Abstract
CNOT1 is a member of the CCR4-NOT complex, which is a master regulator, orchestrating gene expression, RNA deadenylation, and protein ubiquitination. We report on 39 individuals with heterozygous de novo CNOT1 variants, including missense, splice site, and nonsense variants, who present with a clinical spectrum of intellectual disability, motor delay, speech delay, seizures, hypotonia, and behavioral problems. To link CNOT1 dysfunction to the neurodevelopmental phenotype observed, we generated variant-specific Drosophila models, which showed learning and memory defects upon CNOT1 knockdown. Introduction of human wild-type CNOT1 was able to rescue this phenotype, whereas mutants could not or only partially, supporting our hypothesis that CNOT1 impairment results in neurodevelopmental delay. Furthermore, the genetic interaction with autism-spectrum genes, such as ASH1L, DYRK1A, MED13, and SHANK3, was impaired in our Drosophila models. Molecular characterization of CNOT1 variants revealed normal CNOT1 expression levels, with both mutant and wild-type alleles expressed at similar levels. Analysis of protein-protein interactions with other members indicated that the CCR4-NOT complex remained intact. An integrated omics approach of patient-derived genomics and transcriptomics data suggested only minimal effects on endonucleolytic nonsense-mediated mRNA decay components, suggesting that de novo CNOT1 variants are likely haploinsufficient hypomorph or neomorph, rather than dominant negative. In summary, we provide strong evidence that de novo CNOT1 variants cause neurodevelopmental delay with a wide range of additional co-morbidities. Whereas the underlying pathophysiological mechanism warrants further analysis, our data demonstrate an essential and central role of the CCR4-NOT complex in human brain development.
Keywords: CNOT1, neurodevelopment, intellectual disability, Drosophila, genomics, exome sequencing, CCR4-NOT complex, de novo mutations, developmental delay
Main Text
Master regulators controlling development include, but are not limited to, paired box (PAX) proteins,1 SRY-related HMG-box (SOX) proteins,2 and the relatively unknown CCR4-NOT protein complex.3 Although the full spectrum of functional diversity for the CCR4-NOT complex has not yet been established, it has already become apparent that it is active on all levels of gene expression, from accessibility of the DNA to translation and degradation of mRNA.4
The human CCR4-NOT complex contains up to 11 different subunits,3 with each of the subunits having specified functions. For instance, the catalytic activity of CNOT6, CNOT6L, CNOT7, and CNOT8 plays an important role in the deadenylation step leading to mRNA degradation, and the E3 ligase activity5 of CNOT4 is involved in protein substrate recognition and ubiquitination. The residual subunits seem to have a scaffolding function,3,6,7 with CNOT1 being the central scaffolding protein (in)directly binding to all CCR4-NOT partners.8 In addition to its scaffolding function, CNOT1 has been considered as a translational regulator through the binding of nuclear receptors and as a regulator of deadenylase activity.9,10 For the latter, CNOT1 also exhibits the capacity to bind proteins that are not part of the CCR4-NOT complex, but are known to either be involved in general or tissue-specific mRNA degradation pathways.11, 12, 13, 14, 15 Interrogation of gnomAD, as well as visualization of the CNOT1 tolerance landscape using MetaDome16 (Figure S1), has shown that CNOT1 is significantly depleted of loss-of-function (LoF; pLI = 1.00) and missense variation (Z-score = 7.25), suggesting that such variants may lead to genetic disease. Indeed, two recent studies together reported five individuals with a recurrent de novo missense variant in CNOT1 (GenBank: NM_016284.4; c.1603C>T [p.Arg535Cys]) causing a novel syndrome characterized by pancreatic agenesis and holoprosencephaly (MIM: 618500).17,18 De novo variants elsewhere in the gene, however, have not been systematically been reported.
We collected 39 individuals (19 females, 20 males), from 37 nuclear families, with a (likely) pathogenic variant CNOT1 through international collaborations, facilitated by MatchMaker Exchange,19 for further molecular and clinical studies to establish the role of CNOT1 in neurodevelopmental disorders (Figure 1, Table S1). For 31 individuals, it was established that the variant had occurred de novo. For three individuals, the variant was inherited from a mildly affected (n = 2) or mosaic (n = 1) mother. Additional segregation to establish the inheritance of the CNOT1 variant in the two transmitting mothers by study of the maternal grandparents was not performed. For three individuals, inheritance could not be established because of absence of (one of the) parental samples (Table S1). The ages of the index subjects ranged from newborn to 22 years (Table S2). In essence, individuals with a (de novo) CNOT1 variant show a broad phenotypic spectrum (Figures 1 and S2 and Table S2). The most consistent features observed included intellectual disability (ID) (72%) of varying degree, development delay (DD) (92%), speech delay (83%), motor delay (83%), and hypotonia (74%) (Tables 1 and S2). Although facial abnormalities were very common (92%, Figure 1), these did not yield a typical gestalt. Similarly, abnormal growth (74%), behavioral problems (65%), abnormalities of the brain (65%) and skeletal, and muscle and soft tissue abnormalities (67%) were frequently observed, albeit with different features in each individual. From the clinical characteristics of the cohort, we concluded that there is no recognizable “CNOT1 phenotype.” This fits previous observations that genes involved in newly discovered neurodevelopmental disorders are often clinically not recognizable.20
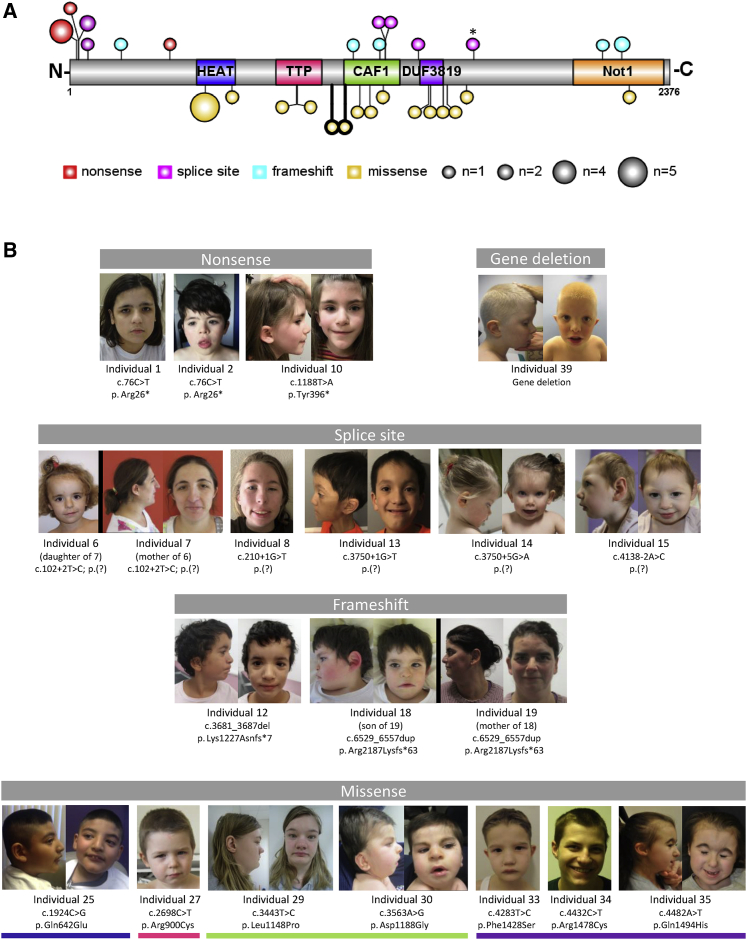
Figure 1.
Schematic Representation of CNOT1 and Facial Photos of Individuals Carrying a (Likely) Pathogenic CNOT1 Variant
(A) Graphical representation of the linear protein structure of CNOT1 with the functional domains indicated by colored boxes (blue, HEAT; pink, TTP; green, CAF; purple, DUF3819; orange, Not1). Variants observed in this study are indicated by circles, in which the size of the circle corresponds to the number of recurrences and the color of the circle to the variant type. Note that the variant in individual 16, affecting the last base of exon 34, creates a splice site (indicated by an asterisk). The variants in individual 28, who was shown to have two de novo missense variants, are outlined in bold.
(B) Facial photos of individuals with a (likely) pathogenic CNOT1 variant, categorized by the type of variant observed. The colored lines underneath the individuals with missense variants correspond to the functional domains indicated in (A). Although shared facial features can be observed between individuals in the cohort, e.g., straight eyebrows (individuals 2, 5, 6, 12, 18, 19, 27, 33), hypertelorism (individuals 18, 25, 29, 33, 39), epicanthus inversus (individuals 8, 18, 30, 33), low nasal bridge (individuals 2, 14, 18, 29, 39), large ears (individuals 15, 18, 25, 39), thickened helix (individuals 10, 14, 18, 25, 29, 35), and protruding helix (individuals 5, 7, 10, 14, 33, 39), a syndromic facial phenotype is lacking. Also, no evident facial resemblance is observed when comparing between individuals based on variant type. Clinical photos of hands and feet are provided in Figure S2 and additional clinical information for all individuals in this study is provided in Table S2 with a summary in Table 1.
Table 1.
Summary of Clinical Characteristics Associated with De Novo CNOT1 Individuals
|
All Individuals (n = 39) |
Individuals with Presumed Loss-of-Function Variants (n = 20) |
Individuals with Missense Variants (n = 19) |
||||
|---|---|---|---|---|---|---|
| % (present/assessed) | % (present/assessed) | % (present/assessed) | ||||
| Neurological Abnormalities | 97 | (38/39) | 100 | (20/20) | 95 | (18/19) |
| Intellectual disability | 72 | (23/32) | 67 | (12/18) | 79 | (11/14) |
| Normal | 13 | (4/32) | 11 | (2/18) | 14 | (2/14) |
| Borderline | 16 | (5/32) | 22 | (4/18) | 7 | (1/14) |
| Mild | 34 | (11/32) | 28 | (5/18) | 43 | (6/14) |
| Moderate | 9 | (3/32) | 17 | (3/18) | 0 | (0/14) |
| Severe | 13 | (4/32) | 6 | (1/18) | 21 | (3/14) |
| Unspecified | 16 | (5/32) | 17 | (3/18) | 14 | (2/14) |
| Developmental delay | 92 | (34/37) | 95 | (18/19) | 89 | (16/18) |
| Motor delay | 83 | (29/35) | 89 | (17/19) | 75 | (12/16) |
| Speech delay | 83 | (29/35) | 79 | (15/19) | 88 | (14/16) |
| Dysarthria | 34 | (10/29) | 28 | (5/18) | 45 | (5/11) |
| Epilepsy | 25 | (9/36) | 22 | (4/18) | 28 | (5/18) |
| Hypotonia | 74 | (26/35) | 79 | (15/19) | 69 | (11/16) |
| Behavioral disturbances | 63 | (20/32) | 61 | (11/18) | 64 | (9/14) |
| Sleep disturbances | 32 | (10/31) | 28 | (5/18) | 38 | (5/13) |
| Abnormal Brain Imaging | 65 | (20/31) | 64 | (9/14) | 65 | (11/17) |
| Holoprosencephaly | 13 | (4/31) | 0 | (0/14) | 24 | (4/17) |
| Other MRI findings | 62 | (18/29) | 64 | (9/14) | 60 | (9/15) |
| Abnormala Growth | 74 | (28/38) | 79 | (15/19) | 68 | (13/19) |
| Abnormal term of delivery | 16 | (6/38) | 21 | (4/19) | 11 | (2/19) |
| Preterm (<37 weeks of gestation) | 16 | (6/38) | 21 | (4/19) | 11 | (2/19) |
| Postterm (>42 weeks of gestation) | 0 | (0/38) | 0 | (0/19) | 0 | (0/19) |
| Abnormal birth weight | 23 | (8/35) | 17 | (3/18) | 29 | (5/17) |
| Small for gestational age | 23 | (8/35) | 17 | (3/18) | 29 | (5/17) |
| Large for gestational age | 0 | (0/35) | 0 | (0/18) | 0 | (0/17) |
| Abnormal head circumference at birth | 24 | (4/17) | 20 | (2/10) | 29 | (2/7) |
| Decreased head circumference | 18 | (3/17) | 10 | (1/10) | 29 | (2/7) |
| Increased head circumference | 6 | (1/17) | 10 | (1/10) | 0 | (0/7) |
| Abnormal height | 61 | (20/33) | 72 | (13/18) | 47 | (7/15) |
| Short stature | 55 | (18/33) | 72 | (13/18) | 33 | (5/15) |
| Tall stature | 6 | (2/33) | 0 | (0/18) | 13 | (2/15) |
| Abnormal head circumference | 26 | (9/34) | 28 | (5/18) | 25 | (4/16) |
| Decreased head circumference | 21 | (7/34) | 17 | (3/18) | 25 | (4/16) |
| Increased head circumference | 6 | (2/34) | 11 | (2/18) | 0 | (0/16) |
| Abnormal weight | 23 | (7/31) | 29 | (5/17) | 14 | (2/14) |
| Underweight | 19 | (6/31) | 24 | (4/17) | 14 | (2/14) |
| Overweight | 3 | (1/31) | 6 | (1/17) | 0 | (0/14) |
| Other Abnormalities | ||||||
| Facial abnormalities | 92 | (36/39) | 100 | (20/20) | 84 | (16/19) |
| Cardiac abnormalities | 33 | (8/24) | 42 | (5/12) | 25 | (3/12) |
| Urogenital abnormalities | 39 | (7/18) | 63 | (5/8) | 20 | (2/10) |
| Gastrointestinal abnormalities | 50 | (9/18) | 57 | (4/7) | 45 | (5/11) |
| Dysphagia/feeding difficulties | 50 | (14/28) | 33 | (5/15) | 69 | (9/13) |
| Pulmonal abnormalities | 24 | (4/17) | 13 | (1/8) | 33 | (3/9) |
| Immunological abnormalities | 14 | (2/14) | 33 | (2/6) | 0 | (0/8) |
| Endocrine abnormalities | 56 | (10/18) | 50 | (3/6) | 58 | (7/12) |
| Skeletal, muscle and soft tissue abnormalities | 67 | (16/24) | 77 | (10/13) | 55 | (6/11) |
| Hearing abnormalities | 12 | (3/25) | 8 | (1/13) | 17 | (2/12) |
| Vision abnormalities | 28 | (7/25) | 38 | (5/13) | 17 | (2/12) |
| Ectodermal abnormalities | 52 | (12/23) | 64 | (9/14) | 33 | (3/9) |
| Hands and feet abnormalities | 64 | (14/22) | 64 | (7/11) | 64 | (7/11) |
Only features present in at least 10% of all individuals are listed.
Abnormal growth parameters defined as > +2 SD and < −2 SD. Clinical details per individual are specified in Table S2.
Given the relative large number of individuals (n = 39) collected, we next reasoned that there may be a more subtle genotype-phenotype correlation, which could be based on the type of variant (truncating versus missense) or the location of the missense variants (e.g., different CNOT1 protein domains; Figure 1). Whereas previously a striking resemblance was observed for individuals with the same de novo p.Arg535Cys variant within the HEAT domain, consisting of holoprosencephaly and absence or insufficient function of the pancreas,17,18 no such other features were observed (Fishers’ exact tests), neither when discriminated by the type of variants nor for missense variants clustering to specific domains (Tables 1 and S1). Of note, individual 25, who had a de novo p.Gln642Glu, which is also located in the HEAT domain, did not have pancreatic agenesis and/or holoprosencephaly (Table S2), suggesting that variants leading to p.Arg535Cys represent a separate clinically recognizable entity within the spectrum of CNOT1 (neuro)developmental disorders.
All types of genetic variation were observed in CNOT1: three different nonsense variants of which one was recurrent in four individuals; six unique splice site variants, of which one was observed twice in an index and mother; five unique frameshifts, one of which was observed twice in an index and mother; 15 different missense variants, one recurrent in five individuals and one individual with two de novo missense variants; and lastly two (partial) gene deletions (Figure 1; Table S1). Interestingly, the missense variants of 16 of 18 individuals suggested clustering to CNOT1 functional domains: six affected the HEAT domain, which modulates substrate specificity; two in the TTP-binding domain involved in deadenylation; four in the CAF1 domain, binding proteins with catalytic properties; four in a domain of unknown function (DUF3819); and one in the Not1 domain, which is associated with interaction of multiple protein partners (Figure S1). Effect prediction of these missense variants shows that they in general occur in evolutionary well-conserved regions (Figure S3) and that they may disturb hydrogen bonds, salt bridges, and structural stability, thereby often affecting interactions with protein binding partners (Table S3). In addition, the position of the missense variants, buried in the core of the protein, may affect protein folding and stability (Figure S3).
To functionally elucidate the pathophysiological effects of the observed de novo variants on CNOT1 scaffolding capacity (Figure S4), we generated eight different CNOT1 variant constructs, including seven missense variants and one loss-of-function variant (Table S4) and transfected these in COS-1 cells. Using a PalmMyr-CFP-tagged construct, targeting CNOT1 to the cell membrane, we confirmed co-localization, as proxy for interaction, of wild-type CNOT1 with its partners CNOT2, CNOT4, and CNOT8 (Figures S5). We next assessed whether the de novo missense variants in CNOT1 impacted these interactions. Hereto, each CNOT1 variant was co-transfected with the interaction partner most likely to be disrupted (Figures S4 and S6; Table S5). Apart from one missense variant (p.Lys1241Arg), none of the variants seemed to affect binding of the respective CCR4-NOT1 interaction partner (Figure S6). Although the effects might be (1) more subtle than quantified here, (2) only identifiable in a different cell type system, or (3) dependent on the sub-unit composition which differs among tissues and during neural development,21 we concluded that in our cell-based assays the functional consequence of de novo CNOT1 variants is not mediated by major changes in the composition of the CCR4-NOT1 complex.
The CCR4-NOT complex has an important function in cell viability, and it has been shown that functional depletion of CNOT1 results in endoplasmic reticulum stress-induced apoptosis.9 We therefore set out to measure the consequence of de novo CNOT1 variants on apoptosis. We used fluorescence-activated cell sorting (FACS) to discriminate viable cells from those in apoptosis, by using 7-AAD and Annexin V as markers. This failed to observe increased apoptosis in three patient-derived Epstein-Barr virus immortalized B-lymphoid cell lines (Figure S7).
With the CCR4-NOT1 complex being involved in mRNA deadenylation, we then reasoned that its role in endonucleolytic nonsense-mediated mRNA decay (NMD) might be impaired. Therefore, we performed QuantSeq 3′mRNA sequencing (Lexogen) for five individuals with a de novo CNOT1 variant and 15 control subjects. As a proof-of-principle, we first evaluated the expression levels of CNOT1 in individuals with a de novo CNOT1 variant. Unexpectedly, the mRNA expression levels of individual 1 were normal, despite her de novo p.Arg26∗ variant (Figures S8A and S8B). This indicates that transcripts harboring the de novo p.Arg26∗ nonsense variant did not undergo NMD. By using allele-specific PCR primers for qPCR (Table S7), we determined that the wild-type and the mutant allele were indeed equally expressed (Figure S8C). While this observation would support the recently suggested non-canonical rule of NMD escape if the premature termination codon is located <150 nt from the start codon (start-proximal rule),22 this rule does not explain the observed normal levels of full-length CNOT1 protein by using CNOT1 antibodies against the C terminus (Figure S8D). Since the second methionine is located 234 amino acid residues downstream of the canonical one, this suggests translational upregulation of the single normal CNOT1 allele. Of note, any CNOT1 protein translated from the premature termination codon-containing allele escaping NMD would be only a maximum of 33 amino acids in size, and in the absence of an antibody directed against this, part of the protein remains undetectable.
There are two NMD pathways by which the cell degrades RNA.23 One is mainly performed by SMG6-dependent endocleavage, which degrades aberrant RNA with a premature stop codon. The other goes by SMG7-dependent exonucleolytic activity, which is usually directed toward transcripts with an upstream open reading frame or a long 3′ untranslated region. The latter is part of normal regulation of RNA expression, also referred to as “regulatory NMD.” We integrated the exome-sequencing data of five individuals with de novo CNOT1 variants with 3′mRNA Quantseq data, where analysis was aimed at the identification of SMG6-dependent NMD failure as a consequence of CCR4-NOT dysfunction. Hence, we focused on nonsense and frameshift variants (Figure S9), which are under normal physiological circumstances degraded by the endonucleolytic NMD pathway, but in individuals with de novo CNOT1 mutations would not. For all variants identified, we cross-referenced with data in GTEX to confirm that indeed the expression in controls with nonsense and frameshift variants in these genes show a reduced expression, fitting our hypothesis of NMD. This led to the identification of only three genes with a nonsense or frameshift variant in control subjects and individuals with de novo CNOT1 variants for which we were able to assess whether these genes showed reduced (proxy for NMD functional expression) or normal expression (proxy for NMD dysfunctional gene expression; Table S7 and Table S8). For these, no differences in endonucleolytic NMD were observed between control subjects and individuals with de novo CNOT1 variants (Figure S10). This suggests no influence of the five CNOT1 variants on SMG6-dependent endonucleolytic NMD, although effects at spatio-temporal level and/or subtle, more global (transcript-dependent) NMD effects cannot be excluded.
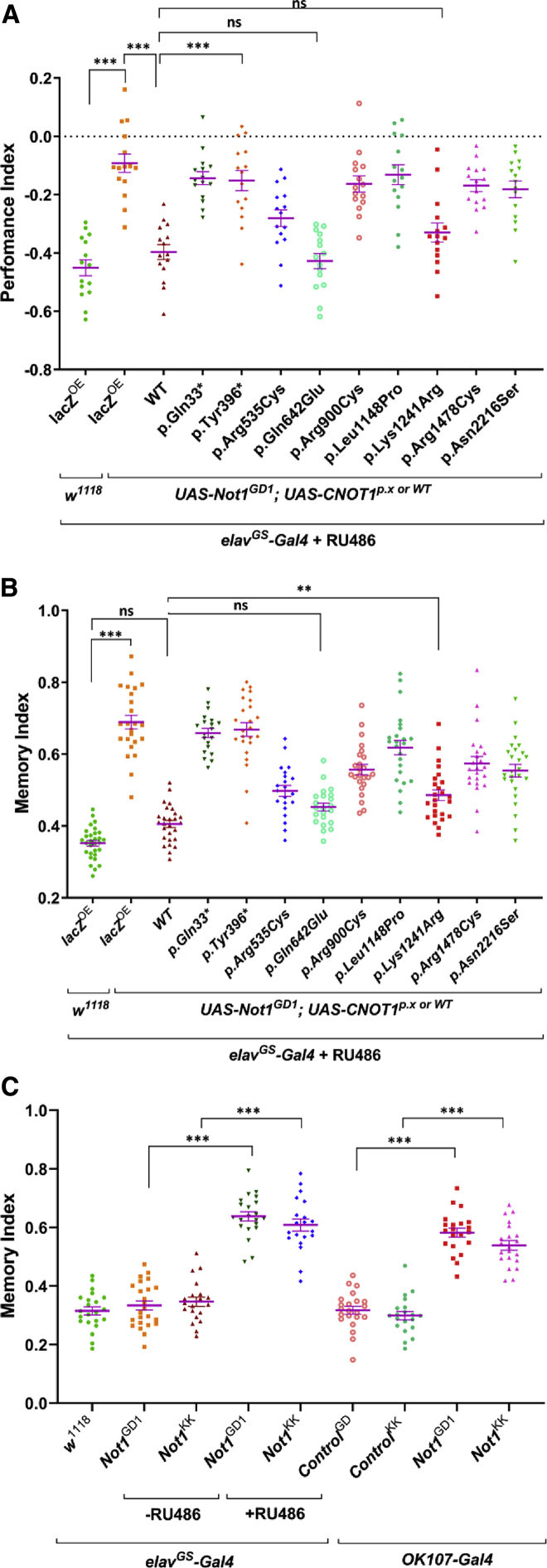
We next set out to study the effects of individual variants in a model organism, as our assays did not yield a convincing pathophysiological mechanism to disease. Hereto, we chose Drosophila melanogaster, in which Not1 is the ortholog of human CNOT1, showing 62% overall identity (Figure S11A). To test the effect of de novo CNOT1 variants on learning and memory, we used the UAS-GAL4 system in two different RNAi lines targeting Not1 (GD12571 for UAS-Not1GD1 and KK196587 for UAS-Not1KK), as well as for two different Gal4-driver lines, elav and OK107, for pan-neuronal or (memory-pivotal) mushroom body-specific knockdown, respectively, and assessed memory in a courtship suppression behavior (Figure S11B). As constitutive Not1 knockdown by elav-Gal4 led to early lethality after eclosion (Figure S12A), we adopted the elav-Gal4 GeneSwitch system (elavGS) inducible by RU486.24 Both RU induced pan-neuronal knockdown in adult flies and mushroom body-specific knockdown (OK107) of Not1 with both RNAi lines, showed a statistically increased memory index indicating impaired learning and memory in those flies (Figures 2A, S12A, and S12C). We next aimed to rescue the learning and memory phenotypes by introducing human wild-type CNOT1 cDNA, as well as by nine of the de novo variants identified in this study, which includes two loss-of-function variants and seven missense variants, all targeting different functional domains (Figure S11C). As both RNAi lines showed similar results upon Not1 knockdown, we only assessed the rescue of memory and learning loss by wild-type and variants of CNOT1 with the RU486-induced (elavGS>Not1GD1) knockdown line. When human wild-type CNOT1 is introduced, a statistically significant rescue of the memory phenotype was observed, whereas for eight of the nine mutants the phenotype could not or only partially be rescued (Figure 2B). The impact of these variants on learning and memory was further substantiated by two additional memory tests performed at larval stages, i.e., aversive and appetitive associative learning (Figures 2C and S12D). These findings suggest that the de novo CNOT1 mutations as observed in this study are likely sufficient to cause a neurodevelopmental disorder.
Figure 2.
Disease-Associated Variants of Human CNOT1 Induce Memory Deficits in Drosophila Model
(A) Depletion of Drosophila Not1 function in all neurons by inducible RNAi knockdown in adults only by using two different UAS-RNAi lines – Not1GD1 and Not1KK with elavGS-Gal4 and exposure to RU (GeneSwitch24), or specifically in the mushroom body with OK107-Gal4, produces memory deficits.
(B and C) RU-induced expression of disease-associated human CNOT1 variant cDNAs (UAS-CNOT1p.X or WT) in a pan-neuronal Not1GD1–RNAi knockdown background (using elavGS-Gal4) induces memory deficits both during adult (B) and larval (C) stages. One-way ANOVA, ∗∗p < 0.01, ∗∗∗p < 0.0001.
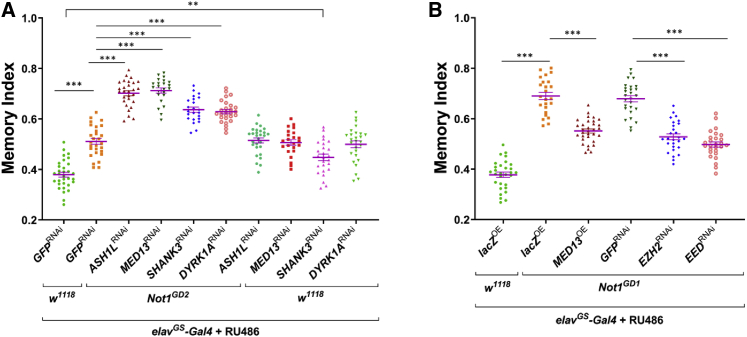
In order to test for a potential link between CNOT1 to genes previously implicated to impact ID/ASD cognitive functions, we used a weaker Not1 RNAi line, which produces only moderate memory defects, in conjunction with knockdown of ash1, MED13, shank, and Dyrk1a for the human genes ASH1L (MIM: 617796), MED13 (MIM: 618009), SHANK3 (MIM: 606232), and DYRK1A (MIM: 614104).25, 26, 27 Whereas we find that pan-neuronal knockdown of ash1, MED13, shank, and Dyrk1a by themselves also caused moderate memory defects, the defects are greatly enhanced by weaker Not1 knockdown (elavGS>Not1GD2; Figure 3A). Conversely, pan-neuronal overexpression of MED13 or knockdown of potential mediators of neurodevelopmental disorders (i.e., Polycomb Repressor Complex 2 [PRC2] components, E(z) and esc, for human EZH2 and EED)28,29 significantly reduces the severe memory defect of strong Not1 knockdown (Figure 3B). These results were further supported by an independent larval olfactory-mediated aversive-associative learning test (Figure S13): knockdown of ash1, MED13, shank, and Dyrk1a exacerbates the moderate larval memory deficits induced by the weaker Not1-RNAi line (Figure S13A). Conversely, overexpression of MED13 or reduction of PRC2 components rescued the memory phenotypes upon Not1 knockdown in larval brains (Figure S13B). Thus, in addition to its role in causing ID/ASD-associated developmental cognitive dysfunctions, our genetic data also implicate CNOT1 as a candidate in contributing to other neurodevelopmental phenotypes.
Figure 3.
Genetic Interactions between Not1 and Known ID/ASD Genes and Rescue of Neuronal Not1 Knockdown-Induced Neurodevelopmental Defects with Transcriptional Modifiers
(A) Neuronal RNAi knockdown of known ID/ASD genes (ASH1, DYRK1A, MED13, SHANK3; in flies ash1, mnb, skd, Prosap, respectively) exacerbates Not1 knockdown-induced memory defects in adults.
(B) Overexpression of MED13 or knockdown of PRC2 components E(z) (EZH2 in humans) and esc (EED in humans) ameliorates Not1 knockdown-induced memory deficits in adults. One-way ANOVA, ∗∗p < 0.01, ∗∗∗p < 0.0001.
In conclusion, we have collected 39 individuals with a neurodevelopmental disorder who have a broad clinical phenotypic spectrum, ranging from individuals with severe ID with co-morbidities such as seizures, hypotonia, and behavioral problems, to individuals with borderline normal IQ and normal everyday functioning. Functional characterization of the (de novo) variants at both the level of RNA and protein of CNOT1, as well as its functional roles in important biological processes such as mRNA decay and cell viability, have not revealed the pathophysiological mechanisms underlying this novel neurodevelopmental disorder. Nonetheless, knockdown and rescue experiments in Drosophila have indicated the plausibility of the pathogenicity of the variants on neurodevelopment, which is further strengthened by the interaction studies with autism-spectrum genes. Given the importance of CNOT1 in many essential biological processes, it is anticipated that de novo variants in CNOT1 impact its normal function in a more complex manner, involving either transcript- and/or tissue-dependent hypomorphic or neomorphic alleles.
Declaration of Interests
A.B. and K.M. are, and M.T.C. was, an employee of GeneDx, Inc.
Acknowledgments
We wish to thank all families participating in this study. In addition, we wish to thank the members of the Genome Technology Center and Cell culture facility, Department of Human Genetics, Radboud University Medical Center, Nijmegen, for data processing and cell culture of patient-derived cell lines, as well as the Radboud Technology Center for Flow Cytometry for help in FACS sorting. This work was financially supported by an Aspasia grant of the Dutch Research Council (015.014.066 to L.E.L.M.V.). D.L.P. is recipient of a CAPES Fellowship (99999.013311/2013-01). F.L.R. is funded from the Cambridge Biomedical Centre. In addition, the collaborations in this study were facilitated by the ERN ITHACA, one of the 24 European Reference Networks (ERNs) approved by the ERN Board of Member States, co-funded by European Commission. For more information about the ERNs and the EU health strategy please visit https://ec.europa.eu/health/ern. The aims of this study contribute to the Solve-RD project (to L.E.L.M.V.) which has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement No 779257.
Published: June 17, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.05.017.
Contributor Information
Lisenka E.L.M. Vissers, Email: lisenka.vissers@radboudumc.nl.
Rolf Bodmer, Email: rolf@sbpdiscovery.org.
Web Resources
OMIM, https://www.omim.org/
YASARA, http://www.yasara.org/
Supplemental Data
References
- 1.Relaix F. Pax genes: Master regulators of development and tissue homeostasis. Semin. Cell Dev. Biol. 2015;44:62–63. doi: 10.1016/j.semcdb.2015.10.036. [DOI] [PubMed] [Google Scholar]
- 2.Kamachi Y., Kondoh H. Sox proteins: regulators of cell fate specification and differentiation. Development. 2013;140:4129–4144. doi: 10.1242/dev.091793. [DOI] [PubMed] [Google Scholar]
- 3.Shirai Y.T., Suzuki T., Morita M., Takahashi A., Yamamoto T. Multifunctional roles of the mammalian CCR4-NOT complex in physiological phenomena. Front. Genet. 2014;5:286. doi: 10.3389/fgene.2014.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ukleja M., Valpuesta J.M., Dziembowski A., Cuellar J. Beyond the known functions of the CCR4-NOT complex in gene expression regulatory mechanisms: New structural insights to unravel CCR4-NOT mRNA processing machinery. BioEssays. 2016;38:1048–1058. doi: 10.1002/bies.201600092. [DOI] [PubMed] [Google Scholar]
- 5.Albert T.K., Hanzawa H., Legtenberg Y.I., de Ruwe M.J., van den Heuvel F.A., Collart M.A., Boelens R., Timmers H.T. Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex. EMBO J. 2002;21:355–364. doi: 10.1093/emboj/21.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collart M.A. The Ccr4-Not complex is a key regulator of eukaryotic gene expression. Wiley Interdiscip. Rev. RNA. 2016;7:438–454. doi: 10.1002/wrna.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Azzouz N., Panasenko O.O., Deluen C., Hsieh J., Theiler G., Collart M.A. Specific roles for the Ccr4-Not complex subunits in expression of the genome. RNA. 2009;15:377–383. doi: 10.1261/rna.1348209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y., Boland A., Kuzuoğlu-Öztürk D., Bawankar P., Loh B., Chang C.T., Weichenrieder O., Izaurralde E. A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol. Cell. 2014;54:737–750. doi: 10.1016/j.molcel.2014.03.034. [DOI] [PubMed] [Google Scholar]
- 9.Ito K., Takahashi A., Morita M., Suzuki T., Yamamoto T. The role of the CNOT1 subunit of the CCR4-NOT complex in mRNA deadenylation and cell viability. Protein Cell. 2011;2:755–763. doi: 10.1007/s13238-011-1092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winkler G.S., Mulder K.W., Bardwell V.J., Kalkhoven E., Timmers H.T. Human Ccr4-Not complex is a ligand-dependent repressor of nuclear receptor-mediated transcription. EMBO J. 2006;25:3089–3099. doi: 10.1038/sj.emboj.7601194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fabian M.R., Cieplak M.K., Frank F., Morita M., Green J., Srikumar T., Nagar B., Yamamoto T., Raught B., Duchaine T.F., Sonenberg N. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat. Struct. Mol. Biol. 2011;18:1211–1217. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki A., Saba R., Miyoshi K., Morita Y., Saga Y. Interaction between NANOS2 and the CCR4-NOT deadenylation complex is essential for male germ cell development in mouse. PLoS ONE. 2012;7:e33558. doi: 10.1371/journal.pone.0033558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murakawa Y., Hinz M., Mothes J., Schuetz A., Uhl M., Wyler E., Yasuda T., Mastrobuoni G., Friedel C.C., Dölken L. RC3H1 post-transcriptionally regulates A20 mRNA and modulates the activity of the IKK/NF-κB pathway. Nat. Commun. 2015;6:7367. doi: 10.1038/ncomms8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fabian M.R., Frank F., Rouya C., Siddiqui N., Lai W.S., Karetnikov A., Blackshear P.J., Nagar B., Sonenberg N. Structural basis for the recruitment of the human CCR4-NOT deadenylase complex by tristetraprolin. Nat. Struct. Mol. Biol. 2013;20:735–739. doi: 10.1038/nsmb.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du H., Zhao Y., He J., Zhang Y., Xi H., Liu M., Ma J., Wu L. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 2016;7:12626. doi: 10.1038/ncomms12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiel L., Baakman C., Gilissen D., Veltman J.A., Vriend G., Gilissen C. MetaDome: Pathogenicity analysis of genetic variants through aggregation of homologous human protein domains. Hum. Mutat. 2019;40:1030–1038. doi: 10.1002/humu.23798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Franco E., Watson R.A., Weninger W.J., Wong C.C., Flanagan S.E., Caswell R., Green A., Tudor C., Lelliott C.J., Geyer S.H. A Specific CNOT1 Mutation Results in a Novel Syndrome of Pancreatic Agenesis and Holoprosencephaly through Impaired Pancreatic and Neurological Development. Am. J. Hum. Genet. 2019;104:985–989. doi: 10.1016/j.ajhg.2019.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kruszka P., Berger S.I., Weiss K., Everson J.L., Martinez A.F., Hong S., Anyane-Yeboa K., Lipinski R.J., Muenke M. A CCR4-NOT Transcription Complex, Subunit 1, CNOT1, Variant Associated with Holoprosencephaly. Am. J. Hum. Genet. 2019;104:990–993. doi: 10.1016/j.ajhg.2019.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaplanis J., Samocha K.E., Wiel L., Zhang Z., Arvai K.J., Eberhardt R.Y., Gallone G., Lelieveld S.H., Martin H.C., McRae J.F. Integrating healthcare and research genetic data empowers the discovery of 49 novel developmental disorders. bioRxiv. 2019 doi: 10.1101/797787. [DOI] [Google Scholar]
- 21.Chen C., Ito K., Takahashi A., Wang G., Suzuki T., Nakazawa T., Yamamoto T., Yokoyama K. Distinct expression patterns of the subunits of the CCR4-NOT deadenylase complex during neural development. Biochem. Biophys. Res. Commun. 2011;411:360–364. doi: 10.1016/j.bbrc.2011.06.148. [DOI] [PubMed] [Google Scholar]
- 22.Hoek T.A., Khuperkar D., Lindeboom R.G.H., Sonneveld S., Verhagen B.M.P., Boersma S., Vermeulen M., Tanenbaum M.E. Single-Molecule Imaging Uncovers Rules Governing Nonsense-Mediated mRNA Decay. Mol. Cell. 2019;75:324–339.e11. doi: 10.1016/j.molcel.2019.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ottens F., Boehm V., Sibley C.R., Ule J., Gehring N.H. Transcript-specific characteristics determine the contribution of endo- and exonucleolytic decay pathways during the degradation of nonsense-mediated decay substrates. RNA. 2017;23:1224–1236. doi: 10.1261/rna.059659.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGuire S.E., Roman G., Davis R.L. Gene expression systems in Drosophila: a synthesis of time and space. Trends Genet. 2004;20:384–391. doi: 10.1016/j.tig.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 25.van Bon B.W., Coe B.P., Bernier R., Green C., Gerdts J., Witherspoon K., Kleefstra T., Willemsen M.H., Kumar R., Bosco P. Disruptive de novo mutations of DYRK1A lead to a syndromic form of autism and ID. Mol. Psychiatry. 2016;21:126–132. doi: 10.1038/mp.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S., DDD Study. Homozygosity Mapping Collaborative for Autism. UK10K Consortium Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snijders Blok L., Hiatt S.M., Bowling K.M., Prokop J.W., Engel K.L., Cochran J.N., Bebin E.M., Bijlsma E.K., Ruivenkamp C.A.L., Terhal P., DDD study De novo mutations in MED13, a component of the Mediator complex, are associated with a novel neurodevelopmental disorder. Hum. Genet. 2018;137:375–388. doi: 10.1007/s00439-018-1887-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moccia A., Martin D.M. Nervous system development and disease: A focus on trithorax related proteins and chromatin remodelers. Mol. Cell. Neurosci. 2018;87:46–54. doi: 10.1016/j.mcn.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuehner J.N., Bruggeman E.C., Wen Z., Yao B. Epigenetic Regulations in Neuropsychiatric Disorders. Front. Genet. 2019;10:268. doi: 10.3389/fgene.2019.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.