Abstract
Understanding of the nephrotoxicity induced by drug candidates is vital to drug discovery and development. Herein, an in situ metabolomics method based on air flow-assisted desorption electrospray ionization mass spectrometry imaging (AFADESI-MSI) was established for direct analysis of metabolites in renal tissue sections. This method was subsequently applied to investigate spatially resolved metabolic profile changes in rat kidney after the administration of aristolochic acid I, a known nephrotoxic drug, aimed to discover metabolites associated with nephrotoxicity. As a result, 38 metabolites related to the arginine–creatinine metabolic pathway, the urea cycle, the serine synthesis pathway, metabolism of lipids, choline, histamine, lysine, and adenosine triphosphate were significantly changed in the group treated with aristolochic acid I. These metabolites exhibited a unique distribution in rat kidney and a good spatial match with histopathological renal lesions. This study provides new insights into the mechanisms underlying aristolochic acids nephrotoxicity and demonstrates that AFADESI-MSI-based in situ metabolomics is a promising technique for investigation of the molecular mechanism of drug toxicity.
Key words: Aristolochic acid, Nephrotoxicity, Mass spectrometry imaging, In situ metabolomics, AFADESI
Abbreviations: AA, aristolochic acids; AAI, aristolochic acids I; AAN, AA-induced nephrotoxicity; AFADESI, air flow-assisted desorption electrospray ionization; ATP, adenosine triphosphate; CPT1, xarnitine palmitoyltransferase 1; DESI, desorption electrospray ionization; DG, diglyceride; GC, gas chromatograph; HDL, high-density lipoprotein; H&E, hematoxylin and eosin; LC, liquid chromatography; LDL, low-density lipoprotein; MALDI, matrix-assisted laser desorption ionization; MG, monoglyceride; MS, mass spectrometry; MSI, mass spectrometry imaging; OPLS-DA, orthogonal projections to the latent structures' discriminant analysis; PC, phosphatidylcholine; PG, phosphatidylglycerol; PE, phosphatidylethanolamine; PS, phosphatidylserine; ROI, region of interest; RSD, relative standard deviation; TG, triglyceride; TIC, total ion current; Ucr, urine creatinine; Upr, urine protein
Graphical abstract
This study investigated the spatially resolved metabolic profile changes in rat kidney after the administration of aristolochic acid I (AAI) by an in situ metabolomics method based on air flow-assisted desorption electrospray ionization mass spectrometry imaging (AFADESI-MSI), providing new insights into the mechanisms underlying aristolochic acids nephrotoxicity.
1. Introduction
Aristolochic acids (AAs) are structurally related nitrophenanthrene carboxylic acid derivatives found primarily in the Aristolochia, Bragantia, and Asarum species1. These acids have been used in obstetrics, and in the treatment of tumors and snake bites since antiquity. AAs exhibit multiple pharmacological effects, such as antiviral, antibacterial, and antineoplastic activities, and have been investigated as lead compounds by pharmaceutical companies in Germany for approximately 20 years until they were found to be carcinogenic in rats2. Their other main adverse effect is nephrotoxicity, which was initially reported in a number of women who had followed a slimming regimen with medicinal herbs containing AAs in Belgium and was subsequently confirmed by extensive studies in animals and humans3, 4, 5, 6. Epidemiological investigations have indicated that intake of herbs containing AAs and grains contaminated by AAs has been associated with the etiology of acute and chronic kidney diseases, which has caused global public health concerns, especially in Asia7, 8, 9.
Great efforts have been made to investigate the pathogenesis of renal injury and dysfunction induced by AAs. The nephrotoxicity of AAs has been found to be manifested by rapidly progressive interstitial nephropathy10, 11, 12 and thought to occur through multiple mechanisms, such as DNA damage, apoptosis, immune inflammation, oxidative stress, and endoplasmic reticulum stress13, 14, 15, 16, 17. However, the underlying mechanism of renal injury and dysfunction associated with AAs remains incompletely understood.
Discovering functional endogenous metabolites is very important for understanding the complex pathological process of AAs-induced nephrotoxicity. Metabolomics is a promising molecular profiling technology that provides high-throughput and unbiased evaluation of the metabolic responses of organisms to toxic stimuli, which makes it especially useful in the discovery of novel toxic biomarkers and elucidation of toxic mechanisms18,19. Various liquid chromatography–mass spectrometry (LC–MS) and gas chromatograph (GC)–MS-based metabolomics methods have been developed to study the nephrotoxicity of AAs20, 21, 22, 23, 24. These studies provide a wealth of information on the perturbation of various metabolites in biological samples for understanding the toxic effects of AAs, but little is known about their spatial distribution within the kidney, which is vital to unveiling the complex pathological processes underlying AAs-induced nephrotoxicity.
Mass spectrometry imaging (MSI) is a novel label-free and sensitive imaging technique that is regarded as most suitable for in situ metabolomics because of its ability to provide molecular and spatial information simultaneously. Desorption electrospray ionization MSI (DESI-MSI) under ambient conditions, matrix-assisted laser desorption ionization MSI (MALDI-MSI), and secondary ion MS have been used for in situ analysis of lipids, metabolites, drugs, and metals in various biological tissue sections25, 26, 27, 28, 29, 30, 31. Previously, our group developed a sensitive and high-coverage air-flow-assisted desorption electrospray ionization mass spectrometry imaging (AFADESI-MSI) technology and AFADESI-MSI based in situ metabolomics method, and demonstrated their outstanding performance and great potential in areas of in situ screening of cancer biomarkers, label-free molecular pathological diagnosis, and action mechanism study of drug candidates32, 33, 34, 35.
In the present study, an in situ metabolomics method was developed by combining AFADESI with an ultra-high resolution Q-OT-qIT mass spectrometer, aimed to investigate the acute nephrotoxicity effect induced by AA exposure. First, an AA-induced nephrotoxicity (AAN) rat model was produced by oral administration of aristolochic acids I (AAI) and verified by biochemical and histopathological examination. Then, various endogenous metabolites in rat kidney sections were detected by using the AFADESI-MSI method. A subsequent orthogonal partial least squares-discriminant analysis was performed to investigate the in situ metabolic changes in AAs’ nephrotoxicity. Finally, the differentiating metabolites were tentatively identified and imaged, providing a new perspective on the nephrotoxicity mechanism of AAI in rat. The research strategy is illustrated in Scheme 1.
Scheme 1.
The research strategy for in situ metabolomics in nephrotoxicity of aristolochic acids I. Scale bar: 2.5 mm.
2. Materials and methods
2.1. Chemicals and reagents
Aristolochic acid I was obtained from the National Institute for Food and Drug Control (Beijing, China). High-performance liquid chromatography-grade acetonitrile and methanol were purchased from Merck (Muskegon, MI, USA). Ultrapure water (Wahaha, Hangzhou, China) was obtained from a local market and used throughout the experiment.
2.2. Animals
Twelve Sprague–Dawley rats (8 weeks old) were purchased from Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). The rats were randomized to the AAN (n = 6) and control groups (n = 6). The AAN group of 100 mg/kg body weight was orally administered a single AAI dose in 0.5% carboxymethyl cellulose-Na. The control group was treated with 0.5% carboxymethyl cellulose-Na instead. On day 7 after administration, the rats from each group were placed in metabolic cages for the researchers to obtain 24 h urine samples. Blood samples were collected through the angular vein and centrifuged at 3000 × g for 10 min at 4 °C to obtain serum. Then, the rats were euthanized with CO2 gas, and their kidneys were immediately removed and snap-frozen in liquid nitrogen. All samples were stored at −80 °C until analysis. The animal experiments were in facilities approved by the Association for Assessment and Accreditation of Laboratory Animal Care International, and the animals were maintained in accordance with the “Guide for the Care and Use of Laboratory Animals”. All the animal experiments were approved by the Animal Care Committee of Peking Union Medical College and Chinese Academy of Medical Sciences.
2.3. Biochemical analysis and histopathological staining
Analyses of urine and serum were performed at the Deyi Diagnostics Corporation (Beijing, China). Concentrations of the urine protein (Upr) and creatinine, serum creatinine, blood urea nitrogen, serum high-density lipoprotein (HDL), and low-density lipoprotein (LDL) were measured. Then, after 24 h, the urine-protein–creatinine ratio (Upr/Ucr) was calculated.
The kidneys were sectioned at a 10 μm thickness using a Leica CM1860 cryostat (Leica Microsystem Ltd., Wetzlar, Germany) at −20 °C and stained with hematoxylin and eosin (H&E) to reveal histopathological lesions.
2.4. Sample preparation and AFADESI-MSI analysis
The kidneys were fixed atop a drop of saline on the cutting stage. Tissues were sectioned at a 10 μm thickness by using a Leica CM1860 cryostat (Leica Microsystem Ltd.) at −20 °C and thaw-mounted onto glass slides. All sections were stored at −80 °C until use and dried in a vacuum desiccator for approximately 30 min prior to analysis.
MSI experiments were performed on an AFADESI-MSI platform equipped with a Q-OT-qIT hybrid mass spectrometer (Orbitrap Fusion Lumos; Thermo Fisher Scientific, San Jose, CA, USA) and a home-made AFADESI ion source. The AFADESI-MSI analysis was carried out by continuously scanning the tissue surface in the x-direction at a constant rate of 200 μm/s, separated by a 200 μm vertical step in y-direction. The mass spectra were acquired in a positive full MS mode, with a scan range of 100–1000 Da, mass resolution of 120,000 at m/z 200, automatic gain control target of 2 × 105, and maximum injection time of 70 ms. The spray voltage and transport tube voltage were set to 7 and 3 kV, respectively. Nitrogen (0.5 MPa) and acetonitrile/water (8:2, v/v, 5 μL/min) was used as the spray gas and spray solvent, respectively. The extracting gas flow was 45 L/min, and the capillary temperature was 350 °C. Data were acquired by using Xcalibur software (Version 2.2, Thermo Scientific).
2.5. Data processing and analysis
Raw data files were converted to the cdf. format by using the Xcalibur software and then imported into custom-developed high-performance imaging software (MassImager 2.0, Beijing, China) for ion image reconstructions. After background subtraction and normalization, MS profiles from regions of interest (ROIs) were precisely extracted by matching H&E staining images of the adjacent section and generating separate two-dimensional data matrixes (m/z, intensity) in .txt format. The separated sample dataset matrixes were then imported into Markerview™ software 1.2.1 (AB SCIEX, Toronto, Ontario, Canada) for background deduction, peak picking, and peak alignment before multivariate statistical analyses. Multivariate analyses of the processed datasets from the ROIs were performed by using SIMCA-P 15.0 (Umetrics AB, Umea, Sweden). The metabolic profiles of the control and AAI-treated groups were compared by performing supervised multivariate OPLS-DA to achieve the maximum separation. Potential metabolic biomarkers were selected on the basis of their contribution to the class separation and variation within the data set. The paired t-test was used to compare the differences between the control group and the AAN group. Variables with P values < 0.05 were considered to be indicative of statistical significance when compared those of the control group.
3. Results
3.1. Optimization of conditions for AFADESI-MSI analysis
The AFADESI ion source was successfully coupled with high mass resolution Q-OT-qIT hybrid mass spectrometer by a home-made interface (Supporting Information Fig. S1). A stainless steel transport tube (i.d. 3 mm, o.d. 4 mm, length 500 mm) was used to connect the AFADESI system with the mass spectrometer and pump back the air flow. A sprayer-to-surface distance of 2.5 mm, a spray impact angle of 65°, and a collection angle of 15° were used to position the AFADESI spray. The parameters for the AFADESI-MSI platform were tuned to optimize the response of endogenous metabolites in kidney tissues. Most of the parameters for desorption/ionization of the AFADESI-MSI system were referred to in our previously reported work32. The instrument was tuned to achieve a mass accuracy of <2 ppm by external calibration, which gave a great advantage for rapid and direct identification of compounds in tissue.
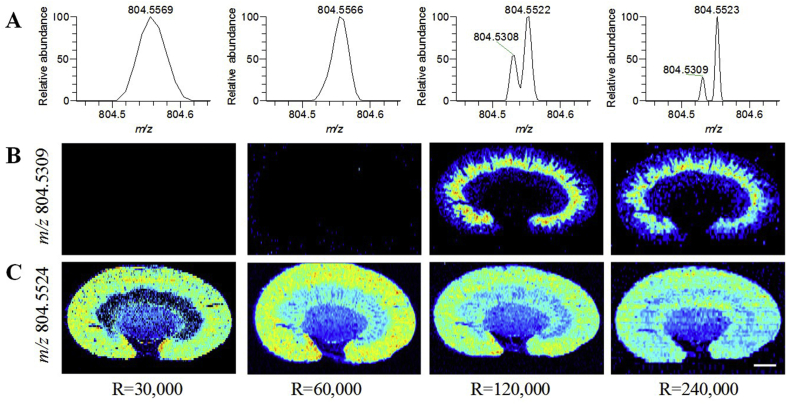
Mass resolution is an important factor that influences the MS image quality of a tissue section. High mass resolution combined with a small bin width is of great advantage for discrimination of a large number of endogenous metabolites with similar exact mass values when complex biological samples are under investigation. The effect of mass resolution on the selectivity, sensitivity, and spatial resolution was investigated by analyzing adjacent tissue sections from the same rat kidney. As illustrated in Fig. 1, when the MS resolution was increased from 30,000 to 240,000, signals that differ in mass by only 0.02 Da could be effectively separated by mass spectrometry, resulting in a significant improvement in the selectivity of the MS image (with a bin width of Δ m/z = 0.005) and a significant increase in the number of ions detected in full-scan MS analyses (Supporting Information Fig. S2A). However, the scanning speed decreased with increasing MS resolution in Orbitrap instruments, which might affect the sensitivity of the AFADESI-MS analysis differently from how it affected the selectivity of this analysis. When an MS resolution of 240,000 was employed, a significant decrease in total ion current was observed (Fig. S2B). In addition, decreased spatial resolution was also observed, which was probably due to the decreased scanning speed at an MS resolution of 240,000. As a result, an MS resolution of 120,000 at m/z 200 was chosen for the AFADESI-MSI analysis of metabolites as a good balance among specificity, sensitivity, and spatial resolution. Other parameters, such as spray solvent, ion-spray needle voltage, vaporizer temperature, sheath gas, aux gas, sweep gas, and isolation window, were also optimized to achieve the maximum MS intensity.
Figure 1.
MS spectra and images of metabolites with close m/z in rat kidney. (A) MS spectra of m/z 804.5309 and m/z 804.5524 ions in rat kidney. (B) MS image of m/z 804.5309. (C) MS image of m/z 804.5524. These two ions cannot be separated by mass spectrometry at MS resolutions of 30,000 and 60,000, but they can be distinguished at MS resolutions of 120,000 and 240,000. Scale bar: 2.5 mm.
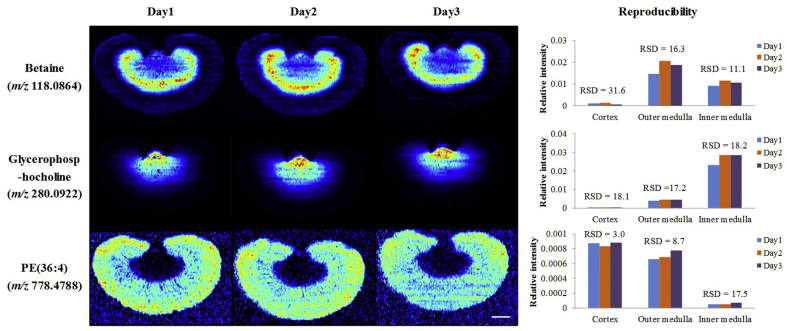
The reproducibility of the developed AFADESI-MSI method was assessed by analyzing adjacent renal slices on three successive days. Positive mode MS images for distribution of nine representative metbolites, including betaine (m/z 118.0864), glycerophosphocholine (m/z 280.0922), phosphatidylethanolamine (PE(36:4), m/z 778.4788), choline (m/z 104.1071), creatinine (m/z 136.0483), spermidine (m/z 146.1653), monoglyceride (MG(20:4), m/z 417.2405), linoleylcarnitine (m/z 424.3425), phosphatidylcholine (PC(38:4), m/z 832.5832), were extracted and compared, and the relative standard deviation (RSD) of the relative intensities of these metabolites in cortex, outer medulla, and inner medulla were also calculated to evaluate the intra-day precision. The reproducibility of MSI of betaine, glycerophosphocholine, and PE(36:4) are shown in Fig. 2, and the reproducibility of MSI of other six metabolites in Supporting Information Fig. S3.
Figure 2.
The reproducibility of the developed AFADESI-MSI method. Positive mode images for the distribution of betaine (m/z 118.0864), glycerophosphocholine (m/z 280.0922), and PE(36:4, m/z 778.4788) in adjacent renal slices analyzed on three successive days, and their relative standard deviation (RSD) of relative intensities in cortex, outer medulla, and inner medulla. Scale bar: 2.5 mm.
MS images of these metabolites with different m/z values in adjacent tissue sections were comparative when using total ion current (TIC) normalization to correct the possible differences in instrumental drift with time, and most of the RSD of the relative intensities of these metabolites were less than 20%, indicating the AFADESI-MSI proposed here can reproducibly provide semi-quantitative data, which enables comparison between sections under different pharmacological treatments analyzed in different times.
3.2. Assessment of renal damage in AAI-treated rats
The body weight and biochemical parameters are summarized in Table 1. Body weight was significantly reduced in the AAI-treated rats relative to that in the control rats. Twenty-four hour Upr, Upr/Ucr, and Ucr were significantly increased in the AAI-treated rats, which indicated that AAI had caused the nephrotoxic effect in rats. However, blood creatinine and blood urea nitrogen were only slightly higher in the AAI-treated groups, probably due to their limitations relating to sensitivity and specificity in the detection of histological injury in preclinical toxicity studies36,37. In addition, serum triglycerides were significantly increased in the AAI-treated group. Serum cholesteryl esters, serum HDL, and serum LDL had no significant differences between the two groups.
Table 1.
Body weight and biochemical parameters from aristolochic acid I (AAI)-treated and control rats on day 7 after administration.
| Parameter | Control | AAI-treateda |
|---|---|---|
| Body weight (g) | 284 ± 9.93 | 233 ± 16.3** |
| Ucr (urine creatinine, mmol/L) | 4.17 ± 0.86 | 4.85 ± 1.32* |
| Upr (24 h urine protein, mg) | 55.7 ± 11.6 | 75.3 ± 23.7* |
| Upr/Ucr | 0.65 ± 0.22 | 1.03 ± 0.14* |
| Bcr (blood creatinine, μmol/L) | 26.0 ± 2.45 | 27.6 ± 2.06 |
| Bun (blood urea nitrogen, mmol/L) | 5.00 ± 0.73 | 5.76 ± 0.45 |
| Serum CE (mmoL/L) | 1.48 ± 0.11 | 1.67 ± 0.29 |
| Serum TG (mmoL/L) | 0.66 ± 0.28 | 1.17 ± 0.45* |
| Serum HDL (mmoL/L) | 1.08 ± 0.05 | 1.17 ± 0.17 |
| Serum LDL (mmoL/L) | 0.28 ± 0.02 | 0.28 ± 0.08 |
P values were calculated by performing the independent Student's t-test. *P < 0.05; **P < 0.01; AAI, aristolochic acid I. CE, cholesteryl ester ; HDL, high-density lipoprotein; LDL, low-density lipoprotein; TG, triglyceride.
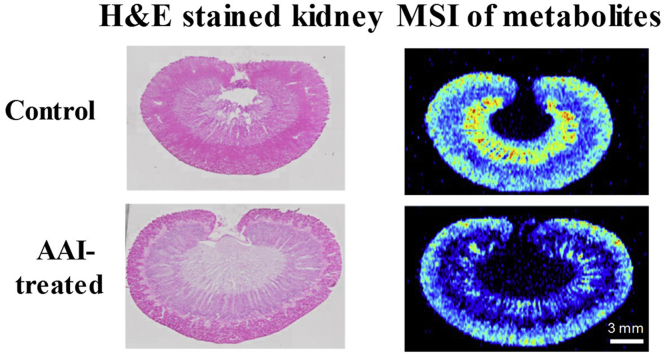
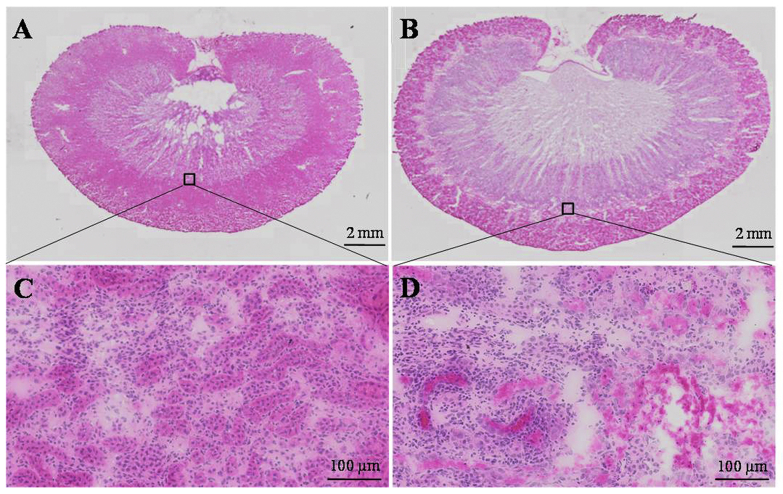
Representative photomicrographs of H&E-stained kidney tissues are presented in Fig. 3. The histological examinations of kidneys indicated swelling and morphological changes in the AAI-treated rats. Extensive tubular necrosis and inflammatory cell infiltration occurred across the cortex and corticomedullary. These pathological changes indicated that AAI caused rapidly progressive tubulo-interstitial lesions in rats.
Figure 3.
Photomicrographs of whole H&E stained renal sections from (A) control and (B) AAI-treated rats. The morphology of renal corticomedullary from control and AAI-treated rats are shown in images (C) and (D) with a 20-fold magnification, respectively.
3.3. AFADESI-MSI analysis of AAI-treated rat kidneys
To investigate the biological effects induced by acute AAI exposure on the kidneys of rats, in situ molecular profiles of frozen sections of kidneys of the controls and dosed (100 mg/kg AAI) rats were acquired by AFADESI-MSI operated in the positive-ion mode with a mass range of m/z 100−1000. Representative AFADESI-MSI spectra from the renal cortex, outer medulla, and inter medulla are presented in Supporting Information Fig. S4. Many peaks, corresponding to all types of metabolites, were observed in the AFADESI-MSI spectra; 512, 368, and 388 biological informative peaks (isotope excluded) were detected in the renal cortex, outer medulla, and inter medulla, respectively. The OPLS-DA scatter plot of the renal cortex, renal outer medulla, and renal inter medulla (Supporting Information Fig. S5A) indicated that the three regions from the control groups could be clearly separated for the AFADESI-MSI analysis, which indicated obvious differences in metabolite profiling of these morphology regions. Compared with the control group, the trend of separation among the renal cortex, renal outer medulla, and renal inter medulla from the AAI-treated group were less significant (Fig. S5B), probably because of tissue-specific effects of AAI exposure on metabolites in different morphology regions of the kidney.
A series of OPLS-DA analyses were performed to further investigate the metabolic effects of AAI exposure on the renal cortex, renal outer medulla, and renal inter medulla, respectively (Supporting Information Fig. S6). An analysis of three latent variables indicated that the R2(X), R2(Y), and Q2(cum) values were 60.1%, 96.5%, and 54.8%, respectively, for the orthogonal projections to the latent structures' discriminant analysis (OPLS-DA) model of the renal cortex; 76.4%, 88.2%, and 33.3%, respectively, for the OPLS-DA model of the renal outer medulla; and 71.8%, 84.7%, and 37.5%, respectively, for the OPLS-DA model of the renal inter medulla. The results indicated that the metabolomic alterations in the renal cortex after AAI treatment were more apparent than those in the renal outer medulla and renal inter medulla, which suggested that the renal cortex is more susceptible to AAI than the medulla. On the basis of a variable importance in projection threshold >1.0 and P < 0.05, we selected 85, 65, and 51 variables that accounted for the class separation as potential metabolic biomarkers for AA-induced toxicity of the renal cortex, renal outer medulla, and renal inter medulla, respectively. The structures of the potential biomarkers were provisionally identified on the basis of our previously described strategy. Briefly, tentative assignment of the structures of these potential biomarkers was first performed by searching the public databases METLIN (http://metlin.scripps.edu/), HMDB (http://hmdb.ca/), Massbank (http://massbank.imm.ac.cn/MassBank), and LIPID MAPS (http://www.lipidmaps.org/) in reference to the exact mass with a criterion of <2 ppm for mass accuracy and isotope patterns, which generated a possible list of endogenous metabolites. Then, the high-resolution MS/MS experiments were conducted directly from renal sections to obtain structural information based on the interpretation of the metabolites’ fragmentation patterns and database searches. As a result, a total of 38 discriminating metabolites were tentatively identified (Table 2). Fourteen metabolites, including creatinine, creatine, arginine, spermidine, choline, betaine, proline betaine, glycerophosphocholine, N-acetylhistamine, phosphoserine, inosine, fructosyl−lysine, carnitine, and linoleylcarnitine were provisionally identified on the basis of exact mass, exact MS/MS data, and databases. Because of the lack of useful fragmentation data, the remaining 27 discriminating metabolites were putatively assigned on the basis of exact mass and databases. The peak intensities of these 38 metabolites and their fold changes between the exposed and control groups are shown in Supporting Information Table S1.
Table 2.
Metabolites tentatively identified in the AFADESI-MSI positive-ion mode with renal distribution information in control and AAI-treated rats.
| Metabolite identification | Elemental composition | Adduct | Theoretical (m/z) | Measured (m/z) | Deltac (ppm) | Tissue distributiond |
|
|---|---|---|---|---|---|---|---|
| Control | AAI-treated | ||||||
| Creatininea | C4H7N3O | [M+Na]+ | 136.0481 | 136.0483 | 1.47 | IM, C | IM (↑), JC (↑) |
| Creatinea | C4H9N3O2 | [M+K]+ | 170.0326 | 170.0328 | 1.18 | C | C, M (↑) |
| Argininea | C6H14N4O2 | [M+H]+ | 175.1190 | 175.1191 | 0.57 | C | IM (↑), JC (↑) |
| Spermidinea | C7H19N3 | [M+H]+ | 146.1652 | 146.1653 | 0.68 | C | JC (↑), M (↑) |
| Cholinea | C5H13NO | [M+H]+ | 104.1070 | 104.1071 | 0.96 | C | C (↑), M (↑) |
| Betainea | C5H11NO2 | [M+H]+ | 118.0863 | 118.0864 | 0.85 | OM | OM (↓) |
| Proline betainea | C7H13NO2 | [M+H]+ | 144.1019 | 144.1021 | 1.39 | OM | OM (↓) |
| Glycerophosphocholinea | C8H20NO6P | [M+Na]+ | 280.0920 | 280.0922 | 0.71 | IM | IM (↑), JC (↑) |
| N-Acetylhistaminea | C7H11N3O | [M+H]+ | 154.0975 | 154.0977 | 1.30 | – | JC (↑) |
| Phosphoserinea | C3H8NO6P | [M+Na]+ | 207.9981 | 207.9980 | −0.48 | C, OM | JC (↑), M (↑) |
| Inosinea | C10H12N4O5 | [M+K]+ | 307.0439 | 307.0441 | 0.65 | C, M | C, M (↓) |
| Fructosyl-lysinea | C12H24N2O7 | [M+H]+ | 309.1656 | 309.1659 | 0.97 | JC | JC (↓) |
| Carnitinea | C7H15NO3 | [M+H]+ | 162.1125 | 162.1126 | 0.62 | OM | OM (↑) |
| Linoleylcarnitinea | C25H45NO4 | [M+H]+ | 424.3421 | 424.3425 | 0.94 | OM | OM (↓) |
| MG(18:2)b | C21H38O4 | [M+K]+ | 393.2402 | 393.2404 | 0.51 | C | JC (↑) |
| MG(20:4)b | C23H38O4 | [M+K]+ | 417.2402 | 417.2405 | 0.72 | C | JC (↑) |
| MG(22:5)b | C25H40O4 | [M+K]+ | 443.2558 | 443.2563 | 1.13 | – | JC (↑) |
| MG(22:6)b | C25H38O4 | [M+K]+ | 441.2402 | 441.2406 | 0.91 | C | JC (↑) |
| DG(36:2)b | C39H72O5 | [M+K]+ | 659.5011 | 659.5015 | 0.61 | C, JC | C, JC (↑), OM (↑) |
| DG(38:4)b | C41H72O5 | [M+K]+ | 683.5011 | 683.5020 | 1.32 | C, JC | C, JC (↑), OM (↑) |
| DG(38:6)b | C41H68O5 | [M+K]+ | 679.4698 | 679.4705 | 1.03 | C, JC | JC (↑) |
| DG(40:6)b | C43H72O5 | [M+K]+ | 707.5011 | 707.5023 | 1.70 | C | C, JC (↑) |
| PC(34:3)b | C42H78NO8P | [M+Na]+ | 778.5357 | 778.5361 | 0.51 | C, M | JC (↑) |
| PC(36:4)b | C44H80NO8P | [M+K]+ | 804.5514 | 804.5517 | 0.37 | C, JC, OM | JC (↑), OM |
| PC(38:4)b | C46H84NO8P | [M+Na]+ | 832.5827 | 832.5832 | 0.60 | C, M | C, JC (↑), M |
| PC(40:4)b | C48H88NO8P | [M+K]+ | 876.5879 | 876.5882 | 0.34 | OM | JC (↑), OM |
| PC(40:5)b | C48H86NO8P | [M+Na]+ | 858.5983 | 858.5981 | −0.23 | OM | JC (↑), OM |
| PC(40:6)b | C48H84NO8P | [M+K]+ | 872.5566 | 872.5572 | 0.69 | C, OM | C, JC (↑) |
| PE(34:2)b | C39H74NO8P | [M+K]+ | 754.4784 | 754.4791 | 0.93 | C, JC | C, JC (↓) |
| PE(P-36:4)b | C41H74NO7P | [M+Na]+ | 746.5095 | 746.5101 | 0.80 | C, JC, OM | C, JC (↓), OM |
| PE(36:4)b | C41H74NO8P | [M+K]+ | 778.4784 | 778.4788 | 0.51 | C, JC,OM | C, JC (↓), OM (↓) |
| PE(P-38:5)b | C43H76NO7P | [M+Na]+ | 788.4991 | 788.4998 | 0.89 | C, JC | C, JC (↓) |
| PE(38:5)b | C43H76NO8P | [M+K]+ | 804.4940 | 804.4947 | 0.87 | C, OM | C, OM (↓) |
| PE(38:6)b | C43H74NO8P | [M+K]+ | 802.4784 | 802.4789 | 0.62 | C, OM | C, OM (↓) |
| PS(39:4)b | C45H80NO10P | [M+H]+ | 826.5593 | 826.5606 | 1.57 | C, JC | C, JC (↓) |
| PS(35:1)b | C41H78NO10P | [M+Na]+ | 798.5256 | 798.5252 | −0.50 | C, JC | C, JC (↓) |
| PG(36:0)b | C42H83O10P | [M+Na]+ | 801.5616 | 801.5615 | −0.12 | C, JC | C, JC (↑) |
| PG(42:8)b | C48H79O10P | [M+Na]+ | 869.5303 | 869.5292 | −1.26 | C | C, JC (↑) |
Metabolites provisionally identified on the basis of exact mass, exact MS/MS data, and databases.
Metabolites putatively assigned on the basis of exact mass and databases with no useful fragmentation data.
Delta=(Measured m/z−Theoretical m/z)/Theoretical m/z × 106.
C, cortex; JC, juxtamedullary cortex; M, medulla; OM, outer medulla; IM, inner medulla. (↑) Indicates significant increase in local abundance; (↓) denotes significant decrease in local abundance. The peak intensities of these 38 metabolites and their fold changes between the exposed and control groups are shown in Supporting Information Table S1. –Not applicable.
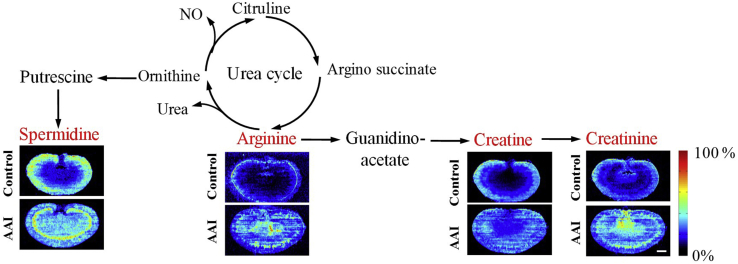
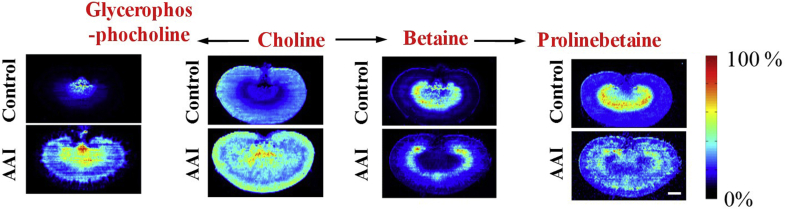
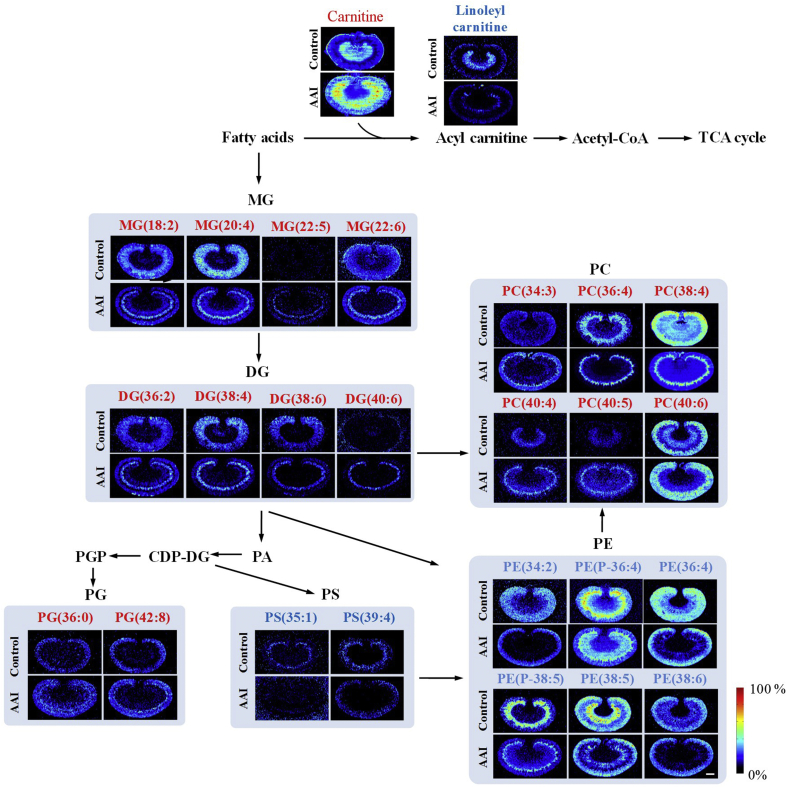
To further evaluate the association of these metabolites with AAI-induced nephrotoxicity, the MS images of these metabolites was extracted and compared with the histological features of AAI-treated kidney obtained by H&E staining of adjacent sections. The results indicated that the distribution of these metabolites showed a good spatial match with histopathological renal lesions (Figure 4, Figure 5, Figure 6, Figure 7).
Figure 4.
Spatial distribution and changes of metabolites involved in arginine–creatinine metabolic pathway in the kidneys from AAI-treated group and control group. The red fonts represent upregulated metabolites. Scale bar: 2.5 mm.
Figure 5.
Spatial distribution and changes of metabolites related to choline metabolism in the kidneys from AAI-treated group and control group. The red fonts represent upregulated metabolites. Scale bar: 2.5 mm.
Figure 6.
Spatial distribution and changes of metabolites related to lipids metabolism in the kidneys from AAI-treated group and control group. The red fonts represent upregulated metabolites and blue fonts represent downregulated metabolites. Scale bar: 2.5 mm.
Figure 7.
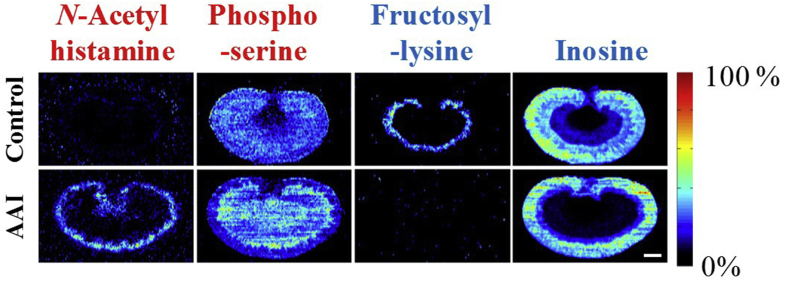
Spatial distribution and changes of N-acetylhistamine, phosphoserine, inosine, and fructosyl-lysine in the kidneys from AAI-treated group and control group. The red fonts represent upregulated metabolites and blue fonts represent downregulated metabolites. Scale bar: 2.5 mm.
4. Discussion
4.1. Perturbation on arginine–creatine metabolic pathway and urea cycle
The spatial distribution and changes in arginine, creatine, and creatinine (Fig. 4) demonstrated that the concentration of arginine, creatine, and creatinine were significantly increased, especially in the juxtamedullary cortex and inner medulla of kidneys, in the AAI-treated rats, respectively. Arginine is an important amino acid involved in multiple physiological processes, such as cell division, wound healing, ammonia removal from the body, immune function, blood pressure regulation, and hormones release38. Creatine and creatinine are downstream metabolites of the arginine–creatinine metabolic pathway. The present study suggested that AAI led to upregulation of the arginine–creatine metabolic pathway, which has been reported in patients with chronic kidney disease39,40.
The concentration of spermidine was significantly increased in the juxtamedullary cortex and medulla of kidneys in the AAI-treated rats (Fig. 4). Spermidine synchronizes an array of biological processes, such as Ca2+, Na+, K+-adenosine triphosphate (ATP)ase, thus maintaining membrane potential and controlling intracellular pH and volume. Spermidine is a precursor of spermine and a downstream metabolite of the urea cycle, which is part of the arginine biosynthesis metabolic pathway39. The increased spermidine and arginine suggested that AAI led to dysfunction of the urea cycle metabolic pathway, which was consistent with the finding of a previous study in which alteration of other metabolites related to the urea cycle, including aspartic acid, citrulline, and spermine, was associated with AAI toxicity22.
4.2. Disturbance of choline metabolism
The spatial distribution of choline and its metabolites in rat kidney is illustrated in Fig. 5. As a component of cell membranes, mitochondrial membranes, and the neurotransmitter acetylcholine, choline is involved in various biological pathways41. Betaine and glycerophosphocholine are metabolites of the oxidation and phosphorylation of choline, respectively. Known as important osmoprotectants, glycerophosphocholine and betaine participate in the formation of the axial osmolality gradient and are able to protect renal medulla cells against damaging influence induced by a high-urea environment41. Furthermore, they contribute to the maintenance of the structure and function of biological macromolecules in cells. Choline and glycerophosphocholine have been found to be increased significantly in the renal cortex and medulla of AAI-treated rats, whereas betaine and prolinebetaine have been found to be decreased significantly in the renal outer medulla; this indicates alterations of organic ion transportation in AAI-treated rats42.
4.3. Disturbance of lipids’ metabolism
The concentrations of carnitine and palmitoylcarnitine were significantly increased in the renal medulla, especially in the juxtamedullary cortex of the kidneys of the AAI-treated rats (Fig. 6). Carnitine and acylcarnitine have an important role in cellular energy distribution by mediating the transportation and metabolism of fatty acids. The binding of fatty acids to carnitine results in formation of acylcarnitine, which is essential for transportation of fatty acids to the mitochondria for energy production. The kidney is rich in mitochondria and has high energy demands. Therefore, mitochondrial dysfunction of the kidney has a critical role in the pathogenesis of kidney diseases43. The accumulation of carnitine and the reduction of linoleylcarnitine suggest that AAI leads to fatty acid metabolism disorder and mitochondrial kidney dysfunction. In addition, because these binding reactions have been found to be catalyzed by carnitine palmitoyltransferase 1 (CPT1)42, the accumulation of carnitine and the reduction of linoleylcarnitine suggested that AAI might lead to fatty acids metabolic disorder by inhibiting the activity of CPT1.
The spatial distribution and changes in lipids are illustrated in Fig. 6. A variety of lipids, including monoglyceride (MG), diglyceride (DG), triglyceride (TG), phosphatidylcholine (PC), phosphatidylglycerol (PG), phosphatidylserine (PS), and phosphatidylethanolamine (PE), were changed significantly in the AAI-treated rats. MG(18:2), MG(20:4), MG(22:5), MG(22:6), DG(36:2), DG(38:4), DG(38:6), DG(40:6), PC(34:3), PC(36:4), PC(38:4), PC(40:4), PC(40:5), PC(40:6), PG(36:0), and PG(42:8) were upregulated in the renal juxtamedullary cortex; PE(34:2), PE(P-36:4), PE(36:4), and PE(38:6) were downregulated in the renal juxtamedullary cortex and medulla; PE(P-38:5) was downregulated in the renal juxtamedullary cortex; and PE(38:5) was downregulated in the renal medulla. These results indicated disturbance of fatty acid metabolism. Lipids have many important biological functions and are involved in regulating a variety of life activities, including energy conversion, material transportation, information recognition and transmission, cellular development, differentiation, and apoptosis44,45. These findings indicated the potential role of lipid mediators in development of AAN, as reported in previous studies20,21.
4.4. Effects on metabolism of N-acetylhistamine, phosphoserine, inosine, and fructosyl–lysine
The spatial distribution and changes in N-acetylhistamine, phosphoserine, inosine, and fructosyl–lysine are illustrated in Fig. 7. N-Acetylhistamine was upregulated in the renal juxtamedullary cortex. N-Acetylhistamine is a metabolite of histamine and an important factor in anaphylactoid reactions46. The results suggested that AAI caused anaphylactoid reactions in the renal juxtamedullary cortex, and N-acetylhistamine may have an important role in the progression of AAI-induced renal injury.
Phosphoserine was increased significantly in the renal juxtamedullary cortex. Phosphoserine is an ester of serine and phosphoric acid, and an important intermediate product of the serine synthesis pathway. The increased phosphoserine probably reflected increased serine synthesis and/or altered filtration by kidneys.
Treatment with AAI caused a significant decrease in inosine in the renal outer medulla. Inosine is typically formed as a breakdown product of ATP and is a known marker of oxidative stress. The kidney is rich in mitochondria and highly demanding of energy. Therefore, mitochondrial dysfunction has a vital role in the pathogenesis of renal diseases47. The decrease may reflect interruption of energy metabolism and mitochondrial dysfunction of the kidney.
Fructosyl–lysine, a conjugate of fructose and lysine, was downregulated in the destroyed area. Lysine is a cationic amino acid and the substrate of organic cationic transporters. The decrease in fructosyl–lysine may reflect the altered activity of renal transporters in the AAI-treated rats42.
The underlying mechanism responsible for the observed changes in different metabolites in the renal tissues of AAI treated rats is presently unclear. But, changes in the content and distribution of most metabolites appear to be associated with mitochondrial dysfunction. One possible explanation for these results is that AAI exposure may induce mitochondrial damage in renal tubular epithelial cell43,47,48, resulting the dysfunction of the mitochondrial lipid metabolism, interruption of energy metabolism, altered activity of ATP-dependent renal transporters, apoptosis, and inflammation. In addition, most of discriminating metabolites in the lesion area were found to be lipids, indicating the lipid metabolism pathway may be closely associated with the nephrotoxicity of aristolochic acids and lipid metabolites may have a potential to be tissue-specific nephrotoxic biomarkers.
5. Conclusions
In summary, an in situ metabolomics method based on AFADESI-MSI was developed to investigate the spatially resolved metabolic effects induced by AAI exposure. The results indicated that the distribution of 38 metabolites related to the arginine–creatine metabolic pathway, the urea cycle, and metabolism of lipids, histamine, serine, lysine, and ATP were changed significantly in the AAI-treated rats. These findings indicated that the proposed AFADESI-MSI-based metabolomics strategy was capable of visualizing unique distributions and alterations of both known and unknown endogenous metabolites with high throughput and high molecular specificity, without the need for labeling and complicated sample pretreatment process. Nevertheless, the molecular images obtained by AFADESI-MSI analysis can be correlated directly with histological staining images, which is helpful for discovery of in situ biomarkers closely related to drug toxicity and for revealing the potential molecular mechanism underlying the toxicity.
Acknowledgments
This research was supported by the National Key Research and Development Program of China (No. 2017YFC1704000) and Outstanding Talent Support Program of Beijing, China (No. 2017000020124G272).
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2019.12.004.
Author contributions
Zeper Abliz and Zhonghua Wang designed and conceptualized this study, Zeper Abliz, Zhonghua Wang, and Jinfeng Wei planned the experiments. Zhonghua Wang, Chunyan Yang, Yaqi Liu, and Wenqing Fu conducted the animal experiments and AFADESI-MSI analysis. Zhonghua Wang, Bingshu He, and Meiling Huo analyzed data. Zhonghua Wang and Bingshu He drafted the manuscript, Zeper Abliz revised the manuscript.
Conflicts of interest
The authors have no conflicts of interest to declare.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Chan W., Cui L., Xu G., Cai Z. Study of the phase I and phase II metabolism of nephrotoxin aristolochic acid by liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2006;20:1755–1760. doi: 10.1002/rcm.2513. [DOI] [PubMed] [Google Scholar]
- 2.Mengs U., Lang W., Poch J.A. The carcinogenic action of aristolochic acid in rats. Arch Toxicol. 1982;51:107–119. [Google Scholar]
- 3.Vanherweghem J.L., Tielemans C., Abramowicz D., Depierreux M., Vanhaelen-Fastre R., Vanhaelen M. Rapidly progressive interstitial renal fibrosis in young women: association with slimming regimen including Chinese herbs. Lancet. 1993;341:387–391. doi: 10.1016/0140-6736(93)92984-2. [DOI] [PubMed] [Google Scholar]
- 4.Liu M.C., Maruyama S., Mizuno M., Morita Y., Hanaki S., Yuzawa Y. The nephrotoxicity of Aristolochia manshuriensis in rats is attributable to its aristolochic acids. J Clin Exp Nephrol. 2003;7:186–194. doi: 10.1007/s10157-003-0229-z. [DOI] [PubMed] [Google Scholar]
- 5.Lord G.M., Tagore R., Cook T., Gower P., Pusey C.D. Nephropathy caused by Chinese herbs in the UK. Lancet. 1999;354:481–482. doi: 10.1016/S0140-6736(99)03380-2. [DOI] [PubMed] [Google Scholar]
- 6.Gökmen M.R., Cosyns J.P., Arlt V.M., Stiborová M., Phillips D.H., Schmeiser H.H. The epidemiology, diagnosis, and management of aristolochic acid nephropathy: a narrative review. Ann Intern Med. 2013;158:469–477. doi: 10.7326/0003-4819-158-6-201303190-00006. [DOI] [PubMed] [Google Scholar]
- 7.Xu X., Nie S., Liu Z., Chen C., Xu G., Zha Y. Epidemiology and clinical correlates of AKI in Chinese hospitalized adults. Clin J Am Soc Nephrol. 2015;10:1510. doi: 10.2215/CJN.02140215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grollman A.P., Shibutani S., Moriya M., Miller F., Wu L., Moll U. Aristolochic acid and the etiology of endemic (Balkan) nephropathy. Proc Natl Acad Sci USA. 2007;104:12129. doi: 10.1073/pnas.0701248104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Debelle F.D., Vanherweghem J.L., Nortier J.L. Aristolochic acid nephropathy: a worldwide problem. Kidney Int. 2008;74:158–169. doi: 10.1038/ki.2008.129. [DOI] [PubMed] [Google Scholar]
- 10.Debelle F.D., Nortier J.L., de Prez E.G., Garbar C.H., Vienne A.R., Salmon I.J. Aristolochic acids induce chronic renal failure with interstitial fibrosis in salt-depleted rats. J Am Soc Nephrol. 2002;13:431. doi: 10.1681/ASN.V132431. [DOI] [PubMed] [Google Scholar]
- 11.Shibutani S., Dong H., Suzuki N., Ueda S., Miller F., Grollman A.P. Selective toxicity of aristolochic acids I and II. Drug Metab Dispos. 2007;35:1217. doi: 10.1124/dmd.107.014688. [DOI] [PubMed] [Google Scholar]
- 12.Yang L., Li X., Wang H. Possible mechanisms explaining the tendency towards interstitial fibrosis in aristolochic acid-induced acute tubular necrosis. Nephrol Dial Transplant. 2006;22:445–456. doi: 10.1093/ndt/gfl556. [DOI] [PubMed] [Google Scholar]
- 13.Randerath K., Reddy M.V., Gupta R.C. 32P-labeling test for DNA damage. Proc Natl Acad Sci USA. 1981;78:6126. doi: 10.1073/pnas.78.10.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prabu S.M., Muthumani M. Silibinin ameliorates arsenic induced nephrotoxicity by abrogation of oxidative stress, inflammation and apoptosis in rats. Mol Biol Rep. 2012;39:11201–11216. doi: 10.1007/s11033-012-2029-6. [DOI] [PubMed] [Google Scholar]
- 15.Pozdzik A.A., Salmon I.J., Husson C.P., Decaestecker C., Rogier E., Bourgeade M.F. Patterns of interstitial inflammation during the evolution of renal injury in experimental aristolochic acid nephropathy. Nephrol Dial Transplant. 2008;23:2480–2491. doi: 10.1093/ndt/gfn140. [DOI] [PubMed] [Google Scholar]
- 16.Hsin Y.H., Cheng C.H., Tzen J.T., Wu M.J., Shu K.H., Chen H.C. Effect of aristolochic acid on intracellular calcium concentration and its links with apoptosis in renal tubular cells. Apoptosis. 2006;11:2167. doi: 10.1007/s10495-006-0289-0. [DOI] [PubMed] [Google Scholar]
- 17.Zhu S., Wang Y., Jin J., Guan C., Li M., Xi C. Endoplasmic reticulum stress mediates aristolochic acid I-induced apoptosis in human renal proximal tubular epithelial cells. Toxicol In Vitro. 2012;26:663–671. doi: 10.1016/j.tiv.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Pan L., Han P., Ma S., Peng R., Wang C., Kong W. Abnormal metabolism of gut microbiota reveals the possible molecular mechanism of nephropathy induced by hyperuricemia. Acta Pharm Sin B. 2020;10:249–261. doi: 10.1016/j.apsb.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi J., Cao B., Wang X.W., Aa J.Y., Duan J.A., Zhu X.X. Metabolomics and its application to the evaluation of the efficacy and toxicity of traditional Chinese herb medicines. J Chromatogr B. 2016;1026:204–216. doi: 10.1016/j.jchromb.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 20.Ni Y., Su M., Qiu Y., Chen M., Liu Y., Zhao A. Metabolic profiling using combined GC–MS and LC–MS provides a systems understanding of aristolochic acid-induced nephrotoxicity in rat. FEBS Lett. 2007;581:707–711. doi: 10.1016/j.febslet.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Y.Y., Wang H.L., Cheng X.L., Wei F., Bai X., Lin R.C. Metabolomics analysis reveals the association between lipid abnormalities and oxidative stress, inflammation, fibrosis, and Nrf2 dysfunction in aristolochic acid-induced nephropathy. Sci Rep. 2015;5:12936. doi: 10.1038/srep12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Y.Y., Tang D.D., Chen H., Mao J.R., Bai X., Cheng X.H. Urinary metabolomics and biomarkers of aristolochic acid nephrotoxicity by UPLC–QTOF/HDMS. Bioanalysis. 2015;7:685–700. doi: 10.4155/bio.14.309. [DOI] [PubMed] [Google Scholar]
- 23.Lin S., Chan W., Li J., Cai Z. Liquid chromatography/mass spectrometry for investigating the biochemical effects induced by aristolochic acid in rats: the plasma metabolome. Rapid Commun Mass Spectrom. 2010;24:1312–1318. doi: 10.1002/rcm.4516. [DOI] [PubMed] [Google Scholar]
- 24.Chen M., Su M., Zhao L., Jiang J., Liu P., Cheng J. Metabolomic study of aristolochic acid-induced nephrotoxicity in rats. J Proteome Res. 2006;5:995–1002. doi: 10.1021/pr050404w. [DOI] [PubMed] [Google Scholar]
- 25.Moreno-Gordaliza E., Esteban-Fernández D., Lázaro A., Humanes B., Aboulmagd S., Tejedor A. MALDI-LTQ-Orbitrap mass spectrometry imaging for lipidomic analysis in kidney under cisplatin chemotherapy. Talanta. 2017;164:16–26. doi: 10.1016/j.talanta.2016.11.026. [DOI] [PubMed] [Google Scholar]
- 26.Römpp A., Spengler B. Mass spectrometry imaging with high resolution in mass and space. Histochem Cell Biol. 2013;139:759–783. doi: 10.1007/s00418-013-1097-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greer T., Sturm R., Li L. Mass spectrometry imaging for drugs and metabolites. J Proteomics. 2011;74:2617–2631. doi: 10.1016/j.jprot.2011.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cornett D.S., Reyzer M.L., Chaurand P., Caprioli R.M. MALDI imaging mass spectrometry: molecular snapshots of biochemical systems. Nat Methods. 2007;4:828. doi: 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- 29.Takáts Z., Wiseman J.M., Gologan B., Cooks R.G. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306:471. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- 30.Liu H., Li W., He Q., Xue J., Wang J., Xiong C. Mass spectrometry imaging of kidney tissue sections of rat subjected to unilateral ureteral obstruction. Sci Rep. 2017;7:41954. doi: 10.1038/srep41954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altelaar A.F.M., Klinkert I., Jalink K., de Lange RPJ, Adan R.A.H., Heeren R.M.A. Gold-enhanced biomolecular surface imaging of cells and tissue by SIMS and MALDI mass spectrometry. Anal Chem. 2006;78:734–742. doi: 10.1021/ac0513111. [DOI] [PubMed] [Google Scholar]
- 32.He J., Sun C., Li T., Luo Z., Huang L., Song X. A sensitive and wide coverage ambient mass spectrometry imaging method for functional metabolites based molecular histology. Adv Sci. 2018;5 doi: 10.1002/advs.201800250. 1800250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun C., Li T., Song X., Huang L., Zang Q., Xu J. Spatially resolved metabolomics to discover tumor-associated metabolic alterations. Proc Natl Acad Sci USA. 2019;116:52. doi: 10.1073/pnas.1808950116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li T., He J., Mao X., Bi Y., Luo Z., Guo C. In situ biomarker discovery and label-free molecular histopathological diagnosis of lung cancer by ambient mass spectrometry imaging. Sci Rep. 2015;5:14089. doi: 10.1038/srep14089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He J., Luo Z., Huang L., He J., Chen Y., Rong X. Ambient mass spectrometry imaging metabolomics method provides novel insights into the action mechanism of drug candidates. Anal Chem. 2015;87:5372–5379. doi: 10.1021/acs.analchem.5b00680. [DOI] [PubMed] [Google Scholar]
- 36.Vaidya V.S., Ozer J.S., Dieterle F., Collings F.B., Ramirez V., Troth S. Kidney injury molecule-1 outperforms traditional biomarkers of kidney injury in preclinical biomarker qualification studies. Nat Biotechnol. 2010;28:478–485. doi: 10.1038/nbt.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonventre J.V., Vaidya V.S., Schmouder R., Feig P., Dieterle F. Next-generation biomarkers for detecting kidney toxicity. Nat Biotechnol. 2010;28:436–440. doi: 10.1038/nbt0510-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tong B.C., Barbul A. Cellular and physiological effects of arginine. Mini Rev Med Chem. 2004;4:823–832. doi: 10.2174/1389557043403305. [DOI] [PubMed] [Google Scholar]
- 39.Benito S., Sánchez A., Unceta N., Andrade F., Aldámiz-Echevarria L., Goicolea M.A. LC–QTOF-MS-based targeted metabolomics of arginine–creatine metabolic pathway-related compounds in plasma: application to identify potential biomarkers in pediatric chronic kidney disease. Anal Bioanal Chem. 2016;408:747–760. doi: 10.1007/s00216-015-9153-9. [DOI] [PubMed] [Google Scholar]
- 40.Benito S., Sánchez-Ortega A., Unceta N., Jansen J.J., Postma G., Andrade F. Plasma biomarker discovery for early chronic kidney disease diagnosis based on chemometric approaches using LC–QTOF targeted metabolomics data. J Pharm Biomed Anal. 2018;149:46–56. doi: 10.1016/j.jpba.2017.10.036. [DOI] [PubMed] [Google Scholar]
- 41.Wang Z.H., He B.S., Sun C.L., Song X.W., He J.M., Zhang R.P. Study on tissue distribution of a variety of endogenous metabolites by air flow assisted ionization-ultra high resolution mass spectrometry imaging. Chin J Anal Chem. 2018;46:406–411. [Google Scholar]
- 42.Lou Y., Li J., Lu Y., Wang X., Jiao R., Wang S. Aristolochic acid-induced destruction of organic ion transporters and fatty acid metabolic disorder in the kidney of rats. Toxicol Lett. 2011;201:72–79. doi: 10.1016/j.toxlet.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 43.Galvan D.L., Green N.H., Danesh F.R. The hallmarks of mitochondrial dysfunction in chronic kidney disease. Kidney Int. 2017;92:1051–1057. doi: 10.1016/j.kint.2017.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wenk M.R. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4:594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- 45.di Paolo G., de Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 46.Xu Y., Guo N., Dou D., Ran X., Liu C. Metabolomics analysis of anaphylactoid reaction reveals its mechanism in a rat model. Asian Pac J Allergy Immunol. 2017;35:224–232. doi: 10.12932/AP0845. [DOI] [PubMed] [Google Scholar]
- 47.Che R.C., Yuan Y.G., Huang S.M., Zhang A.H. Mitochondrial dysfunction in the pathophysiology of renal diseases. Am J Physiol Renal Physiol. 2014;306:367–378. doi: 10.1152/ajprenal.00571.2013. [DOI] [PubMed] [Google Scholar]
- 48.Qi X., Cai Y., Gong L., Liu L., Chen F., Xiao Y. Role of mitochondrial permeability transition in human renal tubular epithelial cell death induced by aristolochic acid. Toxicol Appl Pharmacol. 2007;222:105–110. doi: 10.1016/j.taap.2007.03.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.