Abstract
Genetics researchers and clinical professionals rely on diversity measures such as race, ethnicity, and ancestry (REA) to stratify study participants and patients for a variety of applications in research and precision medicine. However, there are no comprehensive, widely accepted standards or guidelines for collecting and using such data in clinical genetics practice. Two NIH-funded research consortia, the Clinical Genome Resource (ClinGen) and Clinical Sequencing Evidence-generating Research (CSER), have partnered to address this issue and report how REA are currently collected, conceptualized, and used. Surveying clinical genetics professionals and researchers (n = 448), we found heterogeneity in the way REA are perceived, defined, and measured, with variation in the perceived importance of REA in both clinical and research settings. The majority of respondents (>55%) felt that REA are at least somewhat important for clinical variant interpretation, ordering genetic tests, and communicating results to patients. However, there was no consensus on the relevance of REA, including how each of these measures should be used in different scenarios and what information they can convey in the context of human genetics. A lack of common definitions and applications of REA across the precision medicine pipeline may contribute to inconsistencies in data collection, missing or inaccurate classifications, and misleading or inconclusive results. Thus, our findings support the need for standardization and harmonization of REA data collection and use in clinical genetics and precision health research.
Keywords: diversity, clinical genetics, race, ethnicity, ancestry, survey, ClinGen, CSER, precision medicine
Introduction
Different aspects of human diversity are captured by the concepts of “race,” “ethnicity,” and “ancestry” (REA). While these terms are frequently used as prompts in the collection of demographic data, most notably on the United States Census, there are no consistent, universal definitions for REA. Categorizing people by REA is also common in biomedical research, clinical care, and reporting health statistics for funding in the United States.1 However, these terms are not interchangeable, and they are important to define in clinical genetics practice.
We consider race and ethnicity to be socio-cultural factors that provide information about environmental exposures but are not directly indicative of genetic risk factors for disease. The related but distinct concept of ancestry, referring to the genetic inheritance of variants from global ancestral populations, has increasingly been used in genomics research and has implications for certain clinical applications of genetics such as assessing whether a variant is rare or common in a particular ancestral population.2 Ancestry as a genetic concept has also gained prominence in the public sphere due to the rise in popularity of direct-to-consumer genetic testing products.3,4 However, there are no standard definitions of these terms in medical genetics and no consensus on protocols for the collection or estimation of this information across research institutions or healthcare systems. For example, our prior work demonstrated that the categorization of diversity measures on clinical requisition forms varies widely among genetic testing laboratories.5 Importantly, it is unclear to what extent this information is used to interpret genetic test results and whether inconsistent data collection contributes to variation in reporting and delivery of clinical genetics among laboratories, including variant classification, application of screening guidelines, risk elimination, or other measures relevant to clinical care.
Previous studies have examined how race is conceptualized by physicians and genetics professionals.6,7 The ways in which REA influence health outcomes, particularly as they inform health disparities and population-based treatment or interventions, is also an active area of research.8 The literature has shown that definitions of race and ethnicity are historically fluid, context specific, and often used interchangeably.8, 9, 10, 11 The conflation of these concepts, as well as limitations in the type of information available in different settings, also leads to race and/or ethnicity being used as a proxy for genetic ancestry or genomic background. This practice may disproportionately disadvantage people from multiple racial, ethnic, and ancestral backgrounds because self-reported identity measures are often based on incomplete knowledge and are thus inaccurate representations of ancestry, and those with multiple ancestries are not easily grouped into discrete racial or ethnic categories.13 Furthermore, the lack of diversity in genomic databases means that even if self-reported measures serve as a reasonable proxy for ancestral background, this information may have limited utility and actionability in clinical genetics.14
The Ancestry and Diversity Working Group (ADWG) of the NIH-funded Clinical Genome Resource (ClinGen) is a multi-disciplinary group of investigators from academic research institutions and hospitals across the United States. ClinGen is an important knowledgebase for clinical genetics, including a set of FDA-recognized variant-disease expert curations. The ClinGen ADWG is responsible for ensuring that this resource is representative of and responsive to a broad spectrum of human diversity. ADWG members have expertise spanning genomics, epidemiology, public health, bioethics, and social sciences. Positioned at the intersections of these diverse professional perspectives, the ADWG is intended to conduct research and provide evidence-based guidance about the use of REA in clinical genetics.
Another NIH-funded research consortium, Clinical Sequencing Evidence-generating Research (CSER), is a collaborative effort of seven independent research sites conducting studies in clinical translational genetics with a specific focus on diverse populations. CSER investigators are also working to address disparities in clinical genetics and the development of standards for the use of REA. This study is the result of a collaborative effort between the two consortia to determine how REA are conceptualized, utilized, and communicated in clinical genetics research and practice.
There may be some effective and necessary uses of REA data, such as contextualizing the genomic or environmental background of a genetic variant with potential clinical significance, or tracking racial and ethnic health disparities over time. However, continued collection and use of these measures without a clear justification and framework may lead to differential clinical treatment and quality of care among racial and ethnic minority groups with negative implications more broadly for the fair distribution of benefits from precision medicine research. Despite the tradeoffs between the potential utility and harms of racial and ethnic classification,15 no study to date has conducted a comprehensive assessment of how the terms race, ethnicity, and ancestry are understood by clinical genetics professionals and researchers, or how these perceptions inform clinical genetic testing, variant interpretation, reporting, diagnosis, and treatment. The survey described in this paper interrogates both perceptions and reported use of REA among clinical genetics professionals.
Subjects and Methods
The target study population included both non-clinical genetics researchers and clinical genetics professionals (i.e., clinical geneticists, genetic counselors, clinical laboratory directors, and other clinical laboratory employees). Most questions were designed for clinical genetics professionals and related to clinical activities such as test ordering, variant interpretation, and reporting genetic test results to patients. In total, 121 survey questions (see Supplemental Note) were developed and refined through an iterative process by ClinGen ADWG and CSER investigators.
An initial set of survey questions was developed and approved by ADWG members, reviewed and revised based on feedback from CSER investigators, and further revised based on cognitive interviews. In order to refine survey questions for clarity and understanding by the intended respondents, cognitive interviews were conducted by a single interviewer (A.B.P.) with 11 representatives from target participant groups including non-clinical genetics researchers and clinical genetics professionals, most of whom were recruited through CSER. During cognitive interviews, participants were asked to read survey questions aloud and talk through their understanding and interpretation of each question. Their ability to determine or recall the requested information was observed. If a participant had difficulty or was unable to answer a question, the interviewer discussed the question further with the participant to assess interpretation issues and provided alternative phrasing until the intended question was clearly understood. As interview participants read and answered survey questions while speaking aloud their thought processes, the interviewer asked follow-up questions about their interpretation of the questions to ensure consistency with the intended design.
Audio and video were recorded for online interviews (n = 8), and audio was recorded for in-person interviews (n = 3). Every three interviews, the interviewer used notes and recordings to revise survey questions to reflect language that was best understood and interpreted by the majority of participants. As the language and format of questions was refined through cognitive interviews, participants were able to understand more questions without needing clarification, and the ability of participants in later interviews to correctly interpret and answer questions was improved relative to earlier ones. The revised survey was reviewed and approved by ClinGen ADWG members and CSER investigators.
The survey required answers to multiple-choice questions about respondents’ perceived definitions and utility of REA in research and clinical genetics; these questions were developed based on responses to a previous study of physician perspectives on race and ethnicity.16 Optional participant demographic questions included inquiries about personal identity: (1) free-text boxes for “sex,” “gender,” “race,” “ethnicity,” and “ancestry” and (2) multiple-choice options reflecting the categories provided on the U.S. Census and approved by the U.S. Office of Management and Budget (OMB). Self-reported professional demographics (career type, professional experience, and frequency of conducting variant classification for clinical or research purposes, ordering genetic tests, and reporting results to patients) were used to create branching logic to ensure the relevance of each question to each survey respondent. The remaining questions covered the type and source(s) of REA data used in clinical gene and variant curation and interpretation, clinical contextual factors impacting the interpretation and communication of genetic testing results to patients, and opinions on whether guidelines are needed for the clinical genetics community around the use of such diversity measures. These questions included a combination of multiple-choice, true-false, and free-text response options.
Institutional Review
The Stanford University Institutional Review Board (IRB) determined the proposed study did not meet the definition of research or clinical investigation on human subjects and was therefore exempt from IRB review.
Data Collection
The survey was administered from September 2018 through March 2019 and disseminated by targeted emails with links to an online questionnaire (hosted on the platform Qualtrics).Separate recruitment links were sent for each disseminating organization: ClinGen (n = 788), CSER (n = 184), the American Board of Genetic Counselors (ABGC, n = 4,661), the American Society of Human Genetics (ASHG, n = 2,659), and the American College of Medical Genetics and Genomics (ACMG, n = 2,218). A follow-up recruitment email was also sent to the ClinGen and CSER listservs, as well as a manually curated subset of clinical geneticists in the United States whose emails are publicly available (n = 638). Table S1 shows the number of individuals contacted through each organization and their corresponding response rate estimates. After response rates were estimated for each disseminating organization and follow-up emails were sent to the initial solicitations, a snowball approach was adopted in February and March 2019. The purpose of the snowball approach was to reach additional target participants through individual emails to colleagues and professional networks, and social media such as public posts on Facebook and Twitter. There was no incentive provided for completing the survey.
Data Analysis
Data were downloaded from Qualtrics and analyzed using a custom Python script. The majority of questions were designed to assess current practices in research and clinical genetics and as such most data analysis involved descriptive statistics. For certain analyses, responses on a Likert scale were combined to show broad categories of agreement (e.g., “well” or “very well” and “often” or “always”). Due to our focus on hypothesis generation in lieu of proposed a priori hypotheses, no statistical analyses were performed. Selected descriptive data are summarized in the Results.
Results
Study Participants
A total of 448 respondents completed the survey, including non-clinical researchers (n = 87), clinical genetics professionals (n = 268), trainees (n = 12), and others (Table 1). For all types of respondents, we report on demographics (Table 2), perceived definitions of REA (Figure 1), and opinions on the need for guidelines. In order to gain a sense of how REA are used in clinical genetics, we focus on results obtained from clinical genetics professionals, identified by their reported professional roles and activities.
Table 1.
Survey Participant Professional Roles and Affiliations
| n | Percent (%) Total Survey Participants | |
|---|---|---|
| Primary Professional Role | 398 | 88.8 |
| Genetic Counselor | 168 | 37.5 |
| Non-clinical Researcher | 87 | 19.4 |
| Clinical Geneticist | 57 | 12.7 |
| Clinical Lab Director | 43 | 9.6 |
| Other | 31 | 6.9 |
| Trainee | 12 | 2.7 |
| Primary Professional Affiliation(s) | 435 | 97.1 |
| University/Academic Institution | 194 | 43.3 |
| Hospital or Medical Center | 78 | 17.4 |
| Academic Institution and Hospital or Medical Center | 54 | 12.1 |
| Commercial Laboratory | 37 | 8.3 |
| Government Institution | 24 | 5.4 |
| Other | 20 | 4.5 |
| Non-academic Research Institution | 15 | 3.3 |
| Industry or Private Practice | 11 | 2.5 |
| Non-governmental Organization | 2 | 0.4 |
| Years of Experience | 398 | 88.8 |
| 1–5 | 125 | 31.4 |
| 6–10 | 93 | 23.4 |
| 11–15 | 56 | 14.1 |
| 16–20 | 40 | 10.1 |
| 21–25 | 29 | 7.3 |
| 26–30 | 23 | 5.8 |
| 31–35 | 22 | 5.5 |
| >35 | 10 | 2.5 |
| Area(s) of Clinical Expertise [SelectAllThat Apply] | 448 | Totals > 100 |
| Adult | 219 | 48.9 |
| Pediatric | 192 | 42.9 |
| Prenatal | 84 | 18.8 |
| N/A or None of the Above | 113 | 25.2 |
Survey participants were asked (but not required) to provide information about their professional roles, affiliations, years of experience, and clinical areas of expertise. For professional roles and affiliations, participants were asked to select all that apply and/or write in an alternate response. Few respondents reported more than one clinical role (e.g., Genetic Counselor and Clinical Lab Director, n = 2). Those with a clinical role who also identify as non-clinical researchers (n = 21) were assigned to their clinical role for the purpose of summarizing these data. Similarly, there were few respondents who reported more than one affiliation, with the exception of Academic Institution AND Hospital or Medical Center (n = 54). Those reporting more than two professional roles or affiliations were classified as "Other."The total percentage of clinical areas of expertise adds up to >100% as we report all responses from participants with multiple areas of expertise.
Table 2.
Participant Self-Reported Sex, Gender, Race, and Ethnicity
| n | Percent (%) Total Survey Participants | |
|---|---|---|
| Sex and Gender[Free Text Response] | 448 | 100 |
| Female | 281 | 62.7 |
| Male | 98 | 21.9 |
| Transgender | 1 | 0.22 |
| Irrelevant (not related to sex or gender) or Missing Response | 68 | 15.2 |
| Race and Ethnicity[Multiple Choice] | 448 | 100 |
| Single Selection | 373 | 83.3 |
| American Indian/Alaska Native | 0 | 0.0 |
| Asian | 43 | 9.6 |
| Black or African American | 7 | 1.6 |
| Hispanic or Latino | 6 | 1.3 |
| Native Hawaiian or Other Pacific Islander | 0 | 0.0 |
| White | 317 | 70.8 |
| Multiple Selections | 14 | 3.1 |
| American Indian/Alaska Native and White | 3 | 0.7 |
| American Indian/Alaska Native, Hispanic or Latino, and White | 3 | 0.7 |
| Asian and White | 4 | 0.9 |
| Asian and Hispanic or Latino | 1 | 0.22 |
| Black or African American and White | 1 | 0.22 |
| Hispanic or Latino and White | 1 | 0.22 |
| All of the Above | 1 | 0.22 |
| Missing Response (Race and Ethnicity) | 61 | 13.6 |
All survey respondents were asked to write free-text responses describing their identities with regard to sex, gender, race, ethnicity, and ancestry. They were also asked to select their race and ethnicity from multiple-choice options. Shown here are aggregate results of free-text responses about sex and gender, as well as aggregate responses to the multiple-choice race and ethnicity question.
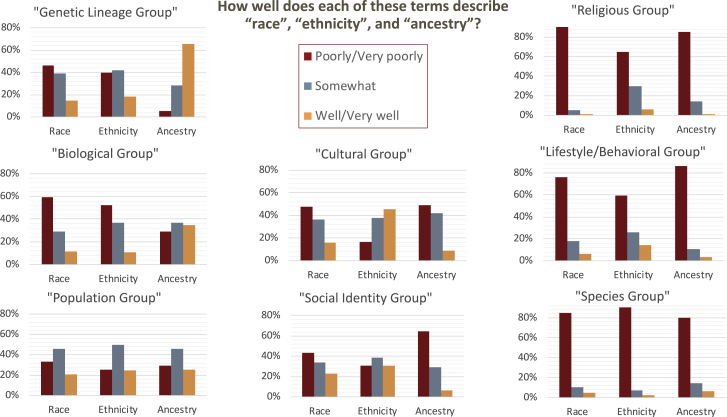
Figure 1.
Perceived Definitions of Race, Ethnicity, and Ancestry
Each cluster of bars corresponds to a complete set of survey responses, such that 100% of participants responded that each description fit each term (race, ethnicity, or ancestry) either very well, well, somewhat, poorly, or very poorly.
Table 1 indicates the primary employment affiliations, professional roles, years of experience, and clinical expertise of respondents. Most respondents were clinical genetics professionals who are employed at academic or commercial diagnostic institutions in the United States. Of the respondents who provided information about their professional experience, 55% (n = 218) had 1–10 years of professional experience, 24% (n = 96) had between 11 and 20 years of experience, and 21% (n = 84) had >20 years. Among those who provided information about their professional roles (n = 398), 42% were genetic counselors (n = 168); others included clinical geneticists (n = 57, 14%), clinical laboratory directors (n = 43, 11%), non-clinical researchers (n = 87, 22%), and trainees (n = 12, 3%).
Sixty percent of survey participants report interpreting or curating genetic variants for a variety of purposes (n = 271), including for research (n = 170, 63%), providing clinical genetic testing reports to healthcare providers (n = 125, 46%), informing the diagnosis of a patient with a genetic disorder (n = 122, 45%), reclassifying variants and/or verifying laboratory results (n = 121, 45%), preparing for a discussion with a patient (n = 120, 44%), and curating genetic variants for large-scale consortium efforts, such as ClinGen (n = 99, 36%).
Among those who provided information about their geographic location (n = 399, 89% of total participants) 93% are in North America, with 347 in the U.S. and 23 in Canada. Outside North America, 19 participants are from Europe, and the remaining individuals are from Australia, Japan, Mexico, Oman, Singapore, and South Korea. The majority of respondents self-identified as white (71%), and 63% identified as female (Table 2).
Adjusted Response Rate
The overall response rate (4.2%) was adjusted for overlap in professional society membership and consortium affiliations. Respondents were asked to indicate all relevant professional affiliations with the organizations sampled, which facilitated the elimination of double counting such that each survey respondent was counted only once in the denominator of all possible respondents. Table S1 indicates response rates that are specific to each disseminating organization, which are also individually adjusted to ensure each study participant was counted in only one targeted recruitment group. Due to residual overlap in organizational membership among survey non-responders, our response rate is likely underestimated. Furthermore, with a snowball approach (employed during the latter portion of the recruitment period), the number of potential respondents cannot be estimated, so these responses are not counted in the overall response rate for the survey.
Collecting REA Data in Practice
Nearly all participants (n = 422, 94%) collect or use information about “population identity, e.g., population allele frequencies, self-reported race or ethnicity, and/or ancestral origins” in their work. The majority of participants reported using this information for the purpose of clinical practice alone (n = 178, 39%) or for the purposes of both clinical practice and research (n = 130, 29%). Among respondents who see patients, order genetic tests, and/or communicate results (n = 268), only 5 individuals (2%) report that no REA information is used in their work, and 4 (2%) were not sure. Race and/or ethnicity is most often obtained directly from the patient (n = 204, 94%) or from the patient’s medical record (n = 87, 40%). Some reported that race and/or ethnicity is recorded by another care provider, “possibly without verifying directly with the patient” (n = 39, 18%).
Conceptualizing REA in Clinical Genetics
Figure 1 illustrates differences among survey respondents in their agreement with select phrases offered to describe the terms race, ethnicity, and ancestry. Nearly two-thirds of survey participants indicated that they felt “ancestry” was well described by the term “genetic lineage group” (n = 295, 66%). “Cultural group” was perceived to describe “ethnicity” well or very well by 46% of participants (n = 205). No term or phrase was considered a good description of “race,” and ∼80% of participants felt that none of the three REA terms were well described by “species group,” “lifestyle/behavioral group,” or “religious group.” Beyond these areas of relative agreement, there was otherwise substantial heterogeneity in respondents’ agreement with the definitions provided.
Respondents were asked about their confidence in describing differences among “race,” “ethnicity,” and “ancestry,” in clinical genetics and in general. More than two-thirds of participants reported being “somewhat” or “not at all confident” in their ability to distinguish among REA terms, both in general and as they relate to genetics and clinical care. However, there was a marked shift in the proportion of survey participants who responded that they are “confident” when asked about their ability to distinguish among terms in the context of genetics and clinical care. Roughly twice as many participants report being “not at all confident” about the terms in general (n = 123, 27%), compared with those who felt “not at all confident” about the terms related to their line of work in genomics and clinical care (n = 62, 14%).
Use of REA in Clinical Genetics
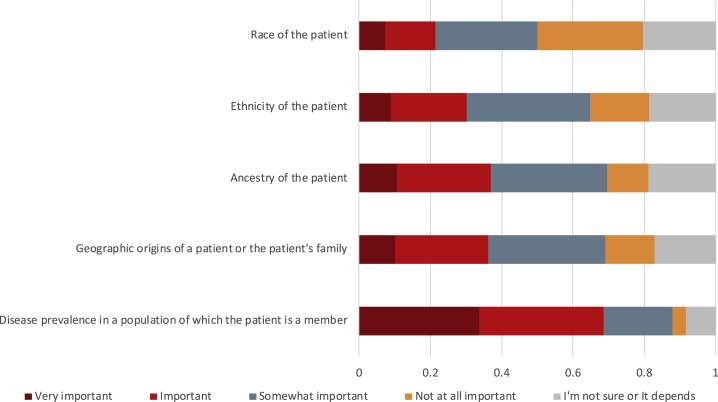
A total of 268 (60%) of survey participants indicated that they order clinical genetic tests and/or return results to patients in their own work and answered a series of questions about patient-facing activities, including perceptions about the relevance of REA for clinical decision making. Patient race (21%), ethnicity (30%), ancestry (37%), geographic origins of a patient or their family (36%), and disease prevalence in a population of which the patient is a member (69%) were all considered important or very important information for the purpose of ordering a genetic test (n = 217) (Figure 2). When asked about other types of clinical decision making, 49% percent of participants agreed that REA would be relevant for obtaining consent.
Figure 2.
Perceived Importance of Patient Data Types in Ordering a Genetic Test
A subset of clinical genetics professionals who work with patients (n = 217) indicated the degree of importance for each type of information (disease prevalence in a population, geographic origins, and REA of a patient) for the purpose of ordering a genetic test.
Patient-facing clinical genetics professionals were also asked about clinical factors and circumstances that would likely motivate them to discuss REA with a patient (Table 3). These included whether test results were positive (48%), revealed a variant of uncertain significance (VUS) (50%), or whether the patient was from a racial or ethnic minority group (53%). A large majority (n = 189, 90%) agreed that REA may be relevant for contextualizing genetic test results for patients. Across all clinical scenarios, there was substantial heterogeneity in the perceived relevance of REA, and for most categories up to a third responded “I’m not sure.”
Table 3.
Perceived Importance of Discussing Race, Ethnicity, and Ancestry with Patients Undergoing Clinical Genetic Testing
| True | False | I’m not sure | |
|---|---|---|---|
| Race, ethnicity, and/or ancestry may be relevant for the following: | n = 210 | ||
| Obtaining consent | 76 (36.2%) | 108 (51.4%) | 26 (12.4%) |
| Contextualizing genetic test results | 189 (90%) | 11 (5.2%) | 10 (4.8%) |
| Tailoring treatment options | 89 (42.4%) | 52 (24.8%) | 69 (32.9%) |
| Factors motivating discussions about race, ethnicity, and ancestry with a patient: | n = 209 | ||
| Positive test result | 101 (48.3%) | 55 (26.3%) | 53 (26.4%) |
| Negative test result | 62 (29.7%) | 71 (34%) | 76 (36.4%) |
| Variant of uncertain significance (VUS) result | 104 (49.8%) | 63 (30.1%) | 41 (19.6%) |
| Patient is from a racial or ethnic minority group | 111 (53.1%) | 70 (33.5%) | 28 (13.4%) |
Survey respondents who see patients responded to true/false questions about the type of clinical functions for which race, ethnicity, or ancestry may be relevant, in addition to factors that might motivate them to discuss these with a patient.
REA in Genetic Variant Interpretation
When asked about the importance of REA for clinical interpretation of genetic variants, respondents perceived these measures as highly important, though practical use and implementation was reportedly much lower. Table 4 shows the degree to which participants reported each of these measures to be important. Some participants pointed out that the type of information most likely to inform interpretation depends on the information that is available. Specifically, ancestry was considered more relevant than race or ethnicity for clinical interpretation but was reportedly less often available and used in practice.
Table 4.
Importance of Race, Ethnicity, and Ancestry in Clinical Variant Interpretation
| n = 271 | Race | Ethnicity | Ancestry |
|---|---|---|---|
| Very important | 22 (8.1%) | 22 (8.1%) | 43 (15.9%) |
| Important | 48 (17.7%) | 67 (24.7%) | 81 (29.9%) |
| Somewhat important | 85 (31.4%) | 70 (25.8%) | 71 (26.2%) |
| It depends | 54 (19.9%) | 58 (21.4%) | 49 (18.1%) |
| Not at all important | 36 (13.3%) | 29 (10.7%) | 10 (3.7%) |
| I’m not sure | 26 (9.6%) | 25 (9.2%) | 17 (6.3%) |
Survey respondents who reported having a professional role in clinical care, such as seeing patients and ordering genetic tests, were asked to evaluate the importance of race, ethnicity, and ancestry for the purpose of clinical variant interpretation. Results are shown here by the multiple-choice (Likert scale) options provided.
Fewer than half of participants reported that any of the REA measures or geographic origin(s) were likely to inform clinical variant interpretation in practice. For example, despite ancestry being reported as most consistent with the concept of a "genetic lineage group" and as the most important to clinical variant interpretation (46% responding that it was important or very important; Table 4), the majority of survey participants (85%) still report that genetic ancestry is rarely (n = 31, 12%) or never (n = 196, 73%) calculated from a patient’s DNA for the purpose of clinical variant interpretation. Importantly, participants (who included clinical lab directors and ordering clinicians) also report that clinical lab reports either rarely (n = 40, 18%) or never (n = 161, 74%) contain ancestry estimates based on genetic data when reporting carrier or diagnostic test results.
Among those who report doing any ancestry analysis for clinical variant interpretation (n = 60), the most common methodological approaches are Principal Component Analysis (PCA, n = 34, 56%) and admixture analysis (n = 16, 26%). Five participants (8%) report using multidimensional scaling, while another 15 (25%) do not know what method is used to calculate ancestry. For relevant methods, the most commonly used reference panels for ancestry inference are 1000 Genomes (n = 30, 73%), HapMap (n = 10, 24%), Human Genome Diversity Project (HGDP, n = 11, 27%), or some combination thereof; some also report using internal reference panels either alone or in combination with others (n = 11, 27%).
Necessity of REA Guidelines for Clinical Genetics
The ACMG/AMP Guidelines for the Interpretation of Sequence Variants call for the use of population databases and rely on allele frequency data to guide the assessment of genetic variants.17 Table 5 shows the proportion of participants who report using each type of REA data to inform clinical variant interpretation. Consistent with the ACMG/AMP recommendations, both population allele frequencies (86%) and the related concept of absence of a variant from “population databases” (83%) are most widely considered informative for interpretation and the majority of participants indicated using multiple publicly available databases to obtain this information.
Table 5.
Data Types Most Likely to Inform Clinical Variant Interpretation
| Data Type[Select All That Apply] | Individuals (n)“Most Likely” to Use Each Type of Data to Inform Variant Interpretation |
|---|---|
| Population allele frequencies | 233 (86%) |
| Whether a variant has been seen before in a population database | 224 (82.7%) |
| Ancestry of the patient or population in which the variant was observed | 120 (44.3%) |
| Geographic origin(s) of the patient’s family or population in which the variant was observed | 108 (40%) |
| Ethnicity of the patient or population in which the variant was observed | 103 (38%) |
| Race of the patient or population in which the variant was observed | 89 (32.8%) |
| All of the above | 49 (18.1%) |
Respondents involved in clinical variant interpretation (n = 271) selected the type(s) of data they are “most likely” to use when interpreting variants.
The majority of respondents who answered questions on the need for new guidelines (n = 425) felt that guidelines would be helpful for the collection and use of REA in clinical genetics (n = 276, 65%). Thirteen participants (3%) disagreed and offered reasons as to why they did not feel new guidelines would be helpful. Common themes include the complexity of issues that cannot be easily digested or distilled, that guidelines would be ineffective or burdensome to implement, and that issues of diversity measures are irrelevant to the field.
Discussion
This study illuminates clinical genetics professionals’ beliefs about the meaning and utility of REA, as well as its reported use in clinical practice. While most respondents considered REA at least somewhat important for clinical interpretation and ordering or communicating the results of genetic tests, there were inconsistent understandings of these concepts and differences of opinion on how these terms should be used in clinical genetics, for what purpose(s). Interestingly, there appeared to be a disconnect between the type of information that is considered most useful by clinical genetics professionals and the type of information they reported having access to (and using) in practice. Highly variable opinions, definitions, and perceived utility of REA for clinical interpretation and the delivery of genomic medicine is consistent with the established view that definitions of race and ethnicity are cultural, dynamic, and change over time.9, 10, 11 These results support the need to standardize the collection and use of REA in clinical genetics.
Perceived Definitions and Utility of REA Vary Widely
Our results indicate that participants are most likely to understand ancestry in biologic terms (i.e., a “genetic lineage group") and in the context of clinical genetics they evaluate the importance of ancestry as higher than that of race or ethnicity. However, this measure is the least available and used most infrequently. Likely due to its lack of availability in practice, survey respondents did not express a greater reliance on ancestry for clinical variant interpretation than on race or ethnicity. Nearly 70% of participants said that disease prevalence in a population of which a patient is a member is important for ordering genetic tests, but without access to genetic ancestry, clinicians must rely on self-reported (or assigned) measures of race and ethnicity in determining to which population a patient should be attributed. Respondents confirm that this information is most often obtained either directly from a patient or an existing medical record and used as a proxy for genetic ancestry. However, as shown previously,5 there is a lack of consistency in how such self-reported measures are collected on clinical laboratory requisition forms (if at all), and some clinical lab directors reported that this information is of limited usefulness. This is particularly problematic for individuals with multiple ancestral backgrounds, as a single ancestral population cannot be assigned even with the ability to calculate or obtain genetic ancestry estimates. A similar challenge exists in the context of clinical variant interpretation, given that the vast majority of participants use population allele frequencies to determine whether a variant is rare and thus considered more likely to be pathogenic. Race and ethnicity are socio-cultural (not biological) in nature, so while they may be useful as a proxy for ancestral background in some cases, this approach is not recommended. In order for this assessment to be relevant, greater representation of diverse populations in allele frequency databases is essential.
There was notable disagreement and uncertainty among survey participants with regard to the clinical scenarios that would be most likely to motivate the use of REA in clinical decision making or discussions with patients. Most survey participants expressed that REA are each at least somewhat important for ordering genetic tests, interpreting results, and communicating with patients. However, participants do not agree about the factors that would contribute to the relevance of REA, which indicates that they may have less practical utility than commonly assumed. It may also suggest that the use of REA is highly contextual such that it becomes important for ordering tests, billing, and medical coverage in some cases and in others it can inform discussions about the limitations of testing in certain populations.
There is evidence to suggest that descriptions of patient race and ethnicity in electronic medical records (EMRs) are often inaccurate,18 which calls into question the utility of this information. Because race, ethnicity, and ancestry each convey and represent a distinct type of information, such as one’s physical or social environment, lived experiences (e.g., racism), cultural traditions (e.g., nutrition, lifestyle), and genomic background, it is not recommended to collapse these measures or use one as a proxy for another. In order to mitigate the continued use of REA in clinical genetics without rigorous scientific evaluation and standardization of this practice, we recommend that use of this information should be justified through formal inquiry on the basis of effectiveness and necessity. It may be that using REA is not appropriate in all clinical scenarios, or that each measure may have specific, unique utility for particular applications. If REA is necessary and effective for the delivery of health care in certain clinical settings, formal and consistent definitions must be developed through a deeply interdisciplinary and deliberative process, then widely disseminated and adopted across clinical laboratories. Future efforts to determine what information about patients can and should be required will need to combine stakeholder engagement, policy analysis, and research on the utility of these measures in specific clinical settings.
Interpreting Variants from Diverse Populations
Respondents reported high perceived utility of REA information in the context of variant interpretation. Nevertheless, there is little guidance regarding how such information should be used in clinical variant interpretation and curation. The ACMG/AMP Guidelines for variant interpretation suggest that absence of a variant from “population databases” is moderate evidence for the pathogenicity of a variant (PM2). However, most databases that clinicians and researchers use to verify the presence or absence of a variant are not representative of the global diversity in human genomic variation.14,19 Further complicating the matter, self-reported race or ethnicity is often used as a proxy for genetic ancestry (since the latter is rarely available), so determining which genomic background or ancestral population is most relevant for the assessment of allele frequencies and the validity of PM2 for a particular patient remains a challenge. Future discussions among interdisciplinary researchers could focus on solutions to this particular challenge as it has a direct impact on the reported pathogenicity of variants.
Study Limitations
Although the results of this survey provide important insights for understanding how REA are used in clinical genetics, there are limitations. Our survey participants were heavily skewed toward individuals who are involved in the U.S.-based research consortia ClinGen and CSER. Additionally, our method of dissemination and recruitment may have biased our sample to individuals who are already interested in the topic of diversity in genetics, and thus may think more deeply about the role of these attributes in both research and clinical care relative to non-responders. Nonresponse error is also possible (with < 5% response rate) such that certain types of individuals may have been less likely to respond to the survey, representing a limitation of the study. Both of these factors could lead to bias such that survey respondents have higher literacy and/or interest in ancestry, diversity, and genetics.
Due to overlap in organizational membership among survey non-responders (which could not be accounted for), the response rate is underestimated. We may have also underestimated the proportion of clinical professionals who make their own determination about a patient’s race or ethnicity, because those who report obtaining this information from a medical record were not asked how this information was placed in the medical record. Finally, while the lack of racial and ethnic diversity among study participants may appear to be a limitation, it reflects the demographics of the clinical genetics professions and is likely a representative sample of our target study population. It may be useful in future studies to actively recruit a more diverse sample of clinical genetics professionals, in order to shed light on how perspectives may differ.
Conclusions and Next Steps
The results of this survey establish a baseline understanding of how REA are perceived and utilized by clinical genetics professionals, and our results show that there is little standardization or consistency. As a result of considerable individual-level variability in beliefs about how REA should inform clinical genetics and a lack of generally accepted understanding or standards, there is ample opportunity for bias to influence the research and implementation of precision medicine. Therefore, we recommend further research and suggest that REA utilization in clinical genetics be standardized, evidence based, and justified, in order for the implementation of genomic medicine to be consistent, scientifically valid, and ethically responsible. Furthermore, increased recognition of the lack of standards may be achieved through greater visibility of these topics at large annual conferences and professional meetings.
The National Academy of Science, Engineering, and Medicine (NASEM) standards for the development of clinical practice guidelines recommend a systematic review of evidence from the literature about a given practice or protocol, and the subsequent assemblage of a multi-disciplinary group of experts to serve as a guideline-development committee.20 Since the appropriate use of REA is a complex issue touching on many areas of clinical genetics, we recommend that diverse stakeholders and representatives from patient populations, clinical laboratories, educational and research institutions, funding agencies, professional organizations, accreditation bodies, scientific journals, and multi-institutional research consortia collaborate to develop a set of standards and recommendations about the utility and use of REA. In addition to the NASEM, other professional organizations might consider participating in the development of these standards, such as the American College of Medical Genetics and Genomics (ACMG), which is in the process of re-evaluating its variant classification guidelines. Among other strategic collaborative goals of the clinical genetics community, the development of consistent definitions, reporting, and appropriate utilization of REA should be highly prioritized as essential to the validity and relevance of genomic medicine for patients of all backgrounds.
Consortia
Additional members of the ClinGen Ancestry and Diversity Working Group who are not listed as authors include Jonathan Berg, Vence Bonham, Christopher Gignoux, Eimear Kenney, Samuel Oh, Manuel Rivas, Sandra Soo-Jin Lee, and Timothy Thornton.
Declaration of Interests
G.W.H. is an employee of Concert Genetics and a member of the Board of Directors of My Gene Counsel. S.E.P. is a member of the Scientific Advisory Board of Baylor Genetics Laboratory. C.D.B. is President and Chairman of CBD Consulting LTD and a venture partner at F-Prime Capital.
Acknowledgments
Funding for this study was provided by the National Human Genome Research Institute (NHGRI) Clinical Genome Resource (ClinGen): 5U41HG009649-03; the Clinical Sequencing Evidence-generating Research (CSER) Coordinating Center: U24HG007307; the UCSF Program in Prenatal and Pediatric Genome Sequencing (P3EGS): U01HG009599; and a Chan-Zuckerberg Investigator Award (J.Y.Z.).
Individuals who contributed to the development of this study through working group discussions and/or participation in refinement of the survey instrument include Vence Bonham, Eimear Kenney, Manuel Rivas, Jonathan Berg, Tim Thornton, Laura Amendola, Kate Foreman, Gail Jarvik, Mary Norton, Sarah Scollon, Jacqueline Odgis, and other anonymous contributors.
Dissemination of the survey was supported by authors and the following individuals, on behalf of the affiliated organizations and consortia: Jeffrey Ou (CSER); Danielle Azzariti (ClinGen); Candice Miller (ABGC); Denise Calvert (ACMG); and Mona Miller (ASHG). Maria Cerezo (GWAS Catalog) and others contributed to targeted social media posts. Lauren Hicks and Catherine Gooch contributed a manually curated email list of publicly listed clinical geneticists.
The authors appreciate feedback on earlier versions of this paper by attendees to the Stanford Center for Biomedical Ethics writing seminar.
Published: June 2, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.05.005.
Web Resources
The Clinical Genome Resource (ClinGen), https://www.clinicalgenome.org
Clinical Sequencing Evidence-generating Research (CSER), https://cser-consortium.org
Qualtrics Software, https://www.qualtrics.com
U.S. Census Bureau, https://www.census.gov/topics/population/race/about.html
Supplemental Information
References
- 1.National Institutes of Health Racial and Ethnic Categories and Definitions for NIH Diversity Programs and for Other Reporting Purposes. 2015. https://grants.nih.gov/grants/guide/notice-files/not-od-15-089.html NOT-OD-15-089.
- 2.Kumar R., Seibold M.A., Aldrich M.C., Williams L.K., Reiner A.P., Colangelo L., Galanter J., Gignoux C., Hu D., Sen S. Genetic ancestry in lung-function predictions. N. Engl. J. Med. 2010;363:321–330. doi: 10.1056/NEJMoa0907897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Royal C.D., Novembre J., Fullerton S.M., Goldstein D.B., Long J.C., Bamshad M.J., Clark A.G. Inferring genetic ancestry: opportunities, challenges, and implications. Am. J. Hum. Genet. 2010;86:661–673. doi: 10.1016/j.ajhg.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jorde L.B., Bamshad M.J. Genetic Ancestry Testing: What is it and why is it important? JAMA. 2020 doi: 10.1001/jama.2020.0517. Published online February 14, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Popejoy A.B., Ritter D.I., Crooks K., Currey E., Fullerton S.M., Hindorff L.A., Koenig B., Ramos E.M., Sorokin E.P., Wand H., Clinical Genome Resource (ClinGen) Ancestry and Diversity Working Group (ADWG) The clinical imperative for inclusivity: Race, ethnicity, and ancestry (REA) in genomics. Hum. Mutat. 2018;39:1713–1720. doi: 10.1002/humu.23644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonham V.L., Sellers S.L., Gallagher T.H., Frank D., Odunlami A.O., Price E.G., Cooper L.A. Physicians’ attitudes toward race, genetics, and clinical medicine. Genet. Med. 2009;11:279–286. doi: 10.1097/GIM.0b013e318195aaf4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson S.C., Yu J.H., Wagner J.K., Harrell T.M., Royal C.D., Bamshad M.J. A content analysis of the views of genetics professionals on race, ancestry, and genetics. AJOB Empir. Bioeth. 2018;9:222–234. doi: 10.1080/23294515.2018.1544177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cunningham B.A., Bonham V.L., Sellers S.L., Yeh H.C., Cooper L.A. Physicians’ anxiety due to uncertainty and use of race in medical decision making. Med. Care. 2014;52:728–733. doi: 10.1097/MLR.0000000000000157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Race E., Race, Ethnicity, and Genetics Working Group The use of racial, ethnic, and ancestral categories in human genetics research. Am. J. Hum. Genet. 2005;77:519–532. doi: 10.1086/491747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smedley A., Smedley B. Fourth Edition. Westview Press; Boulder, CO: 2012. Race in North America: Origins and Evolution of a Worldview. [Google Scholar]
- 11.Omi M., Winant H. Third Edition. Routledge; London, UK: 2014. Racial Formation in the United States. [Google Scholar]
- 12.Roth W. The multiple dimensions of race. Ethn. Racial Stud. 2016;39:1310–1338. [Google Scholar]
- 13.Braun L., Fausto-Sterling A., Fullwiley D., Hammonds E.M., Nelson A., Quivers W., Reverby S.M., Shields A.E. Racial categories in medical practice: how useful are they? PLoS Med. 2007;4:e271. doi: 10.1371/journal.pmed.0040271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landry L.G., Ali N., Williams D.R., Rehm H.L., Bonham V.L. Lack of diversity in genomic databases is a barrier to translating precision medicine research into practice. Health Aff. (Millwood) 2018;37:780–785. doi: 10.1377/hlthaff.2017.1595. [DOI] [PubMed] [Google Scholar]
- 15.Mays V.M., Ponce N.A., Washington D.L., Cochran S.D. Classification of race and ethnicity: implications for public health. Annu. Rev. Public Health. 2003;24:83–110. doi: 10.1146/annurev.publhealth.24.100901.140927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonham V.L., Sellers S.L., Woolford S. Physicians’ knowledge, beliefs, and use of race and human genetic variation: new measures and insights. BMC Health Serv. Res. 2014;14:456. doi: 10.1186/1472-6963-14-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klinger E.V., Carlini S.V., Gonzalez I., Hubert S.S., Linder J.A., Rigotti N.A., Kontos E.Z., Park E.R., Marinacci L.X., Haas J.S. Accuracy of race, ethnicity, and language preference in an electronic health record. J. Gen. Intern. Med. 2015;30:719–723. doi: 10.1007/s11606-014-3102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Institute of Medicine of the National Academies Clinical Practice Guidelines We Can Trust: Standards for Developing Trustworthy Clinical Practice Guidelines (CPGs) 2011. https://www.nap.edu/resource/13058/Clinical-Practice-Guidelines-2011-Report-Brief.pdf
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.