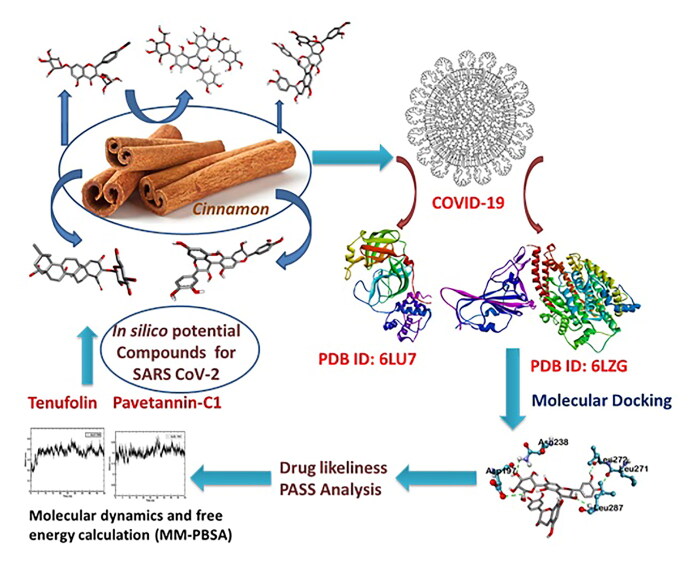
Abstract
Cinnamon has been utilized to remedy a lot of afflictions of humans. Literary works illustrate that it possesses numerous biological activities. Our research study is intended to recognize the phyto-derived antiviral substances from Cinnamon against COVID-19 main protease enzyme and to understand the in silico molecular basis of its activity. In the present study, 48 isolates compounds from Cinnamon retrieved from the PubMed database, are subjected to docking analysis. Docking study was performed using Autodock vina and PyRx software. Afterwards, admetSAR, as well as DruLiTo servers, were used to investigate drug-likeness prophecy. Our study shows that the nine phytochemicals of Cinnamon are very likely against the main protease enzyme of COVID-19. Further MD simulations could identify Tenufolin (TEN) and Pavetannin C1 (PAV) as hit compounds. Utilizing contemporary strategies, these phyto-compounds from a natural origin might establish a reliable medication or support lead identification. Identified hit compounds can be further taken for in vitro and in vivo studies to examine their effectiveness versus COVID-19.
Keywords: Cinnamon, SARS CoV-2, main protease, spike glycoprotein, autodock
Graphical Abstract
Communicated by Ramaswamy H. Sarma
1. Introduction
Several representatives of the Coronaviridae family circulate in the human community and typically induce moderate respiratory illness (Corman et al., 2019). In comparison, severe acute respiratory coronavirus syndrome (SARS-CoV) and Middle East respiratory coronavirus syndrome (MERS-CoV) are introduced from animals to humans and lead to serious respiratory diseases in affected persons, SARS and MERS, respectively (Fehr et al., 2017). SARS originated in Guangdong province, China in 2002 and its eventual global expanded (de Wit et al., 2016; WHO, 2003). Chinese horseshoe bats act as SARS-CoV reservoir hosts (Lau et al., 2005; Li et al., 2005). Intermediate hosts such as civet cats and raccoon puppies, often marketed as food outlets in Chinese wet markets, enabled human transmission (Guan et al., 2003). Currently, no new antivirals or licensed vaccinations are available to counter SARS, although traditional preventive steps, including travel bans although patient isolation, eventually stopped the SARS pandemic in 2002 and 2003.
New respiratory infectious disease appeared in Wuhan, Hubei Province, China (Huang et al., 2020; Wang et al., 2020; Zhu et al., 2020). An initial outbreak epidemic was connected to seafood center, possibly involved in animal exposure. Eventually, human-to-human infection emerged (Chan et al., 2020) and the now-called coronavirus disease 19 (COVID-19) circulated exponentially in China. A unique coronavirus, SARS-coronavirus 2 (SARS-CoV-2), closely associated to SARS-CoV, was found in clinics and is suspected to be the emerging lung disease’s etiologic agent (Chan et al., 2020). On Feb 12, 2020, China recorded 44,730 lab-confirmed disorders, including 8,204 serious cases and 1,114 deaths (WHO, 2020). Illnesses are also identified in 24 non-China nations, correlated with foreign travel. Whether sequencing correlations among SARS-CoV-2 and SARS-CoV translates into related biological properties, namely outbreak possibility, is currently unclear (Munster et al., 2020).
Virus entry in cells is regulated by spike (S) glycoprotein; spike 1 (S1) surface unit helps the virus to be connected to cellular receptors. Cellular proteases cleave the S protein at the S1/S2 and S20 locations to allow viral particle entry. The viral capside is then fused with the cellular membrane, a subunit S2-guided process (Hoffmann et al., 2020). It was established that angiotensin-converting enzyme 2 (ACE2) (15) mediates SARS-CoV entry and that serine protease TMPRSS2 is responsible for S protein cleavage (Glowacka et al., 2011; Li et al., 2003)(Hasan et al., 2020). Analysis of receptor binding motif (RBM) sequences within the receptor binding domain reveals that it is accountable for binding to ACE2 and that residues have been preserved by SARS-CoV and SARS-CoV-2, indicating that binding to ACE2 could be identical while the same residues are absent in other coronaviruses (Ge et al., 2013; Menachery et al., 2019). Some human ACE2 antibodies avoided SARS-CoV and SARS-CoV-2 infections (Boopathi et al., 2020).
Proteases are one of the important and best studied proteins in corona viruses (Khan, Jha, et al., 2020). It is very essential for the processing poly proteins that are translated from viral RNA (Hilgenfeld, 2014). Inhibition of this viral protease enzyme wills results into the blocking of viral replication. In this scenario, the development of inhibitors to the SARS-CoV-2 main protease gains importance in the drug discovery process against COVID-19. Recently Zhang et al solved the crystal structure of inhibitor bound SARS-CoV-2 main protease (Khan, Zia, et al., 2020; Zhang et al., 2020).
The development of new therapeutics is an expensive and time consuming process. Normally it will take years to get the newly developed drug for the treatment (Elfiky, 2020; Enayatkhani et al., 2020; Muralidharan et al., 2020; Pant et al., 2020; Wahedi et al., 2020). Drug repurposing is an efficient strategy in medicinal chemistry to bring faster and effective solutions to the unmet medical needs (Gupta et al., 2020) (Elfiky & Azzam, 2020; Sinha et al., 2020). Repurposing of drugs derived from natural origin is considered as crucial therapeutic approach for the treatment of COVID-19 (Enmozhi et al., 2020), considering the fast pace of its spread around the world (Elmezayen et al., 2020).
In the present research, we have chosen Cinnamon, which is a potent source of antiviral agents (Aanouz et al., 2020). Cinnamon is a traditional Indian medicine which is being used from hundreds of year to relieve various lung-related disorders includes pneumonia, infectious disease, as well as malignant pleural effusion (Lai et al., 2018; Townsend et al., 2013). Recently, several studies also provided scientific data to support and unveil its antiparasitic, antihypertensive, anti-diabetic, anti-hyperlipedimic, anti-oxidant, anti-inflammatory, analgesic, antimicrobial, antiviral, antitumor, anti-hypertension, anti-hyperlipemic, gastro-protective and immunomodulation activities.
Based on the above properties of Cinnamon, this research aimed to show a variety of active compounds across all Cinnamon varities and decide whether and how they interact with proteins i.e. main protease (Joshi et al., 2020) and spike protein, that are essential in the management of SARS- CoV-2.
2. Materials and methods
2.1. Data source
Within this research, a dataset of active phytochemicals were acquired from Indian Medicinal Plants, Phytochemistry, and also Therapeutics data source (Chaudhuri et al., 2018; Mohanraj et al., 2018).
2.2. Docking studies
2.2.1. Preparation of protein
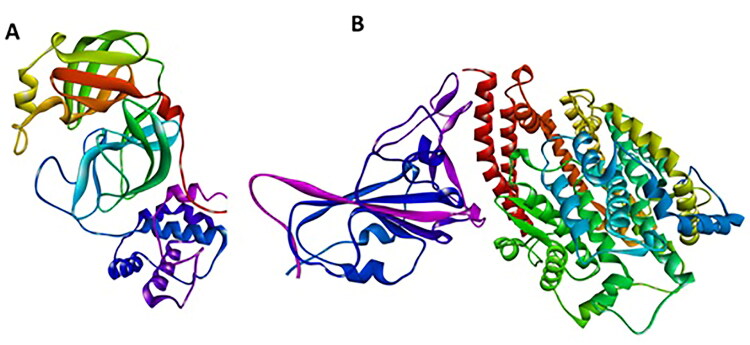
The X-ray crystal structures of Main protease and spike receptor domain complexed with ACE2 (PDB ID: 6LU7, 6LZG) were downloaded from the RCSB PDB (Protein Data Bank) database (Islam et al., 2020; Sarma et al., 2020). The Graphical User Interface program ‘‘Auto-Dock Tools’’ was used to prepare, run, and analyze the docking simulations. Kollman united atom charges, solvation parameters and polar hydrogen’s were added to the receptor for the preparation of protein in docking simulation. Since ligands are not peptides, Gasteiger charge was assigned and then non-polar hydrogens were merged. AutoDock requires pre-calculated grid maps, one for each atom type, present in the ligand being docked as it stores the potential energy arising. This grid must surround the region of interest (active site) in the macromolecule. (Morris et al., 2009) (Figure 1).
Figure 1.
Three dimensional crystal structure of the molecular target, COVID-19. (A) Main protease (6LU7) (B) Spike receptor-binding domain complexed with its receptor ACE2.
2.2.2. Preparation of ligands and analysis of drug likeliness
The crystal 3D structure of the following active compounds of Cinnamon were retrieved from PubChem database (O'Boyle et al., 2011). Drug-likeliness properties of ligands were analyzed for the selected active compounds using DruLiTo software (Pangastuti et al., 2016).
2.2.3. Validation of target protein-ligand complex structures
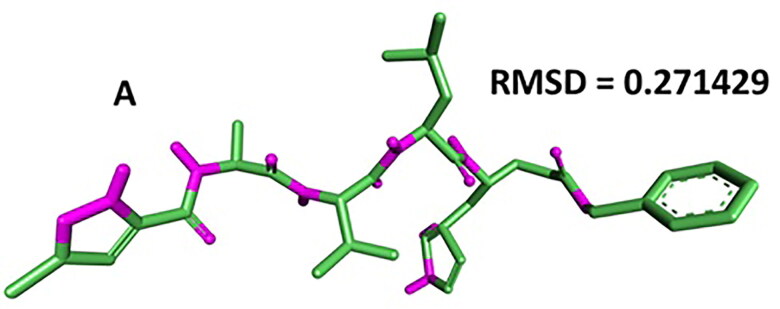
Autodock 4.0 methodology was validated with the respective co-crystallized ligands of target proteins to ensure the virtual screening process. Autodock 4.0 represents valid RMSD score and accurate binding with target receptor. In this context, Mpro (PDB ID: 6LU7) was tested with its co-crystalized inhibitor N3 (n-[(5-methylisoxazol-3- yl)carbonyl]alanyl-l-valyl-n∼1∼-((1r,2z)-4-(benzyloxy)-4-oxo-1-{[(3r)-2-oxopyrrolidin-3-yl]methyl}but-2-enyl)-l-leucinamide) (Agostino et al., 2009; Cosconati et al., 2010).
2.2.4. Prediction of protein active site
Accurate prediction of active sites is an important tool in bioinformatics. In this study, for spike protein (PDB ID: 6LZG), the active site was predicted by using Biovia Drug discovery studio visualizer 2020 (Design, 2014).
2.2.5. Compound screening using PyRx program
Molecular testing of all the compound libraries was performed using PyRx software by autodock wizard as the engine for docking (Dallakyan & Olson, 2015). During the docking period, the ligands were considered to be flexible, and the protein was supposed to be rigid. The configuration file for the grid parameters was generated using Grid box for 6LU7 (x = −17.59, y = 15.81, z = 63.53) and 6LZG (x = −26.73, y = 8.32, z = −14.07) in PyRx respectively (Hall & Ji, 2020). The application was also used to know/predict the amino acids in the active site of the protein that interact with the ligands. The results less than 1.0 Å in positional root-mean-square deviation (RMSD) were considered ideal and clustered together for finding the favourable binding. The highest binding energy (most negative) was recognised as the ligand with maximum binding affinity. Visual examination of the docking site was performed using Biovia Drug discovery studio 2019, and the results were validated using Autodock Vina (Seeliger & de Groot, 2010).
2.3. ADMET analysis
ADMET of the ligands is pharmacokinetic properties calculation that is required to be examined to establish their function inside the body. The ADMET inheritance of the ligands was studied, making use of admetSAR (Cheng et al., 2012; Yang et al., 2019).
2.4. PASS computer program
Prophecy of Cinnamon for antiviral activity was created with the assistance of software, PASS. PASS is a computer system based program utilized for the prognosis of various sorts of physiological actions for multiple compounds consisting of phytoconstituents. The estimated activity of a substance is predicted as probable activity (Pa) and probable inactivity (Pi). The substances revealing Pa higher than Pi are actually the only components thought about as feasible for a specific medical activity (Goel et al., 2011; Khurana et al., 2011; Mittal et al., 2008).
2.5. Molecular dynamics and free energy calculation (MM-PBSA)
The crystal structure of Main protease (6LU7) and Spike receptor-binding domain complexed with its receptor ACE2 (6LZG) with selected top ligands identified from docking analysis such as Tenufolin (TEN) and Pavetannin C1 (PAV) were subjected to molecular dynamics using gromacs GPU enabled package. Ligand topology was selected from Prodrug server. The pdb2gmx, a module of GROMACS was used to add hydrogens to the heavy atoms. Prepared systems were first vacuum minimized for 1500 steps using the steepest descent algorithm. Then the structures were solvated in a cubic periodic box with a water simple point charge (SPCE) water model. The complex systems were subsequently maintained with an appropriate salt concentration of 0.15 M by adding suitable numbers of Na+ and Cl− counter ions. The system preparation was referred on the basis of previously published paper (Gangadharappa et al., 2019). Each resultant structure from the NPT equilibration phase was subjected for final production run in NPT ensemble for 50 ns simulation time. Trajectory analysis was performed by using the gromacs simulation package of RMSD and RMSF (Mohankumar et al., 2020). Molecular Mechanics Poisson-Boltzmann surface area (MM-PBSA) approach was employed to understand the binding free energy (ΔG binding) of an inhibitor with protein over simulation time. A GROMACS utility g_mmpbsa was employed to estimate the binding free energy (Kumari et al., 2014). To obtain an accurate result, we computed ΔG for the last 20 ns in with dt 1000 frames.
3. Results
3.1. Binding site prediction and validation analysis of the target proteins
For the main protease, target structure (PDB- 6LU7) with reported inhibitor as co-crystallized ligand was retrieved from PDB for docking purpose. To further validate the docking methodology, RMSD value was calculated. RMSD score of the co-crystalized (internal) ligand and extracted internal ligand of the docked target protein-ligand complex structure served as control docking model as represented in Figure 2. The docking outcome showed that Autodock 4.0 determined the optimal orientation of the co-crystallized ligand. RMSD value 0.271 indicates the methodology is accurate to predict the binding affinity for unknown ligands.
Figure 2.
Data obtained in the validation of the molecular docking protocol for the receptor (A) 6LU7. Pink – Native ligand; Green and Blue: Docked pose.
For spike protein, target structure with reported inhibitor is not available in the PDB collection or literature. We have utilized the active site prediction option of Discovery Studio Visualizer (DSV) 2020. After loading the protein structure (PDB- 6LZG) to DSV, it reads the protein and highlights the probable active site i.e. residue information in yellow color. For the following spike protein, three residues i.e. HIS374; HIS378; GLU402 were recognized as crucial amino acids at the active pocket. Considering all the three residues, grid file was generated with following dimensions (x = −26.73, y = 8.32, z = −14.07) using PyRx program and progressed for molecular docking. Further Molecular Dynamics integrated with MM-PBSA calculations has validated the hit molecules identified from docking.
3.2. Molecular docking studies
In order to identify a prospective candidate for managing COVID-19, molecular docking was executed over 48 phytoconstituents acquired from different species of Cinnamon on the binding pocket of enzyme COVID-19 (PDB ID: 6LU7; 6LZG). From the literature it was clearly indicated that virus enters into the host cell with the assistance of ACE2 receptor. S-protein binds directly to the Angiotensin Converting Enzyme 2 (ACE2) receptor of the human host cell surface –thus enabling virus entry and replication. We have taken the PDB ID: 6LZG where spike protein is complexed with ACE2 to investigate the binding affinity of cinnamon derivatives with the complex (Sinha et al., 2020).
All these 48 compounds were docked against the target enzyme COVID-19 and ranked based on their dock score. Compounds possessing dock score of −7.0 or even less are thought about a better representative for restraint of the COVID-19. A comprehensive evaluation can be done by referring to table in supplementary file. This table exemplifies the list of active molecules acquired after docking studies. Those active molecules possess dock score value of −7.0 or lower were picked. Total of 7 compounds was selected based on the binding interactions with 6LU7 and 6LZG (Figures 3–10). Out of the seven compounds, Tenufolin exhibited the best-docked score (−8.8 Kcal/mol) with SARS-CoV2 Main Proteases (6LU7) and Pavetannin C1 exhibited the best-docked score (−11.1 Kcal/mol) with SARS-CoV2 spike protein (6LUZ).
Figure 3.
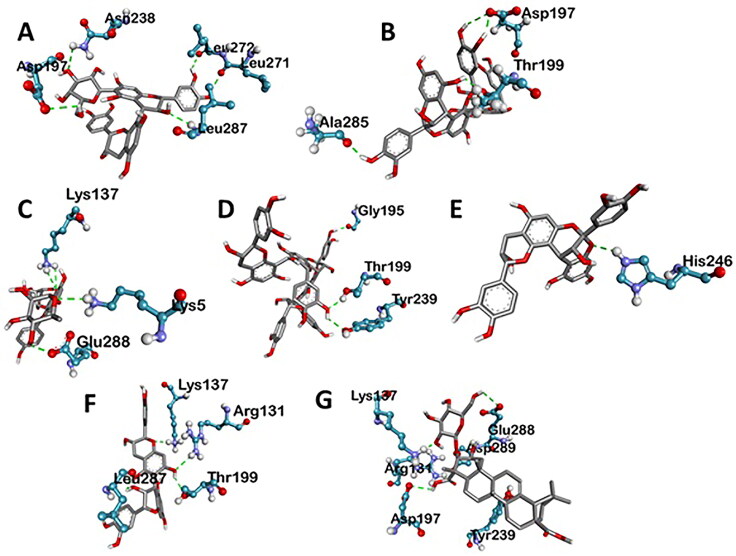
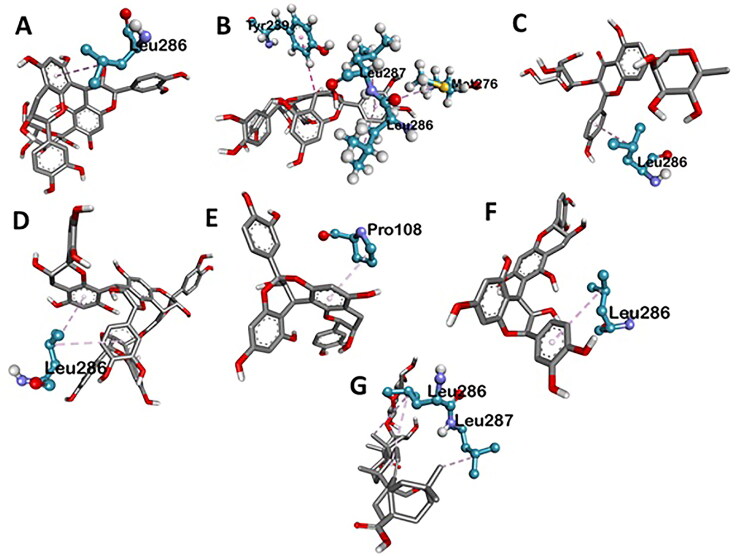
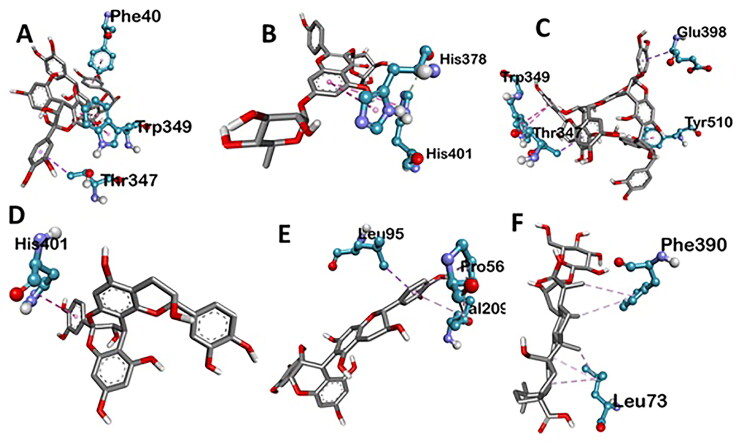
Various three-dimensional interactions of ligands with COVID-19 main protease (6LU7) via Hydrogen Bond. A: 6-Glucopyranosylprocyanidin B1; B: Cinnamtannin B1; C: Kaempferol 3-alpha-L-arabinofuranoside-7-rhamnoside-3; D: Pavetannin C1; E: Proanthocyanidin-A2; F: Procyanidin_B7; G: Tenufolin.
Figure 4.
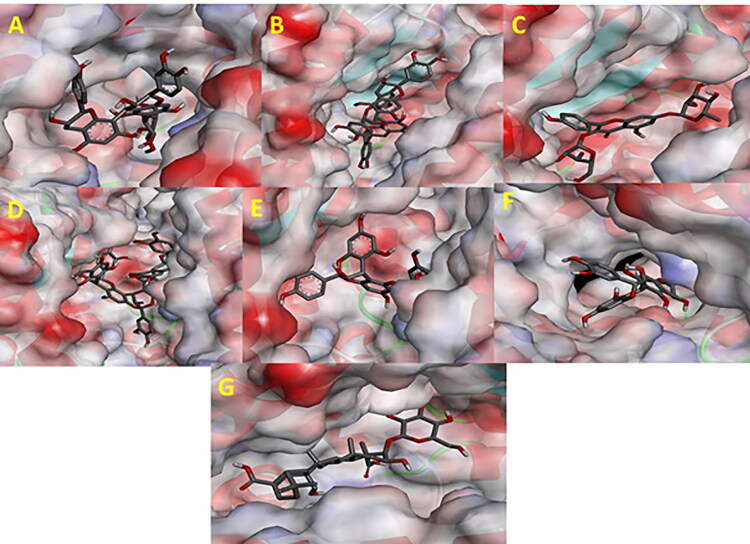
Various three-dimensional interactions of ligands with COVID-19 main protease (6LU7) via Hydrophobic Interactions. A: 6-Glucopyranosylprocyanidin B1; B: Cinnamtannin B1; C: Kaempferol 3-alpha-L-arabinofuranoside-7-rhamnoside-3; D: Pavetannin C1; E: Proanthocyanidin-A2; F: Procyanidin_B7; G: Tenufolin.
Figure 5.
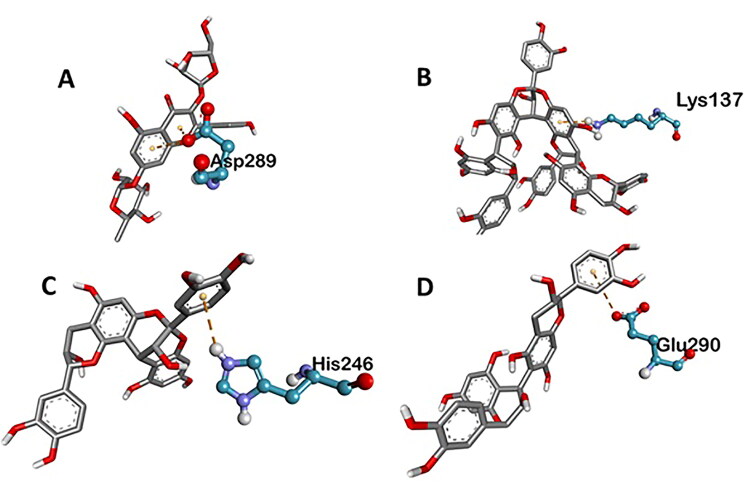
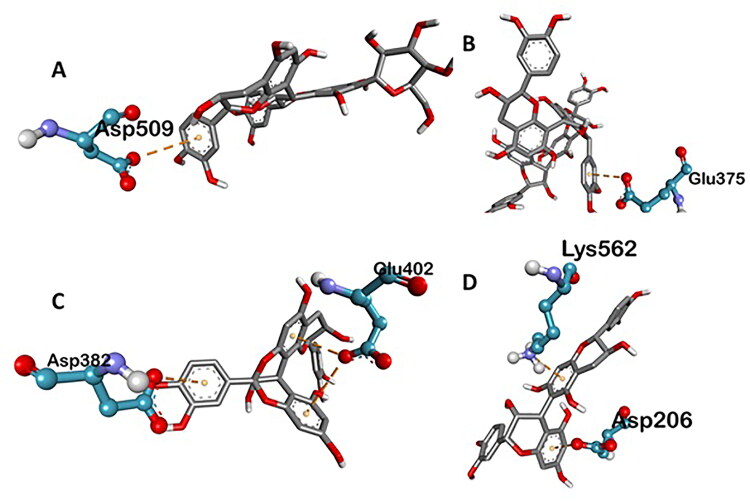
Various three-dimensional interactions of ligands with COVID-19 main protease (6LU7) via electrostatic interactions. A: Kaempferol 3-alpha-L-arabinofuranoside-7-rhamnoside-3; B: Pavetannin-C1; C: Proanthocyanidin-A2; D: Procyanidin_B7.
Figure 6.
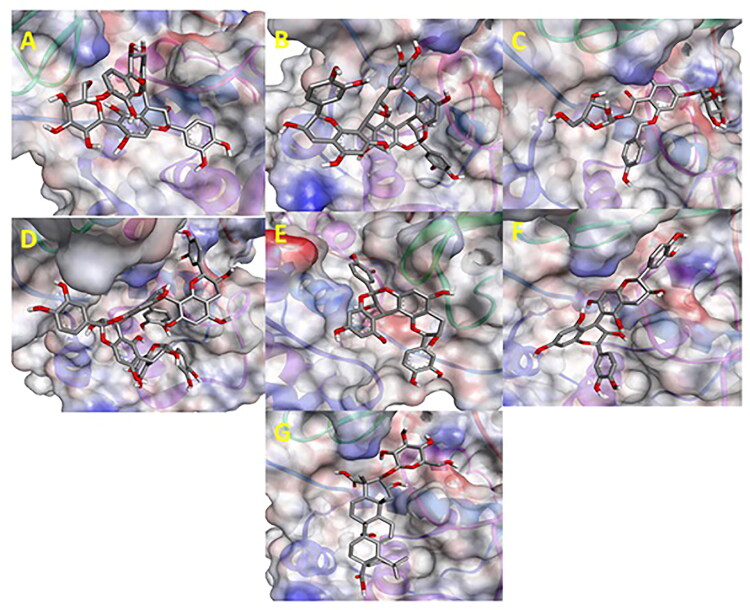
In silico docked complexes of Ligand (Ball and Stick representation) with COVID-19 main protease (6LU7) (Molecular representation) by Biovia Drug Discovery Studio 2019. A: 6-Glucopyranosylprocyanidin B1; B: Cinnamtannin B1; C: Kaempferol 3-alpha-L-arabinofuranoside-7-rhamnoside-3; D: Pavetannin-C1; E: Proanthocyanidin-A2; F: Procyanidin_B7; G: Tenufolin.
Figure 7.
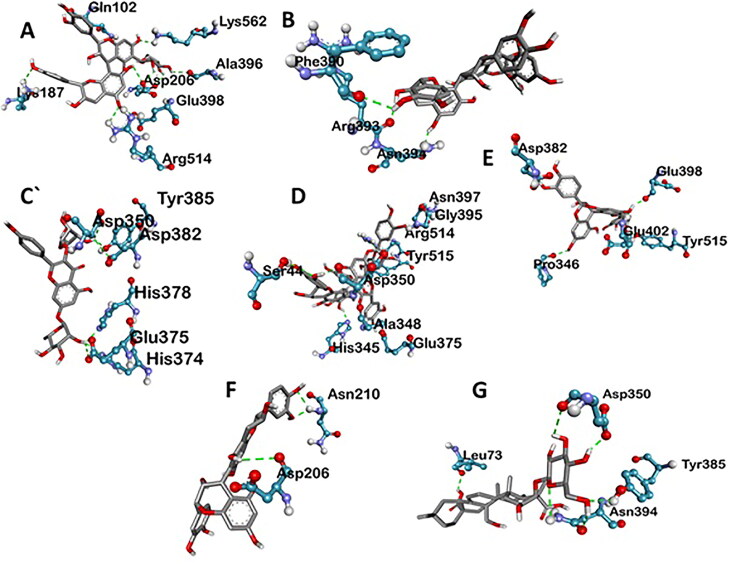
Various three-dimensional Interactions of ligands with COVID-19 spike protein (6LZG) via Hydrogen Bond. A: 6-Glucopyranosylprocyanidin B1; B: Cinnamtannin B1; C: Kaempferol 3-alpha-L-arabinofuranoside-7-rhamnoside-3; D: Pavetannin C1; E: Proanthocyanidin-A2; F: Procyanidin_B7; G: Tenufolin.
Figure 8.
Various three-dimensional interactions of ligands with COVID-19 spike protein (6LZG) via Hydrophobic interactions. A: Cinnamtannin B1; B: Kaempferol 3-alpha-L-arabinofuranoside-7-rhamnoside-3; C: Pavetannin C1; D: Proanthocyanidin-A2; E: Procyanidin_B7; F: Tenufolin.
Figure 9.
Various three-dimensional interactions of ligands with COVID-19 spike protein (6LZG) via electrostatic interactions. A: 6-Glucopyranosylprocyanidin B1; B: Pavetannin C1; C: Proanthocyanidin-A2; D: Procyanidin-B7.
Figure 10.
In silico docked complexes of Ligand (Ball and Stick representation) with COVID-19 spike protein (6LZG) (Molecular representation) by Biovia Drug Discovery Studio 2019. A: 6-Glucopyranosylprocyanidin B1; B: Cinnamtannin B1; C: Kaempferol 3-alpha-L-arabinofuranoside-7-rhamnoside-3; D: Pavetannin C1; E: Proanthocyanidin-A2; F: Procyanidin_B7; G: Tenufolin.
3.2.1. Molecular interaction studies
The rigid docking results were envisioned utilizing Discovery studio for evaluation of communications. The best binding postures of protein-ligand communications were envisioned and charted in Tables 1 and 2.
Table 1.
Physicochemical properties of the active compounds and accordance with the rules of drug-likeness.
| Ligands | MW | logp | Alogp | HBA | HBD | TPSA | AMR | nRB | No. of violations |
|---|---|---|---|---|---|---|---|---|---|
| Tenufolin | 634.02 | 3.077 | −1.812 | 12 | 0 | 52.6 | 172.99 | 6 | 2 |
| 6-Glucopyranosylprocyanidin B1 | 713.99 | −1.91 | −7.417 | 7 | 0 | 27.69 | 84.7 | 5 | 1 |
| Cinnamatannin B1 | 827.91 | 1.337 | −3.554 | 18 | 0 | 36.92 | 236.83 | 4 | 2 |
| Kaempferol 3-alpha-L-arabinofuranoside-7-rhamnoside | 545 | 0.018 | −5.168 | 10 | 0 | 63.22 | 83.68 | 6 | 1 |
| Pavetannin C1 | 1103.88 | 1.612 | −4.88 | 24 | 0 | 46.15 | 315.59 | 6 | 3 |
| Proanthocyanidin A2 | 551.94 | 1.062 | −2.229 | 12 | 0 | 27.69 | 158.07 | 2 | 3 |
| Procyanidin B7 | 551.94 | 1.127 | −2.262 | 12 | 0 | 18.46 | 159.83 | 3 | 3 |
MW: Molecular Weight; HBA: Hydrogen bond acceptor; HBD: Hydrogen bond donor; TPSA: topological polar surface area; AMR: Atom Molar Refractivity; nRB: No. of rotatable bonds.
Table 2.
Interactions of COVID-19 Main Protease (6LU7) amino acid residues with ligands at receptor sites.
| Ligands | Binding affinity, ΔG (Kcal/mol) | Amino acids involved and distance (A°) |
||
|---|---|---|---|---|
| Hydrogen binding interactions | Hydrophobic interactions | Electrostatic interactions | ||
| N-[(5-methylisoxazol-3-yl)carbonyl]alanyl-l-valyl-n∼1∼-((1r,2z)-4-(benzyloxy)-4-oxo-1-{[(3r)-2-oxopyrrolidin-3-yl]methyl}but-2-enyl)-l-leucinamide | −7.4 | HIS A:41 (4.27), GLU A: 166 (3.52), GLY A:143 (3.82) | CYS A:145 (7.03), GLN A:189 (5.51), GLY A:143 (3.76) | – |
| Tenuifolin | −8.8 | ARG A:131 (6.24), ASP A:197 (3.79), TYR A:239 (4.97), GLU A:288 (5.31), LYS A:137 (4.34), ASP A:289 (4.24) | LEU A:286 (5.30, 6.78), LEU A:287 (4.64) | – |
| Cinnamtannin-B1 | −8.4 | ALA A:285 (4.28), THR A:199 (3.69, 4.94), ASP A:197 (3.19, 4.05) | TYR A: 239 (5.92), MET A:276 (6.90), LEU A: 287 (4.86), LEU A:286 (5.06) | – |
| Procyanidin-B7 | −8.2 | ARG A:131 (6.50), LYS A:137 (5.13), THR A:199 (2.94), LEU A:287 (3.54) | LEU A: 286 (5.52) | GLU A:290 (6.55) |
| Kaempferol 3-alpha-L-arabinofuranoside-7-rhamnoside | −8.1 | LYS A:137 (5.91), LYS A:5 (5.96), GLU A:288 (5.47, 5.91) | LEU A:286 (5.12) | ASP A:289 (5.68, 6.17) |
| Proanthocyanidin-A2 | −8 | HIS A:246 (5.78) | PRO A:108 (5.14) | HIS A:246 (5.90) |
| 6-Glucopyranosyl procyanidin B1 | −7.6 | ASN A: 238 (5.42), LEU A:287 (3.76), LEU A:271 (4.86), LEU A:272 (3.15), ASP A:197 (2.95) | LEU A:286 (5.46) | – |
| Pavetannin-C1 | −7.3 | GLY A:195 (3.88), LYS A:5 (5.53, 6.11), TYR A: 239 (5.38), THR A:199 (4.25) | LEU A:286 (6.10, 6.62) | LYS A:137 (8.07) |
Main Protease, involved in severe acute respiratory syndrome (SARS), displayed best docking score of −8.8 kcal/mol with Tenuifolin, among the phytochemicals. Rest of the compounds 6-Glucopyranosylprocyanidin B1, Cinnamtannin-B1, Kaempferol 3-alpha-L-arabinofuranoside-7-rhamnoside, Pavetannin-C1, Proanthocyanidin-A2 and Procyanidin-B7 also interacted well with main Protease showing moderate binding energies of −7.6 kcal/mol, −8.4 kcal/mol, −8.1 kcal/mol, −7.3 kcal/mol, −8 kcal/mol and −8.2 kcal/mol respectively (Table 1, Figures 1–4). Meticulous in silico analysis revealed that all the phytoconstituents interacted decently at the active site of main protease, reflecting probable inhibitory tendencies against the COVID-19.
Tenufolin has the best correlation with main protease (6LU7) protein complexes. The main protease with tenufolin complex formed six hydrogen bond, i.e. ARG A:131; 6.24 A ASP A:197; 3.79 A TYR A:239; 4.97 A GLU A:288; 5.31 A LYS A:137; 4.34 A ASP A:289; 4.24 A and two amino acids are involved in the formation of hydrophobic interactions i.e. LEU A:286; 5.30 A 6.78 A LEU A:287; 4.64 A
Spike protein (6LZG), associated with SARS was found to exhibit the best possible interaction with Pavetannin C1 (−11.1 kcal/mol) among the phytochemicals (Table 2). Rest of the phytoconstituents 6-Glucopyranosylprocyanidin B1, Cinnamtannin-B1, Kaempferol 3-alpha-L-arabinofuranoside-7-rhamnoside, Proanthocyanidin-A2, Procyanidin-B7, Tenuifolin displayed moderate binding with spike protein at the active site with binding energies of −9.9 kcal/mol, −10.2 kcal/mol, −8.7 kcal/mol, −9.4 kcal/mol, −9.6 kcal/mol and −8.7 kcal/mol respectively. Meticulous interface profiling showed promising phytochemicals inhibitory activity against COVID-19’s Spike Protein (Figures 6–9).
Pavetannin C1 has the best correlation with main protease (6LZG) protein complexes. The spike protein with pavetannin complex formed nine hydrogen bond, i.e. ASN A:397; 4.70 A GLY A:395; 3.01 A HIS A:345; 4.46 A TYR A:515; 5.60 A ARG A:514; 5.06 A ALA A:348; 2.90 A ASP A:350; 3.77 A SER A:44; 4.55 A GLU A:375; 5.05 A and four amino acids are involved in the formation of hydrophobic interactions i.e. TRP A:349; 4.55 A TYR A:510; 6.91 A THR A:347; 5.14 A and GLU A:398; 5.10 A (Table 3).
Table 3.
Interactions of COVID-19 Spike Protein amino acid residues with ligands at receptor sites.
| Ligands | Binding affinity, ΔG (Kcal/mol) | Amino acids involved and distance (A°) |
||
|---|---|---|---|---|
| Hydrogen binding interactions | Hydrophobic interactions | Electrostatic interactions | ||
| Pavetannin-C1 | −11.1 | ASN A:397 (4.70), GLY A:395 (3.01), HIS A:345 (4.46), TYR A:515 (5.60), ARG A:514 (5.06), ALA A:348 (2.90), ASP A:350 3.77), SER A:44 (4.55), GLU A:375 (5.05) | TRP A:349 (4.55), TYR A:510 (6.91), THR A:347 (5.14), GLU A:398 (5.10) | GLU A:375 (6.26) |
| Cinnamtannin-B1 | −10.2 | PHE A:390 (4.34), ASN A:394 (3.61), ARG A:393 (4.34) | PHE A:40 (5.80), TRP A:349 (4.71), THR A:347 (5.57) | – |
| 6-Glucopyranosyl procyanidin B1 | −9.9 | ASP A:206 (3.82, 4.17, 4.65), ALA A:396 (5.06), LYS A:562 (5.89), ASN A:103 (4.91), GLN A:102 (5.14), LYS A:187 (6.00), GLU A:398 (4.05), ARG A:514 (4.84) | ASP A:509 (6.50) | |
| Procyanidin-B7 | −9.6 | ASP A:206 (3.11), ASN A:210 (3.52, 4.52) | LEU A:95 (4.95), VAL A:209 (5.03), PRO A:565 (5.76) | ASP A:206 (4.22), LYS A:562 (5.30) |
| Proanthocyanidin-A2 | −9.4 | PRO A:346 (4.75), GLU A:402 (3.62), ASP A:382 (4.87), GLU A:398 (5.02), TYR A:515 (4.71) | HIS A:401 (5.21) | GLU A:402 (5.52, 7.02), ASP A:382 (7.16) |
| Kaempferol 3-alpha-L- arabinofuranoside- 7-rhamnoside | −8.7 | ASP A:350 (4.43), TYR A:385 (5.22), ASP A:382 (4.23), HIS A:345 (5.08), HIA A:374 (4.88), GLU A:375 (4.91), HIS A:378 (4.93) | HIS A:401 (7.11), HIS A:378 (6.01) | – |
| Tenuifolin | −8.7 | LEU A:73 (4.15), ASP A:350 (2.98, 4.00), TYR A:385 (6.97), ASN A:394 (4.57) | PHE A:390 (4.91, 6.25), LEU A:73 (4.41, 4.42, 4.89) | – |
3.3. Drug likeliness
The physicochemical properties of the chosen seven active compounds were studied on DruLiTo software. As all the compounds are of natural source, none of them obeyed Lipinski’s rule (Table 1). Kaempferol 3-alpha-L-arabinofuranoside-7-rhamnoside and Tenufolin shows higher TPSA (63.22, 52.6) and AMR (315.59, 172.9) (Table 1 and Figure 11). TPSA, as well as AMR, are fundamental physicochemical properties mostly entailed in drug absorption, transport and penetration mechanism (Ertl et al., 2000).
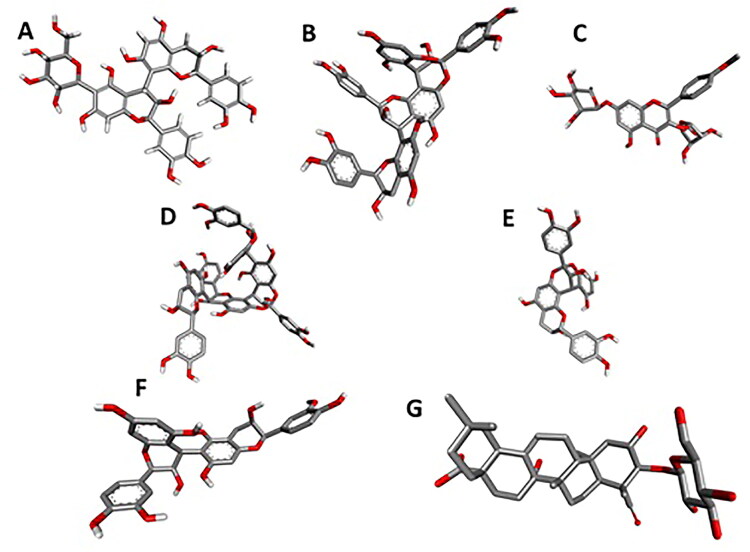
Figure 11.
The three dimensional structures of in silico active ligands. A: 6-Glucopyranosylprocyanidin B1; B: Cinnamtannin B1; C: Kaempferol 3-alpha-L-arabinofuranoside-7-rhamnoside-3; D: Pavetannin_C_1; E: Proanthocyanidin-A2; F: Procyanidin_B7; G: Tenufolin.
3.4. ADME/T evaluation by using admetSAR
The ADMET properties of the ligands were assessed, making use of admetSAR. ADMET properties for the substances in the research study were evaluated, making use of admetSAR. All the substances revealed excellent human intestinal absorption (HIA), blood-brain barrier (B.B.B.) infiltration. None of the compound was found carcinogenic. All the compounds were AMES negative. The results of HIA, B.B.B., LD50 values for the compounds are listed in Table 4.
Table 4.
ADME/T Properties of different compounds from Cinnamon.
| Ligands | HIA | BBB | AMES toxicity | Carcinogenicity | LD50 in rat (mol/kg) |
|---|---|---|---|---|---|
| 6-Glucopyranosylprocyanidin B1 | 0.929 | 0.593 | Non-toxic | Non-carcinogenic | 2.111 |
| Cinnamtannin-B1 | 0.7933 | 0.5685 | Non-toxic | Non-carcinogenic | 2.0438 |
| Kaempferol 3-alpha-L-arabinofuranoside-7-rhamnoside | 0.9353 | 0.744 | Non-toxic | Non-carcinogenic | 2.5152 |
| Pavetannin-C1 | 0.7933 | 0.5685 | Non-toxic | Non-carcinogenic | 2.0438 |
| Proanthocyanidin-A2 | 0.7933 | 0.5685 | Non-toxic | Non-carcinogenic | 2.0438 |
| Procyanidin-B7 | 0.9617 | 0.5434 | Non-toxic | Non-carcinogenic | 1.8446 |
| Tenuifolin | 0.5405 | 0.5396 | Non-toxic | Non-carcinogenic | 3.0142 |
HIA: human intestinal absorption; BBB: Blood-Brain Barrier; LD50: Lethal Dose, 50%.
3.5. PASS predictions for antiviral activity
The biological activity spectra of previously identified phytoconstituents were obtained by online PASS version. These predictions were interpreted and used in a flexible manner and given in Table 5.
Table 5.
Results of PASS calculations for antiviral activity of isolated phyto-constituents from ginger.
| Main predicted activity by PASS online | 6-Glucopyranosyl procyanidin B1 |
Cinnamtannin-B1 |
Kaempferol 3-alpha-L-arabinofuranoside-7-rhamnoside |
Pavetannin-C1 |
Proanthocyanidin-A2 |
Procyanidin-B7 |
Tenuifolin |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pa | Pi | Pa | Pi | Pa | Pi | Pa | Pi | Pa | Pi | Pa | Pi | Pa | Pi | |
| Antiviral (Rhinovirus) | 0,395 | 0,097 | 0,436 | 0,058 | – | – | 0,416 | 0,075 | 0,446 | 0,051 | 0,459 | 0,043 | 0,405 | 0,086 |
| Antiviral (HIV) | 0,203 | 0,029 | 0,161 | 0,054 | 0,146 | 0,070 | 0,170 | 0,047 | 0,180 | 0,040 | 0,261 | 0,014 | – | – |
| Antiviral (Influenza) | 0,722 | 0,004 | 0,348 | 0,065 | 0,717 | 0,005 | 0,265 | 0,117 | 0,400 | 0,046 | 0,474 | 0,027 | 0,769 | 0,003 |
| Antiviral (Herpes) | 0,555 | 0,005 | 0,369 | 0,050 | 0,567 | 0,005 | 0,360 | 0,054 | 0,388 | 0,041 | 0,422 | 0,026 | 0,496 | 0,010 |
| Antiviral (Hepatitis B) | 0,405 | 0,014 | 0,213 | 0,075 | 0,452 | 0,009 | 0,229 | 0,062 | 0,221 | 0,068 | 0,309 | 0,031 | 0,242 | 0,055 |
| Antiviral (Trachoma) | – | – | – | – | 0,121 | 0,028 | 0,099 | 0,044 | – | – | 0,122 | 0,028 | – | – |
PASS = Prediction of Activity Spectra for Substances; Pi = probable inactivity.
3.6. Molecular dynamics simulations
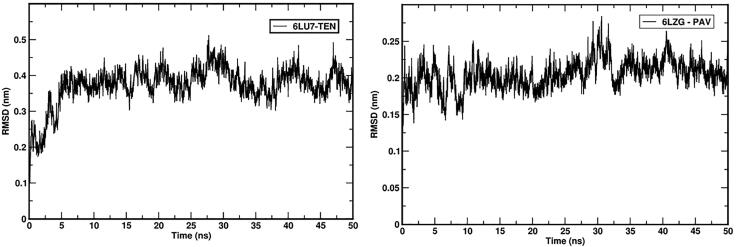
RMSD analysis of the protein gives insights and variations in its structural confirmation during the simulations, which includes the stability of protein and confirms whether simulation has equilibrated. The 6LU7 complex with TEN and 6LZG complex with PAV RMSD plot of backbone atoms was shown in Figures 12 and 13. RMSD was plotted for 6LU7-TEN and 6LGZ-PAV complex structures that converged during the 50 ns MD simulation. Both complex structures were found stable after 10 ns. The average values of 6LU7-TEN was ∼0.45 nm and 6LGZ-PAV was ∼0.24 nm of 50 ns simulation. This indicates the protein complex structures were stable during the md simulation.
Figure 12.
RMSD plots of respective complexes from GROMACS.
Figure 13.
RMSF plots of respective complexes from GROMACS.
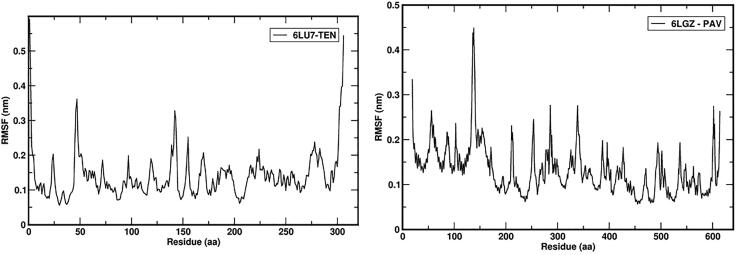
RMSF analysis measures the fluctuations of each residue during simulation. Ligand binding poses energy and interaction is directly dependent on residual fluctuation (RMSF) values. The 6LU7 complex with TEN and 6LZG complex with PAV RMSF plot for each residue was shown in Figure 13. It was observed that residues in loop region are more fluctuated during the simulation. This indicates that the protein was not fluctuated in the 50 ns simulation periods.
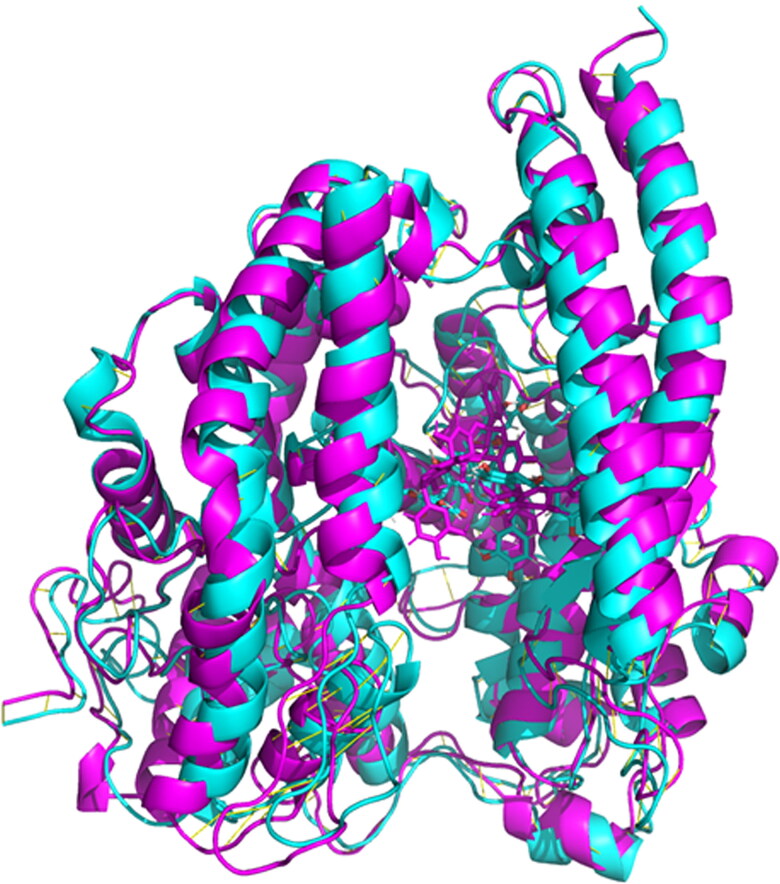
RBD domain superimpose analysis was carried out with open and close confirmation of the protein. Initial and last confirmation of the dynamics trajectory was analyzed using the Pymol software. RMSD = 2.192 (572 to 572 atoms) indicates that there are some structural changes between the protein structure. Superimposed structure was depicted in Figure 14.
Figure 14.
Superimpose analysis of RBD domain during the simulation. Magenta color at 0 ns and cyan color at 50 ns.
We have reported the total binding energy (deltaG value) of complex contributed from various energy terms after MD simulations. The binding free energy value of 6LU7_TEN was found to be −123.949 +/− 16.613 kJ/mol which indicates the inhibitor has higher affinity with the 6LU7 complex and binding free energy value of 6LZG_PAV was found to be −158.870 +/− 30.378 kJ/mol which indicates that the inhibitor has higher affinity with 6LZG complex. The 6LU7_TEN and 6LZG_PAV systems were submitted for residue level MM-PBSA binding energy calculation. Hotspots in interaction with the inhibitor (TEN) complex with 6LU7 are as follows: LYS137 −1.392 kcal/mol, ASP197 −1.088 kcal/mol, TYR239 −2.051 kcal/mol and GLU288 −1.103 kcal/mol. The total of hotspot interaction binding energy was −5.634 kcal/mol. Hotspots in interaction with the inhibitor (PAV) complexed with 6LZG are as follows: SER44 − 1.061 kcal/mol, HIS345 −0.247 kcal/mol, ALA348 −3.580 kcal/mol, ASP350 −9.864 kcal/mol, ARG514 −0.212 kcal/mol and TYR515 −6.672 kcal/mol. The total of hotspot interaction binding energy was −21.636 kcal/mol. MD trajectories analysis and MM-PBSA results revealed that TEN complex with 6LU7 showed moderate binding with key residues and PAV complex with 6LZG formed strong binding with key residues towards COVID-19 targets (Table 6).
Table 6.
MM-PBSA energy values of respective complexes from GROMACS.
| S. No | System | Binding energy | Hotspot interaction binding energy |
|---|---|---|---|
| 1 | 6LU7_TEN | −123.949 +/− 16.613 kJ/mol | −5.634 kcal/mol |
| 2 | 6LZG_PAV | −158.870 +/− 30.378 kJ/mol | −21.636 kcal/mol |
4. Discussion
Coronaviruses have a long history of infecting humans and animals and causing respiratory, digestive, liver and central nervous system diseases (To et al., 2013). A novel emerged SARS-CoV-2 is presenting major threats to human health nowadays (Zhu et al., 2020). The primary focus has been on clinical management which includes the prevention of infection, control measures and supportive care. Currently, no specific clinical therapeutics are available for the treatment of SARS-CoV-2-mediated infections (Zhou et al., 2020). Thus, the need of the hour is to identify and characterize novel drug candidates to overcome the health loses caused by SARS-CoV-2.
With this new breakthrough of Mpro structure in COVID-19, it has offered an astounding possibility to recognize the prospective drug candidates for the effective therapy of coronavirus. In this context, natural products have gained importance as potent anti-viral agents during recent years (Lin et al., 2014; Martinez et al., 2015). Considering the immediate need of therapeutics against COVID-19 and services of natural products in drug discovery, we have screened phytoconstituents from Cinnamon as novel hit molecules against Mpro, of SARS-CoV-2 for the identification of Mpro inhibitors to provide natural scaffolds for drug development.
Generally speaking, after virtual screening, recognizing lead compounds focuses on three main criteria: binding intensity, molecule linkage associations and therapeutic characterization. Lead compounds are more definitely molecules with very small binding energies, intense hydrogen and hydrophobic bond interactions, and relatively good ADMET properties. Therefore, top seven compounds were picked as suitable hit molecules for further study based on their low binding energies and relatively good poses inside the active site pocket after virtual screening.
Out of 48 candidates, seven compounds displayed a higher binding affinity and least binding energy with the main protease enzyme and spike protein. Tenuifolin has exhibited highest dock score with least binding energy of −8.8 Kcal/mol and found to make six hydrogen bonds with six amino acids, i.e. ARG A:105 (6.34), GLN A:107 (4.08), GLN A:110 (3.43, 5.02), THR A:111 (3.39), ASP A:295 (3.97) and one hydrophobic interaction with PHE A:294 (4.35). Except with, ARG A:105, remaining, the bond length of hydrogen bonds is <5 A°, which indicates the bonding is stronger and formed stable complexe. Whereas, Gingerenone A has a binding energy of −6.5 kcal/mol, associates with three hydrogen bonds with GLN A: 107 (4.04), GLN A:110 (3.79), THR A:111 (4.55) and hydrophobic interactions with VAL A:104 (4.98), ILE A:106 (5.48), PHE A:294 (4.73). These two compounds have the least binding affinity in comparison with other ligands due to the formation of more hydrogen bonds with the proteins. All the ligands are involved in hydrophobic interactions; mainly, two amino acids were involved, i.e. PHE A:294, VAL A:104 and ILE A:106.
Physicochemical characteristics were analyzed in light of the traditional drug-like laws that are implemented in the early stages of drug development, namely Lipinski’s Five law (RO5). Lipinski’s rule of Five (RO5) indicates that a medication molecule appears to show good oral bioavailability, smooth membrane permeability, and strong gastrointestinal absorption in human intestine while its log p ≤ 5; MW ≤ 500 dalton; HBAs ≤10 and HBDs ≤5. In our study, all the selected candidates with strong dock score have violated the Lipinski’s rule particularly in two main properties i.e. molecular weight >500 and hydrogen bonding acceptor >10. A significant class of drug molecules omitted from the initial ‘rule-of-five’ study are natural products. The high efficiency of natural product-based drug development can be due to the fact that evolutionary selection, which determines structural prerequisites for protein binding, has biologically pre-validated their chemical structures. It also has the benefit over conventional semi-synthetic methods to refine a structurally diverse natural product lead without the concomitant rise in molecular size. Therefore, synthetic biology may be a great lead optimization method to preserving or growing ligand effectiveness.
In the In silico measurement of intestinal absorption, both phytochemicals were theoretically strongly soluble in the gastrointestinal tract (Table 5). Despite effective distribution of medications in the human body, significant toxic effects such as carcinogenicity (Benigni & Bossa, 2011; Liebler & Guengerich, 2005) became important concerns. Accordingly, in the early phases of drug growth, identification and evaluation of novel product candidates is proposed to be important. None of the compounds have demonstrated in silico toxicity in this analysis.
The biological activity spectra of all selected compounds were determined by using an online version of PASS software. Results obtained (Table 6) were interpreted and used in a flexible manner. Thus, we anticipate that the consumption of Cinnamon has the potential to boost immunity to fight against COVID-19 infections.
In this research study, we have actually made use of Bioinformatics resources, PyRx and also Autodock-Vina and GROMACS to identify the potent molecules from Cinnamon against COVID-19 Main Proteases and spike protein, which participate in a vital part in Corona virus propagation. Final in silico validation by molecular dynamics integrated with free energy calculations and per residue analysis have demonstrated stable complexes with main protease and spike protein. Overall results proposed that tenufolin and Pavetannin C1 are the potent compounds which has good binding efficacy with main protease and spike proteins. Comparatively, Pavetannin C1 has strong affinity towards spike protein and which need to verified experimentally. Natural substances are deemed key targets of drug discovery and lead greatly to innovative product design and production initiatives. As the compounds are bulky in nature with high molecular weight, so further optimization is required. Further, its efficiency can be proved by its in-vitro, in-vivo and clinical studies.
Supplementary Material
Funding Statement
Vivek thank Karnataka Biotechnology and Information Technology Services (KBITS), Bangalore for providing the grant to establish the computational facility under Biotech Policy-II, Biotechnology Finishing School and Biotechnology Skill Enhancement Programme (BiSEP) at the Department of Biotechnology, Siddaganga Institute of Technology, Tumakuru - 572103, Karnataka.
Acknowledgements
Authors are thankful to K L College of Pharmacy, Koneru Lakshmaiah Education Foundation and M.S. Ramaiah University of Applied Sciences for providing necessary support to conduct the research and Siddaganga Institute of Technology, Tumakuru, India, for their encouragement and support in MD simulations.
Disclosure statement
All the authors declare no conflict of interest.
References
- Aanouz I., Belhassan A., El Khatabi K., Lakhlifi T., El Idrissi M., & Bouachrine M. (2020). Moroccan medicinal plants as inhibitors of COVID-19: Computational investigations. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1758790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostino M., Jene C., Boyle T., Ramsland P. A., & Yuriev E. (2009). Molecular docking of carbohydrate ligands to antibodies: Structural validation against crystal structures. Journal of Chemical Information and Modeling, 49(12), 2749–2760. 10.1021/ci900388a [DOI] [PubMed] [Google Scholar]
- Benigni R., & Bossa C. (2011). Mechanisms of chemical carcinogenicity and mutagenicity: A review with implications for predictive toxicology. Chemical Reviews, 111(4), 2507–2536. 10.1021/cr100222q [DOI] [PubMed] [Google Scholar]
- Boopathi S., Poma A. B., & Kolandaivel P. (2020). Novel 2019 Coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. F.-W., Yuan S., Kok K.-H., To K. K.-W., Chu H., Yang J., Xing F., Liu J., Yip C. C.-Y., Poon R. W.-S., Tsoi H.-W., Lo S. K.-F., Chan K.-H., Poon V. K.-M., Chan W.-M., Ip J. D., Cai J.-P., Cheng V. C.-C., Chen H., Hui C. K.-M., & Yuen K.-Y. (2020). A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. The Lancet, 395(10223), 514–523. 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri S., Symons J. A., & Deval J. (2018). Innovation and trends in the development and approval of antiviral medicines: 1987–2017 and beyond. Antiviral Research, 155, 76–88. 10.1016/j.antiviral.2018.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F., Li W., Zhou Y., Shen J., Wu Z., Liu G., & Tang Y. (2012). admetSAR: A comprehensive source and free tool for assessment of chemical ADMET properties. ACS Publications. [DOI] [PubMed] [Google Scholar]
- Corman V., Lienau J., & Witzenrath M. (2019). Coronaviruses as the cause of respiratory infections. Der Internist, 60(11), 1136–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosconati S., Forli S., Perryman A. L., Harris R., Goodsell D. S., & Olson A. J. (2010). Virtual screening with AutoDock: Theory and practice. Expert Opinion on Drug Discovery, 5(6), 597–607. 10.1517/17460441.2010.484460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallakyan S., & Olson A. J. (2015). Small-molecule library screening by docking with PyRx In Chemical biology (pp. 243–250). Springer. [DOI] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., & Munster V. J. (2016). SARS and MERS: Recent insights into emerging coronaviruses. Nature Reviews. Microbiology, 14(8), 523–534. 10.1038/nrmicro.2016.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Design L. (2014). Pharmacophore and ligand-based design with Biovia Discovery Studio®.
- Elfiky A. A. (2020). Natural products may interfere with SARS-CoV-2 attachment to the host cell. Journal of Biomolecular Structure and Dynamics, 1–16. 10.1080/07391102.2020.1761881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A., & Azzam E. B. (2020). Novel guanosine derivatives against MERS CoV polymerase: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1758789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmezayen A. D., Al-Obaidi A., Şahin A. T., & Yelekçi K. (2020). Drug repurposing for coronavirus (COVID-19): In silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1758791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enayatkhani, M., Hasaniazad, M., Faezi, S., Guklani, H., Davoodian, P., Ahmadi, N., Ali Einakian, M., Karmostaji, A., Ahmadi, K. (2020). Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: An in silico study. Journal of Biomolecular Structure and Dynamics, 1–19. 10.1080/07391102.2020.1756411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enmozhi S. K., Raja K., Sebastine I., & Joseph J. (2020). Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: An in silico approach. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1760136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertl P., Rohde B., & Selzer P. (2000). Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. Journal of Medicinal Chemistry, 43(20), 3714–3717. 10.1021/jm000942e [DOI] [PubMed] [Google Scholar]
- Fehr A. R., Channappanavar R., & Perlman S. (2017). Middle East respiratory syndrome: Emergence of a pathogenic human coronavirus. Annual Review of Medicine, 68, 387–399. 10.1146/annurev-med-051215-031152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangadharappa B. S., Sharath R., Revanasiddappa P. D., Chandramohan V., Balasubramaniam M., & Vardhineni T. P. (2019). Structural insights of metallo-beta-lactamase revealed an effective way of inhibition of enzyme by natural inhibitors. Journal of Biomolecular Structure and Dynamics, 1–15. 10.1080/07391102.2019.1667265 [DOI] [PubMed] [Google Scholar]
- Ge X.-Y., Li J.-L., Yang X.-L., Chmura A. A., Zhu G., Epstein J. H., Mazet J. K., Hu B., Zhang W., Peng C., Zhang Y.-J., Luo C.-M., Tan B., Wang N., Zhu Y., Crameri G., Zhang S.-Y., Wang L.-F., Daszak P., & Shi Z.-L. (2013). Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature, 503(7477), 535–538. 10.1038/nature12711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka I., Bertram S., Müller M. A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T. S., He Y., Gnirss K., Niemeyer D., Schneider H., Drosten C., & Pöhlmann S. (2011). Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. Journal of Virology, 85(9), 4122–4134. 10.1128/JVI.02232-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel R. K., Singh D., Lagunin A., & Poroikov V. (2011). PASS-assisted exploration of new therapeutic potential of natural products. Medicinal Chemistry Research, 20(9), 1509–1514. 10.1007/s00044-010-9398-y [DOI] [Google Scholar]
- Guan Y., Zheng B. J., He Y. Q., Liu X. L., Zhuang Z. X., Cheung C. L., Luo S. W., Li P. H., Zhang L. J., Guan Y. J., Butt K. M., Wong K. L., Chan K. W., Lim W., Shortridge K. F., Yuen K. Y., Peiris J. S. M., & Poon L. L. M. (2003). Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science (New York, N.Y.), 302(5643), 276–278. 10.1126/science.1087139 [DOI] [PubMed] [Google Scholar]
- Gupta M. K., Vemula S., Donde R., Gouda G., Behera L., & Vadde R. (2020). In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. Journal of Biomolecular Structure and Dynamics, 1–17. 10.1080/07391102.2020.1751300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D. C. Jr., & Ji H.-F. (2020). A search for medications to treat COVID-19 via in silico molecular docking models of the SARS-CoV-2 spike glycoprotein and 3CL protease. Travel Medicine and Infectious Disease, 101646 10.1016/j.tmaid.2020.101646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan, A., Paray, B. A., Hussain, A., Qadir, F. A., Attar, F., Aziz, F. M., Sharifi, M., Derakhshankhah, H., Rasti, B., Mehrabi, M., Shahpasand, K., Saboury, A. A., Falahati, F. (2020). A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1754293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgenfeld R. (2014). From SARS to MERS: Crystallographic studies on coronaviral proteases enable antiviral drug design. The FEBS Journal, 281(18), 4085–4096. 10.1111/febs.12936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N.-H., Nitsche A., Müller M. A., Drosten C., & Pöhlmann S. (2020). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell, 181(2), 271–280. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., … Cao B. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet, 395(10223), 497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, R., Parves, R., Paul, A. S., Uddin, N., Rahman, M. S., Mamun, A. A., Hossain, M. N., Ali, M. A., Halim, M. A. (2020). A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. Journal of Biomolecular Structure and Dynamics, 1–20. 10.1080/07391102.2020.1761883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, R. S., Jagdale, S. S., Bansode, S. B., Shankar, S. S., Tellis, M. B., Pandya, V. K., Chugh, A., Giri, A. P., Kulkarni, M. J. (2020). Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease$. Journal of Biomolecular Structure and Dynamics, 1–16. 10.1080/07391102.2020.1760137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, R. J., Jha, R. K., Amera, G., Jain, M., Singh, E., Pathak, A., Singh, R. P., Muthukumaran, J., Singh, A. K. (2020). Targeting SARS-Cov-2: A systematic drug repurposing approach to identify promising inhibitors against 3C-like Proteinase and 2’-O-RiboseMethyltransferase. Journal of Biomolecular Structure and Dynamics, 1–40. 10.1080/07391102.2020.1753577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A., Zia K., Ashraf S., Uddin R., & Ul-Haq Z. (2020). Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1751298 [DOI] [PubMed] [Google Scholar]
- Khurana N., Ishar M. P. S., Gajbhiye A., & Goel R. K. (2011). PASS assisted prediction and pharmacological evaluation of novel nicotinic analogs for nootropic activity in mice. European Journal of Pharmacology, 662(1–3), 22–30. 10.1016/j.ejphar.2011.04.048 [DOI] [PubMed] [Google Scholar]
- Kumari R., Open Source Drug Discovery Consortium, Kumar R., & Lynn A. (2014). g_mmpbsa-a GROMACS tool for high-throughput MM-PBSA calculations. Journal of Chemical Information and Modeling, 54(7), 1951–1962. 10.1021/ci500020m [DOI] [PubMed] [Google Scholar]
- Lai K., Shen H., Zhou X., Qiu Z., Cai S., Huang K., Wang Q., Wang C., Lin J., Hao C., Kong L., Zhang S., Chen Y., Luo W., Jiang M., Xie J., & Zhong N. (2018). Clinical practice guidelines for diagnosis and management of cough-Chinese Thoracic Society (CTS) Asthma Consortium. Journal of Thoracic Disease, 10(11), 6314–6351. 10.21037/jtd.2018.09.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S. K. P., Woo P. C. Y., Li K. S. M., Huang Y., Tsoi H.-W., Wong B. H. L., Wong S. S. Y., Leung S.-Y., Chan K.-H., & Yuen K.-Y. (2005). Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proceedings of the National Academy of Sciences of the United States of America, 102(39), 14040–14045. 10.1073/pnas.0506735102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., & Harrison S. C. (2005). Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science (New York, N.Y.), 309(5742), 1864–1868. 10.1126/science.1116480 [DOI] [PubMed] [Google Scholar]
- Li W., Moore M. J., Vasilieva N., Sui J., Wong S. K., Berne M. A., Somasundaran M., Sullivan J. L., Luzuriaga K., Greenough T. C., Choe H., & Farzan M. (2003). Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature, 426(6965), 450–454. 10.1038/nature02145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebler D. C., & Guengerich F. P. (2005). Elucidating mechanisms of drug-induced toxicity. Nature Reviews. Drug Discovery, 4(5), 410–420. 10.1038/nrd1720 [DOI] [PubMed] [Google Scholar]
- Lin L.-T., Hsu W.-C., & Lin C.-C. (2014). Antiviral natural products and herbal medicines. Journal of Traditional and Complementary Medicine, 4(1), 24–35. 10.4103/2225-4110.124335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J., Sasse F., Brönstrup M., Diez J., & Meyerhans A. (2015). Antiviral drug discovery: Broad-spectrum drugs from nature. Natural Product Reports, 32(1), 29–48. 10.1039/C4NP00085D [DOI] [PubMed] [Google Scholar]
- Menachery V. D., Dinnon K. H., Yount B. L., McAnarney E. T., Gralinski L. E., Hale A., Graham R. L., Scobey T., Anthony S. J., Wang L., Graham B., Randell S. H., Lipkin W. I., & Baric R. S. (2019). Trypsin treatment unlocks barrier for zoonotic bat coronavirus infection. Journal of Virology, 94(5), e01774-19. 10.1128/JVI.01774-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal M., Goel R. K., Bhargava G., & Mahajan M. P. (2008). PASS-assisted exploration of antidepressant activity of 1, 3, 4-trisubstituted-β-lactam derivatives. Bioorganic & Medicinal Chemistry Letters, 18(20), 5347–5349. 10.1016/j.bmcl.2008.09.064 [DOI] [PubMed] [Google Scholar]
- Mohankumar T., Chandramohan V., Lalithamba H. S., Jayaraj R. L., Kumaradhas P., Sivanandam M., Hunday G., Vijayakumar R., Balakrishnan R., Manimaran D., & Elangovan N. (2020). Design and molecular dynamic Investigations of 7,8-dihydroxyflavone derivatives as potential neuroprotective agents against alpha-synuclein. Scientific Reports, 10(1), 599 10.1038/s41598-020-57417-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanraj K., Karthikeyan B. S., Vivek-Ananth R., Chand R. B., Aparna S., Mangalapandi P., & Samal A. (2018). IMPPAT: A curated database of Indian medicinal plants, phytochemistry and therapeutics. Scientific Reports, 8(1), 1–17. 10.1038/s41598-018-22631-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., & Olson A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V. J., Koopmans M., van Doremalen N., van Riel D., & de Wit E. (2020). A novel coronavirus emerging in China—key questions for impact assessment. New England Journal of Medicine, 382(8), 692–694. 10.1056/NEJMp2000929 [DOI] [PubMed] [Google Scholar]
- Muralidharan N., Sakthivel R., Velmurugan D., & Gromiha M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–7. 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- O'Boyle N. M., Banck M., James C. A., Morley C., Vandermeersch T., & Hutchison G. R. (2011). Open Babel: An open chemical toolbox. Journal of Cheminformatics, 3(1), 33 10.1186/1758-2946-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pangastuti A., Amin I. F., Amin A. Z., & Amin M. (2016). Natural bioactive compound from Moringa oleiferaagainst cancer based on in silico screening. Jurnal Teknologi, 78(5), 1–4. 10.11113/jt.v78.8328 [DOI] [Google Scholar]
- Pant S., Singh M., Ravichandiran V., Murty U., & Srivastava H. K. (2020). Peptide-like and small-molecule inhibitors against Covid-19. Journal of Biomolecular Structure and Dynamics, 1–15. 10.1080/07391102.2020.1757510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma, P., Sekhar, N., Prajapat, M., Avti, P., Kaur, H., Kumar, S., Singh, S., Kumar, H., Dhibar, D. P., Medhi, B. (2020). In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1753580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeliger D., & de Groot B. L. (2010). Ligand docking and binding site analysis with PyMOL and Autodock/Vina. Journal of Computer-Aided Molecular Design, 24(5), 417–422. 10.1007/s10822-010-9352-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S. K., Shakya A., Prasad S. K., Singh S., Gurav N. S., Prasad R. S., & Gurav S. S. (2020). An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1762741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K. K., Hung I. F., Chan J. F., & Yuen K.-Y. (2013). From SARS coronavirus to novel animal and human coronaviruses. Journal of Thoracic Disease, 5(Suppl 2), S103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend E. A., Siviski M. E., Zhang Y., Xu C., Hoonjan B., & Emala C. W. (2013). Effects of ginger and its constituents on airway smooth muscle relaxation and calcium regulation. American Journal of Respiratory Cell and Molecular Biology, 48(2), 157–163. 10.1165/rcmb.2012-0231OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahedi H. M., Ahmad S., & Abbasi S. W. (2020). Stilbene-based natural compounds as promising drug candidates against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–16. 10.1080/07391102.2020.1762743 [DOI] [PubMed] [Google Scholar]
- Wang C., Horby P. W., Hayden F. G., & Gao G. F. (2020). A novel coronavirus outbreak of global health concern. The Lancet, 395(10223), 470–473. 10.1016/S0140-6736(20)30185-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2003). Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003. World Health Organization. [Google Scholar]
- WHO (2020). Coronavirus disease 2019 (COVID-19): Situation report, 74.
- Yang H., Lou C., Sun L., Li J., Cai Y., Wang Z., Li W., Liu G., & Tang Y. (2019). admetSAR 2.0: Web-service for prediction and optimization of chemical ADMET properties. Bioinformatics (Oxford, England), 35(6), 1067–1069. 10.1093/bioinformatics/bty707 [DOI] [PubMed] [Google Scholar]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science, 368(6489), 409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Hou Y., Shen J., Huang Y., Martin W., & Cheng F. (2020). Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discovery, 6(1), 14–18. 10.1038/s41421-020-0153-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. F., & Tan W. (2020). A novel coronavirus from patients with pneumonia in China, 2019. The New England Journal of Medicine, 382(8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.