Abstract
Polyploidy—or chromosome doubling—plays a significant role in plant speciation and evolution. Much of the existing evidence indicates that fusion of unreduced (or 2n) gametes is the major pathway responsible for polyploid formation. In the early 1900s, a theory was put forward that the mechanism of “hybridization followed by chromosome doubling” would enable the survival and development of the hybrid zygote by providing each chromosome with a homolog with which to pair. However, to date there is only scant empirical evidence supporting this theory. In our previous study, interspecific-interploid crosses between the tetraploid Hylocereus megalanthus, as the female parent, and the diploid H. undatus, as the male parent, yielded only allopentaploids, allohexaploids, and 5x-and 6x-aneuploids instead of the expected allotriploids. No viable hybrids were obtained from the reciprocal cross. Since H. undatus underwent normal meiosis with regular pairing in the pollen mother cells and only reduced pollen grains were observed, the allohexaploids obtained supported the concept of “chromosome doubling.” In this work, we report ploidy level, fruit morphology, and pollen viability and diameter in a group of putative hybrids obtained from an embryo rescue procedure following controlled H. megalanthus × H. undatus crosses, with the aim to elucidate, for the first time, the timing and developmental stage of the chromosome doubling. As in our previous report, no triploids were obtained, but tetraploids, pentaploids, hexaploids, and 5x- and 6x-aneuploids were found in the regenerated plants. The tetraploids exhibited the morphological features of the maternal parent and could not be considered true hybrids. Based on our previous studies, we can assume that the pentaploids were a result of a fertilization event between one unreduced (2n) female gamete from the tetraploid H. megalanthus and a normal (n) haploid male gamete from H. undatus. All the allohexaploids obtained from the embryo rescue technique where those that regenerated from fertilized ovules 10 days after pollination (at the pro-embryo stage), showing that the chromosome doubling event occurred at a very early development stage, i.e., at the zygote stage or shortly after zygote formation. These allohexaploids thus constitute empirical evidence of “hybridization followed by chromosome doubling.”
Keywords: allopolyploidization, flow cytometry, genome duplication, Hylocereus, interspecific-interploid crosses, true hybrids, unreduced gametes
Introduction
Genome doubling or polyploidy – the state of having more than two full sets of chromosomes – has played a major role in the diversification and speciation of the plant kingdom, generating the genetic and epigenetic novelty that has contributed significantly to the diversity prevailing today (Stebbins, 1971; Soltis and Soltis, 1993; Osborn et al., 2003; Van de Peer et al., 2017). Autopolyploids arise within a single species and carry homologous chromosomes, while allopolyploids arise from two different taxa and have homoeologous chromosomes (Leitch and Bennett, 1997; Ramsey and Schemske, 1998; Soltis and Soltis, 2000). More specifically, taxonomically, autopolyploids are formed from within a single species, whereas allopolyploids are formed by hybridization between two or more species. Genetically, autopolyploids are plants with random associations among four (in the tetraploid cases) homologous chromosomes, resulting in tetrasomic segregation, whereas allopolyploids have two sets of homoelogous chromosomes that do not typically pair, leading to disomic segregation (Doyle and Egan, 2010; Doyle and Sherman-Broyles, 2017). As such, allopolyploids can, potentially, generate all the enzymes produced by each parent as well as new hybrid enzymes. This genetic redundancy or “enzyme multiplicity” of allopolyploids is considered to be an advantage, contributing to evolutionary success (Soltis and Soltis, 1993; Soltis et al., 2014; Glover et al., 2016; Lloyd et al., 2018) and facilitating speciation when a new hybrid is both fertile and reproductively isolated from its parental species.
The biological significance of allopolyploids thus derives from their origin, establishment, and persistence. Allopolyploid establishment and persistence have been extensively studied over the past few decades (Jenczewski and Alix, 2004; Feldman and Levy, 2009; Steige and Slotte, 2016; and references therein in all these citations), but what do we know about the pathways to the origin and formation of allopolyploids? It was over 100 years ago that Winge (1917) proposed his theory of “hybridization followed by chromosome doubling” as a mechanism enabling the survival and development of the hybrid zygote by providing each chromosome with a homolog with which to pair. According to Winge, hybridization involving two genetically very different gametes would not produce a viable zygote, since the gametes would not be able to pair (as was then solely in the realm of speculation and not based on scientific examinations). He thus suggested that chromosomes would split longitudinally, thereby allowing to generate a pair of homologous chromosomes that would permit the development of the hybrid with double the parental number of chromosomes. However, to date, most empirical studies have not provided evidence to support Winge's theory. Rather, the findings of most studies support one of the following two mechanisms for allopolyploid origin: (1) sexual polyploidization through the fusion of 2n gametes, where the increase in the chromosome number occurs in the first generation through the union of one or both unreduced gametes (Harlan and deWet, 1975 and references therein, Gao et al., 2019); or (2) chromosome doubling in somatic tissues, i.e., somatic doubling, which is a mitotic rather than a meiotic event. The first report of spontaneous somatic doubling was that for Primula kewensis, a first-generation hybrid between P. floribunda and P. verticillata (Newton and Pellew, 1929). The hybrid diploid plant was sterile, but periodically one of the cuttings produced fertile flowers in branches that were undergoing spontaneous doubling (somatic doubling), a process that apparently restored fertility by providing each chromosome with an identical partner with which to pair (Newton and Darlington, 1929). Today, it is known that somatic doubling in meristem tissues can also be induced by artificial means (Ramsey and Schemske, 1998 and references therein; Sattler et al., 2016 and references therein).
As mentioned above, over the years only scant empirical evidence has been offered in support of Winge's theory of “hybridization followed by chromosome doubling.” To the best of our knowledge, the following examples are the only crosses that support his theory: 1) fertile Nicotiana glutinosa L. × N. tabacum L. tobacco allotetraploids (Clausen and Goodspeed, 1925); 2) two ryegrass genotypes with an unexpected double number of chromosomes from a BC2 progeny [a cross between the diploid Lolium multiflorum and a triploid BC1 (itself the result of a cross between the tetraploid Festuca arundinacea var. glaucescens and a synthetic L. multiflorum tetraploid)] (Morgan et al., 2001); and 3) hexaploid and 6x-aneuploid pitaya hybrids resulting from a cross between the tetraploid Hylocereus megalanthus and the diploid H. undatus (Tel-Zur et al., 2003; Tel-Zur et al., 2004). These three examples suggest that the unexpected high ploidy level of the newly formed hybrids was the result of a process that occurred immediately after or soon after fertilization.
Hylocereus species are night-blooming vine cacti native to the tropical and sub-tropical regions of the Americas (Barthlott and Hunt, 1993). These species are characterized by triple-ribbed stems, large flowers, and attractive edible fruits (Mizrahi and Nerd, 1999). Cytological studies show that Hylocereus species are diploids (Banerji and Sen, 1955; Spencer, 1955; Lichtenzveig et al., 2000; Tel-Zur et al., 2003; Tel-Zur et al., 2004), with the exception of H. megalanthus, which is a tetraploid. The diploid species bear large (250-800 g) flavorless, red-purple fruits, whereas the tetraploid H. megalanthus bears sweeter, but smaller (180-250 g), yellow fruits (Tel-Zur et al., 2003; Tel-Zur et al., 2011). With the aim of producing elite cultivars with improved fruit quality, a long-term breeding program was thus initiated about three decades ago at Ben-Gurion University of the Negev. In this framework, interspecific-interploid crosses were performed with the aim to combine the size and attractiveness of the diploids with the fruit quality of the tetraploid species (Tel-Zur et al., 2004). No true hybrids or chimeras were obtained in the sampled H. megalanthus × H. undatus crosses (Tel-Zur et al., 2003: Tel-Zur et al., 2004), but for the reciprocal cross, tetraploids, pentaploids, hexaploids, and 6x-aneuploids, rather than the expected triploids, were obtained (Tel-Zur et al., 2003), thereby indicating a uni-directional gene flow among these species. The tetraploids exhibited the morphological features of the maternal parent and were therefore not considered true hybrids; rather, they were considered to be either a result of self-pollination due to contamination during the hand-cross pollination process or to be of somatic origin, since polyembryony has been reported in H. megalanthus (Cisneros et al., 2011). It was therefore proposed that zygotic or post-zygotic somatic chromosome doubling constituted the mechanism for the formation of the hexaploid and 6x-aneuploid hybrids (Tel-Zur et al., 2003; Tel-Zur et al., 2004). The above premise was based on our previous work (Lichtenzveig et al., 2000) showing the formation of both unreduced pollen grains and unbalanced gametes due to irregular chromosome disjunction at anaphase. Anaphase I separations such as 22–22, 23–21, and 24–20 were observed in the tetraploid H. megalanthus, suggesting that some degree of aneuploidy could be tolerated, while the diploid species exhibited both the regular chromosome disjunction at anaphase I and a uniform pollen diameter (Lichtenzveig et al., 2000; Tel-Zur et al., 2003). Furthermore, to date, all interspecific diploid Hylocereus × Hylocereus crosses have produced diploid hybrids, which strongly indicates the negligible production of unreduced gametes by the diploid species (Tel-Zur et al., 2004). Consequently, we can assume that the allohexaploid and 6x-aneuploid hybrids obtained from crosses of H. megalanthus and H. undatus occurred at frequencies that were much higher than the frequency that would be expected for the fusion of unreduced gametes from both egg and pollen donor parents. In addition, no chimeras were obtained, i.e., plants with identical fruit and vegetative morphology were observed in the H. megalanthus × H. undatus hybrids studied, which indicates a very low likelihood of somatic doubling later in development and provides further support for our theory.
Against the above background, the main goals of this research were to confirm the occurrence of spontaneous chromosome doubling following interspecific-interploid hybridization, i.e., Winge's theory, and to identify the timing or the developmental stage at which the event occurs. We postulated that genome doubling takes place either in the hybrid zygote or shortly after zygote formation, a process that ensures the viability of the new hybrid embryo. The methodology applied involved estimating the ploidy level by measuring the relative DNA content by means of flow cytometry of the putative allopolyploids regenerated by the embryo rescue technique following H. megalanthus × H. undatus crosses. In addition, the viability and fertility of the resulting true allopolyploids were evaluated in terms of fruit morphology and weight and in terms of the pollen viability and diameter of selected allopolyploids.
Materials and Methods
Plant Material and Growth Conditions
The plant material used in this study comprised 38 plants regenerated from crosses between the tetraploid H. megalanthus [(Schum. ex Vaupel) Moran] Bauer (accession 90-003) as the female parent and the diploid H. undatus (Haw) Br. and Rose (accession 89-024) as the male parent, according to the embryo rescue technique given in Cisneros et al. (2013) and described briefly below. It has previously been shown that the total number of seeds per fruit in H. megalanthus is about 200-300, resulting in 20-35% aborted/empty seeds (Cisneros et al., 2011; Tel-Zur et al., 2011). A detailed description of zygote, embryo, and endosperm formation in H. megalanthus is provided in Cisneros et al. (2011). In particular, that study reported that an 8-cell pro-embryo was observed in H. megalanthus at 11 days after pollination (DAP) and well-developed endosperm was observed 28 DAP. The growing embryo consumed the endosperm, which remained incipient (only a few cells surrounding the embryo) at 42 DAP (Cisneros et al., 2011). Endosperm recovery and/or ploidy determination of endosperm cells is technically very challenging and is thus beyond the scope of our work as discussed below.
The development of the embryo rescue technique in our laboratory started with the rescue of embryos following intraspecific H. undatus and H. megalanthus crosses. All the resulting offspring showed the expected ploidy, i.e., diploid and tetraploid (unpublished data). In parallel, we applied the embryo rescue technique to fertilized ovules carrying embryos at very early pro-embryonic stage following reciprocal interspecific diploid crosses [H. undatus × H. monacanthus (syn. H. polyrhizus)], which resulted in diploid hybrids alone (Cisneros and Tel-Zur, 2010), thereby showing that the technique did not affect the ploidy level of the resulting hybrids. Briefly, the protocol used for embryo rescue technique included half-strength basal Murashige and Skoog (MS) medium containing 680 µM glutamine, 0.54 µM α-naphthaleneacetic acid, 0.45 µM thidiazuron, and 0.17 M sucrose. The regenerated plants were obtained from fertilized ovules collected from developing fruits at 10, 30, and 47 DAP when the embryos were at the pro-embryo (10 DAP), globular (30 DAP), and heart (47 DAP) stages, respectively, according to Cisneros et al. (2011). The 38 putative allopolyploid plants were planted in January 2013 in 10-L pots held under 50% shade in a greenhouse located on the Bergmann Campus, Ben-Gurion University of the Negev, Beer-Sheva, Israel (31°15'N, 34°48'E). Each plant was watered with 200 L year-1 applied via a drip system, with a nutrient concentration of 60 mg L-1 of N, 18 mg L-1 of P and 60 mg L-1 of K fertilizer with trace elements (23-7-23 Deshanim, Israel).
Ploidy Identification
Cytological Observations
Developing floral buds were collected from the putative allopolyploid lines and fixed for 24 h in 3:1 ethanol/glacial acetic acid. The buds were stored in 70% ethanol at 4°C until examination. The chromosomes were stained with 2% acetocarmine by using the standard squash method. Pollen mother cells (PMCs) were examined through an Axio ImagerA1 microscope with LED illumination (Zeiss) and photographed with a ZEISS Axiocam 305 color camera and the ZEN imaging software program.
Flow Cytometry Analysis
Tissue was collected from the tips of newly developed branches of 4- to 8-year-old putative hybrids, of the allohexaploid J-42, and of the parental species. Nuclear suspensions were prepared from this tissue according to the protocol of Li et al. (2017). Samples were analyzed using a iCyt Synergy SY3200 sorter (Sony Biotechnology, San Jose, USA) equipped with a 561-nm laser and a 595/50 band pass filter. Results were analyzed using Winlist 3D software ver 8.0 (Verity Software House). Genome size was assessed by comparing the nuclear DNA content of the tetraploid H. megalanthus with that of the relevant putative allopolyploid. The tissue of each putative hybrid was analyzed at least four times to verify the reproducibility of the results. In addition, a range of nuclear DNA contents indicating each ploidy level was determined on the basis of previously reported nuclear DNA contents for the parental species, i.e., H. megalanthus accession 90-003 with 8.70 pg/2C (Tel-Zur et al., 2011), H. undatus accession 89-024 with 3.86 pg/2C (Tel-Zur et al., 2011), and the allohexaploid J-42 reported to have 66 chromosomes (Tel-Zur et al., 2004) with 13.4 pg/2C (unpublished data); the ranges of nuclear DNA content per ploidy level were determined according to the variance around the means; thus: 8.00 to 9.50, 10.00 to 11.50, and 12.0 to 13.5 pg/2C for 4x, 5x, and 6x, respectively.
Fruit Morphology and Weight
Flowers of the putative allopolyploids were cross pollinated by hand at anthesis with a mixture of fresh pollen collected from diploid Hylocereus spp. Pollen grains were applied to the surface of the stigmata with the aid of a brush early in the morning. Fruits were collected at the ripening stage of full color over five consecutive years (2014-2018). Fruits from each allopolyploid were weighed on the harvesting day.
Pollen Grain Stainability and Diameter
Pollen grains from the allopentaploid N-149 and the allohexaploid N-134 were collected at anthesis and stored at 4°C until evaluation. These particular allopolyploids were chosen for this study due to the high number of flowers they produced every year. Pollen grains were stained with 2% acetocarmine (Belling, 1921), since previous studies showed similar outcomes for staining with acetocarmine, fluorescein diacetate, and Alexander's reagent, but acetocarmine can be used to stain both fresh and stored pollen, while the other two reagents are suitable only for fresh pollen. About 300 to 400. and 100 to 150 pollen grains from four different flowers of each line were evaluated for pollen viability and diameter, respectively. Photomicrographs were taken with an Axio ImagerA1 microscope with LED illumination (Zeiss) and photographed with a ZEISS Axiocam 305 color camera and the ZEN imaging software program. Averages ± se were calculated for pollen diameter and pollen viability. A t-test was used to assess the differences in pollen performances between allopentaploids and allohexaploids.
Results
Ploidy Identification
Cytological Observations
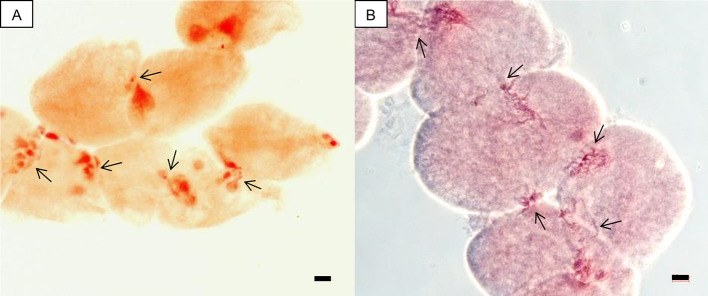
In this study, an attempt was first made to determine ploidy of putative hybrids by chromosome counting. However, cytomixis, i.e., the transfer of chromatin material from one cell to another through intercellular channels (cytomictic channels) during meiosis, proved to be an obstacle that obviated direct counting. Migration of chromatin through cytomictic channels and chains of pollen mother cells (PMCs) united by the migrating chromatin were observed in both allopentaploids and allohexaploids ( Figure 1 ). Despite our efforts to very carefully count chromosomes in the PMCs, our results for the number of chromosomes in different PMCs isolated from the same flower bud were not consistent, probably due to the cytomixis. Consequently, ploidy was estimated by flow cytometry.
Figure 1.
Cytological photographs of groups of PMCs involved in chromatin transfer, i.e., cytomixis (arrow). (A) A chain of PMCs united by channels of transferred chromatin in the allopentaploid N-124. (B) Migration of chromatin through cytomictic channels in the allohexaploid N-121. Scale bar, 10 µm.
Flow Cytometry Analysis
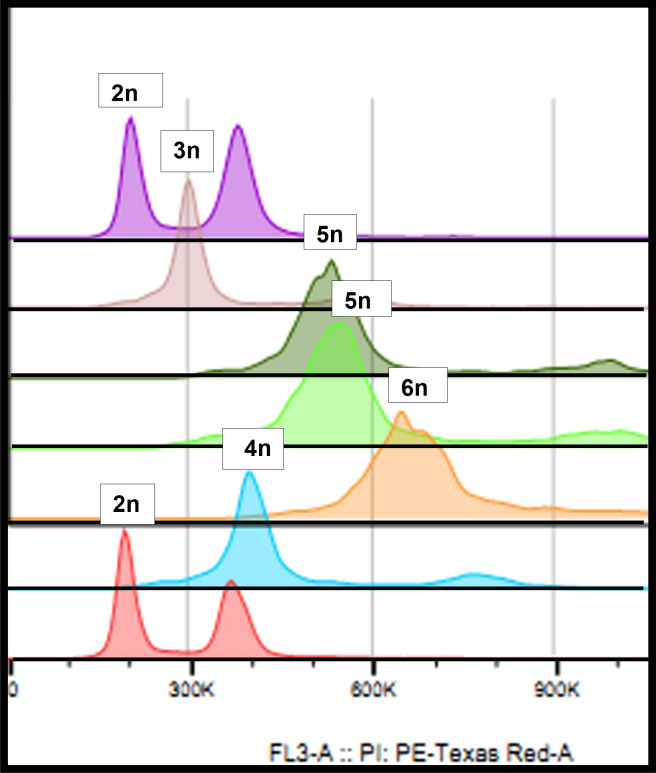
The estimated ploidy – calculated as described in Materials and Methods – indicated that among the 38 putative hybrids studied, 32 were true hybrids, and 6 were tetraploids (not considered true hybrids) like the female H. megalanthus parent ( Table 1 and Figure 2 ). Of the 32 true hybrids, 18 were allopentaploids and 8 allohexaploids. Nuclear DNA content of the allopentaploids ranged from 10.07 to 11.40 pg/2C, and that of the allohexploids ranged from 12.17 to 13.39 pg/2C ( Table 2 ). Estimation of the ploidy level of the other 6 hybrids showed that they exhibited an intermediate ploidy and that they were probably aneuploids, i.e., 3 were 4–5x, and 3 were 5–6x, ranging from 9.55 to 9.78 pg/2C and 11.67 to 11.76 pg/2C, respectively ( Table 2 ). No allotriploids were found in the studied hybrids. Average genome sizes ± se were calculated and are reported for each hybrid line in Table 2 .
Table 1.
Ploidy of the confirmed hybrids* according to cytometric analysis.
| Cross combination ♀ × ♂ |
DAP** | No. of progeny planted | No. of confirmed hybrids | No. of confirmed hybrids according to ploidy | ||||
|---|---|---|---|---|---|---|---|---|
| 3x | 4–5x | 5x | 5–6x | 6x | ||||
| H. megalanthus × H. undatus | 10 | 19 | 18 | 0 | 1 | 8 | 1 | 8 |
| 30 | 15 | 13 | 0 | 2 | 10 | 1 | 0 | |
| 47 | 4 | 1 | 0 | 0 | 0 | 1 | 0 | |
*Total number of putative hybrids studied: 38; true hybrids: 32. **DAP, days after pollination.
Figure 2.
Flow cytometry histogram: violet and dusky pink—the diploid maternal species H. undatus at the beginning and the end of the run, respectively; brown—a triploid hybrid (S-75); olive green and light green—two independent runs of the regenerated allopentaploid N-122; orange—the regenerated allohexaploid N-121; and blue—the paternal species, the tetraploid H. megalanthus.
Table 2.
Ploidy level estimated using flow cytometric analysis and fruit weight in interspecific-interploid hybrids from a cross of H. megalanthus ♀ × H. undatus ♂ rescued 10, 30, or 47 days after pollination (DAP) by hand.
| Hybrid code | DAP | Nuclear DNA content pg/2C ± se |
Ploidy estimated | Fruit weight g ± se |
|---|---|---|---|---|
| N-121 | 10 | 12.98 ± 0.28 | 6x | 112.8 ± 16.5 |
| N-122 | 10.33 ± 0.26 | 5x | 124.0 ± 42.8 | |
| N-123 | 13.20 ± 0.32 | 6x | 84.2 ± 9.8 | |
| N-124 | 11.30 ± 0.12 | 5x | 92.5 ± 12.4 | |
| N-132 | 11.76 ± 0.89 | 5-6x | 70.3 ± 12.4 | |
| N-133 | 11.32 ± 0.43 | 5x | 100.7 ± 9.2 | |
| N-134 | 12.17 ± 0.28 | 6x | 81.8 ± 3.3 | |
| N-135 | 10.72 ± 0.18 | 5x | 123.8 ± 6.1 | |
| N-136 | 11.40 ± 0.43 | 5x | 133.6 ± 7.0 | |
| N-137 | 11.18 ± 0.54 | 5x | 128.8 ± 7.9 | |
| N-138 | 12.69 ± 0.14 | 6x | 90.8 ± 6.0 | |
| N-140 | 10.07 ± 0.33 | 5x | 110.9 ± 3.6 | |
| N-173 | 10.23 ± 0.35 | 5x | 142.7 ± 8.4 | |
| N-174 | 12.65 ± 0.50 | 6x | 72.2 ± 4.4 | |
| N-175 | 12.71 ± 0.38 | 6x | 62.0 ± 12.3 | |
| N-176 | 13.39 ± 0.31 | 6x | 80.9 ± 6.6 | |
| N-177 | 9.55 ± 0.43 | 4-5x | 133.0 ± 15.0 | |
| N-178 | 12.54 ± 0.09 | 6x | 93.0 ± 3.8 | |
| N-125 | 30 | 9.78 ± 0.05 | 4-5x | 87.6 ± 11.4 |
| N-126 | 11.29 ± 0.27 | 5x | 152.1 ± 10.5 | |
| N-127 | 10.31 ± 0.37 | 5x | 154.6 ± 10.8 | |
| N-128 | 11.67 ± 0.32 | 5-6x | 123.9 ± 9.5 | |
| N-129 | 10.74 ± 0.43 | 5x | 171.0 ± 13.1 | |
| N-130 | 11.18 ± 0.07 | 5x | 171.2 ± 10.5 | |
| N-131 | 10.94 ± 0.94 | 5x | 147.2 ± 10.7 | |
| N-144 | 11.19 ± 0.32 | 5x | 128.1 ± 8.3 | |
| N-145 | 10.52 ± 0.45 | 5x | 105.5 ± 7.0 | |
| N-147 | 9.58 ± 0.55 | 4-5x | 121.0 ± 6.0 | |
| N-148 | 10.56 ± 0.51 | 5x | 139.3 ± 11.7 | |
| N-149 | 11.15 ± 0.45 | 5x | 125.9 ± 10.8 | |
| N-150 | 10.89 ± 0.33 | 5x | 133.2 ± 10.4 | |
| N-013 | 47 | 11.74 ± 0.08 | 5-6x | 141.8 ± 13.0 |
Fruit Morphology and Weight
The putative hybrids developed well under growth conditions similar to those for the parental species. Plant morphology of the putative hybrids, i.e., triple-ribbed stems as well as shape and number of thorns at base of the vegetative buds, was identical to that of the parental lines, indicating that it was impossible to identify true hybrids according to the morphology of the vegetative parts. Fruit morphology of the hybrids was compared with that of the parental species ( Tables 2 and 3 ; Figure 3 ). All 32 allopolyploids bore elongated fruits. The fruits were covered by bracts, with thorns at the base of each bract—a trait inherited from the maternal tetraploid species H. megalanthus. The spiny peel of all 32 allopolyploids (allopentaploids, allohexaploids, and the probable aneuploids) was yellow-orange in color, with a pink layer between the peel and the flesh. The fruit flesh was white, as in both parental species. All the allopolyploids set both viable and aborted seeds ( Figures 3C, D ).
Table 3.
Fruit characteristics of the parental species and allopolyploids.
| Plant material | Ploidy | Weight, g ± se | Fruit characteristics | ||
|---|---|---|---|---|---|
| Spiny peel | Peel color | Flesh color | |||
| Parental species | |||||
| H. megalanthus* | 4x | 267 ± 17 | + | Yellow | White |
| H. undatus * | 2x | 372 ± 11 | − | Red | White |
|
Cross combination
♀ × ♂ | |||||
| H. megalanthus × H. undatus | 4-5x | ND** | + | Yellow-orange | White |
| 5x | 133.5 ± 3 | + | Yellow-orange | White | |
| 5-6x | ND** | + | Yellow-orange | White | |
| 6x | 87 ± 3 | + | Yellow-orange | White | |
*Data previously reported by Lichtenzveig et al. (2000) and Tel-Zur et al. (2011). **ND, not determined.
Figure 3.
Fruit morphology. (A) The maternal species, the tetraploid H. megalanthus. (B) The paternal species, the diploid H. undatus. (C) The allopentaploid N-144. (D) The allohexaploid N-176. The average fruit weights were 218, 372, 128, and 81 g for H. megalanthus, H. undatus, allopentaploid N-144 and allohexaploid N-176, respectively.
Average fruit weights ± se were calculated and are reported for each hybrid line in Table 2 . A t-test was used to assess the difference in fruit weights between allopentaploids and allohexaploids. Statistically significant differences were observed for fruit weight, as determined by an unpaired two-tailed t-test (significant at P = 0.0001), between the allopentaploids and the allohexaploids, with the allohexaploid lines bearing smaller fruit (87 ± 3 g) than the allopentaploid lines (133.5 ± 3 g).
Pollen Grain Stainability and Diameter
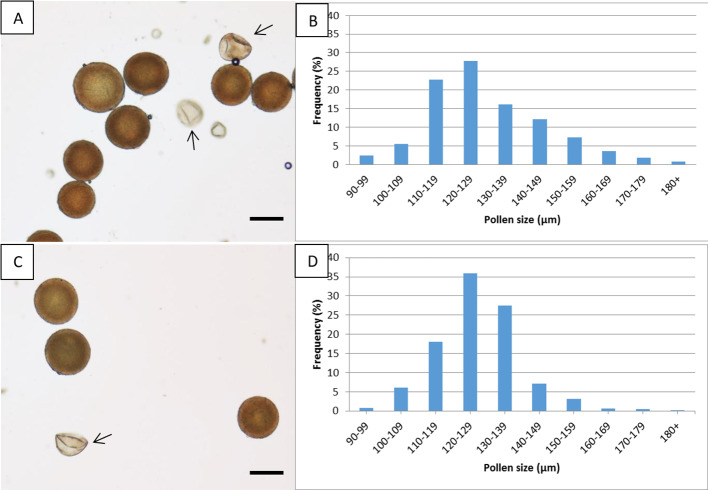
The percent of pollen viability ± se of the allopentaploid N-149 (29.9 ± 0.06%) differed significantly from that of the allohexaploid N-134 (49.5 ± 0.01%), as shown by a t-test (significant at P = 0.019). Images showing viable and aborted pollen grains are presented in Figures 4A, C . The average ± se pollen diameters were 129.5 ± 0.9 and 126.8 ± 0.6 μm for the allopentaploid N-149 and the allohexaploid N-134, respectively. No statistically significant differences were observed for pollen diameter, as determined by an unpaired two-tailed t-test (significant at P = 0.248) between the allopentaploid and allohexaploid lines. The diameters of most of the stainable pollen grains of the allopentaploid N-149 and allohexaploid N-134 (~67% and ~81%, respectively) lay in the range of 110–139 μm, whereas for both allopolyploids, 7% had a smaller diameter than the average, and the remaining diameters exceeded the average range. Pollen diameter frequency distribution is presented in Figures 4B, D .
Figure 4.
Pollen viability and diameter. Images showing viable and aborted (arrow) pollen grains and pollen diameter distribution in the allopentaploid N-149 (A, B) and the allohexaploid N-134 (C, D). Scale bar, 100 µm.
Discussion
The results of this study strongly support Winge's theory of “hybridization followed by chromosome doubling” by showing high ploidy, i.e., 6x instead 3x, following interspecific-interploid hybridization. No hybrids were obtained for the H. undatus × H. megalanthus cross, and in the reciprocal cross only allopentaploids, allohexaploids, and 4-5x and 5-6x aneuploids were obtained. It has been proposed that the main route to polyploidization is through unreduced gametes and the formation of triploids (Bretagnolle and Thompson, 1995). The “triploid block” acts as a reproductive barrier in the endosperm, preventing backcrossing with the parental species. Triploids were not obtained for the H. megalanthus × H. undatus crosses, but crosses between the diploid H. monacanthus as the maternal species and the tetraploid H. megalanthus as the paternal species resulted in triploid and 3x-aneuploid hybrids, while the reciprocal cross yield only pentaploids (Tel-Zur et al., 2003; Tel-Zur et al., 2004). These findings suggest that H. monacanthus is more closely related to H. megalanthus than to H. undatus. Moreover, back-crosses, using triploid H. monacanthus × H. megalanthus hybrids, were indeed obtained (Tel-Zur et al., 2012), thereby showing that the “triploid block” does not exist in Hylocereus species.
As mentioned above, we have previously shown that the tetraploid H. megalanthus produces both normal (reduced) and unbalanced and unreduced gametes, but the diploid H. undatus produces only normally reduced gametes (Lichtenzveig et al., 2000; Tel-Zur et al., 2003). Thus, the presence of allohexaploids and the absence of allotriploids in H. megalanthus × H. undatus cross combinations provide empirical evidence supporting our hypothesis that chromosome doubling occurred at the zygote formation stage or very soon thereafter. We thus suggest that chromosome doubling occurs as a spontaneous process of mitotic cell division without cytokinesis. The sources of the 4x-5x aneuploids are probably unbalanced gametes and unbalanced-unreduced gametes, respectively, from the tetraploid H. megalanthus (Lichtenzveig et al., 2000; Tel-Zur et al., 2003). The 6x aneuploids are probably a result of a fertilization event with one reduced but unbalanced gamete from H. megalanthus followed by chromosome doubling. In this context, it is interesting to note that chromosome doubling at the zygote level can be artificially induced. For example, high temperatures induced the formation of synthetic triploids and tetraploids through embryo sac and zygote embryo chromosome doubling in Populus simonii and its hybrids (Guo et al., 2017). Similarly, corn plants exposed to high temperatures after pollination produced diploid, tetraploid, and octoploid seedlings (Randolph, 1932).
One of the crucial questions in allopolyploidy formation relates to the genetic consequences of combining two genetic systems into a common nucleus (Huxley, 1942; McClintock, 1984; Parisod et al., 2009; Lloyd et al., 2018). Subsequent to the successful formation of a new allopolyploid, its establishment depends on the following factors: 1) the correct control of chromosome pairing, a pre-requisite for the production of viable seeds and pollen grains, 2) epigenetic factors, and 3) ‘homoeologous exchanges,' i.e., exchanges of large chromosomal segments between homoeologous chromosomes via the meiotic homologous recombination pathway (Jenczewski and Alix, 2004; Feldman and Levy, 2009; Madlung and Wendel, 2013; Glover et al., 2016; Steige and Slotte, 2016; Liang and Schnable, 2018). Natural allopolyploids are stable and well adapted, whereas synthetic allopolyploids can exhibit incompatible interactions between parental genomes, i.e., intergenomic incompatibilities (Comai, 2000). Cytomixis, as was observed here in both allopentaploids and allohexaploids ( Figure 1 ), can be the consequence of such intergenomic incompatibilities, but in Hylocereus species—like in other species—the biological and evolutionary significance of cytomixis is not known (Mursalimov and Deineko, 2018).
In a parallel but opposite scenario to that reported here, reciprocal crosses between the diploid Arabidopsis thaliana and the tetraploid A. arenosa resulted in aborted seeds or unsuccessful pollen germination, but crosses between the tetraploid A. thaliana as the maternal parent and the tetraploid A. arenosa restored seed viability (Bushell et al., 2003). Seeds from reciprocal crosses between the diploid A. lyrata and A. arenosa failed to germinate, due to failure of endosperm cellularization in the A. lyrata × A. arenosa cross, while in the reciprocal cross the endosperm cellularized precociously (Lafon-Placette et al., 2017). Interestingly, crosses between the tetraploid A. lyrate and the diploid A. arenosa were fully viable, but crosses between the tetraploid A. arenosa and the diploid A. lyrate did not produce viable seeds (Lafon-Placette et al., 2017). Endosperm defects can explain the failure of the cross and the cross direction, illustrating very clearly the role of the endosperm in hybridization barriers.
In flowering plants, sexual reproduction is characterized by two separate gametic fusion events that form the embryo and the endosperm. In diploid crosses, these events result in a diploid embryo and most commonly a triploid endosperm, with a ratio of 1:1 and 2:1 of maternal-to-paternal genomes, respectively. A fundamental question regarding embryo survival and seed development following interspecific-interploidy crosses centers on maternal or paternal genomic excess and the cases in which the embryo and the endosperm can develop with different maternal/paternal genome ratios. To address this question, Johnston et al. (1980) put forward the endosperm balance number (EBN) theory, which stated that rather than an absolute numerical ploidy, each species has an effective genome ratio, which must be 2:1 maternal/paternal for normal endosperm development. This oversimplification of the very important biological phenomenon of zygote formation and embryo development was useful for plant breeders for predicting the success of a cross, especially among Solanum species (Carputo et al., 1997; Jansky, 2006), but failed to predict crossing success in other plant species.
We note here that ploidy determination in immature embryos was not directly addressed in this study. Hylocereus embryos – even completely mature embryos – are very small, and there is thus not enough tissue to provide sufficient material even for a single flow cytometry run. Nonetheless, even though genome duplication can occur spontaneously in somatic tissues, the probability that such an event would occur in all the studied true hybrids is – in our opinion – extremely low. Moreover, there were no differences between the morphologies of the hybrid plants at the vegetative level and, similarly, no differences in the flow cytometry results for tissues collected from the tips of different newly developed branches; both these findings indicate the low probability of chimera events in the new hybrids.
Determination of endosperm ploidy in the allopolyploids was beyond the scope of this study, but we did calculate the expected endosperm ploidy of the allopentaploid and allohexaploid H. megalanthus × H. undatus crosses described above as 9x (8m:1p) and 5x (4m:1p), respectively, with no production of 3x hybrids with an endosperm ploidy of 5x (4m:1p) or 10x (8m:2p) ( Table 4 ). Likewise, no hybrids were obtained in the reciprocal cross H. undatus × H. megalanthus with an expected endosperm ploidy of 4x (2m:2p)—even when the embryo rescue technique was used ( Table 4 ). Whole genome duplication (hexaploid rather than triploid) did not change the maternal/paternal ratio in the embryo ( Table 4 ), but it did change the maternal-to-endosperm-to-embryo genome ratios by doubling the ploidy of the embryo, which probably allowed normal embryonic development. It therefore seems likely that only the above-described ratios between the endosperm and/or the maternal tissues and/or the embryo permit the survival of the new hybrid embryo in H. megalanthus × H. undatus crosses. Studies focusing on the nature of zygotic genome activation have shown that the maternal and paternal genomes contribute equally to early plant embryo development and that early embryogenesis is mostly under zygotic control (Nodine and Bartel, 2012). In rice, for example, polyploid zygotes with a maternal excess developed normally, whereas most polyploid zygotes with a paternal excess showed developmental arrest, indicating that paternal and maternal genomes act synergistically to allow zygote development but probably with distinct functions for each (Toda et al., 2018). The findings that only allopentaploid and allohexaploids were obtained in the H. megalanthus × H. undatus cross combination and the lack of allotriploids in both reciprocal crosses suggest that there are (still) unknown factor(s) preventing the formation and development of a triploid embryo.
Table 4.
Theoretical outcome of interspecific-interploidy crosses following paternal and maternal excess.
| Parental ploidy | Ploidy (m/p) | ||||
|---|---|---|---|---|---|
| Maternal (m) | Paternal (p) | Predicted endosperm ploidy (m/p) | Hybrid ploidy obtained (m/p) | ||
| Paternal excess (p) | 2x | 4x | 4x (2m:2p) | NH 1 | |
| Maternal excess (m) | 4x | 2x | Normal (reduced) embryo sac | ||
| 5x (4m:1p) | NH 2 | ||||
| Unreduced embryo sac 3 | |||||
| 9x (8m:1p) | 5x (4m:1p) | ||||
| Normal (reduced) embryo sac followed by zygote/embryo chromosome doubling 4 | |||||
| 5x (4m:1p) | 6x (4m:2p) | ||||
| Both endosperm and zygote/embryo chromosome doubling 5 | |||||
| 10x (8m:2p) | 6x (4m:2p) | ||||
2x: H. undatus, 4x: H. megalanthus.
NH - No hybrids were obtained from the H. undatus × H. megalanthus cross, even when we used an embryo rescue technique.
NH- No H. megalanthus × H. undatus allotriploids were obtained.
Assuming that all the embryo sac cells (including the egg cell) result from unreduced gamete formation, since megasporogenesis occurs before megagametogenesis.
Ploidy in the embryo sac cells was assumed to be reduced, while the allohexaploids are a result of chromosome doubling.
Chromosome doubling occurs in the endosperm and zygote/embryo, resulting in 10x and 6x, respectively.
Hylocereus species bear large flowers (30-35 cm diameter) with numerous mega- and microspores, which simplify technical manipulations (Nerd and Mizrahi, 1997), such as hand cross pollination and the embryo rescue technique (Cisneros et al., 2013). Thus, from a technical point of view, Hylocereus species can be used as good model plants in polyploid studies. Among the 32 true hybrids obtained, all eight (25%) allohexaploids were obtained from embryo rescue at 10 DAP (pro-embryo stage), showing that the chromosome doubling event occurs very early in embryo development. We note here that in a previous report (Cisneros et al., 2013) two plants regenerated from embryo rescue 30 DAP following H. megalanthus × H. undatus crossing were reported in error as a diploid and a triploid, whereas they were, in fact, tetraploids. Tissues in Hylocereus species are rich in polysaccharides, making the flow cytometric analysis a challenging task. Subsequent to that report (Cisneros et al., 2013), and in view of the limitations of the published protocols, we developed an accurate protocol (Li et al., 2017) aimed to solve, simplify, and streamline flow cytometry for ploidy determination in polysaccharide-rich tissues; that procedure was used in this work.
One of the key questions following interspecific and interspecific-interploid hybridization is the fertility of the resulting hybrids. Fruit morphology verifies hybrid origin but also provides the essential information that these hybrids (both allopentaploid and allohexaploid) can grow and can set fruit and seeds. Fruit weights in the allohexaploids were lower than those in the allopentaploids, a finding that is in line with previous reports in auto- and allopolyploid Hylocereus species, showing that fruit weight declined as the ploidy level increased (Tel-Zur et al., 2004; Tel-Zur et al., 2012; Cohen et al., 2013). All the allopolyploids set both viable and aborted seeds, and the allopentaploids and allohexaploids produced both viable and aborted pollen grains, showing them to be partially fertile. Pollen viability was about 30% and 50% for the studied allopentaploids and allohexaploids, respectively, with most of the pollen grains presenting – in both cases (67% and 81%, respectively) – a large diameter of 110 to 139 μm. The pollen grains of H. undatus exhibited a uniform diameter of 70–80 μm, but those of H. megalanthus showed a wide variation in diameter, with values lying between 90 and 190 μm. These differences in diameter represent different ploidy levels in the viable pollen grains and are, most probably, due to meiotic abnormalities, such as the formation of unreduced gametes, unbalanced chromosome segregation, or cytomixis (Ramanna and Jacobsen, 2003; Shamina, 2005; Mursalimov and Deineko, 2018). All the above findings indicate that both allopentaploids and allohexaploids can reproduce by sexual reproduction, since both produce a certain percent of viable male and female gametes. Taken as a whole, our data indicate that interspecific-interploid hybridization among Hylocereus does not culminate in a genetic dead-end in the F1 generation.
Conclusions
On the basis of our findings, we concluded that the allohexaploids were produced as a result of “hybridization followed by chromosome doubling” during the very early stage of embryo development, which probably facilitates the survival of hybrids with specific genomic combinations and maternal/paternal ratios in the endosperm.
Interspecific-interploidy Hylocereus hybrids constitute a unique model system for the study of polyploidization mechanisms, providing experimental support for somatic chromosome doubling at the zygote or very early embryo development stages, although the nature of the trigger(s) as well as the details of all the process remain unclear. Questions as to the roles of the endosperm in the control of the doubling and in the initial development of the embryo remain open. Further experimental work, including studies on meiotic stability, synapsis, pairing, and recombination, could provide new insights into the possible establishment of these recently produced hybrids with high ploidy.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
NT-Z conceived and planned the experiments. JM and UZ performed the experiments and analyzed the data. NT-Z wrote the manuscript and YM performed part of the experiments and revised the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Ms. Inez Mureinik for editing the manuscript and Dr. Benyamin Rosental and Ms. Orly Gershoni-Yahalom, Faculty of Health Sciences and Regenerative Medicine and Stem Cell Research Center, Ben-Gurion University of the Negev, for providing flow cytometry assistance.
References
- Banerji I., Sen S. (1955). A contribution to the cytology and embryology of Hylocereus undatus (Haw) Br. & R. Bull. Bot. Soc Bengal 8, 18–23. [Google Scholar]
- Barthlott W., Hunt D. R. (1993). “Cactaceae,” in The Families and the Genera of Vascular Plants, vol. 2 . Ed. Kubitzki K. (Berlin: Springer; ), 161–196. [Google Scholar]
- Belling J. (1921). On counting chromosomes in pollen mother cells. Am. Nat. 55, 573–574. 10.1086/279843 [DOI] [Google Scholar]
- Bretagnolle F., Thompson J. D. (1995). Tansley Review No. 78. Gametes with the somatic chromosome number: Mechanisms of their formation and role in the evolution of autopolyploid plants. New Phytol. 129, 1–22. 10.1111/j.1469-8137.1995.tb03005.x [DOI] [PubMed] [Google Scholar]
- Bushell C., Spielman M., Scott R. J. (2003). The basis of natural and artificial postzygotic hybridization barriers in Arabidopsis species. Plant Cell 15, 1430–1442. 10.1105/tpc.010496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carputo D., Barone A., Cardi T., Sebastiano A., Frusciante L., Peloquin S. J. (1997). Endosperm balance number manipulation for direct in vivo germplasm introgression to potato from a sexually isolated relative (Solanum commersonii Dun.). PNAS 94, 12013–12017. 10.1073/pnas.94.22.12013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisneros A., Tel-Zur N. (2010). Embryo rescue and plant regeneration following interspecific crosses in the genus Hylocereus (Cactaceae). Euphytica 174, 73–82. 10.1007/s10681-010-0135-x [DOI] [Google Scholar]
- Cisneros A., Garcia R. B., Tel-Zur N. (2011). Ovule morphology, embryogenesis and seed development in three Hylocereus species (Cactaceae). Flora 206, 1076–1084. 10.1016/j.flora.2011.07.013 [DOI] [Google Scholar]
- Cisneros A., Garcia R. B., Tel-Zur N. (2013). Creation of novel interspecific-interploid Hylocereus hybrids (Cactaceae) via embryo rescue. Euphytica 189, 433–443. 10.1007/s10681-012-0800-3 [DOI] [Google Scholar]
- Clausen R. E., Goodspeed T. H. (1925). Interspecific hybridization in Nicotiana. II. A tetraploid glutinosa-tabacum hybrid, an experimental verification of Winge's hypothesis. Genetics 10, 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen H., Fait A., Tel-Zur N. (2013). Morphological, cytological and metabolic consequences of autopolyploidization in Hylocereus (Cactaceae) species. BMC Plant Biol. 13, 173. 10.1186/1471-2229-13-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L. (2000). Genetic and epigenetic interactions in allopolyploid plants. Plant Mol. Biol. 43, 387–399. 10.1023/A:1006480722854 [DOI] [PubMed] [Google Scholar]
- Doyle J. J., Egan A. N. (2010). Dating the origins of polyploidy events. New Phytol. 186, 73–85. 10.1111/j.1469-8137.2009.03118.x [DOI] [PubMed] [Google Scholar]
- Doyle J. J., Sherman-Broyles S. (2017). Double trouble: taxonomy and definitions of polyploidy. New Phytol. 213, 487–493. 10.1111/nph.14276 [DOI] [PubMed] [Google Scholar]
- Feldman M., Levy A. A. (2009). Genome evolution in allopolyploid wheat – a revolutionary reprogramming followed by gradual changes. J. Genet. Genomics 36, 511–518. 10.1016/S1673-8527(08)60142-3 [DOI] [PubMed] [Google Scholar]
- Gao S.-m., Yang A.-H., Zhang F., Fan L.-J., Zhou Y. (2019). The strong competitive role of 2n pollen in several polyploidy hybridization in Rosa hybrid . BMC Plant Biol. 9, 127. 10.1186/s12870-019-1696-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover N. M., Redestig H., Dessimoz C. (2016). Homoelogs: what are they and how do we infer them? Trends Plant Sci. 21, 609–621. 10.1016/j.tplants.2016.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Xu W., Zhang Y., Zhang J., Wei Z. (2017). Inducing triploids and tetraploids with high temperatures in Populus sect. Tacamahaca . Plant Cell Rep. 36, 313–326. 10.1007/s00299-016-2081-0 [DOI] [PubMed] [Google Scholar]
- Harlan J. R., deWet J. M. J. (1975). O. Winge and a prayer: the origins of polyploidy. Bot. Rev. 41, 361–390. 10.1007/BF02860830 [DOI] [Google Scholar]
- Huxley J. (1942). Evolution: the modern synthesis (London: Harper and Brothers; ). [Google Scholar]
- Jansky S. (2006). Overcoming hybridization barriers in potato. Plant Breed. 125, 1–12. 10.1111/j.1439-0523.2006.01178.x [DOI] [Google Scholar]
- Jenczewski E., Alix K. (2004). From diploids to allopolyploids: the emergence of efficient pairing control genes in plants. Crit. Rev. Plant Sci. 23, 21–45. 10.1080/07352680490273239 [DOI] [Google Scholar]
- Johnston S. A., den Nijs T. P. M., Peloquin S. J., Hanneman R. E., Jr. (1980). The significance of genetic balance to endosperm development in interspecific crosses. Theor. Appl. Genet. 57, 5–9. 10.1007/BF00276002 [DOI] [PubMed] [Google Scholar]
- Lafon-Placette C., Johannessen I. M., Hornslien K. S., Ali M. F., Bjerkan K. N., Bramsiepe J., et al. (2017). Endosperm-based hybridization barriers explain the pattern of gene flow between Arabidopsis lyrate and Arabidopsis arenosa in Central Europe. PNAS, 114 (6), E1027–E1035. 10.1073/pnas.1615123114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch L. J., Bennett M. D. (1997). Polyploidy in angiosperms. Trends Plant Sci. 2, 470–476. 10.1016/S1360-1385(97)01154-0 [DOI] [Google Scholar]
- Li D., Cisneros A., Tel-Zur N. (2017). An improved protocol for flow cytometry analysis of dragon fruit (Hylocereus spp.) – Species with a high polysaccharide content. Sci. Hortic. 220, 130–133. 10.1016/j.scienta.2017.03.033 [DOI] [Google Scholar]
- Liang Z., Schnable J. C. (2018). Functional divergence between subgenomes and gene pairs after whole genome duplication. Mol. Plant 11, 388–397. 10.1016/j.molp.2017.12.010 [DOI] [PubMed] [Google Scholar]
- Lichtenzveig J., Abbo S., Nerd A., Tel-Zur N., Mizrahi Y. (2000). Cytology and mating systems in the climbing cacti Hylocereus and Selenicereus . Am. J. Bot. 87, 1058–1065. 10.2307/2657005 [DOI] [PubMed] [Google Scholar]
- Lloyd A., Blary A., Charif D., Charpentier C., Tran J., Balzergue S., et al. (2018). Homoelogous exchanges cause extensive dosage-dependent gene expression changes in an allopolyploid crop. New Phytol. 217, 367–377. 10.1111/nph.14836 [DOI] [PubMed] [Google Scholar]
- Madlung A., Wendel J. F. (2013). Genetic and epigenetic aspects of polyploid evolution in plants. Cytogenet. Genome Res. 140, 270–285. 10.1159/000351430 [DOI] [PubMed] [Google Scholar]
- McClintock B. (1984). The significance of responses of the genome to challenge. Science 226, 792–801. 10.1126/science.15739260 [DOI] [PubMed] [Google Scholar]
- Mizrahi Y., Nerd A. (1999). “Climbing and columnar cacti,” in New arid land fruit crops. Perspective in new crops and new uses. Ed. Janick J. (Alexandria, VA: ASHS Press; ), 358–366. [Google Scholar]
- Morgan W. G., King I. P., Kock S., Harper J. A., Thomas H. M. (2001). Introgression of chromosomes of Festuca arundinacea var. glaucescens into Lolium multiflorum revealed by genomic in situ hybridization (GISH). Theor. Appl. Genet. 103, 696–701. 10.1007/s001220100634 [DOI] [Google Scholar]
- Mursalimov S., Deineko E. (2018). Cytomixis in plants: facts and doubts. Protoplasma 255, 719–731. 10.1007/s00709-017-1188-7 [DOI] [PubMed] [Google Scholar]
- Nerd A., Mizrahi Y. (1997). Reproductive biology of cactus fruit crops. Hortic. Rev. 18, 321–346. [Google Scholar]
- Newton W. C. F., Darlington C. D. (1929). Meiosis in polyploids. J. Genet. 21, 1–56. 10.1007/BF02983355 [DOI] [Google Scholar]
- Newton W. C. F., Pellew C. (1929). Primula kewensis and its derivatives. J. Genet. 20, 405–467. 10.1007/BF02983158 [DOI] [Google Scholar]
- Nodine M. D., Bartel D. P. (2012). Maternal and paternal genomes contribute equally to the transcriptome of early plant embryos. Nature 482, 92–U120. 10.1038/nature10756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn T. C., Pires J. C., Auger D. L., Chen Z. J., Lee H.-S., et al. (2003). Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 19, 141–147. 10.1016/s0168-9525(03)00015-5 [DOI] [PubMed] [Google Scholar]
- Parisod C., Salmon A., Zerjal T., Tenaillon M., Grandbastien M.-A., Ainouche M. (2009). Rapid structural and epigenetic reorganization near transposable elements in hybrid and allopolyploid genomes in Spartina . New Phytol. 184, 1003–1015. 10.1111/j.1469-8137.2009.03029.x [DOI] [PubMed] [Google Scholar]
- Ramanna M. S., Jacobsen E. (2003). Relevance of sexual polyploidization for crop improvement – A review. Euphytica 133, 3–18. 10.1023/A:1025600824483 [DOI] [Google Scholar]
- Ramsey J., Schemske D. W. (1998). Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29, 467–501. 10.1146/annurev.ecolsys.29.1.467 [DOI] [Google Scholar]
- Randolph L. F. (1932). Some effects of high temperature on polyploidy and other variations in maize. Proc. Natl. Acad. Sci. U. S. A. 18, 22–229. 10.1073/pnas.18.3.222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler M. C., Carvalho C. R., Clarindo W. R. (2016). The polyploid and its key role in plant breeding. Planta 243, 281–296. 10.1007/s00425-015-2450-x [DOI] [PubMed] [Google Scholar]
- Shamina N. V. (2005). A catalogue of abnormalities in the division spindles of higher plants. Cell Biol. Internat. 29, 384–391. 10.1016/j.cellbi.2005.02.004 [DOI] [PubMed] [Google Scholar]
- Soltis D. E., Soltis P. S. (1993). Molecular data and the dynamic nature of polyploidy. Crit. Rev. Plant Sci. 12, 243–273. 10.1080/07352689309701903 [DOI] [Google Scholar]
- Soltis P. S., Soltis D. E. (2000). The role of genetic and genomic attributes in the success of polyploid. PNAS 97, 7051–7057. 10.1073/pnas.97.13.7051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis P. S., Liu X., Marchant D. B., Visger C. J., Soltis D. E. (2014). Polyploidy and novelty: Gottlieb's legacy. Phil. Trans. R. Soc B. 369, 20130351. 10.1098/rstb.2013.0351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer J. L. (1955). A cytological study of the Cactaceae of Puerto Rico. Bot. Gaz. 117, 33–37. 10.1086/335887 [DOI] [Google Scholar]
- Stebbins G. L. (1971). Chromosomal evolution in higher plants (New York: Columbia University Press; ). [Google Scholar]
- Steige K. A., Slotte T. (2016). Genomic legacies of the progenitors and the evolutionary consequences of allopolyploidy. Curr. Opin. Plant Biol. 30, 88–93. 10.1016/j.pbi.2016.02.006 [DOI] [PubMed] [Google Scholar]
- Tel-Zur N., Abbo S., Bar-Zvi D., Mizrahi Y. (2003). Chromosome doubling in vine cacti hybrids. J. Hered. 94, 329–333. 10.1093/jhered/esg064 [DOI] [PubMed] [Google Scholar]
- Tel-Zur N., Abbo S., Bar-Zvi D., Mizrahi Y. (2004). Genetic relationships among Hylocereus and Selenicereus vine cacti (Cactaceae): evidence from hybridization and cytological studies. Ann. Bot. 94, 527–534. 10.1093/aob/mch183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tel-Zur N., Mizrahi Y., Cisneros A., Mouyal J., Schneider B., Doyle J. J. (2011). Phenotypic and genomic characterization of a vine cactus collection (Cactaceae). Genet. Resour. Crop Evol. 58, 1075–1085. 10.1007/s10722-010-9643-8 [DOI] [Google Scholar]
- Tel-Zur N., Dudai M., Raveh E., Mizrahi Y. (2012). Selection of interspecific hybrids (Hylocereus spp.) for self-compatibility. Plant Breed. 131, 681–685. 10.1111/j.1439-0523.2012.01992.x [DOI] [Google Scholar]
- Toda E., Ohnishi Y., Okamoto T. (2018). An imbalanced parental genome ration affects the development of rice zygotes. J. Exp. Bot. 69, 2609–2619. 10.1093/jxb/ery094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y., Mizrachi E., Marchal K. (2017). The evolutionary significance of polyploidy. Nat. Rev. Genet. 18, 411–424. 10.1038/nrg.2017.26 [DOI] [PubMed] [Google Scholar]
- Winge Ö. (1917). The chromosomes. Their numbers and general importance. Compt. Rend. Trav. Du. Lab. Carlsberg. 13, 131–275. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.