Abstract
The discovery of genetic causes of inherited skin disorders has been pivotal to the understanding of epidermal differentiation, function, and renewal. Here we show via exome sequencing that mutations in ASPRV1 (aspartic peptidase retroviral-like 1) cause a dominant Mendelian disorder featuring palmoplantar keratoderma and lamellar ichthyosis, a phenotype that has otherwise been exclusively recessive. ASPRV1 encodes a mammalian-specific and stratified epithelia-specific protease important in processing of filaggrin, a critical component of the uppermost epidermal layer. Three different heterozygous ASPRV1 missense mutations in four unrelated ichthyosis kindreds segregate with disease and disrupt protein residues within close proximity to each other and autocatalytic cleavage sites. Expression of mutant ASPRV1 proteins demonstrates that all three mutations alter ASPRV1 auto-cleavage and filaggrin processing, a function vital to epidermal barrier integrity.
Keywords: ASPRV1, SASPase, ichthyosis, keratoderma, skin, epidermis, dominant, Mendelian, exome, de novo
Main text
Mendelian disorders of cornification are severe skin disorders that feature localized or generalized scaling and redness, with significant morbidity and mortality. Extensive genetic heterogeneity is evidenced by the observation that mutations in more than 50 genes have been shown to cause disorders of cornification, and yet approximately 15% of cases remain unsolved.
The Yale Human Investigation Committee approved the study protocol, and participants provided verbal and written informed consent. In the course of exome sequencing, a cohort of subjects negative for pathogenic mutations in previously described cornification disorder genes, five affected members of a kindred were found to be heterozygous for the ASPRV1 mutation c.932G>C, encoding p.Arg311Pro (GenBank: NM_152792). This was the only coding mutation that both co-segregated with the disorder in seven members of the extended family (Kindred 292) and was not found in databases of human genetic variation (Figure 1A and Table S1). Subsequently, members of three unrelated kindreds were also found to be heterozygous for mutations in ASPRV1 (Figures 1B–1D and Table S1). In kindred 1055, the proband and his affected mother and brother are heterozygous for c.595A>G, encoding p.Lys199Glu. This mutation is absent from the unaffected maternal grandparents and thus arose de novo in the proband’s mother. In kindreds 630 and 1099, the probands are heterozygous for c.940C>A, encoding p.Pro314Thr. This mutation is absent from the unaffected parents of subject 1099 and thus arose de novo in this subject. The three mutations are not described in the literature, they occur at highly conserved residues (Figure S1), and they are predicted to be damaging (Table S1).1
Figure 1.
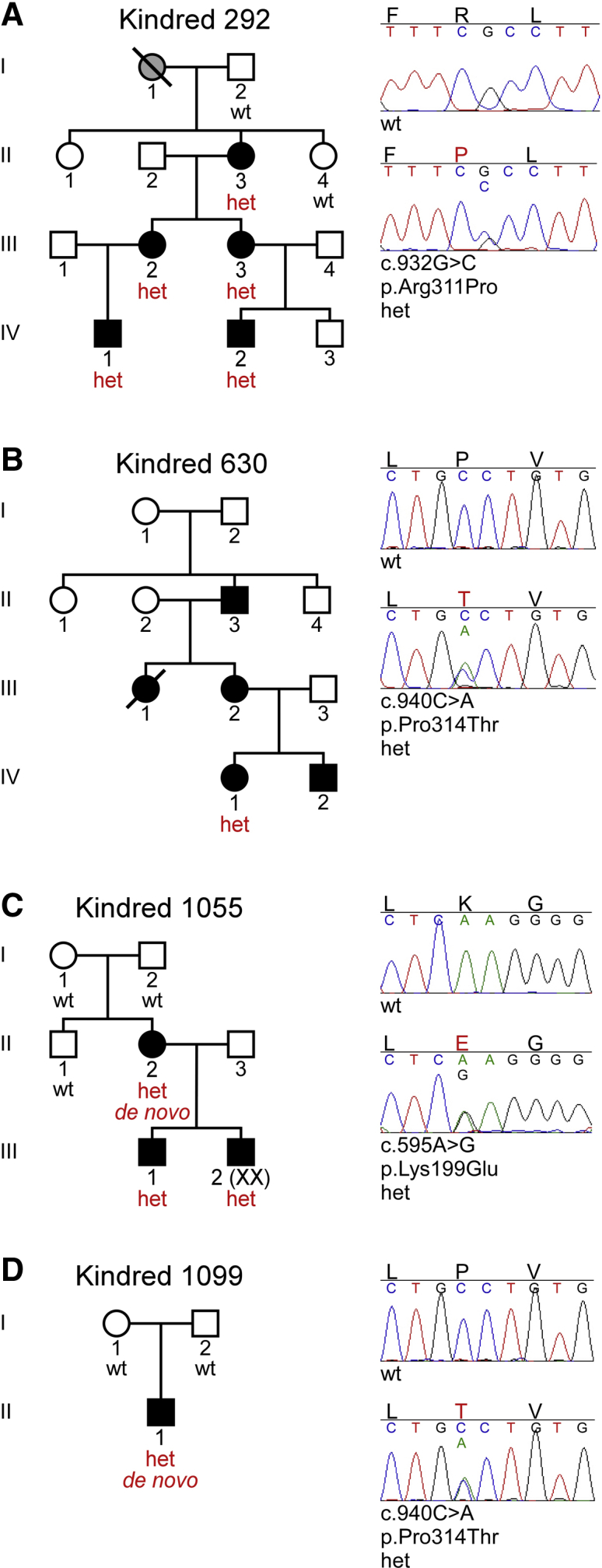
Ichthyosis Subjects Heterozygous for ASPRV1 Mutations
Sanger sequencing was performed subsequent to exome sequencing results (Table S1).
(A) In kindred 292, five subjects (II-3, III-2, III-3, IV-1, IV-2) are heterozygous (het) for c.932G>C, encoding p.Arg311Pro; two unaffected first-degree relatives (I-2, II-4) are wild-type (WT) at this site.
(B) In kindred 630, the proband (IV-1) is heterozygous for c.940C>A, encoding p.Pro314Thr. DNA from other members of the kindred was not available.
(C) In kindred 1055, the proband (III-1) and his affected mother (II-2) and brother (III-2) are heterozygous for c.595A>G, encoding p.Lys199Glu. This mutation is absent from the unaffected maternal uncle (II-1) and grandparents (I-1, I-2), and thus arose de novo in the proband’s mother.
(D) In kindred 1099, the proband (II-1) is heterozygous for c.940C>A, encoding p.Pro314Thr, which is absent from his unaffected parents (I-1, I-2) and thus arose de novo.
All of the subjects heterozygous for deleterious missense mutations in ASPRV1 were found to exhibit a consistent skin phenotype (Figure 2). All subjects presented at birth or within the first months of life with scaling affecting the entire body, including the flexures, palms, and soles. None presented with collodion membrane and there were no complications with pregnancy or delivery. Scales are large and plate-like, most prominent on the arms and legs. Erythema is absent or mild. Scaling improves, but does not completely resolve, during warmer weather. Subjects report inability to perspire when scaling is severe. Palmoplantar keratoderma is present, with prominent scaling and accentuation of the creases. Some subjects report moderate itch. Histology demonstrates acanthosis, compact orthohyperkeratosis, and a slightly expanded granular layer. Members of kindred 630 were previously described in a 1984 report of a new dominant type of lamellar ichthyosis, a phenotype that has otherwise been exclusively recessive.2,3
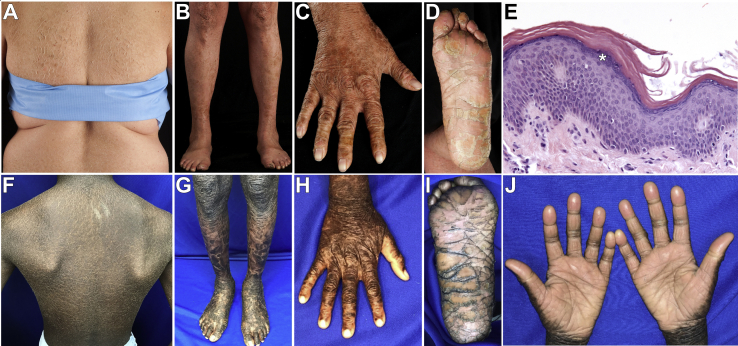
Figure 2.
Subjects Heterozygous for ASPRV1 Mutations Display a Consistent Skin Phenotype
(A–E) Subject 292-2. There are large, plate-like, geometric scales over the back and posterior arms (A) and shins (B), corrugated hyperkeratosis over the extensor wrist and dorsal hand (C), and hyperlinearity of the foot, with large, plate-like, adherent scale and a few fissures of the heel (D). Histology (E) shows acanthosis, compact orthohyperkeratosis, and a prominent stratum granulosum with coarse keratohyalin granules containing filaggrin (asterisk).
(F–J) Subject 1099. There is similar scale on the back (F), with larger plate-like scale on the lower extremities and feet (G), corrugated hyperkeratosis and smoothened scale over the hand (H), marked hyperlinearity of the feet with thick hyperkeratosis and many deep fissures (I), and hyperlinearity and hyperkeratosis of the palms with evidence of constricting bands of hyperkeratosis (pseudoainhum) on multiple digits (J).
ASPRV1 (MIM: 611765) encodes aspartic peptidase retroviral-like 1, also known as skin aspartic protease (SASPase), which was first characterized in 2005 by Bernard et al. in laboratories at L’Oreal and Galderma via analyses of total protein extract from human reconstructed epidermis. It is a single exon gene exclusive to mammals, without apparent orthologs in lower species. The full-length protein is 343 amino acids and 37 kDa, but immunoblot analyses, mass spectrometry, and peptide sequencing revealed expression of both a 28 kDa pro-form (residues 85–343) and a 14 kDa active enzyme generated by auto-cleavage (residues 191–326). There is a putative transmembrane domain spanning residues 57–75. The active site aspartic acid residue at Asp212 was hypothesized based on homology and confirmed by demonstration that mutagenesis of this site abolishes auto-activation. Immunostaining of normal human skin sections displayed strong localization in the granular layer of the epidermis and the inner root sheath of hair follicles.4 The mouse ortholog was later shown to similarly exhibit auto-cleavage and an Asp212 residue essential to proteolytic activity, with expression restricted to suprabasal layers of stratified epithelia.5 Paralogous retroviral-like proteases require homodimerization to form an active site pocket, and it is thought that ASPRV1 also forms homodimers.4,6
One known target of ASPRV1 is filaggrin. Experiments utilizing recombinant purified protein products have demonstrated that ASPRV1 directly cleaves filaggrin,7 a process critical to epidermal integrity.8 Filaggrin is first expressed as a pro-form containing N-terminal and C-terminal domains flanking 10–12 tandem repeats that are processed in a stepwise fashion to form filaggrin monomers. These monomers bind keratin intermediate filaments, resulting in aggregation and alignment of tightly packed parallel arrays. Crosslinking by transglutaminases creates an insoluble keratin matrix that acts as a scaffold for attachment of cornified envelope proteins and lipids, forming the stratum corneum, the uppermost epidermal layer.8
Each of the ASPRV1 mutations observed in our subjects are present in the 14 kDa active enzyme (Figure S1). Missense alteration p.Lys199Glu is eight residues distal to the N-terminal auto-cleavage site. Missense alterations p.Arg311Pro and p.Pro314Thr are tightly clustered within three residues of each other and 12–15 residues from the C-terminal auto-cleavage site. A model of the crystal structure of the active enzyme9 shows that all three mutation sites are on the same surface and tightly clustered within the three-dimensional protein (Figure 3A).
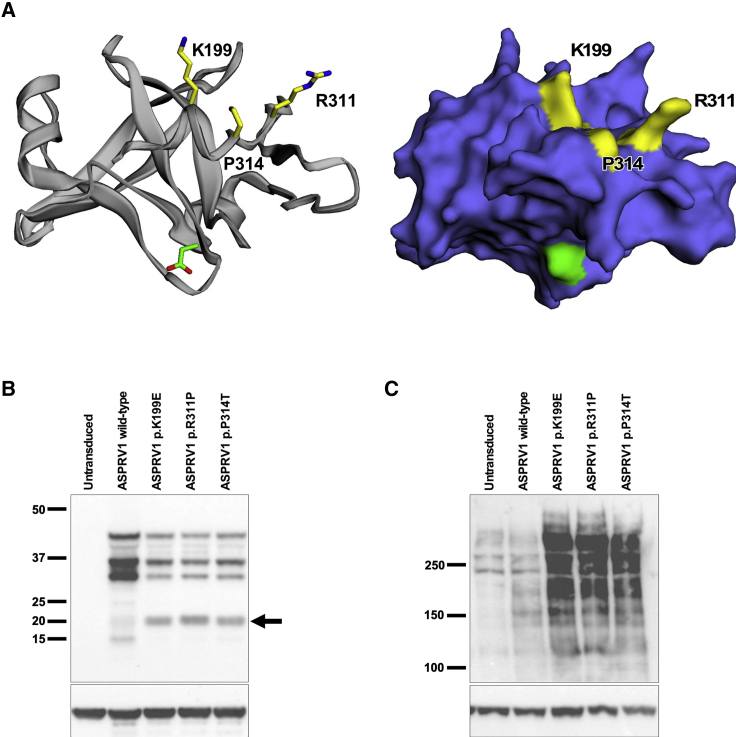
Figure 3.
Clustered ASPRV1 Mutations Generate a Novel ASPRV1 Auto-cleavage Product and Result in Accumulation of High-Molecular-Weight Forms of Filaggrin
(A) A model of the ASPRV1 active enzyme crystal structure9 (ribbon and stick rendering on left, surface rendering on right) shows that all three pathogenic mutation sites (yellow) are tightly clustered within the three-dimensional protein. The catalytic aspartate is shown in green.
(B and C) Human keratinocytes (HK), either nontransduced or transduced with wild-type or mutant ASPRV1, were switched to high calcium (1.2 mM) for 8 days to promote differentiation. Immunoblotting of protein extracts was performed with anti-actin (Sigma SAB4200248, 1:2,000) as a loading control on replicate blots to show equivalent protein loading across all samples. (B) Immunoblotting with an anti-HA antibody (Cell Signaling 3724, 1:500) shows bands at ~37 kDa and ~28 kDa in cells expressing either wild-type or mutant forms of HA-tagged ASPRV1. In addition, cells expressing each of the ASPRV1 mutant forms show a unique ~20 kDa band not seen in nontransduced cells or cells expressing wild-type ASPRV1 (arrow). (C) Immunoblotting with an anti-filaggrin antibody directed against the N-terminal domain (residues 1–261, Thermo Fisher Scientific PA5-79267, 1:500) shows profilaggrin and higher molecular weight filaggrin cleavage products within differentiated keratinocytes. In nontransduced keratinocytes and keratinocytes transduced with wild-type ASPRV1, there is a low quantity of high-molecular-weight filaggrin products, consistent with normal processing of filaggrin to filaggrin repeat monomers. Cells expressing mutant ASPRV1 (encoding p.Lys199Glu, p.Arg311Pro, or p.Pro314Thr) demonstrate an accumulation of high-molecular-weight filaggrin products, suggesting that ASPRV1 mutations impair filaggrin processing.
To assess the effect of ASPRV1 mutations on filaggrin processing, we expressed wild-type or mutant forms of HA-tagged ASPRV1 in human keratinocytes. Immunoblotting of protein extracts with anti-HA and anti-ASPRV1 antibodies demonstrates that cells transduced with each of the ASPRV1 mutant forms express a unique 20 kDa form of ASPRV1 not seen in nontransduced cells or cells transduced with wild-type ASPRV1 (Figures 3B and S2A). We hypothesize that this may be due to derangement of autocatalytic specificity in mutant forms of ASPRV1. Immunoblotting with an anti-filaggrin antibody recognizing the amino terminus of the protein shows low quantities of high-molecular-weight filaggrin in nontransduced keratinocytes and keratinocytes transduced with wild-type ASPRV1, consistent with normal filaggrin processing. Expression of each of the three ASPRV1 mutants results in accumulation of high-molecular-weight filaggrin products (Figures 3C and S2B), suggesting impaired filaggrin processing by mutant forms of ASPRV1.
To further examine the effect of ASPRV1 mutations, we obtained affected skin from subject 292-2 and immunostained with antibodies to ASPRV1, keratin 14 (a marker of basal keratinocytes), keratin 10 (a marker of suprabasal keratinocyte differentiation), and filaggrin. In skin from both the subject and an age-matched control subject, ASPRV1 is localized to the cytoplasm and nuclei of cells within the stratum granulosum. Immunolocalization of keratin 14 shows restriction to the basal layer in affected and control skin, and keratin 10 localizes in suprabasal layers of the epidermis in both. While filaggrin staining is tightly focused within the stratum granulosum of control skin, affected skin shows epidermal thickening with strong expression in an expanded stratum granulosum and weak expression within the upper laryers of the stratum spinosum (Figure 4).
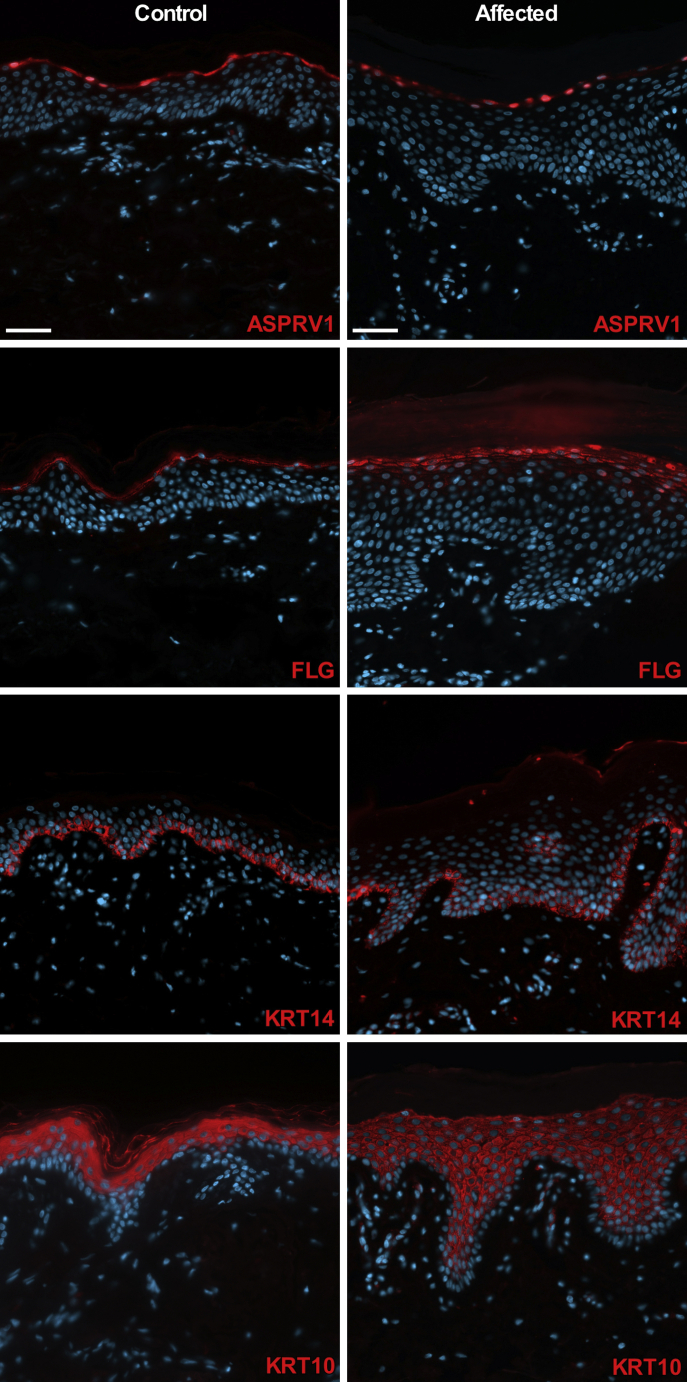
Figure 4.
ASPRV1 Mutant Skin Shows Expansion of Filaggrin Immunolocalization
ASPRV1 is localized to the cytoplasm and nuclei of cells within the stratum granulosum in age-matched control skin and affected tissue from subject 292-2. In contrast to the tightly focused filaggrin (FLG) immunolocalization noted in control tissue, affected tissue shows strong signal within a thickened granular layer of the epidermis and weak expression in the upper layers of stratum spinosum. Immunolocalization of keratin 14 (KRT14, a basal keratinocyte marker) is restricted to the basal layer and keratin 10 (KRT10, a suprabasal keratinocyte marker) is found in suprabasal layers in control and affected tissue. Scale bars are 50 μm. Primary antibodies (red staining) are rabbit anti-ASPRV1 (Sigma HPA034809, 1:200), rabbit anti-FLG (BioLegend 905801, 1:200), mouse anti-KRT14 (Santa Cruz sc-53252, 1:200), and guinea pig anti-KRT10 (Origene AP09544SU-N, 1:300). Nuclei are stained with DAPI (blue staining).
Animal models of ASPRV1 dysfunction have previously been described. Asprv1 knockout mice have increased fine wrinkles on their sides6 and this phenotype was further analyzed by knocking out Asprv1 in hairless mice. Compared to wild-type and Asprv1 ± hairless mice, Asprv1−/− hairless mice have more fine wrinkles and skin that is dry and rough, with raised scales, many horny cells, and thickened stratum corneum.7 Additionally, a single German Shepherd dog with ichthyosis and a de novo missense mutation in ASPRV1 has been reported. This mutation (c.1052T>C, encoding p.Leu351Pro) is within one residue of the C-terminal auto-cleavage site, at a position corresponding to Leu325 in the human protein.10
Immunoblotting and immunofluorescent staining of Asprv1−/− hairless mouse epidermis for differentiation markers showed normal expression and localization with the exception of filaggrin. Immunofluorescent staining showed increased filaggrin-positive layers in the lower stratum corneum, and immunoblotting showed accumulation of premature filaggrin and deficiency of mature filaggrin.7 Human subjects heterozygous for ASPRV1 mutations also show normal localization of differentiation markers within a thickened epidermis, while expression of disease-causing mutants in differentiated keratinocytes leads to accumulation of premature filaggrin.
The phenotypic and molecular similarities between our human subjects and Asprv1 knockout mice implies that the human mutations result in loss of function. Haploinsufficiency is unlikely given that Asprv1+/− mice do not appear phenotypically different from wild-type mice6,7 and heterozygous null mutations are present in genetic databases of unaffected individuals with expected frequency (ASPRV1 probability of loss intolerance 0.01). This and the fact that the human mutations are clustered missense substitutions suggests a dominant-negative mechanism for the mutations in our human subjects, which could be facilitated by homodimerization; additional neomorphic effects are also possible.
Proteases are essential to epidermal differentiation and desquamation. Mutations in genes encoding proteases, protease inhibitors, and protease targets cause a variety of keratinization disorders in which the specific skin phenotype can be understood via the nature of the genetic defect. Ichthyosis vulgaris is due to loss-of-function mutations in filaggrin (FLG [MIM: 135940]), which encodes a protease target, resulting in absence of the granular layer formed from processed filaggrin and concomitant defects in skin moisturization.11 Netherton syndrome and a form of peeling skin syndrome are caused by caused by loss-of-function mutations in serine protease inhibitor Kazal-type 5 (SPINK5 [MIM: 605010]) and cystatin A (CSTA [MIM: 184600]), respectively.12,13 Both encode protease inhibitors, and their loss results in excess of desquamation, observed as redness and peeling. In contrast, a recessive form of ichthyosis is caused by mutations in suppression of tumorigenicity 14 (ST14 [MIM: 606797]), also known as matripase, which encodes a serine protease.14 St14 knockout mice demonstrate aberrant filaggrin processing and hyperkeratotic phenotypes.15 Similarly, subjects with ASPRV1 mutations have aberrant protease function, with a consequent excess of unprocessed filaggrin and a desquamation defect that manifests as thick scale and palmoplantar hyperkeratosis.
While filaggrin is currently the only known target of ASPRV1, there may be others. The phenotype of subjects with ASPRV1 mutations includes lamellar ichthyosis-like manifestations never observed in ichthyosis vulgaris subjects with loss-of-function mutations in FLG, suggesting the possibility of a broader role for ASPRV1 in epidermal barrier function beyond filaggrin processing. Alternatively, the aberrant ASPRV1 auto-cleavage form and/or accumulation of unprocessed or improperly processed filaggrin may result in toxicity, differentiating the phenotype from that of subjects with loss of filaggrin.
Because the skin disorder in subjects with ASPRV1 mutations is a defect of desquamation, pathogenesis-driven therapy might include keratolytic agents such as lactic acid and urea. These agents are currently used by subject 1055 and his affected mother, with almost complete resolution of non-palmoplantar scale.
In a previously described cohort of 196 Japanese atopic dermatitis subjects and 28 control subjects, four heterozygous ASPRV1 missense mutations were reported in subjects (encoding p.Ala24Ser and p.Arg311Cys in one subject and on the same allele, p.Ile186Thr in one subject, p.Val187Ile in three subjects), and two were reported in control subjects (encoding p.Asp232Tyr and p.Val243Ala, each in one subject). Examination of in vitro autoprocessing activity by purified 28 kDa ASPRV1 demonstrated that p.Val187Ile decreased autoprocessing, and p.Val243Ala showed no autoprocessing.7 Notably, the latter was found in a phenotypically normal control subject, which further supports the hypothesis that ASPRV1 haploinsufficency is unlikely to be pathogenic. The p.Arg311Cys missense alteration, affecting the same residue as the p.Arg311Pro missense alteration in kindred 292, was not functionally assessed because the protein purification was unsuccessful,7 but it is present in gnomAD, a database of normal human variation, at an allele frequency of 0.0002 and 0.002 in East Asians. Another variant affecting this residue, p.Arg311His, has an allele frequency of 0.00004. The p.Val187Ile missense alteration shown to decrease ASPRV1 autoprocessing has an allele frequency of 0.0004, and 0.002 in Latinos.16 While these allele frequencies are inconsistent with pathogenicity for a dominant, rare, and severe disorder such as that of our subjects, they don’t exclude the possibility of association with a relatively mild atopic dermatitis that might be considered subclinical in the context of participation in gnomAD. However, a subsequent study from the same group did not detect any of these mutations in a larger Irish, Scottish, and South African cohort and concluded that mutations in ASPRV1 are not associated with atopic eczema or clinically dry skin.17
The identification of four unrelated kindreds and ten affected subjects heterozygous for previously unreported and damaging missense mutations in ASPRV1, all of whom exhibit ichthyosis and palmoplantar keratoderma, two of whom have a mutation which arose de novo, and eight of whom are within extended kindreds in which the mutation and disorder are shown to cosegregate, provides conclusive evidence that mutations in ASPRV1 cause this dominant disorder. Previously described disorders phenotypically indistinguishable from these subjects have heretofore been exclusively recessive. The tight clustering of the mutant sites within the protein and the observation of a recurrent mutation (encoding p.Pro314Thr) suggests that ASPRV1 mutations resulting in this phenotype are likely restricted to specific sites. This discovery highlights the central importance of aspartic proteases in epidermal differentiation.
Acknowledgments
We thank the study subjects, their families, and the health care professionals whose participation made this work possible. We thank Nicholas Theodosakis, Irina Tikhonova, Christopher Castaldi, Kaya Bilguvar, James Knight, Stefanos Koutsoukos, and Richard Presland for technical contributions. This work was supported in part by the Foundation for Ichthyosis and Related Skin Types (FIRST) and the National Institutes of Health (NIH R01 AR068392 to K.A.C. and UM1 HG006504 to the Yale Center for Mendelian Genomics).
Published: June 8, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.05.013.
Web Resources
1000 Genomes, http://www.internationalgenome.org
BWA-MEM, http://bio-bwa.sourceforge.net
Exome Aggregation Consortium (ExAC) Browser, http://exac.broadinstitute.org
ExonPrimer, https://ihg.helmholtz-muenchen.de/ihg/ExonPrimer.html
GenBank, https://www.ncbi.nlm.nih.gov/genbank
Genome Analysis Toolkit (GATK), https://software.broadinstitute.org/gatk
Integrative Genomics Viewer (IGV), http://software.broadinstitute.org/software/igv
OMIM, https://www.omim.org
Protein Homology/anologY Recognition Engine (Phyre2), http://www.sbg.bio.ic.ac.uk/∼phyre2/html/page.cgi?id=index
SNPmasker, http://bioinfo.ebc.ee/snpmasker
UCSC Genome Browser, https://genome.ucsc.edu/index.html
Variant Effect Predictor, http://useast.ensembl.org/info/docs/tools/vep/index.html
Declaration of Interests
The authors declare no competing interests.
Supplemental Information
References
- 1.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Traupe H., Kolde G., Happle R. Autosomal dominant lamellar ichthyosis: a new skin disorder. Clin. Genet. 1984;26:457–461. doi: 10.1111/j.1399-0004.1984.tb01089.x. [DOI] [PubMed] [Google Scholar]
- 3.Kolde G., Happle R., Traupe H. Autosomal-dominant lamellar ichthyosis: ultrastructural characteristics of a new type of congenital ichthyosis. Arch. Dermatol. Res. 1985;278:1–5. doi: 10.1007/BF00412487. [DOI] [PubMed] [Google Scholar]
- 4.Bernard D., Méhul B., Thomas-Collignon A., Delattre C., Donovan M., Schmidt R. Identification and characterization of a novel retroviral-like aspartic protease specifically expressed in human epidermis. J. Invest. Dermatol. 2005;125:278–287. doi: 10.1111/j.0022-202X.2005.23816.x. [DOI] [PubMed] [Google Scholar]
- 5.Rhiemeier V., Breitenbach U., Richter K.H., Gebhardt C., Vogt I., Hartenstein B., Fürstenberger G., Mauch C., Hess J., Angel P. A novel aspartic proteinase-like gene expressed in stratified epithelia and squamous cell carcinoma of the skin. Am. J. Pathol. 2006;168:1354–1364. doi: 10.2353/ajpath.2006.050871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsui T., Kinoshita-Ida Y., Hayashi-Kisumi F., Hata M., Matsubara K., Chiba M., Katahira-Tayama S., Morita K., Miyachi Y., Tsukita S. Mouse homologue of skin-specific retroviral-like aspartic protease involved in wrinkle formation. J. Biol. Chem. 2006;281:27512–27525. doi: 10.1074/jbc.M603559200. [DOI] [PubMed] [Google Scholar]
- 7.Matsui T., Miyamoto K., Kubo A., Kawasaki H., Ebihara T., Hata K., Tanahashi S., Ichinose S., Imoto I., Inazawa J. SASPase regulates stratum corneum hydration through profilaggrin-to-filaggrin processing. EMBO Mol. Med. 2011;3:320–333. doi: 10.1002/emmm.201100140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandilands A., Sutherland C., Irvine A.D., McLean W.H. Filaggrin in the frontline: role in skin barrier function and disease. J. Cell Sci. 2009;122:1285–1294. doi: 10.1242/jcs.033969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bauer A., Waluk D.P., Galichet A., Timm K., Jagannathan V., Sayar B.S., Wiener D.J., Dietschi E., Müller E.J., Roosje P. A de novo variant in the ASPRV1 gene in a dog with ichthyosis. PLoS Genet. 2017;13:e1006651. doi: 10.1371/journal.pgen.1006651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith F.J., Irvine A.D., Terron-Kwiatkowski A., Sandilands A., Campbell L.E., Zhao Y., Liao H., Evans A.T., Goudie D.R., Lewis-Jones S. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat. Genet. 2006;38:337–342. doi: 10.1038/ng1743. [DOI] [PubMed] [Google Scholar]
- 12.Chavanas S., Bodemer C., Rochat A., Hamel-Teillac D., Ali M., Irvine A.D., Bonafé J.L., Wilkinson J., Taïeb A., Barrandon Y. Mutations in SPINK5, encoding a serine protease inhibitor, cause Netherton syndrome. Nat. Genet. 2000;25:141–142. doi: 10.1038/75977. [DOI] [PubMed] [Google Scholar]
- 13.Blaydon D.C., Nitoiu D., Eckl K.M., Cabral R.M., Bland P., Hausser I., van Heel D.A., Rajpopat S., Fischer J., Oji V. Mutations in CSTA, encoding Cystatin A, underlie exfoliative ichthyosis and reveal a role for this protease inhibitor in cell-cell adhesion. Am. J. Hum. Genet. 2011;89:564–571. doi: 10.1016/j.ajhg.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Basel-Vanagaite L., Attia R., Ishida-Yamamoto A., Rainshtein L., Ben Amitai D., Lurie R., Pasmanik-Chor M., Indelman M., Zvulunov A., Saban S. Autosomal recessive ichthyosis with hypotrichosis caused by a mutation in ST14, encoding type II transmembrane serine protease matriptase. Am. J. Hum. Genet. 2007;80:467–477. doi: 10.1086/512487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.List K., Szabo R., Wertz P.W., Segre J., Haudenschild C.C., Kim S.Y., Bugge T.H. Loss of proteolytically processed filaggrin caused by epidermal deletion of Matriptase/MT-SP1. J. Cell Biol. 2003;163:901–910. doi: 10.1083/jcb.200304161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv. 2019 doi: 10.1101/531210. [DOI] [Google Scholar]
- 17.Sandilands A., Brown S.J., Goh C.S., Pohler E., Wilson N.J., Campbell L.E., Miyamoto K., Kubo A., Irvine A.D., Thawer-Esmail F. Mutations in the SASPase gene (ASPRV1) are not associated with atopic eczema or clinically dry skin. J. Invest. Dermatol. 2012;132:1507–1510. doi: 10.1038/jid.2011.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.