Abstract
World Health Organization characterized novel coronavirus disease (COVID-19), caused by severe acute respiratory syndrome (SARS) coronavirus-2 (SARS-CoV-2) as world pandemic. This infection has been spreading alarmingly by causing huge social and economic disruption. In order to response quickly, the inhibitors already designed against different targets of previous human coronavirus infections will be a great starting point for anti-SARS-CoV-2 inhibitors. In this study, our approach integrates different ligand based drug design strategies of some in-house chemicals. The study design was composed of some major aspects: (a) classification QSAR based data mining of diverse SARS-CoV papain-like protease (PLpro) inhibitors, (b) QSAR based virtual screening (VS) to identify in-house molecules that could be effective against putative target SARS-CoV PLpro and (c) finally validation of hits through receptor-ligand interaction analysis. This approach could be used to aid in the process of COVID-19 drug discovery. It will introduce key concepts, set the stage for QSAR based screening of active molecules against putative SARS-CoV-2 PLpro enzyme. Moreover, the QSAR models reported here would be of further use to screen large database. This study will assume that the reader is approaching the field of QSAR and molecular docking based drug discovery against SARS-CoV-2 PLpro with little prior knowledge.
Communicated by Ramaswamy H. Sarma
Keywords: COVID-19, SARS-CoV-2, SARS-CoV PLpro, Monte Carlo based optimization, QSAR based virtual screening, ADME, molecular docking
Introduction
Ligand based and target based drug discoveries are the most applied modern drug discovery outlooks (Hajjo et al., 2012; Polishchuk, 2017). Traditionally medicinal chemists wielded “chemical intuition” for lead optimization, which sometimes rendered biased fingerprint/fragment or scaffold selection (Bandyopadhyay et al., 2019). Now-a-days the comparative learning between the statistical and intelligent approaches enriched lead optimization aspects by combining statistical significance and putative visualizations of fingerprint (or scaffold) devoid of traditional selection-biases (Adhikari et al., 2016; Jain et al., 2020). Significantly, medicinal chemists put their primary focal point on virtual compound libraries rather than industry chemical collections to trigger time- and money-efficiency (Choudhury, 2020; Van Hilten et al., 2019).
The use of such drug discovery approaches in terms of quantitative structure-activity relationship (QSAR), artificial intelligence (AI), virtual screening (VS), drug repurposing etc. demands more when the world faced unwanted and uncontrolled scenario as like current pandemic posted by novel coronavius (2019-nCoV) (Adeoye et al., 2020; Bhardwaj et al., 2020; Elasnaoui & Chawki, 2020; Elfiky, 2020a; Mittal et al., 2020; Paniri et al., 2020; Pant et al., 2020; Patil et al., 2020; Sarma et al., 2020; Wahedi et al., 2020). In this easily accessible world of 21st century coronavirus disease-2019 (COVID-19) has been spreading alarmingly by causing huge social and economic disruption (Aanouz et al., 2020; Arya & Dwivedi, 2020; Basit et al., 2020; Boopathi et al., 2020; Ghosh et al., 2020; Hendaus, 2020; Hendaus & Jomha, 2020; Pillaiyar et al., 2020). COVID-19 respiratory disease is attributed as world pandemic by World Health Organization (https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19—11-march-2020). The 2019-nCoV infection has spread over to 216 countries and territories since its outbreak in the last month of 2019 in China, so far 6 287 771 confirmed cases and 379 941 deaths have been documented as on 3rd June 2020 (https://www.who.int/emergencies/diseases/novel-coronavirus-2019).
2019-nCoV is 3rd human coronavirus (HCoV) as identified in the 21st century (Ahmad et al., 2020; Anwar et al., 2020; Borkotoky & Banerjee, 2020; Chandra et al., 2020; Pillaiyar et al., 2020). Previously, two corona virus diseases severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV) in 2002 and Middle East respiratory syndrome (MERS) coronavirus (MERS-CoV) in 2012 infected at least 8,422 people (fatality rate of ∼10%) and 1,700 people (fatality rate of ∼36%), respectively (http://www.who.int/csr/sars/archive/2003_05_07a/en).
2019-nCoV (also known as SARS-CoV-2; we will use SARS-CoV-2 in rest of the paper) is an envelope virus having a single-stranded positive sense RNA genome (Elfiky & Azzam, 2020; Ghosh et al., 2020; Nejadi Babadaei et al., 2020a, Babadaei et al., 2020b). The spike protein (S) of SARS-CoV-2 regulates viral entry into the host cells (Amin & Abbas, 2020; AP and VS 2020; de Oliveira et al., 2020; Elfiky, 2020b; Enayatkhani et al., 2020; Gupta et al., 2020; Hasan et al., 2020; Sinha et al., 2020; Sk et al., 2020). Two polyproteins i.e. pp1a and pp1ab are promptly translated upon entry into the host cells. Then these are disbanded by two viral proteases, one is 3 C-like protease (3CLpro) and another is papain-like protease (PLpro) (Figure 1) enzymes (Báez-Santos et al., 2015; Elmezayen et al., 2020; Ghosh et al., 2020; Joshi et al., 2020; Khan, Ali, et al., 2020; Khan, Zia, et al., 2020; Lin et al., 2018; Muralidharan et al., 2020). Both proteases are essential for SARS-CoV-2 viral replication and thus, can be is considered as druggable targets (Ghosh et al., 2020).
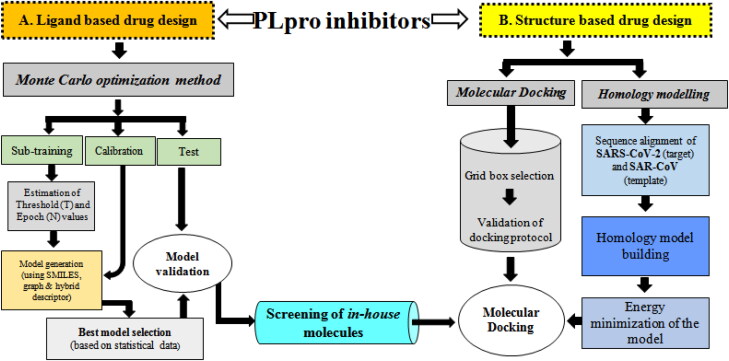
Figure 1.
Schematic representations of current work designwhich are composed of two major aspects- (A) Ligand based and (B) Structure based approaches.
The molecular docking and target based virtual screening studies have moved at a much faster pace (Al-Khafaji et al., 2020; Das et al., 2020; Enmozhi et al., 2020; Gyebi et al., 2020; Islam et al., 2020; Khan, Jha, et al., 2020; Kumar et al., 2020; Lobo-Galo et al., 2020; Mahanta et al., 2020) after deliberation of the first ligand bound SARS-CoV-2 3CLpro crystal structure in February, 2020. However, SARS-CoV-2 PLpro ligand based as well as structure based screening approaching were limited due to little proteomic knowledge.
In response against the social and economic disruptionposted by SARS-CoV-2 outbreak, screening of SARS-CoV PLpro inhibitors is fastest options which offer more strategic and economic benefits. The use of virtual compound libraries already gained in publicity and has achieved some successes. Here, the virtual screening has fostered the application of drug design to the SARS-CoV-2 targets. This current communication, a component of our rational drug design and discovery headway (Adhikari et al., 2017; Amin et al., 2018; Banerjee et al., 2020; Dutta et al., 2019; Halder et al., 2013), we propounded mathematical modelling workflow based on Monte Carlo optimization and other approaches which further leads to the screening of possible SARS-CoV-2 PLpro inhibitors (Figure 1).
The study design was composed of two major aspects-(A) Ligand based approaches: (i) classification QSAR based data mining of diverse SARS-CoV papin-like protease (PLpro) inhibitors, (ii) QSAR based virtual screening (VS) to identify in-house molecules that could be effective against putative target SARS-CoV PLpro; and (B) Structure based approaches: finally validation of hits through receptor-ligand interaction analysis (Figure 1).
Therefore, this study may introduce key concepts, set the stage for molecule identification and QSAR based screening of in-house molecules active against putative SARS-CoV-2 PLpro enzyme.
Methods and materials
Dataset
A set of diverse SARS-CoV PLpro inhibitors were collected with inhibitory activities (Báez-Santos et al., 2014; Cheng et al., 2015; Chou et al., 2008; Frieman et al., 2011; Ghosh et al., 2009, 2010; Park et al., 2012; Ratia et al., 2006, 2008). Compounds with no inhibitory activity and without definite activity were not taken for this study. In addition, duplicate molecules were eliminated. Finally, ninety one molecules were considered for the further molecular modelling study (Table S1). The average SARS-CoV PLpro pIC50 value was considered as the ‘activity threshold’ for the current study. Compounds having the SARS-CoV PLpro pIC50 value less than the ‘activity threshold’ were classified as lower PLpro inhibitors or Inactives and those with PLpro pIC50 value higher than the ‘activity threshold’ were yielded as promising PLpro inhibitors or Actives. Thus, out of 91 molecules, 40 compounds were distinguished as Actives and 51 molecules were considered as Inactives.
Classification based QSAR
The classification modelling assists to discriminate the Active and Inactive molecules in terms of their investigated biological significance. Here, we performed Monte Carlo based Coral QSAR study. Performing this study not only offers a graphical visualization of critical fingerprint or fragments attributed to enhance/decrease the SARS-CoV PLpro inhibitory activity but also it allows the chance of screening external set compounds.
Monte Carlo optimization based QSAR
Descriptors calculation
SMILES-based descriptors
SMILES-based descriptors were calculated by the following equation:
In this equation, T and N represent threshold value and number of epoch, respectively. The correlation weights were represented by CW. The different coefficients like a, b, c, d, α, β and γ were used for descriptor modification. NOSP, HALO, BOND and ATOMPAIR represent global SMILES attributes and the local smile attributes were denoted by Sk, SSk and SSSk (Toropov et al., 2013; Toropova et al., 2015).
Graph-based descriptors
GAO (graph of atomic orbital), HSG (hydrogen-suppressed graph) and HFG (hydrogen-filled graph) represents different graph based descriptors and was calculated by following equation:
Where, 0ECk, 1ECk and 3ECk represent different Morgan’s connectivity indices. Ak denotes different chemical atoms like: C, N, O etc. α, β and γ were the coefficients with 0 and 1 value. The coefficients having value 0 and 1 were denoted by as α, β and γ (Toropov et al., 2013; Worachartcheewan et al., 2014).
Hybrid descriptors
The amalgamation of SMILES and graph-based descriptors forms hybrid descriptors which are represented as:
Model development and validation
In our study, by using balance of correlation method twenty-one classification models were developed from three different splits. The dataset containing of 91PLpro inhibitors were distributed into training (41 compounds), calibration (35 compounds) and test (15 compounds) sets which were used for the study. Further, optimization of T (threshold) and N (epoch) were also performed separately for each model (Toropova et al., 2015). The sensitivity, specificity, accuracy along with the MCC values was recorded as a measure of internal and external validation. Finally, the important structural attributes that were solely answerable for promoting or hindering of PLpro activity were identified.
Target based molecular modelling
Homology modelling was performed which provided 3 D models for SARS-CoV-2 protein structure as the ligand-bound crystal structures are not available till date. The homology model for SARS-CoV-2 was built using Swiss Model web server (https://swissmodel.expasy.org/) and subsequently, validated by Verify3D (https://servicesn.mbi.ucla.edu/Verify3D/), ProSA (https://prosa.services.came.sbg.ac.at/prosa.php) and PROCHECK (https://spdbv.vital-it.ch/). In addition, the model was optimized using Swiss PDB viewer software using GROMOS96 Force-Field followed by determination of RMSD value by the aid of PyMOL software (https://pymol.org/2/). Lastly, the energy minimized model was implemented for the molecular docking analysis. The docking study was conducted by the aid of AutoDock Vina (Trott & Olson, 2009). Notably, grid box was selected by covering the geometric pattern occupied by the prototype in-bound ligand in the crystal structure of PDB: 4OW0. The docked poses of ligands were visualized by PyMOL software (https://pymol.org/2/) and the 2 D-interaction plots were generated by Discovery Studio 3.5 Visualizer (Accelrys Software Inc., San Diego, California, USA).
Result and discussions
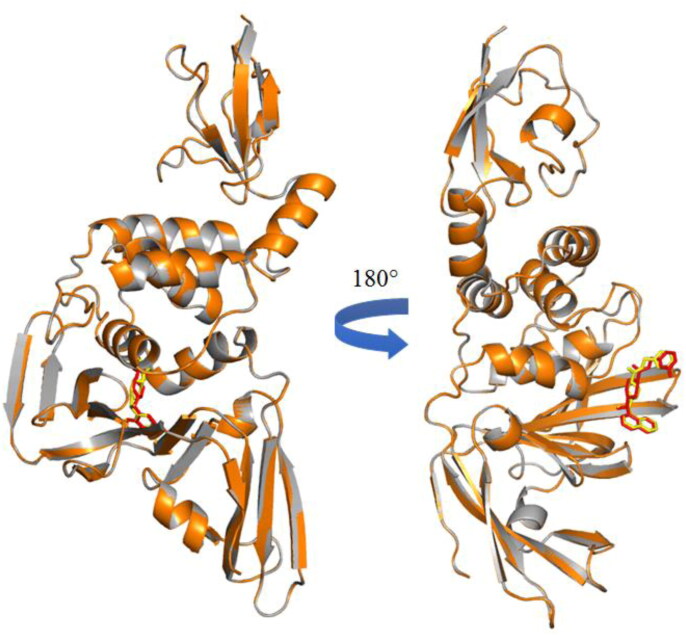
It is already reported that some small molecules exhibited potent SARS-CoV PLpro inhibition (Ghosh et al., 2009, 2010, 2020).SARS-CoV PLpro shares 82.80% sequence similarity with the homologus SARS-CoV-2 strain (Figure 2).
Figure 2.
Comparison of SARS-CoV PLpro and SARS-CoV2 PLpro: A modeled structure of SARS‐CoV‐2 PLpro (orange), the crystal structure of SARS‐CoV (grey, PDB: 4OW0) where PLpro inhibitor binds at the catalytic site [docked ligand (brick red) and in-bound ligand (light yellow)].
Significantly, PLpro active site amino acids (P248, P249, Y269, D165, E168, L163, G164, Q270, Y274, Y265, T302) of both stains are highly conserved (Figure 2). Thus, it may pretend that SARS-CoV PLpro inhibitors would to be potential inhibitors of SARS-CoV-2 PLpro enzyme. As the development of new small molecules against the proteasesof the COVID-19 is challenging as well as time and money consuming, it is better to screen compounds based on the previous ones.
Target based molecular modelling
The Swiss model constructed an excellent homology model of SARS-CoV-2 based on the sequence identity between the PLpro SARS-CoV (PDB: 4OW0) and PLpro SARS-CoV-2. The quality of the model was validated by the aid of Verify3D (96.79% of the residues had average 3 D -1 D score s ≤ 0.2), ProSA server (Z-Score = −8.79), the Ramachandran plot (91.4% and 8.2% of the residues in the most favorable and the additional allowed region, respectively; 0.4% residues in generously allowed regions while no residues were found to be in disallowed region). The RMSD score, as determined by using PyMOL, was recorded 0.090 which confirmed the model acceptability.
The amino acid sequences of SARS-CoV PLpro (PDB: 4OW0) and COVID-19 PLpro (homology modelled) are depicted. Notably, the active site amino acids including P248, P249, Y269, D165, E168, L163, G164, Q270, Y274, Y265, T302 etc. are highly conserved.
Classification QSAR study
The whole set of molecules able to bind to the SARS-CoV PLpro enzyme were taken after extensive literature studies, retrieving only those ligands having an absolute IC50 values. This set consisted of 91 PLpro inhibitors. Depending on the ‘activity threshold’, out of 91 compounds, 40 compounds were identified as Active and 51 molecules were considered as Inactives.
Monte Carlo optimization based coral QSAR
In Monte Carlo optimization (Toropov et al., 2018; Toropova et al., 2015), a total of twenty-one different models from three different splits were generated using SMILES and graph-based descriptors with a combination of different connectivity indices which were calculated for generation of different models (Table S2). Each models were developed after the search for desirable T (threshold) and N (epoch) values as per the test set statistics as suggested by Toropova et al. (2015). The statistical parameters of three best models from three different splits are shown in Table 1.
Table 1.
The statistical performance of three best models from three different splits.
| Parameter | Set | T | N | TP | TN | FP | FN | Total | Sensitivity | Specificity | Accuracy | MCC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M2 SMILES, GAO (0ECk) | Sub-Training | 3 | 7 | 15 | 24 | 2 | 0 | 41 | 1.0000 | 0.9231 | 0.9512 | 0.9025 |
| Calibration | 15 | 18 | 0 | 2 | 35 | 0.8824 | 0.1000 | 0.9429 | 0.8911 | |||
| Test | 4 | 4 | 3 | 4 | 15 | 0.5000 | 0.5741 | 0.5333 | 0.5014 | |||
| M13 SMILES, HSG (0ECk) | Sub-Training | 1 | 7 | 15 | 24 | 0 | 2 | 41 | 0.8824 | 1.0000 | 0.9512 | 0.9025 |
| Calibration | 17 | 17 | 1 | 0 | 35 | 1.0000 | 0.9444 | 0.9714 | 0.9444 | |||
| Test | 5 | 7 | 2 | 1 | 15 | 0.8333 | 0.7778 | 0.8000 | 0.6001 | |||
| M15 SMILES | Sub-Training | 2 | 6 | 10 | 30 | 0 | 1 | 41 | 0.9091 | 1.0000 | 0.9756 | 0.9380 |
| Calibration | 20 | 15 | 0 | 0 | 35 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | |||
| Test | 8 | 4 | 2 | 1 | 15 | 0.8889 | 0.6667 | 0.8000 | 0.5774 |
Where, T = Threshold, N = Epoch;The selected model is shown in bold face,.
Among all the 21 models developed in 3 different splits, model M13 (SMILES and HSG with 0ECk) from split-2 was found to be the best one.
The end point values calculate for M13 is shown below:.
Endpoint = 0.1282 (± 0.0048) + 0.04293 (± 0.00023) * DCW (1,7).
As can be seen from Table 1, the model M13 showed a satisfactory predictive ability. The values of the sensitivity, specificity, accuracy along with the MCC obtained for both the sub-training and calibration sets were highly encouraging. Indeed, the values attained for the test set (i.e. sensitivity, specificity and accuracy of 0.8333, 0.7778 and 0.8000, respectively) suggested the acceptable external predictive power of the classification based model. However, the MCC value of the test set was comparatively poor than the MCC values of the sub-training and calibration sets. Different structural attributes of the best model M13 (SMILES and HSG with 0ECk) from split-2 is depicted in Table S3.
QSAR based virtual screening
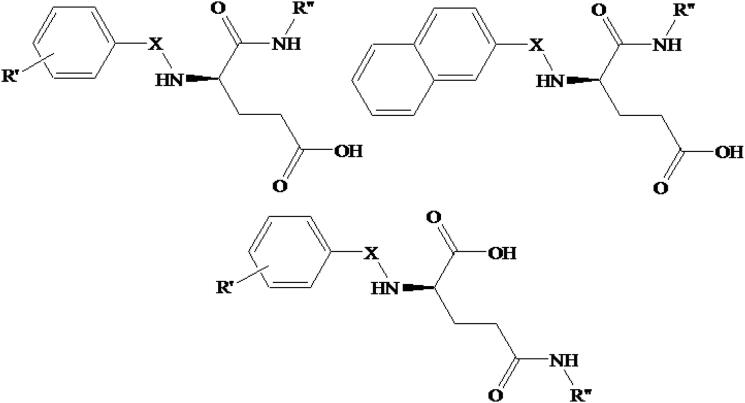
Since the QSAR models proved reasonably accurate, we used the best model to screen our in-house molecules (IH-001 – IH-067) from our previous publications (Adhikari et al., 2016; Halder et al., 2015; Mukherjee et al., 2017). General structures of our in-house compounds (IH-001 – IH-067) are depicted in Figure 3.
Figure 3.
General structures of our in-house compounds (IH-001 – IH-067).
These compounds were already reported as metalloproteinase inhibitors (Adhikari et al., 2016; Halder et al., 2015; Mukherjee et al., 2017). First, we predicted the compounds and then screening as per their potentiality in the Monte Carlo based classification QSAR model. Second, we defined an applicability criterion to choose the best hits.
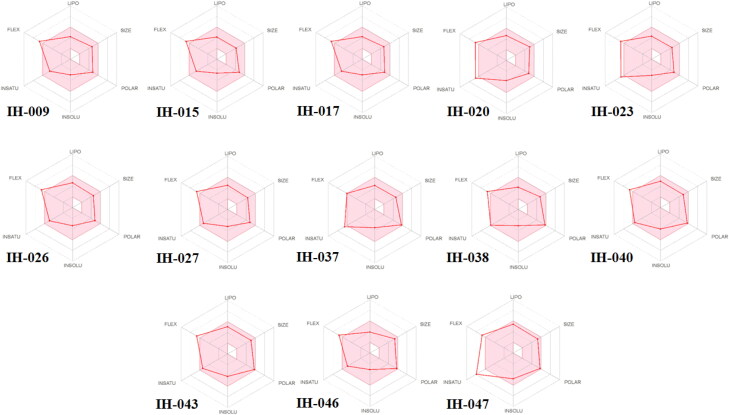
In this regards, we first screened the sixty seven in-house compounds. From the in-house database, a collection of 56 compounds were predicted as Active from the Monte Carlo based QSAR model (Table S4). After screening 56 similar compounds in SwissADME (Daina et al., 2017) − 13 compounds including IH-009, IH-015, IH-017, IH-020, IH-023, IH-027, IH-037, IH-038, IH-040, IH-043, IH-046, IH-047 were passed the ADME criteria (Figure 4).
Figure 4.
Radar plot of the in-house compounds after calculating ADME data by SwissADME server (http://www.swissadme.ch/) suggesting the drug-likeness [the pink area represents the optimal range of each properties.LIPO = Lipophilicity (between −0.7 and +5.0), SIZE = Molecular weight (between 150 and 500 g/mol), POLAR = Polarity (between 20 and 130Å2), INSOLU = Solubility (not higher than 6), INSATU = Saturation (fraction of carbons in the sp3 hybridization not less than 0.25), FLEX = Flexibility (no more than 9 rotatable bonds)].
Lastly, these potential chemotypes were considered for molecular docking study against the putative target SARS-CoV-2 PLpro.
Binding interactions of the in-house VS hits to SARS-CoV-2 PLpro
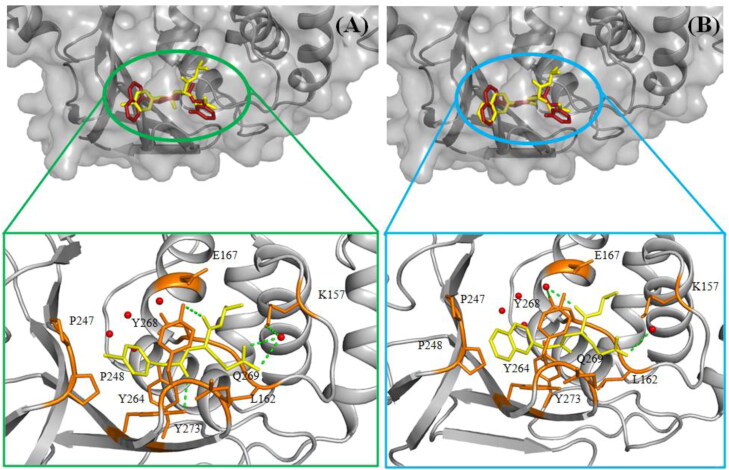
Molecular docking study offers a vital tool to predict the possible structural conformations between ligand and active sites of a receptor/enzyme. Here, molecular docking approach was employed on the in-house VS hits by using the AutoDock Vina to understand the docking/binding mode between in-house VS hits and SARS-CoV-2 PLpro. Meanwhile, the docking modes of two prototype in-house VS hits in the catalytic site amino acid residues of COVID-19 PLpro are illustrated in Figure 5.
Figure 5.
The docking modes of two prototype in-house VS hits (A) IH-009 and (B) IH-027 in the catalytic site amino acid residues of SARS-CoV-2 PLpro (proetin, grey cartoon; active site amino acids, orange stick; in-bound lignad, red stick; in-house molecules, yellow stick; water molecule, red ball; hydrogen bond interactions, light green dashed lines).
Furthermore, the re-dock binding pose of the in-bound PLpro inhibitor (red stick) on the SARS-CoV-2 active site are shown in Figure 5. Notably, the positions of the in-house VS hits in the COVID-19 PLpro catalytic site are basically the same from Figure 5, suggesting that in-house VS hits capture the right position in the PLpro cavity. This phenomenon validated the accuracy of docking study.
The docking poses of the all thirteen in-house VS hits are depicted in Figure S1. The entire in-house VS hits snugly occupied the binding site of SARS-CoV-2 Plpro. In addition, the Figure S1 pinnacle the superimposition of docking poses of thirteen in-house VS hits in the homology modelled SARS-CoV-2 PLpro. This observation justified that these investigated derivatives exhibit potential to be PLpro inhibitor and may be a valid weapon against SARS-CoV-2.
The binding interaction of IH-009 with SARS-CoV-2 PLpro revealed three hydrogen bonds with three amino acids (Leu162, Tyr264 and Tyr268), one π-π T-shaped interaction between phenyl ring and Tyr268, along with additional π-alkyl interaction between phenyl ring and Pro248. Apart from that the bromine molecule of IH-009 interacted with two proline amino acid residues (Pro 247 and 248). The carboxylic acid feature formed water mediated hydrogen bond interactions. These interactions were more or less consistence with other molecules also (Table S5). Notably, the napthyl ring formed two π-π T-shaped and two π-alkyl interactions. An additional π-sigma interaction was noticed where Tyr264 was involved as given in Table S5. However, the exact mechanism of the binding is still sketchy as it required further molecular dynamic simulation study. Moreover, the in vivo effects of these in-house VS hits would be needed to confirm the mechanism.
Conclusion
Here, we have constructed a classification based QSAR model that could be used as a tool for predicting new molecules and/or virtual screening. The model developed by Monte Carlo optimization based QSAR were followed by virtual screening of some in-house chemicals. Then ADME data driven screening was performed by SwissADME and identified compounds with good drug-likeness. Finally, molecular docking study of QSAR derived virtual hits was performed to increase the confidence in the final hypotheses. The molecular docking study performed against putative target SARS-CoV-2 PLpro suggesting potentiality of these investigated in-house molecules. Thus, it can be concluded that the in-house molecules have potential to use as a seed for drug design and optimization against SARS-CoV-2 PLpro. After extensive in vitro and in vivo studies, these in-house VS hits may be emerged as therapeutic options for COVID-19. This study may also motivate medicinal chemists to design similar type of compounds in hopes to trigger biological potency as well as efficacy without accruing toxicities.
Supplementary Material
Funding Statement
Financial assistance from the Council of Scientific and Industrial Research (CSIR), New Delhi, India in the form of a Senior Research Fellowship (SRF) [FILE NO.: 09/096(0967)/2019-EMR-I, Dated: 01-04-2019] to Sk. Abdul Amin is thankfully acknowledged. Tarun Jha is also thankful for the financial support from RUSA 2.0 of UGC, New Delhi, India to Jadavpur University, Kolkata, India. We are very much thankful to the Department of Pharmaceutical Technology, Jadavpur University, Kolkata, India and Department of Pharmaceutical Sciences, Dr. Harisingh Gour University, India for providing the research facilities.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Aanouz I., Belhassan A., El Khatabi K., Lakhlifi T., El Idrissi M., & Bouachrine M. (2020). Moroccan Medicinal plants as inhibitors of COVID-19: Computational investigations. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1758790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeoye A. O., Oso B. J., Olaoye I. F., Tijjani H., & Adebayo A. I. (2020). Repurposing of chloroquine and some clinically approved antiviral drugs as effective therapeutics to prevent cellular entry and replication of coronavirus. Journal of Biomolecular Structure and Dynamics, 1–4. 10.1080/07391102.2020.1765876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari N., Baidya S. K., Saha A., & Jha T. (2017). Structural insight into the viral 3C-like protease inhibitors: Comparative SAR/QSAR approaches In Satya P. Gupta (Ed.), Viral proteases and their inhibitors (pp. 317–409). Academic Press. [Google Scholar]
- Adhikari N., Halder A. K., Mallick S., Saha A., Saha K. D., & Jha T. (2016). Robust design of some selective matrix metalloproteinase-2 inhibitors over matrix metalloproteinase-9 through in silico/fragment-based lead identification and de novo lead modification: Syntheses and biological assays . Bioorganic & Medicinal Chemistry, 24(18), 4291–4309. 10.1016/j.bmc.2016.07.023 [DOI] [PubMed] [Google Scholar]
- Ahmad S., Abbasi H. W., Shahid S., Gul S., & Abbasi S. W. (2020). Molecular docking, simulation and MM-PBSA studies of Nigella Sativa compounds: A computational quest to identify potential natural antiviral for COVID-19 treatment. Journal of Biomolecular Structure and Dynamics, 1–6. 10.1080/07391102.2020.1775129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khafaji K., Al-DuhaidahawiL D., & Tok T. T. (2020). Using integrated computational approaches to identify safe and rapid treatment for SARS -CoV-2. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1764392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin M., & Abbas G. (2020). Docking study of Chloroquine and Hydroxychloroquine interaction with SARS-CoV-2 spike glycoprotein-An in silico insight into the comparative efficacy of repurposing antiviral drugs. Journal of Biomolecular Structure and Dynamics, 1, 1–11. 10.1080/07391102.2020.1775703 [DOI] [PubMed] [Google Scholar]
- Amin S. A., Adhikari N., & Jha T. (2018). Design of aminopeptidase N inhibitors as anti-cancer agents. Journal of Medicinal Chemistry, 61(15), 6468–6490. 10.1021/acs.jmedchem.7b00782 [DOI] [PubMed] [Google Scholar]
- Anwar F., Altayb H. N., Al-Abbasi F. A., Al-Malki A. L., Kamal M. A., & Kumar V. (2020). Antiviral effects of probiotic metabolites on COVID-19. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1775123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ap K., & Vs A. (2020). Design of multi-epitope vaccine candidate against SARS-CoV-2: A in-silico study. Journal of Biomolecular Structure & Dynamics. 10.1080/07391102.2020.1770127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya A., & Dwivedi V. D. (2020). Synergistic effect of Vitamin D and Remdesivir can fight COVID-19. Journal of Biomolecular Structure and Dynamics, 1–2. 10.1080/07391102.2020.1773929 [DOI] [PubMed] [Google Scholar]
- Báez-Santos Y. M., Barraza S. J., Wilson M. W., Agius M. P., Mielech A. M., Davis N. M., Baker S. C., Larsen S. D., & Mesecar A. D. (2014). X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human coronavirus papain-like proteases. Journal of Medicinal Chemistry, 57(6), 2393–2412. 10.1021/jm401712t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Báez-Santos Y. M., John S. E., & Mesecar A. D. (2015). The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds. Antiviral Research, 115, 21–38. 10.1016/j.antiviral.2014.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay D., Kreatsoulas C., Brady P. G., Boyer J., He Z., Scavello G., Peryea T., Jadhav A., Nguyen D.-T., & Guha R. (2019). Scaffold-based analytics: Enabling hit-to-lead decisions by visualizing chemical series linked across large datasets. Journal of Chemical Information and Modeling, 59(11), 4880–4892. 10.1021/acs.jcim.9b00243 [DOI] [PubMed] [Google Scholar]
- Banerjee S., Amin S. A., Baidya S. K., Adhikari N., & Jha T. (2020). Exploring the structural aspects of ureido-amino acid-based APN inhibitors: A validated comparative multi-QSAR modelling study. SAR and QSAR in Environment Research, 31(5), 325–345. 10.1080/1062936X.2020.1734080 [DOI] [PubMed] [Google Scholar]
- Basit A., Ali T., & Rehman S. U. (2020). Truncated human Angiotensin Converting Enzyme 2; a potential inhibitor of SARS-CoV-2 spike glycoprotein and potent COVID-19 therapeutic agent. Journal of Biomolecular Structure and Dynamics, 1–7. 10.1080/07391102.2020.1768150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj V. K., Singh R., Sharma J., Rajendran V., Purohit R., & Kumar S. (2020). Identification of bioactive molecules from Tea plant as SARS-CoV-2 main protease inhibitors. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1766572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boopathi S., Poma A. B., & Kolandaivel P. (2020). Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkotoky S., & Banerjee M. (2020). A computational prediction of SARS-CoV-2 structural protein inhibitors from Azadirachta indica (Neem). Journal of Biomolecular Structure and Dynamics, 1–7. 10.1080/07391102.2020.1774419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra A., Gurjar V., Qamar I., & Singh N. (2020). Identification of potential inhibitors of SARS-COV-2 endoribonuclease (EndoU) from FDA approved drugs: A drug repurposing approach to find therapeutics for COID19. Journal of Biomolecular Structure and Dynamics, 1–6. 10.1080/07391102.2020.1775127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K. W., Cheng S. C., Chen W. Y., Lin M. H., Chuang S. J., Cheng I. H., Sun C. Y., & Chou C. Y. (2015). Thiopurine analogs and mycophenolic acid synergistically inhibit the papain-like protease of Middle East respiratory syndrome coronavirus . Antiviral Research, 115, 9–16. 10.1016/j.antiviral.2014.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C.-Y., Chien C.-H., Han Y.-S., Prebanda M. T., Hsieh H.-P., Turk B., Chang G.-G., & Chen X. (2008). Thiopurine analogues inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biochemical Pharmacology, 75(8), 1601–1609. 10.1016/j.bcp.2008.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury C. (2020). Fragment tailoring strategy to design novel chemical entities as potential binders of novel corona virus main protease. Journal of Biomolecular Structure and Dynamics, 1–5. 10.1080/07391102.2020.1771424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A., Michielin O., & Zoete V. (2017). SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Reports, 7, 42717 10.1038/srep42717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Sarmah S., Lyndem S., & Roy A. S. (2020). An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1763201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira O. V., Rocha G. B., Paluch A. S., & Costa L. T. (2020). Repurposing approved drugs as inhibitors of SARS-CoV-2 S-protein from molecular modeling and virtual screening. Journal of Biomolecular Structure and Dynamics, 1–4. 10.1080/07391102.2020.1772885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DS Visualizer 3.5 , Accelrys Software Inc. [Google Scholar]
- Dutta S., Halder A. K., Adhikari N., Amin S. A., Das S., Saha A., & Jha T. (2019). Synthesis, anticancer activity, structure-activity relationship and binding mode of interaction studies of substituted pentanoic acids. Future Medicinal Chemistry, 11(14), 1679–1702. 10.4155/fmc-2018-0361 [DOI] [PubMed] [Google Scholar]
- Elasnaoui K., & Chawki Y. (2020). Using X-ray images and deep learning for automated detection of coronavirus disease. Journal of Biomolecular Structure and Dynamics, 1–22. 10.1080/07391102.2020.1767212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A. (2020. a). SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An insilico perspective. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1761882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A. (2020. b). Natural products may interfere with SARS-CoV-2 attachment to the host cell. Journal of Biomolecular Structure and Dynamics, 1–6. 10.1080/07391102.2020.1761881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A., & Azzam E. B. (2020). Novel Guanosine Derivatives against MERS CoV polymerase: An in silico perspective. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1758789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmezayen A. D., Al-Obaidi A., Şahin A. T., & Yelekçi K. (2020). Drug repurposing for coronavirus (COVID-19): In silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1758791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enayatkhani M., Hasaniazad M., Faezi S., Guklani H., Davoodian P., Ahmadi N., Einakian M. A., Karmostaji A., & Ahmadi K. (2020). Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: An in silico study. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1756411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enmozhi S. K., Raja K., Sebastine I., & Joseph J. (2020). Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: An in silico approach. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1760136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Basu D., Matthews K., Taylor J., Jones G., Pickles R., Baric R., & Engel D. A. (2011). Yeast based small molecule screen for inhibitors of SARS-CoV. PLoS One, 6, e28479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A. K., Brindisi M., Shahabi D., Chapman M. E., & Mesecar A. D. (2020). Drug development and medicinal chemistry efforts toward SARS-coronavirus and Covid-19 therapeutics. ChemMedChem, 15(11), 907–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A. K., Takayama J., Aubin Y., Ratia K., Chaudhuri R., Baez Y., Sleeman K., Coughlin M., Nichols D. B., Mulhearn D. C., Prabhakar B. S., Baker S. C., Johnson M. E., & Mesecar A. D. (2009). Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors for the severe acute respiratory syndrome-coronavirus papain-like protease. Journal of Medicinal Chemistry, 52(16), 5228–5240. 10.1021/jm900611t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A. K., Takayama J., Rao K. V., Ratia K., Chaudhuri R., Mulhearn D. C., Lee H., Nichols D. B., Baliji S., Baker S. C., Johnson M. E., & Mesecar A. D. (2010). Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: Design, synthesis, protein-ligand X-ray structure and biological evaluation . Journal of Medicinal Chemistry, 53(13), 4968–4979. 10.1021/jm1004489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M. K., Vemula S., Donde R., Gouda G., Behera L., & Vadde R. (2020). In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1751300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyebi G. A., Ogunro O. B., Adegunloye A. P., Ogunyemi O. M., & Afolabi S. O. (2020). Potential Inhibitors of Coronavirus 3-Chymotrypsin-Like Protease (3CLpro): An in-silico screening of Alkaloids and Terpenoids from African medicinal plants. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1764868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjo R., Setola V., Roth B. L., & Tropsha A. (2012). Chemocentric informatics approach to drug discovery: Identification and experimental validation of selective estrogen receptor modulators as ligands of 5-hydroxytryptamine-6 receptors and as potential cognition enhancers. Journal of Medicinal Chemistry, 55(12), 5704–5719. 10.1021/jm2011657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder A. K., Mallick S., Shikha D., Saha A., Saha K. D., & Jha T. (2015). Design of dual MMP-2/HDAC-8 inhibitors by pharmacophore mapping, molecular docking, synthesis and biological activity. RSC Advances, 5(88), 72373–72386. 10.1039/C5RA12606A [DOI] [Google Scholar]
- Halder A. K., Saha A., & Jha T. (2013). Exploration of structural and physicochemical requirements andsearch of virtual hits for aminopeptidase N inhibitors. Molecular Diversity, 17(1), 123–137. 10.1007/s11030-013-9422-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A., Paray B. A., Hussain A., Qadir F. A., Attar F., Aziz F. M., Sharifi M., Derakhshankhah H., Rasti B., Mehrabi M., & Shahpasand K. (2020). A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1754293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendaus M. A. (2020). Remdesivir in the treatment of Coronavirus Disease 2019 (COVID-19): A simplified summary. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1767691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendaus M. A., & Jomha F. A. (2020). Covid-19 induced superimposed bacterial infection. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1772110 [DOI] [PubMed] [Google Scholar]
- http://www.swissadme.ch/ (as accessed on 24th May 2020)
- http://www.who.int/csr/sars/archive/2003_05_07a/en (as accessed on 10th May 2020)
- https://prosa.services.came.sbg.ac.at/prosa.php (as accessed on 10th May 2020)
- https://pymol.org/2/ (as accessed on 10th May 2020)
- https://servicesn.mbi.ucla.edu/SAVES/ (as accessed on 10th May 2020)
- https://servicesn.mbi.ucla.edu/Verify3D/ (as accessed on 10th May 2020)
- https://spdbv.vital-it.ch/ (as accessed on 10th May 2020)
- https://swissmodel.expasy.org/ (as accessed on 10th May 2020)
- https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19—11-march-2020 (as accessed on 17th May 2020)
- https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (as accessed on 3rd June 2020)
- Islam R., Parves R., Paul A. S., Uddin N., Rahman M. S., Mamun A. A., Hossain M. N., Ali M. A., & Halim M. A. (2020). A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. Journal of Biomolecular Structure and Dynamics, 1–20. 10.1080/07391102.2020.1761883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Amin S. A., Adhikari N., Jha T., & Gayen S. (2020). Good and bad molecular fingerprints for human rhinovirus 3C protease inhibition: Identification, validation, and application in designing of new inhibitors through Monte Carlo-based QSAR study. Journal of Biomolecular Structure & Dynamics, 38(1), 66–77. 10.1080/07391102.2019.1566093 [DOI] [PubMed] [Google Scholar]
- Joshi R. S., Jagdale S. S., Bansode S. B., Shankar S. S., Tellis M. B., Pandya V. K., Chugh A., Giri A. P., & Kulkarni M. J. (2020). Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1760137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. T., Ali A., Wang Q., Irfan M., Khan A., Zeb M. T., Zhang Y. J., Chinnasamy S., & Wei D. Q. (2020). Marine natural compounds as potents inhibitors against the main protease of SARS-CoV-2. A molecular dynamic study. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1769733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan R. J., Jha R. K., Amera G. M., Jain M., Singh E., Pathak A., Singh R. P., Muthukumaran J., & Singh A. K. (2020). Targeting SARS-CoV-2: A systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2′-O-ribose methyltransferase. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1753577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A., Zia K., Ashraf S., Uddin R., & Ul-Haq Z. (2020). Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1751298 [DOI] [PubMed] [Google Scholar]
- Kumar D., Kumari K., Jayaraj A., Kumar V., Kumar R. V., Dass S. K., Chandra R. & Singh P. (2020). Understanding the binding affinity of noscapines with protease of SARS-CoV-2 for COVID-19 using MD simulations at different temperatures. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1752310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M. H., Moses D. C., Hsieh C. H., Cheng S. C., Chen Y. H., Sun C. Y., & Chou C. Y. (2018). Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes. Antiviral Research, 150, 155–163. 10.1016/j.antiviral.2017.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo-Galo N., Terrazas-López M., Martínez-Martínez A., & Díaz-Sánchez Á. G. (2020). FDA-approved thiol-reacting drugs that potentially bind into the SARS-CoV-2 main protease, essential for viral replication. Journal of Biomolecular Structure and Dynamics, 1–2. 10.1080/07391102.2020.1764393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahanta S., Chowdhury P., Gogoi N., Goswami N., Borah D., Kumar R., Chetia D., Borah P., Buragohain A. K., & Gogoi B. (2020). Potential anti-viral activity of approved repurposed drug against main protease of SARS-CoV-2: An in silico based approach. Journal of Biomolecular Structure and Dynamics, 1–5. 10.1080/07391102.2020.1768902 [DOI] [PubMed] [Google Scholar]
- Mittal L., Kumari A., Srivastava M., Singh M., & Asthana S. (2020). Identification of potential molecules against COVID-19 main protease through structure-guided virtual screening approach. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1768151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A., Adhikari N., & Jha T. (2017). A pentanoic acid derivative targeting matrix metalloproteinase-2 (MMP-2) induces apoptosis in a chronic myeloid leukemia cell line. European Journal of Medicinal Chemistry, 141, 37–50. 10.1016/j.ejmech.2017.09.052 [DOI] [PubMed] [Google Scholar]
- Muralidharan N., Sakthivel R., Velmurugan D., & Gromiha M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 Protease against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–6. 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- Babadaei N., Hasan M. M., Vahdani A., Haj Bloukh Y., Sharifi S., Kachooei M., Haghighat E., S., & Falahati M. (2020. b). Development of Remdesivir repositioning as a nucleotide analog against COVID-19 RNA dependent RNA polymerase. Journal of Biomolecular Structure and Dynamics, 1–2. 10.1080/07391102.2020.1767210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejadi Babadaei M. M., Hasan A., Haj Bloukh S., Edis Z., Sharifi M., Kachooei E., & Falahati M. (2020. a). The expression level of angiotensin-converting enzyme 2 determine the severity of COVID-19: Lung and heart tissue as targets. Journal of Biomolecular Structure and Dynamic, 1–3. 10.1080/07391102.2020.1767211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paniri A., Hosseini M. M., & Akhavan-Niaki H. (2020). First comprehensive computational analysis of functional consequences of TMPRSS2 SNPs in susceptibility to SARS-CoV-2 among different populations. Journal of Biomolecular Structure and Dynamics, 1–8. 10.1080/07391102.2020.1767690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant S., Singh M., Ravichandiran V., Murty U. S. N., & Srivastava H. K. (2020). Peptide-like and small-molecule inhibitors against Covid-19. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1757510Z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-Y., Kim J. H., Kim Y. M., Jeong H. J., Kim D. W., Park K. H., Kwon H.-J., Park S.-J., Lee W. S., & Ryu Y. B. (2012). Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases. Bioorganic & Medicinal Chemistry, 20(19), 5928–5935. 10.1016/j.bmc.2012.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil V. M., Singhal S., & Masand N. (2020). A systematic review on use of aminoquinolines for the therapeutic management of COVID-19: Efficacy, safety and clinical trials. Life Sciences, 254, 117775 10.1016/j.lfs.2020.117775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillaiyar T., Meenakshisundaram S., & Manickam M. (2020). Recent discovery and development of inhibitors targeting coronaviruses. Drug Discovery Today. [DOI] [PMC free article] [PubMed]
- Polishchuk P. (2017). Interpretation of quantitative structure-activity relationship models: Past, present, and future. Journal of Chemical Information and Modeling, 57(11), 2618–2639. 10.1021/acs.jcim.7b00274 [DOI] [PubMed] [Google Scholar]
- Ratia K., Pegan S., Takayama J., Sleeman K., Coughlin M., Baliji S., Chaudhuri R., Fu W., Prabhakar B. S., Johnson M. E., Baker S. C., Ghosh A. K., & Mesecar A. D. (2008). A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proceedings of the National Academy of Sciences of the United States of America, 105(42), 16119–16124. 10.1073/pnas.0805240105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratia K., Saikatendu K. S., Santarsiero B. D., Barretto N., Baker S. C., Stevens R. C., & Mesecar A. D. (2006). Severe acute respiratory syndrome coronavirus papain-like protease: Structure of a viral deubiquitinating enzyme. Proceedings of the National Academy of Sciences, 103(15), 5717–5722. 10.1073/pnas.0510851103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma P., Sekhar N., Prajapat M., Avti P., Kaur H., Kumar S., Singh S., Kumar H., Prakash A., Dhibar D. P., & Medhi B. (2020). In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1753580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S. K., Shakya A., Prasad S. K., Singh S., Gurav N. S., Prasad R. S., & Gurav S. S. (2020). An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. Journal of Biomolecular Structure and Dynamics, 1–3. 10.1080/07391102.2020.1762741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sk M. F., Roy R., Jonniya N. A., Poddar S., & Kar P. (2020). Elucidating biophysical basis of binding of inhibitors to SARS-CoV-2 main protease by using molecular dynamics simulations and free energy calculations. Journal of Biomolecular Structure and Dynamics, 1–21. 10.1080/07391102.2020.1768149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toropova A. P., Toropov A. A., & Benfenati E. (2015). A quasi-QSPR modelling for the photocatalytic decolourization rate constants and cellular viability (CV%) of nanoparticles by CORAL. SAR and QSAR in Environmental Research, 26(1), 29–40. 10.1080/1062936X.2014.984327 [DOI] [PubMed] [Google Scholar]
- Toropov A. A., Toropova A. P., Puzyn T., Benfenati E., Gini G., Leszczynska D., & Leszczynski J. (2013). QSAR as a random event: Modeling of nanoparticles uptake in PaCa2 cancer cells. Chemosphere, 92(1), 31–37. 10.1016/j.chemosphere.2013.03.012 [DOI] [PubMed] [Google Scholar]
- Toropov A. A., Toropova A. P., Raitano G., & Benfenati E. (2018). CORAL: Building up QSAR models for the chromosome aberration test. Saudi Journal of Biological Sciences, 26(6), 1101–1106. 10.1016/j.sjbs.2018.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., & Olson A. J. (2009). AutoDockVina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31, 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hilten H. N., Chevillard F., & Kolb P. (2019). Virtual compound libraries in computer-assisted drug discovery. Journal of Chemical Information and Modeling, 59, 644–651. [DOI] [PubMed] [Google Scholar]
- Wahedi H. M., Ahmad S., & Abbasi S. W. (2020). Stilbene-based natural compounds as promising drug candidates against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–6. 10.1080/07391102.2020.1762743 [DOI] [PubMed] [Google Scholar]
- Worachartcheewan A., Mandi P., Prachayasittikul V., Toropova A. P., Toropov A. A., & Nantasenamat C. (2014). Large-scale QSAR study of aromatase inhibitors using SMILES-based descriptors. Chemometrics and Intelligent Laboratory Systems, 138, 120–126. 10.1016/j.chemolab.2014.07.017 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.