Abstract
Partitiviruses (dsRNA viruses, family Partitiviridae) are ubiquitously detected in plants and fungi. Although previous surveys suggested their omnipresence in the white root rot fungus, Rosellinia necatrix, only a few of them have been molecularly and biologically characterized thus far. We report the characterization of a total of 20 partitiviruses from 16 R. necatrix strains belonging to 15 new species, for which “Rosellinia necatrix partitivirus 11–Rosellinia necatrix partitivirus 25” were proposed, and 5 previously reported species. The newly identified partitiviruses have been taxonomically placed in two genera, Alphapartitivirus, and Betapartitivirus. Some partitiviruses were transfected into reference strains of the natural host, R. necatrix, and an experimental host, Cryphonectria parasitica, using purified virions. A comparative analysis of resultant transfectants revealed interesting differences and similarities between the RNA accumulation and symptom induction patterns of R. necatrix and C. parasitica. Other interesting findings include the identification of a probable reassortment event and a quintuple partitivirus infection of a single fungal strain. These combined results provide a foundation for further studies aimed at elucidating mechanisms that underly the differences observed.
Keywords: partitivirus, dsRNA virus, phytopathogenic fungus, Rosellinia necatrix, Cryphonectria parasitica, diversity, reassortment, horizontal transfer
Introduction
Plant pathogenic fungi provide platforms for identifying eukaryotic viruses (Pearson et al., 2009; Xie and Jiang, 2014; Ghabrial et al., 2015; Suzuki, 2017; Hillman et al., 2018). Research performed in previous decades have revealed that the diversity of fungal viruses or mycoviruses is much greater than previously imagined (Yu et al., 2010; Kondo et al., 2013a; Liu et al., 2014; Kanhayuwa et al., 2015; Zhang et al., 2016). Further, studies involving particular fungal-viral systems have provided interesting insights into virus-virus and virus-host interactions (Cho et al., 2013; Jiang et al., 2013; Xie and Jiang, 2014; Hillman et al., 2018). Antagonistic, mutualistic and neutral virus/virus interactions, and beneficial, harmful, or no effects on their hosts have been reported. Among fungal hosts commonly used for the study of viruses is the phytopathogenic ascomycete fungus, Rosellinia necatrix, which infects over 400 plant species and causes white root rot in perennial crops worldwide, and particularly in Japan (Pliego et al., 2012; Kondo et al., 2013b). A survey of over 1,000 field isolates showed that ~20% of field isolates harbor diverse populations of viruses (Arakawa et al., 2002; Ikeda et al., 2004; Kondo et al., 2013b; Zhang et al., 2014, 2016; Arjona-Lopez et al., 2018). Importantly, the characterization of some viruses has led to the approval of proposals to create new viral families by the International Committee on Taxonomy of Viruses (ICTV) (Chiba et al., 2009; Lin et al., 2012). The majority of viruses detected in R. necatrix have been partitiviruses (Yaegashi et al., 2013) (Kanematsu and Sasaki, unpublished results), which was also the case for the fungal forest inhabitant, Heterobasidion spp. (Vainio and Hantula, 2016).
Partitiviruses are bisegmented, biparticulate viruses with double-stranded RNA (dsRNA) genomes that range from 3.0 to 4.8 kbp (Nibert et al., 2014; Vainio et al., 2018a). With a few exceptions, RNA-dependent RNA polymerase (RdRP) is encoded within the large segment, while capsid protein (CP) is encoded within the smaller segment. Partitiviruses typically form icosahedral particles that are 30–35 nm in diameter. The near-atomic resolution of ~5 Å has been achieved for the T = 1 capsids of partitiviruses via cryo-electron microscopy (Nibert et al., 2013). Based on phylogenetic relationships and host ranges, partitiviruses are now classified into 5 genera: Alphapartitivirus, Betapartitivirus, Gammapartitivirus, Deltapartitivirus, and Cryspovirus (Nibert et al., 2014; Vainio et al., 2018a). The first two genera contain members that infect both plant and fungal species, while the genera Gammapartitivirus and Deltapartitivirus are composed of members that infect exclusively fungal and plant species, respectively. In this regard, it should be noted that fungus-infecting, betapartitivirus-related sequences are detected in various plant genomes, a paleological record of their capacity to infect plant species (viral fossils or endogenous virus elements) and their association with plants several tens of millions of years ago (Liu et al., 2010; Chiba et al., 2011). Cryspoviruses have been isolated from protozoa of the genus Cryptosporidium that infect mammals as a parasite. Recently, the novel genera “Epsilonpartitivirus” and “Zetapartitivirus” have been proposed for some novel viruses or virus-like agents associated with fungi and arthropods (Nerva et al., 2017a; Jiang et al., 2019). Partitiviruses, in most cases, produce symptomless infections. Exceptions include growth-reducing Heterobasidion partitivirus 3 (an alphapartitivirus) (Vainio et al., 2010) and Aspergillus fumigatus partitivirus 1 (a gammapartitivirus) (Bhatti et al., 2011), and hypovirulence-conferring partitiviruses such as Rhizoctonia solani partitivirus 2 (an alphapartitivirus, RsPV2) (Zheng et al., 2014), Sclerotinia sclerotiorum partitivirus 1 (a betapartitivirus, SsPV1) (Xiao et al., 2014), Aspergillus flavus partitivirus 1 (a zetapartitivirus) (Jiang et al., 2019) and Heterobasidion partitivirus 13 (an alphapartitivirus) (Vainio et al., 2018b). In other cases, some partitiviruses have been shown to contribute to hypovirulence and/or growth defects induced by other co-infecting fungal dsRNA viruses such as megabirnavirus and chrysovirus (Wang et al., 2014; Sasaki et al., 2016).
Despite the large number of partitiviruses identified thus far (Nibert et al., 2014; Vainio et al., 2018a), only a few including R. necatrix-infecting partitiviruses (Rosellinia necatrix partitiviruses: RnPV1, RnPV2, RnPV3, RnPV6, RnPV7, RnPV8, RnPV9, and RnPV10) have been identified (Sasaki et al., 2006, 2007, 2016; Chiba et al., 2013a, 2016; Yaegashi and Kanematsu, 2016; Arjona-Lopez et al., 2018). Biological characterization of fungal viruses has been facilitated by the development of improved virion transfection methods (Hillman et al., 2004). Fungal partitiviruses, unlike plant partitiviruses, can be experimentally introduced into both original and experimental hosts. The method has previously been used to study some partitiviruses including RnPV1, RnPV2, RnPV6, SsPV1, and RsPV2 (Sasaki et al., 2007; Chiba et al., 2013a, 2016; Xiao et al., 2014; Zheng et al., 2014). In this regard, it is important to note that the phytopathogenic ascomycete that causes chestnut blight, Cryphonectria parasitica, is a model filamentous host used to explore virus-virus and virus-host interactions. The fungus is an attractive host because it is biologically tractable, genetically manipulatable, and can support many homologous and heterologous fungal viruses (Eusebio-Cope et al., 2015). In addition, a number of biological resources and molecular tools are available for the fungus (Nuss, 2005; Eusebio-Cope et al., 2015; Chiba et al., 2018). Therefore, the use of C. parasitica as a viral host has facilitated the study of viral symptom induction via host antiviral RNA silencing to and counter defense of both homologous and heterologous fungal viruses (Faruk et al., 2008; Andika et al., 2017, 2019). Recent advancements made by using this fungus as a model system include the characterization of mechanisms of induction of and susceptibility to mechanisms of antiviral RNA silencing of different fungal viruses (Chiba and Suzuki, 2015; Andika et al., 2017, 2019). It has also been shown that a fungal gammapartitivirus replicates in plant cells (Nerva et al., 2017b).
Here, we report the characterization of a total of 20 partitiviruses belonging to 15 new and 5 previously-identified species placed in the genus Alphapartitivirus or Betapartitivirus. Some tested partitiviruses differ with respect to viral content in both R. necatrix and C. parasitica, and symptom induction in the experimental host. This study has enhanced our understanding of the molecular and biological diversity of fungal partitiviruses.
Materials and Methods
Fungal and Viral Materials
A total of 16 field isolates of R. necatrix listed in Table 1 were used in the study. Most fungal isolates were collected from Japanese pear or apple trees in Saga, Fukuoka, Hyogo, Nagano, and Gunma Prefectures, Japan, while Rn459 was collected from an avocado tree in Malaga, Spain (Arakawa et al., 2002; Ikeda et al., 2004; Yaegashi et al., 2013; Arjona-Lopez et al., 2018). The Japanese fungal strains had earlier been shown to harbor partiti-like dsRNAs of ~2.0–2.5 kbp by a conventional dsRNA extraction method (Arakawa et al., 2002; Ikeda et al., 2004; Sasaki and Kanematsu, unpublished data). The partitivirus isolate from the Spanish strain, Rn459, was previously partially sequenced (Arjona-Lopez et al., 2018). The standard strain, W97, of R. necatrix has been described in previous reports (Kanematsu et al., 2010; Shimizu et al., 2018). The standard C. parasitica strain, EP155, and its dicer-like 2 (dcl2) knockout (KO) mutant strain, Δdcl2, were a generous gift from Dr. Donald L. Nuss (University of Maryland, College Park, MD). The Δdcl2 strain carries deletion of a key gene used for antiviral RNA silencing, dcl2 (Segers et al., 2007). All fungal materials were cultured either on potato dextrose agar (PDA, BD Difco Laboratories, Detroit, MI, USA) plates or potato dextrose broth (PDB, BD Difco Laboratories).
Table 1.
List of Rosellinia necatrix isolates used.
| Isolatea | Collection locality | Host plants | Collection year | MCGb | Group of NGS | Virus detectede | Source |
|---|---|---|---|---|---|---|---|
| W98 | Saga | Japanese pear | 1998 | 80 | Pool-3/2 | RnPV11 | Ikeda et al., 2004 |
| W118 | Saga | Japanese pear | 1998 | 86 | Pool-3/1 | RnPV3, 12, 13 | Ikeda et al., 2004 |
| W129 | Fukuoka | Japanese pear | 1998 | 88 | Pool-2 | RnPV25 | Ikeda et al., 2004 |
| W442 | Hyogo | Unknown | 2000 | 169 | Pool-3/1 | RnPV18, 19 | This study |
| W558 | Saga | Japanese pear | 1998 | 85 | Pool-1 | RnPV6 | Ikeda et al., 2004 |
| W662 | Gunma | Apple | 2000 | 301 | Pool-1 | RnPV23, 24 | Ikeda et al., 2004 |
| W744 | Saga | Japanese pear | 2001 | 325 | Pool-3/2 | RnPV1, 14–17 | Ikeda et al., 2004 |
| W1030 | Nagano | Apple | 2009 | 139 | NAc | RnPV4 | Yaegashi et al., 2013 |
| W1031 | Nagano | Apple | 2009 | 139 | Pool-4 | RnPV3, 4 | Yaegashi et al., 2013 |
| W1040 | Nagano | Apple | 2009 | 139 | Pool-1 | RnPV5 (+one)f | Yaegashi et al., 2013 |
| W1041 | Nagano | Apple | 2009 | 139 | NAc | RnPV5 | Yaegashi et al., 2013 |
| W1050 | Nagano | Apple | 2009 | 139 | Pool-4 | RnPV22 | Yaegashi et al., 2013 |
| W1126 | Iwate | Lacquer tree | 2011 | U1 | NAc | RnPV22 | From Dr. M. Tabata |
| W1134 | Kagawa | Lacquer tree | 2011 | U13 | Pool-3/1 | RnPV20, 21 | From Dr. M. Tabata |
| W1135 | Kagawa | Lacquer tree | 2011 | U14 | NAc | RnPV20 | From Dr. M. Tabata |
| Rn459 | Malaga | Avocado | 2016 | unknown | NAd | RnPV10 | Arjona-Lopez et al., 2018 |
All isolates were collected in Japan, except for Rn459 isolated from Spain.
Mycelial compatibility group.
Not applicable. For these viruses, conventional cDNA libraries were constructed and subjected to Sanger sequencing.
Partial genomic sequence was reported by Arjona-Lopez et al. (2018).
Viral infection was confirmed by RT-PCR.
The partial genome sequence derived from an additional potential novel partitivirus besides RnPV5 was detected in the W1040 strain.
Partitivirus Purification, Electron Microscopy, and Transfection
Partitivirus particles were semi-purified following the method described by Chiba et al. (2016). After differential centrifugation, viral fractions were subjected to sucrose/cesium chloride gradient ultracentrifugation. Purified virus preparations were examined by transmission electron microscopic (TEM, H-7650 Hitachi, Tokyo, Japan) observation after EM staining (the EM stainer, an alternative for uranyl acetate, Nissin EM Co., Tokyo, Japan) (Nakakoshi et al., 2011). Virus preparations were loaded on sodium dodecyl sulfate (SDS)—polyacrylamide (10%) gel and electrophoreses, and then stained with Rapid stain CBB kit (Nacalai tesque inc., Kyoto Japan). Semi-purified particle fractions were used to transfect virus-free protoplasts derived from either strain W97 from R. necatrix, or EP155 and Δdcl2 strains from C. parasitica. Transfection of strain W97 was conducted according to the method described by Kanematsu et al. (2010), and transfection of C. parasitica-derived strains was performed according to the method described by Salaipeth et al. (2014). After regeneration, partitiviral infection of candidate transfectants were assessed using the one-tube RT-PCR method using PrimeScript™ One Step RT-PCR Kit ver.2 (Takara Bio Inc., Shiga Japan) and toothpicks described by Urayama et al. (2015).
RNA Analyses
Viral genomic dsRNA sequences were determined using two approaches: conventional complimentary DNA (cDNA) library construction using purified viral dsRNA and subsequent Sanger sequencing (Chiba et al., 2016), and next-generation high-throughput sequencing (NGS) of total RNA from fractions obtained from infected mycelia (Shamsi et al., 2019). dsRNA was isolated from mycelia cultured in PDB media for 1 week as previously described (Chiba et al., 2013a). Total RNA, dsRNA or single-stranded RNA (ssRNA) fractions were obtained from R. necatrix strains as described by Chiba et al. (2013a).
For RNA-Seq analyses, RNA samples were pooled into four groups: two were comprised of dsRNA samples and were named Pool-1 and -2 (1.2 and 0.9 μg, respectively) and other two, Pool-3 and -4, contained total RNA samples (14.1 and 67.5 μg, respectively) (see Table 1). Each RNA pool with/without ribosomal RNA depletion treatment (the Ribo-Zero kit, Illumina, San Diego, CA, USA) was subjected to cDNA library construction (the TruSeq RNA Sample Preparation kit v2, Illumina) and pair-end deep sequencing (100 bp pair-end reads) using the Illumina HiSeq 2500/4000 platforms (Illumina) performed by Macrogen Inc. (Tokyo, Japan). After deep sequencing, adapter-trimmed sequence reads (Pool-1: 28,827,612; Pool-2: 29,248,296, Pool-3: 46,288,894, and Pool-4: 47,084,068 raw reads) were de novo assembled using the CLC Genomics Workbench (version 11, CLC Bio-Qiagen, Aarhus, Demark). To verify the virus infection in the fungal strains, we performed RT-PCR using the specific primer sets for each of the partitivirus (Supplementary Table S1). Viral genomic sequences were completed by RLM-RACE (RNA ligase mediated rapid amplification of complementary DNA ends) and gap-filling RT-PCR (Suzuki et al., 2004; Chiba et al., 2009) (Supplementary Table S1). Sequences obtained in this study were deposited in EMBL/GenBank/DDBJ databases with accession numbers LC517370–LC517399, as described in Table 2.
Table 2.
Properties of newly discovered partitiviruses from Rosellinia necatrix.
| Virus namea | Original host strain | Contig No. | Total read count | Genus | Segment lengthb | Blastp | Protein size | Symptomc | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| dsRNA1/(accession) | Hit with highest score in | Identity | RdRp (aa) | R.n. | C.p. | ||||||
| dsRNA2 (accession) | Blastp | (%) | Cp (aa) | Δdcl2 | EP155 | ||||||
| RnPV11 | W98 | 4 262 |
61129 26713 |
Beta- | 2445 (LC517370) 2326 (LC517371) |
Rosellinia necatrix partitivirus 20 Fusarium poae partitivirus 2 |
71 35 |
757 652 |
–~+ | ++ | +++ |
| RnPV12 | W118 | 148 779 |
47804 1197 |
Alpha- |
1845 LC517372
1925 (LC517373) |
Botrytis cinerea partitivirus 2 Erysiphe necator partitivirus 1 |
63 38 |
551 533 |
–~+ | ||
| RnPV13 | W118 | 610 529 |
6211 4558 |
Alpha- | 1965 (LC517374)*
1822 (LC517375) |
Rhizoctonia oryzae-sativae partitivirus 1 Rhizoctonia oryzae-sativae partitivirus 1 |
59 54 |
469 488 |
|||
| RnPV3 f | W118 | 317 347 |
Beta- | 2246 (LC010950) 2065 (LC010951) |
Rosellinia necatrix partitivirus 3 Rosellinia necatrix partitivirus 3 |
99 96 |
709 613 |
– | – | ||
| RnPV14 | W744 | 118 750 |
46844 2477 |
Beta- |
2412 (LC517376)
2427 (LC517377) |
Rosellinia necatrix partitivirus 4 Rosellinia necatrix partitivirus 1-W8 |
72 47 |
743 706 |
–~+ | ++ | |
| RnPV15 | W744 | 826 2267 |
7917 3491 |
Beta- | 2517 (LC517378) 2358 (LC517379) |
Rosellinia necatrix partitivirus 14 Rosellinia necatrix partitivirus 16 |
59 49 |
747 663 |
|||
| RnPV16 | W744 | 509 3216 |
20228 944 |
Beta- | 2372 (LC517380) 2344 (LC517381) |
Podosphaera prunicola partitivirus 4 Rosellinia necatrix partitivirus 15 |
61 49 |
730 662 |
++ | +++ | |
| RnPV17 | W744 | 671 237 |
4185 8985 |
Beta- | 2292 (LC517382) 2235 (LC517383) |
Heterobasidion partitivirus 7 Rhizoctonia solani partitivirus 7 |
63 71 |
725 670 |
|||
| RnPV1f | W744/ W1134 |
43 82/69 |
Beta- | 2374 (AB113347) 2263 (AB113348) |
Rosellinia necatrix partitivirus 1-W8 Rosellinia necatrix partitivirus 1-W8 |
98 99/94 |
709 686 |
||||
| RnPV18d | W442 | 110 874 |
30695 | Beta- | 2410 (LC517384) 2337 (LC517385) |
Enteoleuca partitivirus 2 Dill cryptic virus 2 |
99.7 43 |
740 670 |
– | – | – |
| RnPV19d | W442 | 26 379 |
40899 5244 |
Alpha- | 2013 (LC517386) 1842 (LC517387) |
Entoleuca partitivirus 1 Grosmannia clavigera partitivirus 1 |
99.7 42 |
606 520 |
+ | – | |
| RnPV20 | W1134 | 5 7 |
186156 55670 |
Beta- | 2417 (LC517388) 2318 (LC517389) |
Rosellinia necatrix partitivirus 11 Ceratobasidium partitivirus CP-h |
71 61 |
755 643 |
–~+ | +++ | +++ |
| RnPV21 | W1134 | 161 69 |
49075 3784 |
Beta- |
2352 (LC517390)
2361 (LC517391) |
Podosphaera prunicola partitivirus 4 Rosellinia necatrix partitivirus 1-W8 |
65 94 |
721 686 |
|||
| RnPV22 | W1050/ W1126 |
27 707 |
Alpha- |
2012 (LC517392)
2037 (LC517393) |
Trichoderma atroviride partitivirus 1 Trichoderma atroviride partitivirus 1 |
76 43 |
613 584 |
||||
| RnPV23 | W662 | 127 25 |
Alpha- | 1831 (LC517394)**
1791 (LC517395)** |
Heterobasidion partitivirus 13 Heterobasidion partitivirus 13 |
49 37 |
571 514 |
||||
| RnPV24 | W662 | 77 125 |
Alpha- | 1946 (LC517396)**
1771 (LC517397)** |
Oyster mushroom isometric virus II Medicago sativa alphapartitivirus 1 |
75 28 |
598 498 |
||||
| RnPV25 | W129 | 51 188 |
Beta- | 2374 (LC517398)**
2049 (LC517399)** |
Trichoderma citrinoviride partitivirus 1 Trichoderma citrinoviride partitivirus 1 |
65 54 |
733 642 |
||||
| RnPV10 | Rn459 | Alpha- | 1896 (LC333736) 1911 (LC333737) |
Rosellinia necatrix partitivirus 10 Rosellinia necatrix partitivirus 10 |
100 100 |
573 539 |
|||||
| RnPV4e | W1030/ W1031 |
4 270 |
Alpha- | 2342 (AB698493) 2295 (LC521312) |
Rosellinia necatrix partitivirus 4 Sclerotinia sclerotiorum partitivirus 1 |
100 48 |
744 683 |
||||
| RnPV5e | W1040/ W1041 |
10/522 27/62 |
Beta- | 2046 (AB698494) 1906 (LC521313) |
Rosellinia necatrix partitivirus 5 Trichoderma atroviride partitivirus 1 |
99.7 44 |
647 576 |
||||
| RnPV6f | W558 | 52 137 |
Beta- | 2499 (LC010952) 2462 (LC010953) |
Rosellinia necatrix partitivirus 6 Rosellinia necatrix partitivirus 6 |
100 100 |
756 729 |
–~+ | +++ | +++ | |
RnPV22 was also detected from the W1126 strain.
5′-terminal sequence of dsRNA1 is incomplete;
5′- and 3′-terminal sequences of both segments are incomplete. Underlined: RdRp is encoded by the smaller dsRNA segments.
R.n., R. necatrix W97 strain; C.p., C. parasitica EP155 or Δdcl2 strain. Growth reduction: –, No; +: Slight; ++, Moderate; +++, Great. See Figure 4 and Supplementary Figure S4 for colony morphology of virus-infected fungal strains.
Near-complete or complete dsRNA1 sequences of these viruses from Spanish Entoleuca sp. (d) or R. necatrix (e) have already been deposited in GenBank.
Partitiviruses belonging to the same species have previously been reported from different Japanese strains of R. necatrix.
Bioinformatics
Viral sequences were analyzed with online bioinformatics tools as described by Kondo et al. (2015). After de novo assembly, contigs [Pool-1: 23,341 (~9.9 kb), Pool-2: 23,130 (~9.8 kb), Pool-3: 10,485 (~13.2 kb), and Pool-4: 11,016 (~13.8 kb)] were subjected to local BLAST searches against the viral reference sequence (RefSeq) dataset from the National Center for Biotechnology Information (NCBI). Viral-like sequences were analyzed using Enzyme X v3.3.31 or GENETYX-MAC (Genetyx Co., Tokyo, Japan). Database searches of viral sequences were performed using the BLAST (BLASTn and BLASTp) programs available from NCBI. Pairwise amino acid identity was calculated using SDT v1.2 (Muhire et al., 2014).
Phylogenetic reconstruction was carried out using the maximum-likelihood (ML) method as described previously (Kondo et al., 2019). Deduced amino acid sequences from virus RNA sequences were aligned using MAFFT version 7 (Katoh and Standley, 2013) and unreliably aligned regions were eliminated using Gblocks 0.91b (Talavera and Castresana, 2007). ML phylogenetic trees were constructed by PhyML 3.0 (Guindon et al., 2010) using automatic model selection via smart model selection (SMS) (Lefort et al., 2017). Support for branches was examined via bootstrapping with 1,000 repetitions. The phylogenetic trees (mid-point rooted) were visualized and refined using FigTree version 1.3.1 software2.
Results
dsRNA Profiles of R. necatrix Isolates
Field-collected strains of R. necatrix have previously been screened for mycoviruses by Japanese research groups (Arakawa et al., 2002; Ikeda et al., 2004; Yaegashi and Kanematsu, 2016). We selected a total of 16 fungal strains harboring ~1.5–2.5 kbp dsRNAs for the study (Table 1). dsRNA profiles of some these strains have been reported previously by Ikeda et al. (2004), Yaegashi et al. (2013), and Arjona-Lopez et al. (2018). Bands were confirmed to be dsRNA following double digestion using RQ1 RNase free DNase I (Promega) and S1 Nuclease (Takara) enzymes (Eusebio-Cope and Suzuki, 2015). dsRNA-enriched or total RNA fractions and their mixed pools (Poo1-1–4) were then subjected to either conventional Sanger sequencing of cDNA clones or NGS analyses, respectively (Table 2; see Figure 1 for the dsRNA profile of Pool-3 from R. necatrix-derived strains). It is worth noting that some dsRNA bands of the sequence analysis turned out to be doublets (or more).
Figure 1.
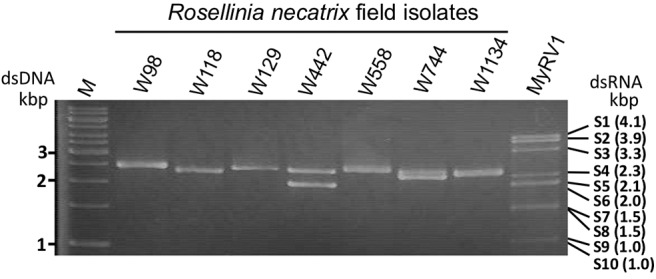
Agarose gel electrophoresis of dsRNA isolated from field isolates of Rosellinia necatrix. dsRNA fractions were obtained from fungal isolates as indicated above each lane, and electrophoresed through a 1% agarose gel in 1 × TAE (40 mM Tris-acetate−1 mM EDTA, pH 7.8). See Table 1 for information regarding fungal isolates. A 1-kb DNA ladder (Thermo Fisher Scientific., Inc., Waltham, MA, USA) and MyRV1 genomic dsRNA (Supyani et al., 2007) were used as size markers.
Sequence Analysis
Properties of fully and near-fully sequenced partitiviruses are summarized in Table 2. Some partially characterized partitiviruses, i.e., RnPV1, RnPV3, RnPV4, RnPV5, RnPV6, and RnPV10, were previously isolated from R. necatrix strains (Sasaki et al., 2005; Chiba et al., 2013b, 2016; Yaegashi et al., 2013; Arjona-Lopez et al., 2018). The partial genomic sequences of RnPV4, RnPV5, and RnPV10, and the complete genomic sequences of RnPV1 and RnPV6 were previously determined. The remainder of the partitivirus isolates for which novel sequence information was obtained in this study were designated Rosellinia necatrix partitivirus 11–25 (RnPV11–RnPV25) (Table 2).
NGS and conventional sequencing, followed by subsequent RT-PCR analyses, suggested that tested fungal strains were either singly (W98, W129, W558, W1030, W1041, W1050, W1126, W1135, and Rn459), doubly (W442, W662, W1031, W1040, and W1134), triply (W118) or quintuply (W744) infected by partitiviruses (Tables 1, 2). Partitivirus infection was confirmed by RT-PCR using specific primer sets designed from NGS contigs or sequences by conventional methods (Table 2 and Supplementary Table S1). Particularly, it is noteworthy that W744 was infected by five distinct partitiviruses, RnPV1, RnPV14, RnPV15, RnPV16, and RnPV17, as has also been reported in some other fungal hosts multiply infected by mitoviruses (Hillman et al., 2018). When the nucleotide sequences of these co-infecting viruses were compared, the highest sequence identity is 59% detected between RnPV14 dsRNA1 and RnPV15 dsRNA1 (see Supplementary Figure S3 for amino acid sequence comparison), which is low enough to avoid possible RNA silencing-mediated inter-virus antagonism. Fifteen novel partitiviruses, including those identified by conventional sequencing methods, from seven fungal strains such as W98 (RnPV11), W118 (RnPV12 and 13), W442 (RnPV18 and 19), W744 (RnPV14–17), W1050/1126 (dsRNA segments with conventional sequencing, RnPV22), W1134 (RnPV20 and 21), and Rn459 (two reported dsRNA contigs for RnPV10, Arjona-Lopez et al., 2018) had two segments that were completely sequenced using the RLM-RACE method (Table 2). In addition, the only fully-sequenced coding regions of the three other novel partitiviruses were from the fungal strains W662 (RnPV23 and 24) and W129 (RnPV25) (Table 2).
Sequence analysis revealed that each segment encoded one open reading frame (ORF), and BlastP analysis confirmed that the ORF from dsRNA1 (larger dsRNA segments, in general) encoded RdRP, which was closely related to alpha- or betapartitiviruses of the Partitiviridae family (Table 2, see below). The ORF from dsRNA2 (smaller dsRNA segments, in general) encoded CP, which was also related to other known alpha- or betapartitiviruses. Viral sequences from R. necatrix from W744 (RnPV1), W118/1031 (RnPV3), W1030/1031 (RnPV4, dsRNA1 segment determined by conventional sequencing), W1041/1040 (RnPV5) and W558 (RnPV6) strains were 85–100% identical to known R. necatrix partitiviruses RnPV1/W8, RnPV3/W1029, RnPV4/W1028 (dsRNA1), RnPV5/W1028 (dsRNA1), and RnPV6/W113, respectively (Table 2). For RnPV4 and RnPV5, previously undescribed dsRNA2 segments were newly sequenced (Table 2). Intriguingly, two dsRNA1 sequence for RnPV18 and 19, respectively, from the single W442 strain were 99% identical to two reported partitivirus sequences (Entoleuca partitivirus 2 and 1, EnPV2 and 1), respectively, which were both obtained from a Spanish fungal isolate (E97-14) of Entoleuca sp. entirely different from, but sympatric to, R. necatrix that infest avocado orchards in Spain (Velasco et al., 2019, 2020) (Table 2). Note that the dsRNA2 sequences of EnPV1 and 2 from the GenBank/EMBL/DDBJ databases were unavailable. The 5′-terminus of EnPV1 remains incomplete. Thus, we report the full-length sequence of the genome of RnPV18 and RnPV19, respectively (Table 2).
Genome Organization, Protein Sequence Similarities, and Phylogenetic Analysis
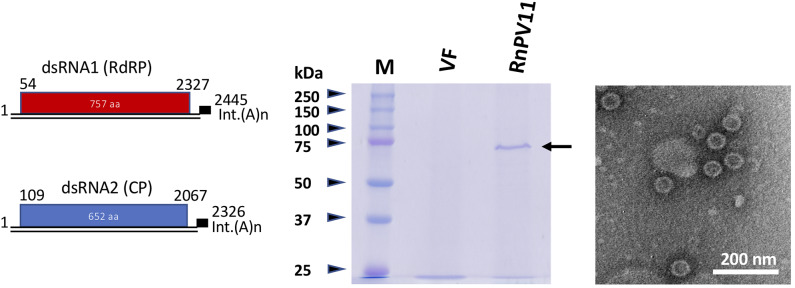
All novel partitivirus strains from R. necatrix characterized molecularly have been determined to belong to either the genus Alphapartitivirus or Betapartitivirus (Table 2). No gammapartitiviruses, which exclusively infect fungi, were detected within R. necatrix as a part of this or previous studies (Sasaki et al., 2005; Chiba et al., 2013a; Yaegashi et al., 2013; Yaegashi and Kanematsu, 2016; Arjona-Lopez et al., 2018). Their sequence characteristics are similar to those of previously reported partitivirus members (Nibert et al., 2014). Namely, the size ranges of their segments, 5′- and 3′-untranslated regions (UTRs), and encoded proteins of these viruses mostly fall within ranges reported previously, whereas a few expand the reported size distributions within alpha- and betapartitiviruses (Nibert et al., 2014). The genome sizes of most reported alphapartitiviruses range from 3.6 to 3.9 kbp (Nibert et al., 2014), but RnPV22 isolate extended the range from 3.6 to 4.0 kbp. Similarly, an expansion of genome segment and coding protein size is also observed for betapartitiviruses RnPV6, RnPV14, and RnPV15. dsRNA1 segments of partitiviruses are usually longer than dsRNA2 segments, but we encountered few exceptions in RnPV12, RnPV14, RnPV21, and RnPV22, which had longer dsRNA2 than dsRNA1 segments (Table 2), as also has been reported for RnPV6 (Chiba et al., 2016). As an example, the genome organization, protein component, and virion morphology of a novel R. necatrix virus betapartitivirus, RnPV11, isolated from the strain W98, is shown in Figure 2. The RnPV11 genomic segments of ~2.4 and 2.3 kbp were shown to be encased in spherical particles of ~30 nm in diameter composed of the CP of ~72 kDa.
Figure 2.
Schematic of the genome organization, particle protein composition, and morphology of a newly discovered partitivirus of R. necatrix. (Left) Genome organization of a representative betapartitivirus, RnPV11/W98. Sequence information for RnPV11 is summarized in Table 2. (Middle) SDS-PAGE analysis of purified preparations of RnPV11. Virus particles were purified from the mycelia of the W98 strain (lane RnPV11) and analyzed in 10% polyacrylamide gel electrophoresis. The arrow indicates the migration of RnPV11/W98 CP. A preparation was also obtained in parallel from virus-free W97 (VF). Protein size standards used (lane M, Precision Plus Protein Dual Color Standards) were purchased from Bio-Rad Laboratories, Inc., Hercules, CA, USA. (Right) Electron microscopy of RnPV11 virions. A purified virus preparation of RnPV11 was examined using a Hitachi electron microscope model H-7650.
Partitiviruses analyzed in this study had terminal sequences similar to those described previously (Nibert et al., 2014; Vainio et al., 2018a). For several partitiviruses from R. necatrix isolates, such as RnPV19, RnPV20, and RnPV22, “CAA” repeats were present at the 5′-UTR region, which has been assumed to be a translation enhancer (Jiang and Ghabrial, 2004) (Supplementary Figure S1). The 20–30 nucleotides located at the 5′-terminal regions are highly conserved between genome segments, and the 5′-terminal-most nucleotides are considered consensus sequences and are shown in bold in Supplementary Table S2. The 5′-terminal G residue is strictly conserved at the terminal end or +1 from the end (Supplementary Figure S2). For 3′-termini, the plus strands of the genome segments of some betapartitiviruses, such as RnPV11, RnPV14, and RnPV15, end with interrupted poly(A) tracts, while those of other partitiviruses contain additional nucleotides, which include the di- or tri-nucleotide “UC,” “CC,” or “CU” that follow interrupted poly(A) tracts (Supplementary Table S2).
The BlastP (or BlastN as mentioned above) results are summarized in Table 2. The species demarcation criteria set by the ICTV are ≤ 90% and ≤ 80% amino acid identities for RdRP and CP, respectively (Vainio et al., 2018a). Of 20 newly sequenced alpha- or betapartitivirus isolates, 15 (including EnPV dsRNA segments) showed much smaller amino acid identities with known partitiviruses, which ranged 28–76% for RdRP or CP, than the above-mentioned species demarcation criteria (Table 2). In addition, pairwise comparisons of proteins encoded by new partitiviruses revealed moderate levels of amino acid sequence identity (less than above criteria) among RdRPs and CPs (Supplementary Figure S3). These results indicated that the 15 partitiviruses that were newly identified belong to new species, which were named Rosellinia necatrix partitivirus 11 to Rosellinia necatrix partitivirus 25. Notably, the nucleotide sequence identities of dsRNA1 and dsRNA2 of RnPV21/W1134 were most similar to Podosphaera prunicola partitivirus 4 (45%) and RnPV1/W8 or W744 (94%) (Table 2), respectively. This strongly suggests that a reassortment event likely occurred between RnPV1 and another partitivirus (see Discussion).
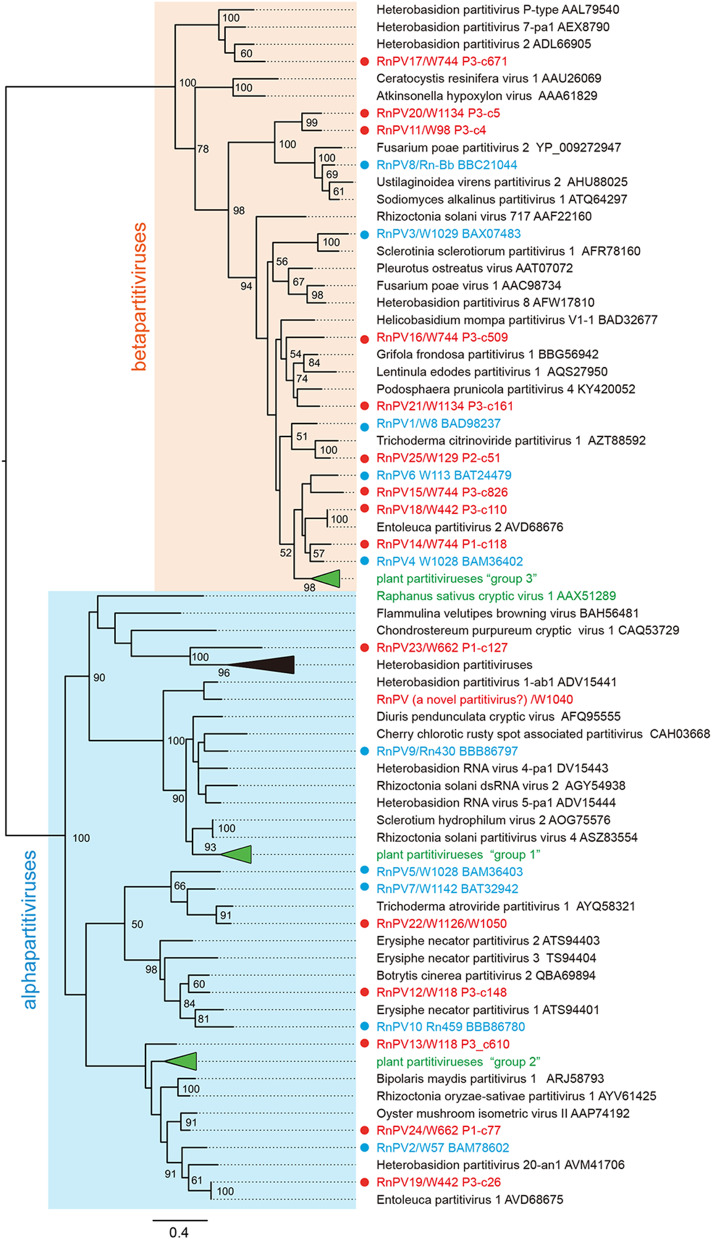
Members of the Alphapartitivirus and Betapartitivirus genera have been characterized from 14 to 17 species of fungi (ascomycetous and basidiomycetous) and plants (largely dicot plants), respectively (Vainio et al., 2018a). Phylogenetic analysis based on RdRP, encoded by dsRNA1, of the novel 15 R. necatrix partitiviruses using selected alpha- and betapartitiviruses that included approved members of both genera revealed that six novel R. necatrix partitiviruses clustered with alphapartitiviruses with a bootstrap value of 100 (Figure 3). These included RnPV12, RnPV13, RnPV19, RnPV22, RnPV23, and RnPV24. The remaining nine novel R. necatrix partitiviruses (RnPV11, RnPV14, RnPV15, RnPV16, RnPV17, RnPV18, RnPV20, RnPV21, and RnPV25) were clustered with betapartitiviruses with bootstrap values of 100. Note that some inter-subgroup relationships within the alphapartitivirus branch were not well supported with high bootstrap values. Notably, most R. necatrix partitiviruses were discretely placed within the tree, while some viruses (RnPV4, RnPV6, RnPV14, RnPV15, and RnPV18) were clustered together and nested with a clade of plant betapartitiviruses (namely group 3) (Figure 3 and Supplementary Table S4).
Figure 3.
Phylogenetic analysis of novel partitiviruses from field-collected isolates of R. necatrix. Maximum likelihood (ML) phylogenetic trees based on amino acid alignments of RdRPs were constructed using PhyML 3.0 with a best fit model RtREV with +G +I +F. Sequences of members of Alphapartitivirus and Betapartitivirus genera were analyzed together with novel R. necatrix partitiviruses. Virus names are followed by GenBank accession numbers. Red and blue circles indicate novel or reported R. necatrix partitiviruses, respectively. The numbers at the nodes indicate bootstrap values. The virus names and accession numbers in the collapsed triangles are described in Supplementary Table S4.
Phenotypic Effects of Novel Partitiviruses on Both Original and Experimental Hosts
The investigation of the biological properties of any virus requires comparing sets of isogenic virus-free and -infected strains. However, when assessing R. necatrix-partitivirus interactions, this was difficult because mixed infections occurred frequently and virus curing via hyphal tipping presented difficulties. To this end, we used a virion transfection approach now available for various virus/host combinations.
Of the 15 sequenced, novel partitiviruses of R. necatrix identified in this study, 13 (in 5 fungal strains) were tested for their effects on three virus-free strains of two fungal species: the standard strain, W97, of the original host species, R. necatrix, and two strains (EP155 wild-type and its Δdcl2 KO mutant, an antiviral RNA silencing deficient strain) of the experimental host, C. parasitica. Note that many of the fungal strains tested were co-infected by multiple partitiviruses (see Table 1). Obtained transfectants are summarized in Supplementary Table S3. RnPV11 was singly transfected in W97 (Table 2). The five, three and two viruses coinfecting W744, W118, and W1134, respectively, could not be separated via transfection of W97. No significant phenotypic change was observed in W97 transfectants, except W97 co-infected with five viruses (RnPV1 and RnPV14 to RnPV17) originally harbored in W744. Initially, the W97 transfectant infected by the five viruses displayed considerably reduced growth, its mycelia appeared to be deep white in color and were fluffy (Supplementary Figure S4A). Subsequent hyphal fusion with virus free W97 led to the presentation of a milder growth defect (Supplementary Figure S4C).
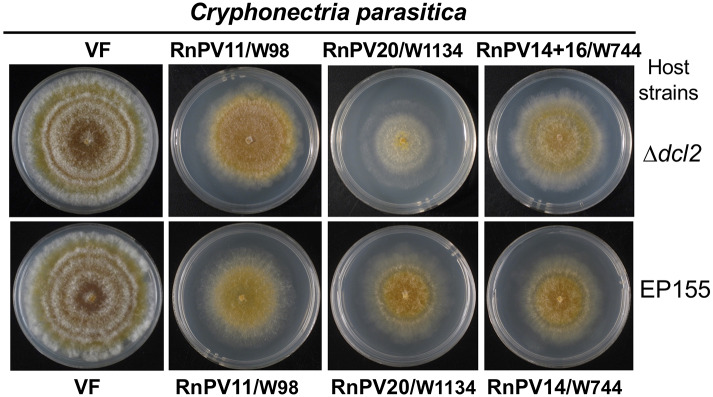
Multiple partitiviruses, RnPV18 and RnPV19, coinfecting a single strain W442, segregated and their single infectants could be obtained in C. parasitica Δdcl2 (Supplementary Figure S4B). These Δdcl2 transfectants as well as other partitivirus transfectants (RnPV11, RnPV20 and RnPV14–16) (Supplementary Table S3) were then anastomosed with RNA silencing-proficient C. parasitica EP155 to move the viruses. Even after repeated coculturing, EP155 stably maintained partitiviruses (RnPV11, RnPV20, or RnPV14–16) and their infectants showed reduction in growth rates, pigmentation, and growth of aerial hyphae (Figure 4). On the other hand, EP155 strains singly infected by RnPV18 and RnPV19 were either infected asymptomatically or displayed only slightly reduced growth rates in EP155 (Supplementary Figure S4B). In Δdcl2, most partitiviruses (RnPV11, RnPV20, RnPV14–16, and probably RnPV19) produced symptomatic infections and induced reduced growth rates and irregular margins (Figure 4 and Supplementary Figure S4B). Similar symptoms have previously been attributed to other partitiviruses from R. necatrix, i.e., RnPV2 and RnPV6 (Chiba et al., 2013a, 2016) (Table 2).
Figure 4.
The morphology of C. parasitica colonies infected with betapartitiviruses from different R. necatrix fungal strains. C. parasitica Δdcl2 singly infected by RnPV11/W98 or RnPV20/W1134, and doubly infected by RnPV14, and RnPV16 from the W744 strain were grown in PDA for 1 week on a benchtop and photographed. C. parasitica EP155 colonies singly infected with the viruses are shown. VF refers to virus free strains.
Different Virus Accumulation Levels in Between Virus Strains and in Between Different Fungal Host Strains
We attempted to compare genomic dsRNA accumulation in the standard R. necatrix strain W97. To this end, RnPV6/W558, RnPV10/Rn459, and RnPV11/W98 were selected, because their W97 single transfectants could readily be obtained. For RnPV25/W129, the original filed-collected strain W129 was used. Total RNA fractions were isolated from two biological replicates for each strain, and levels of viral dsRNA accumulation were compared by normalizing to host ribosomal RNAs (Figure 5). RnPV11/W98 dsRNA1 and dsRNA2 of 2.4 and 2.3 kbp accumulated in W97 at a level slightly greater than or comparable to, RnPV10/Rn459 (1.9 kbp + 1.9 kbp), or RnPV25/W129 (2.4 kbp + <2.0 kbp). The band intensity of RnPV6/W558 dsRNA (2.5 kbp + 2.5 kbp) was much fainter than those of dsRNA from the other three partitiviruses. Note that dsRNA1 and dsRNA2 of the four tested partitiviruses, except for RnPV25/W129, were completely sequenced and co-migrated in the agarose gel. Considering a second dsRNA band for RnPV25/W129, the size of partially sequenced dsRNA2 appears to be similar to dsRNA1 and have comigrated with it (Figure 5). These results suggest variability in partitiviral dsRNA accumulation within the same R. necatrix host strain.
Figure 5.
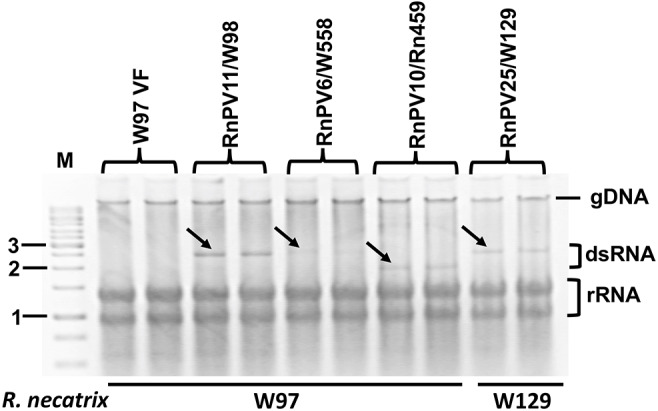
Virus accumulation of four partitivirus strains in R. necatrix host strains. One alphapartitivirus (RnPV10) and three betapartitiviruses (RnPV6, RnPV11, and RnPV25) were used for this analysis. Total nucleic acid fractions were isolated from W97 mycelia singly infected by RnPV11/W98, RnPV6/W558, or RnPV10/Rn549, and analyzed via 1.2% agarose gel electrophoresis. These W97 single infectants were obtained by transfection of protoplasts with respective partitivirus virions. For RnPV25/W129, the original host strain W129 was used. Black arrows indicate the migration positions of dsRNA genomic segments of the respective partitiviruses. M refers to the 1-kb DNA ladder. rRNA was used as a loading control. gDNA indicates the migration position of the host genomic DNA.
Generally, partitiviruses from R. necatrix accumulate to the greater levels in the original host, rather than the experimental host strain, C. parasitica (Chiba et al., 2013a, 2016). Next, we compared the accumulation of a single partitivirus (RnPV11 or RnPV6) in the three host strains, which included the standard R. necatrix strain W97, C. parasitica EP155 and Δdcl2. Inspection of Figure 6 clearly shows that RnPV11/W98 and RnPV6/558 accumulated more in the RNA silencing-deficient Δdcl2 than in RNA silencing-competent EP155. However, both viruses accumulated more highly in W97 than in wild-type EP155 and Δdcl2. We obtained a similar accumulation profile using three biological replicates (data not shown).
Figure 6.
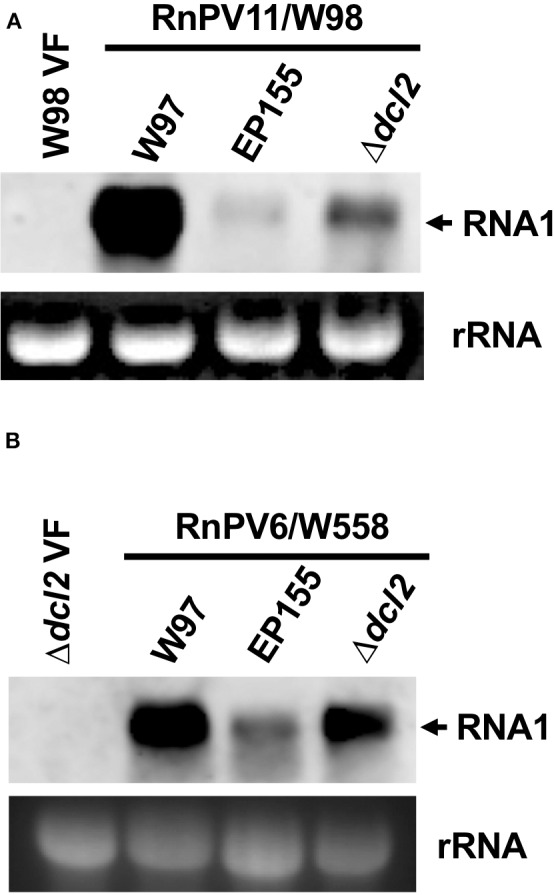
Comparison of virus accumulation in three different fungal strains R. necatrix W97 and C. parasitica wild-type EP155 and its mutant Δdcl2. A. Equal amounts of total RNA was obtained from three fungal strains each singly infected by RnPV11/W98 (A) or RnPV6/W558 (B), and subjected to Northern blotting. The probes used are DIG-labeled PCR fragments specific to RnPV11 dsRNA1 (A) and RnPV6 dsRNA1 (B), respectively, that were prepared using specific primer sets, W98-RdRp-F and W98-RdRp-R, and W558-RdRp-F and W558-RdRp-R. See Supplementary Table S1 for the sequences of the primers used in probe preparation.
Discussion
In this study, a total of 20 partitiviruses were isolated from 16 field isolates of the white root rot fungus, R. necatrix. The isolates were characterized molecularly and a subset was characterized biologically. Interestingly, 15 (RnPV11 to RnPV25) may belong to new species, Rosellinia necatrix partitivirus 11 to Rosellinia necatrix partitivirus 25, and six other partitiviruses (RnPV1, RnPV3–RnPV6) belong to known or previously proposed partitiviral species. This study has shown that all of the characterized partitiviruses should be placed within the two genera of Alphapartitivirus and Betapartitivirus, which are members of the family Partitiviridae. However, they show great variability with respect to their biological and virological properties. This study also provided a platform for the further exploration of molecular mechanisms that underly differences in viral symptoms and replication patterns.
The 15, completely sequenced, novel R. necatrix partitiviruses (6 alphapartitiviruses and 9 betapartitiviruses) provide interesting insights into partitivirus biology. Partitiviruses that infect fungi are currently classified into three genera: Alphapartitivirus, Betapartitivirus, and Gammapartitivirus (Nibert et al., 2014; Vainio et al., 2018a). This and previous studies have revealed the prevalence of alpha- and betapartitiviruses, but not gammapartitiviruses, in R. necatrix (Chiba et al., 2013a, 2016; Kondo et al., 2013b; Arjona-Lopez et al., 2018). Our failure to detect gammapartitiviruses in R. necatrix is surprising, because gammapartitiviruses have been reported only in ascomycetes thus far (Nibert et al., 2014; Vainio et al., 2018a). Among the other most extensively surveyed fungi for virus hunting are ascomycetous C. parasitica (class: Sordariomycetes), and Sclerotinia sclerotiorum (class: Leotiomycetes), and basidiomycetous Heterobasidion spp. The fact that no gammapartitiviruses have been reported from these fungi (Jiang et al., 2013; Vainio and Hantula, 2016) may suggest that gammapartitiviruses are restricted to specific ascomycetes and/or narrow host ranges, although a gammapartitivirus has been shown to replicate in plant protoplasts (Nerva et al., 2017b). Alphapartitivirus and Betapartitivirus each include both plant and fungal partitiviruses. It is presumed that those viruses, or their ancestors, may have been transferred between the two kingdoms, which eventually lead to their higher prevalence relative to gammapartitiviruses.
An interesting relation was found between the newly discovered betapartitivirus, RnPV21/W1134, and a previously characterized betapartitivirus, RnPV1/W8 (Sasaki et al., 2005), which was also discovered in the W744 strain in this study. The two segments of RnPV21 possessed the conserved terminal sequences, indicating that they represented the genome of the virus. However, the two viruses shared 56 and 94% amino acid sequence identities for RdRP and CP, respectively. While the highest degree of CP amino acid sequence identify was detected between RnPV1 and RnPV21, the highest degree of RdRP amino acid sequence identity (65%) was observed with an another betapartitivirus (Podosphaera prunicola partitivirus 4) from the sweet cherry powdery mildew fungus. Generally, partitivirus CP genes are less conserved than RdRP (Nibert et al., 2014). These facts strongly suggest that there might have been a reassortment event between two betapartitiviruses. Similar reassortment events have recently been proposed among isolates of different partitiviruses by Petrzik (2019).
Co-infection of the Japanese R. necatrix strain, W442, by two novel partitiviruses, RnPV18 (Betapartitivirus) and RnPV19 (Alphapartitivirus), was detected (Table 2). The Japanese fungal strain W442 was isolated in Hyogo Prefecture (Table 1). Interestingly, the same combination of partitiviruses (EnPV2 and EnPV1, closely related to RnPV18 and RnPV19, respectively) have been reported from Spanish isolates of Entoleuca sp. as well as R. necatrix that infested or colonized avocado soil of southern coastal area of Spain (Velasco et al., 2019, 2020). These observations suggest that interspecific horizontal transfer of the two partitiviruses may have occurred between R. necatrix and Entoleuca sp., as previously suggested or demonstrated for different viruses (Deng et al., 2003; Liu et al., 2003; Yaegashi et al., 2013; Vainio and Hantula, 2016; Khalifa and MacDiarmid, 2019). The coinfection of two fungal hosts by the same set of two taxonomically different partitiviruses (alpha- and betapartitiviruses) suggests the possibility of mutualistic or commensal interactions between the coinfecting partitiviruses. At present, we cannot rule out the possibility that the two viruses accidentally co-transferred from an originally co-infected fungal strain. Further, these partitiviruses were isolated from geographically distinct locations, Japan and Spain. The fact that RnPV18 and RnPV19, and EnPV2 and EnPV1 were isolated from two different sympatric fungal species suggests that contamination in the laboratory was highly unlikely. It is difficult to determine the origin of the fungus carrying the viruses, which may have been brought together via crop plants and/or host fungi as a result of human activities such as tourism and trade (see below).
Horizontal transfer of fungal viruses between two strains of single fungal species, and also between different fungal species, have been suggested by several research groups (Yaegashi et al., 2013). The same subset of partitiviruses, megabirnaviruses (dsRNA viruses), hypoviruses, and fusagraviruses (ssRNA viruses) were detectable within two different fungal species, R. necatrix and Entoleuca sp., collected from avocado fields in Andalusia, Spain (Velasco et al., 2018, 2019). Previous viral identification and characterization studies compared genomic sequences of viruses that belonged to the same species, but were isolated from different continents. For example, 95 and 94% amino acid sequence identities were detected between the isolates of positive-sense (a mitovirus, family Narnaviridae) and negative-sense (a mymonovirus, family Mymonavirudae) ssRNA viruses from Sclerotinia sclerotiorum collected from the USA and Australia (Mu et al., 2017). The sequence identities determined in this study were greater than those previously reported. This may suggest that the virus-harboring fungi were imported relatively recently with crop seedlings. In Spain, R. necatrix is an endemic soil-borne inhabitant in soils in which previous rainfed susceptible crops such as olive, almond, and grapes were cultivated. Later (1970's) these crops were replaced by high-value avocado irrigated crop (Lopez-Herrera and Zea-Bonilla, 2007), in which soils are highly infested by this pathogen with some colonized by the antagonistic fungi Entoleuca sp. (Arjona-Girona and Lopez-Herrera, 2018). EnPV1 and EnPV2 and these have been also detected in a Fusarium isolate collected from the same avocado orchards in addition to Entoleuca sp. and R. necatrix (Velasco et al., 2019, 2020). Presumably, these fungal viruses have been present in these soils for many years and their horizontal transmission between different fungal species inhabiting the same soils is likely to happen. However, it remains unknown which of them has been the initial source of transmission of these fungal viruses to the rest of fungal species or how the viruses have been horizontally transferred.
Of the 16 fungal isolates tested, five produced mixed infections, as exemplified in W744 that was infected with five different betapartitiviruses. There have been reported cases of virus/virus interactions in mixed infections (Hillman et al., 2018). Thus, there is a need to obtain single infectants when assigning two genomic segments to single viruses and characterizing each partitivirus. To this end, we used the transfection approach previously developed for assessing various virus-host combinations (Sasaki et al., 2006; Kanematsu et al., 2010; Chiba et al., 2013a, 2016; Salaipeth et al., 2014). We obtained transfectants either in the R. necatrix W97, C. parasitica EP155 genetic background, or both W97 and EP155, which were singly infected partitiviruses. For these viruses, we confirmed that the two segments matched that of the genome of specified partitiviruses. For other coinfecting partitiviruses, our conclusions were based on the highly conserved terminal sequences found between segments (Supplementary Table S2). Furthermore, this facilitated the comparison of virus titers in EP155 and its Δdcl2 disruptant.
Some of the characterized partitiviruses differed from one another with respect to their accumulation levels and degree of symptom induction (Figures 4–6 and Table 2). These characteristics are related to antiviral/counter-defense responses incited by hosts and viruses. Previous studies have revealed that many homologous and heterologous viruses are targeted by antiviral RNA silencing (Segers et al., 2007; Sun et al., 2009; Chiba et al., 2013a,b, 2016; Salaipeth et al., 2014; Chiba and Suzuki, 2015). Many of these viruses induce antiviral RNA silencing via transcriptional up-regulation of the genes including a dicer (dcl2) and an argonaute (agl2) (Sun et al., 2009; Chiba and Suzuki, 2015). Previous reports showed that a victorivirus (dsRNA virus, family Totiviridae), RnVV1, was sensitive to, and targeted by, RNA silencing, but did not induce antiviral RNA silencing (Chiba and Suzuki, 2015). CHV1 is tolerant to RNA silencing and does not induce dcl2 transcription, which was largely attributed to the activity of the RNA silencing suppressor, p29 (Segers et al., 2007; Chiba and Suzuki, 2015). Andika et al. (2017) developed an antiviral RNA silencing monitoring system through the dcl2 transcription using a GFP reporter construct driven by the dcl2 promoter with C. parasitica genetic background. Interestingly, partitiviruses isolated from R. necatrix showed great variability in induction levels of dcl2 (Aulia et al., unpublished data). It is known that some partitiviruses tolerate RNA silencing while others are sensitive to silencing (Chiba et al., 2016) (Chiba and Suzuki, unpublished data). This study provided researchers with the materials necessary to obtain insights into mechanisms controlling diversity of host-partitivirus interactions.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found here: https://www.ddbj.nig.ac.jp/, LC517370-LC517399, LC521312-LC521313 (32 entries).
Author Contributions
NS designed the experiments and wrote the manuscript. PT, SH, CM, KH, and JA-L performed the experimental work. SK collected samples. PT, SK, CL-H, HK, and NS analyzed the data. PT and HK were involved in discussion and manuscript revision. All authors have given approval to the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer EV declared a past co-authorship with one of the authors NS.
Acknowledgments
The authors sincerely thank Dr. Donald L. Nuss (IBBR, University of Maryland) and Dr. Masanobu Tabata (Forestry & Forest Products Research Institute) for their generous gift of the C. parasitica strains EP155, Δdcl2, and Δagl2 and some R. necatrix field isolates used in this study.
Funding. This work was supported in part by Yomogi Inc. (to NS), the Program for the Promotion of Basic and Applied Research for Innovation in Bio-Oriented Industries (to SK and NS), and Grants-in-Aid for Scientific Research (A) and on Innovative Areas from the Japanese Ministry of Education, Culture, Sports, Science, and Technology (MEXT) (17H01463 and 16H06436, 16H06429, and 16K21723 to NS). The funder, Yomogi Inc., was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01064/full#supplementary-material
References
- Andika I. B., Jamal A., Kondo H., Suzuki N. (2017). SAGA complex mediates the transcriptional up-regulation of antiviral RNA silencing. Proc. Natl. Acad. Sci. U.S.A. 114, E3499–E3506. 10.1073/pnas.1701196114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andika I. B., Kondo H., Suzuki N. (2019). Dicer functions transcriptionally and post-transcriptionally in a multilayer antiviral defense. Proc. Natl. Acad. Sci. U.S.A. 116, 2274–2281. 10.1073/pnas.1812407116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa M., Nakamura H., Uetake Y., Matsumoto N. (2002). Presence and distribution of double-stranded RNA elements in the white root rot fungus Rosellinia necatrix. Mycoscience 43, 21–26. 10.1007/s102670200004 [DOI] [Google Scholar]
- Arjona-Girona I., Lopez-Herrera C. J. (2018). Study of a new biocontrol fungal agent for avocado white root rot. Biol. Control 117, 6–12. 10.1016/j.biocontrol.2017.08.018 [DOI] [Google Scholar]
- Arjona-Lopez J. M., Telengech P., Jamal A., Hisano S., Kondo H., Yelin M. D., et al. (2018). Novel, diverse RNA viruses from mediterranean isolates of the phytopathogenic fungus, Rosellinia necatrix: insights into evolutionary biology of fungal viruses. Environ. Microbiol. 20, 1464–1483. 10.1111/1462-2920.14065 [DOI] [PubMed] [Google Scholar]
- Bhatti M. F., Jamal A., Petrou M. A., Cairns T. C., Bignell E. M., Coutts R. H. (2011). The effects of dsRNA mycoviruses on growth and murine virulence of Aspergillus fumigatus. Fungal. Genet. Biol. 48, 1071–1075. 10.1016/j.fgb.2011.07.008 [DOI] [PubMed] [Google Scholar]
- Chiba S., Jamal A., Suzuki N. (2018). First evidence for internal ribosomal entry sites in diverse fungal virus genomes. MBio 9:e02350-17. 10.1128/mBio.02350-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S., Kondo H., Tani A., Saisho D., Sakamoto W., Kanematsu S., et al. (2011). Widespread endogenization of genome sequences of non-retroviral RNA viruses into plant genomes. PLoS Pathog. 7:e1002146. 10.1371/journal.ppat.1002146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S., Lin Y. H., Kondo H., Kanematsu S., Suzuki N. (2013a). Effects of defective-interfering RNA on symptom induction by, and replication of a novel partitivirus from a phytopathogenic fungus Rosellinia necatrix. J. Virol. 87, 2330–2341. 10.1128/JVI.02835-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S., Lin Y. H., Kondo H., Kanematsu S., Suzuki N. (2013b). A novel victorivirus from a phytopathogenic fungus, Rosellinia necatrix is infectious as particles and targeted by RNA silencing. J. Virol. 87, 6727–6738. 10.1128/JVI.00557-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S., Lin Y. H., Kondo H., Kanematsu S., Suzuki N. (2016). A novel betapartitivirus RnPV6 from Rosellinia necatrix tolerates host RNA silencing but is interfered by its defective RNAs. Virus Res. 219, 62–72. 10.1016/j.virusres.2015.10.017 [DOI] [PubMed] [Google Scholar]
- Chiba S., Salaipeth L., Lin Y. H., Sasaki A., Kanematsu S., Suzuki N. (2009). A novel bipartite double-stranded RNA mycovirus from the white root rot fungus Rosellinia necatrix: molecular and biological characterization, taxonomic considerations, and potential for biological control. J. Virol. 83, 12801–12812. 10.1128/JVI.01830-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S., Suzuki N. (2015). Highly activated RNA silencing via strong induction of dicer by one virus can interfere with the replication of an unrelated Virus Proc. Natl. Acad. Sci. U.S.A. 112, E4911–E4918. 10.1073/pnas.1509151112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W. K., Lee K. M., Yu J., Son M., Kim K. H. (2013). Insight into mycoviruses infecting fusarium species. Adv. Virus Res. 86, 273–288. 10.1016/B978-0-12-394315-6.00010-6 [DOI] [PubMed] [Google Scholar]
- Deng F., Xu R., Boland G. J. (2003). Hypovirulence-associated double-stranded RNA from Sclerotinia homoeocarpa is conspecific with Ophiostoma novo-ulmi mitovirus 3a-Ld. Phytopathology 93, 1407–1414. 10.1094/PHYTO.2003.93.11.1407 [DOI] [PubMed] [Google Scholar]
- Eusebio-Cope A., Sun L., Tanaka T., Chiba S., Kasahara S., Suzuki N. (2015). The chestnut blight fungus for studies on virus/host and virus/virus interactions: from a natural to a model host. Virology 477, 164–175. 10.1016/j.virol.2014.09.024 [DOI] [PubMed] [Google Scholar]
- Eusebio-Cope A., Suzuki N. (2015). Mycoreovirus genome rearrangements associated with RNA silencing deficiency. Nucl. Acids Res. 43, 3802–3813. 10.1093/nar/gkv239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faruk M. I., Eusebio-Cope A., Suzuki N. (2008). A host factor involved in hypovirus symptom expression in the chestnut blight fungus, Cryphonectria parasitica. J. Virol. 82, 740–754. 10.1128/JVI.02015-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghabrial S. A., Caston J. R., Jiang D., Nibert M. L., Suzuki N. (2015). 50-plus years of fungal viruses. Virology 479–480, 356–368. 10.1016/j.virol.2015.02.034 [DOI] [PubMed] [Google Scholar]
- Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Hillman B. I., Aulia A., Suzuki N. (2018). Viruses of plant-interacting fungi. Adv. Virus Res. 100, 99–116. 10.1016/bs.aivir.2017.10.003 [DOI] [PubMed] [Google Scholar]
- Hillman B. I., Supyani S., Kondo H., Suzuki N. (2004). A reovirus of the fungus Cryphonectria parasitica that is infectious as particles and related to the coltivirus genus of animal pathogen. J. Virol. 78, 892–898. 10.1128/JVI.78.2.892-898.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K., Nakamura H., Arakawa M., Matsumoto N. (2004). Diversity and vertical transmission of double-stranded RNA elements in root rot pathogens of trees, helicobasidium mompa and Rosellinia necatrix. Mycol. Res. 108, 626–634. 10.1017/S0953756204000061 [DOI] [PubMed] [Google Scholar]
- Jiang D., Fu Y., Guoqing L., Ghabrial S. A. (2013). Viruses of the plant pathogenic fungus Sclerotinia sclerotiorum. Adv. Virus Res. 86, 215–248. 10.1016/B978-0-12-394315-6.00008-8 [DOI] [PubMed] [Google Scholar]
- Jiang D., Ghabrial S. A. (2004). Molecular characterization of Penicillium chrysogenum virus: reconsideration of the taxonomy of the genus chrysoVirus J. Gen. Virol. 85, 2111–2121. 10.1099/vir.0.79842-0 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Wang J., Yang B., Wang Q., Zhou J., Yu W. (2019). Molecular characterization of a debilitation-associated partitivirus Infecting the pathogenic fungus aspergillus flavus. Front. Microbiol. 10:626. 10.3389/fmicb.2019.00626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanematsu S., Sasaki A., Onoue M., Oikawa Y., Ito T. (2010). Extending the fungal host range of a partitivirus and a mycoreovirus from Rosellinia necatrix by inoculation of protoplasts with virus particles. Phytopathology 100, 922–930. 10.1094/PHYTO-100-9-0922 [DOI] [PubMed] [Google Scholar]
- Kanhayuwa L., Kotta-Loizou I., Ozkan S., Gunning A. P., Coutts R. H. (2015). A novel mycovirus from Aspergillus fumigatus contains four unique dsRNAs as its genome and is infectious as dsRNA. Proc. Natl. Acad. Sci. U.S.A. 112, 9100–9105. 10.1073/pnas.1419225112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalifa M. E., MacDiarmid R. M. (2019). A novel totivirus naturally occurring in two different fungal genera. Front. Microbiol. 10:2318. 10.3389/fmicb.2019.02318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo H., Chiba S., Maruyama K., Andika I. B., Suzuki N. (2019). A novel insect-infecting virga/nege-like virus group and its pervasive endogenization into insect genomes. Virus Res. 262, 37–47. 10.1016/j.virusres.2017.11.020 [DOI] [PubMed] [Google Scholar]
- Kondo H., Chiba S., Sasaki A., Kanematsu S., Suzuki N. (2013a). Evidence for negative-strand RNA virus infection in fungi. Virology 435, 201–209. 10.1016/j.virol.2012.10.002 [DOI] [PubMed] [Google Scholar]
- Kondo H., Chiba S., Suzuki N. (2015). Detection and analysis of non-retroviral RNA virus-like elements in plant, fungal, and insect genomes. Methods Mol. Biol. 1236, 73–88. 10.1007/978-1-4939-1743-3_7 [DOI] [PubMed] [Google Scholar]
- Kondo H., Kanematsu S., Suzuki N. (2013b). Viruses of the white root rot fungus, Rosellinia necatrix. Adv. Virus Res. 86, 177–214. 10.1016/B978-0-12-394315-6.00007-6 [DOI] [PubMed] [Google Scholar]
- Lefort V., Longueville J. E., Gascuel O. (2017). SMS: smart model selection in PhyML. Mol. Biol. Evol. 34, 2422–2424. 10.1093/molbev/msx149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. H., Chiba S., Tani A., Kondo H., Sasaki A., Kanematsu S., et al. (2012). A novel quadripartite dsRNA virus isolated from a phytopathogenic filamentous fungus, Rosellinia necatrix. Virology 426, 42–50. 10.1016/j.virol.2012.01.013 [DOI] [PubMed] [Google Scholar]
- Liu H., Fu Y., Jiang D., Li G., Xie J., Cheng J., et al. (2010). Widespread horizontal gene transfer from double-stranded RNA viruses to eukaryotic nuclear genomes. J. Virol. 84, 11876–11887. 10.1128/JVI.00955-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Xie J., Cheng J., Fu Y., Li G., Yi X., et al. (2014). Fungal negative-stranded RNA virus that is related to bornaviruses and nyaviruses. Proc. Natl. Acad. Sci. U.S.A. 111, 12205–12210. 10.1073/pnas.1401786111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. C., Linder-Basso D., Hillman B. I., Kaneko S., Milgroom M. G. (2003). Evidence for interspecies transmission of viruses in natural populations of filamentous fungi in the genus Cryphonectria. Mol. Ecol. 12, 1619–1628. 10.1046/j.1365-294X.2003.01847.x [DOI] [PubMed] [Google Scholar]
- Lopez-Herrera C. J., Zea-Bonilla T. (2007). Effects of benomyl, carbendazim, fluazinam and thiophanate methyl on white root rot of avocado. Crop. Protect. 26, 1186–1192. 10.1016/j.cropro.2006.10.015 [DOI] [Google Scholar]
- Mu F., Xie J., Cheng S., You M. P., Barbetti M. J., Jia J., et al. (2017). Virome characterization of a collection of S. sclerotiorum from Australia. Front. Microbiol. 8:2540. 10.3389/fmicb.2017.02540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhire B. M., Varsani A., Martin D. P. (2014). SDT: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 9:e0108277. 10.1371/journal.pone.0108277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakakoshi M., Nishioka H., Katayama E. (2011). New versatile staining reagents for biological transmission electron microscopy that substitute for uranyl acetate. J. Electron. Microsc. 60, 401–407. 10.1093/jmicro/dfr084 [DOI] [PubMed] [Google Scholar]
- Nerva L., Silvestri A., Ciuffo M., Palmano S., Varese G. C., Turina M. (2017a). Transmission of penicillium aurantiogriseum partiti-like virus 1 to a new fungal host (Cryphonectria parasitica) confers higher resistance to salinity and reveals adaptive genomic changes. Environ. Microbiol. 19, 4480–4492. 10.1111/1462-2920.13894 [DOI] [PubMed] [Google Scholar]
- Nerva L., Varese G. C., Falk B. W., Turina M. (2017b). Mycoviruses of an endophytic fungus can replicate in plant cells: evolutionary implications. Sci. Rep. 7:1908. 10.1038/s41598-017-02017-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibert M. L., Ghabrial S. A., Maiss E., Lesker T., Vainio E. J., Jiang D., et al. (2014). Taxonomic reorganization of family partitiviridae and other recent progress in partitivirus research. Virus Res. 188C, 128–141. 10.1016/j.virusres.2014.04.007 [DOI] [PubMed] [Google Scholar]
- Nibert M. L., Tang J., Xie J., Collier A. M., Ghabrial S. A., Baker T. S., et al. (2013). 3D structures of fungal partitiviruses. Adv. Virus Res. 86, 59–85. 10.1016/B978-0-12-394315-6.00003-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuss D. L. (2005). Hypovirulence: mycoviruses at the fungal-plant interface. Nat. Rev. Microbiol. 3, 632–642. 10.1038/nrmicro1206 [DOI] [PubMed] [Google Scholar]
- Pearson M. N., Beever R. E., Boine B., Arthur K. (2009). Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol. Plant. Pathol. 10, 115–128. 10.1111/j.1364-3703.2008.00503.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrzik K. (2019). Evolutionary forces at work in partitiviruses. Virus Genes 55, 563–573. 10.1007/s11262-019-01680-0 [DOI] [PubMed] [Google Scholar]
- Pliego C., Lopez-Herrera C., Ramos C., Cazorla F. M. (2012). Developing tools to unravel the biological secrets of Rosellinia necatrix, an emergent threat to woody crops. Mol. Plant. Pathol 13, 226–239. 10.1111/j.1364-3703.2011.00753.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salaipeth L., Chiba S., Eusebio-Cope A., Kanematsu S., Suzuki N. (2014). Biological properties and expression strategy of Rosellinia necatrix megabirnavirus 1 analyzed in an experimental host, Cryphonectria parasitica. J. Gen. Virol. 95, 740–750. 10.1099/vir.0.058164-0 [DOI] [PubMed] [Google Scholar]
- Sasaki A., Kanematsu S., Onoue M., Oikawa Y., Nakamura H., Yoshida K. (2007). Artificial infection of Rosellinia necatrix with purified viral particles of a member of the genus mycoreovirus reveals its uneven distribution in single colonies. Phytopathology 97, 278–286. 10.1094/PHYTO-97-3-0278 [DOI] [PubMed] [Google Scholar]
- Sasaki A., Kanematsu S., Onoue M., Oyama Y., Yoshida K. (2006). Infection of Rosellinia necatrix with purified viral particles of a member of Partitiviridae (RnPV1-W8). Arch. Virol. 151, 697–707. 10.1007/s00705-005-0662-2 [DOI] [PubMed] [Google Scholar]
- Sasaki A., Miyanishi M., Ozaki K., Onoue M., Yoshida K. (2005). Molecular characterization of a partitivirus from the plant pathogenic ascomycete Rosellinia necatrix. Arch. Virol. 150, 1069–1083. 10.1007/s00705-005-0494-0 [DOI] [PubMed] [Google Scholar]
- Sasaki A., Nakamura H., Suzuki N., Kanematsu S. (2016). Characterization of a new megabirnavirus that confers hypovirulence with the aid of a co-infecting partitivirus to the host fungus, Rosellinia necatrix. Virus Res. 219, 73–82. 10.1016/j.virusres.2015.12.009 [DOI] [PubMed] [Google Scholar]
- Segers G. C., Zhang X., Deng F., Sun Q., Nuss D. L. (2007). Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc. Natl. Acad. Sci. U.S.A. 104, 12902–12906. 10.1073/pnas.0702500104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamsi W., Sato Y., Jamal A., Shahi S., Kondo H., Suzuki N., et al. (2019). Molecular and biological characterization of a novel botybirnavirus identified from a Pakistani isolate of alternaria alternata. Virus Res. 263, 119–128. 10.1016/j.virusres.2019.01.006 [DOI] [PubMed] [Google Scholar]
- Shimizu T., Kanematsu S., Yaegashi H. (2018). Draft genome sequence and transcriptional analysis of Rosellinia necatrix infected with a virulent mycoVirus Phytopathology 108, 1206–1211. 10.1094/PHYTO-11-17-0365-R [DOI] [PubMed] [Google Scholar]
- Sun Q., Choi G. H., Nuss D. L. (2009). A single argonaute gene is required for induction of RNA silencing antiviral defense and promotes viral RNA recombination. Proc. Natl. Acad. Sci. U.S.A. 106, 17927–17932. 10.1073/pnas.0907552106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supyani S., Hillman B. I., Suzuki N. (2007). Baculovirus expression of the 11 mycoreovirus-1 genome segments and identification of the guanylyltransferase-encoding segment. J. Gen. Virol. 88, 342–350. 10.1099/vir.0.82318-0 [DOI] [PubMed] [Google Scholar]
- Suzuki N. (2017). Frontiers in fungal virology. J. Gen. Plant Pathol. 83, 419–423. 10.1007/s10327-017-0740-9 [DOI] [Google Scholar]
- Suzuki N., Supyani S., Maruyama K., Hillman B. I. (2004). Complete genome sequence of Mycoreovirus-1/Cp9B21, a member of a novel genus within the family reoviridae, isolated from the chestnut blight fungus Cryphonectria parasitica. J. Gen. Virol. 85, 3437–3448. 10.1099/vir.0.80293-0 [DOI] [PubMed] [Google Scholar]
- Talavera G., Castresana J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577. 10.1080/10635150701472164 [DOI] [PubMed] [Google Scholar]
- Urayama S., Katoh Y., Fukuhara T., Arie T., Moriyama H., Teraoka T. (2015). Rapid detection of Magnaporthe oryzae chrysovirus 1-A from fungal colonies on agar plates and lesions of rice blast. J. Gen. Plant Pathol. 81, 97–102. 10.1007/s10327-014-0567-6 [DOI] [Google Scholar]
- Vainio E. J., Chiba S., Ghabrial S. A., Maiss E., Roossinck M., Sabanadzovic S., et al. (2018a). ICTV virus taxonomy profile: partitiviridae. J. Gen. Virol. 99, 17–18. 10.1099/jgv.0.000985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainio E. J., Hantula J. (2016). Taxonomy, biogeography and importance of heterobasidion viruses. Virus Res. 219, 2–10. 10.1016/j.virusres.2015.10.014 [DOI] [PubMed] [Google Scholar]
- Vainio E. J., Jurvansuu J., Hyder R., Kashif M., Piri T., Tuomivirta T., et al. (2018b). Heterobasidion partitivirus 13 mediates severe growth debilitation and major alterations in the gene expression of a fungal forest pathogen. J. Virol. 92, e01744–e01717. 10.1128/JVI.01744-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainio E. J., Korhonen K., Tuomivirta T. T., Hantula J. (2010). A novel putative partitivirus of the saprotrophic fungus Heterobasidion ecrustosum infects pathogenic species of the heterobasidion annosum complex. Fungal. Biol. 114, 955–965. 10.1016/j.funbio.2010.09.006 [DOI] [PubMed] [Google Scholar]
- Velasco L., Arjona-Girona I., Ariza-Fernandez M. T., Cretazzo E., Lopez-Herrera C. (2018). A novel hpovirus species from Xylariaceae fungi infecting avocado. Front. Microbiol. 9:778. 10.3389/fmicb.2018.00778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco L., Arjona-Girona I., Cretazzo E., Lopez-Herrera C. (2019). Viromes in Xylariaceae fungi infecting avocado in Spain. Virology 532, 11–21. 10.1016/j.virol.2019.03.021 [DOI] [PubMed] [Google Scholar]
- Velasco L., Lopez Herrera C. J., Cretazzo E. (2020). Two novel partitiviruses that accumulate differentially in Rosellinia necatrix and Entoleuca sp. infecting avocado. Virus. Res. 285. 10.1016/j.virusres.2020.198020 [DOI] [PubMed] [Google Scholar]
- Wang L., Jiang J., Wang Y., Hong N., Zhang F., Xu W., et al. (2014). Hypovirulence of the phytopathogenic fungus Botryosphaeria dothidea: association with a coinfecting chrysovirus and a partitiVirus J. Virol. 88, 7517–7527. 10.1128/JVI.00538-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Cheng J., Tang J., Fu Y., Jiang D., Baker T. S., et al. (2014). A novel partitivirus that confers hypovirulence on plant pathogenic fungi. J. Virol. 88, 10120–10133. 10.1128/JVI.01036-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Jiang D. (2014). New insights into mycoviruses and exploration for the biological control of crop fungal diseases. Annu. Rev. Phytopathol. 52, 45–68. 10.1146/annurev-phyto-102313-050222 [DOI] [PubMed] [Google Scholar]
- Yaegashi H., Kanematsu S. (2016). Natural infection of the soil-borne fungus Rosellinia necatrix with novel mycoviruses under greenhouse conditions. Virus Res. 219, 83–91. 10.1016/j.virusres.2015.11.004 [DOI] [PubMed] [Google Scholar]
- Yaegashi H., Nakamura H., Sawahata T., Sasaki A., Iwanami Y., Ito T., et al. (2013). Appearance of mycovirus-like double-stranded RNAs in the white root rot fungus, Rosellinia necatrix, in an apple orchard. FEMS Microbiol. Ecol. 83, 49–62. 10.1111/j.1574-6941.2012.01454.x [DOI] [PubMed] [Google Scholar]
- Yu X., Li B., Fu Y., Jiang D., Ghabrial S. A., Li G., et al. (2010). A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc. Natl. Acad. Sci. U.S.A. 107, 8387–8392. 10.1073/pnas.0913535107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Hisano S., Tani A., Kondo H., Kanematsu S., Suzuki N. (2016). A capsidless ssRNA virus hosted by an unrelated dsRNA virus Nat. Microbiol. 1:15001. 10.1038/nmicrobiol.2015.1 [DOI] [PubMed] [Google Scholar]
- Zhang R., Liu S., Chiba S., Kondo H., Kanematsu S., Suzuki N. (2014). A novel single-stranded RNA virus isolated from a phytopathogenic filamentous fungus, Rosellinia necatrix, with similarity to hypo-like viruses. Front. Microbiol. 5:360. 10.3389/fmicb.2014.00360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Zhang M., Chen Q., Zhu M., Zhou E. (2014). A novel mycovirus closely related to viruses in the genus alphapartitivirus confers hypovirulence in the phytopathogenic fungus Rhizoctonia solani. Virology 456–457, 220–226. 10.1016/j.virol.2014.03.029 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found here: https://www.ddbj.nig.ac.jp/, LC517370-LC517399, LC521312-LC521313 (32 entries).