Abstract
Partial or complete loss-of-function variants in SCN5A are the most common genetic cause of the arrhythmia disorder Brugada syndrome (BrS1). However, the pathogenicity of SCN5A variants is often unknown or disputed; 80% of the 1,390 SCN5A missense variants observed in at least one individual to date are variants of uncertain significance (VUSs). The designation of VUS is a barrier to the use of sequence data in clinical care. We selected 83 variants: 10 previously studied control variants, 10 suspected benign variants, and 63 suspected Brugada syndrome-associated variants, selected on the basis of their frequency in the general population and in individuals with Brugada syndrome. We used high-throughput automated patch clamping to study the function of the 83 variants, with the goal of reclassifying variants with functional data. The ten previously studied controls had functional properties concordant with published manual patch clamp data. All 10 suspected benign variants had wild-type-like function. 22 suspected BrS variants had loss of channel function (<10% normalized peak current) and 22 variants had partial loss of function (10%–50% normalized peak current). The previously unstudied variants were initially classified as likely benign (n = 2), likely pathogenic (n = 10), or VUSs (n = 61). After the patch clamp studies, 16 variants were benign/likely benign, 45 were pathogenic/likely pathogenic, and only 12 were still VUSs. Structural modeling identified likely mechanisms for loss of function including altered thermostability and disruptions to alpha helices, disulfide bonds, or the permeation pore. High-throughput patch clamping enabled reclassification of the majority of tested VUSs in SCN5A.
Keywords: SCN5A, NaV1.5, Brugada syndrome, high-throughput, variant of uncertain significance, patch clamp
Introduction
Variants in the cardiac sodium channel gene SCN5A (which encodes the protein NaV1.5) have been linked to many arrhythmia and cardiac conditions, including Brugada syndrome (BrS [MIM: 601144]),1 long QT syndrome (LQT [MIM: 603830]),2 dilated cardiomyopathy (MIM: 601154),3 cardiac conduction disease (MIM: 113900),4 and sick sinus syndrome (MIM: 608567).5 BrS is diagnosed by a characteristic ECG pattern (ST-segment elevation in the right precordial leads) at baseline or upon drug challenge and is associated with an increased risk for sudden cardiac death.6 SCN5A loss-of-function variants are the most common (and in fact, the only ClinGen-validated)7 genetic cause of BrS, and are present in approximately 30% of case subjects (BrS1).1,8,9 Variants that destabilize fast inactivation of the channel and generate a persistent (“late”) current are associated with LQT.10,11 The risk of sudden cardiac death in individuals with Brugada syndrome can be partially prevented, for example with implantable defibrillators. Therefore, incidentally discovered SCN5A variants that are pathogenic or likely pathogenic for BrS are considered actionable and reportable.12
A multi-center study of 2,111 unrelated individuals with Brugada syndrome discovered 293 distinct SCN5A variants, including 225 variants present in only one individual.1 This result indicates that Brugada syndrome risk is distributed among many different, rare SCN5A variants. A large body of work reporting phenotypes of SCN5A variant carriers and studying these variants by in vitro patch clamping has established the link between partial or total loss of NaV1.5 function and Brugada syndrome.1,13, 14, 15, 16 In a recent literature curation, we identified 1,712 SCN5A variants that have been observed to date in at least one individual in the literature or in the genome aggregation database (gnomAD).10 The strongest predictor of a variant’s association with Brugada syndrome was a partial or total reduction in peak current recorded during voltage clamp experiments in heterologous expression systems (mainly HEK293 cells).10 Additionally, a weaker association was observed between a variant’s Brugada syndrome association and its V1/2 activation, the voltage that elicits half-maximal channel activation.
Because SCN5A variants linked to Brugada syndrome have incomplete penetrance, assessment of variant risk using phenotypic data in single variant carriers can be minimally informative.17 Nevertheless, an important challenge is to accurately classify variants, especially when only limited phenotype data are available.18 The American College of Medical Genetics and Genomics (ACMG) classification scheme uses 28 criteria to classify variants into 5 categories: pathogenic, likely pathogenic, benign, likely benign, or variant of uncertain significance (VUS).19 Two “strong evidence” ACMG criteria rely on in vitro functional data: pathogenic strong criterion 3 (PS3, well-established functional variant studies show a deleterious effect) and benign strong criterion 3 (BS3, well-established functional variant studies show no deleterious effect).
Conventionally, variants in ion channels such as SCN5A have been assessed by time-intensive manual patch clamping of variants in heterologous expression. Recently, automated high-throughput patch clamp devices have allowed the rapid evaluation of dozens of variants in ion channel genes, including the arrhythmia-associated genes KCNQ1 and KCNH2.20, 21, 22 In this study, we use automated patch clamping to study 83 SCN5A variants. We identify 44 new partial or total loss-of-function variants and reclassify 49/61 variants of uncertain significance. We further use structural modeling to identify likely mechanisms of NaV1.5 loss of function, including altered thermostability and disruptions to critical protein features.
Material and Methods
Selection of Variants
83 variants were studied by automated patch clamping (Figure 1): 10 previously studied controls, 10 suspected benign variants, and 63 suspected BrS variants. The previously studied variants were chosen because they had a spectrum of normal and abnormal channel function, representing multiple types of perturbations to the channel. The suspected BrS variants were selected because they appeared in at least one case of BrS in the literature and were relatively rare or absent in the gnomAD database of putative controls (minor allele frequency of 0 to 5e−5, except for p.Asp1243Asn which had 5 BrS cases and an allele frequency of 1.4e−4; Figure 2B, Table S1).10,23 In contrast, the suspected benign variants were observed in multiple unaffected or gnomAD individuals (minor allele frequency of 5e−5 to 3e−4), but never in cases of BrS or LQT in the literature (Figure 2B, Table S1). p.Arg1958Ter was also classified as a suspected BrS variant because it is a nonsense variant. However, to our knowledge, it has not been observed in any individuals with BrS (Figure 2B, Table S1).
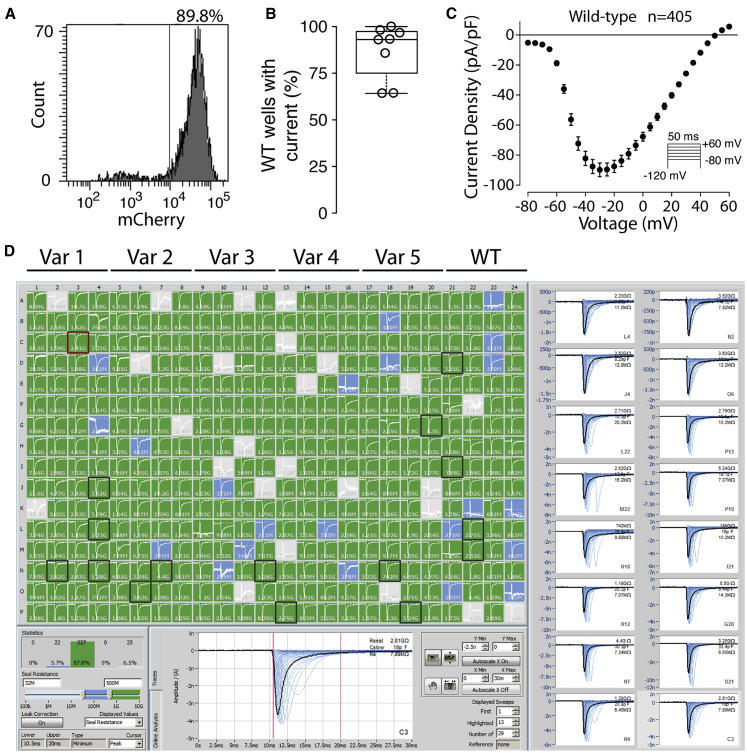
Figure 1.
Molecular and Functional Expression of SCN5A
(A) Flow cytometry plot of representative wild-type stable cell line. Histogram of mCherry signal is shown. 89.8% of cells expressed a high-level of mCherry, indicating successful plasmid integration.
(B) Boxplot of percentage of wild-type wells with NaV1.5-like current across eight independent transfections. Only wells with a cell passing seal resistance and capacitance criteria (see Material and Methods) were considered.
(C) Wild-type current density-voltage plot, averaged across 405 wild-type cells passing quality control (seal resistance, capacitance, in voltage control, and minimum peak current criteria, see Material and Methods). The predicted reversal potential given the internal and external solutions used in this study is 45.3 mV. Error bars indicate SEM.
(D) Example SyncroPatch experiment. A typical experiment studied 5 variants and wild-type on a single 384-well chip. In this experiment all 5 variants had wild-type-like currents. Green wells indicate a cell with seal resistance >0.5 GΩ, blue wells indicate a cell with seal resistance 0.2–0.5 GΩ (not included in analysis), and gray wells indicate no cell present or a cell with seal resistance <0.2 GΩ (not included in analysis). Randomly selected cells with seals >0.5GΩ are highlighted with a black square and displayed on the right.
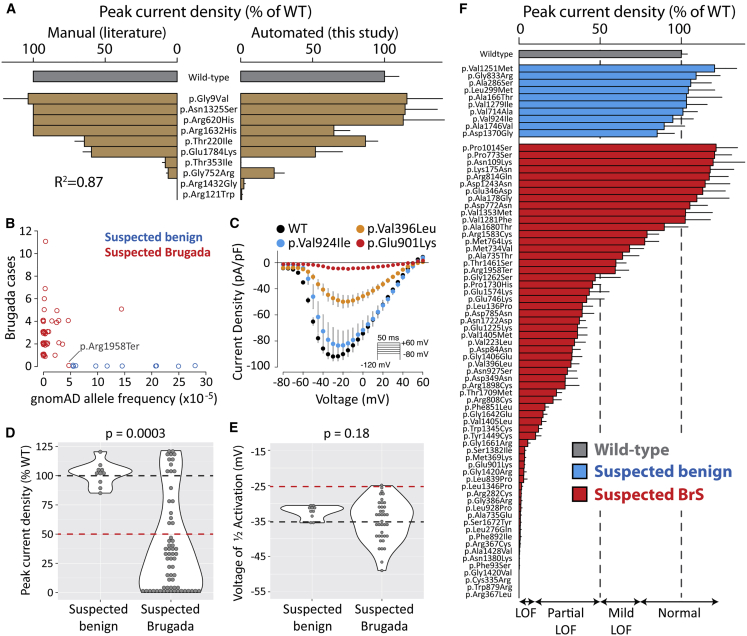
Figure 2.
Suspected Brugada Syndrome Variants Have Reduced Peak Current
(A) Peak current density of ten variants previously studied by manual patch clamp. Mean ± standard errors. Left: literature values. For some variants, standard errors were not reported. Right: automated patch clamp values (this study).
(B) Selection of suspected benign and suspected BrS-associated variants. Each data point is a variant. Points were “jittered” in the x and y axes with a small random number so that points with identical values would be visible. The x axis is the minor allele frequency in the gnomAD database. The y axis indicates the number of individuals with Brugada syndrome in the literature.10 Values are taken from a recent collation of SCN5A data from the literature.10 p.Arg1958Ter was classified as a suspected BrS variant because it is a nonsense variant, even though it has not been observed in any cases of BrS.
(C) Current-voltage curves for selected variants, showing an example of a normal variant (p.Val924Ile), a partial loss-of-function variant (p.Val396Leu), and a loss-of-function variant (p.Glu901Lys). Error bars indicate standard error of the mean.
(D and E) Violin plots comparing peak current density (D) or voltage of 1/2 activation (E) for suspected Brugada syndrome versus suspected benign variants. Wild-type values are indicated with a dashed black line, and cutoffs for deleteriousness are indicated with a red line (50% peak current or 10 mV rightward shift in V1/2 activation). p values are from a two-tailed Mann-Whitney U test. For a complete list of measured parameters, see Table S7 and Data S1.
(F) Peak current density (normalized to wild-type) for all previously unstudied variants. Error bar indicates standard error of the mean.
SCN5A Mutagenesis
To facilitate mutagenesis, a “zone” system was developed that divided the 6 kilobase SCN5A cDNA into 8 zones of ∼750 bp each (Table S2). These zones were separated by restriction enzyme sites already present in the SCN5A coding sequence or introduced with synonymous mutations. Mutagenesis primers were designed with the online QuikChange primer design tool. All mutagenesis primers are listed in Table S3. The QuikChange Lighting Multi kit (Agilent) was used to perform the mutagenesis with one primer/mutation, following manufacturer’s instructions. Mutants were created in a small (<4 kilobase) plasmid containing the zone of interest. Following sequence verification, mutant zones were moved with the appropriate restriction enzymes into a plasmid containing an AttB recombination site and a full-length SCN5A:IRES:mCherry-blasticidinR (Figure S1). This plasmid was modified from a previously published promoterless dsRed-Express-derivative plasmid designed for rapid stable line generation.24 This modification introduced SCN5A into the plasmid and included the generation of a mCherry-blasticidinR fusion protein. For all plasmids, the entire SCN5A coding region was Sanger sequenced to confirm that the variant of interest was included and to verify that there were no off-target variants. The “wild-type” SCN5A used in this study had the more common His allele of the p.His558Arg polymorphism (rs1805124) and the most common splice isoform in the heart, a 2,015-amino acid sequence (not counting the stop codon) that includes deletion of the Gln1077 residue (ENST00000443581). However, the variants are named according to the 2,016-amino acid Gln1077-included isoform (ENST00000333535), as is standard in the literature.
Stable SCN5A-Expressing Cell Lines
Human Embryonic Kidney HEK293T “negative selection” landing pad cells were used, which express Blue Fluorescent Protein and an activatable iCasp caspase from a tetracycline-sensitive promoter.25 In these cells, an AttP integration site is located in between the promoter and the BFP/iCasp. Bxb1 integrase-mediated integration of a plasmid containing an AttB site at the genomic AttP location disrupts the expression of BFP/iCasp. After integration of an AttB-containing SCN5A:IRES:mCherry:blasticidinR plasmid, these cells switch to expression of SCN5A and an mCherry-blasticidinR fusion protein. Cells were cultured at 37°C in a humidified 95% air/5% CO2 incubator in “HEK media”: Dulbecco’s Eagle’s medium supplemented with 10% fetal bovine serum, 1% non-essential amino acids, and 1% penicillin/streptomycin. On day −1, cells were plated in 12-well plates. On day 0, each well was transfected with 2.1 μl FuGENE 6 (Promega), 600 ng of the mutant SCN5A plasmid, and 100 ng of a plasmid expressing Bxb1 recombinase (a gift from Pawel Pelczar,26 Addgene plasmid #51271). On day 1, cells were washed with HEK media to remove the FuGENE reagent. On day 3, the cells were exposed to 1 μg/mL doxycycline (Sigma) in HEK media to induce expression of either the SCN5A mutant or iCasp9 in non-integrated cells from the Tet-sensitive promoter. On day 5, cells were exposed to 1 μg/mL doxycycline, 100 μg/mL blasticidin S (Sigma), and 10 nM AP1903 (MedChemExpress) in HEK media. The blasticidin selected for expression of the blasticidin resistance gene present on the SCN5A plasmid. The AP1903 activated the iCasp caspase protein expressed from non-integrated cells; this resulted in selection against non-integrated cells. Cells were exposed to doxycycline+blasticidin+AP1903 in HEK media for 4–7 days to select for integrated cells, passaging cells as needed. Less than 48 h prior to the SyncroPatch experiment, cells were analyzed by analytical flow cytometry on a BD Fortessa 4-laser instrument to determine the percentage of cells that were mCherry-positive, Blue Fluorescent Protein-negative, indicating cells with successful integration of the SCN5A plasmid. BFP was measured by excitation at 405 and emission at 450/50 and mCherry was measured by excitation at 561 and emission at 610/20.
Automated Patch Clamping
Cells were patch clamped with the SyncroPatch 384PE automated patch clamping device (Nanion). To prepare cells for patch clamping, cells were washed in PBS, treated with Accutase (Millipore-Sigma) for 3 min at 37°C, then recovered in CHO-S-serum free media (GIBCO). Cells were pelleted and resuspended in divalent-free reference solution (DVF) at ∼200,000–400,000 cells/mL. DVF contained (mM) NaCl 140, KCl 4, alpha-D(+)-glucose 5, HEPES 10 (pH 7.4) adjusted with NaOH. Cells were then added to a medium resistance (4–6 MΩ) 384-well recording chamber with 1 patch aperture per well (NPC-384, Nanion), which contained DVF and internal solution: CsCl 10, NaCl 10, CsF 110, EGTA 10, HEPES 10 (pH 7.2) adjusted with CsOH. Next, to enhance seal resistance, 50% of the DVF was exchanged with a calcium-containing seal enhancing solution: NaCl 80, NMDG 60, KCl 4, MgCl2 1, CaCl2 10, alpha-D(+)-glucose 5, HEPES 10 (pH 7.4) adjusted with HCl. The cells were washed three times in external recording solution: NaCl 80, NMDG 60, KCl 4, MgCl2 1, CaCl2 2, alpha-D(+)-glucose 5, HEPES 10 (pH 7.4) adjusted with HCl. Currents elicited in response to activation, inactivation, and recovery from inactivation protocols were then recorded (Figure S2). A late current measurement was captured every 5 s. After 1 min, 50% of the external solution was exchanged with external solution containing 200 μM tetracaine hydrochloride (Sigma; effective concentration 100 μM tetracaine). After tetracaine addition, late current measurements were obtained every 5 s for 1 additional minute. At least 10 cells expressing wild-type SCN5A were included for comparison in each SyncroPatch experiment (Figure 1), and at least 2 independent transfections and at least 10 replicate cells were studied per mutant. Recordings were performed at room temperature.
We also conducted experiments to assess the effects of incubation at low temperature or mexiletine (a sodium channel blocker), interventions reported to increase cell surface expression of mistrafficked channels.27, 28, 29, 30, 31 For these experiments, cells stably expressing loss-of-function variants were generated as described above. The cells were incubated for 24 h at 30°C, or at 37°C with or without 500 μM mexiletine hydrochloride (Sigma), washed with HEK media, and were patch clamped as described above.
Data Analysis
Cells with a seal resistance of 0.5–10 GΩ and a cell capacitance between 5 and 30 pF were analyzed. Built-in SyncroPatch windowing methods were used to calculate currents in peak/inactivation/recovery from inactivation protocols (maximum current from 0.5 to 5 ms post voltage shift) or late current (mean current from 190 to 200 ms). These values were exported to .csv files and analyzed in custom R scripts. Current-voltage plots were created for each well, and each well was manually analyzed and classified as normal/in voltage control (voltage-dependent current with peak current near −20 mV), out of voltage control (voltage-dependent current that rapidly jumped around −60 mV), or having no current (<25 pA peak current). Example current-voltage curves of these three classes are shown in Figure S3.
Only wells that were in voltage control (Figure S3) with peak currents between 100 and 2,000 pA were used to assess additional features, such as the voltage dependence of activation, voltage dependence of inactivation, inactivation time, or recovery from inactivation (Table S4). Additional details on peak current averaging are presented in the Supplemental Methods. Only wells with a peak current above 500 pA and wells where the seal resistance and capacitance did not change by more than 10% during addition of tetracaine (see below) were used to measure late current. Activation and inactivation best-fit curves were calculated for each well by fitting Boltzmann equations using the R function nls (nonlinear least-squares). Recovery from inactivation data and inactivation time data were fitted with exponential curves with the nls function. Wells with high noise for which the best-fit curve did not fit the data well (data points had >10% average deviation from the best-fit line) were removed from the analysis. Tetracaine-sensitive late current was calculated as the mean of 5 post-tetracaine raw late current values subtracted from the mean of 5 pre-tetracaine values and was normalized to tetracaine-sensitive peak current. Outlier values exceeding 3 standard deviations from the mean were excluded (1.5% of all values, Table S4). Per-well V1/2 activation (voltage at which half the channels are activated), V1/2 inactivation (voltage at which half the channels are inactivated), time of 50% recovery from inactivation, inactivation time constant, and late current parameters were averaged across all wells meeting the above inclusion criteria. All measured parameters with at least 5 cells meeting inclusion criteria are reported. For many severe loss-of-function variants, there were fewer than 5 qualifying wells to accurately quantify channel parameters other than peak current density. All comparisons of variant parameters between groups were made in R with two-tailed t tests (t.test) or with two-tailed Mann-Whitney U tests (wilcox.test with the parameter paired = FALSE) when the distributions were non-normally distributed. Differences in dispersion between groups were tested with Levene’s Test (levene.test, car package). Violin plots were made with geom_violin (ggplot2).
Variants were classified according to American College of Medical Genetics and Genomics (ACMG) criteria (Figure S4).19 A custom R script was used to implement these criteria. Variant classifications pre- and post-functional data are presented in Table S1. A cutoff of 6/∼250,000 alleles in gnomAD v.2.123 was used to determine criteria BS1 and PM2, following a previous recommended cutoff for Brugada syndrome.32 BP4 and PP3 were determined from the consensus of PROVEAN33 and PolyPhen234 classifications. PS4 was interpreted to mean at least 5 observed individuals with Brugada syndrome and an estimated Brugada syndrome penetrance from literature reports whose 95% confidence interval excluded 0.10 Variants with peak current densities between 75% and 125% of wild-type, <10 mV shifts in activation or inactivation, <2-fold shifts in recovery from inactivation, and <1% late current (% of peak) were considered to have normal in vitro functional data (BS3). Variants with peak current densities <50% of wild-type or a >10 mV rightward shift in V1/2 activation were considered to have abnormal loss-of-function functional data (PS3). Variants with >75% peak current and a late current >1% (normalized to peak current) were considered to have abnormal gain-of-function functional data (PS3). These cutoffs were determined from a previous analysis of the correlation between functional parameters and Brugada syndrome and long QT syndrome risk.10 These cutoffs reflect the observation that variants that are not linked to disease often have mild perturbations to their patch clamp parameters. ClinVar classifications35 were used to determine criteria BP6 and PP5. All literature and gnomAD case/control counts, peak current densities, and classifications pre- and post-patch clamp data are presented in Table S1, and ACMG criteria used for variant classification are presented in Table S5. All variants were considered to meet PP2 (missense variant in gene with low rate of benign missense variants and pathogenic missense variants common) except p.Arg1958Ter, which was considered to meet PM4 (protein length changing variant). For gnomAD counts, literature counts, and classifications, variants with the same outcome on the protein sequence (due to the redundant genetic code) were grouped together. Therefore, the PS1 criterion (same amino acid change as established pathogenic variant) was not used. No variants satisfied the PP1 criterion (statistical co-segregation with disease in multiple affected family members) due to the low numbers of carriers in the literature and lack of large published pedigrees for these variants. We performed an initial round of classification without using PM5 (missense variant at a position where a different variant is pathogenic/likely pathogenic). Then, for each variant, we determined the PM5 criterion by searching for variants at the same amino acid position that were initially classified as pathogenic/likely pathogenic. Finally, classifications were recalculated including PM5.
Homology Model and Structural Calculations
All computational modeling was conducted in parallel to and blinded from the experimental characterizations. Structural models of human SCN5A (UniProtKB: Q14524-1, modeled residues: 30–440, 685–957, 1174–1887) bound with SCN1B (UniProtKB: Q07699-1, modeled residues: 20–192) were generated by homology modeling using the protein structure prediction software Rosetta (v.3.10).36 The cryo-EM structure of human SCN9A bound with SCN1B and the Ig domain of SCN2B resolved to 3.2 Å (PDB: 6J8H)37 was used as the primary template while the cryo-EM structure of NavPaS from American Cockroach resolved to 2.6 Å (PDB: 6A95)38 was used as a secondary template. The percent identity between the aligned positions of SCN9A and SCN5A sequences was 76.7%. While the percent identity between NavPaS and SCN5A was only moderate (45.6%), the N-terminal and C-terminal domains in the NavPaS structure were partially resolved, providing coordinates for modeling the corresponding domains of SCN5A. The final model (Figures 4 and S5) covers 70 of the 73 functionally characterized variants. Three variants—p.Pro1014Ser, p.Arg1898Cys, and p.Arg1958Ter—fall outside of the set of modeled residues, and are therefore not covered. Additional information about the homology model and ΔΔG (thermostability) calculations are presented in the Supplemental Methods and Table S6.
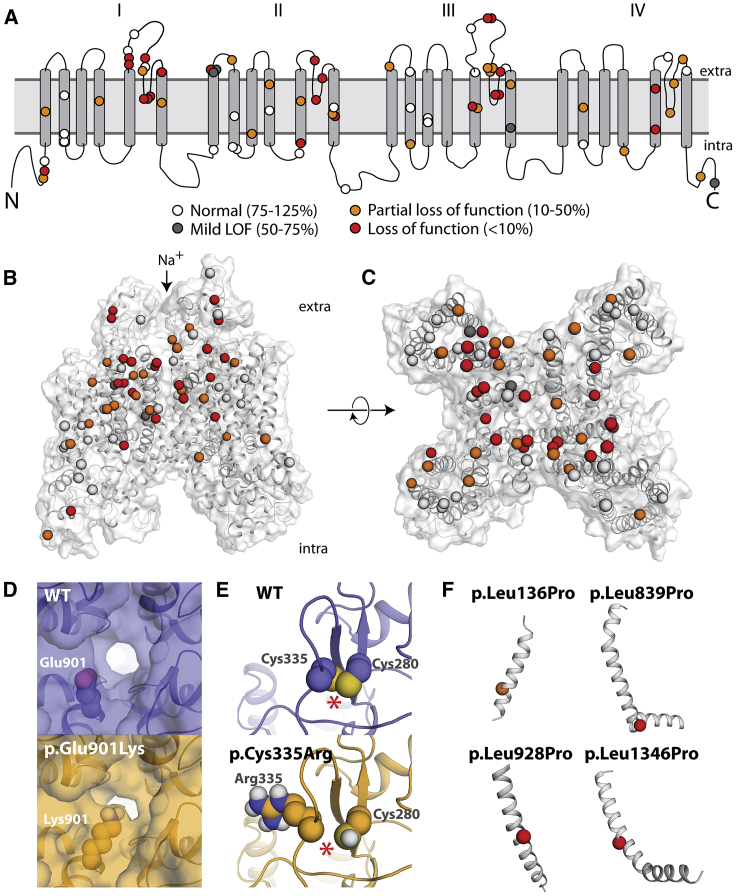
Figure 4.
Structural Basis of SCN5A Loss-of-Function Variants
(A) Two-dimensional schematic of NaV1.5 structure. All previously unstudied variants are shown and color-coded based on peak current density (white 75%–125%, gray 50%–75%, orange 10%–50%, red < 10%).
(B and C) Three-dimensional homology model of NaV1.5. Variants are colored as in (A).
(D) Top-down view of WT (top) and p.Glu901Lys (bottom), as modeled using Rosetta. The lysine residue projects into the pore, likely disrupting sodium passage.
(E) View of WT and p.Cys335Arg, as modeled using Rosetta. The WT protein has a disulfide bond between Cys335 (left) and Cys280 (right), which was inferred from the spatial proximity of these two residues and the fact that the corresponding residues in the template structures also form a disulfide bond; this bond is disrupted by p.Cys335Arg. The disulfide bond is indicated with an asterisk (∗).
(F) Four leucine -> proline variants in this study. p.Leu136Pro is a partial loss-of-function variant and p.Leu839P, p.Leu928Pro, and p.Leu1340Pro are loss of function. The structures of these four variants were not modeled because modeling drastic structural changes involving prolines that are part of a helix usually cannot be reliably modeled in Rosetta. However, these variants likely cause loss of protein function by causing a kink in the alpha helix and protein misfolding.
Results
Automated Patch Clamping of NaV1.5 Variants
Across multiple transfections and variants, a high percentage (85.4% ± 12.3% SEM) of HEK293 cells were mCherry positive, indicating successful integration of an SCN5A expression plasmid (Figures 1A and S1). Wild-type and mutant cells were patch clamped with the SyncroPatch 384PE device (Figure 1). For cells expressing wild-type SCN5A, most wells that passed seal resistance and capacitance quality control cutoffs had voltage-dependent sodium currents (86.9% ± 14.6% SEM, Figures 1B and 1C). Wild-type cells had typical NaV1.5 properties (Figures 1 and S2).
To validate the automated patch clamping method, we patch clamped 10 previously studied variants chosen because they had a range of functional properties. The previously published manual patch clamp properties were concordant with the automated patch clamp data (Figure 2A, Table S7). Peak current is the major parameter of interest for assessing Brugada syndrome risk.10 We observed a strong correlation (R2 = 0.87) between the automated peak current density measurements and previously published manual patch clamp peak current density values (Figure 2A). p.Arg1632His and p.Gly752Arg had differences in peak current compared to previous manual patch clamp data (65% peak current density versus no reported difference from wild-type for p.Arg1632His;39 23% peak current density versus 6% previously reported for p.Gly752Arg40). These differences might represent differences in cell lines,40 differences in data analysis pipelines, or experimental measurement error inherent in these measurements. These two variants had previously published changes to other properties39,40 that were recapitulated by automatic patch clamping (a dramatic increase in time to recover from inactivation for p.Arg1632His [Figure S9] and a right-shift in activation voltage for p.Gly752Arg [Figure S6]).
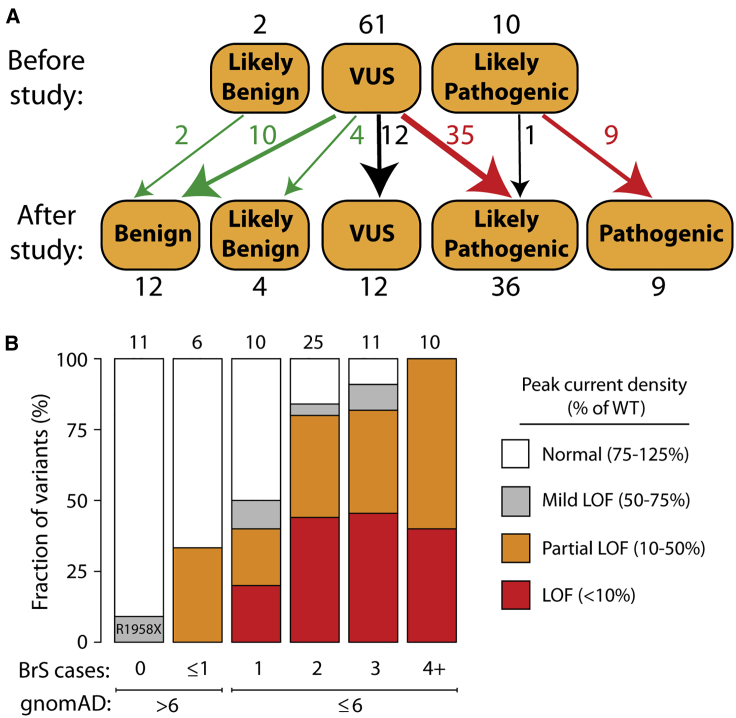
From our previous curation of the SCN5A variant literature,10 we selected 73 previously unstudied variants: 63 suspected Brugada syndrome variants and 10 suspected benign variants. The suspected benign and suspected Brugada syndrome variants were selected based on their frequency in the gnomAD database and in the literature in individuals with Brugada syndrome (Figure 2B, Table S1).10,23 Applying ACMG classification criteria, without patch clamping data, 61 variants were classified as VUS, 2 as likely benign, and 10 as likely pathogenic (Figure 3A, Table S1).
Figure 3.
Reclassifications with Patch Clamp Data
(A) Variants were classified according to the American College of Medical Genetics and Genomics criteria.19 Above: classifications before the patch clamp data in this study. Below: classifications with the patch clamp data in this study. All classifications indicate Brugada syndrome, except for p.Arg814Gln, which was reclassified from likely pathogenic for Brugada syndrome to likely pathogenic for long QT syndrome.
(B) Patch clamp results varied depending on the number of observed individuals with Brugada syndrome in the literature10,18 and the gnomAD allele frequency.23 LoF, loss of function.
Most Suspected Brugada Syndrome Variants Had (Partial) Loss of Function
There was a striking difference in peak current density between the suspected BrS and suspected benign variants (p = 0.0003, two-tailed Mann-Whitney U test, Figure 2D). Overall, 23/63 suspected BrS variants had partial loss of function (10%–50% peak current density, normalized to wild-type) and 22/63 had loss of function (<10%), while 10/10 suspected benign variants had normal peak currents (75%–125%) (Figure 2F, Table 1). We next examined whether large changes to other parameters were a common cause of channel loss of function. There was a statistically significant increase in width of the distribution of V1/2 activations in the suspected BrS set (p = 0.0028, Levene’s test). However, only 2/39 variants with measured V1/2 activation had a >10 mV rightward shift (WT: −36.7 ± 0.6 mV, n = 279; D785N: −26.4 ± 1.3 mV, n = 16; D349N: −26.5 ± 3.1 mV, n = 5). p.Asp785Asn and p.Asp349Asn both also had a partial loss of function in peak current (38.9% and 28.5% of WT, respectively; Figure 2F). One suspected BrS variant, p.Gly1262Ser, had a combination of a partial loss of function of peak current density (47% of wild-type) and a gain of function leftward shift in V1/2 activation (−12.2 mV; Figure S6). Overall, there was no significant difference in V1/2 activation between suspected BrS and suspected benign variants (p = 0.18, Mann-Whitney U test, Figure 2E). Therefore, in this dataset, large shifts in activation gating were not a common cause of Brugada syndrome. Surprisingly, one suspected BrS-associated variant, p.Arg814Gln, had near wild-type-like peak current density (117% of wild-type) and increased late current (1.4% of peak). In addition to Brugada syndrome cases, p.Arg814Gln has been observed in two cases of long QT syndrome,41 consistent with its gain-of-function late current phenotype. Besides these variants, no major differences between suspected BrS and suspected benign variants were observed for V1/2 inactivation, inactivation time, recovery from inactivation, or late current (Figures S6–10, Table S7). Despite being a nonsense variant, p.Arg1958Ter generated substantial current (peak current density of 59.3% of wild-type), likely due to the fact that the stop codon is in the distal C terminus of the protein (Figure 2F).
Table 1.
Functional Class of Suspected Benign and Brugada Syndrome Variants
| Functional Class | Peak Current Density | Suspected Benign | Suspected BrS |
|---|---|---|---|
| Normal | 75%–125% | 10 (100%) | 14 (22%) |
| Mild loss of function | 50%–75% | 0 (0%) | 4 (6%) |
| Partial loss of function | 10%–50% | 0 (0%) | 23 (37%) |
| Loss of function | <10% | 0 (0%) | 22 (35%) |
| Total | ∗ | 10 | 63 |
Some suspected BrS variants have only been observed in only a single individual with BrS, whereas other variants have been observed in multiple individuals. In this study, the patch clamp phenotypes varied according to the strength of the phenotypic evidence for Brugada syndrome (Figure 3B). For example, 4/10 variants that have been observed in exactly 1 case of Brugada syndrome and ≤6 in gnomAD had partial or complete loss of function (0%–50% peak current). In contrast, 10/10 variants that have been observed in at least 4 BrS1 cases and ≤6 in gnomAD had partial or complete loss of function. 2/6 variants seen in ≥1 cases of Brugada syndrome but also in >6 counts in gnomAD had partial loss-of-function defects. Therefore, variants that were more commonly observed in Brugada syndrome cases and less frequently observed in the population were more likely to have loss of channel function.
Reclassification of Variants with Functional Data
When we implemented the ACMG classification criteria19 to classify all studied variants without the automated patch clamp data (Figure 3A), 60/73 variants were classified as VUS. Based on our previous literature curation study, which showed an elevated BrS risk for variants with peak current density <50% of wild-type,10 we defined cutoffs for ACMG functional criteria PS3 and BS3. Variants with <50% peak current were considered to meet criterion PS3 (well-established functional assays show a change), and variants with 75%–125% peak current were considered to meet criterion BS3 (well-established functional assays show no change). Since the phenotypic consequences of mild LoF variants (50%–75% peak current) to BrS risk is unclear,10 these variants were not considered to satisfy either PS3 or BS3. Because of its elevated late current and normal peak current, p.Arg814Gln was considered to meet the PS3 criterion for abnormal gain of function, and was reclassified from likely pathogenic for Brugada syndrome to likely pathogenic for long QT syndrome. p.Gly1262Ser, which had peak current density of 47% of wild-type and a gain of function 12.2 mV leftward shift in V1/2 activation, was considered to meet neither the PS3 nor the BS3 criteria because of the uncertain impact of these features on BrS risk. After patch clamping data were incorporated, 36/61 VUSs were reclassified as likely pathogenic and 14 were reclassified as likely benign or benign. Overall, classifications were changed for 49/61 VUSs (80%) and 61/73 (84%) previously unstudied variants. Therefore, for this set of variants, functional data led to reclassification of the great majority of VUSs.
Partial Rescue of Some Loss-of-Function Variants
Previous studies have shown that pre-incubation of cells at a lower temperature or with a sodium channel blocker can partially rescue the function of some loss-of-function SCN5A variants, typically by improving protein folding and trafficking to the cell surface.27,29, 30, 31 Therefore, we tested whether pre-incubating cells at 30°C or with the sodium channel blocker mexiletine could partially rescue sodium current for the 22 loss-of-function variants (<10% normalized peak current density). Cells expressing these variants were cultured for 24 h in usual conditions at 37° with no added drug, at 30°C, or with 500 μM mexiletine. Compared to usual conditions, 8/22 variants had significantly increased peak current at 30°C and 2/22 variants had significantly increased peak current when treated with mexiletine (Figure S11). p.Phe892Ile and p.Met369Lys had the largest responses to 30°C incubation, with normalized peak current density increasing from 8.7% at 37°C to 34.8% at 30°C for p.Phe892Ile (p = 0.006, two-tailed t test) and from 3.9% to 42.5% for p.Met369Lys (p = 0.0003; Figure S11).
Structural Basis of Loss of Function
In this study, 22 previously unstudied variants had <10% peak current and an additional 23 variants had 10%–50% current. We explored the structural basis of these variants’ decreased channel function. Although the suspected BrS variants were selected for study independent of their position in the protein, 42/45 (partial) loss-of-function variants were located in the four structured transmembrane domains, as opposed to unstructured linker regions or the N and C termini (Figures 4A–4C). 33/45 (73%) loss/partial loss-of-function variants were located in the pore-forming or pore-adjacent S5, S5-S6 linker, or S6 regions, areas that we have previously identified as a hotspot for Brugada variants.10,17 Indeed, variant distance from the pore in the protein structure was strongly correlated with normalized peak current density (Pearson’s r = 0.54, p = 1.3e–6, Figure S12).
Many disease-causing variants cause amino acid substitutions that lead to a significant perturbation to native thermostability (|ΔΔG|) of protein structure.42,43 To investigate the extent to which the perturbation to the native thermostability of NaV1.5 is correlated with molecular function, we evaluated the impact of each variant on estimated thermostability relative to the wild-type structure with a Rosetta ΔΔG protocol.44 Normalized peak currents and estimated |ΔΔG| values were significantly negatively correlated (Pearson’s r = −0.31, p = 0.0092, Figure S13). The estimated |ΔΔG| of variants with functional effects on peak current (peak current density < 50%, n = 44, median |ΔΔG| = 2.00 kcal/mol) was significantly larger than normal/mild loss-of-function variants (peak current density ≥ 50%, n = 26, median |ΔΔG| = 0.88 kcal/mol, Mann-Whitney U test, p = 0.0031, Figure S13B). This result suggests that variant-induced disruption of native thermostability of NaV1.5 may be a major factor contributing to compromised function.
Variants can cause complete or partial loss of function through mechanisms other than affecting thermostability, such as disrupting the native topology or electrostatic environment of the pore. Using a homology model of NaV1.5, several variants had plausible mechanisms that could explain their loss-of-function phenotype (Figures 4D–4F). Seven pore-lining residues in this study, p.Asp349Asn, p.Arg367Cys, p.Arg367Leu, p.Trp879Arg, p.Phe892Ile, p.Glu901Lys, and p.Thr1709Met, caused either complete or partial loss of function while inducing negligible or only minor perturbations to native thermostability (Figures 4C, 4D, and S14). In particular, the residue Glu901 lines the channel pore, and in the homology model, the loss-of-function variant p.Glu901Lys projects into the pore, likely disrupting sodium permeation (Figure 4D). p.Cys335Arg disrupts a disulfide bond, likely destabilizing the tertiary structure of the protein and explaining its loss-of-function phenotype (Figure 4E). Three loss-of-function leucine→proline variants (p.Leu839Pro, p.Leu928Pro, and p.Leu1346Pro) and a fourth partial loss-of-function variant (p.Leu136Pro) likely disrupt alpha helices, as is seen in other proteins with proline variants (Figure 4F).45 In contrast, p.Ala166Thr, a suspected benign variant located distally from the pore and not predicted to alter protein structure, had wild-type-like electrophysiological properties (Figures S5B and S5C, Table S1). These data indicate that structural features can help predict or explain channel loss of function in SCN5A variants.
Discussion
Reclassification of Brugada Syndrome Variants with Patch Clamping Data
This study identified 44 novel (partial) loss-of-function variants by high-throughput, automated patch clamping. In addition, this nearly doubles (from 24 to 46) the number of known loss-of-function missense SCN5A variants with <10% peak current (Table S8). As a result of the patch clamp data, 35 novel pathogenic/likely pathogenic variants and 14 novel benign/likely benign variants were identified. Overall, 61/73 variants, including 49/61 VUSs, were reclassified with our patch clamp data. A nonsense variant, p.Arg1958Ter, has been observed in 13 individuals in the gnomAD database and never in a published case of Brugada syndrome. p.Arg1958Ter generated substantial current (peak current density of 59.3% of wild-type) and likely escapes nonsense-mediated decay because it located is in the last exon of SCN5A. p.Arg1958Ter was not considered to meet the PVS1 criterion (null variant) and was classified as a VUS both pre- and post-study due to its mild loss-of-function in vitro phenotype. Eight other nonsense and frameshift variants in the C terminus of SCN5A are present in the gnomAD database;23 these variants may also generate sodium current and not have complete loss of channel function.
A recent guide from the ClinGen consortium recommended that in vitro assays that are well validated with clearly pathogenic and benign controls and predict disease risk well be used as PS3/BS3 at the strong level in the ACMG classification scheme.46 Since patch clamping is the gold standard method to assay ion channel function and has been performed for hundreds of SCN5A variants including (for the most part) correctly predicting the phenotypic impact of dozens of clearly benign and pathogenic variants, we implemented the PS3 and BS3 criteria at the strong level for variants between 0%–50% peak current density (PS3) and 75%–125% (BS3). These ranges were chosen based on the correlation between peak current and variant-specific BrS risk.10 Because of their uncertain impact on Brugada syndrome risk, mild loss-of-function variants (50%–75% peak current) were not considered to meet BS3 or PS3. As a result, the classification of these variants was not changed due to this study.
While each variant is rare, the collective allele frequency of the reclassified variants in this study is 0.2% in gnomAD (∼0.4% of individuals). Although the majority of the 1,390 observed SCN5A missense variants remain VUSs,10 this study suggests that variant properties can be used to identify a subset of SCN5A variants that are highly enriched for altered in vitro properties and disease association. In this study, two properties predict a high rate of in vitro loss of function: (1) the observation of the variant in at least one individual with BrS and (2) ultra-rare frequency in the population (≤6 in gnomAD—concordant with a previous computational estimate32). Including this study, 135 of the 276 variants with these properties have been now studied in vitro. The remaining 141 variants are excellent candidates for future high-throughput functional characterization and possible reclassification. An important open question is the rate of deleterious in vitro phenotypes in the 742 ultra-rare variants (≤6 in gnomAD) that have not been observed in any BrS cases to date. Some of these variants might not have appeared in published BrS cases despite a true BrS risk, or carriers may present with other SCN5A-associated arrhythmia phenotypes.47
It is important to note that SCN5A variant classification does not completely predict Brugada syndrome risk in individual patients. BrS is an incompletely penetrant disease, and its presentation is influenced by demographic factors such as age and sex,48 as well as common genetic variants—including multiple noncoding haplotypes near SCN5A.49 Individuals with loss-of-function SCN5A variants can present with other arrhythmias besides Brugada syndrome, including sick sinus syndrome,5 atrial standstill,50 or other conduction disease.51 Therefore, carriers of the loss-of-function variants identified in this study may present with those conditions instead of Brugada syndrome. In addition, some “overlap” SCN5A variants can have increased risk of both Brugada syndrome and long QT syndrome.52 One suspected BrS-associated variant in this study, p.Arg814Gln, had late current above our cutoff (>1% of peak current) and has appeared in the literature in both BrS and LQT cases.41,53 Carriers of incidental pathogenic or likely pathogenic SCN5A variants should have follow-up ECG screening and a clinical and family history taken to determine each individual’s phenotype and sudden cardiac death risk. A common variant in SCN5A, p.His558Arg, is present at a minor allele frequency of 22.3% in the gnomAD database.23 This variant has been shown to modulate the effect of some SCN5A variants, although there is still a strong correlation between variant properties in the His558 and Arg558 backgrounds.54 Future work will investigate the role of the Arg558 background on the variants in this study, which may enable more precise predictions of disease risk from SCN5A genotype.
Mechanisms of NaV1.5 Loss of Function
In this set of variants, the most common cause of channel loss of function was a partial or total reduction in peak current. This is consistent with previous clamp studies13, 14, 15 and our previous literature analysis which found that SCN5A peak current was the largest single predictor of Brugada syndrome risk.10 Two variants, p.Asp785Asn and p.Asp349Asn, had a >10 mV rightward shift in V1/2 activation; however, these two variants also had <50% peak current. Thus, while variants with large gating defects have been previously described (e.g., p.Arg1632His39), the major mechanism of NaV1.5 loss of function is a reduction in peak current. Previous studies have found that channel misfolding and a resulting cell-surface trafficking deficiency is a common mechanism of loss-of-function variants in SCN5A5,27 and other ion channel genes.55 We observed a negative correlation between peak current density and computationally predicted |ΔΔG|, an estimate of variant-induced perturbations to native thermostability, consistent with other studies that showed that thermostability perturbations are a major cause of altered protein function.56, 57, 58, 59 In addition, structural modeling identified several probable modes of channel dysfunction, including pore-lining variants that likely disrupt sodium permeation, removal of a disulfide bond, or creation of prolines that likely disrupt alpha helices. Although these structural analyses were enlightening, structural features do not yet fully predict channel function, highlighting the complementarity of structural modeling and empirical electrophysiological measurements. Consistent with previous studies of SCN5A variants,27, 28, 29, 30, 31 some but not all (8/22) SCN5A loss-of-function variants were partially rescued by 30° treatment or mexiletine. Although these treatments are not practical in the clinic for treatment of Brugada syndrome, this result suggests that other drugs or interventions may help rescue some SCN5A loss-of-function variants.
High-Throughput Approaches to Variant Classification
High-throughput patch clamping is a promising method for reclassifying the thousands of variants of uncertain significance in Mendelian arrhythmia genes. Previously, Vanoye et al.22 reclassified 23/35 “uninformative” KCNQ1 variants (variants of uncertain significance, variants with conflicting interpretations, or no available data). Ng et al.21 reclassified 13 uninformative variants in KCNH2 with patch clamping and cell surface abundance assays. This work combines high-throughput patch clamping and structural modeling, and explicitly incorporates ACMG classification criteria to reclassify 49/61 SCN5A VUSs, 35 to likely pathogenic, and 14 to benign/likely benign. Our approach may be extended to study additional SCN5A variants, including gain-of-function long QT-associated variants or additional variants observed in population sequencing efforts. Accurate classification of the large number of variants in arrhythmia-associated genes will require integrating data from multiple different model systems, such as patch clamping,21,22 induced pluripotent stem cell-derived cardiomyocytes,60, 61, 62 structural and computational models,17,63,64 and ultra-high-throughput multiplexed assays.65, 66, 67
Limitations
This study assays variants in a heterologous expression system. While peak current as measured in this system is the strongest available in vitro predictor of Brugada syndrome risk,10 it is possible for some variants to show different properties in HEK293T cells compared to cardiomyocytes, e.g., because of other proteins that interact with or modify NaV1.5.68, 69, 70 This study examined only the most common haplotype (H558) and the most common cardiac splice isoform of SCN5A; for some variants, it has been shown that alternate haplotypes/isoforms can modulate channel properties.54
Conclusion
This study used automated patch clamping to study 73 previously unstudied SCN5A variants, resulting in the reclassification of 49/61 variants of uncertain significance. This approach can help reclassify variants in this important disease gene and improve the accuracy and scope of genetic medicine.
Data and Code Availability
Additional data and code from this study is available upon reasonable request from the corresponding author.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank Kenneth Matreyek and Douglas Fowler for sharing HEK293 cell lines and Tim Strassmaier and Carlos Vanoye for helpful advice. The Nanion SyncroPatch 384PE is housed and managed within the Vanderbilt High-Throughput Screening center, an institutionally supported core, and was funded by NIH Shared Instrumentation Grant 1S10OD025281. This research was funded by NIH grants K99 HG010904 (A.M.G.), K99 HL135442 (B.M.K.), R01 HL149826 (D.M.R.), and P50 GM115305 (D.M.R.) and American Heart Association fellowships 20POST35220002 (B.L.) and 20PRE35180088 (A.M.).
Published: June 12, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.05.015.
Web Resources
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
Supplemental Information
Summary of Patch Clamp Data for Each Variant. For each variant, peak current density (normalized to wild-type), voltage of 1/2 activation, inactivation time, voltage of 1/2 inactivation, recovery from inactivation, and late current (% of peak) are presented. Means, standard errors of the mean, and the number of qualifying cells are presented for each parameter. Some parameters have “normalized” values also included which indicates the difference in the parameter value from wild-type. Only variants with at least 5 qualifying cells are included, so most severe loss of function variants are not included for most parameters. In addition, variants are classified into categories based on their functional properties, and each variant’s BS3 and PS3 ACMG criteria for Brugada Syndrome (loss of function) or Long QT syndrome (gain of function) is presented.
References
- 1.Kapplinger J.D., Tester D.J., Alders M., Benito B., Berthet M., Brugada J., Brugada P., Fressart V., Guerchicoff A., Harris-Kerr C. An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm. 2010;7:33–46. doi: 10.1016/j.hrthm.2009.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapplinger J.D., Tester D.J., Salisbury B.A., Carr J.L., Harris-Kerr C., Pollevick G.D., Wilde A.A.M., Ackerman M.J. Spectrum and prevalence of mutations from the first 2,500 consecutive unrelated patients referred for the FAMILION long QT syndrome genetic test. Heart Rhythm. 2009;6:1297–1303. doi: 10.1016/j.hrthm.2009.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moreau A., Gosselin-Badaroudine P., Delemotte L., Klein M.L., Chahine M. Gating pore currents are defects in common with two Nav1.5 mutations in patients with mixed arrhythmias and dilated cardiomyopathy. J. Gen. Physiol. 2015;145:93–106. doi: 10.1085/jgp.201411304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bezzina C.R., Rook M.B., Groenewegen W.A., Herfst L.J., van der Wal A.C., Lam J., Jongsma H.J., Wilde A.A., Mannens M.M. Compound heterozygosity for mutations (W156X and R225W) in SCN5A associated with severe cardiac conduction disturbances and degenerative changes in the conduction system. Circ. Res. 2003;92:159–168. doi: 10.1161/01.res.0000052672.97759.36. [DOI] [PubMed] [Google Scholar]
- 5.Gui J., Wang T., Jones R.P., Trump D., Zimmer T., Lei M. Multiple loss-of-function mechanisms contribute to SCN5A-related familial sick sinus syndrome. PLoS ONE. 2010;5:e10985. doi: 10.1371/journal.pone.0010985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brugada J., Campuzano O., Arbelo E., Sarquella-Brugada G., Brugada R. Present Status of Brugada Syndrome: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2018;72:1046–1059. doi: 10.1016/j.jacc.2018.06.037. [DOI] [PubMed] [Google Scholar]
- 7.Hosseini S.M., Kim R., Udupa S., Costain G., Jobling R., Liston E., Jamal S.M., Szybowska M., Morel C.F., Bowdin S., National Institutes of Health Clinical Genome Resource Consortium Reappraisal of Reported Genes for Sudden Arrhythmic Death: Evidence-Based Evaluation of Gene Validity for Brugada Syndrome. Circulation. 2018;138:1195–1205. doi: 10.1161/CIRCULATIONAHA.118.035070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brugada P., Brugada J. Right bundle branch block, persistent ST segment elevation and sudden cardiac death: a distinct clinical and electrocardiographic syndrome. A multicenter report. J. Am. Coll. Cardiol. 1992;20:1391–1396. doi: 10.1016/0735-1097(92)90253-j. [DOI] [PubMed] [Google Scholar]
- 9.Milman A., Andorin A., Postema P.G., Gourraud J.B., Sacher F., Mabo P., Kim S.H., Maeda S., Takahashi Y., Kamakura T. Ethnic differences in patients with Brugada syndrome and arrhythmic events: New insights from Survey on Arrhythmic Events in Brugada Syndrome. Heart Rhythm. 2019;16:1468–1474. doi: 10.1016/j.hrthm.2019.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Kroncke B.M., Glazer A.M., Smith D.K., Blume J.D., Roden D.M. SCN5A (NaV1.5) Variant Functional Perturbation and Clinical Presentation: Variants of a Certain Significance. Circ Genom Precis Med. 2018;11:e002095. doi: 10.1161/CIRCGEN.118.002095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang D.W., Yazawa K., George A.L., Jr., Bennett P.B. Characterization of human cardiac Na+ channel mutations in the congenital long QT syndrome. Proc. Natl. Acad. Sci. USA. 1996;93:13200–13205. doi: 10.1073/pnas.93.23.13200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalia S.S., Adelman K., Bale S.J., Chung W.K., Eng C., Evans J.P., Herman G.E., Hufnagel S.B., Klein T.E., Korf B.R. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet. Med. 2017;19:249–255. doi: 10.1038/gim.2016.190. [DOI] [PubMed] [Google Scholar]
- 13.Chen Q., Kirsch G.E., Zhang D., Brugada R., Brugada J., Brugada P., Potenza D., Moya A., Borggrefe M., Breithardt G. Genetic basis and molecular mechanism for idiopathic ventricular fibrillation. Nature. 1998;392:293–296. doi: 10.1038/32675. [DOI] [PubMed] [Google Scholar]
- 14.Deschênes I., Baroudi G., Berthet M., Barde I., Chalvidan T., Denjoy I., Guicheney P., Chahine M. Electrophysiological characterization of SCN5A mutations causing long QT (E1784K) and Brugada (R1512W and R1432G) syndromes. Cardiovasc. Res. 2000;46:55–65. doi: 10.1016/s0008-6363(00)00006-7. [DOI] [PubMed] [Google Scholar]
- 15.Kapplinger J.D., Giudicessi J.R., Ye D., Tester D.J., Callis T.E., Valdivia C.R., Makielski J.C., Wilde A.A., Ackerman M.J. Enhanced Classification of Brugada Syndrome-Associated and Long-QT Syndrome-Associated Genetic Variants in the SCN5A-Encoded Na(v)1.5 Cardiac Sodium Channel. Circ Cardiovasc Genet. 2015;8:582–595. doi: 10.1161/CIRCGENETICS.114.000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Priori S.G., Napolitano C., Gasparini M., Pappone C., Della Bella P., Giordano U., Bloise R., Giustetto C., De Nardis R., Grillo M. Natural history of Brugada syndrome: insights for risk stratification and management. Circulation. 2002;105:1342–1347. doi: 10.1161/hc1102.105288. [DOI] [PubMed] [Google Scholar]
- 17.Kroncke B.M., Mendenhall J., Smith D.K., Sanders C.R., Capra J.A., George A.L., Blume J.D., Meiler J., Roden D.M. Protein structure aids predicting functional perturbation of missense variants in SCN5A and KCNQ1. Comput. Struct. Biotechnol. J. 2019;17:206–214. doi: 10.1016/j.csbj.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kroncke B.M., Smith D.K., Glazer A.M., Roden D.M., Blume J.D. A Bayesian method using sparse data to estimate penetrance of disease-associated genetic variants. bioRxiv. 2019 doi: 10.1101/571158. [DOI] [Google Scholar]
- 19.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang S.K., Vanoye C.G., Misra S.N., Echevarria D.M., Calhoun J.D., O’Connor J.B., Fabre K.L., McKnight D., Demmer L., Goldenberg P. Spectrum of KV2.1 dysfunction in KCNB1-associated neurodevelopmental disorders. Ann. Neurol. 2019;86:899–912. doi: 10.1002/ana.25607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng C.A., Perry M.D., Liang W., Smith N.J., Foo B., Shrier A., Lukacs G.L., Hill A.P., Vandenberg J.I. High-throughput phenotyping of heteromeric human ether-a-go-go-related gene potassium channel variants can discriminate pathogenic from rare benign variants. Heart Rhythm. 2020;17:492–500. doi: 10.1016/j.hrthm.2019.09.020. [DOI] [PubMed] [Google Scholar]
- 22.Vanoye C.G., Desai R.R., Fabre K.L., Gallagher S.L., Potet F., DeKeyser J.M., Macaya D., Meiler J., Sanders C.R., George A.L., Jr. High-Throughput Functional Evaluation of KCNQ1 Decrypts Variants of Unknown Significance. Circ Genom Precis Med. 2018;11:e002345. doi: 10.1161/CIRCGEN.118.002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matreyek K.A., Stephany J.J., Fowler D.M. A platform for functional assessment of large variant libraries in mammalian cells. Nucleic Acids Res. 2017;45:e102. doi: 10.1093/nar/gkx183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matreyek K.A., Stephany J.J., Chiasson M.A., Hasle N., Fowler D.M. An Improved Platform for Functional Assessment of Large Protein Libraries in Mammalian Cells. Nucleic Acids Res. 2020;48:e1. doi: 10.1093/nar/gkz910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hermann M., Stillhard P., Wildner H., Seruggia D., Kapp V., Sánchez-Iranzo H., Mercader N., Montoliu L., Zeilhofer H.U., Pelczar P. Binary recombinase systems for high-resolution conditional mutagenesis. Nucleic Acids Res. 2014;42:3894–3907. doi: 10.1093/nar/gkt1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clatot J., Ziyadeh-Isleem A., Maugenre S., Denjoy I., Liu H., Dilanian G., Hatem S.N., Deschênes I., Coulombe A., Guicheney P., Neyroud N. Dominant-negative effect of SCN5A N-terminal mutations through the interaction of Na(v)1.5 α-subunits. Cardiovasc. Res. 2012;96:53–63. doi: 10.1093/cvr/cvs211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makiyama T., Akao M., Tsuji K., Doi T., Ohno S., Takenaka K., Kobori A., Ninomiya T., Yoshida H., Takano M. High risk for bradyarrhythmic complications in patients with Brugada syndrome caused by SCN5A gene mutations. J. Am. Coll. Cardiol. 2005;46:2100–2106. doi: 10.1016/j.jacc.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 29.Pfahnl A.E., Viswanathan P.C., Weiss R., Shang L.L., Sanyal S., Shusterman V., Kornblit C., London B., Dudley S.C., Jr. A sodium channel pore mutation causing Brugada syndrome. Heart Rhythm. 2007;4:46–53. doi: 10.1016/j.hrthm.2006.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valdivia C.R., Ackerman M.J., Tester D.J., Wada T., McCormack J., Ye B., Makielski J.C. A novel SCN5A arrhythmia mutation, M1766L, with expression defect rescued by mexiletine. Cardiovasc. Res. 2002;55:279–289. doi: 10.1016/s0008-6363(02)00445-5. [DOI] [PubMed] [Google Scholar]
- 31.Valdivia C.R., Tester D.J., Rok B.A., Porter C.B., Munger T.M., Jahangir A., Makielski J.C., Ackerman M.J. A trafficking defective, Brugada syndrome-causing SCN5A mutation rescued by drugs. Cardiovasc. Res. 2004;62:53–62. doi: 10.1016/j.cardiores.2004.01.022. [DOI] [PubMed] [Google Scholar]
- 32.Whiffin N., Minikel E., Walsh R., O’Donnell-Luria A.H., Karczewski K., Ing A.Y., Barton P.J.R., Funke B., Cook S.A., MacArthur D., Ware J.S. Using high-resolution variant frequencies to empower clinical genome interpretation. Genet. Med. 2017;19:1151–1158. doi: 10.1038/gim.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi Y., Chan A.P. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leaver-Fay A., Tyka M., Lewis S.M., Lange O.F., Thompson J., Jacak R., Kaufman K., Renfrew P.D., Smith C.A., Sheffler W. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen H., Liu D., Wu K., Lei J., Yan N. Structures of human Nav1.7 channel in complex with auxiliary subunits and animal toxins. Science. 2019;363:1303–1308. doi: 10.1126/science.aaw2493. [DOI] [PubMed] [Google Scholar]
- 38.Shen H., Li Z., Jiang Y., Pan X., Wu J., Cristofori-Armstrong B., Smith J.J., Chin Y.K.Y., Lei J., Zhou Q. Structural basis for the modulation of voltage-gated sodium channels by animal toxins. Science. 2018;362:362. doi: 10.1126/science.aau2596. [DOI] [PubMed] [Google Scholar]
- 39.Benson D.W., Wang D.W., Dyment M., Knilans T.K., Fish F.A., Strieper M.J., Rhodes T.H., George A.L., Jr. Congenital sick sinus syndrome caused by recessive mutations in the cardiac sodium channel gene (SCN5A) J. Clin. Invest. 2003;112:1019–1028. doi: 10.1172/JCI18062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Potet F., Mabo P., Le Coq G., Probst V., Schott J.J., Airaud F., Guihard G., Daubert J.C., Escande D., Le Marec H. Novel brugada SCN5A mutation leading to ST segment elevation in the inferior or the right precordial leads. J. Cardiovasc. Electrophysiol. 2003;14:200–203. doi: 10.1046/j.1540-8167.2003.02382.x. [DOI] [PubMed] [Google Scholar]
- 41.Itoh H., Berthet M., Fressart V., Denjoy I., Maugenre S., Klug D., Mizusawa Y., Makiyama T., Hofman N., Stallmeyer B. Asymmetry of parental origin in long QT syndrome: preferential maternal transmission of KCNQ1 variants linked to channel dysfunction. Eur. J. Hum. Genet. 2016;24:1160–1166. doi: 10.1038/ejhg.2015.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yue P., Li Z., Moult J. Loss of protein structure stability as a major causative factor in monogenic disease. J. Mol. Biol. 2005;353:459–473. doi: 10.1016/j.jmb.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 43.Stein A., Fowler D.M., Hartmann-Petersen R., Lindorff-Larsen K. Biophysical and Mechanistic Models for Disease-Causing Protein Variants. Trends Biochem. Sci. 2019;44:575–588. doi: 10.1016/j.tibs.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park H., Bradley P., Greisen P., Jr., Liu Y., Mulligan V.K., Kim D.E., Baker D., DiMaio F. Simultaneous Optimization of Biomolecular Energy Functions on Features from Small Molecules and Macromolecules. J. Chem. Theory Comput. 2016;12:6201–6212. doi: 10.1021/acs.jctc.6b00819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim M.K., Kang Y.K. Positional preference of proline in alpha-helices. Protein Sci. 1999;8:1492–1499. doi: 10.1110/ps.8.7.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brnich S.E., Abou Tayoun A.N., Couch F.J., Cutting G.R., Greenblatt M.S., Heinen C.D., Kanavy D.M., Luo X., McNulty S.M., Starita L.M., Clinical Genome Resource Sequence Variant Interpretation Working Group Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. Genome Med. 2019;12:3. doi: 10.1186/s13073-019-0690-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilde A.A.M., Amin A.S. Clinical Spectrum of SCN5A Mutations: Long QT Syndrome, Brugada Syndrome, and Cardiomyopathy. JACC Clin. Electrophysiol. 2018;4:569–579. doi: 10.1016/j.jacep.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 48.Milman A., Gourraud J.B., Andorin A., Postema P.G., Sacher F., Mabo P., Conte G., Giustetto C., Sarquella-Brugada G., Hochstadt A. Gender differences in patients with Brugada syndrome and arrhythmic events: Data from a survey on arrhythmic events in 678 patients. Heart Rhythm. 2018;15:1457–1465. doi: 10.1016/j.hrthm.2018.06.019. [DOI] [PubMed] [Google Scholar]
- 49.Bezzina C.R., Barc J., Mizusawa Y., Remme C.A., Gourraud J.B., Simonet F., Verkerk A.O., Schwartz P.J., Crotti L., Dagradi F. Common variants at SCN5A-SCN10A and HEY2 are associated with Brugada syndrome, a rare disease with high risk of sudden cardiac death. Nat. Genet. 2013;45:1044–1049. doi: 10.1038/ng.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takehara N., Makita N., Kawabe J., Sato N., Kawamura Y., Kitabatake A., Kikuchi K. A cardiac sodium channel mutation identified in Brugada syndrome associated with atrial standstill. J. Intern. Med. 2004;255:137–142. doi: 10.1046/j.0954-6820.2003.01247.x. [DOI] [PubMed] [Google Scholar]
- 51.Probst V., Allouis M., Sacher F., Pattier S., Babuty D., Mabo P., Mansourati J., Victor J., Nguyen J.M., Schott J.J. Progressive cardiac conduction defect is the prevailing phenotype in carriers of a Brugada syndrome SCN5A mutation. J. Cardiovasc. Electrophysiol. 2006;17:270–275. doi: 10.1111/j.1540-8167.2006.00349.x. [DOI] [PubMed] [Google Scholar]
- 52.Remme C.A., Wilde A.A., Bezzina C.R. Cardiac sodium channel overlap syndromes: different faces of SCN5A mutations. Trends Cardiovasc. Med. 2008;18:78–87. doi: 10.1016/j.tcm.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 53.Frigo G., Rampazzo A., Bauce B., Pilichou K., Beffagna G., Danieli G.A., Nava A., Martini B. Homozygous SCN5A mutation in Brugada syndrome with monomorphic ventricular tachycardia and structural heart abnormalities. Europace. 2007;9:391–397. doi: 10.1093/europace/eum053. [DOI] [PubMed] [Google Scholar]
- 54.Makielski J.C., Ye B., Valdivia C.R., Pagel M.D., Pu J., Tester D.J., Ackerman M.J. A ubiquitous splice variant and a common polymorphism affect heterologous expression of recombinant human SCN5A heart sodium channels. Circ. Res. 2003;93:821–828. doi: 10.1161/01.RES.0000096652.14509.96. [DOI] [PubMed] [Google Scholar]
- 55.Anderson C.L., Delisle B.P., Anson B.D., Kilby J.A., Will M.L., Tester D.J., Gong Q., Zhou Z., Ackerman M.J., January C.T. Most LQT2 mutations reduce Kv11.1 (hERG) current by a class 2 (trafficking-deficient) mechanism. Circulation. 2006;113:365–373. doi: 10.1161/CIRCULATIONAHA.105.570200. [DOI] [PubMed] [Google Scholar]
- 56.Casadio R., Vassura M., Tiwari S., Fariselli P., Luigi Martelli P. Correlating disease-related mutations to their effect on protein stability: a large-scale analysis of the human proteome. Hum. Mutat. 2011;32:1161–1170. doi: 10.1002/humu.21555. [DOI] [PubMed] [Google Scholar]
- 57.DePristo M.A., Weinreich D.M., Hartl D.L. Missense meanderings in sequence space: a biophysical view of protein evolution. Nat. Rev. Genet. 2005;6:678–687. doi: 10.1038/nrg1672. [DOI] [PubMed] [Google Scholar]
- 58.Tokuriki N., Tawfik D.S. Stability effects of mutations and protein evolvability. Curr. Opin. Struct. Biol. 2009;19:596–604. doi: 10.1016/j.sbi.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 59.Zhou Y., Bowie J.U. Building a thermostable membrane protein. J. Biol. Chem. 2000;275:6975–6979. doi: 10.1074/jbc.275.10.6975. [DOI] [PubMed] [Google Scholar]
- 60.Chavali N.V., Kryshtal D.O., Parikh S.S., Wang L., Glazer A.M., Blackwell D.J., Kroncke B.M., Shoemaker M.B., Knollmann B.C. Patient-independent human induced pluripotent stem cell model: A new tool for rapid determination of genetic variant pathogenicity in long QT syndrome. Heart Rhythm. 2019;16:1686–1695. doi: 10.1016/j.hrthm.2019.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fatima A., Kaifeng S., Dittmann S., Xu G., Gupta M.K., Linke M., Zechner U., Nguemo F., Milting H., Farr M. The disease-specific phenotype in cardiomyocytes derived from induced pluripotent stem cells of two long QT syndrome type 3 patients. PLoS ONE. 2013;8:e83005. doi: 10.1371/journal.pone.0083005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Selga E., Sendfeld F., Martinez-Moreno R., Medine C.N., Tura-Ceide O., Wilmut S.I., Pérez G.J., Scornik F.S., Brugada R., Mills N.L. Sodium channel current loss of function in induced pluripotent stem cell-derived cardiomyocytes from a Brugada syndrome patient. J. Mol. Cell. Cardiol. 2018;114:10–19. doi: 10.1016/j.yjmcc.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heyne H.O., Baez-Nieto D., Iqbal S., Palmer D., Brunklaus A., the Epi25 Collaborative, Johannesen K.M., Lauxmann S., Lemke J.R., Moller R.S. Predicting functional effects of missense variants in the voltage-gated sodium and calcium channels. bioRxiv. 2019 doi: 10.1101/671453. [DOI] [PubMed] [Google Scholar]
- 64.Li B., Mendenhall J.L., Kroncke B.M., Taylor K.C., Huang H., Smith D.K., Vanoye C.G., Blume J.D., George A.L., Jr., Sanders C.R., Meiler J. Predicting the Functional Impact of KCNQ1 Variants of Unknown Significance. Circ Cardiovasc Genet. 2017;10:10. doi: 10.1161/CIRCGENETICS.117.001754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Findlay G.M., Daza R.M., Martin B., Zhang M.D., Leith A.P., Gasperini M., Janizek J.D., Huang X., Starita L.M., Shendure J. Accurate classification of BRCA1 variants with saturation genome editing. Nature. 2018;562:217–222. doi: 10.1038/s41586-018-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Glazer A.M., Kroncke B.M., Matreyek K.A., Yang T., Wada Y., Shields T., Salem J.E., Fowler D.M., Roden D.M. Deep Mutational Scan of an SCN5A voltage sensor. Circ Genom Precis Med. 2020;13:e002786. doi: 10.1161/CIRCGEN.119.002786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matreyek K.A., Starita L.M., Stephany J.J., Martin B., Chiasson M.A., Gray V.E., Kircher M., Khechaduri A., Dines J.N., Hause R.J. Multiplex assessment of protein variant abundance by massively parallel sequencing. Nat. Genet. 2018;50:874–882. doi: 10.1038/s41588-018-0122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aiba T., Farinelli F., Kostecki G., Hesketh G.G., Edwards D., Biswas S., Tung L., Tomaselli G.F. A mutation causing Brugada syndrome identifies a mechanism for altered autonomic and oxidant regulation of cardiac sodium currents. Circ Cardiovasc Genet. 2014;7:249–256. doi: 10.1161/CIRCGENETICS.113.000480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Casini S., Albesa M., Wang Z., Portero V., Ross-Kaschitza D., Rougier J.S., Marchal G.A., Chung W.K., Bezzina C.R., Abriel H., Remme C.A. Functional Consequences of the SCN5A-p.Y1977N Mutation within the PY Ubiquitylation Motif: Discrepancy between HEK293 Cells and Transgenic Mice. Int. J. Mol. Sci. 2019;20:11. doi: 10.3390/ijms20205033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Clatot J., Hoshi M., Wan X., Liu H., Jain A., Shinlapawittayatorn K., Marionneau C., Ficker E., Ha T., Deschênes I. Voltage-gated sodium channels assemble and gate as dimers. Nat. Commun. 2017;8:2077. doi: 10.1038/s41467-017-02262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of Patch Clamp Data for Each Variant. For each variant, peak current density (normalized to wild-type), voltage of 1/2 activation, inactivation time, voltage of 1/2 inactivation, recovery from inactivation, and late current (% of peak) are presented. Means, standard errors of the mean, and the number of qualifying cells are presented for each parameter. Some parameters have “normalized” values also included which indicates the difference in the parameter value from wild-type. Only variants with at least 5 qualifying cells are included, so most severe loss of function variants are not included for most parameters. In addition, variants are classified into categories based on their functional properties, and each variant’s BS3 and PS3 ACMG criteria for Brugada Syndrome (loss of function) or Long QT syndrome (gain of function) is presented.
Data Availability Statement
Additional data and code from this study is available upon reasonable request from the corresponding author.