Abstract
Zygotic cleavage failure (ZCF) is a unique early embryonic phenotype resulting in female infertility and recurrent failure of in vitro fertilization (IVF) and/or intracytoplasmic sperm injection (ICSI). With this phenotype, morphologically normal oocytes can be retrieved and successfully fertilized, but they fail to undergo cleavage. Until now, whether this phenotype has a Mendelian inheritance pattern and which underlying genetic factors play a role in its development remained to be elucidated. B cell translocation gene 4 (BTG4) is a key adaptor of the CCR4-NOT deadenylase complex, which is involved in maternal mRNA decay in mice, but no human diseases caused by mutations in BTG4 have previously been reported. Here, we identified four homozygous mutations in BTG4 (GenBank: NM_017589.4) that are responsible for the phenotype of ZCF, and we found they followed a recessive inheritance pattern. Three of them—c.73C>T (p.Gln25Ter), c.1A>G (p.?), and c.475_478del (p.Ile159LeufsTer15)—resulted in complete loss of full-length BTG4 protein. For c.166G>A (p.Ala56Thr), although the protein level and distribution of mutant BTG4 was not altered in zygotes from affected individuals or in HeLa cells, the interaction between BTG4 and CNOT7 was abolished. In vivo studies further demonstrated that the process of maternal mRNA decay was disrupted in the zygotes of the affected individuals, which provides a mechanistic explanation for the phenotype of ZCF. Thus, we provide evidence that ZCF is a Mendelian phenotype resulting from mutations in BTG4. These findings contribute to our understanding of the role of BTG4 in human early embryonic development and provide a genetic marker for female infertility.
Keywords: BTG4, mutation, zygotic cleavage failure, female infertility
Introduction
Female infertility is a world-wide health problem that affects an estimated 48 million women.1 With the help of in vitro fertilization (IVF) and intracytoplasmic sperm injection (ICSI) techniques, many infertile couples have successfully given birth to their own children. However, a number of women cannot establish pregnancy because of recurrent IVF and/or ICSI failure due to oocyte maturation arrest or fertilization failure.2, 3, 4 Several genes have been found to be responsible for oocyte maturation arrest (TUBB8 [MIM: 616768] and PATL2 [MIM: 614661])5,6 and fertilization failure (TLE6 [MIM: 612399] and WEE2 [MIM: 614084]),7,8 thus demonstrating the critical role of genetic factors in these diseases. For human reproduction, an oocyte and a sperm will first form a zygote after fertilization and then undergo “zygotic cleavage” to begin embryonic development. For some individuals, morphologically normal oocytes can be retrieved and can be fertilized to form pronuclei. However, most of these zygotes cannot complete the first cleavage and finally arrest at the one-cell stage, thus showing the phenotype of zygotic cleavage failure (ZCF). Until now, there has been no convincing evidence to show the causative gene responsible for this phenotype.
During oocyte growth, numerous maternal mRNAs are synthesized and stored. After meiotic resumption, protein translation is activated to support oocyte maturation.9 Upon fertilization, male and female pronuclei develop in the zygote, then the zygote undergoes a series of cell cycle regulatory processes and the first cleavage occurs.10 During this period, maternal mRNAs are dramatically decayed to facilitate maternal-zygotic transition (MZT).9 The first cleavage of the zygote, also known as zygotic cleavage, relies on several key proteins that are translated during oocyte maturation. For example, polo-like kinase 1 (PLK1) and heat shock protein 90 beta family member 1 (HSP90B1) are associated with spindle assembly and chromosome arrangement during the prometaphase stage of the first cleavage in mouse zygotes.11,12 RAD9 checkpoint clamp component A (RAD9A) plays an important role in zygotic chromatin reprogramming and is also essential for the first cleavage event in mice,13 and tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein epsilon (YWHAE) is required for mitotic entry because it regulates maturation-promoting factor in fertilized mouse eggs.14 In 2016, we showed that B cell translocation gene 4 (BTG4) is a meiotic-cell-cycle-coupled MZT-licensing factor in mouse oocytes.15 BTG4 bridges CCR4-NOT transcription complex subunit 7 (CNOT7) to eukaryotic translation initiation factor 4E (EIF4E) and facilitates the decay of maternal mRNAs, and Btg4 null female mice produce morphologically normal oocytes that, due to the accumulation of maternal mRNAs, arrest at the one-cell stage after fertilization.15 Despite the pivotal role of Btg4 and other maternal genes in the zygotic cleavage in mice, none of them have been associated with the phenotype of ZCF in humans.
In the present study, we used whole-exome sequencing (WES) to identify four homozygous pathogenic variants in BTG4 (MIM: 605673; GenBank: NM_017589.4) that are responsible for human ZCF in four independent families. All affected individuals had homozygous frameshift, nonsense, or missense loss-of-function variants that followed a Mendelian recessive inheritance pattern. Our single zygote transcriptome analysis showed that ZCF in these affected individuals could be attributed to the impaired decay of maternal mRNAs. Our findings thus show that ZCF is a Mendelian phenotype and establish the causal relationship between BTG4 and the phenotype, and this might provide a diagnostic genetic marker for these affected individuals and a target for future therapeutic strategies.
Material and Methods
Clinical Samples
We recruited infertile individuals diagnosed with ZCF and controls from the Reproductive and Genetic Hospital of CITIC-Xiangya, the Reproductive Hospital of Guangxi Zhuang Autonomous Region, the Shanghai Ji Ai Genetics and IVF Institute affiliated with the Obstetrics and Gynecology Hospital of Fudan University, and the Shaanxi Maternal and Child Care Service Center. All blood samples were donated for the investigation after informed consent was obtained. Studies of human subjects were approved by the Fudan University Medicine Institutional Review Board and the Ethics Committee of the Reproductive and Genetic Hospital of CITIC-Xiangya.
Whole-exome Sequencing and Variant Analysis
The blood samples from the affected individuals were subjected to WES. Functional prediction was assessed using SIFT and the MutationTaster program. Variants were filtered using the following criteria:16 (1) variants with minor allele frequencies less than 1% in the gnomAD, 1000 Genomes Project, and ExAC databases; (2) exonic nonsynonymous or splice-site variants or coding indels; (3) homozygous or compound-heterozygous mutations in the proband and homozygous variants were considered as priority in consanguineous families; (4) mRNAs that were highly expressed or specifically expressed in oocytes.
Sanger Sequencing
Specific primers flanking the variants in the BTG4 gene were used for amplification via polymerase chain reaction (PCR) with an ABI 3100 DNA analyzer (Applied Biosystems).
Molecular Modeling and Evolutionary Conservation Analysis
The three-dimensional structure of wild-type BTG4 (NP_060059.1) was predicted using the SWISS-MODEL web server (PDB ID:5CI8). Molecular graphics and analysis were performed with PyMol software. Evolutionary conservation analysis was performed with MultiAlin software.
Immunofluorescent Staining
Oocytes and zygotes were first treated with acidic Tyrode’s solution to remove the zona pellucida. After thorough washing, the oocytes were fixed with 4% paraformaldehyde (Sangon Biotech, Cat#E672002) for 20 min at room temperature, washed three times in PBS supplemented with 0.1% bovine serum albumin (BSA) (Sigma, Cat#B2064), and then subjected to membrane permeabilization with 1% Triton X-100 (Sigma, Cat#T8787) for 30 min. The oocytes were washed again, blocked in a solution containing 5% donkey serum albumin (Jackson ImmunoResearch, Cat#017-000-121) and 2% BSA in PBS for 1 h at room temperature, and then incubated with rabbit anti-BTG4 (1:100 dilution; Abcam, Cat#ab235085) at 4°C overnight. After being washed three times, the oocytes were incubated with Alexa Fluor 555 donkey anti-rabbit IgG (1:1,000 dilution, Thermo Fisher Scientific, Cat#A31572) and 4′,6-diamidino-2-phenylindole (DAPI; 1:100 dilution, Thermo Fisher Scientific, Cat#62247) for 1 h at room temperature. Confocal images were obtained through the use of an Olympus microscope.
Construction of Expression Plasmids and Transfection
Human or mouse BTG4 and CNOT7 (MIM: 604913) cDNA were ligated into eukaryote expression vectors, and site-directed mutagenesis using PrimerSTAR Max DNA Polymerase (Takara, Cat#R045A) was performed for the generation of the BTG4 mutants. HeLa cells obtained from the American Type Culture Collection (ATCC) were grown at 37°C in 5% CO2 to 70%–80% confluence in DMEM medium (GIBCO, Cat#11965092) supplemented with 10% fetal bovine serum (Hyclone, Cat#SH30070). Transient transfections were performed with Lipofectamine 2000 (Thermo Fisher Scientific, Cat#11668019) in accordance with the manufacturer’s protocol. The compound was added directly to the well with the growing cells, and the transiently transfected cells were harvested at 48 h for immunoblot analysis.
Immunoblots and Immunoprecipitation
Transfected cells were lysed in protein loading buffer and heated at 95°C for 10 min. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblots were performed following standard procedures using a Mini-PROTEAN Tetra Cell System (Bio-Rad). For immunoprecipitation, the protocol was as previously described.17 HeLa cells were harvested in the lysis buffer, and after centrifugation, the supernatant was subjected to immunoprecipitation with affinity beads (Sigma). After incubation at 4°C for 4 h, the beads were washed with lysis buffer three times. The bead-bound proteins were eluted using sodium dodecyl sulfate (SDS) sample buffer for immunoblot analysis.
Single-cell RNA-seq Library Preparation, Sequencing, and RNA Analysis
RNA-seq libraries were prepared using the SMARTSeq2 protocol as we described previously,18 and these were then sequenced on a BGISEQ500 platform (BGI-Shenzhen, China) in the 100 bp paired-end mode. Sequencing data were quality controlled and filtered using SOAPnuke (v1.5.2), and we mapped the sequences to the human reference genome of GRCh38 (without masking repeats) and analyzed the protein-coding genes using the RSEMs (v1.2.12) software. The expression levels of the genes were quantified using normalized fragments per kilobase million (FPKM).
Quantitative Real-time Polymerase Chain Reaction (qRT-PCR)
qRT-PCR analysis was performed using the Power SYBR Green PCR Master Mix (Roche, Cat#42352720) and an Applied Biosystems 7500 Real-Time PCR System. Relative mRNA levels were calculated by normalizing to the levels of endogenous GAPDH mRNA. The relative transcript levels of BTG4 mutant zygotes were regarded as 1, and the fold changes in other mutant zygotes were compared to this. Primer sequences are listed in Table S1.
Results
Clinical Characteristics of the Affected Individuals
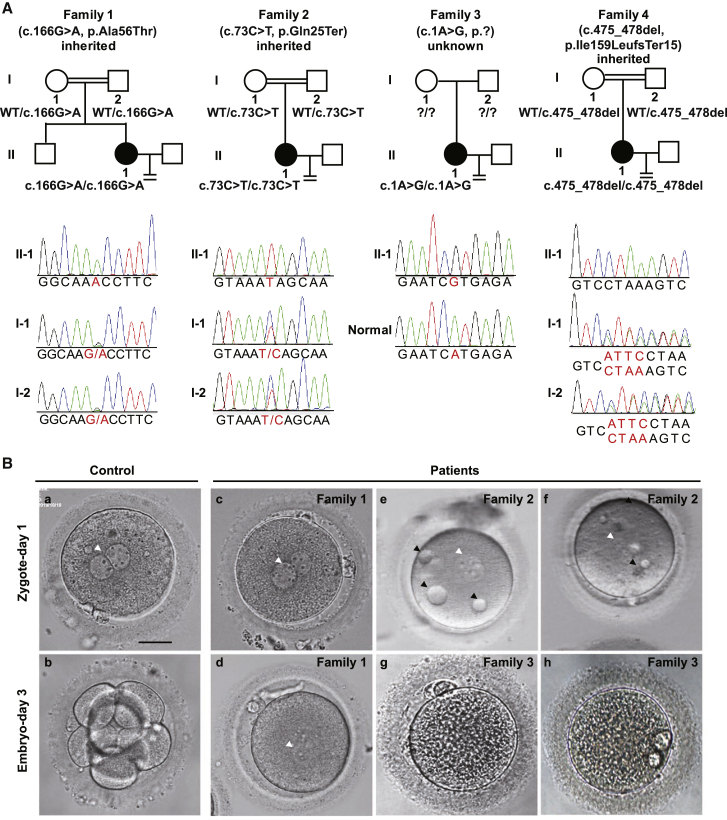
We recruited four independent female affected individuals with infertility of unknown cause. These affected individuals had normal menstrual cycles and sex hormone levels. Three of them (families 1, 2, and 4) were from consanguineous families (Figure 1A), and all of them had undergone two or three failed IVF and/or ICSI attempts each (Table 1). Affected individual II-1 in family 1 had undergone two different controlled ovarian hyperstimulation protocols in separate IVF and ICSI attempts, with similar outcomes. Altogether, 12 firstt polar body (PB1) oocytes were retrieved, and 11 could be fertilized, but all of the embryos were arrested before the first cleavage even after another 2 days of cultivation (Figure 1B and Table 1). The affected individual in family 2 had undergone one failed IVF attempt and two failed ICSI attempts. Overall, 11 PB1 oocytes were retrieved, of which 10 two-pronuclear (2PN) zygotes were formed, and with some, small vacuoles were distributed throughout the cytoplasm (Figure 1B). The images of these zygotes were not recorded on day 3, but according to the description of the clinician, none of them were cleaved (Table 1). The proband in family 3 had undergone two IVF attempts, and 18 PB1 oocytes were retrieved, half of which could be fertilized, but none could undergo cleavage (Figure 1B and Table 1). The proband in family 4 had undergone two IVF attempts in which 17 PB1 oocytes were retrieved, and 12 could be fertilized, but none could undergo cleavage (Table 1).
Figure 1.
Pedigree-based Identification of the BTG4 Pathogenic Variants and the ZCF Phenotype
(A) Genetic analysis in four families affected by ZCF. Black circles indicate the affected individuals. Sanger sequencing confirmation is shown below the pedigrees. The variants c.166G>A (p.Ala56Thr) (family 1), c.73C>T (p.Gln25Ter) (family 2), and c.475_478del (p.Ile159LeufsTer15) (family 4) in BTG4 were inherited from the parents of the affected individuals, but the inheritance pattern of family 3 was unknown due to lack of information about the parents (indicated by question marks).
(B) The morphology of a control zygote and the affected individuals’ zygotes on day 1 and/or day 3. The white arrowheads indicate the pronuclei, and the black arrowheads indicate the small vacuoles. Scale bar = 20 μm.
Table 1.
Oocyte and Embryo Characteristics of IVF and ICSI Attempts for the Affected Individual
| Individual | Age (Years) | Duration of Infertility (Years) | IVF and ICSI Attempts | Retrieved Oocytes | Immature Oocytes | PB1 Oocytes | Abnormally Fertilized Oocytes | Normally Fertilized Oocytes | Cleaved Fertilized Oocytes |
|---|---|---|---|---|---|---|---|---|---|
| II-1 in Family 1 | 25 | 4 | first IVF | 6 | 1 | 5 | 1 | 4 | 0 |
| II-1 in Family 1 | 25 | 4 | second ICSI | 7 | 0 | 7 | 0 | 7 | 0 |
| II-1 in Family 2 | 33 | 1 | first IVF | 4 | 0 | 4 | 1 | 3 | 0 |
| II-1 in Family 2 | 33 | 1 | second ICSI | 5 | 2 | 3 | 0 | 3 | 0 |
| II-1 in Family 2 | 33 | 1 | third ICSI | 7 | 3 | 4 | 0 | 4 | 0 |
| II-1 in Family 3 | 29 | 8 | first IVF | 15 | 3 | 12 | 5 | 7 | 0 |
| II-1 in Family 3 | 29 | 8 | second IVF | 9 | 3 | 6 | 4 | 2 | 0 |
| II-1 in Family 4 | 29 | 7 | first IVF | 14 | 1 | 13 | 4 | 9 | 0 |
| II-1 in Family 4 | 29 | 7 | second IVF | 6 | 2 | 4 | 1 | 3 | 0 |
IVF— in vitro fertilization; ICSI—intracytoplasmic sperm injection (ICSI); PB1—first polar body.
Identification of BTG4 Pathogenic Variants
WES was used for the identification of the pathogenic variants. Initially, we focused on two consanguineous families (families 1 and 2), and homozygous variants were considered a priority. After we filtered by frequency (<1%), expression level in oocytes, and corresponding function, BTG4 (GenBank: NM_017589.4) was the most likely candidate gene for the disease. Subsequently, we identified pathogenic homozygous variants in BTG4 in another two independent families (families 3 and 4). No homozygous or compound heterozygous variants in BTG4 were found in our control database, and this further confirmed the genetic contribution of BTG4 to human ZCF. The affected individual in family 1 had a homozygous missense variant, c.166G>A (p.Ala56Thr), and the affected individual in family 2 had a homozygous nonsense variant, c.73C>T (p.Gln25Ter) (Figure 1A). The affected individual in family 3 had a homozygous start-lost variant, c.1A>G (p.?), and the affected individual in family 4 had a homozygous frameshift variant, c.475_478del (p.Ile159LeufsTer15) (Figure 1A). All variants were confirmed using Sanger sequencing. The inheritance pattern in family 3 was unknown due to the unavailability of DNA samples from her parents, whereas the other three families followed a recessive inheritance pattern (Figure 1A). The variants c.166G>A (p.Ala56Thr), c.73C>T (p.Gln25Ter), and c.475_478del (p.Ile159LeufsTer15) have not been reported in any public databases, and variant c.1A>G (p.?) has a low frequency of 0.0000123 in the gnomAD exome database (Table 2). The variants c.166G>A (p.Ala56Thr), c.73C>T (p.Gln25Ter), and c.1A>G (p.?) were located in the BTG domain, and c.166G>A (p.Ala56Thr) was located in the Box A motif of the BTG domain, whereas the variant c.475_478del (p.Ile159LeufsTer15) was located in the C-terminal domain (Figure 2A).
Table 2.
Overview of the BTG4 Mutations Observed in the Families
| Probands in Families | Genomic Position on chr11 (bp) | cDNA Change | Protein Change | Mutation Type | Genotype | SIFTa | MutTasa | gnomAD AFb |
|---|---|---|---|---|---|---|---|---|
| Family 1 | 111,369,336 | c.166G>A | p.Ala56Thr | missense | homozygous | D | D | NA |
| Family 2 | 111,369,429 | c.73C>T | p.Gln25Ter | nonsense | homozygous | NA | D | NA |
| Family 3 | 111,369,501 | c.1A>G | p.? | start-lost | homozygous | D | D | 1.23 × 10−5 |
| Family 4 | 111,367,968 | c.475_478del | p.Ile159LeufsTer15 | frameshift deletion | homozygous | NA | D | NA |
BTG4—B cell translocation gene 4
Mutation assessment by SIFT and MutationTaster (MutTas). D—damaging; NA—not available.
Allele frequency of corresponding mutations in the gnomAD database. NA—not available.
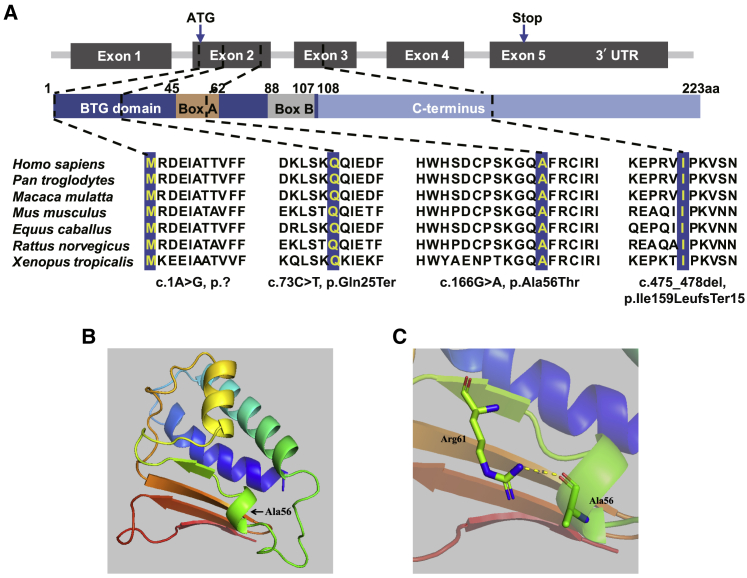
Figure 2.
Location of Variants, Amino Acid Conservation across Species, and Structural Implications of BTG4 Amino Acid Substitution.
(A) Location of the variants in the gene and protein structure and the conservation of the mutated amino acids in seven different species. All four residues are highly conserved across species.
(B) Overview of the predicted structure of the wild-type BTG4 protein. Arrow indicates the location of Ala56.
(C) Magnified view of the predicted structure surrounding Ala56. The yellow dashed line represents the predicted hydrogen bond.
Impact of the BTG4 Variants on Protein Structure
The amino acids at positions p.Met1, p.Gln25, p.Ala56, and p.Ile159 are highly conserved across species (Figure 2A). Except for c.166G>A (p.Ala56Thr), the variants were predicted to result in protein loss or a truncated protein. According to the three-dimensional structure prediction, Ala56 is located in one of the helical motifs, and Ala56 can form hydrogen bonds with nearby residues (Figure 2B). The substitution of Ala56 by Thr56 may affect the corresponding helix or the hydrogen bond between residues, which may further affect the function of the protein.
Localization and Level of Mutant Protein in Oocytes and HeLa cells
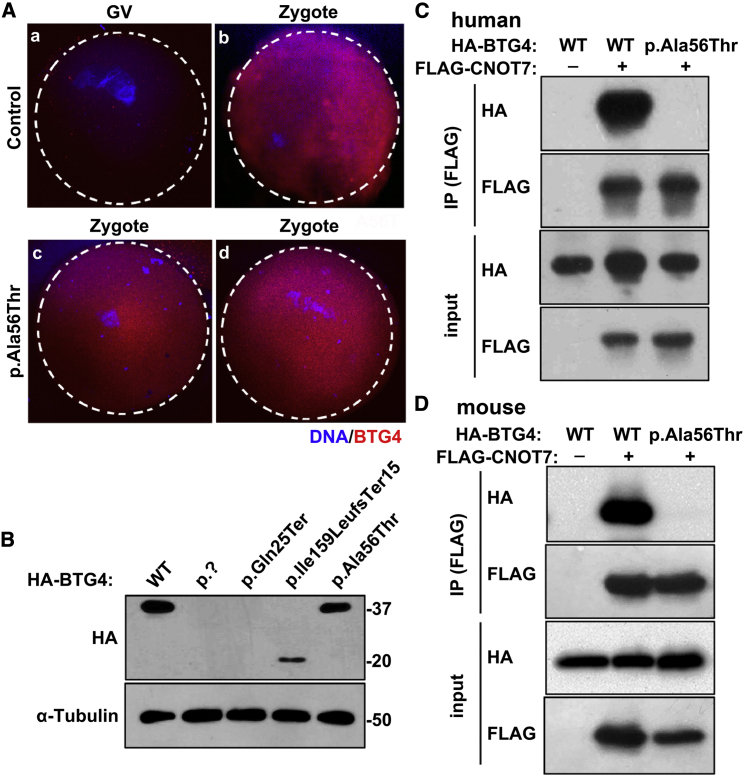
The results of immunofluorescence staining in control oocytes suggested that the BTG4 protein is normally present only after germinal vesicle (GV) breakdown (Figure 3A). Compared to controls, the c.166G>A (p.Ala56Thr) variant did not affect the localization or temporal distribution of BTG4 protein in the zygotes from affected individual (Figure 3A). To further evaluate the protein levels of the pathogenic variants in vitro, we performed immunoblot analysis with HeLa cells transfected with wild-type or mutant BTG4 plasmids. The variants c.1A>G (p.?) and c.73C>T (p.Gln25Ter) resulted in undetectable protein levels, while the variant c.475_478del (p.Ile159LeufsTer15) produced a truncated protein (∼20 kDa). For the variant c.166G>A (p.Ala56Thr), there was no change in protein level (Figure 3B), and this was consistent with the immunofluorescence staining in the mutant zygotes (Figure 3A).
Figure 3.
Localization and Level of the Mutant Protein in Oocytes and HeLa cells and the Effect of the c.166G>A (p.Ala56Thr) Variant on the BTG4-CNOT7 Interaction
(A) Immunofluorescence of control and c.166G>A (p.Ala56Thr) GV oocytes and zygotes.
BTG4 is present after the GV stage (A and B), and zygotes with the c.166G>A (p.Ala56Thr) variant exhibit similar BTG4 protein localization as the controls (C and D).
(B) Immunoblot analysis of HeLa cell extracts after transfection with wild-type or mutant HA-tagged BTG4 expression plasmids. The wild-type protein and the c.166G>A (p.Ala56Thr) variant were both about 37 kDa, the truncated c.475_478del (p.Ile159LeufsTer15) variant was about 20 kDa, and the c.1A>G (p.?) and c.73C>T (p.Gln25Ter) variants showed no detectable bands. α-Tubulin was used as the internal control.
(C) Co-IP assay showing the loss of the BTG4 and CNOT7 interaction by the c.166G>A (p.Ala56Thr) variant in humans. Cell lysates were immunoprecipitated with anti-FLAG antibody, and the resulting protein samples were separated by SDS-PAGE and analyzed via immunoblot using antibodies against the HA tag.
(D) Co-IP assay showing the loss of the BTG4 and CNOT7 interaction by the c.166G>A (p.Ala56Thr) variant in mice. Cell lysates were immunoprecipitated with anti-FLAG antibody and analyzed via immunoblot using antibodies against HA.
Variant c.166G>A (p.Ala56Thr) Abolished the Interaction Between BTG4 and CNOT7
It has been reported that BTG4 functions as an adaptor by recruiting the CCR4-NOT catalytic subunits CNOT7 and CNOT8, which are involved in mRNA decay.15 Because the c.166G>A (p.Ala56Thr) variant had no effect on BTG4 protein level and distribution pattern, we speculated that this variant might impair the interaction between BTG4 and CNOT7. Coimmunoprecipitation (Co-IP) experiments showed that wild-type BTG4 interacted with CNOT7, while the interaction was completely abolished in the BTG4 c.166G>A (p.Ala56Thr) variant (Figure 3C). Due to the high conservation of the Ala56 residue across species (Figure 2A), we repeated the experiment using mouse constructs and obtained similar results (Figure 3D). These findings demonstrated that the conserved Ala56 residue is essential for BTG4 to bind CNOT7.
The BTG4 Variants Impaired the Decay of Maternal mRNA in Zygotes of the Affected Individual
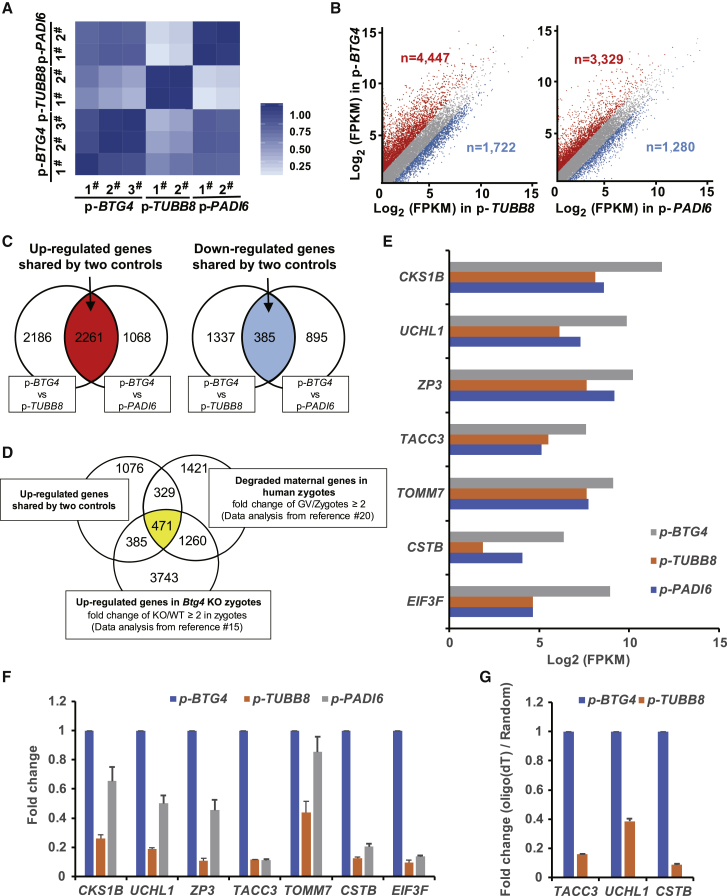
Because BTG4 facilitates the decay of maternal mRNAs in mice,15 we next investigated the effects of the BTG4 pathogenic variants on mRNA metabolism in zygotes of the affected individual. Because variants in TUBB8 and PADI6 (MIM: 610363) have no influence on maternal mRNA decay,6,19 we used zygotes from other affected individuals with the TUBB8 (GenBank: NM_177987.3) variant (c.922G>A, p.Gly308Ser, likely pathogenic, unpublished) or the PADI6 (GenBank: NM_207421.4) pathogenic variant (c.1521dup [p.Ser508GlnfsTer5])16 as controls. The individual with the TUBB8 variant has a mixed phenotype of fertilization problem and ZCF, whereas the phenotype of the individual with the PADI6 variant was typically early embryonic arrest. A total of 18,324 genes (FPKM > 0.1) were analyzed from all samples (Table S2), and all replicates showed high correlations (Figure 4A). As indicated in Figure 4B, a large number of genes exhibited higher expression levels in zygotes with the BTG4 c.166G>A (p.Ala56Thr) variant compared to zygotes with the TUBB8 variant (NBTG4 versus TUBB8 = 4,447) or the PADI6 variant (NBTG4 versus PADI6 = 3,329) (Figure 4B). In contrast, the numbers of downregulated genes were much smaller (NBTG4 versus TUBB8 = 1,722; NBTG4 versus PADI6 = 1,280) (Figure 4B). The number of upregulated genes shared by the two controls was 2,261, whereas the number of downregulated genes was only 385 (Figure 4C); this indicates impairment of mRNA decay in the zygotes with the c.166G>A (p.Ala56Thr) pathogenic variant. Generally, previous studies indicated that during the process of meiotic maturation and fertilization in humans, about 3,481 maternal genes need to be degraded in zygotes (fold change ≥ 2 between GV and zygote stage).20 In zygotes of Btg4 knockout mice, about 5,859 maternal genes were upregulated, and these genes should have been degraded in the wild-type zygotes (fold change ≥ 2 between Btg4 knockout and wild-type zygotes).15 When overlapping the 2,261 upregulated genes in zygotes of the affected individual with the BTG4 c.166G>A (p.Ala56Thr) variant (Figure 4C), with the 3,481 degraded maternal genes in normal human zygotes (data analysis from Wu et al.20), and with the 5,859 upregulated maternal genes in zygotes of Btg4 knockout mice (data analysis from Yu et al.15), we finally focused on 471 genes (Figure 4D). We thereby inferred that the 471 genes could be functionally conservative as well as key in both human and mouse, and these genes need to be degraded for normal zygotic cleavage (Figure 4D and Table S2). Therefore, we then randomly chose seven of the top fold-changed genes from the 471 genes, and we analyzed them in terms of the FRKM ratio of the RNA-seq data. The results were similar to those of the qRT–PCR, and these maternal transcripts were indeed maintained at higher expression levels in zygotes with BTG4 pathogenic variants compared with the controls (Figure 4E-F). Furthermore, in the control group with TUBB8 variants, the efficacy of oligo-dT–mediated reverse transcription for the selected genes (TACC3 [MIM: 605303], UCHL1 [MIM: 191342], and CSTB [MIM: 601145]) decreased significantly compared to the random primer-mediated transcription (Figure 4G), implying that there were longer poly(A) tails in the affected zygotes. These results demonstrated the impairment of mRNA decay following BTG4 dysfunction, which ultimately resulted in maternal mRNA accumulation in the zygotes of the affected individual.
Figure 4.
The Pathogenic Variants in BTG4 Resulted in Impaired Maternal mRNA Decay in Zygotes with the c.166G>A (p.Ala56Thr) Variant
(A) Heatmap of the Spearman correlation coefficients among zygotes with the three different variants: BTG4 (c.166G>A [p.Ala56Thr]), TUBB8 (c.922G>A [p.Gly308Ser]), and PADI6 (c.1521dup [p.Ser508GlnfsTer5]).
(B) Scatterplot comparing transcripts between BTG4 variant (c.166G>A, p.Ala56Thr) and TUBB8 (c.922G>A [p.Gly308Ser]) and/or PADI6 (c.1521dup [p.Ser508GlnfsTer5]) variant zygotes. Transcripts that were increased or decreased by more than 2-fold in the BTG4 variant zygotes are highlighted in red or blue, respectively.
(C) Venn diagrams showing the shared upregulated (red region pointed by arrow) or downregulated (blue region pointed by arrow) genes between the BTG4 variant (c.166G>A [p.Ala56Thr]) and the TUBB8 variant (c.922G>A [p.Gly308Ser]) and between the BTG4 variant and the PADI6 variant (c.1521dup [p.Ser508GlnfsTer5]).
(D) Venn diagrams showing the overlap of upregulated genes in zygotes harboring the BTG4 variant (c.166G>A [p.Ala56Thr]) with the reported human (GV/Zygote ≥ 2) and mouse (Btg4 knockout [KO] mice/wild type [WT] mice ≥ 2 in zygotes) mRNA sequencing data.
(E) RNA-seq results confirming the relative expression of representative genes.
(F) qRT-PCR verification of the expression of representative genes relative to GAPDH in zygotes with variants in different genes. The error bar denotes SD.
(G) qRT-PCR results obtained from oligo-dT versus random primer-mediated reverse transcription reactions; these results reflect changes in the poly(A) tail length of the given transcripts from the above representative genes. The error bar denotes SD.
Discussion
Several genetic factors associated with female infertility characterized by abnormalities in oocyte maturation, fertilization, and early embryo cleavage have been identified in recent years, including TUBB8,6 PATL2,5 WEE2,8 PADI6,19 PANX1 (MIM: 608420),21 and TLE6.7 In this study, we show that human ZCF is a Mendelian phenotype, and we report four different homozygous mutations in BTG4 that are responsible for the disease, indicating that human ZCF has a recessive inheritance pattern. All four pathogenic variants resulted in the functional loss of BTG4, including protein loss, truncation, and impairment of the interaction between BTG4 and CNOT7, and our in vivo studies showed that loss of maternal mRNA decay in the zygotes explained the failure to undergo the first cleavage. Our study demonstrates that mutations in BTG4 cause a human disease characterized by female infertility due to ZCF.
In mammalian oogenesis, maternal mRNAs are transcribed and stored during oocyte growth, and these mRNAs support oocyte meiotic maturation and the oocyte-to-embryo transition. These mRNAs undergo massive degradation as early as the meiotic resumption stage, and about 90% of maternal mRNA is eliminated during the oocyte-to-embryo transition.22 BTG4 acts as a meiotic cell cycle-coupled MZT-licensing factor in mouse oocytes, and Btg4 null females produce otherwise morphologically normal oocytes that arrest at the one-cell stage due to the accumulation of maternal mRNAs.15 Our present findings confirm the role of BTG4 and the phenotype of BTG4 dysfunction in humans, and this result suggests the important and conserved function of BTG4 during female reproduction.
The BTG4 protein contains two conserved regions—the N-terminal BTG domain that contains the Box-A and Box-B motifs and the functionally unclear C-terminal domain (Figure 2A). The c.1A>G (p.?) variant altered the first amino acid of BTG4 protein, whereas the c.73C>T (p.Gln25Ter) variant resulted in a premature stop of the BTG4 protein, and the c.475_478del (p.Ile159LeufsTer15) variant located in the C-terminal domain produced a truncated protein. All three of these variants caused severe impairment of the BTG4 protein (Figure 3B). As for the c.166G>A (p.Ala56Thr) variant, although the protein level and distribution pattern were unchanged (Figure 3A and 3B), it totally abolished the interaction of BTG4 with CNOT7 (Figure 3C–3D). Previous studies suggested that BTG4 primarily interacts with the CNOT7 and CNOT8 catalytic subunits of the CCR4-NOT deadenylase complex through multiple conserved amino acid residues.15,22 Tryptophan-95 is a key residue in mediating the interaction between BTG4 and CNOT7 and CNOT8, and the homologous mutation of tryptophan-95 to alanine (Btg4W95A) in mice causes a similar phenotype to that of Btg4-knockout mice.15 The present study suggests that alanine-56 is another key residue in BTG4 function, and mutating this residue to threonine-56 abolished the BTG4-CNOT7 interaction in both humans and mice. All of these findings suggest the important role of the BTG domain.
It has been reported that variants in TUBB8 cause oocyte maturation defect 2 (OOMD2 [MIM: 616780]) because of abnormal spindle assembly6 and that mice with variants of PADI6—which is a member of the subcortical maternal complex23,24—and Padi6-knockout mice exhibit early embryonic arrest due to failure to undergo zygotic genome activation, but this is not due to disruption of maternal mRNA decay.19,25 Compared to zygotes with TUBB8 or PADI6 variants, a large number of maternal mRNAs remain intact in the zygotes of the affected individual described here, thus demonstrating the specific function of BTG4 in maternal mRNA decay. It should be noted that there are more differentially expressed genes in the group of BTG4 versus TUBB8 zygotes than those in the group of BTG4 versus PADI6 zygotes. One possibility is that the secondary consequence of the PADI6 variant might further cause impairment of mRNA metabolism. In addition, although single-cell RNA sequencing has been successfully pursued in several recent studies,26,27 we admit that because of paucity of human zygotes, only limited zygotes were used for RNA sequencing in the current study, and this may have led to some variation for numbers of differentiated expressed genes in transcriptomic analysis. Besides, the impairment of mRNA decay in vivo was only demonstrated in the affected individual from family 1 (variant c.166G>A [p.Ala56Thr]), whereas such an effect could not be investigated and confirmed in affected individuals from other three families due to unavailability of their zygotes. Further analysis needs to be performed in more cases or in Btg4 knock-in mice. Overall, our present findings emphasize the importance of MZT in human zygotes. Until now, little was known about the pathophysiological mechanism of human MZT, and further studies are needed to identify other critical factors that participate in the MZT process in humans.
In conclusion, homozygous mutations in BTG4 prevent maternal mRNA decay and lead to female infertility characterized by ZCF. These findings will facilitate genetic diagnoses for clinical infertility patients with recurrent failure of IVF, and they lay the foundation for developing possible therapeutic treatments for these affected individuals in the future.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2017YFC1001500, 2016YFC1000600, 2016YFC1000200, 2018YFC1003100, and 2018YFC1003800), the National Natural Science Foundation of China (81725006, 81822019, 81771581, 81971450, and 81971382), the Natural Science Foundation of Hunan Province (2018JJ3893), the Scientific Research Foundation of Reproductive and Genetic Hospital of China International Trust Investment Corporation (CITIC) Xiangya (YNXM-201913), the Shanghai Municipal Science and Technology Major Project (2017SHZDZX01), the Project of Shanghai Municipal Science and Technology Commission (19JC1411001), the Shanghai Rising-Star Program (17QA1400200), the Natural Science Foundation of Shanghai (19ZR1444500 and 17ZR1401900), Shuguang Program of Shanghai Education Development Foundation and Shanghai Municipal Education Commission (18SG03), the Strategic Collaborative Research Program of the Ferring Institute of Reproductive Medicine, Ferring Pharmaceuticals and Chinese Academy of Sciences (FIRMC200507), the Capacity Building Planning Program for Shanghai Women and Children’s Health Service, the Collaborative Innovation Center Project Construction for Shanghai Women and Children’s Health, and Start-up Funding of Guangdong Second Provincial General Hospital (YY2019-001).
Published: June 4, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.05.010.
Contributor Information
Ge Lin, Email: linggf@hotmail.com.
Qing Sang, Email: sangqing@fudan.edu.cn.
Web Resources
1000 Genomes Project, https://www.internationalgenome.org/home
ExAC Browser, http://exac.broadinstitute.org/
GeneBank, https://www.ncbi.nlm.nih.gov/genbank/
Genome Aggregation Database (gnomAD), https://gnomad.broadinstitute.org/
MultiAlin, http://multalin.toulouse.inra.fr/multalin/multalin.html
OMIM, https://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
PyMol software, https://pymol.org/2/
Swiss Model web server, https://swissmodel.expasy.org/
Supplemental Data
References
- 1.Mascarenhas M.N., Flaxman S.R., Boerma T., Vanderpoel S., Stevens G.A. National, regional, and global trends in infertility prevalence since 1990: a systematic analysis of 277 health surveys. PLoS Med. 2012;9:e1001356. doi: 10.1371/journal.pmed.1001356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hart R.J. Physiological aspects of female fertility: role of the environment, modern lifestyle, and genetics. Physiol. Rev. 2016;96:873–909. doi: 10.1152/physrev.00023.2015. [DOI] [PubMed] [Google Scholar]
- 3.Matzuk M.M., Lamb D.J. The biology of infertility: research advances and clinical challenges. Nat. Med. 2008;14:1197–1213. doi: 10.1038/nm.f.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yatsenko S.A., Rajkovic A. Genetics of human female infertility. Biol. Reprod. 2019;101:549–566. doi: 10.1093/biolre/ioz084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen B., Zhang Z., Sun X., Kuang Y., Mao X., Wang X., Yan Z., Li B., Xu Y., Yu M. Biallelic mutations in PATL2 cause female infertility characterized by oocyte maturation arrest. Am. J. Hum. Genet. 2017;101:609–615. doi: 10.1016/j.ajhg.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng R., Sang Q., Kuang Y., Sun X., Yan Z., Zhang S., Shi J., Tian G., Luchniak A., Fukuda Y. Mutations in TUBB8 and human oocyte meiotic arrest. N. Engl. J. Med. 2016;374:223–232. doi: 10.1056/NEJMoa1510791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alazami A.M., Awad S.M., Coskun S., Al-Hassan S., Hijazi H., Abdulwahab F.M., Poizat C., Alkuraya F.S. TLE6 mutation causes the earliest known human embryonic lethality. Genome Biol. 2015;16:240. doi: 10.1186/s13059-015-0792-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sang Q., Li B., Kuang Y., Wang X., Zhang Z., Chen B., Wu L., Lyu Q., Fu Y., Yan Z. Homozygous mutations in WEE2 cause fertilization failure and female infertility. Am. J. Hum. Genet. 2018;102:649–657. doi: 10.1016/j.ajhg.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sha Q.-Q., Zhang J., Fan H.-Y. A story of birth and death: mRNA translation and clearance at the onset of maternal-to-zygotic transition in mammals. Biol. Reprod. 2019;101:579–590. doi: 10.1093/biolre/ioz012. [DOI] [PubMed] [Google Scholar]
- 10.Clift D., Schuh M. Restarting life: fertilization and the transition from meiosis to mitosis. Nat. Rev. Mol. Cell Biol. 2013;14:549–562. doi: 10.1038/nrm3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z., Chen C., Cui P., Liao Y., Yao L., Zhang Y., Rui R., Ju S. Plk1 inhibition leads to a failure of mitotic division during the first mitotic division in pig embryos. J. Assist. Reprod. Genet. 2017;34:399–407. doi: 10.1007/s10815-016-0864-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Audouard C., Le Masson F., Charry C., Li Z., Christians E.S. Oocyte-targeted deletion reveals that hsp90b1 is needed for the completion of first mitosis in mouse zygotes. PLoS ONE. 2011;6:e17109. doi: 10.1371/journal.pone.0017109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang L., Meng T.-G., Ma X.-S., Wang Z.-B., Qi S.-T., Chen Q., Zhang Q.-H., Liang Q.-X., Wang Z.-W., Hu M.-W. Rad9a is involved in chromatin decondensation and post-zygotic embryo development in mice. Cell Death Differ. 2019;26:969–980. doi: 10.1038/s41418-018-0181-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui C., Ren X., Liu D., Deng X., Qin X., Zhao X., Wang E., Yu B. 14-3-3 epsilon prevents G2/M transition of fertilized mouse eggs by binding with CDC25B. BMC Dev. Biol. 2014;14:33. doi: 10.1186/s12861-014-0033-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu C., Ji S.-Y., Sha Q.-Q., Dang Y., Zhou J.-J., Zhang Y.-L., Liu Y., Wang Z.-W., Hu B., Sun Q.-Y. BTG4 is a meiotic cell cycle-coupled maternal-zygotic-transition licensing factor in oocytes. Nat. Struct. Mol. Biol. 2016;23:387–394. doi: 10.1038/nsmb.3204. [DOI] [PubMed] [Google Scholar]
- 16.Zheng W., Chen L., Dai J., Dai C., Guo J., Lu C., Gong F., Lu G., Lin G. New biallelic mutations in PADI6 cause recurrent preimplantation embryonic arrest characterized by direct cleavage. J. Assist. Reprod. Genet. 2020;37:205–212. doi: 10.1007/s10815-019-01606-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sha Q.Q., Yu J.L., Guo J.X., Dai X.X., Jiang J.C., Zhang Y.L., Yu C., Ji S.Y., Jiang Y., Zhang S.Y. CNOT6L couples the selective degradation of maternal transcripts to meiotic cell cycle progression in mouse oocyte. EMBO J. 2018;37:e99333. doi: 10.15252/embj.201899333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leng L., Sun J., Huang J., Gong F., Yang L., Zhang S., Yuan X., Fang F., Xu X., Luo Y. Single-Cell Transcriptome Analysis of Uniparental Embryos Reveals Parent-of-Origin Effects on Human Preimplantation Development. Cell Stem Cell. 2019;25:697–712.e6. doi: 10.1016/j.stem.2019.09.004. [DOI] [PubMed] [Google Scholar]
- 19.Xu Y., Shi Y., Fu J., Yu M., Feng R., Sang Q., Liang B., Chen B., Qu R., Li B. Mutations in PADI6 cause female infertility characterized by early embryonic arrest. Am. J. Hum. Genet. 2016;99:744–752. doi: 10.1016/j.ajhg.2016.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu J., Xu J., Liu B., Yao G., Wang P., Lin Z., Huang B., Wang X., Li T., Shi S. Chromatin analysis in human early development reveals epigenetic transition during ZGA. Nature. 2018;557:256–260. doi: 10.1038/s41586-018-0080-8. [DOI] [PubMed] [Google Scholar]
- 21.Sang Q., Zhang Z., Shi J., Sun X., Li B., Yan Z., Xue S., Ai A., Lyu Q., Li W. A pannexin 1 channelopathy causes human oocyte death. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aav8731. [DOI] [PubMed] [Google Scholar]
- 22.Horiuchi M., Takeuchi K., Noda N., Muroya N., Suzuki T., Nakamura T., Kawamura-Tsuzuku J., Takahasi K., Yamamoto T., Inagaki F. Structural basis for the antiproliferative activity of the Tob-hCaf1 complex. J. Biol. Chem. 2009;284:13244–13255. doi: 10.1074/jbc.M809250200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L., Zheng P., Dean J. Maternal control of early mouse development. Development. 2010;137:859–870. doi: 10.1242/dev.039487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu X., Gao Z., Qin D., Li L. A maternal functional module in the mammalian oocyte-to-embryo transition. Trends Mol. Med. 2017;23:1014–1023. doi: 10.1016/j.molmed.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 25.Yurttas P., Vitale A.M., Fitzhenry R.J., Cohen-Gould L., Wu W., Gossen J.A., Coonrod S.A. Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo. Development. 2008;135:2627–2636. doi: 10.1242/dev.016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou F., Wang R., Yuan P., Ren Y., Mao Y., Li R., Lian Y., Li J., Wen L., Yan L. Reconstituting the transcriptome and DNA methylome landscapes of human implantation. Nature. 2019;572:660–664. doi: 10.1038/s41586-019-1500-0. [DOI] [PubMed] [Google Scholar]
- 27.Li L., Dong J., Yan L., Yong J., Liu X., Hu Y., Fan X., Wu X., Guo H., Wang X. Single-Cell RNA-Seq Analysis Maps Development of Human Germline Cells and Gonadal Niche Interactions. Cell Stem Cell. 2017;20:858–873.e4. doi: 10.1016/j.stem.2017.03.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.