequenced to scan for mutations.
TABLE 1.
Primers used in this study.
| Gene | Primer sequences (5′→3′) | Product size (bp) | References |
| PCR primers | |||
| fosA | F:GCTGCACGCCCGCTGGAATA | 217 | Chen et al., 2014 |
| R:CGACGCCCCCTCGCTTTTGT | |||
| fosB | F:CAGAGATATTTTAGGGGCTGACA | 312 | Chen et al., 2014 |
| R:CTCAATCTATCTTCTAAACTTCCTG | |||
| fosC | F:GGGTTACATGCCCTTGCTCA | 354 | Chen et al., 2014 |
| R:AACCCGCACAACGACAGATG | |||
| fosD | F: AACTCTAACTTGTGTCCGTCAG | 220 | Liu et al., 2017 |
| R: GTGGCTTATGGGTTGCGTTA | |||
| fosX | F: ATGATCAGTCATATGACATTTATCG | 243 | Zhang et al., 2020 |
| R: ATTTAGCCCCTTGTCGATAACG | |||
| murA | F:GCCCTTGAAAGAATGGTTCGT | 1600* | NC_002745.2** |
| R:GTTACAATACTCGACGCAGGT | |||
| glpT | F:TGAATAAAACAGCAGGGCAA | 1699* | NC_002745.2** |
| R:CACAGCTAGTATGTATAACGAC | |||
| uhpT | F:TGTGTTTATGTTCAGTATTTTGGA | 1571* | NC_002745.2** |
| R:TCTTTCATCTCTTCACGCAC | |||
| tet38 | F:GCGGATACAACAGCGAGTGA | 1353 | Truong-Bolduc et al., 2005 |
| R:TCGACGCACCTAATGGGAAT | |||
| qRT-PCR primers | |||
| gmk | F:ACTAGGGATGCGTTTGAAGC | 122 | Chen and Hooper, 2018 |
| R:TCATGACCTTCGTCCATTGT | |||
| tet38 | F:TGACAGGTGTGGCTATTGGT | 112 | Chen and Hooper, 2018 |
| R:TTGCCTGGGAAATTTAATGC | |||
| murA | F:TGTGCACCTTGCAATTGACT | 102 | G et al., 2019 |
| R:CCGTTTTATGCATGTTGCAG | |||
*PCR product including surrounding sequences adjacent to the target gene. **GenBank-EMBL-DDBL accession number.
Fosfomycin Treatment and Total RNA Isolation
Actively growing S. aureus specimens were treated with increasing concentrations of fosfomycin (1/8 MIC, 1/4 MIC, and 1/2 MIC) for 2 h, after which the cells were harvested, and total RNA was extracted using a Bacterial RNA Miniprep Kit (Biomiga, Shanghai, China) according to the manufacturer’s instructions. Then, 1000 ng RNA was used as the template for reverse transcription using a RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, United States) to obtain cDNA.
Quantitative Real-Time PCR (qRT-PCR)
qPCR was performed on a CFX-96 TouchTM Real-Time PCR system (Bio-Rad, Hercules, CA, United States) using TB Green Premix Ex Taq II (Tli RNase H Plus) (2×) (Takara, Japan), specific primers (Table 1), and 100 ng cDNA as the template. Cycling conditions were as follows: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 20 s. A melting curve was performed after each run (raising the temperature by 0.5°C/s, from 65 to 95°C). Each sample was run in triplicate, and the means of the Ct values were used for analysis. The relative expression levels of tet38 and murA genes were normalized to the gmk reference gene (Chen and Hooper, 2018). The quantification of the target genes was analyzed using the comparative threshold cycle 2–ΔΔCt method. All experiments were repeated in triplicate independently. The relative expression of the mRNA of the target gene was normalized to that of S. aureus ATCC 29213.
Multilocus Sequence Typing (MLST)
Isolates were screened using a previously described method to detect the following seven housekeeping genes: carbamate kinase (arcC), shikimate dehydrogenase (aroE), glycerol kinase (glp), guanylate kinase (gmk), phosphate acetyltransferase (pta), triosephosphate isomerase (tpi), and acetyl coenzyme A acetyltransferase (yqiL) (Enright and Spratt, 1999). The sequences of the PCR products were compared with those available from the MLST website2 for S. aureus. Also, the allelic number was determined for each sequence.
Planktonic Growth Assay
The planktonic growth rates of 8 tet38-overexpressed S. aureus isolates were determined as described previously (Wijesinghe et al., 2019), with some modifications. Briefly, 8 tet38-overexpressed isolates and ATCC 29213 standard cell suspensions were prepared by adjusting the turbidity of suspension to 0.5 McFarland standard in sterile saline. Then, 200 μL of each suspension was inoculated in 20 mL sterile LB broth containing fosfomycin in 0, 1/8 MIC, 1/4 MIC, and 1/2 MIC, respectively, for growth at 37°C and 180 rpm for 24 h. The growth rate of the planktonic bacteria was determined by measuring the optical density (OD) of the suspension in each well of the 96-well plate at 600 nm at 2-h intervals for 24 h using a microtiter plate reader (BioTek, United States). The growth curve was generated in triplicate for each experiment. ATCC 29213 served as the control strain.
Statistical Analysis
The relative expression of murA and tet38 was compared using Student’s t-test, and P-value < 0.05 was considered to be statistically significant.
Results
Susceptibility to Fosfomycin and Other Types of Antibiotics
The susceptibility to fosfomycin of 200 S. aureus isolates was determined by the agar dilution method using glucose-6-phosphate (25 mg/L). The results showed that 5.5% (11/200) of the isolates were resistant to fosfomycin. Also, resistance to other antibiotics was determined (Table 2); 81.8% (9/11) of the isolates displayed resistance to erythromycin, while 72.7% (8/11) belonged to MDR S. aureus.
TABLE 2.
Characteristics and resistance spectrum of fosfomycin-resistant S. aureus strains.
| Isolates | ST type | FOM | OXA | ERY | CIP | LVX | GEN | RIF | LNZ | VAN | TEC |
| JP3187 | 5 | 256R | >128R | 32R | >256R | 32R | 4 | <0.03 | 2 | 2 | 4 |
| JP3189 | 4539 | 64R | >128R | 64R | 64R | 32R | 16R | >16R | 1 | 1 | 4 |
| JP3212 | 5 | 256R | >128R | 64R | 128R | 32R | 4 | <0.03 | 2 | 2 | 8 |
| JP3235 | 5708 | 64R | >128R | 1 | 128R | 32R | 64R | >16R | 2 | 1 | 4 |
| JP3244 | 7 | 128R | 0.5 | 64R | 0.25 | 0.25 | <0.125 | <0.03 | 2 | 1 | 0.5 |
| JP3505 | 4739 | 512R | 0.5 | 1 | 2 | 1 | <0.125 | <0.03 | 4 | 2 | 0.5 |
| JP3535 | 5 | 64R | >128R | 16R | 128R | 16R | 0.25 | <0.03 | 4 | 1 | 2 |
| JP3539 | 59 | 64R | 8R | 64R | 0.5 | 0.25 | 0.25 | <0.03 | 4 | 2 | 1 |
| JP3589 | 1 | 64R | 0.25 | 64R | 2 | 1 | 2 | <0.03 | 2 | 2 | 1 |
| JP3592 | 239 | 256R | >128R | 64R | 128R | 64R | <0.125 | >8R | 4 | 1 | 1 |
| JP3600 | 965 | 64R | 0.5 | 64R | 4R | 1 | 0.5 | <0.03 | 2 | 2 | 2 |
FOM, fosfomycin; OXA, oxacillin; ERY, erythromycin; CIP, ciprofloxacin; LVX, levofloxacin; GEN, gentamicin; RIF, rifampicin; LNZ, linezolid; VAN, vancomycin; TEC, teicoplanin. Superscript “R” indicates resistance.
Molecular Mechanisms of Fosfomycin-Resistant Isolates
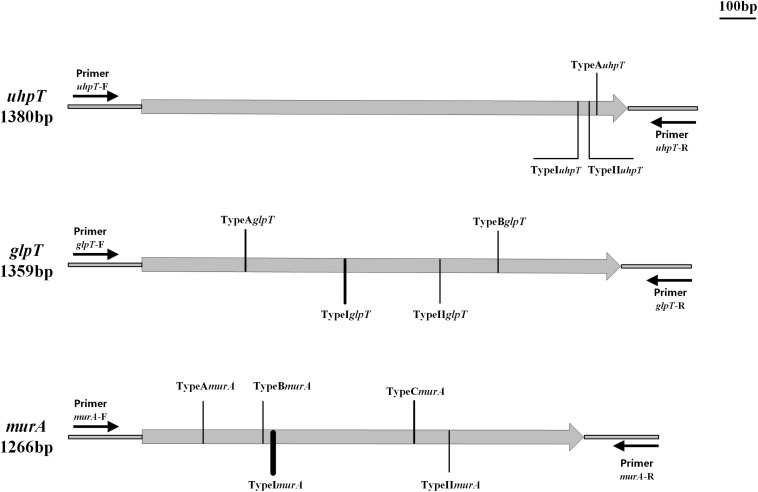
Strains carrying the fosA, fosB, fosC, fosD, or fosX gene were not found in the current study (Table 3). Based on the classification method of Fu et al. (2015) we named the sense mutations as TypeA, TypeB, and TypeC according to the amino acid sequence, and the nonsense mutations were named as TypeI, TypeII, and TypeIII; the subscripts represent different genes (Fu et al., 2015). Three distinct mutations were detected in the uhpT gene of the 11 fosfomycin-resistant S. aureus isolates and the corresponding sensitive strains. Mutation TypeAuhpT, found in JP3212, resulted in an amino acid substitution at position 457 (Leu457Val) of UhpT. Conversely, the other two mutations (TypeI–IIuhpT), which resulted in distinct amino acid substitutions within the UhpT protein, were identified in fosfomycin-sensitive isolates, although one mutation (TypeIIuhpT) was also found in fosfomycin-resistant S. aureus (Figure 1 and Table 3).
TABLE 3.
Characteristics and amino acid substitutions in fosfomycin-resistant and fosfomycin-sensitive S. aureus.
| Strains | Type | tet38 | fos | Amino acid substitution | ||||||
| fosA | fosB | fosC | fosD | fosX | uhpT | glpT | murA | |||
| JP3187 | R | + | – | – | – | – | – | None | TypeAglpT | TypeImurA |
| JP3189 | R | + | – | – | – | – | – | None | None | TypeCmurA |
| JP3212 | R | + | – | – | – | – | – | TypeAuhpT | None | TypeImurA |
| JP3235 | R | + | – | – | – | – | – | None | None | TypeImurA TypeCmurA |
| JP3244 | R | + | – | – | – | – | – | None | None | TypeImurA TypeCmurA |
| JP3505 | R | + | – | – | – | – | – | None | TypeIglpT | TypeAmurA TypeImurA |
| JP3535 | R | + | – | – | – | – | – | None | TypeIglpT TypeBglpT | TypeImurA |
| JP3539 | R | + | – | – | – | – | – | None | TypeAglpT TypeIglpT | TypeImurA TypeIImurA |
| JP3589 | R | + | – | – | – | – | – | None | None | TypeImurA TypeBmurA |
| JP3592 | R | + | – | – | – | – | – | None | None | TypeImurA TypeCmurA |
| JP3600 | R | + | – | – | – | – | – | TypeIIuhpT | TypeIglpT | TypeImurA |
| JP3200 | S | + | – | – | – | – | – | None | None | None |
| JP3203 | S | + | – | – | – | – | – | TypeIuhpT | None | None |
| JP3277 | S | – | – | – | – | – | None | None | None | |
| JP3230 | S | + | – | – | – | – | – | None | None | None |
| JP3240 | S | + | – | – | – | – | – | TypeIIuhpT | TypeIglpT TypeIIglpT | None |
| JP3245 | S | + | – | – | – | – | – | None | None | None |
| JP3502 | S | + | – | – | – | – | – | None | None | TypeImurA TypeIImurA |
| JP3512 | S | + | – | – | – | – | – | None | None | None |
| JP3518 | S | + | – | – | – | – | – | None | None | TypeImurA |
| JP3520 | S | + | – | – | – | – | – | None | TypeIglpT | TypeImurA |
| JP3522 | S | + | – | – | – | – | – | None | None | TypeImurA |
+, carries the corresponding gene; –, does not carry the corresponding gene; None: nonsense mutation; TypeAuhpT: T1369 G; TypeIuhpT: G1364A; TypeIIuhpT: T1368G; TypeAglpT: C299T; TypeBglpT: G1064A; TypeIglpT: G583A; TypeIIglpT: T829G; TypeAmurA: G187A; TypeBmurA: G349T; TypeCmurA: G770A; TypeImurA: C371G; TypeIImurA: A873T.
FIGURE 1.
Types and positions of mutations in uhpT, glpT, and murA genes. TypeAuhpT: T1369 G; TypeIuhpT: G1364A; TypeIIuhpT: T1368G. TypeAglpT: C299T; TypeBglpT: G1064A; TypeIglpT: G583A; TypeIIglpT: T829G. TypeAmurA: G187A; TypeBmurA: G349T; TypeCmurA: G770A; TypeImurA: C371G; and TypeIImurA: A873T.
Moreover, four different mutations were detected in the glpT gene (TypeA–BglpT and TypeI–IIglpT). Notably, TypeBglpT, found in the fosfomycin-resistant isolates JP3535, produced a premature stop codon within the glpT coding sequence at position 355 (Trp335Ter), thereby resulting in truncated proteins. In addition, TypeIIglpT was detected only in the fosfomycin-sensitive isolates (Figure 1 and Table 3).
Of the 11 fosfomycin-resistant isolates, 6 contained one of the three different mutations (TypeA–CmurA) in the murA gene. TypeA–CmurA, which resulted in distinct amino acid substitutions within the MurA protein at positions 63 (Ala63Thr), 117 (Gly117Trp), and 257 (Gly257Asp), could only be found in fosfomycin-resistant isolates, and two mutations (TypeI–IImurA) could be found in both fosfomycin-resistant and fosfomycin-susceptible S. aureus (Figure 1 and Table 3). Moreover, only one sense mutation was present in each fosfomycin-resistant S. aureus isolate.
Expression Analysis of murA and tet38
qRT-PCR revealed significant differences in the expression of murA between resistant and susceptible groups of S. aureus as compared with S. aureus ATCC 29213 (P < 0.05) (Table 4). The data showed that the average expression level of murA gene decreased by 0.7-fold in fosfomycin-resistant and fosfomycin-susceptible S. aureus isolates. In addition, compared with the fosfomycin-susceptible S. aureus, the expression of murA in the resistance isolates was not significantly higher.
TABLE 4.
Relative expression of target enzyme gene murA and efflux pump gene tet38 in fosfomycin-susceptible and fosfomycin-resistant S. aureus.
| Strains | Relative expression level of murAa (mean ± SD) | Relative expression level of tet38a (mean ± SD) |
| S1 | 0.77 ± 0.15 | 0.77 ± 0.13 |
| S2 | 1.45 ± 0.10 | 1.48 ± 1.02 |
| S3 | 0.40 ± 0.05 | 0.71 ± 0.21 |
| S4 | 0.48 ± 0.01 | 1.55 ± 0.33 |
| S5 | 0.48 ± 0.06 | 0.98 ± 0.35 |
| JP3187 | 0.75 ± 0.09 | 1.71 ± 0.93 |
| JP3189 | 0.35 ± 0.01 | 0.71 ± 0.25 |
| JP3212 | 0.47 ± 0.01 | 21.6 ± 5.75 |
| JP3235 | 1.47 ± 0.14 | 0.92 ± 0.34 |
| JP3244 | 0.21 ± 0.04 | 1.24 ± 0.15 |
| JP3505 | 0.23 ± 0.04 | 2.65 ± 1.04 |
| JP3535 | 0.34 ± 0.03 | 2.74 ± 0.37 |
| JP3539 | 0.39 ± 0.05 | 1.71 ± 0.48 |
| JP3589 | 0.42 ± 0.08 | 0.54 ± 0.23 |
| JP3592 | 0.40 ± 0.07 | 143.36 ± 2.05 |
| JP3600 | 1.52 ± 0.23 | 24.59 ± 0.17 |
S1–S5 represent the 5 fosfomycin-susceptible S. aureus strains. ATCC 29213 served as the control strain. aThe relative gene expression with more than 2-fold change compared with ATCC 29213 after fosfomycin induction is shown in bold.
However, the results (Table 4) indicated that compared with that in ATCC29213 and susceptible isolates, the expression level of efflux pump gene tet38 in JP3212, JP3535, JP359, and JP3600 was elevated. Notably, the level of the tet38 gene in JP3212, JP3535, JP3592, and JP3600 was altered markedly (21. 60-, 2. 74-, 143. 36-, and 24.59-fold) as compared with that in ATCC29213 (Table 4).
Exposure to Fosfomycin Resulted in Increased Expression of tet38 Efflux Pump Genes Among Some Resistant Isolates
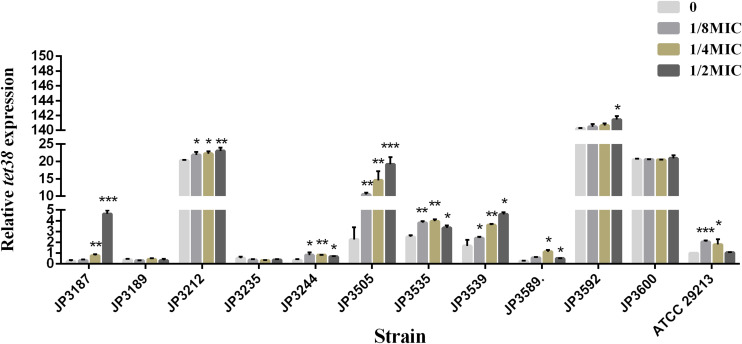
The expression of tet38 in the presence of increasing amounts of fosfomycin with 0, 1/8 MIC, 1/4 MIC, and 1/2 MIC concentrations was determined by qRT-PCR. Notably, the expression of the tet38 gene in JP3187, JP3212, JP3244, JP3505 JP3535, JP3539, P3589, JP3592, and ATCC 29213 was upregulated with the increase in fosfomycin concentration as compared with the 0 MIC strains (Figure 2 and Table 4). Also, 4.63-fold and 6.42-fold increases were noted in the expression of tet38 in JP3505 cells treated with 1/8 MIC (64 mg/L) and 1/4 MIC (128 mg/L) fosfomycin, respectively, as compared with that in cells that were not treated with fosfomycin (Table 5). A further 8.46-fold increase was observed in those treated with 1/2 MIC (256 mg/L) fosfomycin.
FIGURE 2.
The relative expression of efflux pump gene tet38 in fosfomycin-resistant S. aureus exposed to 0, 1/8 MIC, 1/4 MIC, and 1/2 MIC concentrations. Bars indicate the mean values, and asterisks denote the significant difference of expression (P < 0.05).
TABLE 5.
Relative expression of efflux pump gene tet38 in fosfomycin-resistant S. aureus exposed to different concentrations of fosfomycin.
| Strains | The relative expression level of tet38a (mean ± SD) | |||
| 0 MIC | 1/8 MIC | 1/4 MIC | 1/2 MIC | |
| JP3187 | 0.250.09 | 0.320.08 | 0.77 ± 0.10 | 4.64 ± 0.25 |
| JP3189 | 0.380.08 | 0.320.01 | 0.430.07 | 0.320.13 |
| JP3212 | 20.320.10 | 21.890.69 | 22.220.56 | 23.020.8 |
| JP3235 | 0.530.12 | 0.360.08 | 0.330.03 | 0.370.06 |
| JP3244 | 0.350.06 | 0.81 ± 0.20 | 0.83 ± 0.01 | 0.680.03 |
| JP3505 | 2.270.91 | 10.51 ± 0.38 | 14.57 ± 2.12 | 19.21 ± 1.65 |
| JP3535 | 2.500.12 | 3.820.12 | 3.950.14 | 3.340.17 |
| JP3539 | 1.670.45 | 2.400.08 | 3.60 ± 0.08 | 4.60 ± 0.14 |
| JP3589 | 0.270.02 | 0.57 ± 0.06 | 1.13 ± 0.14 | 0.510.01 |
| JP3592 | 140.270.05 | 140.480.30 | 140.650.23 | 141.470.36 |
| JP3600 | 20.580.20 | 20.620.04 | 20.470.08 | 20.930.72 |
aThe relative gene expression with more than 2-fold change compared with 0 MIC after fosfomycin induction is shown in bold.
Molecular Typing
The 11 fosfomycin-resistant S. aureus specimens were categorized into 9 STs (Table 2): ST1 (n = 1), ST5 (n = 3), ST59 (n = 1), ST7 (n = 1), ST239 (n = 1), ST965 (n = 1), ST4539 (n = 1), ST4739 (n = 1), and a new ST that was found in the current study (ST 5708) (n = 1).
Growth Rate
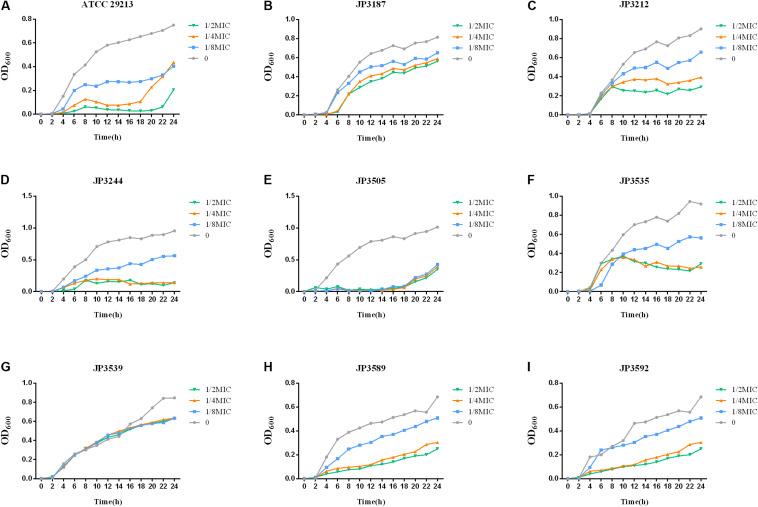
In order to gain quantitative insight into the fitness cost imposed by tet38-overexpressed isolates, the growth curves of 8 tet38-overexpressed S. aureus were recorded. We identified a fitness cost after fosfomycin induction. The growth of 8 tet38-overexpression strains was inhibited in LB at a subinhibitory concentration of fosfomycin (Figure 3).
FIGURE 3.
Growth curves at different fosfomycin concentrations in tet38-overexpression S. aureus. The values shown are the average of three independent experiments. Different colors of lines represent different concentrations of fosfomycin. (A) Growth of control strain ATCC 29213. (B–I) Growth of 8 tet38-overexpression isolates.
Discussion
Due to the unique mechanisms of action, fosfomycin exhibits significant antimicrobial activity against a broad spectrum of pathogens, including S. aureus (Goto, 1977). A review described that the susceptibility of S. aureus to fosfomycin ranged from 33.2% to 100% in the nine available studies [odds ratio (OD) = 91.7%, 95% confidence interval (CI): 88.7–94.9%] (Vardakas et al., 2016). In the current study, the susceptibility rate of fosfomycin in S. aureus was 94.5% (189/200). However, the prevalence of fosfomycin resistance in clinical isolates of S. aureus has been reported with increasing frequency in many areas (Del Rio et al., 2014; Mihailescu et al., 2014; Shi et al., 2014).
The resistance mechanism of fosfomycin in Gram-negative bacteria has been widely reported; also, in a previous study, we reported the resistance of fosfomycin in ESBL-producing Escherichia coli (Bi et al., 2017). Fosfomycin enters the cell via two transporters, GlpT and UhpT, and mutations or insertions in glpT and/or uhpT genes result in the loss of function (Takahata et al., 2010). According to the study by Castaneda-Garcia et al. (2009), glpT inactivation played an essential role in the resistance to fosfomycin in Pseudomonas aeruginosa (Castaneda-Garcia et al., 2009). The murA gene is also closely related to fosfomycin resistance (Takahata et al., 2010; Couce et al., 2012). In addition, fosfomycin activity can be inhibited via the catalytic activity of FosA, FosB, and FosC, respectively (Garcia et al., 1995; Lee et al., 2012).
Among Gram-positive bacteria, the resistance mechanism of fosfomycin is rarely reported. In the current study, none of the resistant strains carried the fosA, fosB, fosC, fosD, or fosX gene, indicating that these genes might not be the primary factors mediating the resistance of S. aureus against fosfomycin. Other studies have shown that the prevalence of fosfomycin resistance genes (fosA, fosB, and fosC) was not the predominant factor contributing to resistance in S. aureus (Xu et al., 2017). In addition, a total of 12 mutations were found in 11 strains of fosfomycin-resistant S. aureus by sequencing analysis. Of these, 3 were detected in uhpT, and TypeAuhpT was carried only by fosfomycin-resistant strain JP3212, while TypeIuhpT and TypeIIuhpT were found in both fosfomycin-resistant and fosfomycin-sensitive strains, which likely did not contribute to fosfomycin resistance. Within the glpT gene of 11 drug-resistant strains, 2 mutations TypeA–BglpT were observed only in the drug-resistant strains. On the other hand, mutation TypeIglpT was widely detected in both drug-resistant and susceptible strains. Intriguingly, TypeBglpT, which generated a stop codon (TA1064G) at position 335 (Figure 1), was harbored in JP3535. Also, we found a mutation (TypeIIglpT) merely in the fosfomycin-sensitive strains. Of the five murA gene mutations found in the drug-resistant strains, TypeImurA and TypeIImurA could also be found in susceptible strains, and the remaining three mutations (TypeA–CmurA) were found only in drug-resistant strains (TypeAmurA: JP3505; TypeBmurA: JP3589; TypeCmurA: JP3189, JP3235, JP3244; JP3592). In addition, TypeImurA was also found in fosfomycin-resistant strains (10/11). Among the above mutations, four mutation sites, TypeAglpT, TypeBglpT, TypeCmurA, and TypeIImurA, were consistent with those reported (Fu et al., 2015). We also found that the frequency of murA mutation in S. aureus was high, which might play a major role in mediating fosfomycin resistance, which needs an in-depth investigation.
Although several studies have mentioned that the overexpression of the murA gene can greatly increase the MICs of fosfomycin, the difference in murA expression between fosfomycin-sensitive and fosfomycin-resistant S. aureus has not been reported (Garcia et al., 1995; Olesen et al., 2014). In the current study, the results of qRT-PCR revealed a significant difference between the fosfomycin-resistant and fosfomycin-susceptible S. aureus with respect to the expression of murA as compared with that of S. aureus ATCC 29213. However, statistical differences could not be detected between two types of strains (Table 4), indicating that the overexpression of target gene murA has no role in conferring fosfomycin resistance in the strains identified in this study. Interestingly, some resistant strains showed a downward trend in the expression of murA, suggesting that the role of the murA gene in fosfomycin needs to be studied further.
Recent studies have shown that the tet38 gene exerts a specific effect on fosfomycin resistance. According to the study by Truong-Bolduc, the overexpression of tet38 resulted in a fourfold increase in the MIC of fosfomycin compared with that of the parent strain (Truong-Bolduc et al., 2018). The results of the current study showed that the expression of the tet38 efflux pump gene in fosfomycin-resistant strains JP3212, JP3535, JP3592, and JP3600 was significantly higher than that in the control strain ATCC29213 and the susceptible strains (P = 0.007, P = 0.002, P < 0.001, P < 0.001, respectively). Furthermore, under the treatment of 0, 1/8 MIC, 1/4 MIC, and 1/2 MIC with fosfomycin, we found that the expression of efflux pump gene tet38 was upregulated in most resistant isolates, even in the reference strain ATCC 29213. Although nonsense mutation was detected in uhpT, glpT, and murA genes, the level of tet38 in JP3600 was high even without the drug, which might explain the resistance to fosfomycin. This phenomenon suggests that the tet38 efflux pump plays a critical role in mediating fosfomycin resistance. Reportedly, abscess and other factors can promote the expression of tet38 (Chen and Hooper, 2018), and the current study has shown that the stimulation of the drug also enhances the expression of the efflux pump, albeit the specific mechanism remains to be studied further. Moreover, the overexpression of tet38 can also lead to changes in the cost of bacterial fitness. Some studies have demonstrated that the global regulator MgrA functions as a direct regulator of tetR21, which is a TetR-like regulator of the tet38 efflux pump gene. TetR21 acts as a repressor of tet38 expression and may also regulate the expression of other bacterial resistance determinants (Truong-Bolduc et al., 2005, 2015). We speculated that the high expression of the tet38 gene in S. aureus might be related to the regulation of TetR-like regulator TetR21 and the global regulator MgrA. We will also continue to focus on these phenomena in other bacteria in future studies.
MLST analyses designated three fosfomycin-resistant S. aureus isolated to ST5. Combined with drug sensitivity, we found that the ST5 resistant strains were resistant to at least five antibiotics. Among 11 fosfomycin-resistant strains, 72.7% were MDR strains, and further follow-up treatment was essential. Wu et al. (2018) reported that ST5 and ST239 strains were usually resistant to fosfomycin and constituted the predominant HA-MRSA clones in China. The new sequence type found in the resistant strain has been submitted to the repository (see text footnote 2).
Conclusion
A total of 11 fosfomycin-resistant strains were screened out from 200 S. aureus isolates, and the mechanism was explored. Our findings indicated that fosA, fosB, fosC, fosD, and fosX genes might not be the major resistant mechanism of S. aureus to fosfomycin. The mutations within the glpT, uhpT, and murA genes might play a critical role in conferring fosfomycin resistance. However, the role of overexpression of murA in fosfomycin resistance needs to be discussed further in S. aureus. Also, the phenomenon of overexpression in the tet38 gene under a subinhibitory concentration of fosfomycin needs to be investigated further.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
WX conducted the experiments, analyzed the data, and wrote the manuscript. TC participated in experiments and writing. HW and WZ provided fosfomycin-resistant strains and participated in the analysis of results. KY and QW participated in the analysis of the results. TZ helped to design the study. YX and XZ designed the study and corrected the manuscript. All authors read and approved the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This study was supported by a research grant from the National Natural Science Foundation of China (no. 81171614).
References
- Bi W., Li B., Song J., Hong Y., Zhang X., Liu H., et al. (2017). Antimicrobial susceptibility and mechanisms of fosfomycin resistance in extended-spectrum beta-lactamase-producing Escherichia coli strains from urinary tract infections in Wenzhou. China. Int. J. Antimicrob. Agents 50 29–34. 10.1016/j.ijantimicag.2017.02.010 [DOI] [PubMed] [Google Scholar]
- Castaneda-Garcia A., Rodriguez-Rojas A., Guelfo J. R., Blazquez J. (2009). The glycerol-3-phosphate permease GlpT is the only fosfomycin transporter in Pseudomonas aeruginosa. J. Bacteriol. 191 6968–6974. 10.1128/jb.00748-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Hooper D. C. (2018). Effect of Staphylococcus aureus Tet38 native efflux pump on in vivo response to tetracycline in a murine subcutaneous abscess model. J. Antimicrob. Chemother. 73 720–723. 10.1093/jac/dkx432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Xu X., Qu T., Yu Y., Ying C., Liu Q., et al. (2014). Prevalence of the fosfomycin-resistance determinant, fosB3, in Enterococcus faecium clinical isolates from China. J. Med. Microbiol. 63 1484–1489. 10.1099/jmm.0.077701-0 [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute [CLSI] (2018). Performance Standards Forantimicro-Bial Susceptibility Testing: Twenty Eighth Informational Supplement M100-S28. Wayne, PA: CLSI. [Google Scholar]
- Couce A., Briales A., Rodriguez-Rojas A., Costas C., Pascual A., Blazquez J. (2012). Genomewide overexpression screen for fosfomycin resistance in Escherichia coli: MurA confers clinical resistance at low fitness cost. Antimicrob. Agents Chemother. 56 2767–2769. 10.1128/aac.06122-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rio A., Gasch O., Moreno A., Pena C., Cuquet J., Soy D., et al. (2014). Efficacy and safety of fosfomycin plus imipenem as rescue therapy for complicated bacteremia and endocarditis due to methicillin-resistant Staphylococcus aureus: a multicenter clinical trial. Clin. Infect. Dis. 59 1105–1112. 10.1093/cid/ciu580 [DOI] [PubMed] [Google Scholar]
- Enright M. C., Spratt B. G. (1999). Multilocus sequence typing. Trends Microbiol. 7 482–487. [DOI] [PubMed] [Google Scholar]
- Etienne J., Gerbaud G., Fleurette J., Courvalin P. (1991). Characterization of staphylococcal plasmids hybridizing with the fosfomycin resistance gene fosB. FEMS Microbiol. Lett. 68 119–122. 10.1111/j.1574-6968.1991.tb04580.x [DOI] [PubMed] [Google Scholar]
- Fu Z., Ma Y., Chen C., Guo Y., Hu F., Liu Y., et al. (2015). Prevalence of Fosfomycin Resistance and Mutations in murA, glpT, and uhpT in Methicillin-Resistant Staphylococcus aureus strains isolated from blood and cerebrospinal fluid samples. Front. Microbiol. 6:1544. 10.3389/fmicb.2015.01544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- G C. B., Sahukhal G. S., Elasri M. O. (2019). Role of the msaABCR Operon in Cell Wall Biosynthesis, Autolysis, Integrity, and Antibiotic Resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 63:e00680-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia P., Arca P., Evaristo Suarez J. (1995). Product of fosC, a gene from Pseudomonas syringae, mediates fosfomycin resistance by using ATP as cosubstrate. Antimicrob. Agents Chemother. 39 1569–1573. 10.1128/aac.39.7.1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatadi S., Madhavi Y. V., Chopra S., Nanduri S. (2019). Promising antibacterial agents against multidrug resistant Staphylococcus aureus. Bioorg. Chem. 92:103252. 10.1016/j.bioorg.2019.103252 [DOI] [PubMed] [Google Scholar]
- Goto S. (1977). Fosfomycin, antimicrobial activity in vitro and in vivo. Chemotherapy 23(Suppl. 1), 63–74. 10.1159/000222028 [DOI] [PubMed] [Google Scholar]
- Lee S. Y., Park Y. J., Yu J. K., Jung S., Kim Y., Jeong S. H., et al. (2012). Prevalence of acquired fosfomycin resistance among extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae clinical isolates in Korea and IS26-composite transposon surrounding fosA3. J. Antimicrob. Chemother. 67 2843–2847. 10.1093/jac/dks319 [DOI] [PubMed] [Google Scholar]
- Liu B. H., Lei C. W., Zhang A. Y., Pan Y., Kong L. H., Xiang R., et al. (2017). Colocation of the Multiresistance Gene cfr and the Fosfomycin Resistance Gene fosD on a Novel Plasmid in Staphylococcus arlettae from a Chicken Farm. Antimicrob. Agents Chemother. 61:e01388-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehraj J., Witte W., Akmatov M. K., Layer F., Werner G., Krause G. (2016). Epidemiology of Staphylococcus aureus nasal carriage patterns in the community. Curr. Top. Microbiol. Immunol. 398 55–87. [DOI] [PubMed] [Google Scholar]
- Michalopoulos A. S., Livaditis I. G., Gougoutas V. (2011). The revival of fosfomycin. Int. J. Infect. Dis. 15 e732–e739. 10.1016/j.ijid.2011.07.007 [DOI] [PubMed] [Google Scholar]
- Mihailescu R., Furustrand Tafin U., Corvec S., Oliva A., Betrisey B., Borens O., et al. (2014). High activity of Fosfomycin and Rifampin against methicillin-resistant Staphylococcus aureus biofilm in vitro and in an experimental foreign-body infection model. Antimicrob. Agents Chemother. 58 2547–2553. 10.1128/aac.02420-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaminami H., Noguchi N., Nishijima S., Kurokawa I., Sasatsu M. (2008). Characterization of the pTZ2162 encoding multidrug efflux gene qacB from Staphylococcus aureus. Plasmid 60 108–117. 10.1016/j.plasmid.2008.04.003 [DOI] [PubMed] [Google Scholar]
- Olesen S. H., Ingles D. J., Yang Y., Schonbrunn E. (2014). Differential antibacterial properties of the MurA inhibitors terreic acid and fosfomycin. J. Basic Microbiol. 54 322–326. 10.1002/jobm.201200617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Mao N. F., Wang L., Zhang H. B., Chen Q., Liu H., et al. (2014). Efficacy of combined vancomycin and fosfomycin against methicillin-resistant Staphylococcus aureus in biofilms in vivo. PLoS One 9:e113133. 10.1371/journal.pone.0113133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorr A. F., Pogue J. M., Mohr J. F. (2017). Intravenous fosfomycin for the treatment of hospitalized patients with serious infections. Expert Rev. Anti Infect. Ther. 15 935–945. 10.1080/14787210.2017.1379897 [DOI] [PubMed] [Google Scholar]
- Silver L. L. (2017). Fosfomycin: mechanism and resistance. Cold Spring Harb. Perspect. Med. 7:a025262. 10.1101/cshperspect.a025262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata S., Ida T., Hiraishi T., Sakakibara S., Maebashi K., Terada S., et al. (2010). Molecular mechanisms of fosfomycin resistance in clinical isolates of Escherichia coli. Int. J. Antimicrob. Agents 35 333–337. 10.1016/j.ijantimicag.2009.11.011 [DOI] [PubMed] [Google Scholar]
- Truong-Bolduc Q. C., Bolduc G. R., Medeiros H., Vyas J. M., Wang Y., Hooper D. C. (2015). Role of the Tet38 Efflux Pump in Staphylococcus aureus internalization and survival in epithelial cells. Infect. Immun. 83 4362–4372. 10.1128/iai.00723-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong-Bolduc Q. C., Dunman P. M., Strahilevitz J., Projan S. J., Hooper D. C. (2005). MgrA is a multiple regulator of two new efflux pumps in Staphylococcus aureus. J. Bacteriol. 187 2395–2405. 10.1128/jb.187.7.2395-2405.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong-Bolduc Q. C., Wang Y., Hooper D. C. (2018). Tet38 efflux pump contributes to fosfomycin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 62:e00927-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushanov L., Lasareishvili B., Janashia I., Zautner A. E. (2020). Application of Campylobacter jejuni phages: challenges and perspectives. Animals 10:279. 10.3390/ani10020279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardakas K. Z., Legakis N. J., Triarides N., Falagas M. E. (2016). Susceptibility of contemporary isolates to fosfomycin: a systematic review of the literature. Int. J. Antimicrob. Agents 47 269–285. 10.1016/j.ijantimicag.2016.02.001 [DOI] [PubMed] [Google Scholar]
- Wang Y., Lin J., Zhang T., He S., Li Y., Zhang W., et al. (2020). Environmental contamination prevalence, antimicrobial resistance and molecular characteristics of methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis isolated from secondary schools in Guangzhou, China. Int. J. Environ. Res. Public Health 17:623. 10.3390/ijerph17020623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijesinghe G., Dilhari A., Gayani B., Kottegoda N., Samaranayake L., Weerasekera M. (2019). Influence of laboratory culture media on in vitro growth, adhesion, and biofilm formation of Pseudomonas aeruginosa and Staphylococcus aureus. Med. Princ. Pract. 28 28–35. 10.1159/000494757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Chen Y., Sun L., Qu T., Wang H., Yu Y. (2018). Prevalence of Fosfomycin Resistance in Methicillin-Resistant Staphylococcus aureus isolated from patients in a University Hospital in China from 2013 to 2015. Jpn. J. Infect. Dis. 71 312–314. 10.7883/yoken.jjid.2018.013 [DOI] [PubMed] [Google Scholar]
- Xu S., Fu Z., Zhou Y., Liu Y., Xu X., Wang M. (2017). Mutations of the Transporter Proteins GlpT and UhpT confer fosfomycin resistance in Staphylococcus aureus. Front. Microbiol. 8:914. 10.3389/fmicb.2017.00914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Bi W., Chen L., Zhang Y., Fang R., Cao J., et al. (2020). Molecular mechanisms and epidemiology of fosfomycin resistance in enterococci isolated from patients at a teaching hospital in China, 2013-2016. J. Glob. Antimicrob. Resist. 20 191–196. 10.1016/j.jgar.2019.08.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.