Abstract
Secondary genomic findings are increasingly being returned to individuals as opportunistic screening results. A secondary finding offers the chance to identify and mitigate disease that may otherwise be unrecognized in an individual. As a form of screening, secondary findings must be considered differently from sequencing results in a diagnostic setting. For these reasons, clinicians should employ an evaluation and long-term management strategy that accounts for both the increased disease risk associated with a secondary finding and the lower positive predictive value of a screening result compared to an indication-based testing result. Here we describe an approach to the clinical evaluation and management of an individual who presents with a secondary finding. This approach enumerates five domains of evaluation—(1) medical history, (2) physical exam, (3) family history, (4) diagnostic phenotypic testing, and (5) variant correlation—through which a clinician can distinguish a molecular finding from a clinicomolecular diagnosis of genomic disease. With this framework, both geneticists and non-geneticist clinicians can optimize their ability to detect and mitigate genomic disease while avoiding the pitfalls of overdiagnosis. Our goal with this approach is to help clinicians translate secondary findings into meaningful recognition, treatment, and prevention of disease.
Secondary genomic findings are increasingly being returned to individuals as opportunistic screening results. A secondary finding offers the chance to identify and mitigate disease that may otherwise be unrecognized in an individual. As a form of screening, secondary findings must be considered differently from sequencing results in a diagnostic setting. For these reasons, clinicians should employ an evaluation and long-term management strategy that accounts for both the increased disease risk associated with a secondary finding and the lower positive predictive value of a screening result compared to an indication-based testing result. Here we describe an approach to the clinical evaluation and management of an individual who presents with a secondary finding. This approach enumerates five domains of evaluation—(1) medical history, (2) physical exam, (3) family history, (4) diagnostic phenotypic testing, and (5) variant correlation—through which a clinician can distinguish a molecular finding from a clinicomolecular diagnosis of genomic disease. With this framework, both geneticists and non-geneticist clinicians can optimize their ability to detect and mitigate genomic disease while avoiding the pitfalls of overdiagnosis. Our goal with this approach is to help clinicians translate secondary findings into meaningful recognition, treatment, and prevention of disease.
Main Text
Introduction
A secondary finding is defined as a genomic variant of potential medical value that is unrelated to the primary reason for testing. The American College of Medical Genetics and Genomics (ACMG) has issued recommendations for the reporting of secondary findings in clinical exome and genome sequencing.1,2 The current recommendations include a minimum list of 59 genes associated with genetic disease (ACMG59) in which variants identified by clinical sequencing are recommended to be returned to tested individuals.3 The overarching rationale for returning these variants is that they can be associated with an unrecognized risk for serious heritable disorders and are typically poorly ascertained by the health care system, yet there are medical interventions that should reduce the associated morbidity and mortality.4, 5, 6 While there is some variability among clinical laboratories, standard practice is that only pathogenic or likely pathogenic variants, and not variants of uncertain significance (VUS),7 are returned as secondary findings. An estimated 1%–4% of all individuals harbor a secondary finding.8, 9, 10, 11, 12, 13, 14, 15 Beyond clinical exome or genome sequencing, returning variants identified in clinical research sequencing is becoming more common16 and some research-based sequencing programs opt to return secondary findings in more extended gene lists.15,17 With the increasing use of exome and genome sequencing, the number of people who have a secondary finding returned to them is expected to rise. Clinicians across different specialties will increasingly need to manage individuals with these findings.
While the disorders represented in the ACMG59 list are all well characterized, there are no published guidelines for how to best manage an individual who presents with a secondary finding. Clinical decision aids for certain conditions have been developed by the ACMG in the form of ACT sheets (Table 1), which were modeled after the ACT sheets available for newborn screening results. These straightforward, rapid-decision-making tools are necessary for newborn screening scenarios because time to diagnosis and initiation of intervention is critical.18 For secondary findings, additional guidance geared toward comprehensive evaluation and management is needed. The assumptions of the original ACMG secondary findings recommendations were that a clinician would employ a generalization of the approach typically used for family-based cascade testing.19,20 However, this is complicated by the fact that in family-based cascade testing, the genomic variant is originally identified in a relative affected with the disease, which increases the likelihood that the individual who undergoes the cascade testing actually has the disorder. Secondary findings, by contrast, are described as “opportunistic screening”21 based on the assumption that the individual may not have a recognized personal or family history of the associated disorder. As a general principle, the positive predictive value of a screening test result is often much lower than the same result in a diagnostic testing setting.22 This concept has been addressed quantitatively regarding genetic testing results.23 If a secondary finding is identified in an individual, the clinician has gained information on disease risk and the key next step is to perform a judicious evaluation that properly recognizes the probability of disease. In doing so, the clinician will also minimize overtreatment or overtesting of individuals.24,25 For these reasons, a standardized approach to a person presenting with a secondary genomic finding is needed.
Table 1.
Key Resources for Providers of Individuals with Secondary Findings
| Resource | URL | Description |
|---|---|---|
| ACMG ACT Sheet | https://www.acmg.net/ACMG/Medical-Genetics-Practice-Resources/ACT_Sheets_and_Algorithms.aspx | algorithmic description of short-term actions a provider can take in the management of an individual with a subset of ACMG59 disorders; prepared by ACMG working group |
| Clinical Genome Resource (ClinGen), Actionability | https://clinicalgenome.org/curation-activities/clinical-actionability/ | summaries of actionability assessments for genetic disorders, including evidence pertaining to penetrance of disease features and outcomes associated with specific interventions; reports and scoring provided by the ClinGen pediatric and adult actionability working groups |
| Clinical Genome Resource (ClinGen), Gene-Disease Validity | https://clinicalgenome.org/curation-activities/gene-disease-validity/ | aggregation of evidence available for the association of a gene with a given disorder; strength of evidence assessed by the ClinGen gene-disease validity working group |
| GeneReviews | http://www.genereviews.org | point-of-care resource for clinicians, written by experts on the specific condition and subject to peer review prior to publication; focus is on genetic disorders |
| National Comprehensive Cancer Network (NCCN) | https://www.nccn.org/ | practice guidelines from alliance of cancer centers to guide decision making in the management of cancer, including suspected cancer predisposition syndromes |
| UpToDate | https://www.uptodate.com/ | point-of-care resource for clinicians, written by experts on the specific condition and subject to peer review prior to publication |
The approach focuses on an adult who presents with a chief complaint of having had a secondary finding in an ACMG59 gene identified by a clinical genetic testing lab (see Secondary Findings in Children for a discussion of nuances related to the pediatric setting). This scenario may be encountered by a primary care physician, geneticist, or other specialist and we have outlined a process by which clinicians can decide what portions of the evaluation to do themselves and when to refer. This is meant to suggest a thoughtful, measured approach that balances the risks of overdiagnosis and overutilization of healthcare resources with the opportunity to accomplish the end objective of secondary findings return: to avoid preventable serious harms or death from genetic disease.
Definitions
Thinking about a secondary finding requires an appreciation of the distinction between the mere presence of a genomic variant and the clinical diagnosis of disease. There is a growing understanding of the difference between the pathogenicity of a genetic variant and the presence of a genetic disease.26,27 This is a familiar concept in medicine. A hematocrit or hemoglobin result below the reference range is not equivalent to a clinical diagnosis of anemia, but rather a data point that can be used and contextualized to help make a clinical diagnosis. Sometimes, a single result from a CBC is sufficient to make the diagnosis, while in other instances an abnormal result from a CBC is only a risk factor and is not a diagnosis. While there are many differences between a CBC and a genomic sequencing test, this same concept of contextualism exists for both. Clinicians are accustomed to considering the context in which any clinical result is obtained, but genomic results can pose a challenge. A lack of familiarity with genomics among non-geneticist clinicians may make them feel unprepared to use these results in their practice.28 Even among clinicians with genetics expertise, there is a lack of consensus as to how to interpret genetic information, which makes the application of genetic information obtained via opportunistic screening challenging.29 In an effort to clarify the distinction between a genomic finding and a disease, we propose to define the following terms as applied to the ACMG59:
-
•
Molecular finding: the presence of a secondary genomic finding (regardless of the clinical presentation).
-
•
Clinical finding: the presence of a detectable phenotype compatible with the disease associated with a given gene.
-
•
Clinicomolecular diagnosis: the presence of a correlated clinical and molecular finding by the above definitions.
Domains of Clinical Evaluation
A secondary finding is initially a molecular finding that requires a thorough clinical evaluation, the results of which determine further management (Figure 1). This evaluation can be organized into five domains. The entire evaluation, which may not necessarily be completed by a single clinician, includes the information necessary for risk stratification and further management. The five domains of evaluation mostly overlap with those that are used every day by clinicians:
-
•
Medical history
-
•
Physical examination
-
•
Family history
-
•
Diagnostic phenotypic testing
-
•
Variant correlation
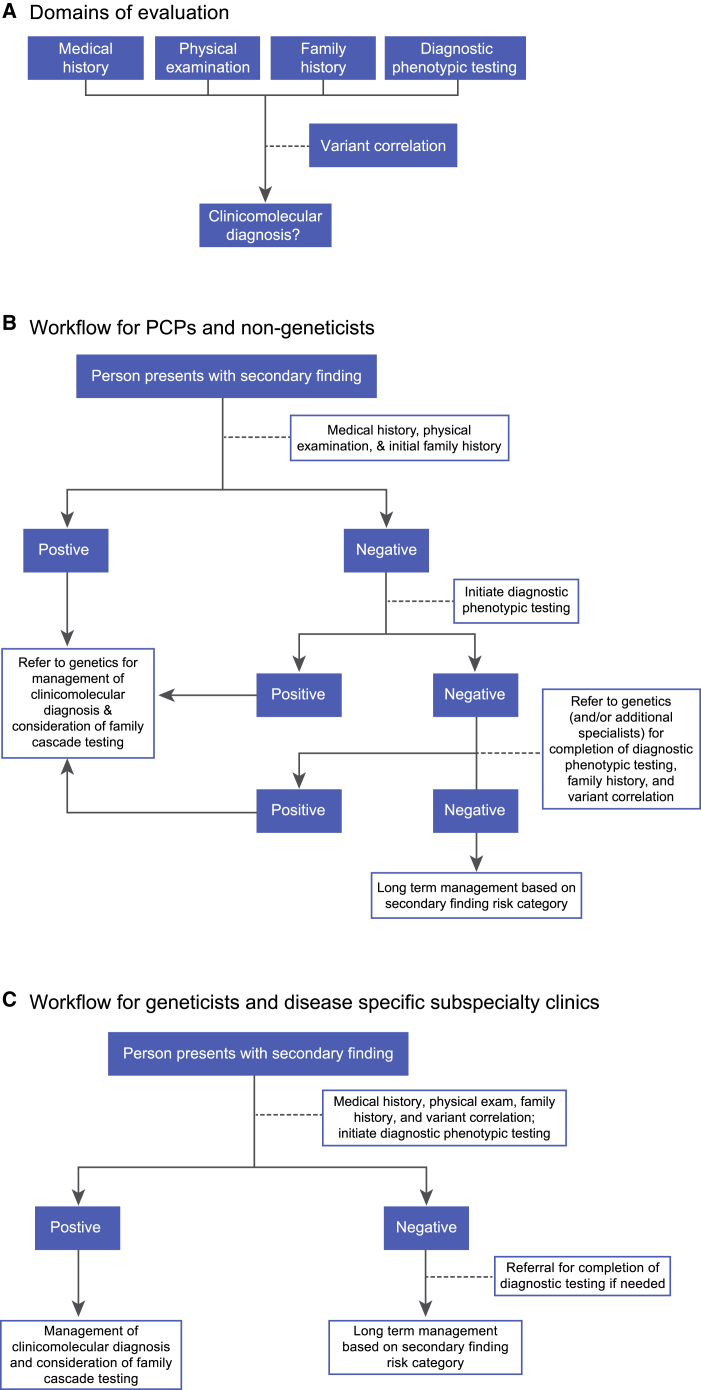
Figure 1.
Domains of Evaluation and Workflow for Clinicians
(A) The five domains of evaluation to be addressed by clinicians for an individual with a secondary genomic finding. Findings suggestive of the phenotype in the personal history, physical exam, or diagnostic phenotypic testing make a clinicomolecular diagnosis highly likely. A family history consistent with the expected phenotype makes a clinicomolecular diagnosis highly likely in the affected family member, but confirmation may require an in-depth family evaluation with cascade-based targeted variant testing by a geneticist. The variant correlation is not sufficient by itself to establish a clinicomolecular diagnosis but is crucial to the ultimate risk assessment. The long-term management of individuals is dependent on whether a clinicomolecular diagnosis is present. If no clinicomolecular diagnosis is made, then the clinician will make a risk assessment guided by the disease categories (Table S1).
(B) When the initial presentation is to a primary care provider (PCP) or other non-geneticist specialist, the clinician can perform an evaluation encompassing the medical history, physical examination, and family history and can in many cases initiate the process of diagnostic phenotypic testing. Once this has been completed, the appropriate next step is to refer the person to a clinician with genetics expertise. This may be a general genetics clinic or a relevant subspecialty clinic (e.g., a hypertrophic cardiomyopathy clinic). The exact timing of the referral to a geneticist or subspecialist depends on the results of the initial evaluation performed by the referring clinician and practical considerations of finding and accessing specialty care. The geneticist or genetically expert subspecialist should perform the variant correlation and complete any necessary phenotypic testing. A genetics clinic is often best equipped to address complexities regarding the inheritance of a variant (including issues pertaining to potential mosaicism, de novo occurrence, parent-of-origin effect, or non-paternity) and initiate the process of family-based cascade testing for the presence or absence of the variant among family members.
(C) If a person presents to a geneticist or disease-specific subspecialty clinic, the clinician can perform the five domains of evaluation (including the variant correlation). Subspecialty consultation may be required for some of the recommended diagnostic phenotypic testing, but this process can be initiated by the clinician. For a person who initially presents to a subspecialist, the recommended workflow will depend on the subspecialist’s genetics expertise and experience with the disorder associated with the secondary finding. A general cardiologist encountering an individual with a secondary finding in MYH7 will likely choose to refer that individual to a geneticist or to a hypertrophic cardiomyopathy clinic (where such clinics are available). If that person seeks care directly with a hypertrophic cardiomyopathy clinic then a referral to a geneticist may not be necessary.
Any clinician should be able to initiate the evaluation process, while some of the steps, such as variant correlation, aspects of the family history, or diagnostic phenotypic testing, may require genetics or other specialty expertise. Several resources are available to guide clinicians in disease-specific aspects of each domain of clinical evaluation (for examples, see Table 1). As discussed below, the consideration of a referral will depend on the clinician’s comfort and the results of the initial evaluation.
Medical History
The medical history refers to the personal medical history and is similar to that which a clinician would perform if the patient presented with a history of a close relative with the associated disorder: targeted questions based on the expected phenotype, such as a detailed cardiovascular system history for an individual with a secondary finding in a gene associated with hypertrophic cardiomyopathy. The medical history in this context includes current or previous signs, symptoms, and diagnoses and the relevant results of any prior evaluations the individual may have had. Resources are available to help any clinician determine which targeted questions should be asked (Table 1).
Physical Examination
The physical examination is a standard part of a clinician encounter. Particular attention should be given to the system(s) most likely to be affected by the disorder in question. A clinician can refer a patient to a specialist to complete an aspect of the physical examination if needed (e.g., a referral to dermatology for closer evaluation of a skin lesion in an individual with a secondary finding in TSC1 [MIM: 191100]).
It is important for the clinician to recognize that a history and/or physical examination that is positive for findings of the disorder is highly predictive of a clinicomolecular diagnosis. In contrast, for many genetic disorders a negative history and physical examination is poorly predictive of the absence of a clinicomolecular diagnosis. This is a consequence of the fact that most of the disorders that were selected for the ACMG59TM often exist in asymptomatic or unrecognized forms for decades in affected individuals, which highlights the value of the opportunistic genomic screen.
Family History
A detailed family history (beyond what is typically included in a general history and physical examination) is a key element of the evaluation. As is standard practice during a genetics evaluation, the family history should, at a minimum, ask for what is known about each first- and second-degree family member of the presenting individual (typically represented on a three-generation pedigree that includes the person undergoing evaluation). The family history should also include targeted questions about family members, where the specific questions asked depend on the suspected disorder. A molecular finding may prompt identification of previously unrecognized or underappreciated diseases or unclear incidents in family members, such as a drowning accident in the family history of a person with a secondary finding associated with sudden cardiac death susceptibility.8
For all adult first-degree relatives (regardless of clinical history) and any additional family member with signs or symptoms corresponding to the expected phenotype, an effort should be made to verify the presence or absence of the variant. While there are often logistic challenges involved, it is important for the clinician to convey the importance of the family history and variant status of family members. In-depth evaluations of large families are likely beyond the remit of the primary care provider or non-geneticist specialist (see Referrals). As is true for the history and physical examination, the clinician must be aware that the family history is often negative for overt manifestations of these disorders, even in families where the disorders are present.
Diagnostic Phenotypic Testing
For each secondary finding gene, there is recommended diagnostic testing (Table S1). Additional testing beyond the options provided here can be considered based on specific signs and symptoms uncovered during the evaluation. However, even in the case of a completely asymptomatic individual with no relevant medical or family history, there is a recommended minimum amount of testing that should be obtained. The tests listed include some redundancy and overlap (such as both echocardiography and cardiac MRI for cardiomyopathies) and should be interpreted as the options available to obtain the necessary information for this domain of the clinical evaluation.
The recommended diagnostic testing is an aggregation of our assessment of best practices based on available practice guidelines and expert reviews (Table 1). It is important to emphasize that practice guidelines focus on individuals expressing signs and symptoms of disease, and therefore the diagnostic testing options are derived from a generalization of recommendations for individuals who have one or more unambiguously affected family members. Evidence for best practices regarding genomic variants identified in an unselected population is limited, and therefore the recommendations presented here are meant to be reviewed and updated as more data about the clinical yield of secondary findings are collected. The presenting individual may require additional testing as appropriate based on the specific personal or family history. While certain phenotypic tests require specialty consultation (such as electrophysiology-guided pharmacologic provocation testing), the decision for which tests are ordered or interpreted by a primary care provider, geneticist, or specialist is generally dependent on the clinician’s own expertise, familiarity, and comfort with the applicable genetic disorder.
Variant Correlation
The variant correlation is a domain of the secondary findings evaluation that requires genetics expertise (not expected to be done by a primary care provider or other clinician not trained in genetics). While the clinical testing laboratory provides a formal pathogenicity assessment, the genetics clinician needs to perform their own review taking into account the patient’s complete medical and family history (to which the lab may not have access). Formal pathogenicity assessments have become more standardized, but discordance among clinical labs persists.30, 31, 32 Based on current tools and data availability, a clinician’s analysis should include three key steps. First, the clinician should review assessments of that variant in the ClinVar database, the most widely accepted database for submission of variant interpretations. If the variant is present in ClinVar, the clinician can review the quality of evidence used by different groups assessing pathogenicity, particularly if there are conflicting interpretations of that variant. Second, the clinician should review what is known about the penetrance of the disorder (Table 1). GeneReviews contains a section on penetrance for all ACMG59 disorders. The Clinical Genome Resource (ClinGen) contains evidence for gene-disease associations and actionability assessments including the penetrance of specific disease features. Third, the clinician should appreciate the difference between a likely pathogenic and a pathogenic secondary finding. A likely pathogenic interpretation implies a small but significant degree of uncertainty regarding the evidence base. A likely pathogenic variant is defined as having a 90%–99% probability of pathogenicity compared to >99% probability for a pathogenic variant.33 While only a small proportion of existing likely pathogenic variants have been re-classified to uncertain significance to date, a clinician should be cognizant of this possibility as well as the variant reclassification process in general.34
Reviewing evidence and literature about a specific gene or variant is a daunting task that requires specific expertise and knowledge and it is unreasonable to expect clinicians to conduct a comprehensive review. However, the three steps listed above are enough for a general sense of what is known about the variant, particularly if there is alternative evidence or reason to question the disease-causing nature of the variant in the presenting individual. The primary goal of this domain of evaluation is not to question the clinical lab’s formal pathogenicity assessment, but rather for the clinician to correlate that assessment with the clinical implications of the gene, variant, and disorder with the presenting individual to make a clinicomolecular diagnosis. It is also important to relay that information back to the laboratory to maintain an updated pathogenicity assessment of the variant.
Referrals
A person with a secondary finding may present to a geneticist, to a primary care provider, or to a subspecialist (Figure 1). Any clinician can initiate the evaluation, though a referral to a clinician with genetics expertise will ultimately be appropriate. The exact timing of the referral to a geneticist or subspecialist depends on the results of the initial evaluation performed by the referring clinician and practical considerations of finding and accessing specialty care. Diagnostic phenotypic testing may require specialty consultation as well. It is our expectation that individuals will require a multi-disciplinary approach for an adequate evaluation across all five domains. Following this evaluation, longitudinal follow-up should generally be coordinated by a clinical geneticist but some primary physicians or specialists with genetics expertise may be able to fulfill this role as well.
Risk Assessment and Longitudinal Care
A comprehensive evaluation that addresses all five domains is necessary for all individuals with secondary findings, with the results considered collectively to make an overall assessment of the clinicomolecular diagnosis and determine a follow-up plan. The ACMG59 diseases are well characterized with available practice recommendations (Table 1). A person with a secondary finding who is determined to have a clinicomolecular diagnosis should be managed according to these recommendations by a geneticist or disease-specific subspecialist.
If the medical history, physical examination, and clinical diagnostics are negative but there are positive findings in the family history (with these findings either verified by targeted variant segregation analysis or highly specific for the disease if variant segregation is not able to be done), then the person would be followed longitudinally as an “at-risk” family member of an affected individual according to the disease-specific guidelines. For example, heterozygous pathogenic variants in ACTA2 (MIM: 611788) are associated with familial thoracic aortic aneurysm. Consider a person presenting with an ACTA2 secondary finding and normal aortic dimensions on echocardiography, but whose father is found to harbor the same variant and previously had a thoracic aortic aneurysm requiring surgical repair. That presenting individual has a molecular finding but no clinical finding. He should be considered an at-risk family member and undergo cardiovascular imaging biannually to assess for any aortic growth (see GeneReviews, Milewicz and Regalado, in Web Resources). A critical aspect of this scenario is that the question of heritable thoracic aortic aneurysm and dissection in this family was raised by detection of a secondary finding in an asymptomatic individual. When prompted by the discovery of a secondary finding, it is not uncommon for the reporting of a family history to change from negative to positive.8 This is a key indicator of the utility of the secondary findings paradigm—to raise questions about the presence of heritable diseases when routine personal and family history has failed to do so.
Negative Evaluation
Individuals in whom there are no clear findings indicating genetic disease in the medical history, physical examination, family history, and diagnostic testing represent a challenging scenario for a clinician. In such instances, the variant correlation offers important information for determining a risk assessment and follow-up plan. Our approach includes four general categories in which secondary findings can be grouped (Table S1). The categories are not meant to be discrete entities nor to offer specific recommendations for any one gene, variant, or disease, but rather to serve as a guide for longitudinal follow-up plans, particularly in the setting of a negative evaluation. We consider “disorders with high or near complete penetrance” to mean that an individual with a truly pathogenic variant will almost certainly exhbit some manifestation of disease during their expected lifetime. Highly studied cancer predisposition syndromes such as hereditary breast and ovarian cancer and Lynch syndrome have penetrance estimates that still allow for a reasonable probability that an individual with a pathogenic variant will not develop any malignancy during their lifetime are therefore included in “disorders with low or moderate penetrance” (even when considering the upper estimate of 87% for females with a pathogenic BRCA1 [MIM: 604370] or BRCA2 [MIM: 612555] variant to develop an associated malignancy; see GeneReviews, Petrucelli et al., in Web Resources). For adenomatous polyposis coli (MIM: 175100) and MUTYH-associated polyposis (MIM: 608456), not all affected individuals will develop colorectal cancer, particularly if the disorder is present in an attenuated form (see GeneReviews, Jasperson et al. and Nielsen et al., in Web Resources), but these disorders are categorized as “high or near complete penetrance” due to the ability to detect polyps as a disease manifestation. Our categorization of penetrance is based on the strategy a clinician would employ in evaluating for the presence or absence of a clinicomolecular diagnosis. This does not necessarily correlate with how these disorders are chacterized in other contexts (for example, the National Comprehensive Cancer Network appropriately refers to BRCA1 and BRCA2 as high penetrance genes in familial high-risk assessment guidelines).35
None of the individuals in our hypothetical examples have clinicomolecular diagnoses, but the long-term management of each differs based on the nature of their specific molecular finding. Ultimately, the clinician’s management of an individual is dependent on several factors, including balancing the certainty of a clinicomolecular diagnosis with the nature of interventions offered for that disorder (Figure 2). As is appreciated in cancer screening, and based on current scientific and clinical evidence, a less interventional approach is often reasonable and appropriate.36 In determining the need for periodic reevaluation of an individual in the absence of a clinicomolecular diagnosis, the clinician must be mindful of the risk category of the disorder in question and the completeness of the evaluation performed. The crucial concept to recognize here is that there is often a point in the evaluation where the clinician should decline to make a clinicomolecular diagnosis and adopt a watch and wait approach.
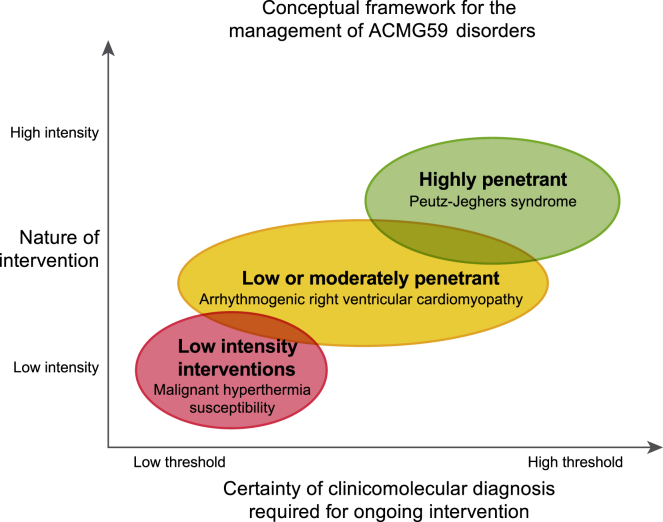
Figure 2.
Conceptual Framework for the Management of ACMG59 Disorders
The results of the clinical evaluation enable the clinician to determine whether or not an individual has a clinicomolecular diagnosis, but uncertainty may still exist. There is an overall positive correlation between the certainty of clinicomolecular diagnosis required for ongoing intervention (beyond the initial clinical evaluation) and the nature of that intervention. The nature of intervention includes consideration of medical risk, cost, and overall inconvenience. Malignant hyperthermia susceptibility can be managed with relatively low burden (avoiding triggering agents during surgery and communicating risk to clinicians) and therefore, even in the setting of an uncertain clinicomolecular diagnosis, these measures should be implemented. By contrast, an individual with Peutz-Jeghers syndrome would, at a minimum, undergo regular moderately invasive screening exams. Therefore, in this latter scenario, a higher degree of certainty is required for an individual to be considered to have a clinicomolecular diagnosis. Given both the catastrophic threat and diagnostic challenges associated with arrhythmogenic right ventricular cardiomyopathy, there are moderate intensity interventions that are acceptable for an individual even when there is uncertainty regarding a clinicomolecular diagnosis. The disorders noted are illustrative examples but are not the only disorders that fall into a given category. For any individual with a secondary finding, a clinician would weigh these factors for the given disorder and determine whether the results of the clinical evaluation meet a threshold for ongoing intervention. The categories are purposefully overlapping as they are not discrete entities but rather concepts for the clinician to be mindful of when establishing a long-term plan for an individual.
Disorders with Typical Childhood Presentation
For adults who present with a secondary finding in one of these genes (Table S1) along with a negative evaluation (that includes adequate personal history and physical, family history with variant segregation analysis, and clinical testing evaluation), it would be appropriate to consider this person to have a molecular finding that is not causing clinical disease. Follow up can be symptom-driven only, and in the absence of new signs or symptoms there may be no need to repeat diagnostic testing. The variant correlation in such cases may add insight. A likely pathogenic secondary finding may not truly be pathogenic or a pathogenic variant with reduced penetrance would be not be expected to cause disease in some individuals who harbor this variant. See Box 1 for an example scenario in this category.
Box 1. RB1 Hypothetical Example.
A 40-year-old man presents with a likely pathogenic secondary finding in RB1, a gene associated with retinoblastoma. He has no personal history of ocular problems and no history of cancer. A physical examination is unremarkable and he has normal visual acuity. The family history is also negative for ocular concerns, retinal tumors, or sarcomas in his parents and siblings; to the best of his knowledge, this is true for all of his grandparents as well. The man’s parents have provided samples to the genetic testing lab, and the variant is present in his father (effectively confirming that this is not a de novo variant). He is referred to an ophthalmologist and undergoes retinal exam with ophthalmoscopy and is found to have no abnormalities.
This person has had an adequate evaluation across all clinical domains with no findings of the expected phenotype of retinoblastoma. The variant is likely pathogenic, which allows a reasonable probability (as high as 10% before phenotyping33) that this variant does not cause disease. For this individual, no specific follow up with an ophthalmologist beyond routine population recommendations would be necessary. If he were to have new relevant signs or symptoms (such as vitreous hemorrhage), the secondary finding may help guide the clinical evaluation and differential diagnosis at that time, which may prompt a reanalysis of the clinical relevance of the secondary finding. For young children with a finding in one of these genes (e.g., a neonate sequenced for multiple anomalies), the approach would be quite different. Because such a child presents with the secondary finding before the typical age of onset of one of these disorders, the absence of clinical findings at this young age does not confidently exclude its development in the future—this is very different from the situation for the adult.
Disorders for Which Low-Intensity Interventions Are Recommended
For secondary findings in these genes (Table S1), the risk-to-benefit ratio of the recommended interventions favor such interventions, even in cases in which the evaluation cannot firmly establish a clinicomolecular diagnosis. This is because the interventions recommended are not overly burdensome or costly.37,38 For example, malignant hyperthermia susceptibility is a disease susceptibility phenotype and therefore does not reliably manifest detectable signs and symptoms (even in the setting of a known volatile anesthetic). The clinical diagnostic test is invasive and expensive (caffeine-halothane contraction test, which requires muscle biopsy). However, most surgical procedures can be performed avoiding known triggering agents, and effective measures (including stocking the reversing agent, dantrolene) can be taken if knowledge of the risk is communicated to the anesthesiologist. The same concept holds true for a secondary finding associated with familial hypercholesterolemia. While lipid levels are easy to measure for a given time point, evidence suggests that there is a genetic risk of cardiovascular disease—independent of lipid levels—due to pathogenic LDLR variants.6 Any person with a molecular finding associated with familial hypercholesterolemia should therefore have their lipids monitored and managed to a risk-appropriate goal (typically through pharmacotherapy) (see GeneReviews, Youngblom et al., in Web Resources). See Box 2 for an example scenario in this category.
Box 2. RYR1 Hypothetical Example.
A 30-year-old woman presents with a pathogenic variant in RYR1, a gene associated with malignant hyperthermia susceptibility. She has no significant medical history and has never had surgery or required anesthesia. No one in her family has ever experienced complications during surgery indicative of malignant hyperthermia and to the best of her knowledge no family members have experienced any notable intolerance to heat or exercise. The woman declines to undergo a muscle biopsy necessary for diagnostic contracture testing.
There is nothing in the clinical history suggestive of malignant hyperthermia. While contracture testing is considered the gold standard diagnostic test, the invasive nature of the required muscle biopsy (and potential lack of availability) will often preclude its use. Despite the lack of evidence of a clinicomolecular diagnosis, this individual should be counseled that her risk of malignant hyperthermia susceptibility is uncertain. Therefore, it would be prudent to treat her as at risk, and, consistent with best practices, any surgery she requires in the future should be done in a center familiar with the disease and potential triggering pharmacologic agents should be avoided.
Disorders with Low or Moderate Penetrance
The ACMG59 includes cardiovascular disorders and cancer susceptibility disorders that exhibit incomplete penetrance. The evidence for this penetrance has primarily come from studying family members of affected individuals in whom a causative variant had been identified. It is reasonable to expect that in the setting of a secondary finding, the penetrance may be somewhat less than the previously observed penetrance from family ascertainment-based studies.39, 40, 41 For this category of disorders (Table S1), an individual with an adequate negative evaluation can be considered unaffected. However, the clinician must still be cautious about excluding the clinicomolecular diagnosis given that the penetrance can be age dependent. The family-based cascade testing is crucial. If a parent or other older individuals in the family is found to also have the molecular finding, they should be highly encouraged to undergo the complete clinical evaluation. The presence or absence of a clinicomolecular diagnosis in older individuals with the same variant will inform the care of the presenting individual. If the older individuals undergo a complete evaluation and are not found to have a clinicomolecular diagnosis, then the presenting individual can be more confidently considered to be unaffected and would not require repeat diagnostic testing. We recognize that confirming variant status and having older individuals undergo complete medical evaluation is not always feasible and that secondary findings are not necessarily familial. Because these disorders are associated with significant morbidity and mortality and the nature of incomplete penetrance makes predicting disease outcomes difficult, in the absence of a comprehensive familial evaluation, it is most prudent to monitor these individuals similarly to how one would monitor an at-risk family member for a period of time after the initial presentation. Such an approach necessitates periodic reevaluation with repeat diagnostic testing performed, even in the absence of new symptoms. Repeat negative evaluations over time will significantly decrease the probability of a clinicomolecular diagnosis. See Box 3 for an example scenario in this category.
Box 3. PKP2 Hypothetical Example.
A 35-year-old man presents with a pathogenic variant in PKP2, a gene associated with arrhythmogenic right ventricular cardiomyopathy. There is no personal history of cardiovascular symptoms, a physical exam is unremarkable, and the family history is negative for cardiomyopathy, ventricular arrhythmias, sudden cardiac death, or incidents suspicious for undiagnosed cardiac death. Targeted variant analysis establishes that this variant was maternally inherited. Clinical evaluation has included a 12 lead ECG, 48 h Holter monitor, transthoracic echocardiogram, and cardiac MRI, which are negative for abnormalities. Detailed review of this variant is consistent with multiple lines of evidence supporting a pathogenic interpretation, but the disorder is known to show incomplete penetrance.
Despite this individual’s molecular finding, his presentation is not consistent with a clinicomolecular diagnosis and he can therefore be considered unaffected at the time of evaluation. Because disorders in this category often have age-dependent penetrance, clinical evaluations of family members who share the same variant are particularly important. Older family members with the variant should undergo the same evaluation and the results will inform the care of the presenting 35-year-old man. If no family members, including the mother, have evidence of a clinicomolecular diagnosis, the provider can take a watch-and-wait approach in the long-term management, with follow-up testing triggered only by new symptoms. However, a thorough evaluation in multiple family members with the same molecular finding will not always be feasible. In such instances, given the risk of later-onset disease, a repeat evaluation of cardiac rhythm and function should be performed periodically.
Disorders with High or Near Complete Penetrance for Some Disease Manifestation
The high penetrance of these disorders (Table S1) makes it likely that an individual with a pathogenic variant will have some indication of disease in the personal history and physical examination, family history, or diagnostic phenotyping evaluation. If an individual has a negative evaluation, the clinician should have a high index of suspicion that the variant identified is not actually pathogenic. For secondary findings in genes in this category, the variant correlation will help guide the follow-up plan. If the secondary finding is likely pathogenic, a thorough negative evaluation across all domains makes the probability of a clinicomolecular diagnosis very low and repeat diagnostic testing in the future would only be triggered by the development of new signs or symptoms. See Box 4 for an example scenario in this category.
Box 4. STK11 Hypothetical Example.
A 45-year-old woman presents with a likely pathogenic variant in STK11, a gene associated with Peutz-Jeghers syndrome. She has no personal history of cancer, benign tumors, or intussusception. A detailed skin examination does not identify any hyperpigmented macules or skin lesions, but she remembers having freckles as a child. The woman was adopted as an infant and does not know anything about her biological parents. She undergoes an evaluation which includes colonoscopy, upper endoscopy, capsule endoscopy, transvaginal US, serum CA 125 level, MRI-MRCP, and breast MRI which are all negative for abnormalities (including polyps).
The results of this evaluation are not consistent with a clinicomolecular diagnosis and the woman in this example should be considered unaffected. Although there is no family history information available, the near-complete penetrance and rarity of the disorder is such that the probability of disease is low in this setting. The significance of her molecular finding can be reassessed in the future if the woman develops concerning signs or symptoms that may be associated with Peutz-Jeghers syndrome (such as bowel obstruction or rectal prolapse), but in the absence of such signs or symptoms, no specific diagnostic follow-up testing is warranted. The provider here should understand that the nature of a highly penetrant disorder is such that a negative evaluation makes a molecular finding highly unlikely to lead to a clinicomolecular diagnosis. It is also important that this information be relayed to the laboratory to incorporate into an updated pathogenicity assessment of the variant, potentially to revise it for the presenting individual and for future reporting.
Secondary Findings in Children
The approach discussed above is tailored for the adult presenting with a secondary finding. Here we define adult as an individual who is responsible for his or her own medical decisions, which in the United States typically refers to someone who is 18 years or older or is considered an emancipated minor.42, 43, 44 The risks and benefits of returning secondary findings to children is complex and beyond the scope of this article. For a child who presents with a secondary finding, we recommend determining parental origin of the variant. For most adult-onset disorders, the parent who harbors the secondary finding would then become the index case subject and be evaluated in accordance with our approach (and the results of the parent’s evaluation would then be informative for the child’s long-term care plan). If a child presents with a secondary finding and the disease has pediatric onset, parental origin cannot be identified, the variant is confirmed de novo, or the secondary finding is in a disease with an autosomal-recessive inheritance pattern, we recommend that the child have an evaluation that is consistent with the established practices of pediatric genetics, specifically considering the typical childhood presentation and likelihood of benefit for interventions prior to adulthood.45,46 In many cases, the age-dependent penetrance of these disorders would support deferring these evaluations for several years.
Conclusion
This approach to the evaluation and clinical care of an individual with a secondary finding is based on a reasonable extrapolation of guidelines for cascade testing in unambiguously affected families, guided by rational risk assessment. It is meant to offer a guide for a physician caring for such an individual. As is true for all such recommendations, the framework we present should be tailored to an individual’s specific needs. The framework and gene categories should also be adjusted as needed as the field of medical genetics advances (specifically with regard to gene-disease association and penetrance).
Our approach does not address certain important aspects of genetic testing in general. These include the need for genetic counseling, ethical and legal considerations of genetic testing, the increasing availability of limited direct-to-consumer and consumer-initiated testing for certain genes on the ACMG59 list, and family planning implications of genetic results. The management of individuals with secondary findings is also complicated by the observation that participants in genome-sequencing studies may be unaware that the return of these results was a possibility (even if a consent process was documented).47 Such considerations underscore the importance of a referral to a genetics clinic or subspecialty clinic with genetic counselors available for a nuanced discussion of the molecular finding and the results from the evaluation, even in the absence of a clinicomolecular diagnosis. The approach presented here is deliberately focused on the clinical evaluation and management of a hypothetical adult with a secondary finding and is meant to be balanced with the important considerations applicable to genetic testing that fall outside the purview of this paper.
One of the promises of genomic medicine is the ability to predict phenotype based on genotype. The potential for secondary findings to identify previously unrecognized disease risk is one way in which genomic medicine can be predictive of disease and meaningfully impact patient care. Before this promise can be a reality, a standard clinical approach to the evaluation and longitudinal care for individuals with secondary findings must be implemented. As the scale of population sequencing studies increases, the return of secondary findings will become more common in the general population. A seven-gene ACMG59 subset associated with risk for hereditary breast and ovarian cancer, Lynch syndrome, and familial hypercholesterolemia—referred to as “Tier 1 genomic tests” by the Centers for Disease Control and Prevention—has been proposed as a screening tool for the general population, which would massively increase the number of people seeking care specifically because of a molecular finding.48 While our approach is focused on ACMG59 genes and disorders, the return of secondary findings from extended gene lists or in a research context is often considered at the discretion of the clinicians and the institution in which an individual is seeking care or engaged in research.15, 16, 17 Individuals with secondary findings outside of the ACMG59 can be managed using the framework we propose. In such instances, clinicians will consider the likelihood of a clinicomolecular diagnosis based on the same domains of evaluation we delineate herein. A clinician will need to determine the specific diagnostic phenotypic testing in order to establish the presence or absence of a clinicomolecular diagnosis, and what is known about the variant and disorder in question will guide long-term management.
Our goal is that all people with secondary findings are adequately evaluated and cared for. As a part of this process, it is essential that clinical outcomes be collected and aggregated to inform modifications and amendments to what we propose, which will also change the secondary findings gene list. The creation of a learning health system model to gather evidence for precision health has been proposed49 and the need for harmonization of outcomes50 to accelerate the accumulation of experience has been demonstrated. These efforts will be enhanced by the standardized approach advocated here. This has the immense benefit of reducing data heterogeneity, which in turn will facilitate rapid learning.49,50 Given current knowledge, a clinician following our approach will optimize the opportunity to mitigate disease while also remaining cognizant of the uncertainties inherent in genomic prediction. Ultimately, predictive genomic medicine has the potential to greatly improve health and healthcare. The implementation of a standardized approach to a secondary finding evaluation is one step toward genomics realizing this potential and future research will refine and improve the the initial approach we propose here.
Declaration of Interests
A.E.K. reports no competing interests. R.L.N. reports receiving salary and holding equity in Invitae and being a paid consultant for Pfizer, a compensated member and equity holder of the scientific advisory board of Genome Medical, an equity holder of the scientific advisory board of Maze Therapeutics, and an unpaid consultant for OMIM (based at Johns Hopkins School of Medicine). B.D.S. reports grant support from the intramural program of the National Human Genome Research Institute, receiving payments for being Deputy Editor-in-Chief at the American Journal of Medical Genetics, and receiving royalties from Oxford University Press. H.L.R. reports grant support from the National Institutes of Health outside the submitted work, salary support from the Broad Institute of MIT and Harvard which offers clinical sequencing, and being a compensated member and equity holder of the scientific advisory board of Genome Medical. M.S.W. reports grant support from the National Human Genome Research Institute outside the submitted work. L.G.B. reports grant support from the intramural research program of the National Human Genome Research Institute, being an uncompensated member of the medical ethics advisory board for Illumina, and receiving honoraria from Cold Spring Harbor Press.
Acknowledgments
This work was supported by the intramural research program of the National Human Genome Research Institute, grant HG200387 04 and the National Institutes of Health Director’s Challenge Award. We thank Allison De Moya of the National Human Genome Research Institute for compilations of resources and Julia Fekecs and Darryl Leja of the National Human Genome Research Institute for assistance with the graphics.
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.05.002.
Web Resources
Centers for Disease Control and Prevention genomic application toolkit, https://www.cdc.gov/genomics/implementation/toolkit/tier1.htm
GeneReviews, Jasperson, K.W., Patel, S.G., and Ahnen, D.J. (1993). APC-Associated Polyposis Conditions. https://www.ncbi.nlm.nih.gov/books/NBK1345/
GeneReviews, Milewicz, D.M., and Regalado, E. (1993). Heritable Thoracic Aortic Disease Overview. https://www.ncbi.nlm.nih.gov/books/NBK1120/
GeneReviews, Nielsen, M., Infante, E., and Brand, R. (1993). MUTYH Polyposis. https://www.ncbi.nlm.nih.gov/books/NBK107219/
GeneReviews, Petrucelli, N., Daly, M.B., and Pal, T. (1993). BRCA1- and BRCA2-Associated Hereditary Breast and Ovarian Cancer. https://www.ncbi.nlm.nih.gov/pubmed/20301425
GeneReviews, Youngblom, E., Pariani, M., and Knowles, J.W. (1993). Familial Hypercholesterolemia. https://www.ncbi.nlm.nih.gov/books/NBK174884/
OMIM, https://www.omim.org/
Supplemental Information
References
- 1.ACMG Board of Directors ACMG policy statement: updated recommendations regarding analysis and reporting of secondary findings in clinical genome-scale sequencing. Genet. Med. 2015;17:68–69. doi: 10.1038/gim.2014.151. [DOI] [PubMed] [Google Scholar]
- 2.Green R.C., Berg J.S., Grody W.W., Kalia S.S., Korf B.R., Martin C.L., McGuire A.L., Nussbaum R.L., O’Daniel J.M., Ormond K.E., American College of Medical Genetics and Genomics ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet. Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalia S.S., Adelman K., Bale S.J., Chung W.K., Eng C., Evans J.P., Herman G.E., Hufnagel S.B., Klein T.E., Korf B.R. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet. Med. 2017;19:249–255. doi: 10.1038/gim.2016.190. [DOI] [PubMed] [Google Scholar]
- 4.Neben C.L., Zimmer A.D., Stedden W., van den Akker J., O’Connor R., Chan R.C., Chen E., Tan Z., Leon A., Ji J. Multi-Gene Panel Testing of 23,179 Individuals for Hereditary Cancer Risk Identifies Pathogenic Variant Carriers Missed by Current Genetic Testing Guidelines. J. Mol. Diagn. 2019;21:646–657. doi: 10.1016/j.jmoldx.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Manickam K., Buchanan A.H., Schwartz M.L.B., Hallquist M.L.G., Williams J.L., Rahm A.K., Rocha H., Savatt J.M., Evans A.E., Butry L.M. Exome Sequencing-Based Screening for BRCA1/2 Expected Pathogenic Variants Among Adult Biobank Participants. JAMA Netw. Open. 2018;1:e182140. doi: 10.1001/jamanetworkopen.2018.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abul-Husn N.S., Manickam K., Jones L.K., Wright E.A., Hartzel D.N., Gonzaga-Jauregui C., O’Dushlaine C., Leader J.B., Lester Kirchner H., Lindbuchler D.M. Genetic identification of familial hypercholesterolemia within a single U.S. health care system. Science. 2016;354:354. doi: 10.1126/science.aaf7000. [DOI] [PubMed] [Google Scholar]
- 7.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hart M.R., Biesecker B.B., Blout C.L., Christensen K.D., Amendola L.M., Bergstrom K.L., Biswas S., Bowling K.M., Brothers K.B., Conlin L.K. Secondary findings from clinical genomic sequencing: prevalence, patient perspectives, family history assessment, and health-care costs from a multisite study. Genet. Med. 2019;21:1100–1110. doi: 10.1038/s41436-018-0308-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnston J.J., Rubinstein W.S., Facio F.M., Ng D., Singh L.N., Teer J.K., Mullikin J.C., Biesecker L.G. Secondary variants in individuals undergoing exome sequencing: screening of 572 individuals identifies high-penetrance mutations in cancer-susceptibility genes. Am. J. Hum. Genet. 2012;91:97–108. doi: 10.1016/j.ajhg.2012.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Natarajan P., Gold N.B., Bick A.G., McLaughlin H., Kraft P., Rehm H.L., Peloso G.M., Wilson J.G., Correa A., Seidman J.G. Aggregate penetrance of genomic variants for actionable disorders in European and African Americans. Sci. Transl. Med. 2016;8:364ra151. doi: 10.1126/scitranslmed.aag2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amendola L.M., Dorschner M.O., Robertson P.D., Salama J.S., Hart R., Shirts B.H., Murray M.L., Tokita M.J., Gallego C.J., Kim D.S. Actionable exomic incidental findings in 6503 participants: challenges of variant classification. Genome Res. 2015;25:305–315. doi: 10.1101/gr.183483.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorschner M.O., Amendola L.M., Turner E.H., Robertson P.D., Shirts B.H., Gallego C.J., Bennett R.L., Jones K.L., Tokita M.J., Bennett J.T., National Heart, Lung, and Blood Institute Grand Opportunity Exome Sequencing Project Actionable, pathogenic incidental findings in 1,000 participants’ exomes. Am. J. Hum. Genet. 2013;93:631–640. doi: 10.1016/j.ajhg.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y., Muzny D.M., Xia F., Niu Z., Person R., Ding Y., Ward P., Braxton A., Wang M., Buhay C. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olfson E., Cottrell C.E., Davidson N.O., Gurnett C.A., Heusel J.W., Stitziel N.O., Chen L.S., Hartz S., Nagarajan R., Saccone N.L., Bierut L.J. Identification of Medically Actionable Secondary Findings in the 1000 Genomes. PLoS ONE. 2015;10:e0135193. doi: 10.1371/journal.pone.0135193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dewey F.E., Murray M.F., Overton J.D., Habegger L., Leader J.B., Fetterolf S.N., O’Dushlaine C., Van Hout C.V., Staples J., Gonzaga-Jauregui C. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science. 2016;354:354. doi: 10.1126/science.aaf6814. [DOI] [PubMed] [Google Scholar]
- 16.National Academies of Sciences, Engineering, and Medicine . 2018. Returning Individual Research Results to Participants: Guidance for a New Research Paradigm (Washington, DC) [PubMed] [Google Scholar]
- 17.Schwartz M.L.B., McCormick C.Z., Lazzeri A.L., Lindbuchler D.M., Hallquist M.L.G., Manickam K., Buchanan A.H., Rahm A.K., Giovanni M.A., Frisbie L. A Model for Genome-First Care: Returning Secondary Genomic Findings to Participants and Their Healthcare Providers in a Large Research Cohort. Am. J. Hum. Genet. 2018;103:328–337. doi: 10.1016/j.ajhg.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simopoulos A.P., Committee for the Study of Inborn Errors of Metabolism (SIEM) Genetic screening: programs, principles, and research--thirty years later. Reviewing the recommendations of the Committee for the Study of Inborn Errors of Metabolism (SIEM) Public Health Genomics. 2009;12:105–111. doi: 10.1159/000156114. [DOI] [PubMed] [Google Scholar]
- 19.Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group Recommendations from the EGAPP Working Group: genetic testing strategies in newly diagnosed individuals with colorectal cancer aimed at reducing morbidity and mortality from Lynch syndrome in relatives. Genet. Med. 2009;11:35–41. doi: 10.1097/GIM.0b013e31818fa2ff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knowles J.W., Rader D.J., Khoury M.J. Cascade Screening for Familial Hypercholesterolemia and the Use of Genetic Testing. JAMA. 2017;318:381–382. doi: 10.1001/jama.2017.8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brothers K.B., Vassy J.L., Green R.C. Reconciling Opportunistic and Population Screening in Clinical Genomics. Mayo Clin. Proc. 2019;94:103–109. doi: 10.1016/j.mayocp.2018.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Black W.C., Welch H.G. Screening for disease. AJR Am. J. Roentgenol. 1997;168:3–11. doi: 10.2214/ajr.168.1.8976910. [DOI] [PubMed] [Google Scholar]
- 23.Biesecker L.G. Genomic screening and genomic diagnostic testing-two very different kettles of fish. Genome Med. 2019;11:75. doi: 10.1186/s13073-019-0696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohane I.S., Masys D.R., Altman R.B. The incidentalome: a threat to genomic medicine. JAMA. 2006;296:212–215. doi: 10.1001/jama.296.2.212. [DOI] [PubMed] [Google Scholar]
- 25.Christensen K.D., Bernhardt B.A., Jarvik G.P., Hindorff L.A., Ou J., Biswas S., Powell B.C., Grundmeier R.W., Machini K., Karavite D.J. Anticipated responses of early adopter genetic specialists and nongenetic specialists to unsolicited genomic secondary findings. Genet. Med. 2018;20:1186–1195. doi: 10.1038/gim.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biesecker L.G., Nussbaum R.L., Rehm H.L. Distinguishing Variant Pathogenicity From Genetic Diagnosis: How to Know Whether a Variant Causes a Condition. JAMA. 2018;320:1929–1930. doi: 10.1001/jama.2018.14900. [DOI] [PubMed] [Google Scholar]
- 27.Murray M.F. Your DNA is not your diagnosis: getting diagnoses right following secondary genomic findings. Genet. Med. 2016;18:765–767. doi: 10.1038/gim.2015.134. [DOI] [PubMed] [Google Scholar]
- 28.Christensen K.D., Vassy J.L., Jamal L., Lehmann L.S., Slashinski M.J., Perry D.L., Robinson J.O., Blumenthal-Barby J., Feuerman L.Z., Murray M.F., MedSeq Project Team Are physicians prepared for whole genome sequencing? a qualitative analysis. Clin. Genet. 2016;89:228–234. doi: 10.1111/cge.12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garrison N.A., Brothers K.B., Goldenberg A.J., Lynch J.A. Genomic Contextualism: Shifting the Rhetoric of Genetic Exceptionalism. Am. J. Bioeth. 2019;19:51–63. doi: 10.1080/15265161.2018.1544304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang S., Lincoln S.E., Kobayashi Y., Nykamp K., Nussbaum R.L., Topper S. Sources of discordance among germ-line variant classifications in ClinVar. Genet. Med. 2017;19:1118–1126. doi: 10.1038/gim.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrison S.M., Dolinsky J.S., Knight Johnson A.E., Pesaran T., Azzariti D.R., Bale S., Chao E.C., Das S., Vincent L., Rehm H.L. Clinical laboratories collaborate to resolve differences in variant interpretations submitted to ClinVar. Genet. Med. 2017;19:1096–1104. doi: 10.1038/gim.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amendola L.M., Jarvik G.P., Leo M.C., McLaughlin H.M., Akkari Y., Amaral M.D., Berg J.S., Biswas S., Bowling K.M., Conlin L.K. Performance of ACMG-AMP Variant-Interpretation Guidelines among Nine Laboratories in the Clinical Sequencing Exploratory Research Consortium. Am. J. Hum. Genet. 2016;98:1067–1076. doi: 10.1016/j.ajhg.2016.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tavtigian S.V., Greenblatt M.S., Harrison S.M., Nussbaum R.L., Prabhu S.A., Boucher K.M., Biesecker L.G., ClinGen Sequence Variant Interpretation Working Group (ClinGen SVI) Modeling the ACMG/AMP variant classification guidelines as a Bayesian classification framework. Genet. Med. 2018;20:1054–1060. doi: 10.1038/gim.2017.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harrison S.M., Rehm H.L. Is ‘likely pathogenic’ really 90% likely? Reclassification data in ClinVar. Genome Med. 2019;11:72. doi: 10.1186/s13073-019-0688-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daly M.B., Pilarski R., Yurgelun M.B., Berry M.P., Buys S.S., Dickson P., Domchek S.M., Elkhanany A., Friedman S., Garber J.E. NCCN Guidelines Insights: Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic, Version 1.2020. J. Natl. Compr. Canc. Netw. 2020;18:380–391. doi: 10.6004/jnccn.2020.0017. [DOI] [PubMed] [Google Scholar]
- 36.Chapa J., Haslam A., Prasad V. Interpreting the Effectiveness of Cancer Screening From National Population Statistics: Is It Sound Practice? Mayo Clin. Proc. 2019;94:951–956. doi: 10.1016/j.mayocp.2019.02.016. [DOI] [PubMed] [Google Scholar]
- 37.Aderibigbe T., Lang B.H., Rosenberg H., Chen Q., Li G. Cost-effectiveness analysis of stocking dantrolene in ambulatory surgery centers for the treatment of malignant hyperthermia. Anesthesiology. 2014;120:1333–1338. doi: 10.1097/ALN.0000000000000257. [DOI] [PubMed] [Google Scholar]
- 38.Ademi Z., Watts G.F., Pang J., Sijbrands E.J., van Bockxmeer F.M., O’Leary P., Geelhoed E., Liew D. Cascade screening based on genetic testing is cost-effective: evidence for the implementation of models of care for familial hypercholesterolemia. J. Clin. Lipidol. 2014;8:390–400. doi: 10.1016/j.jacl.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 39.Begg C.B. On the use of familial aggregation in population-based case probands for calculating penetrance. J. Natl. Cancer Inst. 2002;94:1221–1226. doi: 10.1093/jnci/94.16.1221. [DOI] [PubMed] [Google Scholar]
- 40.Haggerty C.M., James C.A., Calkins H., Tichnell C., Leader J.B., Hartzel D.N., Nevius C.D., Pendergrass S.A., Person T.N., Schwartz M. Electronic health record phenotype in subjects with genetic variants associated with arrhythmogenic right ventricular cardiomyopathy: a study of 30,716 subjects with exome sequencing. Genet. Med. 2017;19:1245–1252. doi: 10.1038/gim.2017.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ranola J.M.O., Tsai G.J., Shirts B.H. Exploring the effect of ascertainment bias on genetic studies that use clinical pedigrees. Eur. J. Hum. Genet. 2019;27:1800–1807. doi: 10.1038/s41431-019-0467-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alderman E.M., Breuner C.C., COMMITTEE ON ADOLESCENCE Unique Needs of the Adolescent. Pediatrics. 2019;144:144. doi: 10.1542/peds.2019-3150. [DOI] [PubMed] [Google Scholar]
- 43.Greydanus D.E., Patel D.R. Consent and confidentiality in adolescent health care. Pediatr. Ann. 1991;20:80–84. doi: 10.3928/0090-4481-19910201-09. [DOI] [PubMed] [Google Scholar]
- 44.English A. Treating adolescents. Legal and ethical considerations. Med. Clin. North Am. 1990;74:1097–1112. doi: 10.1016/s0025-7125(16)30504-1. [DOI] [PubMed] [Google Scholar]
- 45.American College of Medical Genetics and Genomics Incidental findings in clinical genomics: a clarification. Genet. Med. 2013;15:664–666. doi: 10.1038/gim.2013.82. [DOI] [PubMed] [Google Scholar]
- 46.Ross L.F., Saal H.M., David K.L., Anderson R.R., American Academy of Pediatrics. American College of Medical Genetics and Genomics Technical report: Ethical and policy issues in genetic testing and screening of children. Genet. Med. 2013;15:234–245. doi: 10.1038/gim.2012.176. [DOI] [PubMed] [Google Scholar]
- 47.Mackley M.P., Blair E., Parker M., Taylor J.C., Watkins H., Ormondroyd E. Views of rare disease participants in a UK whole-genome sequencing study towards secondary findings: a qualitative study. Eur. J. Hum. Genet. 2018;26:652–659. doi: 10.1038/s41431-018-0106-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murray M.F., Evans J.P., Angrist M., Uhlmann W.R., Lochner Doyle D., Fullerton S.M., Ganiats T.G., Hagenkord J., Imhof S., Rim S.H. NAM Perspectives; 2018. A Proposed Approach for Implementing Genomics-Based Screening Programs for Healthy Adults. [Google Scholar]
- 49.Williams M.S., Buchanan A.H., Davis F.D., Faucett W.A., Hallquist M.L.G., Leader J.B., Martin C.L., McCormick C.Z., Meyer M.N., Murray M.F. Patient-Centered Precision Health In A Learning Health Care System: Geisinger’s Genomic Medicine Experience. Health Aff. (Millwood) 2018;37:757–764. doi: 10.1377/hlthaff.2017.1557. [DOI] [PubMed] [Google Scholar]
- 50.Williams J.L., Chung W.K., Fedotov A., Kiryluk K., Weng C., Connolly J.J., Harr M., Hakonarson H., Leppig K.A., Larson E.B. Harmonizing Outcomes for Genomic Medicine: Comparison of eMERGE Outcomes to ClinGen Outcome/Intervention Pairs. Healthcare (Basel) 2018;6:6. doi: 10.3390/healthcare6030083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.