Abstract
Imbalances of cellular proteostasis are linked to ageing and human diseases, including neurodegenerative and neuromuscular diseases. Heat shock proteins (HSPs) and small heat shock proteins (sHSPs) together form a crucial core of the molecular chaperone family that plays a vital role in maintaining cellular proteostasis by shielding client proteins against aggregation and misfolding. sHSPs are thought to act as the first line of defence against protein unfolding/misfolding and have been suggested to act as “sponges” that rapidly sequester these aberrant species for further processing, refolding, or degradation, with the assistance of the HSP70 chaperone system. Understanding how these chaperones work at the molecular level will offer unprecedented insights for their manipulation as therapeutic avenues for the treatment of ageing and human disease. The evolution in single-molecule force spectroscopy techniques, such as optical tweezers (OT) and atomic force microscopy (AFM), over the last few decades have made it possible to explore at the single-molecule level the structural dynamics of HSPs and sHSPs and to examine the key molecular mechanisms underlying their chaperone activities. In this paper, we describe the working principles of OT and AFM and the experimental strategies used to employ these techniques to study molecular chaperones. We then describe the results of some of the most relevant single-molecule manipulation studies on HSPs and sHSPs and discuss how these findings suggest a more complex physiological role for these chaperones than previously assumed.
Keywords: Heat shock proteins, Small heat shock proteins, Single-molecule manipulation, Structural dynamics, Mechanism of action
Introduction
The proper folding of proteins is essential for life (Klaips et al. 2018). However, protein folding into their native state is constantly challenged by several errors that affect the fidelity in the flow of genetic information from DNA to RNA and, finally, to proteins. These errors can take place during DNA replication or transcription, as well as during translation, and they result in the synthesis of mutated and/or truncated polypeptides that cannot fold into their native state, and therefore are unstable and prone to aggregation (Kapur and Ackerman 2018). If not readily detected by the molecular chaperones and destroyed by the degradation systems, such as the ubiquitin proteasome system (UPS) and autophagy, these aberrant polypeptides or fragments of them can impair protein homeostasis (proteostasis). Loss of proteostasis, in turn, leads to cell dysfunction and toxicity and is considered as an important player in ageing and diseases, ranging from cancer to neurodegeneration and myopathies (Shibata and Morimoto 2014; Klaips et al. 2018).
Molecular chaperones such as the heat shock proteins (HSPs) and the degradation systems together constitute the so-called protein quality control (PQC) system. The PQC surveys the cellular proteome and avoids the accumulation of unfolded/misfolded proteins by assisting the refolding of aberrant peptides into the native state or, when this is not possible, by hiding interactive surfaces on protein aggregates and, finally, by promoting the proteasome or autophagy-mediated clearance of aberrant polypeptides; these concerted actions avoid the irreversible aggregation and accumulation of aberrant polypeptides inside the cells (Bukau et al. 2006; Chen et al. 2011; Hartl et al. 2011; Balchin et al. 2016). Due to its essential functions in the maintenance of the cellular proteome, it is not surprising that defects and imbalances of the PQC, including HSPs, are associated with the development of neurodegenerative diseases; conversely, boosting the activity of chaperones and degradative machineries has proven to be efficient in delaying disease progression in several cellular and animal models of neurodegenerative diseases (Labbadia and Morimoto 2015; Klaips et al. 2018). Thus, understanding at the molecular level how the PQC works holds promise for the design of therapeutic strategies against ageing and neurodegeneration (Hipp et al. 2019).
In this paper, we will focus our attention on the experimental attempts that have been made in the past decades to understand at the molecular level how HSPs, in particular the small HSPs, avoid protein misfolding and aggregation with the ultimate goal of finding strategies to exploit their chaperone activity for cell health and fitness. We will put emphasis on the principle and application of single-molecule manipulation using optical tweezers and atomic force microscopy for the study of the chaperone activity of HSPs and small HSPs.
HSPs and small HSPs
HSPs were discovered more than 40 years ago by Dr. Alfred Tissieres (Tissiéres et al. 1974). After their identification in Drosophila melanogaster, HSPs were found in archaea, bacteria and eukaryotes. The HSPs identified have been classified according to their molecular weight into families: HSP100, HSP90, HSP70, HSP60, HSP40 and small HSPs, the latter characterised by a molecular weight ranging from 15 to 30 kDa (Caspers et al. 1995; Bult et al. 1996; Hartl et al. 2011). In the last decades, the combination of several experimental approaches, ranging from in vitro studies with recombinant purified HSPs to gene expression profiling, knockdown and overexpression screening in cells and animal models, such as for example D. melanogaster and C. elegans allowed us to gain a deeper understanding of the mechanisms of action of HSPs and the identification of the cellular processes that are regulated by HSPs. Advanced mass spectrometry studies that revealed the existence of extensive chaperone–client networks in cells, combined with knockdown/knockout studies, were particularly instrumental in gathering information on the biological processes that are modulated by HSPs both in resting cells and following exposure to stress. Thanks to these studies, it became clear that HSPs regulate, directly or indirectly, nearly all aspects of cell biology in a large variety of organisms, from bacterial cells, to yeast and human cells, but also in plants and mammals (Bukau et al. 2006; Hartl et al. 2011; Schopf et al. 2017). This is because HSPs can bind to many different proteins and regulate protein function and activity by facilitating protein folding, the assembly of multiprotein complexes, as well as interaction of ligands with their targets or receptors, such as in the case of HSP90 (Schopf et al. 2017). In parallel, thanks to the use of nuclear magnetic resonance spectroscopy, Förster resonance energy transfer (FRET), electron microscopy and crystallography, as well as cross-linking experiments followed by mass spectrometric analysis, we gained detailed knowledge on how HSPs work at the molecular level, as well as on how their conformational changes and their binding affinities to specific clients are regulated. For example, these techniques enabled to describe the conformational transitions of the HSP90 cycle and how specific combination of co-chaperones associate with specific HSP90 conformational states (Hessling et al. 2009); alternatively, they enabled to demonstrate the cooperation between chaperones such as HSP70 and HSP90 and how, at the molecular level, this is stabilised by the binding of the substrate, enabling its transfer from one chaperone to another (Genest et al. 2015). Cross-linking experiments followed by mass spectrometry analysis shed light on the mechanisms regulating substrate recognition by the small HSPs that, in contrast to higher molecular weight chaperones such as HSP70 and HSP90, are ATP-independent (de Jong et al. 1993; Kappé et al. 2003).
The fact that small HSP chaperone activity does not require ATP might become extremely important under conditions of energy deprivation and upon stress conditions, when cells need to prioritize use the energy towards cell stress response/adaptation and survival. The term caloristasis has been coined to complement proteostasis and these are considered as two critical components of cellular metabolic homeostasis (Bonorino et al. 2018). Caloristasis, a concept that emphasizes the integrative regulatory interactions needed to understand cellular energy homeostasis, is discussed in more detail in the chapter herein by Tezgin et al. Against this background, the ATP-independent chaperone functions of small HSPs during periods of depleted cellular energy reserves take on even greater significance.
One essential feature of small HSPs is that their monomers associate to form large oligomers with a dimeric substructure; thus, the homodimers formed via the association of the conserved alpha-crystallin domain of small HSPs are considered as the building blocks for the assembly of larger oligomers (Haslbeck et al. 2019). The oligomers of small HSPs are thought to act as a reservoir of the chaperone power; in fact, it is the dynamic dissociation of small HSPs into monomers and dimers that governs their binding affinities for the substrates, and therefore their chaperone function (Haslbeck et al. 2019). The interaction between small HSPs and their substrates involves several binding sites that are located in the conserved alpha-crystallin domain but extend to the less conserved N-terminal region (van Montfort et al. 2001). This would explain how different small HSPs may specifically bind to different substrates (reviewed in Haslbeck et al. 2019). Concerning the chaperone activity of small HSPs, the use of relaxation dispersion and high-pressure nuclear magnetic resonance spectroscopy recently contributed to shed light on the enigmatic mechanism of action of small HSPs. These experiments showed that upon dissociation of the oligomers, the monomers are released and undergo partial unfolding of the conserved alpha-crystallin domain interface; this event seems to be critical to enhance the chaperone activity of small HSPs. Although so far this model has been demonstrated for human Hsp27/HSPB1, considering the high degree of conservation of the alpha-crystallin domain interface between the various small HSPs, it is likely that increased disorder in the small HSP monomers represents a general mechanism that increases their chaperone activity (Alderson et al. 2019).
Although the combination of different techniques allowed us to tremendously improve our understanding of the function of chaperones at the molecular level, the finer mechanisms underlying the structural dynamics and the regulatory actions of HSPs and small HSPs are still partly unresolved. This lack of information is mostly due to the inability of traditional ensemble methods to describe in detail the phenomena that are intrinsically, highly heterogeneous. The structural dynamics and the functions of proteins, such as HSPs and small HSPs, are ultimately determined by their conformational equilibria. By diffusing on a rugged energy landscape, the structure of a protein fluctuates at equilibrium between a large variety of conformations that collectively define its interactome and thus, its functional profile. Some of these conformations are highly populated and thermodynamically stable; others are transient and less probable; others originates when the protein interacts with its own substrate. Regardless of their origin, structural properties and energetics, any of these conformations might play a key role in the functional cycle of a protein and deserves an accurate characterization. Unfortunately, a detailed description of the structural dynamics of a protein with bulk techniques is a daunting task as these methods provide a signal that is the ensemble average of the contributions of a large and often de-phased population of molecules, where the many different conformational transition pathways and transient molecular states are not resolved. The application of single-molecule manipulation methods, such as optical tweezers and atomic force microscopy, to the study of molecular chaperones has recently opened new avenues of research, allowing us to go beyond the ensemble average and uncover information that is not accessible to more traditional techniques. In these experiments, individual proteins are tethered between movable surfaces and watched fluctuating at equilibrium between different conformations along a well-defined reaction coordinate, namely the molecular end-to-end distance (Cecconi et al. 2005; Bertz et al. 2008; Stigler et al. 2011; Ritchie and Woodside 2015; Choudhary et al. 2019). These studies allow a detailed description of the energy landscapes underlying folding and unfolding transitions and an accurate estimate of the kinetics and thermodynamics of these reactions. Providing non-averaged information, these single-molecule techniques also permit the identification and characterisation of less probable molecular species and a detailed description of the role they play in a protein’s functional cycle.
In this paper, we will illustrate the results of single-molecule manipulation studies aimed at elucidating the structural dynamics and the mechanisms of action of different HSPs and sHSPs.
Structural, mechanical and functional properties of HSPs
The experimental approach typically used in optical tweezers experiments to mechanically manipulate a single protein is depicted in Fig. 1a (Jahn et al. 2014). The protein is connected to two optically trapped beads by means of DNA molecular handles that act as spacers to avoid non-specific interactions between the tethering surfaces (Wen et al. 2007; Cecconi et al. 2008; Cecconi et al. 2011; Pfitzner et al. 2013). Each DNA molecule is covalently attached to a cysteine residue of the protein at one end and to a bead (typically made of polystyrene or glass) at the other end through biotin/streptavidin or digoxygenine/antibodies interactions. During the experiments, the protein is stretched and relaxed by varying the distance between the beads while recording the applied force, the molecular extension and the time. The applied force, which acts as denaturant in these experiments, can be varied almost linearly with time (constant-speed experiments) (Heidarsson et al. 2012; Caldarini et al. 2014; Mandal et al. 2017), or it can be kept constant at a pre-set value through a feedback mechanism (force-constant experiments) (Shank et al. 2010; Heidarsson et al. 2013). In other cases, the distance between the two optical traps is kept constant, and the molecule is watched fluctuating between different molecular states at a constant average force (constant-distance measurements) (Gao et al. 2011; Neupane et al. 2016). Under tension, a denaturation or renaturation event is accompanied by an increase or a decrease in the end-to-end distance of the protein; this is because unfolded molecular states are more compliant and thus more extended than folded molecular states. These changes in extension generate detectable discontinuities in the recorded traces that allow us to follow the structural changes of a protein.
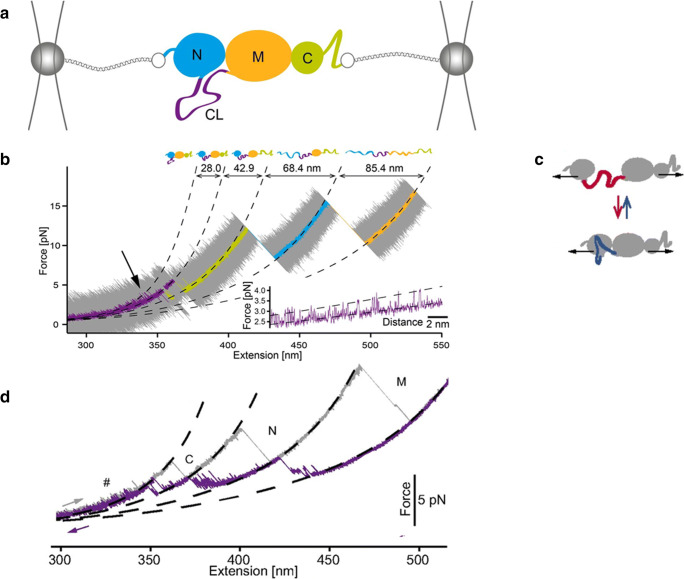
Fig. 1.
Experimental set-up used to manipulate single Hsp82 monomers with optical tweezers (Jahn et al. 2014; Jahn et al. 2016; Jahn et al. 2018). a The monomer comprises a N-terminal ATP–binding domain (N, 211 residues), a middle domain (M, 266 residues) responsible for client binding and a C-terminal domain (C, 172 residues) responsible mainly for dimerization. The N- and M-domains are connected to each other by a charged linker (CL), shown in purple. For mechanical manipulation, the protein is attached between two glass beads by means of two DNA molecular handles. By varying the position of one of the two traps, the force applied on Hsp82 monomer can be varied. b Stretching trace obtained by pulling on a Hsp82 monomer at constant speed. The original data (20 kHz) were filtered to obtain smooth a trace showing three major transitions (rips) corresponding to the sequential unfolding of the C-, N- and M-domain, respectively. The inset shows fluctuations of Hsp82 monomer at low forces due to hopping of the CL between a compact and an extended conformation, as schematically shown in panel c). d Stretching (in grey) and relaxation (in purple) cycle acquired by pulling and relaxing a Hsp82 monomer. The relaxation trace shows various fluctuations corresponding to an ensemble of intermediate states. Reprinted with permission from PNAS, (Jahn et al. 2014; Jahn et al. 2016)
Optical tweezers were employed to decipher structural and dynamics aspects of the chaperone Hsp82 (Jahn et al. 2014; Jahn et al. 2018), a eukaryote orthologue of HSP90. Hsp82 is a dimer with each monomer comprising three domains (N-, M- and C-domain) connected by linkers (Fig. 1a). In particular, the N- and M-domains are connected via a 61-amino acid charged linker (CL) that has so far eluded any structural characterization, suggesting a flexible and disordered conformation for this protein region (Ali et al. 2006; Hainzl et al. 2009). When a monomer of Hsp82 is mechanically stretched, it unfolds through three major steps, corresponding to the sequential unfolding of the C-, N- and M-domain respectively (Fig. 1b). Interestingly, and importantly, the denaturation of the C domain is preceded at low forces by fluctuations of Hsp82 between two molecular states that differ in length by about 28 nm; this length is very similar to that of CL (purple line and inset in Fig. 1b). To better understand the origin of these fluctuations and the role played by CL, mutants were generated in which extended parts of CL were replaced by unstructured glycine-glycine-serine repetitions. No substitution mutants displayed low force fluctuations, suggesting that they originate from reversible folding and unfolding transitions of CL. Performing further experiments with mutants where different protein regions were deleted, the authors showed that indeed CL hops at low forces between a close and an open conformation, interacting with the N-domain (Fig. 1c). When the CL folds, the D-domain docks to the N-domain, and when it opens up, the two domains separate. By analysing the effect of force on the docking–undocking equilibrium, the authors showed that at zero force the molecule spends 75% of the time in the docked state and 25% of the time in the undocked state. What the biological function is of these structural fluctuations at the interface between the N- and C domains is not clear yet; surely, they increase the structural flexibility of the protein, and thus, they might play an important role during the functional cycle of the chaperone.
After its mechanical denaturation, as the applied force is lowered, Hsp82 regains its native structure through a complicated folding process comprising an ensemble of intermediate states (Fig. 1d), (Jahn et al. 2016). One of these partially folded conformations is an on-pathway intermediate state populated by the M-domain, likely corresponding to the so-called smaller alpha/beta/alpha subdomain. All the others are off-pathways (misfolded) intermediate states that both the M- and N-domain populate during their journey to the native state. These misfolding events hinder proper folding, slowing down the productive folding rate constant of Hsp82 by an order of magnitude. The smallest of the three domains instead, the C-domain, folds last through a simple two-state process that involves no detectable intermediate states. These results highlight the complexity of the folding process of a large protein such as Hsp82 and the multitude of conformations that it can populate.
Building on these intriguing results, the folding and unfolding processes of other two HSP90 orthologues, i.e. the bacterial HtpG and the eukaryotic Grp94, were characterised with optical tweezers and compared with Hsp82 (Jahn et al. 2018). Overall, the structural dynamics of HtpG and Grp94 are similar to that of Hsp82, but some important differences exist. The mechanical denaturation of HtpG and Grp94 also involve three major events corresponding to the sequential unfolding of the C-, N- and M-domain respectively, with similar unfolding forces and changes in contour length to those measured for Hsp82. Similar to Hsp82, during folding, these two chaperones populate an ensemble of misfolded states that collectively slow down productive folding. However, some important differences exist in the behaviour of the charged linkers, especially that of Grp94. In fact, this liker, unlike that of Hsp82, at zero force does not fluctuate between an open and a close conformation. Instead, it folds into a compact and thermodynamically stable structure (8.7 kBT) that under tension unfolds through a distinct transition at ~ 7–8 pN, which immediately precedes the denaturation of the N-domain. Through constant-distance measurements, the authors show that at zero force this linker is almost always in its folded state and, thus, the N- and M-domains of Grp94 are always very close to each other with limited rotational freedom. This more rigid N/M-domain interface in Grp94 might have functional consequences and might prevent this chaperone from interacting with some Hsp82 protein clients. Concerning HtpG, it should be noticed that its CL is only 7-amino acid long, and so, the conformational freedom of the N- and M-domain of this chaperone is also quite limited compared with that of Hsp82.
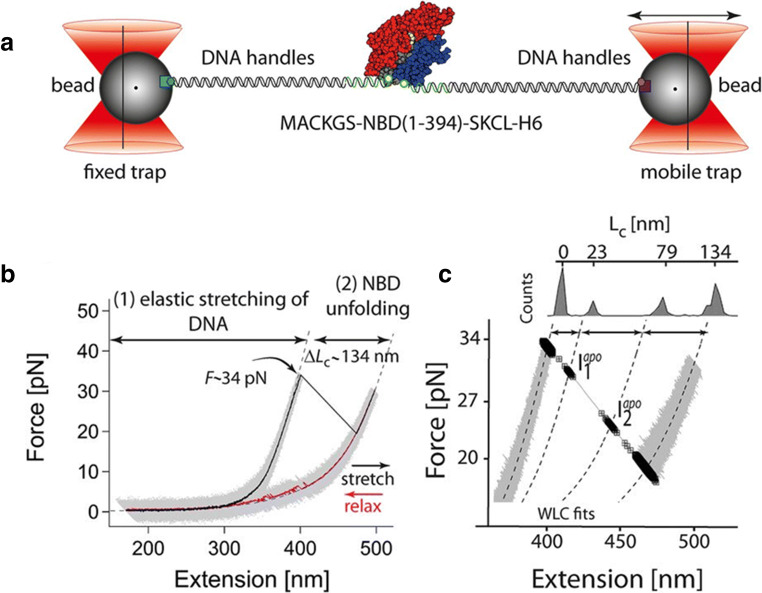
In the same vein, Bauer et al. investigated the mechanical stability of the nucleotide-binding domain (NBD) of the DnaK chaperone from E. coli, a bacterial homologue of human HSP70, in the presence and in the absence of ATP (Fig. 2) (Bauer et al. 2015). The NBD of DnaK is primarily sub-divided into two lobes, termed lobe I and lobe II, connected together by a C-terminal helix. Each lobe is composed of two sub-domains—Ia, Ib for lobe I and IIa, IIb for lobe II. Upon mechanical manipulation and in the absence of ATP, the NBD of DnaK denatures into a completely unfolded polypeptide chain by populating two short-lived intermediate states, named Iapo1 and Iapo2 (Fig. 2 b and c). By selectively varying the lengths of the different subdomains of NBD through loop insertions, the authors showed that the first unfolding event corresponds to the unravelling of the C-terminal helix and the concomitant separation of lobe I from lobe II, leading to Iapo1. As stretching proceeds, the first lobe I unfolds, generating Iapo2, followed by the denaturation of lobe II that completes NBD denaturation. In the presence of ATP, the unfolding pathway of NBD changes dramatically, as lobe II becomes mechanically more stable and the lobe-lobe interactions are stabilised. In fact, under tension, the holo-NBD unfolds by populating three intermediates states that all differ from those observed with the apo-form. The first unfolding event, leading to the intermediate state Iholo1, originates from the unravelling of the C-terminal helix that, in this case, does not induce the concomitant separation of the two lobes, which instead split at higher extensions through a transition that generates the intermediate state Iholo2. As stretching proceeds, firstly, lobe II unfolds (leading to Iholo3), and then lobe I unfolds, completing the mechanical denaturation of the protein. Nucleotide binding thus reverses the mechanical hierarchy between the two lobes of NBD, making lobe II the most mechanically resistant. This is not so surprising if we consider that most of the NBD residues interacting with ATP belong to lobe II (13 total) and only a few residues belong to lobe I (Kityk et al. 2012). Moreover, ATP binding strengthens the interaction between the two domains that, in the holo-form, remain connected after the denaturation of the C-terminal helix; their splitting requires further pulling. In summary, the results of this study provide unique insights into the structural and energetic changes that nucleotide binding can induce in a heat shock protein and provide hints on how a signal can get transmitted through a protein structure.
Fig. 2.
Mechanical manipulation of DnaK NBD in the absence of ATP (Bauer et al. 2015). As the applied force is increased, the apo-form of NBD unfolds at ~ 35 pN (b), through a process involving two short lived intermediate states, Iapo1and Iapo2, (c). Reprinted with permission from PNAS, (Bauer et al. 2015)
In a follow-up study, Bauer and co-authors (Bauer et al. 2018) elucidated the folding mechanism of the NBD of E. coli DnaK. Through single-molecule optical tweezers experiments, the authors show that, upon relaxation of the applied force, a denatured NBD molecule refolds into its native structure through a hierarchical multistep process. This multistep process involves the sequential folding of subdomain IIb, then of whole lobe II and, eventually, of lobe I. Through a careful analysis of the recorded traces, the authors show that along the NBD-folding pathway, the fully folded lobe II represents an obligatory on-pathway intermediate state that acts as an internal chaperone for the folding of lobe I; if lobe II does not reach its native state, lobe I folding fails. While lobe I cannot fold independently, lobe II can fold into a stable protein even in isolation, displaying nucleotide-binding ability and native-like folding behaviour. To better understand the role played by lobe II in the driving of the folding process of E. coli NBD, the authors pulled on the yeast mitochondrial NBD that, unlike its prokaryotic counterpart, does not fold in vitro unless it is helped by dedicated chaperones. They showed that replacement of the yeast lobe II with the E. coli lobe II rescues the folding-incompetent yeast NBP, which now can fold effectively even in the absence of chaperones. The results described in (Bauer et al. 2018) clearly show the importance of lobe II for the folding and nucleotide binding of the E. coli NBP and shed light on the molecular basis of the different folding behaviours displayed by the various members of the sugar kinase family.
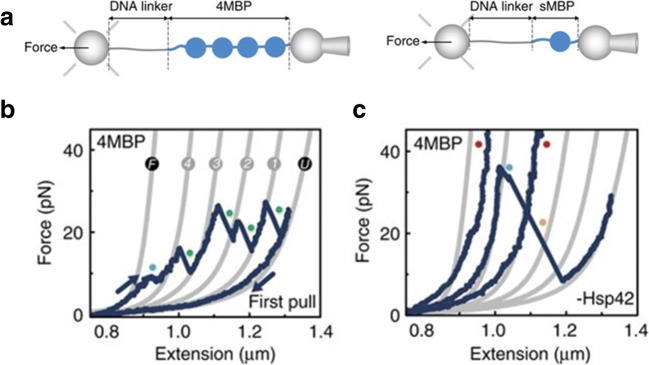
Optical tweezers assays are also well suited to discern the functional activity of chaperone proteins in vitro at the single-molecule level. The experimental approach that has so far been adopted for these studies is illustrated in Fig. 3. A homotetramer of maltose-binding protein from E. coli (4MBP) has been identified as a viable and versatile substrate for these experiments (Bechtluft et al. 2007; Mashaghi et al. 2013; Mashaghi et al. 2016); this is because MBP has a strong propensity to aggregate in vitro and is a client to a wide variety of chaperones, including, e.g. SecB, GroEL and the yeast small HSP Hsp42 (Scholz et al. 1996; Bechtluft et al. 2007). The homotetrameric structure mimics a condition of molecular crowding, where neighbouring domains can interact with each other and aggregate into aberrant molecular structures. In these experiments, a single 4MBP is subject to sequential stretching-relaxation cycles, where the force is increased to denature the molecule and then decreased to allow refolding. During refolding, the neighbouring MBP domains interact with each other and give rise to aggregates that in the subsequent stretching trace generate transitions of variable size and unfolding forces. Figure 3a shows the first stretching-relaxation cycle for a 4MBP (Ungelenk et al. 2016). The stretching trace show discontinuities at 8–9 pN (first blue dot), due to unfolding of mechanically weak helical segments, followed by four unfolding peaks, in a force range between 15 and 30 pN; the latter correspond to the sequential denaturation of the core structure of each MBP domain (subsequent 4 blue dots). This native-like unfolding pattern is visible only in the first stretching trace, as the subsequent stretching traces show non-reproducible discontinuities that correspond to the mechanical denaturation of aggregates of variable size and mechanical resistance. Rarely, in ~ 10% of cases, a MBD domain avoids aggregation and folds into its native state giving rise to a native-like unfolding transition in the subsequent stretching trace. The folding and aggregation processes of individual 4MBPs can be studied in the absence or in the presence of a HSP, and the corresponding results can then be compared to decipher the molecular mechanisms underlying the chaperone activity.
Fig. 3.
Experimental set-up to probe the functional activity of chaperone proteins (Ungelenk et al. 2016). a Four MBP molecules connected end to end and the single MBP molecule are tethered in between two beads via a DNA linker. One bead is kept in a stationary spot by a micro-pipette while the other bead is trapped in the optical trap allowing for the induction of denaturation in the MBP molecule and measurement of applied force. b First unfolding and refolding cycle of the MBP homotetramer. The grey lines represent the WLC fitting of the unfolding rips. The first observed unfolding at low forces (F to 4) represents the untangling of the alpha helical structures from the MBP core. This is followed by four distinct rips (4 to 3, 3 to 2, 2 to 1 and 1 to U) corresponding to the unfolding of the core of each MBP molecule with U representing the completely unstructured homotetramer. The 4MBP molecule is then relaxed by gradually reducing the applied force (last blue line) giving the molecule a chance to fold back in their native states. (C) Subsequent pulling cycles in the absence of chaperone showing aggregated structures unfolding at higher forces (blue dot) or not unfolding at all (red dots). Adapted with permission from (Ungelenk et al. 2016) via Creative Commons Attribution 4.0 International License
This experimental approach was employed by Mashaghi et al. to investigate the chaperone mechanism of the bacterial Hsp70 homologue DnaK (Mashaghi et al. 2016). In Mashaghi et al. (2016), the authors show that when 4MBP is pulled in the presence of the DnaK chaperone system (DnaK, DnaJ, GrpE, ADP), the homotetramer displays far less tendency for aggregation than in the absence of the chaperone. Rather, an increased presence of unfolding events corresponding to the denaturation of single MBP cores and minor aggregates was observed. These results indicate a strong holdase action of DnaK, as well as its propensity to promote native folding, which is in line with previous data obtained from bulk assays (Skowyra et al. 1990; Schröder et al. 1993; Szabo et al. 1994). To decipher the underlying chaperone mechanisms, the authors simplified the experimental assay and characterised the effect of DnaK on the folding process of single MBP domains (sMBP). The results of these experiments show that DnaK binds both to denatured polypeptide chains, preventing protein refolding, and to partially folded native-like protein structures, stabilizing them mechanically. OT studies can be performed using the full-length protein or truncated versions of it, allowing us to study the functions performed by various parts of the chaperone. In addition to the nucleotide-binding domain described above, DnaK comprises a substrate-binding domain (SBD)) containing an α-helical subdomain that acts as a lid and a β-sheet subdomain that accommodates the peptide binding pocket (Mayer et al. 2000). To shed light on the roles played by the lid and the peptide-binding pocket, experiments similar to those described above were performed using a truncated lid variant, DnaK (2–538), and a binding groove mutant, DnaK (V436F). The lid-truncated variant could bind to an unfolded sMBP and prevent its refolding but could not stabilise folded structures. By contrast, the groove-mutated variant was unable to hinder refolding of sMBP, but managed to stabilise (although weakly) near-native conformations. These data indicate that while the groove plays a key role in the DnaK-substrate interaction, the lid is necessary for the stabilization of folded structures. In light of these results, the authors surmised that DnaK counters aggregation by binding not only to extended peptides but also to partially folded, near-native conformations, stabilizing them. They also extended upon the canonical model of DnaK action by suggesting that DnaK can help guide partially folded structures to native conformations by limiting inter-domain interactions, staying bound to them until they are close to a native conformation.
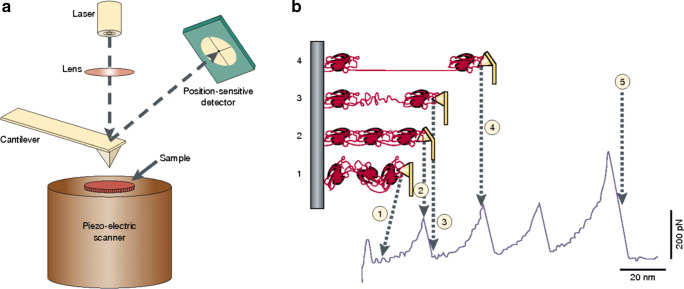
The structural and functional properties of proteins can also be studied with atomic force microscopy. An atomic force microscope is typically used to image the topography of a sample. However, it can also be employed for other purposes, for example to measure molecular forces. In this mode of operation, called “force spectroscopy”, single molecules or pairs of interacting molecules are stretched between an AFM tip and a substrate and the forces generated by their elongation measured at pN level (Fig. 4a) (Bustamante et al. 2000). Specifically, the mechanical properties of proteins are usually studied by pulling on a polyprotein, a molecule made of a linear array of a globular domain, which is stretched between a flat surface, typically made of gold, and a silicon nitrite AFM tip (Rief et al. 1997; Carrion-Vazquez et al. 1999; Yang et al. 2000; Dietz and Rief 2004; Sandal et al. 2008; Perales-Calvo et al. 2018). The need for polymeric proteins in these experiments comes from the necessity of keeping the tethering surfaces far enough from each other in order to avoid short-range tip-surface interactions. As the tip is pulled away from the surface, the tension along the polymer increases and the single domains start unfolding sequentially, producing a characteristic saw tooth-like pattern in the force-extension curves, where each peak corresponds to the unfolding of one domain (Fig. 4b). A careful analysis of the recorded trace allows us to characterise the unfolding pathway of the stretched protein in terms of different parameters, including (i) unfolding forces of tertiary and secondary structures, (ii) unfolding activation barriers and their position along the reaction coordinate and (iii) presence of intermediate states and estimation of their structural features.
Fig. 4.
Mechanical manipulation of a polyprotein with an atomic force microscope (Bustamante et al. 2000). a Schematic representation of an atomic force microscope. The sample is mounted on a piezo-electric scanner that can change the position of the sample relative to the AFM tip, which is integrated at the end of a flexible cantilever. The force applied on the tip is measured by monitoring the deflection of the cantilever through an “optical lever” made of a laser and a position-sensitive detector. b Mechanical denaturation of a polyprotein. As the distance between the tip and the surface increases (from state 1 to state 2), the molecule extends and generates a restoring force that bends the cantilever. At a certain point, one domain stochastically unfolds generating an increase of the molecule contour length that makes the cantilever relax and the force drop (state 3). As the stretching of the polymer continues, the force raises again until another domain unfolds generating another drop in force. At the end of the mechanical denaturation of the polymer, the corresponding stretching trace will be characterised by a saw tooth-like pattern where each peak corresponds to the denaturation of one domain. Reprinted with permission from Springer Nature Customer Service Centre GmbH: Springer Nature, Nature Reviews Molecular Cell Biology (Bustamante et al. 2000), Copyright 2000
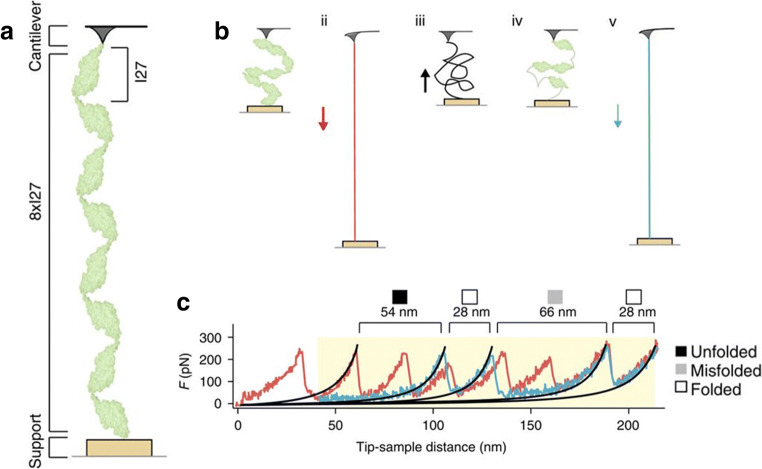
Atomic force microscopy was employed by Nunes et al. to study the chaperone activities of DnaJ and DnaK (Nunes et al. 2015). The authors used an octomer of I27 domains (8xI27) as a substrate for these experiments since 8xI27 is prone to misfolding and aggregation when manipulated with an AFM tip (Fig. 5a) (Wright et al. 2005; Borgia et al. 2011). In these studies, a single I27 octomer is subject to multiple stretching-relaxation cycles in which first the polymer is stretched and mechanically denatured, then the applied tension is decreased to low force values for 5 s to allow refolding and, finally, the polymer is stretched and denatured again (Fig. 5b). During the 5-s relaxation time, a denatured I27 domain can fold into its native state, misfold into an aberrant structure or remain unfolded. Each of these three possible conformations can be easily recognised in the subsequent stretching trace (Fig. 5c), thereby allowing for a statistical analysis of the refolding process of I27 (Fig. 6).
Fig. 5.
Mechanical manipulation of a I27 octomer (8xI27), (Nunes et al. 2015). a A I27 octomer is attached between an AFM tip and a gold surface. b Schematic of the experimental strategy used to study the refolding process of mechanically denatured I27 domains. The I27 octomer is first stretched and mechanically denatured by moving the gold surface away from the tip (I and ii). Then, the surface is approached back towards the tip to lower the applied tension and allow refolding (iii and iv). Finally, the polymer is mechanically unfolded again to probe the refolding process of the denatured I27 domains (v). c The first stretching trace, showing a characteristic saw tooth-like pattern corresponding to the mechanical denaturation of I27 native states, is shown in red. The second stretching trace (panel v in B), acquired after force relaxation, is instead shown in blue and presents different types of transitions corresponding to (i) stretching of unfolded I27 domains (filled square), denaturation of I27 native states (empty square) and denaturation of I27 misfolded states (grey square). Reprinted with permission from Springer Nature Customer Service Centre GmbH: Springer Nature, Nature Communications (Nunes et al. 2015), Copyright 2015
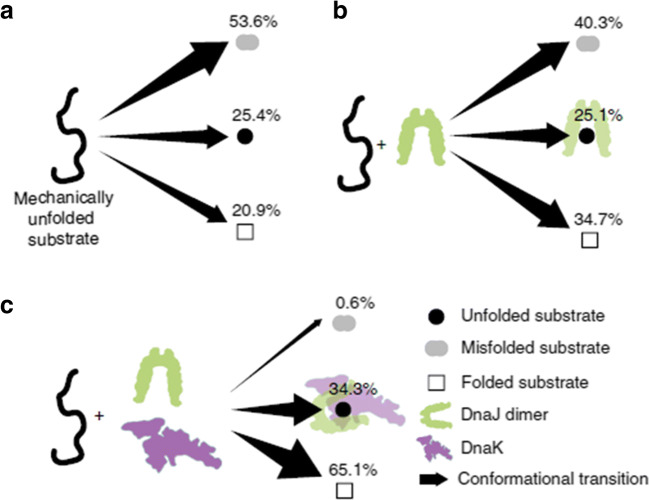
Fig. 6.
Refolding process of a denatured I27 domain upon relaxation of the applied force, under different experimental conditions (Nunes et al. 2015). a In the absence of chaperones, the unfolded domain mostly misfolds. b In the presence of DnaJ, the I27 domain misfolds less and transits into its native state more often, while the probability of remaining unfolded does not change. c In the presence of DnaJ and DnaK (1:2 M ratio), the I27 domain folds into its native state most of the time, or alternatively remains unfolded. Misfolding is almost abolished. Reprinted with permission from Springer Nature Customer Service Centre GmbH: Springer Nature, Nature Communications (Nunes et al. 2015), Copyright 2015
In the absence of a chaperone, ~ 54% of I27 domains misfolds, ~ 21% folds into their native state and the rest remain unfolded. In the presence of DnaJ, these percentages change. In fact, denatured I27 domains misfold less (~ 40%) and transit into their native states more often (~ 35%), while the fraction of unfolded conformers remains the same. These data reveal that DnaJ, under these experimental conditions, presents an unexpected foldase activity. If we consider that DnaJ binds to denatured polypeptide chains (REF) and that its presence does not change the size of the misfolded states observed in the stretching traces, it seems unlikely that it interacts directly with misfolded states facilitating their direct conversion into native states. Instead, the hypothesis that after binding to denatured I27 domains DnaJ limits their conformational space, guiding them preferentially towards their native states seems more likely. In cells, DanJ cooperates with DnaK. It is thus possible to extend the study from one single chaperone to chaperone complexes. When DnaJ and DnaK are both present in a 1:2 M ratio, the folding behaviour of I27 is affected even more: under these conditions, almost no proteins misfold (~ 1%), and the rest either transit into the native state (~ 65%) or remain unfolded (~ 34%). Thus, compared with the no chaperone condition, the chaperone pair DnaJ/DnaK, through synergic mechanisms, stabilises significantly both the native and the unfolded states, thereby, hindering misfolding and aggregation of the substrate protein (Fig. 6).
Structural, mechanical and functional properties of small HSPs
Small HSPs are characterised by their ability to dynamically change their structure and oligomerization state, which has been reported to influence their chaperone activity (Ito et al. 1997; Delbecq and Klevit 2013; Peschek et al. 2013; Haslbeck et al. 2019). As a consequence, experimental techniques, such as optical tweezers and atomic force microscopy that allow us to monitor in real time the conformational changes that a molecule undergoes spontaneously or as a consequence of the interaction with its own substrate, are particularly indicated for the study of these chaperones.
Expanding upon their previous optical tweezers studies on DnaK, the group of Prof. Tans characterised the chaperone action of the small heat shock protein HSP42 (Ungelenk et al. 2016). Strong holdase, as well as some foldase action for HSP42 was reported by analysing its impact on 4MBP. The authors noticed that large and mechanically resistant aggregates of MBP disappeared in the presence of HSP42. Instead, they observed an appreciable increase in the number of native-like core unfolding events, along with transitions corresponding to the denaturation of small and weakly aggregated structures. From these results, the authors hypothesised that HSP42 hinders non-native interactions in 4MBP by forming a complex with near-natively folded MBP structures, and shielding them from aggregation partners; alternatively, but not mutually exclusive, HSP42 disrupts large aggregated structure and allows the protein to fold back in its native conformation, or to aggregate weakly. To learn more about the molecular mechanism underlying its chaperone action, the authors characterised the effect of HSP42 on single MBP domains (sMBP). These studies revealed that HSP42 does not interact with the native state, nor with the denatured state of sMBP (the substrate protein), but its presence increases the probability of observing native-like core unfolding. These unfolding events, however, as in the case of 4MBP, occurred at lower forces than in the absence of HSP42 and hence correspond to the denaturation of native-like molecular structures that are less stable than the native state. These results prompted the authors to conclude that HSP42 carries out its chaperone activity by binding to partially folded, close to native conformations and prevent them from aggregating any further. It can then propel these bound structures towards their native conformations by hindering non-native interactions. It is also possible that this chaperone actively discourages aggregation by guiding the folding protein in a less mechanically stable native-like state, which is not available to the MBP protein in the absence of HSP42. This will require that HSP42 alters the energy landscape of the target protein during the folding state. Although no conclusive evidence is provided for this hypothesis, further single-molecule studies of HSP42 could help us better understand the molecular mechanisms that mediate the biological function of this chaperone. These data paint a more complicated mechanism of action for small HSPs, which extends well-beyond small HSPs acting as “sponges” that promiscuously bind to heterogeneous substrates to avoid their aggregation. Although they are considered as “holdases”, a term that implies a rather passive function, these data suggest that small HSPs may actively favour protein folding in an ATP-independent manner. Future studies using OT and extending the characterization to different small HSPs from several species should unravel this hypothesis and may provide surprising data that can potentially change the way the scientific community looks at small HSPs.
Atomic force microscopy was employed by Bertz et al. (2010) to characterise the mechanical stability of the α-crystallin domain dimer of the small heat shock protein Hsp16.5, from the hyperthermophilic archaeon Methanocaldococcus jannaschii. The α-crystallin domain dimer represents the building block of the three-dimensional structure of Hsp16.5, comprising 24 subunits arranged into a spherical homo-oligomer. Bertz et al. characterised the disassembly and unfolding of the α-crystallin domain dimer through both single-molecule force spectroscopy and fluorescence bulk experiments, where they monitored GdmCl-induced equilibrium unfolding transitions. The results of these latter studies indicate that the disassembly and assembly processes of the dimer are strictly coupled to the unfolding and refolding of the monomers, so that, through an all-or-none process, the system goes directly from a structured dimer to denatured polypeptide chains and vice versa, with no population of intermediate folded monomeric species. Single-molecule manipulation studies provided different information. A first series of experiments were performed by pulling on a molecular construct where the C terminus of each 16.5α was connected to a molecular handle comprising three filamin domains (ddFLN3–5) (Schwaiger et al. 2004) and three titin domains (I27–29) (Li et al. 2002). These domains can easily be distinguished one from another in a force-extension curve by both their contour lengths and their unfolding forces, with the ddFLN3–5 unfolding below 120 pN and I27–29 unfolding at ~ 230 pN. A typical stretching trace recorded in these experiments is characterised by a sawtooth-like pattern, corresponding to the sequential unfolding of filamin domains, followed by a final force peak at ~ 180 pN, which was interpreted as the rupture force of the dimer that dissociates before force could unfold the titin domains. The interpretation of these data was corroborated by the results of similar experiments in which force was applied to different molecular constructs, where the two 16.5α monomers are flanked by three ubiquitin domains, acting as handles for mechanical manipulation, and are connected to each other through an ubiquitin trimer linker that prevents dissociation of the stretched molecular construct upon disassembly of the dimer. The resulting force-extension curves showed the characteristic sawtooth-like unfolding signature of the ubiquitin domains interrupted by a longer unfolding event, which produces an increment in contour length that is consistent with that expected for the complete unfolding of the 16.5α dimer. A closer inspection of the recorded traces shows that the longer unfolding event in reality is a three-step transition process, where two intermediate states are populated. The first transition takes place at a force (183 ± 4 pN) that is quite similar to that of the final force peak observed in the abovementioned experiments, and produces an increment in contour length (9.2 ± 0.4 nm) that corresponds well to that expected for the dissociation of the 16.5α dimer. The second and third transitions generate increments in contour length that are consistent with the sequential denaturation of the two isolated α monomers. Thus, these results indicate that a 16.5α dimer can be split into two stable fully folded monomers, which subsequently can unfold independently of each other. These findings are in contrast to those emerging from chemical unfolding bulk experiments that suggest that no folded monomeric intermediates are populated during dimer disassembly. This may appear surprising, but in reality, discrepancies between chemical and mechanical denaturation have already been reported (Bertz and Rief 2008). Surely, the force spectroscopy data collected by Bertz et al. demonstrate the efficacy of single-molecule manipulation studies in spotting and characterizing molecular species that are difficult to detect with ensemble methods. On the basis of the data illustrated in this study, we can hypothesise that assembly of the three-dimensional structure of Hsp16.5 begins with the folding of polypeptide chains into stable monomeric species that later associate into stable dimers that eventually assemble into the final oligomeric conformation.
Conclusions
Recently, optical tweezers and atomic force microscopy have been successfully applied to the study of structural and functional properties of molecular chaperones. Some of the most significant experimental results obtained with HSPs and sHSPs using these techniques are illustrated in this paper. Optical tweezers have been effectively employed to decipher the intricacy of the multistate conformational dynamics of Hsp82, HtpG and Grp94, revealing an ensemble of on- and off-pathways intermediate states that these chaperones populate during their journey towards the native state (Jahn et al. 2014, 2016, 2018). Particular attention was paid to the characterization of the structural dynamics of the charged linker (CL) connecting the N- and M-domains of these chaperones. The results of these studies show that the CL of Hsp82, unlike that of HtpG and Grp94, fluctuates at equilibrium between a closed and an open conformation, providing a structural flexibility to Hsp82 that might play a key role during its interaction with the client proteins. Using a similar experimental strategy, Bauer and co-authors characterised in great detailed the unfolding and refolding pathways of the nucleotide-binding domain (NBD) of DnaK chaperone, assessing also the effect of nucleotide binding to the energy landscape of the chaperone (Bauer et al. 2015). These studies reveal a mechanical hierarchy between NBD structural lobes that is reversed in the presence of ATP, and disclose the role played by lobe II in the driving of the productive folding process of NBD. OT studies were also applied to decipher the mechanism of action of the small heat shock protein HSP42, revealing an unexpected foldase activity that the chaperone seems to carry out by binding to partially folded close to native conformations of the substrate, rather than to its unfolded state (Bauer et al. 2018). Similarly, AFM was used to study the structural and functional properties of different chaperones. In particular, AFM was employed to characterise the mechanical properties of the α-crystallin domain dimer of the small heat shock protein Hsp16.5, revealing that it can be split into two stable fully folded monomers, in contrast to what was suggested by chemical denaturation bulk studies (Bertz et al. 2010). Moreover, through AFM force spectroscopy the molecular mechanisms underlying the chaperone activities of DnaJ and DnaK were explored in great detail (Nunes et al. 2015).
While excellent details have been extracted through the application of these techniques to the study of biomolecules, OT and AFM are to a certain extent limited in their approach by the fact that they probe a molecular process along a single reaction coordinate, which is determined by the points of force application (Choudhary et al. 2019). As a consequence, any structural transition of the molecule under examination that does not produce an appreciable change in molecular extension along the pulling axis is not detected in these experiments. One way to move past this hurdle is to perform multiple pulling assays with varied attachment points to extract structural dynamics information across different reactions coordinates (Elms et al. 2012; Heidarsson et al. 2012). Also, in case of optical tweezers, multi-trap systems are now available, which can overcome the single-dimensional probing limit (Dame et al. 2006). Alternatively, novel hybrid approaches of combining fluorescence-based imaging techniques with single-molecule manipulation methods can be employed to provide a multidimensional picture of the molecular processes under study (Sirinakis et al. 2012; Lee and Hohng 2013).
In conclusion, key aspects of the structural and functional properties of HSPs and small HSPs have been revealed recently through single-molecule manipulation experiments. However, this field of research is still in its infancy and great room for improvement exists both for the instrumentation and for the methods used to prepare biological samples. It is therefore reasonable to assume that these types of experiments will produce even more surprising and relevant discoveries in the near future.
Finally, the comparison at the single-molecule level of the function of wild-type small HSPs with their disease-causing mutation counterparts may illuminate on how they affect their functionality, via a gain or loss-of-function. This knowledge will be essential for the design of any potential therapeutic approach for the treatment of neuromuscular diseases, myopathies, cardiomyopathies and congenital cataract that have been so far associated with mutations in human genes encoding for small HSPs, such as, e.g. HSPB1, HSPB3, HSPB4, HSPB5 and HSPB8 (Dierick et al. 2005; Boncoraglio et al. 2012; Benndorf et al. 2014).
Funding information
S.C and C.C. acknowledge funding by Italian Ministry of University and Research (MIUR), Departments of excellence 2018–2022; E91I18001480001, PRIN—Progetti di Ricerca di Interesse Nazionale (2017 EX_ALS) and University of Modena and Reggio Emilia (FAR 2016).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Serena Carra, Email: serena.carra@unimore.it.
Ciro Cecconi, Email: ciro.cecconi@unimore.it.
References
- Alderson TR, Roche J, Gastall HY, Dias DM, Pritišanac I, Ying J, Bax A, Benesch JL, Baldwin AJ. Local unfolding of the HSP27 monomer regulates chaperone activity. Nat Commun. 2019;10(1):1068. doi: 10.1038/s41467-019-08557-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH. Crystal structure of an Hsp90–nucleotide–p23/Sba1 closed chaperone complex. Nature. 2006;440(7087):1013. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balchin D, Hayer-Hartl M, Hartl FU. In vivo aspects of protein folding and quality control. Science. 2016;353(6294):aac4354. doi: 10.1126/science.aac4354. [DOI] [PubMed] [Google Scholar]
- Bauer D, Merz DR, Pelz B, Theisen KE, Yacyshyn G, Mokranjac D, Dima RI, Rief M, Žoldák G. Nucleotides regulate the mechanical hierarchy between subdomains of the nucleotide binding domain of the Hsp70 chaperone DnaK. Proc Natl Acad Sci. 2015;112(33):10389–10394. doi: 10.1073/pnas.1504625112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer D, Meinhold S, Jakob RP, Stigler J, Merkel U, Maier T, Rief M, Žoldák G. A folding nucleus and minimal ATP binding domain of Hsp70 identified by single-molecule force spectroscopy. Proc Natl Acad Sci. 2018;115(18):4666–4671. doi: 10.1073/pnas.1716899115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtluft P, Van Leeuwen RG, Tyreman M, Tomkiewicz D, Nouwen N, Tepper HL, Driessen AJ, Tans SJ. Direct observation of chaperone-induced changes in a protein folding pathway. Science. 2007;318(5855):1458–1461. doi: 10.1126/science.1144972. [DOI] [PubMed] [Google Scholar]
- Benndorf R, Martin JL, Pond SLK, Wertheim JO. Neuropathy-and myopathy-associated mutations in human small heat shock proteins: characteristics and evolutionary history of the mutation sites. Mutat Res Rev Mutat Res. 2014;761:15–30. doi: 10.1016/j.mrrev.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertz M, Rief M. Mechanical unfoldons as building blocks of maltose-binding protein. J Mol Biol. 2008;378(2):447–458. doi: 10.1016/j.jmb.2008.02.025. [DOI] [PubMed] [Google Scholar]
- Bertz M, Kunfermann A, Rief M. Navigating the folding energy landscape of green fluorescent protein. Angew Chem Int Ed. 2008;47(43):8192–8195. doi: 10.1002/anie.200802987. [DOI] [PubMed] [Google Scholar]
- Bertz M, Chen J, Feige MJ, Franzmann TM, Buchner J, Rief M. Structural and mechanical hierarchies in the α-crystallin domain dimer of the hyperthermophilic small heat shock protein Hsp16. 5. J Mol Biol. 2010;400(5):1046–1056. doi: 10.1016/j.jmb.2010.05.065. [DOI] [PubMed] [Google Scholar]
- Boncoraglio A, Minoia M, Carra S. The family of mammalian small heat shock proteins (HSPBs): implications in protein deposit diseases and motor neuropathies. Int J Biochem Cell Biol. 2012;44(10):1657–1669. doi: 10.1016/j.biocel.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Bonorino C, Sistonen L, Eriksson J, Mezger V, Santoro G, Hightower LE (2018). The VIII international congress on stress proteins in biology and medicine: täynnä henkeä. Cell Stress Chaperones 23(2): 171–177 [DOI] [PMC free article] [PubMed]
- Borgia MB, Borgia A, Best RB, Steward A, Nettels D, Wunderlich B, Schuler B, Clarke J. Single-molecule fluorescence reveals sequence-specific misfolding in multidomain proteins. Nature. 2011;474(7353):662. doi: 10.1038/nature10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125(3):443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, Gocayne JD. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273(5278):1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- Bustamante C, Macosko JC, Wuite GJ. Grabbing the cat by the tail: manipulating molecules one by one. Nat Rev Mol Cell Biol. 2000;1(2):130–136. doi: 10.1038/35040072. [DOI] [PubMed] [Google Scholar]
- Caldarini M, Sonar P, Valpapuram I, Tavella D, Volonté C, Pandini V, Vanoni M, Aliverti A, Broglia R, Tiana G. The complex folding behavior of HIV-1-protease monomer revealed by optical-tweezer single-molecule experiments and molecular dynamics simulations. Biophys Chem. 2014;195:32–42. doi: 10.1016/j.bpc.2014.08.001. [DOI] [PubMed] [Google Scholar]
- Carrion-Vazquez M, Oberhauser AF, Fowler SB, Marszalek PE, Broedel SE, Clarke J, Fernandez JM. Mechanical and chemical unfolding of a single protein: a comparison. Proc Natl Acad Sci. 1999;96(7):3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspers G-J, Leunissen JA, de Jong WW. The expanding small heat-shock protein family, and structure predictions of the conserved “α-crystallin domain”. J Mol Evol. 1995;40(3):238–248. doi: 10.1007/BF00163229. [DOI] [PubMed] [Google Scholar]
- Cecconi C, Shank EA, Bustamante C, Marqusee S. Direct observation of the three-state folding of a single protein molecule. Science. 2005;309(5743):2057–2060. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- Cecconi C, Shank EA, Dahlquist FW, Marqusee S, Bustamante C. Protein-DNA chimeras for single molecule mechanical folding studies with the optical tweezers. Eur Biophys J. 2008;37(6):729–738. doi: 10.1007/s00249-007-0247-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi C, Shank EA, Marqusee S, Bustamante C (2011) DNA molecular handles for single-molecule protein-folding studies by optical tweezers. DNA Nanotechnology, Springer 255–271 [DOI] [PubMed]
- Chen B, Retzlaff M, Roos T, Frydman J. Cellular strategies of protein quality control. Cold Spring Harb Perspect Biol. 2011;3(8):a004374. doi: 10.1101/cshperspect.a004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary D, Mossa A, Jadhav M, Cecconi C. Bio-molecular applications of recent developments in optical tweezers. Biomolecules. 2019;9(1):23. doi: 10.3390/biom9010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame RT, Noom MC, Wuite GJ. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature. 2006;444(7117):387–390. doi: 10.1038/nature05283. [DOI] [PubMed] [Google Scholar]
- de Jong WW, Leunissen JA, Voorter C. Evolution of the alpha-crystallin/small heat-shock protein family. Mol Biol Evol. 1993;10(1):103–126. doi: 10.1093/oxfordjournals.molbev.a039992. [DOI] [PubMed] [Google Scholar]
- Delbecq SP, Klevit RE. One size does not fit all: the oligomeric states of αB crystallin. FEBS Lett. 2013;587(8):1073–1080. doi: 10.1016/j.febslet.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierick I, Irobi J, De Jonghe P, Timmerman V. Small heat shock proteins in inherited peripheral neuropathies. Ann Med. 2005;37(6):413–422. doi: 10.1080/07853890500296410. [DOI] [PubMed] [Google Scholar]
- Dietz H, Rief M. Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proc Natl Acad Sci. 2004;101(46):16192–16197. doi: 10.1073/pnas.0404549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elms PJ, Chodera JD, Bustamante C, Marqusee S. The molten globule state is unusually deformable under mechanical force. Proc Natl Acad Sci. 2012;109(10):3796–3801. doi: 10.1073/pnas.1115519109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Sirinakis G, Zhang Y. Highly anisotropic stability and folding kinetics of a single coiled coil protein under mechanical tension. J Am Chem Soc. 2011;133(32):12749–12757. doi: 10.1021/ja204005r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genest O, Hoskins JR, Kravats AN, Doyle SM, Wickner S. Hsp70 and Hsp90 of E. coli directly interact for collaboration in protein remodeling. J Mol Biol. 2015;427(24):3877–3889. doi: 10.1016/j.jmb.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainzl O, Lapina MC, Buchner J, Richter K. The charged linker region is an important regulator of Hsp90 function. J Biol Chem. 2009;284(34):22559–22567. doi: 10.1074/jbc.M109.031658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475(7356):324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Haslbeck M, Weinkauf S, Buchner J. Small heat shock proteins: simplicity meets complexity. J Biol Chem. 2019;294(6):2121–2132. doi: 10.1074/jbc.REV118.002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidarsson PTO, Valpapuram I, Camilloni C, Imparato A, Tiana G, Poulsen FM, Kragelund BB, Cecconi C. A highly compliant protein native state with a spontaneous-like mechanical unfolding pathway. J Am Chem Soc. 2012;134(41):17068–17075. doi: 10.1021/ja305862m. [DOI] [PubMed] [Google Scholar]
- Heidarsson PO, Otazo MR, Bellucci L, Mossa A, Imparato A, Paci E, Corni S, Di Felice R, Kragelund BB, Cecconi C. Single-molecule folding mechanism of an EF-hand neuronal calcium sensor. Structure. 2013;21(10):1812–1821. doi: 10.1016/j.str.2013.07.022. [DOI] [PubMed] [Google Scholar]
- Hessling M, Richter K, Buchner J. Dissection of the ATP-induced conformational cycle of the molecular chaperone Hsp90. Nat Struct Mol Biol. 2009;16(3):287–293. doi: 10.1038/nsmb.1565. [DOI] [PubMed] [Google Scholar]
- Hipp MS, Kasturi P, Hartl FU (2019) The proteostasis network and its decline in ageing. Nat Rev Mol Cell Biol 1 [DOI] [PubMed]
- Ito H, Okamoto K, Nakayama H, Isobe T, Kato K. Phosphorylation of αB-crystallin in response to various types of stress. J Biol Chem. 1997;272(47):29934–29941. doi: 10.1074/jbc.272.47.29934. [DOI] [PubMed] [Google Scholar]
- Jahn M, Rehn A, Pelz B, Hellenkamp B, Richter K, Rief M, Buchner J, Hugel T. The charged linker of the molecular chaperone Hsp90 modulates domain contacts and biological function. Proc Natl Acad Sci. 2014;111(50):17881–17886. doi: 10.1073/pnas.1414073111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn M, Buchner J, Hugel T, Rief M. Folding and assembly of the large molecular machine Hsp90 studied in single-molecule experiments. Proc Natl Acad Sci. 2016;113(5):1232–1237. doi: 10.1073/pnas.1518827113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn M, Tych K, Girstmair H, Steinmaßl M, Hugel T, Buchner J, Rief M. Folding and domain interactions of three orthologs of Hsp90 studied by single-molecule force spectroscopy. Structure. 2018;26(1):96–105. doi: 10.1016/j.str.2017.11.023. [DOI] [PubMed] [Google Scholar]
- Kappé G, Franck E, Verschuure P, Boelens WC, Leunissen JA, de Jong WW. The human genome encodes 10 α-crystallin–related small heat shock proteins: HspB1–10. Cell Stress Chaperones. 2003;8(1):53–61. doi: 10.1379/1466-1268(2003)8<53:thgecs>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur M, Ackerman SL. mRNA translation gone awry: translation fidelity and neurological disease. Trends Genet. 2018;34(3):218–231. doi: 10.1016/j.tig.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kityk R, Kopp J, Sinning I, Mayer MP. Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol Cell. 2012;48(6):863–874. doi: 10.1016/j.molcel.2012.09.023. [DOI] [PubMed] [Google Scholar]
- Klaips CL, Jayaraj GG, Hartl FU. Pathways of cellular proteostasis in aging and disease. J Cell Biol. 2018;217(1):51–63. doi: 10.1083/jcb.201709072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J, Morimoto RI. The biology of proteostasis in aging and disease. Annu Rev Biochem. 2015;84:435–464. doi: 10.1146/annurev-biochem-060614-033955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Hohng S. An optical trap combined with three-color FRET. J Am Chem Soc. 2013;135(49):18260–18263. doi: 10.1021/ja408767p. [DOI] [PubMed] [Google Scholar]
- Li H, Linke WA, Oberhauser AF, Carrion-Vazquez M, Kerkvliet JG, Lu H, Marszalek PE, Fernandez JM. Reverse engineering of the giant muscle protein titin. Nature. 2002;418(6901):998–1002. doi: 10.1038/nature00938. [DOI] [PubMed] [Google Scholar]
- Mandal SS, Merz DR, Buchsteiner M, Dima RI, Rief M, Žoldák G. Nanomechanics of the substrate binding domain of Hsp70 determine its allosteric ATP-induced conformational change. Proc Natl Acad Sci. 2017;114(23):6040–6045. doi: 10.1073/pnas.1619843114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashaghi A, Kramer G, Bechtluft P, Zachmann-Brand B, Driessen AJ, Bukau B, Tans SJ. Reshaping of the conformational search of a protein by the chaperone trigger factor. Nature. 2013;500(7460):98. doi: 10.1038/nature12293. [DOI] [PubMed] [Google Scholar]
- Mashaghi A, Bezrukavnikov S, Minde DP, Wentink AS, Kityk R, Zachmann-Brand B, Mayer MP, Kramer G, Bukau B, Tans SJ. Alternative modes of client binding enable functional plasticity of Hsp70. Nature. 2016;539(7629):448–451. doi: 10.1038/nature20137. [DOI] [PubMed] [Google Scholar]
- Mayer MP, Schröder H, Rüdiger S, Paal K, Laufen T, Bukau B. Multistep mechanism of substrate binding determines chaperone activity of Hsp70. Nat Struct Mol Biol. 2000;7(7):586. doi: 10.1038/76819. [DOI] [PubMed] [Google Scholar]
- Neupane K, Foster DA, Dee DR, Yu H, Wang F, Woodside MT. Direct observation of transition paths during the folding of proteins and nucleic acids. Science. 2016;352(6282):239–242. doi: 10.1126/science.aad0637. [DOI] [PubMed] [Google Scholar]
- Nunes JM, Mayer-Hartl M, Hartl FU, Müller DJ. Action of the Hsp70 chaperone system observed with single proteins. Nat Commun. 2015;6:6307. doi: 10.1038/ncomms7307. [DOI] [PubMed] [Google Scholar]
- Perales-Calvo J, Giganti D, Stirnemann G, Garcia-Manyes S. The force-dependent mechanism of DnaK-mediated mechanical folding. Sci Adv. 2018;4(2):eaaq0243. doi: 10.1126/sciadv.aaq0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschek J, Braun N, Rohrberg J, Back KC, Kriehuber T, Kastenmüller A, Weinkauf S, Buchner J. Regulated structural transitions unleash the chaperone activity of αB-crystallin. Proc Natl Acad Sci. 2013;110(40):E3780–E3789. doi: 10.1073/pnas.1308898110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfitzner E, Wachauf C, Kilchherr F, Pelz B, Shih WM, Rief M, Dietz H. Rigid DNA beams for high-resolution single-molecule mechanics. Angew Chem Int Ed. 2013;52(30):7766–7771. doi: 10.1002/anie.201302727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276(5315):1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- Ritchie DB, Woodside MT. Probing the structural dynamics of proteins and nucleic acids with optical tweezers. Curr Opin Struct Biol. 2015;34:43–51. doi: 10.1016/j.sbi.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandal M, Valle F, Tessari I, Mammi S, Bergantino E, Musiani F, Brucale M, Bubacco L, Samorì B. Conformational equilibria in monomeric α-synuclein at the single-molecule level. PLoS Biol. 2008;6(1):e6. doi: 10.1371/journal.pbio.0060006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz C, Zarnt T, Kern G, Lang K, Burtscher H, Fischer G, Schmid FX. Autocatalytic folding of the folding catalyst FKBP12. J Biol Chem. 1996;271(22):12703–12707. doi: 10.1074/jbc.271.22.12703. [DOI] [PubMed] [Google Scholar]
- Schopf FH, Biebl MM, Buchner J. The HSP90 chaperone machinery. Nat Rev Mol Cell Biol. 2017;18(6):345–360. doi: 10.1038/nrm.2017.20. [DOI] [PubMed] [Google Scholar]
- Schröder H, Langer T, Hartl F, Bukau B. DnaK, DnaJ and GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage. EMBO J. 1993;12(11):4137–4144. doi: 10.1002/j.1460-2075.1993.tb06097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger I, Kardinal A, Schleicher M, Noegel AA, Rief M. A mechanical unfolding intermediate in an actin-crosslinking protein. Nat Struct Mol Biol. 2004;11(1):81–85. doi: 10.1038/nsmb705. [DOI] [PubMed] [Google Scholar]
- Shank EA, Cecconi C, Dill JW, Marqusee S, Bustamante C. The folding cooperativity of a protein is controlled by its chain topology. Nature. 2010;465(7298):637–640. doi: 10.1038/nature09021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Morimoto RI. How the nucleus copes with proteotoxic stress. Curr Biol. 2014;24(10):R463–R474. doi: 10.1016/j.cub.2014.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirinakis G, Ren Y, Gao Y, Xi Z, Zhang Y. Combined versatile high-resolution optical tweezers and single-molecule fluorescence microscopy. Rev Sci Instrum. 2012;83(9):093708. doi: 10.1063/1.4752190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Georgopoulos C, Zylicz M. The E. coli dnaK gene product, the hsp70 homolog, can reactivate heat-inactivated RNA polymerase in an ATP hydrolysis-dependent manner. Cell. 1990;62(5):939–944. doi: 10.1016/0092-8674(90)90268-j. [DOI] [PubMed] [Google Scholar]
- Stigler J, Ziegler F, Gieseke A, Gebhardt JCM, Rief M. The complex folding network of single calmodulin molecules. Science. 2011;334(6055):512–516. doi: 10.1126/science.1207598. [DOI] [PubMed] [Google Scholar]
- Szabo A, Langer T, Schröder H, Flanagan J, Bukau B, Hartl FU. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc Natl Acad Sci. 1994;91(22):10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissiéres A, Mitchell HK, Tracy UM. Protein synthesis in salivary glands of Drosophila melanogaster: relation to chromosome puffs. J Mol Biol. 1974;84(3):389–398. doi: 10.1016/0022-2836(74)90447-1. [DOI] [PubMed] [Google Scholar]
- Ungelenk S, Moayed F, Ho C-T, Grousl T, Scharf A, Mashaghi A, Tans S, Mayer MP, Mogk A, Bukau B. Small heat shock proteins sequester misfolding proteins in near-native conformation for cellular protection and efficient refolding. Nat Commun. 2016;7:13673. doi: 10.1038/ncomms13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Montfort RL, Basha E, Friedrich KL, Slingsby C, Vierling E. Crystal structure and assembly of a eukaryotic small heat shock protein. Nat Struct Mol Biol. 2001;8(12):1025. doi: 10.1038/nsb722. [DOI] [PubMed] [Google Scholar]
- Wen J-D, Manosas M, Li PT, Smith SB, Bustamante C, Ritort F, Tinoco I., Jr Force unfolding kinetics of RNA using optical tweezers. I. Effects of experimental variables on measured results. Biophys J. 2007;92(9):2996–3009. doi: 10.1529/biophysj.106.094052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CF, Teichmann SA, Clarke J, Dobson CM. The importance of sequence diversity in the aggregation and evolution of proteins. Nature. 2005;438(7069):878–881. doi: 10.1038/nature04195. [DOI] [PubMed] [Google Scholar]
- Yang G, Cecconi C, Baase WA, Vetter IR, Breyer WA, Haack JA, Matthews BW, Dahlquist FW, Bustamante C. Solid-state synthesis and mechanical unfolding of polymers of T4 lysozyme. Proc Natl Acad Sci. 2000;97(1):139–144. doi: 10.1073/pnas.97.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]