Abstract
The typical plastid genome (plastome) of photosynthetic angiosperms comprises a pair of Inverted Repeat regions (IRs), which separate a Large Single Copy region (LSC) from a Small Single Copy region (SSC). The independent losses of IRs have been documented in only a few distinct plant lineages. The majority of these taxa show uncommonly high levels of plastome structural variations, while a few have otherwise conserved plastomes. For a better understanding of the function of IRs in stabilizing plastome structure, more taxa that have lost IRs need to be investigated. We analyzed the plastomes of eight species from two genera of the putranjivoid clade of Malpighiales using Illumina paired-end sequencing, the de novo assembly strategy GetOrganelle, as well as a combination of two annotation methods. We found that all eight plastomes of the putranjivoid clade have lost their IRB, representing the fifth case of IR loss within autotrophic angiosperms. Coinciding with the loss of the IR, plastomes of the putranjivoid clade have experienced significant structural variations including gene and intron losses, multiple large inversions, as well as the translocation and duplication of plastome segments. However, Balanopaceae, one of the close relatives of the putranjivoid clade, exhibit a relatively conserved plastome organization with canonical IRs. Our results corroborate earlier reports that the IR loss and additional structural reorganizations are closely linked, hinting at a shared mechanism that underpins structural disturbances.
Keywords: plastome evolution, Inverted Repeat region loss, genomic rearrangement, Lophopyxidaceae, Putranjivaceae
Introduction
Plastids, such as chloroplasts, chromoplasts, and leucoplasts, are the place for photosynthesis and the major organelle for organic product storage in plants. Plastids retain a semi-autonomous genetic system with their own genome (plastome). Typically, the plastome of a photosynthetic angiosperm is a circular molecule, with a length of 120–160 kb (Wicke et al., 2011). Structurally, such plastome comprises a pair of Inverted Repeat regions (hereafter called IRs; ~25 kb), a Large Single Copy region (LSC; ~85 kb), and a Small Single Copy region (SSC; ~15 kb) (Ruhlman and Jansen, 2014; Mower and Vickrey, 2018). IRs may play an important role in maintaining plastome stability (Maréchal and Brisson, 2010), which might be one of the reasons why most autotrophic angiosperms possess canonical IRs. However, IR losses have been documented in a few distinct angiosperm lineages, namely the IR-Lacking Clade (IRLC) of Leguminosae (Palmer and Thompson, 1981; Palmer and Thompson, 1982; but see Choi et al., 2019), two Erodium lineages of Geraniaceae (Guisinger et al., 2011; Ruhlman et al., 2017), Carnegiea gigantea of Cactaceae, and Tahina spectabilis of Arecaceae (Choi et al., 2019). Plastomes of the IRLC, C. gigantea (Sanderson et al., 2015), Tahina spectabilis (Barrett et al., 2016) showed significant higher rearrangement degrees compared to their sister clade, while species in a lineage of Erodium that has lost one IR exhibit an otherwise conserved plastome structure (Blazier et al., 2016). Hence, further comparative study is needed to elucidate the function of IRs in stabilizing plastome structure.
Malpighiales are one of the largest orders of flowering plants. Plants in this order exhibit a remarkable morphological and ecological diversity, with many species of great ecological and economic importance (Xi et al., 2012). Previous studies have revealed significant structural variations in the plastomes of multiple taxa in this order. Rabah et al. (2019) compared plastomes of 15 species of the genus Passiflora (Passifloraceae) and found that this genus has experienced widespread genomic changes, including inversions, gene and intron losses along with multiple independent IR expansions and contractions. Lopes et al. (2018) revealed the contraction and expansion of the IRs altering the size, gene content, and gene order of SC and IRs in the plastome of Linum usitatissimum (Linaceae). Tangphatsornruang et al. (2011) reported a 30-kb inversion between trnE-UUC—trnS-GCU and trnT-GGU—trnR-UCU in Hevea brasiliensis (Euphorbiaceae). Two recent studies detected an inversion in the LSC, significant variation in length reduction of the IRs, gene loss and pseudogenization events in plastomes of Podostemaceae (Bedoya et al., 2019; Jin et al., 2020). An inversion over 50 kb spanning from trnK-UUU to rbcL in the LSC is shared by Cratoxylum cochinchinense (Hypericaceae), Tristicha trifaria, and Marathrum foeniculaceum (Podostemaceae) (Jin et al., 2020). Previous studies suggested that multiple lineages of Malpighiales have experienced plastome structural variations, but knowledge of plastomes evolution in this large order is still limited.
The putranjivoid clade in Malpighiales consists of two families: Lophopyxidaceae and Putranjivaceae (Wurdack and Davis, 2009). Lophopyxidaceae have a single genus, whereas Putranjivaceae contain three genera and ca. 216 species. Containing 209 species, Drypetes is the largest genus in Putranjivaceae. The species in this clade are perennial trees or shrubs, growing primarily in tropical and subtropical areas (Kubitzki, 2014).
As it is unknown to date how plastid genomes evolve in the putranjivoid clade, we here assembled the complete plastome sequences for eight species, as well as two species from the closely related family Balanopaceae, representing one genus each from each family. Our analyses focused on exploring the structural variation of plastomes and revealed that all plastomes of the putranjivoid calde have lost the IRB entirely and experienced extensive additional structural rearrangements. In contrast, the plastomes of the two Balanopaceae species retain a relatively conserved plastome structure, indicating an evolutionary shift after the split of both lineages.
Materials and Methods
Taxon Sampling, DNA Extraction and Sequencing
We sampled seven species from the largest genus Drypetes of Putranjivaceae, one species from Lophopyxidaceae, and two species from Balanopaceae as outgroups. Total genomic DNA of all samples was isolated from herbarium specimens or silica gel-dried leaves using the DNeasy Plant Mini Kit (Tiangen Biotech Co., LTD., Beijing, China) or a standardized CTAB-protocol (Doyle and Doyle, 1987). Following quantity checks and library preparations, paired-end sequencing was carried out on Illumina HiSeq 2000 or HiSeq X TEN at the Plant Germplasm and Genomics Center (Kunming Institute of Botany, Chinese Academy of Sciences). A genome skimming sequencing approach was employed. Table S1 provides original collection location, herbarium voucher information, GenBank accession numbers, as well as the read characteristics for all taxa discussed in this study.
Plastome Assembly and Annotation
Plastomes were assembled using GetOrganelle v1.6.1a with default settings, which filtered plastid-like reads, conducted the de novo assembly, purified the assembly graph, and generated the complete plastomes (Camacho et al., 2009; Bankevich et al., 2012; Langmead and Salzberg, 2012; Jin et al., 2019). K-mer gradients were set according to the sequenced read lengths as “-k 21,31,41,51,65,85,91,95,99,101,111,121,127” for 150 bp reads; “-k 21,31,41,51,61,71,81,85,87” were used for 90 bp reads. Final assembly graphs were visualized in Bandage (Wick et al., 2015) to confirm the automatically generated plastomes. Two configurations of each plastome caused by the flip-flop recombination mediated by the IR or the ~1.2 kb sIR (short Inverted Repeat regions) were obtained, and one of them was arbitrarily selected for downstream analysis (Walker et al., 2015). All plastomes were initially annotated using PGA (Qu et al., 2019) and GeSeq (Tillich et al., 2017), with the annotated plastome of Amborella trichopoda (NC_005086) (Goremykin et al., 2003) selected as the reference. For confirmation, all annotations were compared with the previously published plastome of Byrsonima coccolobifolia (NC_037191; Malpighiaceae; Menezes et al., 2018) and manually examined in Geneious Prime (https://www.geneious.com). All newly sequenced plastomes were deposited in GenBank under accession numbers MN504788–MN504797.
Phylogenetic Analysis
Phylogenetic analysis was performed using 71 protein-coding genes, which were shared by all study species ( Table S2 ). Gene sequences were extracted using get_annotated_regions_from_gb.py (https://github.com/Kinggerm/PersonalUtilities, accessed on July 30, 2019; Zhang et al., 2020), aligned individually using prank v.140603 (Loytynoja and Goldman, 2008), then concatenated into a single aligned dataset using concatenate_fasta.py (https://github.com/Kinggerm/PersonalUtilities, accessed on July 30, 2019; Zhang et al., 2020). To reconstruct the phylogenetic relationships among our taxa, we employed RAxML v.8.2.11 (Stamatakis, 2014) with “-m GTRGAMMA”, which performs tree searches and optimization under the maximum likelihood paradigm. For statistical support, we ran 1,000 bootstrap replicates, and visualized the results in FigTree v.1.4.4 (http://tree.bio.ed.ac.uk). We mapped the events manually, facilitated by the small size of the data set, assuming that pseudogenization, gene loss, and IR loss are irreversible events.
Plastome Structural Rearrangements
To build whole plastome alignments for the putranjivoid clade, and the two Balanops species, we used the progressiveMauve algorithm in Mauve v2.3.1 (Darling et al., 2010) with default settings. The IRB was removed from plastid genomes with two copies of the large inverted repeats to allow for an optimal homology assessment (Wicke et al., 2013). Based on the strand orientation of the Locally Collinear Blocks (LCBs) identified by the progressiveMauve alignment, strand orientation determines the sign (+/-). Compared with the references, each LCB was numbered. Subsequently, we used GRIMM (Tesler, 2002) to calculate genome rearrangement distances.
Number of Repeats
Dispersed repeats (including forward, reverse, complement, and palindromic repeats) were identified by REPuter (Kurtz et al., 2001) based on the following criteria: minimum repeat size ≥ 30 bp; sequence identities ≥ 90%; Hamming distance = 3. Again, the IRB was removed, where present. REPuter overestimates the number of repetitive elements in a given sequence by recognizing nested or overlapping repeats within a given region containing multiple repeats (Wang et al., 2018). The FindRepeats plugin of Geneious Prime was also used to identify repeated regions using a minimum repeat length of 30 bp and zero mismatches.
Confirmation of 271 bp sIR-Induced Isomers
sIR range from 11 bp to several kbs in plastomes and are capable of inducing plastomic inversions and isomer (Martin et al., 2014; Wang et al., 2018). As sIR can potentially induce isomers, we used the library information of paired-end reads to confirm the existence of each potential isomers in Lophopyxis maingayi. We mapped the paired-end reads to the plastome sequence of each isomer, visually inspected the mapped read pairs in Geneious, and verified the existence of properly-mapped read pairs spanning the entire sIR. An isomer with read pairs spanning the entire sIR was supported to exist. Specifically, we firstly conducted read mapping using the evaluate_assembly_using_mapping.py script from the GetOrganelle toolkit, which calls Bowtie2 (Langmead and Salzberg, 2012). Because of the relatively short average insert size ( Table S1 ), most read pairs are too short in insert size for providing confirmation and hampered visual inspection. For better visualization, we filtered the alignment using SAMtools (Li et al., 2009) by keeping records with an insert size between 330 and 600. Finally, we imported the filtered alignment file (*.sam) into Geneious Prime, turn on the “Layout-Link paired reads” mode and checked whether there are read pairs spanning the entire sIR.
Results and Discussion
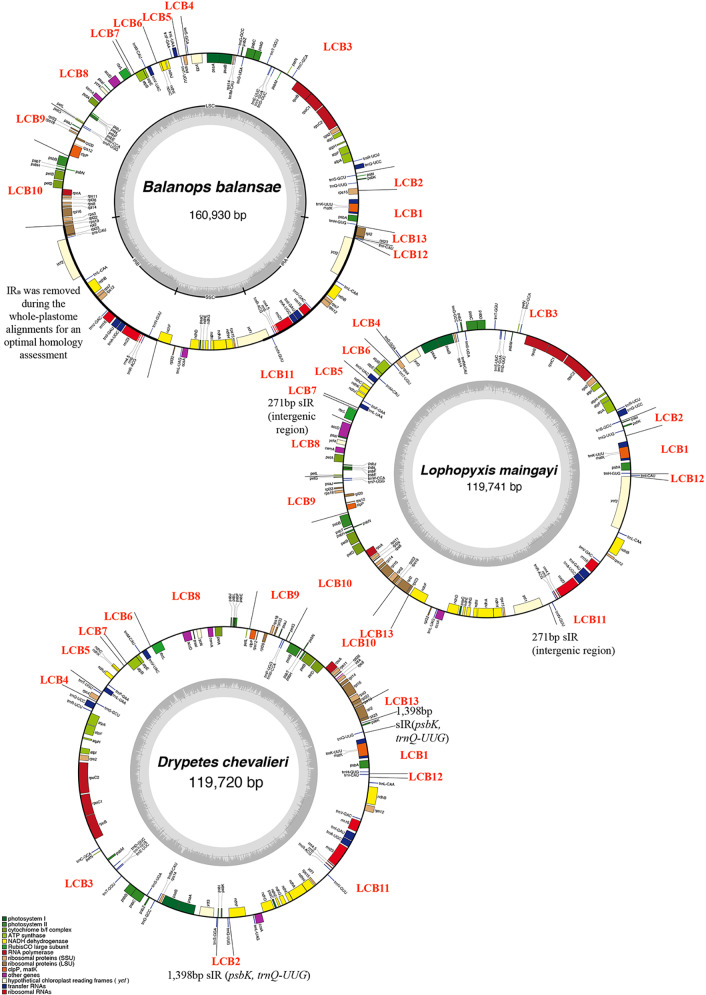
Due to the differences in plant materials, the average base coverages of plastomes varied from 72 x to 640 x ( Table S1 ). However, all ten newly assembled plastomes were complete. Plastomes from the putranjivoid clade are relatively small compared to their sister family Balanopaceae ( Figure 1 ; Table 1 ). Variation in plastome size of the sampled putranjivoid sepcecies was small: Drypetes hainanensis has the smallest plastome with a length of 119,105 bp, while Drypetes lateriflora has the largest plastome with a length of 120,800 bp.
Figure 1.
Plastid genomes of three species of Malpighiales representing three genera involved in this study. GC content graphs are shown as dark gray bars toward the center of each diagram. Each Locally Collinear Block (LCB) was indicated in the circular map, as well as the ~12kb short Inverted Repeat (sIR) and 271bp sIR.
Table 1.
Plastome features of 10 Malpighiales species.
| Species | Plastome size (bp) | IR size (bp) | Number of unique genes* | sIR (bp) | Estimated rearrangement distance† |
|---|---|---|---|---|---|
| Drypetes chevalieri | 119,720 | n.a. | 106 | 1,398 | 7 |
| Drypetes diopa | 119,299 | n.a. | 106 | 1,357 | 7 |
| Drypetes hainanensis | 119,105 | n.a. | 106 | 1,191 | 7 |
| Drypetes indica | 120,596 | n.a. | 106 | 1,047 | 7 |
| Drypetes lateriflora | 120,800 | n.a. | 107 | 1,484 | 7 |
| Drypetes longifolia | 119,268 | n.a. | 106 | 1,260 | 7 |
| Drypetes similis | 119,507 | n.a. | 106 | 1,221 | 7 |
| Lophopyxis maingayi | 119,741 | n.a. | 109 | 271 | 3 |
| Balanops balansae | 160,930 | 26,748 | 112 | np | – |
| Balanops pedicellata | 160,765 | 26,738 | 112 | np | – |
*Number of unique genes refers to unique gene number within the whole plastome.†Gene order changes were calculated relative to references (Balanops).bp, basepair; IR, inverted repeat; n.a., not applicable due to no IR pair; sIR, short inverted repeat that might have induced isomers; np, not present.
Across autotrophic flowering plants, the content of IRs nearly universally includes all 4 rRNA genes, 7 tRNA genes, and a small number of protein genes (Mower and Vickrey, 2018). Plastomes of all studied putranjivoid species have lost a copy of the inverted repeat, namely IRB ( Figure 1 , Figure 2 ; Table 1 ), which led to the observed significant reduction of their overall plastome size. All sampled putranjivoid species have lost the same segment of IRB including 4 rRNA genes, 7 tRNA genes, and several protein coding genes (rps12, rps7, ndhB, ycf2, rpl23, and rpl2). Their plastome sizes were slightly varied due to the differences in intergenic regions. However, not all inversions are shared by L. maingayi and Drypetes species ( Figure 1 , Table 2 ).
Figure 2.
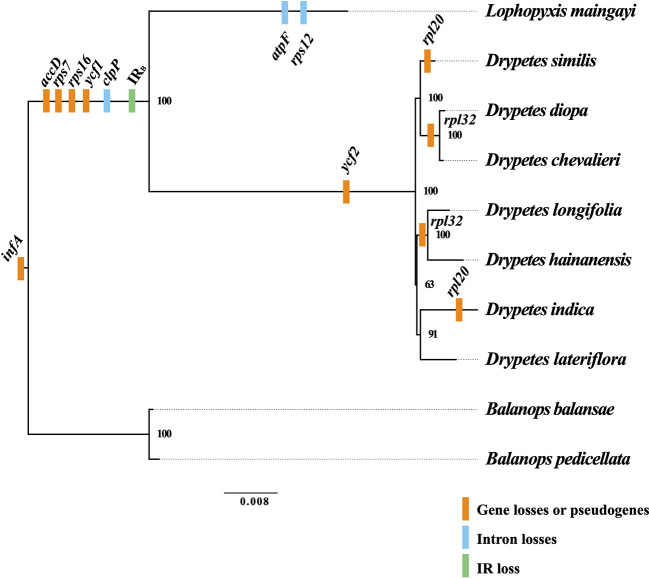
Gene losses or pseudogenes in the plastomes of the putranjivoid clade. Based on the phylogenetic tree, we mapped the events of gene and intron losses, as well as the event of the inverted repeat (IR) loss. Branch lengths correspond to substitutions per site.
Table 2.
Permutations and Locally Collinear Blocks (LCB).
| Species | Gene order |
|---|---|
| Balanops balansae | 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13 |
| Balanops pedicellata | 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13 |
| Lophopyxis maingayi | 1, 2, 3, 4, -6, -5, 7, 8, 9, 10, -13, 11, 12 |
| Drypetes chevalieri | 1, 13, -10, 9, -8, 6, 7, -5, -4, 3, -2, 11, -12 |
| Drypetes diopa | 1, 13, -10, 9, -8, 6, 7, -5, -4, 3, -2, 11, -12 |
| Drypetes hainanensis | 1, 13, -10, 9, -8, 6, 7, -5, -4, 3, -2, 11, -12 |
| Drypetes indica | 1, 13, -10, 9, -8, 6, 7, -5, -4, 3, -2, 11, -12 |
| Drypetes lateriflora | 1, 13, -10, 9, -8, 6, 7, -5, -4, 3, -2, 11, -12 |
| Drypetes longifolia | 1, 13, -10, 9, -8, 6, 7, -5, -4, 3, -2, 11, -12 |
| Drypetes similis | 1, 13, -10, 9, -8, 6, 7, -5, -4, 3, -2, 11, -12 |
Negative numbers indicate a change of strand orientation.
To our knowledge, the IR loss event in the putranjivoid clade represents the fifth reported IR loss of autotrophic flowering plants. Among the five IR losses, the putranjivoid clade and Tahina spectabilis have lost IRB (Barrett et al., 2016), while the IR-lacking legumes (Palmer and Thompson, 1981; Palmer and Thompson, 1982), C. gigantea (Sanderson et al., 2015), and some Erodium species (Guisinger et al., 2011; Ruhlman et al., 2017) all have lost their IRA. Which copy of IR has been lost seems to be a stochastic phenomenon. The two identical copies of the IR contain the same genes. None of the IR-lacking lineages, including all putranjivoid species, exhibits an impaired phenotype or habits (Blazier et al., 2016). Therefore, we may conclude that for those lineages one copy per IR-gene seems to be sufficient to support the overall function of the plastid.
The plastomes of Balanopaceae, one of the closest relatives of the putranjivoid clade, possess a canonical IR structure and a relatively conserved gene content and organization, which resembles those of the supposed ancestral angiosperm plastome (Ruhlman and Jansen, 2014). However, the plastomes of the putranjivoid clade have experienced significant gene content changes ( Table 1 ; Figure 2 ). All examined plastomes from the putranjivoid clade lack intact accD, rps7, rps16, and ycf1 genes ( Figure 2 ), and all examined Putranjivaceae plastomes have one copy of the ycf2 gene lost or became a pseudogene. The rpl20 gene was inferred to be a pseudogene due to the presence of internal stop codons in the plastomes of D. similis and D. indica ( Figure S1 ). Drypetes diopa, D. chevalieri, and D. longifolia lost the rpl32 gene independently, and the rpl32 gene of D. hainanensis was a pseudogene due to internal stop codons ( Figure S2 ). The loss of rps16 is common in angiosperm plastomes (Jansen et al., 2007). A study in Medicago truncatula (Leguminosae) and Populus alba (Salicaceae) showed that the rps16 gene was lost in both species. However, the function of the plastid rps16 was compensated by a nuclear-encoded rps16 in both species (Ueda et al., 2008). The loss of accD in Trifolium species has been achieved by relocation to the nucleus (Magee et al., 2010). Two previous studies (Bedoya et al., 2019; Jin et al., 2020) suggested the uncommon loss or pseudogenization of ycf1 and ycf2 in Podostemaceae. Our results also suggested the loss or pseudogenization of ycf1 in the putranjivoid clade, and the loss or pseudogenization of ycf2 in Putranjivaceae. Moreover, all putranjivoid species lack both clpP introns, and L. maingayi lacks the typical introns in atpF and rps12 ( Figure 2 ). Previous studies indicated the loss of rps12 and clpP introns in various legume lineages (Jansen et al., 2008; Wang et al., 2018). Recent studies on Podostemaceae also found the loss of both introns of clpP in riverweeds (Bedoya et al., 2019; Jin et al., 2020). The loss of the atpF intron was found not only in Lophopyxis maingayi, but also in members of Euphorbiaceae, Phyllanthaceae, Elatinaceae, and Passifloraceae of Malpighiales (Daniell et al., 2008). However, the mechanisms responsible for the intron losses remain elusive.
Plastomes of the putranjivoid clade have experienced notable structural reorganizations. Our progressiveMauve plastomes alignment of the putranjivoid clade with Balanops as references identified 13 syntenic regions ( Figures 1 and 3 , Figures S3 and S4 ; Table 2 ). Genes or intergenic regions located in each LCB were identified ( Table 3 ). Plastomic rearrangement distances were estimated based on the LCB orientations. The plastome of L. maingayi showed fewer rearrangements than those of Putranjivaceae species ( Figure 3 ), as reflected in a lower genome rearrangement distance of 3 for L. maingayi but a higher genome rearrangement distance of 7 for the Drypetes species ( Table 1 ). In L. maingayi, an inversion altered the syntenic blocks (4) (5) (6) (7) into (4) (-6) (-5) (7). LCB (5) and (6) corresponded to a 7.5-kb region between atpB and trnL-UAA. The order of the LCBs (10) (-13), (-13) (11), and the disruption of the adjacency of blocks (12) (13) were also the results of a translocation of LCB (13). LCB (13) corresponds to a 2-kb region spanning from the rpl23 to the rpl2 gene. Alternatively, a reasonable explanation for the changes around LCB (13) is that the rpl23 and rpl2 genes located in IRA were lost, while the identical though inverted copies of these two genes from IRB remained intact. Plastomes of all Drypetes species shared all inversions ( Figure 3 , Figure S3 , and Figure S4 ). One optimal reversal (means rearrangement event such as inversion) scenario included 7 inversion events, which means the minimum number of inversions required for transforming in gene order from a Drypetes plastome to a Balanops plastome is 7.
Figure 3.
Plastid genome variation in the putranjivoid clade. Whole-plastome alignments divide the plastid genome of our study taxa into 13 Locally Collinear Blocks (LCB), which are shown as color-coded representations of syntenic regions. The IRB was removed from plastid genomes with two copies of the large inverted repeats to allow for an optimal homology assessment. Blocks below the horizontal central line represent inversions relative to the references, shown as the upper two taxa. The height of the colored region within a block reflects the average sequence identity relative to the reference. Species names are color-coded to indicate their family: Balanopaceae (black), Lophopyxidaceae (blue), and Putranjivaceae (purple). The pink blocks in both Balanops species indicate the IR regions. Red blocks represent rrn5, rrn4.5, rrn23, and rrn16 genes, green blocks represent trnA-UGC and trnI-GAU genes.
Table 3.
Genes in Locally Collinear Blocks (LCB) identified using ProgressiveMauve alignment for plastomes of the putranjivoid clade.
| LCB | Genes |
|---|---|
| 1 | trnH-GUG, psbA, trnK-UUU, matK |
| 2 | psbI, psbK, trnQ-UUG |
| 3 | trnS-GCU, trnG-UCC, trnR-UCU, atpA, atpF, atpH, atpI, rps2, rpoC2, rpoC1, rpoB, trnC-GCA, petN, psbM, trnD-GUC, trnY-GUA, trnE-UUC, trnT-GGU, psbD, psbC, trnS-UGA, psbZ, trnG-UCC, trnM-CAU, rps14, psaB, psaA, ycf3, trnS-GGA |
| 4 | trnT-UGU, rps4 |
| 5 | ndhC, ndhK, ndhJ, trnF-GAA, trnL-UAA |
| 6 | trnV-UAC, trnM-CAU, atpE, atpB |
| 7 | Intergenic region |
| 8 | petL, psbE, psbF, psbL, psbJ, petA, cemA, ycf4, psaI, accD, rbcL |
| 9 | petG, trnW-CCA, trnP-UGG, psaJ, rpl33, rps18, rpl20, rps12_5’exon, clpP |
| 10 | rps19_fragment, rpl22, rps3, rpl16, rpl14, rps8, rpl36, rps11, rpoA, petD, petB, psbH, psbN, psbT, psbB |
| 11 | ndhF, rpl32, trnL-UAG, ccsA, ndhD, psaC, ndhE, ndhG, ndhI, ndhA, ndhH, rps15, trnN-GUU, trnR-ACG, rrn5, rrn4.5, rrn23, trnA-UGC, trnI-GAU, rrn16, trnV-GAC, rps12_3’exon, ndhB, trnL-CAA, ycf2 |
| 12 | trnI-CAU |
| 13 | rpl23, rpl2, rps19_fragment |
infA, rps7, rps16and ycf1were not included in the LCBs as they were not present in all of the plastomes of the putranjivoid clade.
IRs are thought to play a role in stabilizing the plastome (Maréchal and Brisson, 2010). This hypothesis is based on the fact that legume and conifer plastomes, which have no IRs, also show more rearrangements than plastomes containing canonical IRs (Palmer and Thompson, 1982; Hirao et al., 2008; Mower and Vickrey, 2018). The putranjivoid clade is another solid example that increased structural variations coincide with the loss of the IRs. However, species in a lineage of Erodium, which also have no IRs, still exhibit a conserved overall plastome structure, resembling those of IR-containing species (Blazier et al., 2016). In contrast, many species of Geranium and Pelargonium (Chumley et al., 2006; Guisinger et al., 2011; Röschenbleck et al., 2016; Weng et al., 2016) and Campanulaceae (Haberle et al., 2008), some of which have canonical though expanded IRs, possess highly rearranged plastomes. These cases suggest that further comparative study is needed to elucidate the function of IRs in stabilizing plastome structure.
An emerging consensus is that the presence of smaller repeats, rather than the loss of the IRs, is a major driver of plastome rearrangements (Mower and Vickrey, 2018). In the putranjivoid clade, we observed an obvious tendency that plastomes with more genomic rearrangements were also richer in repeats of 30 bp or more ( Table 4 ). The number of short repeats are the largest in the Drypetes plastomes. While the Balanops plastomes, which are the most conserved ones have the fewest number of repeats. Furthermore, more rearrangement events also coincide with the presence of longer repeats ( Table 4 ). Being the most rearranged, all Drypetes plastomes do possess a pair of sIRs with the length of more than 1,000 bp. As the only case that sIR induced gene duplication found in our study, all Drypetes species have two copy of two genes, psbK and trnQ-UUG, due to the ~1.2kb sIR. Typical IRs in plastomes trigger intra-plastomic homologous recombination, which generates two isomeric plastomes in equimolar abundance (Palmer, 1983; Martin et al., 2014). Multiple studies have detected isomeric plastome structures caused by sIR in several conifers and legumes (Tsumura et al., 2000; Wu et al., 2011; Yi et al., 2013; Qu et al., 2017; Wang et al., 2018). We also confirmed the existence of isomers induced by a pair of 271 bp sIRs in L. maingayi ( Figure S5 ). Based on our findings, we conclude that smaller repeats indeed have played a role in enhancing plastome structural variation in the putranjivoid clade.
Table 4.
Number of repeats.
| Number of Repeats | Species | Repeat Length/Numbers | |||
|---|---|---|---|---|---|
| 30-60bp | 60-100bp | 100-500bp | >1000bp | ||
| 8 | Balanops balansae | 6 | 2 | 0 | 0 |
| 4 | Balanops pedicellata | 4 | 0 | 0 | 0 |
| 18 | Lophopyxis maingayi | 13 | 3 | 2 | 0 |
| 25 | Drypetes chevalieri | 19 | 3 | 2 | 1 |
| 23 | Drypetes diopa | 18 | 3 | 1 | 1 |
| 25 | Drypetes hainanensis | 18 | 3 | 3 | 1 |
| 37 | Drypetes indica | 20 | 10 | 6 | 1 |
| 26 | Drypetes lateriflora | 19 | 4 | 2 | 1 |
| 18 | Drypetes longifolia | 14 | 0 | 3 | 1 |
| 18 | Drypetes similis | 11 | 3 | 3 | 1 |
Data Availability Statement
The datasets generated for this study can be found in the GenBank Database, MN504788–MN504797.
Author Contributions
T-SY, J-JJ, and D-MJ designed the study. T-SY, J-BY, and D-MJ contributed to tissue sample collections, experiments, and sequences. D-MJ, J-JJ, and LG assembled the plastomes. D-MJ and J-JJ conducted the analysis. D-MJ, T-SY, SW, and J-JJ wrote and edited the manuscript; all authors commented on the manuscript.
Funding
This project was funded by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB31010000); the Large-scale Scientific Facilities of the Chinese Academy of Sciences (No. 2017-LSF-GBOWS-02); the National Natural Science Foundation of China [key international (regional) cooperative research project No. 31720103903]; and the open research project of “Cross-Cooperative Team” of the Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Missouri Botanical Garden for providing specimens and the Molecular Biology Experiment Center, Germplasm Bank of Wild Species in Southwest China for skillful laboratory assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00942/full#supplementary-material
References
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19 (5), 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett C. F., Baker W. J., Comer J. R., Conran J. G., Lahmeyer S. C., Leebens-Mack J. H., et al. (2016). Plastid genomes reveal support for deep phylogenetic relationships and extensive rate variation among palms and other commelinid monocots. New Phytol. 209 (2), 855–870. 10.1111/nph.13617 [DOI] [PubMed] [Google Scholar]
- Bedoya A. M., Ruhfel B. R., Philbrick C. T., Madriñán S., Bove C. P., Mesterházy A., et al. (2019). Plastid genomes of five species of riverweeds (Podostemaceae): structural organization and comparative analysis in Malpighiales. Front. Plant Sci. 10, 1035. 10.3389/fpls.2019.01035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazier J. C., Jansen R. K., Mower J. P., Govindu M., Zhang J., Weng M.-L., et al. (2016). Variable presence of the inverted repeat and plastome stability in Erodium . Ann. Bot. 117 (7), 1209–1220. 10.1093/aob/mcw065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. (2009). BLAST+: architecture and applications. BMC Bioinf. 10, 421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi I. S., Jansen R., Ruhlman T. (2019). Lost and found: return of the inverted repeat in the legume clade defined by its absence. Genome Biol. Evol. 11 (4), 1321–1333. 10.1093/gbe/evz076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumley T. W., Palmer J. D., Mower J. P., Fourcade H. M., Calie P. J., Boore J. L., et al. (2006). The complete chloroplast genome sequence of Pelargonium x hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 23 (11), 2175–2190. 10.1093/molbev/msl089 [DOI] [PubMed] [Google Scholar]
- Daniell H., Wurdack K. J., Kanagaraj A., Lee S. B., Saski C., Jansen R. K. (2008). The complete nucleotide sequence of the cassava (Manihot esculenta) chloroplast genome and the evolution of atpF in Malpighiales: RNA editing and multiple losses of a group II intron. Theor. Appl. Genet. 116 (5), 723–737. 10.1007/s00122-007-0706-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A. E., Mau B., Perna N. T. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PloS One 5 (6), e11147. 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. J., Doyle J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15. [Google Scholar]
- Goremykin V. V., Hirsch-Ernst K. I., Wolfl S., Hellwig F. H. (2003). Analysis of the Amborella trichopoda chloroplast genome sequence suggests that Amborella is not a basal angiosperm. Mol. Biol. Evol. 20 (9), 1499–1505. 10.1093/molbev/msg159 [DOI] [PubMed] [Google Scholar]
- Guisinger M. M., Kuehl J. V., Boore J. L., Jansen R. K. (2011). Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol. Biol. Evol. 28 (1), 583–600. 10.1093/molbev/msq229 [DOI] [PubMed] [Google Scholar]
- Haberle R. C., Fourcade H. M., Boore J. L., Jansen R. K. (2008). Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J. Mol. Evol. 66 (4), 350–361. 10.1007/s00239-008-9086-4 [DOI] [PubMed] [Google Scholar]
- Hirao T., Watanabe A., Kurita M., Kondo T., Takata K. (2008). Complete nucleotide sequence of the Cryptomeria japonica D. Don. chloroplast genome and comparative chloroplast genomics: diversified genomic structure of coniferous species. BMC Plant Biol. 8, 70. 10.1186/1471-2229-8-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R. K., Cai Z., Raubeson L. A., Daniell H., dePamphilis C. W., Leebens-Mack J., et al. (2007). Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. U. S. A. 104 (49), 19369–19374. 10.1073/pnas.0709121104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R. K., Wojciechowski M. F., Sanniyasi E., Lee S. B., Daniell H. (2008). Complete plastid genome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps12 and clpP intron losses among legumes (Leguminosae). Mol. Phylogenet. Evol. 48 (3), 1204–1217. 10.1016/j.ympev.2008.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J.-J., Yu W.-B., Yang J.-B., Song Y., dePamphilis C. W., Yi T.-S., et al. (2019). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. bioRxiv. 265470. 10.1101/256479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D.-M., Jin J.-J., Yi T.-S. (2020). Plastome structural conservation and evolution in the clusioid clade of Malpighiales. Sci. Rep. 10 (1), 9091. 10.1038/s41598-020-66024-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubitzki K. (2014). Flowering Plants Eudicots: Malpighiales (Heidelberg: Springer: Springer-Verlag Berlin; ), 247–276. [Google Scholar]
- Kurtz S., Choudhuri J. V., Ohlebusch E., Schleiermacher C., Stoye J., Giegerich R. (2001). REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29 (22), 4633–4642. 10.1093/nar/29.22.4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 (4), 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25 (16), 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes A. D., Pacheco T. G., dos Santos K. G., Vieira L. D., Guerra M. P., Nodari R. O., et al. (2018). The Linum usitatissimum L. plastome reveals atypical structural evolution, new editing sites, and the phylogenetic position of Linaceae within Malpighiales. Plant Cell Rep. 37 (2), 307–328. 10.1007/s00299-017-2231-z [DOI] [PubMed] [Google Scholar]
- Loytynoja A., Goldman N. (2008). Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science 320 (5883), 1632–1635. 10.1126/science.1158395 [DOI] [PubMed] [Google Scholar]
- Magee A. M., Aspinall S., Rice D. W., Cusack B. P., Semon M., Perry A. S., et al. (2010). Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Res. 20 (12), 1700–1710. 10.1101/gr.111955.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal A., Brisson N. (2010). Recombination and the maintenance of plant organelle genome stability. New Phytol. 186 (2), 299–317. 10.1111/j.1469-8137.2010.03195.x [DOI] [PubMed] [Google Scholar]
- Martin G. E., Rousseau-Gueutin M., Cordonnier S., Lima O., Michon-Coudouel S., Naquin D., et al. (2014). The first complete chloroplast genome of the Genistoid legume Lupinus luteus: evidence for a novel major lineage-specific rearrangement and new insights regarding plastome evolution in the legume family. Ann. Bot. 113 (7), 1197–1210. 10.1093/aob/mcu050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes A. P. A., Resende-Moreira L. C., Buzatti R. S. O., Nazareno A. G., Carlsen M., Lobo F. P., et al. (2018). Chloroplast genomes of Byrsonima species (Malpighiaceae): comparative analysis and screening of high divergence sequences. Sci. Rep. 8 (1), 2210. 10.1038/s41598-018-20189-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower J. P., Vickrey T. L. (2018). “Structural diversity among plastid genomes of land plants,” in Advances in botanical research. Ed. J.R. Chaw S. M. (Cambridge, MA, USA: Academic Press; ), 263–292. [Google Scholar]
- Palmer J. D., Thompson W. F. (1981). Rearrangements in the chloroplast genomes of mung bean and pea. Proc. Natl. Acad. Sci. U. S. A. 78 (9), 5533–5537. 10.1073/pnas.78.9.5533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J. D., Thompson W. F. (1982). Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell 29 (2), 537–550. 10.1016/0092-8674(82)90170-2 [DOI] [PubMed] [Google Scholar]
- Palmer J. D. (1983). Chloroplast DNA exists in two orientations. Nature 301, 92–93. 10.1038/301092a0 [DOI] [Google Scholar]
- Qu X.-J., Wu C.-S., Chaw S.-M., Yi T.-S. (2017). Insights into the existence of isomeric plastomes in Cupressoideae (Cupressaceae). Genome Biol. Evol. 9 (4), 1110–1119. 10.1093/gbe/evx071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X.-J., Moore M. J., Li D.-Z., Yi T.-S. (2019). PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 15, 50. 10.1186/s13007-019-0435-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röschenbleck J., Wicke S., Weinl S., Kudla J., Müller K. F. (2016). Genus-wide screening reveals four distinct types of structural plastid genome organization in Pelargonium (Geraniaceae). Genome Biol. Evol. 9, 64–76. 10.1093/gbe/evw271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabah S. O., Shrestha B., Hajrah N. H., Sabir M. J., Alharby H. F., Sabir M. J., et al. (2019). Passiflora plastome sequencing reveals widespread genomic rearrangements. J. Syst. Evol. 57 (1), 1–14. 10.1111/jse.12425 [DOI] [Google Scholar]
- Ruhlman T. A., Jansen R. K. (2014). “The Plastid Genomes of Flowering Plants,” in Chloroplast Biotechnology: Methods and Protocols. Ed. Maliga P. (New York: Springer; ), 3–38. [Google Scholar]
- Ruhlman T. A., Zhang J., Blazier J. C., Sabir J. S. M., Jansen R. K. (2017). Recombination-dependent replication and gene conversion homogenize repeat sequences and diversify plastid genome structure. Am. J. Bot. 104 (4), 559–572. 10.3732/ajb.1600453 [DOI] [PubMed] [Google Scholar]
- Sanderson M. J., Copetti D., Burquez A., Bustamante E., Charboneau J. L. M., Eguiarte L. E., et al. (2015). Exceptional reduction of the plastid genome of saguaro cactus (Carnegiea gigantea): loss of the ndh gene suite and inverted repeat. Am. J. Bot. 102 (7), 1115–1127. 10.3732/ajb.1500184 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30 (9), 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangphatsornruang S., Uthaipaisanwong P., Sangsrakru D., Chanprasert J., Yoocha T., Jomchai N., et al. (2011). Characterization of the complete chloroplast genome of Hevea brasiliensis reveals genome rearrangement, RNA editing sites and phylogenetic relationships. Gene 475 (2), 104–112. 10.1016/j.gene.2011.01.002 [DOI] [PubMed] [Google Scholar]
- Tesler G. (2002). GRIMM: genome rearrangements web server. Bioinformatics 18 (3), 492–493. 10.1093/bioinformatics/18.3.492 [DOI] [PubMed] [Google Scholar]
- Tillich M., Lehwark P., Pellizzer T., Ulbricht-Jones E. S., Fischer A., Bock R., et al. (2017). GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45 (W1), W6–W11. 10.1093/nar/gkx391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumura Y., Suyama Y., Yoshimura K. (2000). Chloroplast DNA inversion polymorphism in populations of Abies and Tsuga . Mol. Biol. Evol. 17 (9), 1302–1312. 10.1093/oxfordjournals.molbev.a026414 [DOI] [PubMed] [Google Scholar]
- Ueda M., Nishikawa T., Fujimoto M., Takanashi H., Arimura S., Tsutsumi N., et al. (2008). Substitution of the gene for chloroplast rps16 was assisted by generation of a dual targeting signal. Mol. Biol. Evol. 25 (8), 1566–1575. 10.1093/molbev/msn102 [DOI] [PubMed] [Google Scholar]
- Walker J. F., Jansen R. K., Zanis M. J., Emery N. C. (2015). Sources of inversion variation in the small single copy (SSC) region of chloroplast genomes. Am. J. Bot. 102 (11), 1751–1752. 10.3732/ajb.1500299 [DOI] [PubMed] [Google Scholar]
- Wang Y.-H., Wicke S., Wang H., Jin J.-J., Chen S.-Y., Zhang S.-D., et al. (2018). Plastid genome evolution in the early-diverging legume subfamily Cercidoideae (Fabaceae). Front. Plant Sci. 9, 138. 10.3389/fpls.2018.00138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M.-L., Ruhlman T. A., Jansen R. K. (2016). Expansion of inverted repeat does not decrease substitution rates in Pelargonium plastid genomes. New Phytol. 214 (2), 842–851. 10.1111/nph.14375 [DOI] [PubMed] [Google Scholar]
- Wick R. R., Schultz M. B., Zobel J., Holt K. E. (2015). Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31 (20), 3350–3352. 10.1093/bioinformatics/btv383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S., Schneeweiss G. M., dePamphilis C. W., Muller K. F., Quandt D. (2011). The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 76 (3-5), 273–297. 10.1007/s11103-011-9762-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S., Müller K. F., de Pamphilis C. W., Quandt D., Wickett N. J., Zhang Y., et al. (2013). Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell 25 (10), 3711–3725. 10.1105/tpc.113.113373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-S., Lin C.-P., Hsu C.-Y., Wang R.-J., Chaw S.-M. (2011). Comparative chloroplast genomes of Pinaceae: insights into the mechanism of diversified genomic organizations. Genome Biol. Evol. 3, 309–319. 10.1093/gbe/evr026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurdack K. J., Davis C. C. (2009). Malpighiales phylogenetics: gaining ground on one of the most recalcitrant clades in the angiosperm tree of life. Am. J. Bot. 96 (8), 1551–1570. 10.3732/ajb.0800207 [DOI] [PubMed] [Google Scholar]
- Xi Z.-X., Ruhfel B. R., Schaefer H., Amorim A. M., Sugumaran M., Wurdack K. J., et al. (2012). Phylogenomics and a posteriori data partitioning resolve the Cretaceous angiosperm radiation Malpighiales. Proc. Natl. Acad. Sci. U. S. A. 109 (43), 17519–17524. 10.1073/pnas.1205818109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X., Gao L., Wang B., Su Y.-J., Wang T. (2013). The complete chloroplast genome sequence of Cephalotaxus oliveri (Cephalotaxaceae): evolutionary comparison of Cephalotaxus chloroplast DNAs and insights into the loss of inverted repeat copies in gymnosperms. Genome Biol. Evol. 5 (4), 688–698. 10.1093/gbe/evt042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Wang Y.-H., Jin J.-J., Stull G. W., Bruneau A., Cardoso D., et al. (2020). Exploration of plastid phylogenomic conflict yields new insights into the deep relationships of Leguminosae. Syst. Biol. 69 (4), 613–622. 10.1093/sysbio/syaa013 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in the GenBank Database, MN504788–MN504797.