Abstract
Background
Pharmaceutically important curcuminoid synthesis in C. longa is controlled by CURS1, CURS2, and CURS3 genes. The present study detected the physicochemical properties and structural characteristics including the secondary and 3D structure of CURS proteins. The primary, secondary, and tertiary structure of the CURS proteins were modeled and characterized using multiple bioinformatics tools such as ExPasy ProtParam tools, self-optimized prediction method with alignment (SOPMA), PSIPRED, and SWISS-MODEL. The predicted secondary structure of curcumin synthase provided an α-helix and random coil as the major components. The reliability of the modeled structure was confirmed using PROCHECK and QMEAN programs.
Results
The molecular weight of CURS1 is 21093.19 Da, theoretical pI as 4.93, and an aliphatic index of 99.19. Molecular weight of CURS2 and CURS3 proteins are 20266.13 Da and 20629.52 Da, theoretical pI as 5.28 and 4.96, and an aliphatic index of 89.30 and 86.37, respectively. In the predicted secondary structure of CURS proteins, alpha helices and random coils of CURS1, CUR2, and CURS3 were 42.72, 41.38, and 44.74% and 24.87, 31.03, and 17.89, respectively. The extended strands were 16.24, 19.40, and 17.89. QMEAN Z-score is − 0.83, − 0.89, and − 1.09 for CURS1, CURS2, and CURS3, respectively.
Conclusion
Prediction of the 3D model of a protein by in silico analysis is a highly challenging aspect to confirm the NMR or X-ray crystallographic data. This report can contribute to the understanding of the structure, physicochemical properties, structural motifs, and protein-protein interaction of CURS1, CUR2, and CURS3.
Keywords: CURS, Curcuminoids, In silico analysis, Bioinformatic tools, Homology modeling
Background
The major class of secondary metabolites from C. longa contains a mixture of curcumin (60–80%), demethoxycurcumin (15–30%), and bisdemethoxycurcumin (2–6%) [1], soluble in methanol, ethanol, or dimethyl sulfoxide and insoluble in water [2]. Curcuminoids have anti-inflammatory, antimutagenic, anti-diabetic, anti-bacterial, and hepatoprotective activities [3]. It is also known for its free-radical scavenging antioxidant activity [4], healing of the dermal wound [5], and prevention of Alzheimer’s disease [6]. Most importantly, curcumin inhibits the cell growth of various cancer cell lines and induces apoptosis in cancer cells [7] and also in the regulation of cancer cell growth [8].
Curcumin synthesis is mediated by curcumin synthase, (CURS), the gene family has three members; curcumin synthase 1 (CURS1, the first identified CURS) and type III polyketide synthases (PKSs), Viz. CURS2 and CURS3, having CURS-like activity with the substrate specificity slightly different from that of CURS1 [9] involved in curcumin synthesis pathway. Type III polyketide synthases (PKSs) consists of structurally simple homodimers of ketosynthase that are involved in the biosynthesis of most of the plant polyketides [10].
The elucidation of protein structure is one of the key features for understanding the biological processes at a molecular level. However, very little is known about the structure of CURS (CURS1, CURS2, and CURS3) proteins. Identification of the 3D structure of a protein is very difficult and complex. X-ray crystallography or NMR spectroscopy methods were used to determine the protein structure, but it is time-consuming and not successful with all proteins, particularly in membrane proteins [11]. A viable alternative approach developed to predict the in silico 3D structure of proteins based on homology modeling using an unknown protein sequence with more than 35% of similarity [12] serves the purpose with better validation.
The present study was aimed at modeling curcumin synthase genes of C. longa using in silico analysis including physicochemical properties of the designed secondary structure, modeling CURS protein 3-D structure, evaluation, and analysis of the modeled structures using different standard computational tools.
Methods
Plant material
Curcuma longa rhizomes were collected and identified using flora and conserved as field germplasm, a voucher specimen was submitted to the herbarium and a voucher number (6949) was provided by the curator of the herbarium. The rhizomes were harvested after 10 months of cultivation and used for extraction. Analytical grade chemicals purchased from Hi-Media Laboratories, Mumbai, India, were used for the extraction.
Cloning and annotation of putative CURS gene
Total RNA from C. longa rhizome was isolated using modified SDS method [13]. Purified RNA was converted into cDNA using Takara PrimeScriptTM RT reagent kit (Cat. # RR037A, Takara Bio Inc., Japan) according to the manufacturer’s instructions. The cDNA synthesis reaction mixture contained 5X Primescript buffer (2.0 μl), Primescript RT enzyme mix (0.5 μl), 0.5 μl Oligo dT primer (50 μM), 2.0 μl random hexamers (100 μM), template RNA (10 μl), and the final volume was adjusted to 20 μl by adding RNase-free water. The reaction conditions were reverse transcription at 37 °C for 15 min, inactivation of reverse transcription at 85 °C for 5 s, and hold at 4 °C. Primers for cloning the CURS gene were designed from the conserved regions of available C. amada, C. longa, and C. zedoaria CURS genes retrieved from GenBank (Accession Nos. CURS1—KM880189.1 C. longa CURS1, AB495007.1 C. longa CURS1 and MF402846.1 C. zedoaria CURS1; CURS2—KF980981.1 C. amada isolate CURS2-XI CURS2, KF980982.1 C. amada isolate CURS2-XII CURS2, LC064068.1 C. longa CURS2, AB506762.1 C. longa CURS2; and CURS3—KX154461.1 C. amada CURS3, AB506763.1 C. longa CURS3, KM880190.1 C. longa CURS3, and MF987835.1 C. zedoaria CURS3) using Multalin and Primer-BLAST. The primers designed were CURS1 (F:5′-ATGGTGAAGA AGCGGTACCTG-3′; R: 5′-TGTTGCCGTACTCTGTGAAGA-3′), CURS2 (F:5′-GCTAATC AGTCAATCCAGA TGG-3′; R: 5′- CGTCTATCGATTGATCGATC GT-3′), and CURS3 (F:5′-GTCAACCG CCTCATG CTCTACA-3′; R:5′-TCACCTCGTCCAT CACGAAGTAC-3′). PCR was carried out using 10× PCR buffer (2 μl), 25 mM MgCl2 (2 μl), 100 mM dNTPs, forward primer 1 μl, reverse primer1 μl, ~ 50 ng cDNA template, and .25 μl Taq DNA polymerase (5 U/μl) and the final volume was made up to 25 μl with sterile double distilled water. The reaction conditions were initial denaturation at 95 °C for 15 min and 35 cycles comprising: 95 °C for 20 s, gradient annealing temperature at (51.5, 52.1, 53.4, 54.0, 55.4 55.9, 58.6 and 59.6 °C) for 40 s, 72 °C for 1 min, and final extension at 72 °C for 10 min. Amplified PCR products were visualized on a 1% (w/v) agarose gel and molecular weight was detected using standard 1 kb DNA ladder. The PCR product was purified and sequenced. The obtained sequence was analyzed using BLAST (http://www.ncbi.nlm.nih.gov) program to find out the homology of the sequence and submitted in NCBI (MK515083, MG386668, and MK511334) translated to corresponding proteins. The Open Reading Frame (ORF) Finder program was used to determine the coding regions of the sequences and the sequences were annotated.
Physicochemical characteristics
The physical and chemical attributes, such as molecular weight, theoretical pI, amino acid composition, atomic composition, extinction coefficient, estimated half-life, instability index, aliphatic index, and grand average of hydropathy (GRAVY) of the CURS proteins, were computed using Expasy ProtParam tool [14].
Secondary structure prediction
The secondary structure properties like the α-helix, β-sheet, and turn of amino acid sequences of CURS proteins were predicted using PSI-blast-based secondary structure PREDiction (PSIPRED) [15] and self-optimized prediction method with alignment (SOPMA) [16].
Protein 3D model prediction
The derived CURS protein sequences were used as query sequences for comparative modeling. SWISS-MODEL (http://swissmodel.expasy.org) was used for the 3D structure prediction of CURS1, CURS2, and CURS3 and its integrated external resources, such as UniProt, InterPro, STRING, and Nature PSI SBKB were also used for analysi s[17].
Model evaluation
Different tools were used to evaluate the internal consistency and reliability of the modeled structure of the CURS1, CURS2, and CURS3. PROCHECK and MolProbity programs were used to assess the stereochemical quality of the model by quantifying the residues in the allowed zones of Ramachandran plot [18]. The obtained protein structure was re-assessed for its reliability and model quality using QMEAN Z-scores from QMEAN server http://swissmodel.expasy.org/docs/structure_assessment [19].
Results
Cloning and annotation of putative CURS gene
PCR-assisted cloning using the designed primers amplified the CURS1, 900 bp; CURS2, 1100 bp; and CURS3, 590 bp, genes and the homology determination provided similarity with the existing CURS genes from the genebank. The ORF finder demarcated a putative 588 bp, 675 bp, and 570 bp ORF for the three cloned CURS nucleotide sequences translated to CURS proteins with 195, 224, and 190 amino acid residues for CURS1, CURS2, and CURS3 with ATG as the initiation codon.
Physicochemical properties
Different physicochemical properties of the CURS proteins were examined using ExPASy ProtParam tool (Table 1). The molecular weight of CURS1 is 21093.19 Da, theoretical pI 4.93, and an aliphatic index of 99.19. The instability index was 32.10 and GRAVY was 0.199. Molecular weight of CURS2 and CURS3 proteins are 20266.13 Da and 20629.52 Da, theoretical pI as 5.28 and 4.96, and an aliphatic index of 89.30 and 86.37, respectively. The instability index was 37.84 and 31.33 and GRAVY was 0.118 and 0.058 for CURS2 and CURS3. Phosphorylation sites were predicted using NetPhos 2.0 server. The CURS1 protein has 3 Ser, 2 Thr, and 1 Tyr; CURS2 has 4 Ser, 5 Thr, and 2 Tyr; and CURS3 showed 4 Ser, 1 Thr, and 3 Tyr.
Table 1.
Physicochemical properties of CURS proteins
| S. no. | Name of the proteins | M. wt. (Da) | Seq. length | pI | EC (assuming all pairs of Cys residues form cystine) | EC (assuming all Cys residues are reduced) | Half-life (h) | II | GRAVY | −R | +R | AI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | CURS1 | 21093.19 | 197 | 4.93 | 28,085 | 27,960 | 30 | 32.10 | 0.199 | 22 | 14 | 99.19 |
| 2 | CURS2 | 20266.13 | 224 | 5.28 | 21,095 | 20,970 | 30 | 37.84 | 0.118 | 20 | 13 | 89.30 |
| 3 | CURS3 | 28629.52 | 190 | 4.95 | 28,085 | 27,960 | 30 | 31.33 | 0.058 | 23 | 14 | 86.37 |
M. wt. molecular weight, pI isoelectric point. −R, number of negative residues, +R number of positive residues, EC extinction coefficient at 280 nm, II instability index, AI aliphatic index, GRAVY grand average hydropathy
Secondary structure prediction
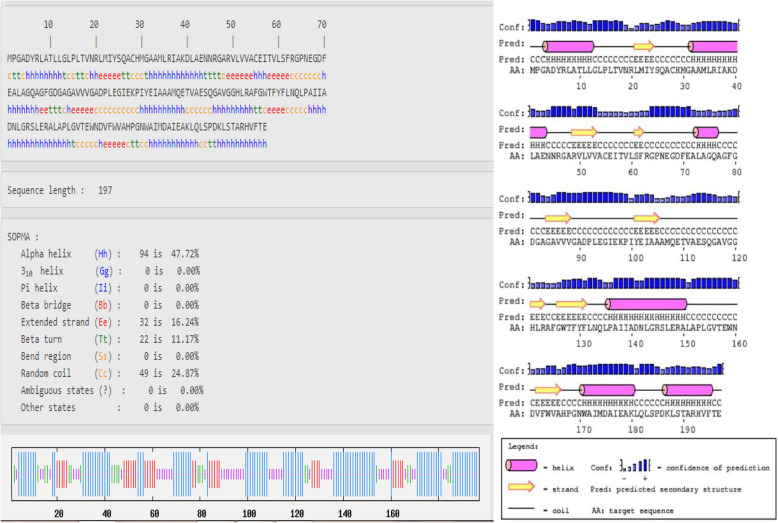
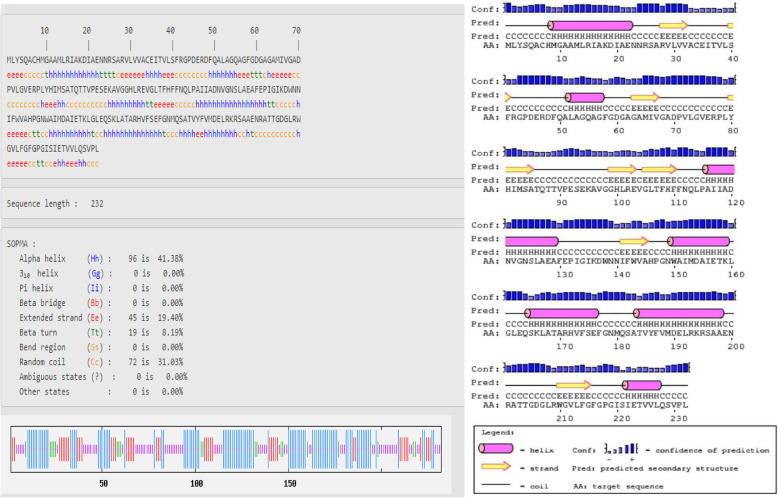
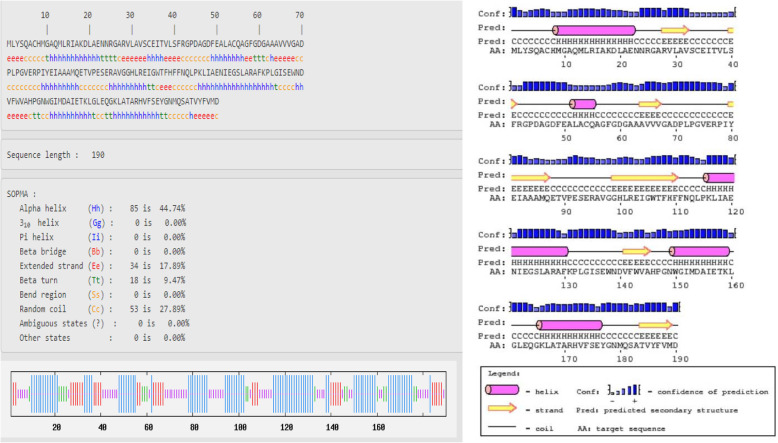
The secondary structure of protein chains was analyzed by SOPMA that predicted the alpha helix, extended strand, beta turn, and random coil (Figs. 1, 2, and 3). In the designed secondary structure of CURS proteins, alpha helices showed 42.72, 41.38, and 44.74% in CURS1, CUR2, and CURS3, respectively. It is followed by random coils 24.87, 31.03, 17.89 and extended strands 16.24, 19.40, 17.89 (Table 2). The CURS proteins revealed the predominant nature of helix and coiling underlining the more compact and strongly bonded and transmembrane position of the CURS protein.
Fig. 1.
Secondary structure analysis of C. longa CURS1
Fig. 2.
Secondary structure analysis of C. longa CURS2
Fig. 3.
Secondary structure analysis of C. longa CURS3
Table 2.
Prediction of secondary structure of CURS proteins by SOPMA
| Parameters | CURS1 | CURS2 | CURS3 |
|---|---|---|---|
| α-Helix | 47.72% | 41.38% | 44.74% |
| Extended β-strand | 16.24% | 19.40% | 17.89% |
| Random coil | 24.87% | 31.03% | 27.89% |
| Ambiguous state | 0.00% | 0.00% | 0.00% |
Model validation
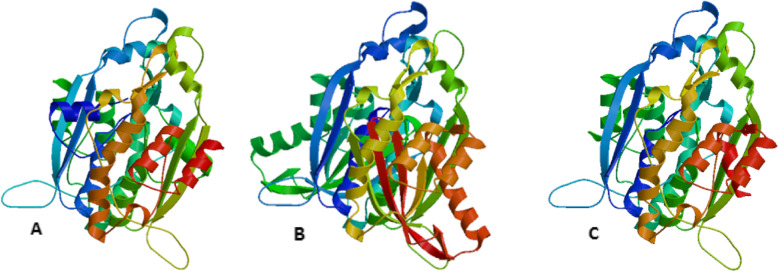
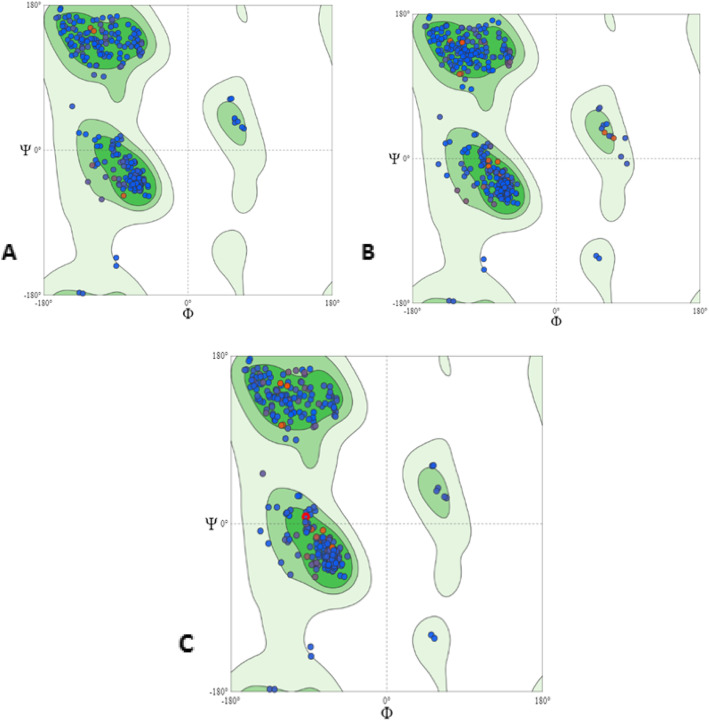
Homology modeling of the CURS proteins was done using the automated homology protein modeling server of SWISS-MODEL, based on ProMod3, an open structure comparative modeling engine (Fig. 4a–c). The CURS1, CURS2, and CURS3 protein models were verified using the Ramachandran plot from the MolProbity program and validated all the amino acid residues of the modeled protein fit in the allowed regions of the Ramachandran plot. The CURS1 protein showed 1.3% MolProbity score, 97.67% residues were in the favored residues, 0% in the outliers regions; and the Clash score was 0.68%. The MolProbity score of CURS2 was 1.6%, favored residues were 95.45%, outliers regions with 0.22%; and Clash Score was 1.85%. In the CURS3 protein, the MolProbity score was 1.33%, 96.01% of the amino acids were in the favored regions, 0% in the outliers regions, and 0.52% Clash Score (Fig. 5a–c). The modeled proteins were submitted to PMDB and accession numbers were provided (PM0082212, PM0082213, and PM0082214).
Fig. 4.
Model 3D structure of protein from Curcuma longaa CURS1, b CURS2, and c CURS3
Fig. 5.
The stereochemical validation of the hypothetical model using Ramachandran plot of a CURS1, b CURS2, and c CURS3 proteins
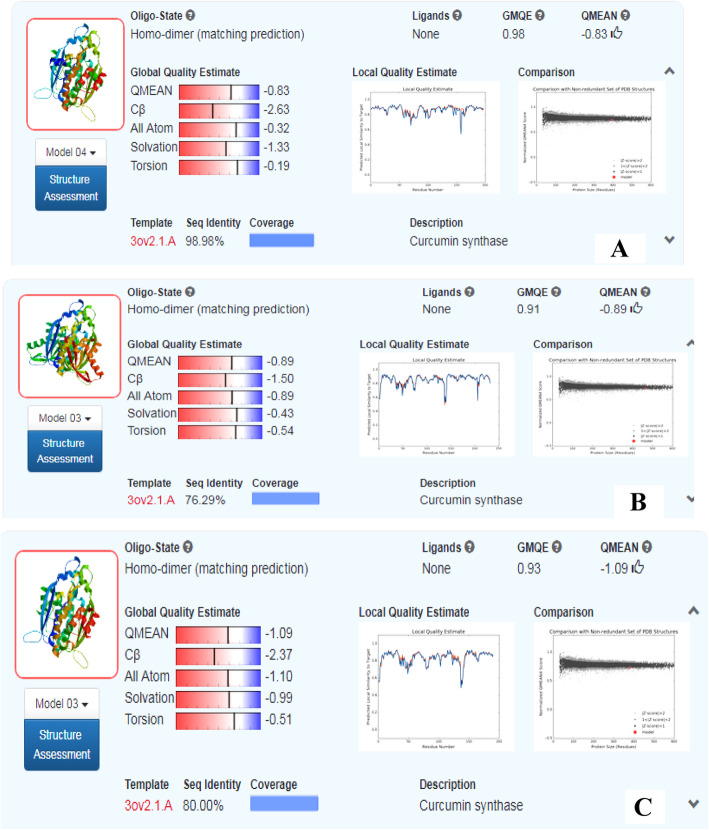
QMEAN Z-score was − 0.83, − 0.89, and − 1.09 for CURS1, CURS2, and CURS3, respectively. The individual Z-scores compared the interaction potential between Cβ atoms only. All atoms with the resolution potential and the torsion angle potential are shown in Fig. 6a–c. The “Local Quality” was estimated for each residue of the model (reported on the x-axis) and the expected similarity to the native structure (y-axis). Usually, residues showing a score below 0.6 are expected to be of low quality. In the “Comparison” plot (Fig. 6a–c), the model quality scores of individual models are related to scores obtained for experimental structures of similar size.
Fig. 6.
Quality estimation (GMQE, QMEAN, local quality estimate, and comparison plot) of a CURS1, b CURS2, and c CURS3 proteins
The QMEAN Z-score provided an estimate of the “degree of nativeness” of the structural features observed in the model on a global scale. It indicates whether the QMEAN score of the model is comparable to the expected score from experimental structures of similar size. QMEAN Z-score value of approximately zero specifies superior quality between the modeled structure and experimental structures. The obtained scores of − 4.0 or below indicate that the models with low quality. The QMEAN Z-scores of the CURS1, CURS2, and CURS3 proteins showed − 0.83, − 0.89, and − 1.09, respectively, and these results indicate that the proposed homology model is reliable and acceptable.
Post-translational modifications
The process of post-translational modification mainly includes phosphorylation, glycosylation, ubiquitination, nitrosylation, methylation, acetylation, lipidation, and proteolysis. The CURS1 protein has 3 Ser, 2 Thr, and 1Tyr residues. S-146 has a score of 0.989 indicates its candidacy for a phosphorylation site than the other. CURS2 has 4 Ser, 5 Thr, and 2Tyr and S-196 has a score of 0.994, and CURS3 has 4 Ser, 1 Thr, and 3Tyr and S-33 with an overall score of 0.959.
Discussion
The cloned putative sequences of CURS1, CURS2, and CURS3 showed better homology with the database CURS sequences and the ORF determination specified the protein characteristics. The aliphatic index of the protein is defined as the relative volume occupied by aliphatic side chains, which include alanine, valine, isoleucine, and leucine, and contribute to protein thermostability [20]. The predicted aliphatic index of CURS1 protein was 99.19%; CURS2, 89.30%; and CURS3, 86.37%. The isoelectric point is the condition where the amino acid maintains the same level of positive and negative charges and the net charge will be zero. Isoelectric points (pI) of CURS1, CURS2, and CURS3 were 4.93, 5.28, and 4.96 suggesting a moderately acidic nature of the protein. Approximately neutral pH is required in in vivo condition compared to in vitro for the optimum activity of the alkaline phosphatase enzyme [21]. The total number of positively charged and negatively charged residues refers to the total no. of lysine (K), arginine (R) and aspartate (D), and glutamate (E), respectively [22]. The instability indices were between 32.10, 37.84, and 31.33. The obtained instability indices for CURS1, CURS2, and CURS3 were lesser than 40, suggesting the stability of the proteins [23, 24]. GRAVY is used for the computational analysis of various physicochemical parameters for a given amino acid sequence [25]. Low range GRAVY value of 0.199, 1.118, and 0.058 indicates its high affinity for water that improves the solubility of a protein [25, 26].
Alpha helical structure is composed of methionine (M), alanine (A), leucine (L), glutamate (E), and lysine (K) amino acids, whereas the beta strand is composed of tryptophan (W), tyrosine (Y), phenylalanine (F), valine (V), isoleucine (I), and threonine (T); furthermore, glycine (G) and proline (P) amino acids help to build the relevant turns [23]. Such findings suggest that the numbers of amino acids are solely responsible for constructing the respective secondary structure of proteins. The percentage score of amino acid distribution infers that alpha helix is dominated over other secondary structures followed by the random coil, extended strand, and beta turn; Figs. 1, 2, and 3 represent the secondary structure of CURS proteins where the alpha helix is maximum than other structures.
The QMEAN quality estimations are based on different geometrical properties and provide both global (i.e., for the entire structure) and local (i.e., per residue) absolute quality estimates on the basis of one single model and its scoring function consists of a linear combination of six structural descriptors [27, 28]. The CASP experiment showed the optimization of weightage factors for the terms contributing to QMEAN has been performed on models from the seventh round of the (CASP7) [29]. QMEAN Z-scores are applied for the experimental structures from the PDB database [30]. The CURS proteins showed the highest phosphorylation sites, higher scores reflect the confidence of the prediction and similarity to one or more of the phosphorylation sites used in the method [31, 32]. Phosphorylation regulates innate inflammatory responses through the activation, cellular translation, and interaction of innate receptors, adaptors, and downstream signaling of molecules in response to infectious and dangerous signals [33].
Conclusion
In the present study, bioinformatics tools were used to model the CURS (CURS1, CURS2, and CURS3) proteins of Curcuma longa. Multiple sequence alignment with CURS proteins had higher homologies with other CURS proteins. Primary structure analysis revealed that CURS proteins are acidic in nature and stable. The secondary structure analysis confirmed that in all three CURS proteins, the alpha helix dominated followed by random coil, extended strand, and beta turns. Tertiary structure predictions were analyzed by Swiss-model and the models were validated using PROCHECK’S Ramachandran plot. The models were validated and submitted in the PMDB server. Prediction of the 3D model of a protein by in silico analysis is a highly challenging aspect to confirm the data obtained from the NMR or X-ray crystallographic-based methods. Consequently, in silico analysis of protein structure is one of the very useful methods for studying the structural and functional aspects of the protein. Our results indicate that future studies with the quaternary structure of CURS proteins will provide a better insight into the exact or most probable molecular mechanisms involved in curcumin synthase. This report can throw light into the protein structure, physicochemical properties, structural motifs, and protein-protein interactions.
Acknowledgements
The authors acknowledge the facilities provided by the Director, Interuniversity Centre for Plant Biotechnology, University of Calicut, for providing facilities.
Abbreviations
- cDNA
Complementary deoxyribonucleic acid
- NCBI
National Centre for Biotechnology Information
- NMR
Nuclear magnetic resonance
- ORF
Open Reading Frame
- PSIPRED
Position-specific iterative blast-based secondary structure prediction
- RNA
Ribonucleic acid
- SOPMA
Self-optimized prediction method with alignment
Authors’ contributions
Both authors contributed equally to the manuscript. YA conceived the idea and corrected the manuscript; SKR executed the work and wrote the preliminary manuscript. Both authors have read the manuscript and approved it for publication in the Journal of Genetic Engineering and Biotechnology.
Authors’ information
Studies involving plants must include a statement specifying the local, national, or international guidelines and legislation and the required or appropriate permissions and/or licenses for the study: The plant material is submitted in the herbaria of the University of Calicut herbarium (CALI) and provided voucher number by the curator of the herbarium.
Funding
SKR received funding in the form of Rajiv Gandhi Fellowship, Govt. of India, as a research fellowship.
Availability of data and materials
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Ethics approval and consent to participate
No animal models were used for the study.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wichitnithad W, Jongaroonngamsang N, Pummangura S, Rojsitthisak P. A simple isocratic HPLC method for the simultaneous determination of curcuminoids in commercial turmeric extracts. Phytochem Anal. 2009;20(4):314–319. doi: 10.1002/pca.1129. [DOI] [PubMed] [Google Scholar]
- 2.Tønnesen HH, Karlsen J. Studies on curcumin and curcuminoids. Z Lebensm Unters Forsch. 1985;180(5):402–404. doi: 10.1007/BF01027775. [DOI] [PubMed] [Google Scholar]
- 3.Krup V, Prakash LH, Harini A. Pharmacological activities of turmeric (Curcuma longa Linn): a review. J Homeop Ayurv Med. 2013;2(133):2167–1206. [Google Scholar]
- 4.Kalpravidh RW, Siritanaratkul N, Insain P, Charoensakdi R, Panichkul N, Hatairaktham S, Srichairatanakool S, Phisalaphong C, Rachmilewitz E, Fucharoen S. Improvement in oxidative stress and antioxidant parameters in β-thalassemia/Hb E patients treated with curcuminoids. Clin Biochem. 2010;43(4-5):424–429. doi: 10.1016/j.clinbiochem.2009.10.057. [DOI] [PubMed] [Google Scholar]
- 5.Gopinath D, Ahmed MR, Gomathi K, Chitra K, Sehgal PK, Jayakumar R. Dermal wound healing processes with curcumin incorporated collagen films. Biomaterials. 2004;25(10):1911–1917. doi: 10.1016/S0142-9612(03)00625-2. [DOI] [PubMed] [Google Scholar]
- 6.Shen L, Ji HF. The pharmacology of curcumin: is it the degradation products? Trends Mol Med. 2012;18(3):138–144. doi: 10.1016/j.molmed.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 7.Weir NM, Selvendiran K, Kutala VK, Tong L, Vishwanath S, Rajaram M, Tridandapani S, Anant S, Kuppusamy P. Curcumin induces G2/M arrest and apoptosis in cisplatin-resistant human ovarian cancer cells by modulating Akt and p38 MAPK. Cancer Biol Ther. 2007;6(2):178–184. doi: 10.4161/cbt.6.2.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu E, Wu J, Cao W, Zhang J, Liu W, Jiang X, Zhang X. Curcumin induces G2/M cell cycle arrest in a p53-dependent manner and upregulates ING4 expression in human glioma. J Neurooncol. 2007;85(3):263–270. doi: 10.1007/s11060-007-9421-4. [DOI] [PubMed] [Google Scholar]
- 9.Katsuyama Y, Kita T, Funa N, Horinouchi S. Curcuminoid biosynthesis by two type III polyketide synthases in the herb Curcuma longa. J Biol Chem. 2009;284(17):11160–11170. doi: 10.1074/jbc.M900070200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Austin MB, Noel JP. The chalcone synthase superfamily of type III polyketide synthases. Nat Prod Rep. 2003;20(1):79–110. doi: 10.1039/b100917f. [DOI] [PubMed] [Google Scholar]
- 11.Johnson MS, Srinivasan N, Sowdhamini R, Blundell TL. Knowledge-based protein modeling. Crit Rev Biochem Mol Biol. 1994;29(1):1–68. doi: 10.3109/10409239409086797. [DOI] [PubMed] [Google Scholar]
- 12.Fiser A. Template-based protein structure modeling. Methods Mol Biol. 2010;673:73–94. doi: 10.1007/978-1-60761-842-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deepa K, Sheeja TE, Santhi R, Sasikumar B, Cyriac A, Deepesh PV, Prasath D. A simple and efficient protocol for isolation of high quality functional RNA from different tissues of turmeric (Curcuma longa L.) Physiol Mol Biol Plants. 2014;20:263–271. doi: 10.1007/s12298-013-0218-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.ProtParam tool. Available from: http://web.expasy.org/protparam/.
- 15.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16(4):404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 16.Geourjon C, Deleage G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics. 1995;11(6):681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- 17.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2000;31(13):3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 20.Ikai A. Thermostability and aliphatic index of globular proteins. J Biochem. 1980;88(6):1895–1898. doi: 10.1093/oxfordjournals.jbchem.a133168. [DOI] [PubMed] [Google Scholar]
- 21.Aminfar Z, Tohidfar M. In silico analysis of squalene synthase in Fabaceae family using bioinformatics tools. J Genet Eng Biotechnol. 2018;16(2):739–747. doi: 10.1016/j.jgeb.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filiz E, Koç İ. In silico sequence analysis and homology modeling of predicted beta-amylase 7-like protein in Brachypodium distachyon. L. J.BioSci Biotechnol. 2014;3(1):61–67. [Google Scholar]
- 23.Lee MH, Jeong JH, Seo JW, Shin CG, Kim YS, In JG, Yang DC, Yi JS, Choi YE. Enhanced triterpene and phytosterol biosynthesis in Panax ginseng overexpressing squalene synthase gene. Plant Cell Physiol. 2004;45(8):976–984. doi: 10.1093/pcp/pch126. [DOI] [PubMed] [Google Scholar]
- 24.Kim YS, Cho JH, Park S, Han JY, Back K, Choi YE. Gene regulation patterns in triterpene biosynthetic pathway driven by overexpression of squalene synthase and methyl jasmonate elicitation in Bupleurum falcatum. Planta. 2011;233(2):343–355. doi: 10.1007/s00425-010-1292-9. [DOI] [PubMed] [Google Scholar]
- 25.Pramanik K, Ghosh PK, Ray S, Sarkar A, Mitra S, Maiti TK. An in silico structural, functional and phylogenetic analysis with three dimensional protein modeling of alkaline phosphatase enzyme of Pseudomonas aeruginosa. J Genet Eng Biotechnol. 2017;15(2):527–537. doi: 10.1016/j.jgeb.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verma A, Singh VK, Gaur S. Computational based functional analysis of Bacillus phytases. Comput Biol Chem. 2016;60:53–58. doi: 10.1016/j.compbiolchem.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 27.Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2010;27(3):343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benkert P, Künzli M, Schwede T. QMEAN server for protein model quality estimation. Nucleic Acids Res. 2009;37(suppl_2):W510–W514. doi: 10.1093/nar/gkp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moult J, Fidelis K, Kryshtafovych A, Rost B, Hubbard T, Tramontano A. Critical assessment of methods of protein structure prediction—Round VII. Proteins. 2007;69(S8):3–9. doi: 10.1002/prot.21767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294(5):1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 32.Ahmad S, Hewett PW, Wang P, Al-Ani B, Cudmore M, Fujisawa T, Haigh JJ, Le Noble F, Wang L, Mukhopadhyay D, Ahmed A. Direct evidence for endothelial vascular endothelial growth factor receptor-1 function in nitric oxide–mediated angiogenesis. Circ Res. 2006;99:715–722. doi: 10.1161/01.RES.0000243989.46006.b9. [DOI] [PubMed] [Google Scholar]
- 33.Liu H, Carvalhais LC, Kazan K, Schenk PM (2016) Development of marker genes for jasmonic acid signaling in shoots and roots of wheat. Plant Signal Behav 11(5). 10.1080/15592324.2016.1176654 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.