Abstract
In the present study, we have explored the interaction of the active components from 10 different medicinal plants of Indian origin that are commonly used for treating cold and respiratory-related disorders, through molecular docking analysis. In the current scenario, COVID-19 patients experience severe respiratory syndromes, hence it is envisaged from our study that these traditional medicines are very likely to provide a favourable effect on COVID-19 infections. The active ingredients identified from these natural products are previously reported for antiviral activities against large group of viruses. Totally 47 bioactives identified from the medicinal plants were investigated against the structural targets of SARS-CoV-2 (Mpro and spike protein) and human ACE2 receptor. The top leads were identified based on interaction energies, number of hydrogen bond and other parameters that explain their potency to inhibit SARS-CoV-2. The bioactive ligands such as Cucurbitacin E, Orientin, Bis-andrographolide, Cucurbitacin B, Isocucurbitacin B, Vitexin, Berberine, Bryonolic acid, Piperine and Magnoflorine targeted the hotspot residues of SARS-CoV-2 main protease. In fact, this protease enzyme has an essential role in mediating the viral replication and therefore compounds targeting this key enzyme are expected to block the viral replication and transcription. The top scoring conformations identified through docking analysis were further demonstrated with molecular dynamics simulation. Besides, the stability of the conformation was studied in detail by investigating the binding free energy using MM-PBSA method. Overall, the study emphasized that the proposed hit Cucurbitacin E and orientin could serve as a promising scaffold for developing anti-COVID-19 drug.
Communicated by Ramaswamy H. Sarma
Keywords: Ethnomedicine, drug discovery, molecular docking, MD simulation, Binding energy, SARS-CoV-2 inhibitors
Introduction
At the peak of COVID19 pandemic which shudder the whole world through human to human transmission of SARS-CoV-2, devouring numerous lives, medical professionals and researchers all over the globe are in pursuit for the discovery of prophylactic or anaphylactic remedy to surmount the condition (Al-Khafaji et al., 2020; Boopathi et al., 2020; Pant et al., 2020). Thus far, no specific vaccine or antiviral therapeutic has been either discovered or anticipated as a conventional medication for this viral infection (Zumla et al., 2016). Current treatment strategy includes supportive care which is extended with supplementation of combination of broad-spectrum antibiotics, antivirals, cortecosteroids and convalescent plasma (Chen et al., 2020; Habibzadeh & Stoneman, 2020). SARS-CoV-2 was found to be an enveloped single-stranded RNA-type beta coronavirus which shares 79.5% sequence similarity with its antecedent class SARS-CoV (Chen et al., 2020; Wu et al., 2020). During the previous outbreak of SARS crisis due to SARS-CoV, several research groups have conducted in vitro analysis and randomized trials on use of natural herbal medicines to control the adverse effects of infection and evidenced their use as effective (Hsu et al., 2008; Jang et al., 2009; Kim et al., 2008; Lau et al., 2005a, 2005b; Li et al., 2005; Wen et al., 2011). As novel coronavirus is closely related to SARS coronavirus, it can be suggested that natural herbal medicine may have potential use in existing health crisis (Aanouz et al., 2020; Elfiky, 2020b; Islam et al., 2020; Umesh et al., 2020; Wahedi et al., 2020). In addition to this, for decades, natural herbal medicine has been expended for prevention and cure of numerous ailments concerning human wellbeing.
The WHO estimates that, 80% of population from underdeveloped realm rely mostly on the traditional medicines. In addition to this, WHO listed around 21,000 plants which are known for their therapeutic potential around the world among which 2500 varieties are found around India including 150 species with large scale commercial usage (Shukla et al., 2019). In India, Siddha is one among the three traditional medical systems practiced and the other two being Ayurveda and Unani. In Siddha medicine Nilavembu Kudineer Chooranam (NKC) has been recommended for the preclusion and control of all the types of viral infectious diseases. NKC is an herbal formulation comprising nine varieties of plant materials viz. Nilavembu (Andrographis paniculata), Vetiver (Vetiveria zizanioides), Vilamiccam ver (Plectranthus vettiveroides), Santanam (Santalum album), Pei pudal (Trichosanthes dioica), Korai kizhangu (Cyperus rotandus), Chukku (Zingeber officinale), Milagu (Piper nigrum) and Parpatakam (Mollugo cerviana) (Kavinilavan et al., 2017). NKC has been reported to have therapeutic applications such as antiviral, antipyretic, antibacterial, antiulcer, antioxidant, analgesic etc (Jain et al., 2019). On the other hand, it has also been stated to act as an immunostimulant and immunomodulant which enhances the host immune system and modulates the defence response that helps to protect the complications of infections (Kamalarajan et al., 2019; Nakkeeran et al., 2016; Ramanathan et al., 2019). Previously, NKC has been shown to alleviate the symptoms of dengue and chikungunya (Anbarasu et al., 2011; Mattummal et al., 2018). In fact, Government of Tamil Nadu have taken initiatives in supplying decoction of NKC to patients visiting the primary health care centres and also promoted the drink for prevention and control of morbidity level during outbreak of viral fever (Ramanathan et al., 2019). Owing to their medicinal properties and specifically antiviral potential, the present study aims at investigating the therapeutic efficacy of active components of NKC and natural constituents from other known medicinal plants (Tinospora cordifolia and Anethumsowa) which are also used as combinations to treat respiratory diseases. Phytomolecules from the components of NKC and the aforementioned medicinal plants against the two major viral targets spike glycoprotein and protease and a host target human angiotensin-converting enzyme 2 (ACE2) receptor, which is the entry point for viral encounter were examined with the prospects of identifying potential drug candidate(s) against COVID19 infection. Thus, ethnomedicines identified from the study through in silico approaches can combat the global health crisis instigated by SARS-CoV-2 (Abdelli et al., 2020; Elfiky & Azzam, 2020; Elmezayen et al., 2020; Enayatkhani et al., 2020; Enmozhi et al., 2020, Elfiky, 2020a; Gupta et al., 2020; Sarma et al., 2020; Sinha et al., 2020). Unlike modern medicines, the antique herbal remedies do not possess side effects, indeed they establish natural resistance to alleviate the viral infection by boosting the immune system. Thus, the main objective of the study is to identify a potent phytocompound as inhibitor against SARS-CoV-2 drug targets from vintage medicinal plants through computational investigations.
Materials and methods
Bioactive ligands
The study focuses predominantly on bioactive ligands from ten traditional medicinal plants that are primarily used in the treatment of respiratory illness. The active components were selected and screened against SARS-CoV-2 main protease, spike (S) protein and host cell ACE2 receptor. The list of active leads used in the present study is shown in Table 1. Ligand 3D structure was obtained in .sdf format from PubChem database and converted into .pdb format through online program (https://cactus.nci.nih.gov/translate/). PubChem ID of bioactives are provided in Supplementary Table 1.
Table 1.
List of natural products that are commonly used to treat fever, cold and respiratory related ailments in traditional South Indian medicine system. The active ingredients extracted from these natural products are used for docking with SARS-CoV-2 drug targets.
| S. No | Natural products Tamil name | Scientific name | Accepted Scientific name | Active ingredients |
|---|---|---|---|---|
| 1. | Nilavembu | Andrographis paniculata | Andrographis paniculata (Burm.f.) Nees | Andrographolide, Bis-andrographolide, Caffeic acid |
| 2. | Vetiver | Chrysopogon zizanoides | Chrysopogon zizanioides (L.) Roberty | Vetivone, α-cadinene, α-calacorene |
| 3. | Korai kizhangu | Cyperus rotundus | Cyperus rotundus L. | Cyperene, β-selinene |
| 4. | Cittapiryan | Mollugo cerviana | Mollugo cerviana (L.) Ser. | Vitexin, Orientin |
| 5. | Milagu | Piper nigrum | Piper nigrum L. | Piperine, p-cymene, D-limonene |
| 6. | Santanam | Santalum album | Santalum album L. | α, β-santalol, Vanillic acid |
| 7. | Pei pudal | Trichosanthes cucumerina | Trichosanthes cucumerina L. | Bryonolic acid, Cucurbitacin B, Cucurbitacin E, Isocucurbitacin B, β-sitosterol, Stigmasterol |
| 8. | Chukku | Zingiber officinale Roscoe | Zingiber officinale Roscoe | 6-shogaol, 6-ginerol, zingiberol, alpha pinene, beta pinene, camphene, Limonene, myrcene |
| 9. | Seenthilkodi | Tinospora cordifolia | Tinospora cordifolia (Willd.) Hook.f. & Thomson | Tinosporin, Berberine, Palmatine, Magnoflorine, |
| 10. | Sathakuppai | Anethum sowa | Anethum sowa Roxb. | α-phellandrene,Carvone and Limonene |
Molecular docking
Crystal Structures of SARS-CoV-2 main protease (PDB ID: 5R82.pdb), spike protein (PDB ID: 6VYB.pdb) and human entry receptor ACE2 (PDB ID:1R42.pdb) were retrieved from RCSB PDB database (http://www.rcsb.org/pdb). In order to minimize the energy, PDB structures of the target proteins were refined through ModRefiner algorithm (Xu & Zhang, 2011). Ligands were optimized by adding hydrogen atoms and energy minimization was performed with MMFF94 Force Field in Autodock v4.2 (Norgan et al., 2011). Grid box was defined on binding pocket of target protein and the grid points were extended in all directions to encompass the binding region. Molecular docking was performed through Autodock v4.2 by tethering ligand to target protein and the binding affinity was determined through kcal mol−1. The pose with highest binding energy was extracted and analysed in Biovia Discovery Studio 4.5 software (Systemes, 2015).
Molecular dynamics (MD) simulation
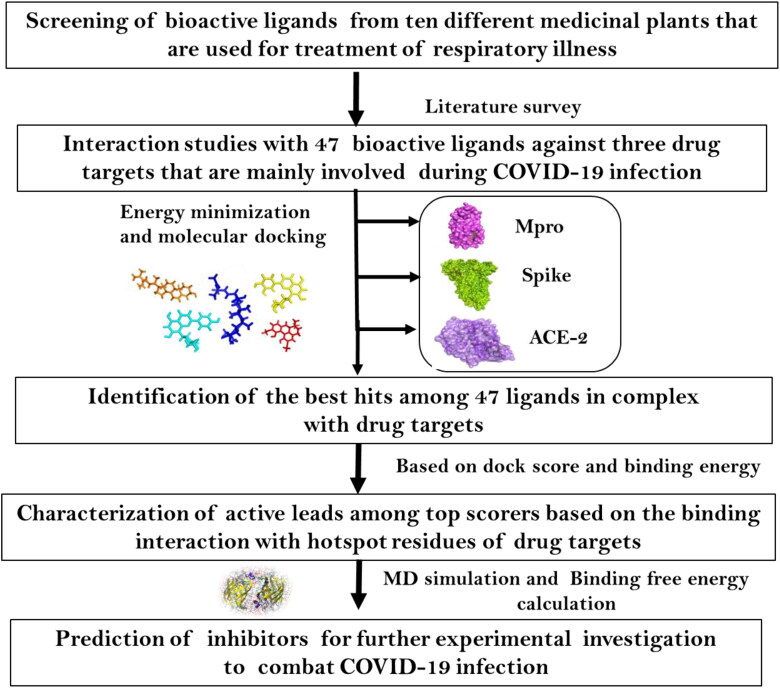
The top two ligands showing interaction with Mpro (revealed through docking analysis) were further subjected to 20 ns MD simulation studies. The energy and structural properties of the ligand-receptor complexes were determined through Gromacs 5.1.4 simulation package using CHARMM force field. The protein-ligand complex was inserted into a cubic box of 10 Å dimension and solvated using SPCE water model and the system was neutralized by adding counter ions (0.15 M NaCl). An initial minimization of 1000-step was carried out by applying steepest descent algorithm followed by 2000-step using conjugate gradient minimization. The system was gradually heated to 300 K for 100 ps and then equilibrated using NVT ensemble for 100 ps and retained at constant temperature. Sequentially, the system was equilibrated using NPT ensemble for 1000 ps. MD production run for 20 ns was performed with a time step of 2 ps and the trajectories were recorded at every step (Beema Shafreen et al., 2014). The structural behaviour of Mpro protein in the presence and absence of the drug molecules were calculated through Root mean square deviation (RMSD) and Root mean square fluctuation (RMSF). Overall methodology is represented as a flowchart in Figure 1 (Borgio et al., 2020).
Figure 1.
Flow chart representing the summary of the work.
Binding energy calculation
The binding free energy between the protein and drug molecules were calculated using molecular mechanics/Poisson-Boltzmann surface area (MM-PBSA) method in Gromacs (g_mmpbsa) software. The following equation was used to calculate the binding free energy.
Where ΔGBind is the binding free energy; ΔEMM is the interaction energies such as ΔEvdw (van der Waals interaction energy) and ΔEelec (electrostatic interaction energy) between the protein and drug complex; ΔGsol is the solvation energy comprising ΔGGB (polar components) and ΔGSA (non-polar components); TΔS is the entropy change at temperature T.
Results
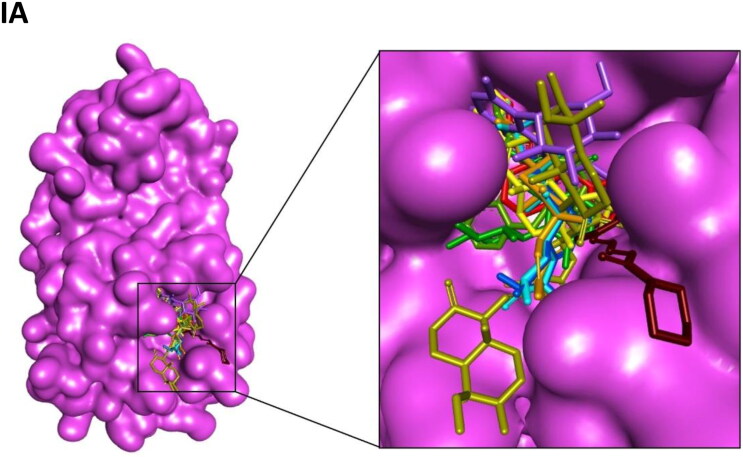
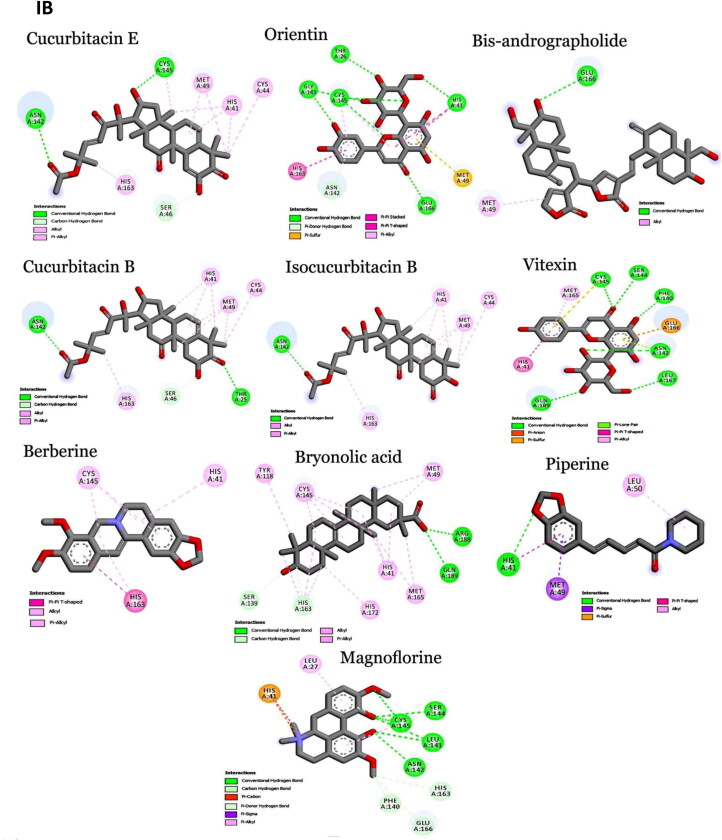
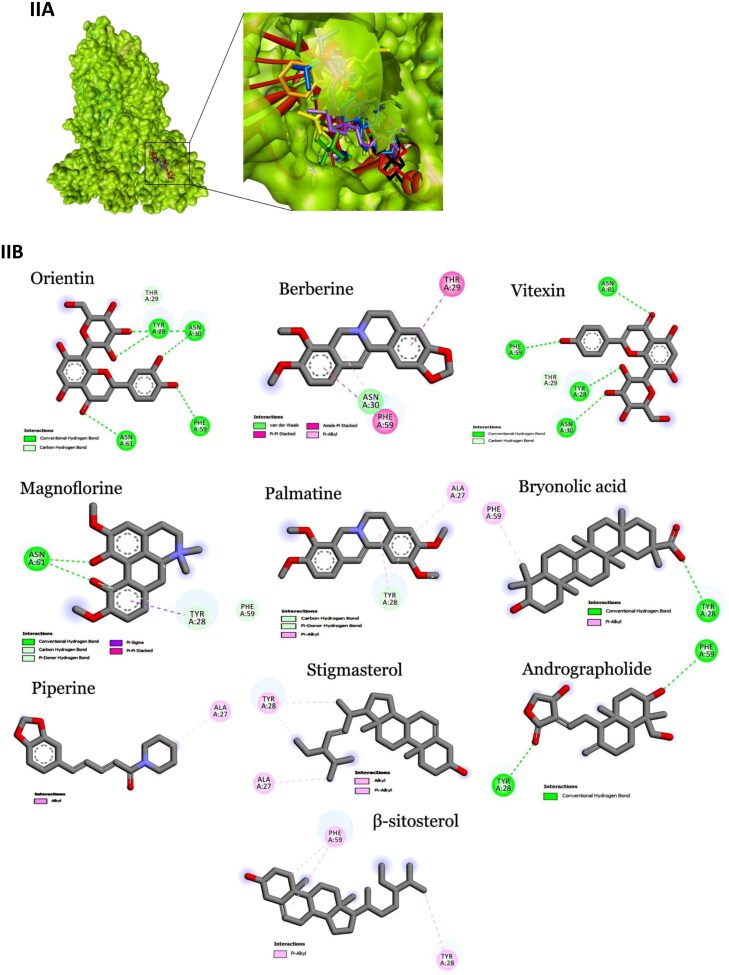
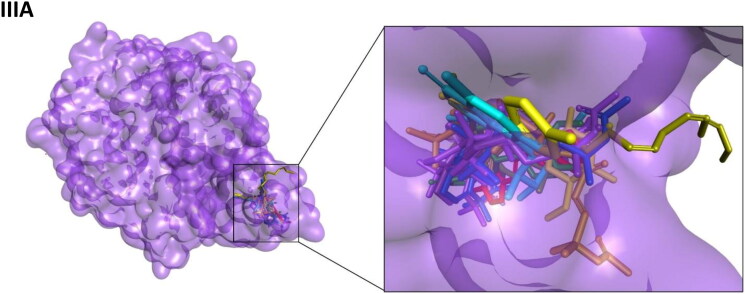
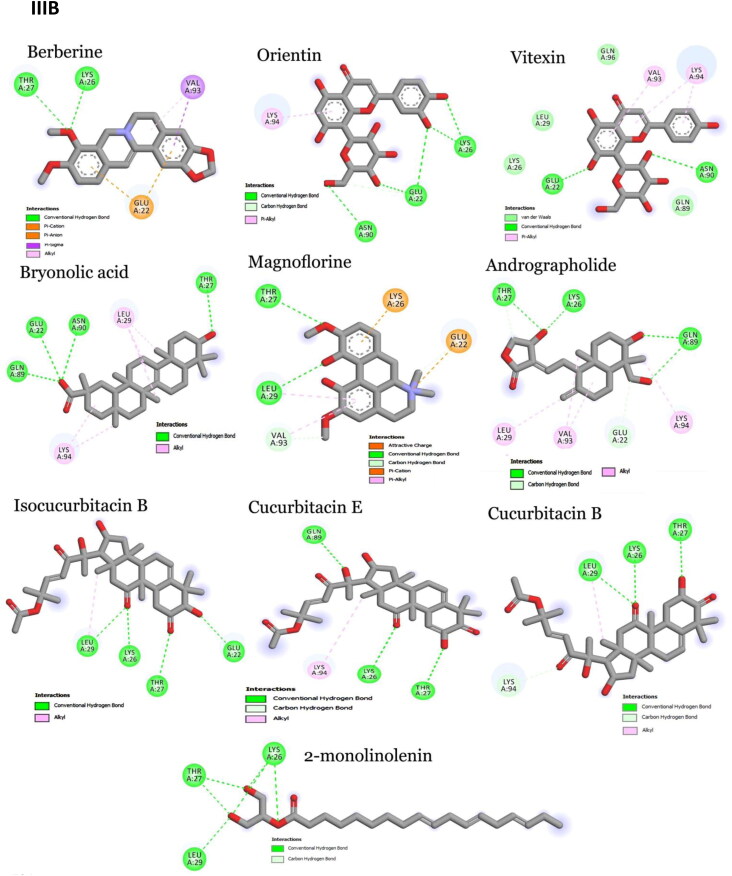
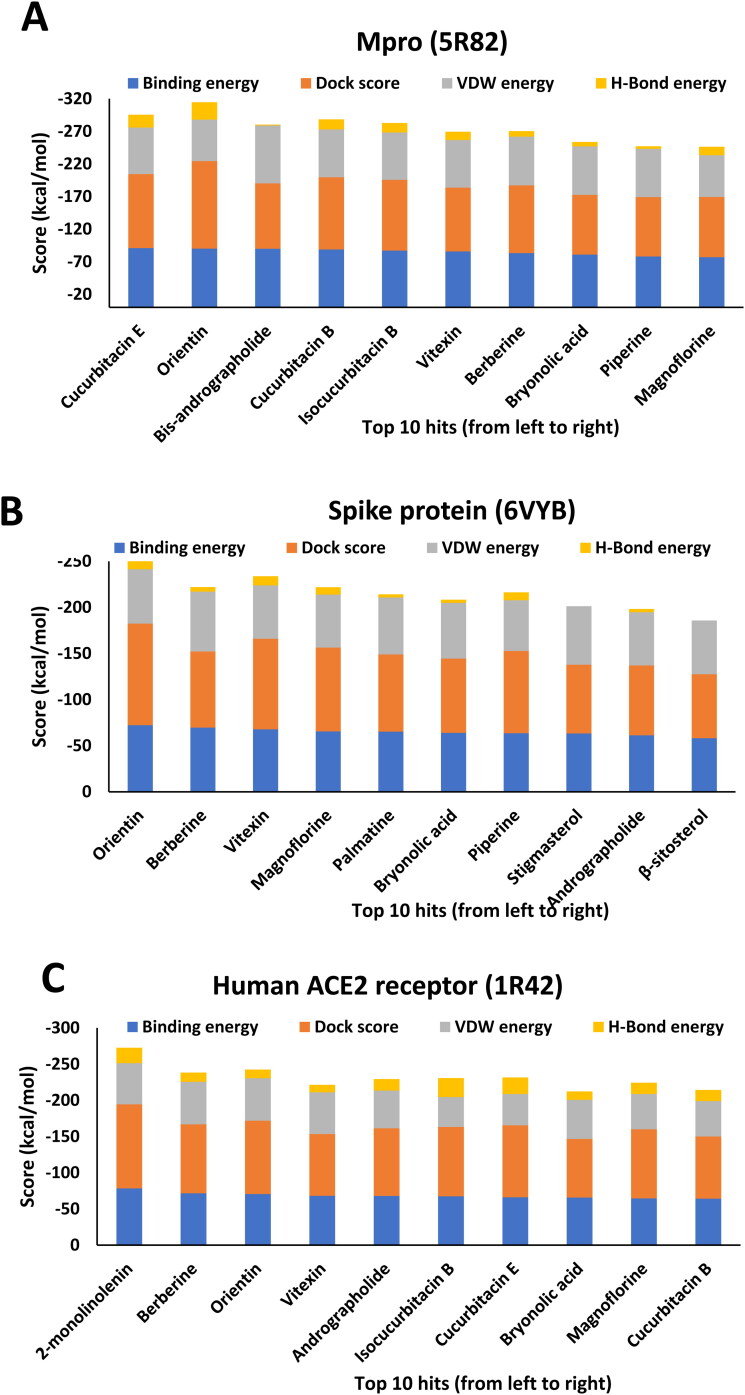
Crystal structures of main protease, spike protein and human entry receptor (ACE2) were used to define the molecular interactions (Meng et al., 2011) with the scrutinized bioactives. The top ten scoring bioactive ligands are represented in Table 2 with binding energy, dock score, van der Waals and H-bond energy. Interestingly, among the bioactives screened, the compounds Orientin, Vitexin, Berberine, Bryonolic acid and Magnoflorine were found to interact with all the drug targets of SARS-CoV-2 including the human entry receptor ACE2 (Figure 2). Although the aforesaid compounds had interaction with all the drug targets of SARS-CoV-2, interaction with active site residues of main protease was found to be much stronger than the other two drug targets (spike protein and ACE2) used in the study (Hasan et al., 2020). The highest binding affinity between the ligands-receptor complex was confirmed through binding energy and other possible non-covalent interactions. Based on the scoring values it is confirmed that these compounds can significantly inhibit SARS-CoV-2 viral replication by binding strongly with the main protease.
Table 2.
The top ten scoring bioactive ligands with binding energy (BE), dock score (DS), van der Waals (VDW) and H-bond (HB) energy.
| Compounds | 5R82 (SARS-Cov-2 Main protease) |
Compounds | 6VYB (SARS-Cov-2 spike Protein) |
Compounds | 1R42 (Human ACE2 receptor) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BE | DS | VDW | HB | BE | DS | VDW | HB | BE | DS | VDW | HB | |||
| Cucurbitacin E | −91.00 | −113.40 | −71.54 | −19.48 | Orientin | −72.30 | −110.05 | −59.02 | −13.24 | 2-monolinolenin | −78.30 | −116.12 | −56.82 | −21.48 |
| Orientin | −90.20 | −134.14 | −63.66 | −26.58 | Berberine | −69.70 | −82.48 | −64.85 | −4.90 | Berberine | −71.50 | −95.33 | −58.76 | −12.73 |
| Bis-andrographolide | −90.00 | −100.30 | −88.61 | −1.39 | Vitexin | −67.70 | −98.34 | −57.79 | −9.93 | Orientin | −70.60 | −101.17 | −58.84 | −11.76 |
| Cucurbitacin B | −88.80 | −110.75 | −73.57 | −15.28 | Magnoflorine | −65.50 | −90.88 | −57.45 | −8.02 | Vitexin | −68.10 | −85.24 | −57.61 | −10.50 |
| Isocucurbitacin B | −87.10 | −108.56 | −72.49 | −14.56 | Palmatine | −65.20 | −83.71 | −61.76 | −3.48 | Andrographolide | −68.00 | −93.29 | −52.22 | −15.77 |
| Vitexin | −85.70 | −98.03 | −72.98 | −12.72 | Bryonolic acid | −63.90 | −80.57 | −60.38 | −3.50 | Isocucurbitacin B | −67.40 | −95.87 | −41.24 | −26.19 |
| Berberine | −83.20 | −103.89 | −74.85 | −8.35 | Piperine | −63.50 | −89.19 | −55.10 | −8.42 | Cucurbitacin E | −66.00 | −99.46 | −43.40 | −22.63 |
| Bryonolic acid | −81.00 | −91.63 | −74.26 | −6.71 | Stigmasterol | −63.30 | −74.57 | −63.32 | 0.00 | Bryonolic acid | −65.60 | −80.99 | −54.03 | −11.60 |
| Piperine | −78.10 | −90.95 | −73.98 | −4.16 | Andrographolide | −61.30 | −75.71 | −57.77 | −3.50 | Magnoflorine | −64.40 | −95.46 | −48.91 | −15.52 |
| Magnoflorine | −77.00 | −92.41 | −64.03 | −12.98 | β-sitosterol | −58.20 | −69.35 | −58.25 | 0.00 | Cucurbitacin B | −64.20 | −85.81 | −49.02 | −15.23 |
Figure 2.
Interaction map of top ten scoring ligands with SARS-CoV-2 drug targets (I) Main protease (Mpro, 5R82) (II) Spike protein (6VYB) (III) ACE-2 (1R42) (A) Crystal structure of drug targets (represented in space-filling model) displaying the druggable pocket of ligands (stick model) (B) 2D interaction map showing the top ten ligand interactions with receptor. Active site binding pockets residues are represented in three letter amino acid code and the type of interactions are mentioned in different colors.
Among 47 bioactives used in the study, Orientin, a C-glycosyl compound, showed the highest binding affinity with main protease with a dock score of −134.14 and binding energy of −90.2 kcal mol−1 (Figure 2-IB). It formed seven conventional hydrogen bonds with five residues (Thr26, His41, Gly143, Cys145 and Glu 166). These hydrogen bond interactions indeed enhance the binding affinity of the complex by stabilizing the ligand at the active site of the target and thereby help in increasing the biological activity of the complex. Notably, among the active site residues the catalytic dyad of main protease (His41 and Cys145) was mainly involved in hydrogen bond interactions. These interactions are expected to prevent the virus from processing polyprotein which consequently hinder the viral replication. The other interactions that stabilizes the Orientin and main protease complex includes pi-pi stacked interaction with His163, pi-pi T-shaped interaction with His41, pi-alkyl interaction with Cys145, pi-sulfur interaction with Met49. In addition, the residue Asn142 formed a pi-donor hydrogen bond with the ligand molecule. The conformation had the van der Waals and H-bond energy of −63.65 and −26.58 kcal mol−1, respectively. All these crucial interactions facilitate in augmenting the efficacy of the drug molecules by stabilizing the binding structures. Orientin docked with spike protein (6VYB) revealed the docking and binding energy score of −110.05 and −72.3 kcal mol−1, respectively. Besides, it formed six hydrogen bond interactions with four amino acids (Tyr28, Asn30, Phe59 and Asn61) and one carbon-hydrogen bond with Thr29. H-bond and van der Waals interaction energies were found to be −13.23 and −59.02 kcal mol−1, respectively. Similarly, Orientin docked with ACE2 receptor revealed the docking and binding energy score of −101.17 and −70.6 kcal mol−1, respectively. The binding affinity was associated with five hydrogen bond interactions formed by three residues (Glu22, Lys26 and Asn90), one carbon hydrogen bond interaction with Glu22 and pi-alkyl interaction with Lys94. The structure possesses van der Waals and H-bond interactions energy of −58.84 and −11.76 kcal mol−1, respectively.
Vitexin, an apigenin flavone glycoside, was found to have the binding energy of −85.7 kcal mol−1 and the dock score of −98.03 during the interaction with main protease. The strong affinity was explicable from six hydrogen bonds formed with six residues of main protease (Phe140, Ser144, Cys145, Asn142, Leu167 and Gln189). Vitexin-Mpro complex was stabilized by pi-anion (Cys145), pi-sulfur (Glu166), pi-lone pair (Asn142), pi-pi T-shaped (His41) and pi-alkyl (Met165) interactions. The van der Waals and H-bond energy was found to be −72.97 and −12.71 kcal mol−1, respectively. Vitexin docked with spike protein reported −98.34 and −67.7 kcal mol−1 as docking and binding energy score, respectively. It formed four hydrogen bonds with four residues (Tyr28, Asn30, Phe59 and Asn61) and one carbon hydrogen bond with Thr29. The conformation had the van der Waals and H-bond energy of −57.78 and −9.93, respectively. ACE2 receptor with Vitexin had a docking and binding energy score of −85.24 and −68.1 kcal mol−1, respectively. Conformational stability of the complex (Vitexin-ACE2) is maintained by hydrogen bond (two hydrogen bonds from Glu22 and Asn90), van der Waals (Lys26, Leu29, Gln89 and Gln96) and pi-alkyl interactions (Val93 and Lys 94). H-bond and van der Waals energy was found to be −57.61 and −10.5 kcal mol−1, respectively.
Main protease and Berberine (an organic heteropentacyclic compound) interaction showed a binding energy of −83.2 kcal mol−1 and a dock score of −103.89. The interaction between ligand with the receptor binding site of Mpro was stabilized by pi-pi T-shaped interaction with His163, Pi-alkyl interaction with Cys145 & His41 and alkyl interaction with His163. The conformation showed van der Waals and H-bond energy of −74.84 and −8.34 kcal mol−1, respectively. Berberine with spike protein revealed the docking score, binding and interaction energies such as H-bond and van der Waals as −82.48, −69.7, −4.89 and −64.84 kcal mol−1, respectively. The other stabilizing interactions includes van der Waals (Asn30), amide pi-stacked (Phe59), pi-pi stacked (Thr29) and pi-alkyl (Phe59) interactions. Berberine with ACE2 receptor had a docking score of −95.33, binding energy of −71.5 kcal mol−1 and H-bond and van der Waals interaction energies of −58.759 and −12.728, respectively. It formed two conventional hydrogen bond interactions with Lys26 and Thr27. Further stabilization occurred with pi-cation (Glu22), pi-anion (Glu22), pi-sulfur (Val93) and alkyl (Val93) interactions.
Bryonolic acid formed a strong interaction with the receptor binding sites of main protease with a binding energy score of −81.0 kcal mol−1 and dock score of −91.63. Bryonolic acid formed two hydrogen bond interactions with Arg188 and Gln189. Further conformational interactions such as carbon hydrogen bond interaction was observed with Ser139 and His163, alkyl interaction with Tyr118, Cys145, His163 and Met49 and pi-alkyl interaction with His41, His163, His172 and Met165 which stabilized the binding of the bryonolic with Mpro. The van der Waals and H-bond interaction energy was −74.25 and −6.71 kcal mol−1, respectively. Bryonolic acid docked with spike protein reported −80.57, −63.9, −60.37 and −3.5 kcal mol−1as docking, binding, H-bond and van der Waals energies, respectively. Key interactions between Bryonolic acid and spike protein includes one conventional hydrogen bond with Tyr28 and pi-alkyl interaction with Phe58. Interaction with ACE2 receptor was found to have a docking score of −80.99, binding energy score of −65.6 kcal mol−1 and van der Waals and H-bond interaction score of −54.032 and −11.596 kcal mol−1, respectively. Energetic contributions to the ligand include conventional hydrogen bonds (four bonds with Glu22, Thr27, Gln89 and Asn 90) and pi-alkyl (Leu29 and Lys94) interactions.
Magnoflorine docked with main protease showed a binding energy score of −77 kcal mol−1 and a dock score of −92.41. It formed six hydrogen bonds with four amino acids (Leu141, Asn142, Ser144 and Cys145). Besides, it also forms carbon hydrogen bond with Phe140, His163 and Glu166 and pi-donor hydrogen bond with Cys145. The extra stabilizing interaction associated with ligand and main protease complex includes pi-cation interaction with His41, pi-alkyl interaction with Leu27 and Cys145 and pi-sigma interaction with His41. The van der Waals and H-bond interaction energy was −64.02 and −12.98 kcal mol−1, respectively. Magnoflorine with spike protein has a dock score of −90.88, binding energy score of −65.5 kcal mol−1and van der Waals and H-bond interactions score of −57.44 and −8.02 kcal mol−1, respectively. It formed conventional hydrogen bonds with Asn61 and carbon hydrogen and pi-donor hydrogen bond with Tyr28. Other significant interactions comprise pi-sigma (Tyr28) and pi-pi stacked (Tyr28) interactions. Magnoflorine with ACE2 receptors exhibited a docking score of −95.46, binding energy score of −64.4 kcal mol−1, H-bond and van der Waals interaction scores of −15.52 and −48.90 kcal mol−1, respectively. It formed two hydrogen bond interactions with Thr27 and Leu29 and carbon hydrogen bond with Val93 and Glu22. Other energetic contribution includes pi-cation (Lys26 and Glu22) and pi-alkyl (Leu29 and Val23) interactions. In addition, it formed electrostatic interaction with Lys26 and Glu22, thereby stabilizing the complex formed.
Besides, the compound Cucurbitacin E, Bis-andrographolide, Cucurbitacin B, Isocucurbitacin B and Piperine had strong binding affinity to main protease of SARS-CoV-2. Cucurbitacin E was the top scorer among the main protease inhibitors. It had the highest binding energy score of −91.0 kcal mol−1 with a dock score of −113.4. The H-bond and van der Waals interactions energy was found to be −19.48 and −71.53 kcal mol−1, respectively. It formed two conventional hydrogen bond interactions with Asn142 and Cys145 and one carbon hydrogen bond with Ser46. Besides, it had alkyl and pi-alkyl interaction with Met49, His41, Cys44 and His163. The compounds Cucurbitacin B and Isocucurbitacin B had similar kind of pi-alkyl and alkyl interactions as Cucurbitacin E. But in case of conventional hydrogen bond interactions both lacked Cys145 interactions. In addition, Isocucurbitacin B lacked carbon hydrogen bond interaction with Ser46. Notably, Cucurbitacin B formed a specific conventional hydrogen bond with Thr25.
Bis-andrographolide had the docking and binding energy score of −100.3 and −90 kcal mol−1, respectively. It formed conventional hydrogen bond with Glu166 and alkyl interaction with Met49. The conformation had the van der Waals and H-bond energy score of −88.60 and −1.38 kcal mol−1, respectively. Piperine interacted with main protease with a dock score of −90.95 and binding energy score of −78.10 kcal mol−1. It formed one conventional hydrogen bond with hotspot residue His41 and other stabilizing interactions include pi-sulfur, pi-sigma, pi-pi T-shaped and alkyl interactions.
The compounds Palmatine, Piperine, Stigmasterol, Andrographolide and β-sitosterol and the components 2-monolinolenin, Andrographolide, Isocucurbitacin B, Cucurbitacin E and B had strong binding affinity to spike and ACE2 receptor, respectively. The schematic representation summarizing the overall scoring functions of top ten bioactive molecules is shown in Figure 3.
Figure 3.
A schematic representation summarizing the top ten scoring functions of ligands in complex with (A) 5R82.pdb (B) 6VYB.pdb (C) 1R42.pdb.
MD simulation and binding energy
Among the ligands screened through docking studies, Cucurbitacin E and Orientin revealed significant interaction with most critical residues of Mpro with high binding affinities. Therefore, these two compounds were selected as the best inhibitors and used for further analysis. MD simulation of 20 ns was performed for the two selected drug molecules to get deep insights into the stability of the Mpro-drug complex. The overall stability was determined through RMSD and RMSF analysis. The energy profile in the presence and absence of ligands were computed during 20 ns MD simulation. Wherein, the potential energy profile of the system was found to be stable throughout the simulation and the lowest energy profile in the presence of ligands indicates that the system is considerably stable than in the absence of ligand (Figure 4(A)). Similarly, RMSD analysis during each time step unveiled the stability of the system. Mpro in the absence of drug molecule maintained the steady state throughout the simulation with an average RMSD value of 3.71 Å, indicating the convergence of the system during 20 ns simulation. However, Mpro in complex with drug molecules exhibited little variations during the simulation run (Figure 4(A)). Orientin presented an average RMSD of 4.6 Å whereas Cucurbitacin E presented an average RMSD of 4.9 Å. Although, the system started with increasing RMSD profile, it favored to equilibrium state after 2 ns and successfully maintained the stable state throughout the remaining simulation run (Figure 4(B)). RMSF provides the visibility for most flexible and mobile regions in the protein during the simulation. In fact, these structural fluctuations reveal the binding and interaction of the drug molecules with the protein motif. RMSF analysis of Mpro in the presence of ligand unveiled the fluctuating residues at 1–12, 128–146 and 185–192 amino acid regions corresponding to the loop region (Figure 4(C)) of Mpro. The average RMSF value for Mpro was found to be 1.53 Å revealing that there was little fluctuation in the loop region during MD run. However, Mpro in complex with Cucurbitacin E and Orientin was found to have high fluctuation in the druggable pocket with an average RMSF value of 2.10 Å and 1.79 Å, respectively. The Mpro-Cucurbitacin E complex was found to have significant fluctuation with the catalytic residues (Cys145) of Mpro in comparison to Orientin. Since, Cys145 resides in the loop region, the fluctuations are favorable and advantageous in accommodating the drug molecule at the receptor site.
Figure 4.
Analysis of MD trajectories (A) Potential energy during 20 ns MD simulation (B) RMSD of protein backbone (C) RMSF.
MM-PBSA is the most reliable method for predicting the binding interaction between protein-drug complexes. The binding free energy for Cucurbitacin E and Orientin was −152.96 and −123.62 kcal/mol respectively. The binding energies obtained from MD trajectories further confirms the accommodation of inhibitors into the druggable pocket of Mpro. Moreover, the binding pocket with polar microenvironment reinforces the strong hydrogen and hydrophobic interactions with the drug molecule and seizes them in the active site of Mpro. Overall, the results were suggestive that Mpro maintains a stable binding state with the inhibitors (Cucurbitacin E and Orientin) for plausible interactions.
Discussion
Siddha, one of the oldest traditional systems of medicine in India, has been shown to be effective in treating respiratory-related infections (Ram et al., 2009). Several natural bioactives isolated from medicinal plants of Siddha system are reported for antiviral property against respiratory viruses. It is thus likely that these compounds may exert an antiviral activity against SARS-CoV-2 (Adeoye et al., 2020; Oliveira et al., 2020). Hence, in the present study anti-COVID-19 drug candidates were identified from Siddha medicinal plant, with special attention to NKC combination plants, due to its antiviral, immunostimulant and immunomodulatory activities (Kavinilavan et al., 2017). The potent anti-COVID-19 drug candidates were screened from 47 bioactives, among them 5 (Orientin, Vitexin, Berberine, Bryonolic acid and Magnoflorine) bioactives had potential interactions with all drug targets (mentioned in the present study). All these drug candidates exhibited strong affinity towards the target site of the receptor molecule with high binding energy. The top scoring receptor-ligand complex was stabilized by non-covalent interactions such as hydrogen-bonding, van der Waals and electrostatics interactions (Chen & Kurgan, 2009). These interactions are indeed prerequisites for biological functions and successful drug developments. Moreover, the top two scoring ligands Cucurbitacin E and Orientin were further substantiated through 20 ns MD simulation studies.
Mpro, one of the best characterized drug targets of SARS-CoV-2, is essential for polyprotein processing (translated from viral RNA) that enables viral replication and gene expressions. The catalytic dyad His41 and Cys145 is essential for substrate hydrolysis. Thus, compounds inhibiting the active site of this protease are expected to prevent replication and transcription of viral components (Das et al., 2020; Gyebi et al., 2020; Joshi et al., 2020; Khan et al., 2020a; 2020b; Kumar et al., 2020; Muralidharan et al., 2020; Zhang et al., 2020a; Zhang et al., 2020b). Other drug targets of SARS-CoV-2 include spike glycoprotein and host cell receptor ACE2. In general, SARS-CoV-2 makes an entry into host cell through ACE2 receptors and it is mediated by receptor binding domain of homo trimeric spike glycoprotein. The crystal structure of spike protein in complex with human ACE2 receptor revealed that residue Ser19, Gln24,Thr27, Phe28, Asp30, Lys31, His34, Glu35,Glu37, Asp38, Tyr41, Gln42, Leu45, Leu79, Met82, Tyr83,Asn330,Lys353, Gly354, Asp355, Arg357 and Arg393 of human ACE2 interacted with residues Ala475/Gly476, Ala474/G476/Asn487, Phe56/Tyr473/Ala475/Tyr489, Tyr489, Lys417/Leu455/Phe456, Leu455/Phe456/Glu484/Tyr489/Phe490/Gln493, Tyr453/Leu455/Gln493, Gln493, Tyr505, Tyr449/Gly496/Gln498, Gln498/Thr500/Asn501, Gly446/Tyr449/Gln498, Gln498/Thr500, Phe486, Phe486, Phe486/Asn487/Tyr489, Thr500, Gly496/Asn501/Gly502/Tyr505, Tyr502/Tyr505, Thr500,Gly502, Thr500, Tyr505, respectively, of spike glycoprotein (Wang et al., 2020). Therefore, bioactives targeting these hotspot residues are likely to become a potent inhibitor of SARS-CoV-2. However, spike protein and ACE2 receptor showed no significant inhibition. In case of ACE2 drug target, only Thr27 (hotspot residue) of ACE2 had interactions with bioactives (Veeramachaneni et al., 2020).
Orientin possesses wide range of pharmacological effects such as antioxidant, antiaging, antibacterial, anti-inflammation, free radical scavenging, cardioprotective, radioprotective, neuroprotective, antidepressant, anticancer and thrombocytopenia activities (Lam et al., 2016; Sharma et al., 2016; Yadav et al., 2018). It has also been documented for antiviral activity against parainfluenza type 3 virus (PIV-3) (Li et al., 2002). Like Orientin, the compound Vitexin was also reported for multiple drug targets including antioxidant, anticancer, anti-inflammatory, antihyperalgesic and neuroprotective effects (He et al., 2016). It has also been reported for antiviral activity against several viruses such as herpes simplex virus type 1 (HSV-1), Hepatitis A Virus (HAV-H10), PIV-3 and rotavirus (Fahmy et al., 2020; Knipping et al., 2012; Li et al., 2002). Bryonolic acid, a pentacyclic triterpenoids, exhibits biological activities such as antiallergic, antitumor, antioxidant and anti-inflammatory (Gatbonton-Schwager et al., 2012; Khallouki et al., 2018; Tanaka et al., 1991). An aporphine alkaloid, Magnoflorine, has been reported for antidiabetic and anticancer property (Patel & Mishra, 2012). In addition, Magnoflorine has been previously reported for their antiviral effects against HSV-1 and poliovirus type-1 (Mohamed et al., 2010). Berberine, an organic hetero pentacyclic compound has been reported for numerous therapeutic values such as antioxidant, anti-inflammatory, antidiarrhea, antilipemic, antibiofilm, immunomodulatory, cardioprotective, nephroprotective, hepatoprotective and glucose metabolism (Neag et al., 2018; Zhang & Shen, 1989). Most importantly, it has been reported for antiviral activity against numerous viruses such as influenza virus (Wu et al., 2011), enterovirus 71 (Wang et al., 2017), human cytomegalovirus (Hayashi et al., 2007), hepatitis C virus (Hung et al., 2019), HSV-1 (Dkhil & Al-Quraishy, 2014) and chikungunya virus (Varghese et al., 2016). Besides, it has been proven to be an antibacterial agent against multiple drug resistance bacteria such as methicillin-resistant Staphylococcus aureus and Streptococcus agalactiae (Chu et al., 2016; Peng et al., 2015; Tan et al., 2019; Yu et al., 2005) and antifungal agent against Candida spp. (Xie et al., 2020).
The compounds Piperine, Cucurbitacin B, Cucurbitacin E, Isocucurbitacin B and Bis-andrographolide have also been reported for numerous pharmacological effects (Chao & Lin, 2010; Derosa et al., 2016; Kaushik et al., 2015). In addition, Piperine has been reported for antiviral activity against Hepatitis B (Jiang et al., 2013). Cucurbitacin B showed antiviral activity against HSV-1 (Hassan et al., 2017) and Bovine viral diarrhea virus (BVDV) (Alsayari et al., 2012). Cucurbitacin E was reported against BVDV (Alsayari et al., 2012) and Bis-andrographolide against HIV virus (Reddy et al., 2005).
The multiple pharmacological effects exhibited by these components strongly suggest that these phytochemicals would certainly find their way into the arsenal of antiviral drugs. Moreover, the active components Vitexin, Berberine, Bryonolic acid, Magnoflorine, Cucurbitacin, Piperine, Palmatine, Stigmasterol, Andrographolide, Sitosterol and 2-monolinolenin were FDA approved drugs. Hence, these phytochemicals can be further taken for preclinical studies to treat COVID-19 infection (Lobo-Galo et al., 2020). The docking studies expounded with MD simulation and binding affinity analyses strongly suggest that the compound Cucurbitacin E and Orientin are promising drug candidate to combat COVID-19 infections. However, in vitro and in vivo experimental evidences are further required to substantiate the postulations observed from the present study. In conclusion, consumption of NKC will helps in developing an immune response, elimination of toxins from the system, soothes the body and improves the overall health condition.
Conclusion
The present study explored the important bioactive constituents of Siddha medicine, especially NKC through molecular docking and simulation analysis for the prevention and cure of COVID19 infection. The compounds Cucurbitacin E, Orientin, Bis-andrographolide, Cucurbitacin B, Isocucurbitacin B, Vitexin, Berberine, Bryonolic acid, Piperine and Magnoflorine were identified as potential lead molecules that have been shown to possess the ability to interact with the components that block the viral replication in SARS-CoV-2. Moreover, the immune-enhancing properties of these compounds without any adverse side effects could provide natural immune power to resist COVID-19 infections. However, preclinical studies would provide deep insight into mechanism of actions and also drugability of these bioactives for the treatment of COVID19 infection.
Supplementary Material
Funding Statement
The authors sincerely acknowledge the computational and bioinformatics analysis provided by the Bioinformatics Infrastructure Facility (funded by DBT, GOI; File No. BT/BI/25/012/2012,BIF). The authors also thankfully acknowledge DST-FIST (Grant No. SR/FST/LSI-639/2015(C)), UGC-SAP (Grant No. F.5-1/2018/DRS-II(SAP-II)) and DST-PURSE (Grant No. SR/PURSE Phase 2/38 (G)). SKP is thankful to UGC for Mid-Career Award [F.19-225/2018(BSR)]. RBS is thankful to RUSA 2.0 [F.24-51/2014-U, Policy (TN Multi-Gen), Dept. of Edn, GoI].
Disclosure statement
The authors declare that they have no conflicts of interest.
Data availability statement
The data that support the findings of this study are openly available in GenBank database.
References
- Aanouz I., Belhassan A., El Khatabi K., Lakhlifi T., El Idrissi M., & Bouachrine M. (2020). Moroccan Medicinal plants as inhibitors of COVID-19: Computational investigations. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1758790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelli I., Hassani F., Bekkel Brikci S., & Ghalem S. (2020). In silico study the inhibition of Angiotensin converting enzyme 2 receptor of COVID-19 by Ammoides verticillata components harvested from western Algeria. Journal of Biomolecular Structure and Dynamics, 1–17. 10.1080/07391102.2020.1763199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeoye A. O., Oso B. J., Olaoye I. F., Tijjani H., & Adebayo A. I. (2020). Repurposing of chloroquine and some clinically approved antiviral drugs as effective therapeutics to prevent cellular entry and replication of coronavirus. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1765876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khafaji K., Al-DuhaidahawiL D., & Taskin Tok T. (2020). Using Integrated Computational Approaches to Identify Safe and Rapid Treatment for SARS-CoV-2. Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1764392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsayari A., Darweesh M., Halaweish F., & Chase C. C. L. (2012). Anti-bovine viral diarrhea virus activity of cucurbitacins as new potential antiviral agents. Planta Medica, 78(11), PD142 10.1055/s-0032-1320500 [DOI] [Google Scholar]
- Anbarasu K., Manisenthil K. K., & Ramachandran S. (2011). Antipyretic, anti-inflammatory and analgesic properties of nilavembu kudineer choornam: A classical preparation used in the treatment of chikungunya fever. Asian Pacific Journal of Tropical Medicine, 4(10), 819–823. 10.1016/S1995-7645(11)60201-0 [DOI] [PubMed] [Google Scholar]
- Beema Shafreen R. M., Selvaraj C., Singh S. K., & Karutha Pandian S. (2014). In silico and in vitro studies of cinnamaldehyde and their derivatives against LuxS in Streptococcus pyogenes: Effects on biofilm and virulence genes. Journal of Molecular Recognition: JMR, 27(2), 106–116. 10.1002/jmr.2339 [DOI] [PubMed] [Google Scholar]
- Boopathi S., Poma A. B., & Kolandaivel P. (2020). Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgio J. F., Alsuwat H. S., Al Otaibi W. M., Ibrahim A. M., Almandil N. B., Al Asoom L. I., Salahuddin M., Kamaraj B., & AbdulAzeez S. (2020). State-of-the-art tools unveil potent drug targets amongst clinically approved drugs to inhibit helicase in SARS-CoV-2. Archives of Medical Science : Ams, 16(3), 508–518. 10.5114/aoms.2020.94567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao W., & Lin B. (2010). Isolation and identification of bioactive compounds in Andrographis paniculata (Chuanxinlian). Chinese Medicine, 5(1), 17 10.1186/1749-8546-5-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., & Kurgan L. (2009). Investigation of atomic level patterns in protein—small ligand interactions. PLoS One, 4(2), e4473 10.1371/journal.pone.0004473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., & Zhang L. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet (London, England), 395(10223), 507–513. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu M., Zhang M.-B., Liu Y.-C., Kang J.-R., Chu Z.-Y., Yin K.-L., Ding L.-Y., Ding R., Xiao R.-X., Yin Y.-N., Liu X.-Y., & Wang Y.-D. (2016). Role of berberine in the treatment of methicillin-resistant Staphylococcus aureus infections. Scientific Reports, 6, 24748 10.1038/srep24748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Sarmah S., Lyndem S., & Singha Roy A. (2020). An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. Journal of Biomolecular Structure and Dynamics, 1–18. 10.1080/07391102.2020.1763201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derosa G., Maffioli P., & Sahebkar A. (2016). Piperine and its role in chronic diseases. Advances in Experimental Medicine and Biology, 928, 173–184. 10.1007/978-3-319-41334-1_8 [DOI] [PubMed] [Google Scholar]
- Dkhil M. A., & Al-Quraishy S. (2014). Evaluation of antiviral activity of berberine against herpes simplex viruses. Journal of Pure and Applied Microbiology, 8, 155–159. [Google Scholar]
- Elfiky A. A. (2020. a). SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 1–15. 10.1080/07391102.2020.1761882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A. (2020. b). Natural products may interfere with SARS-CoV-2 attachment to the host cell. Journal of Biomolecular Structure and Dynamics, 1–16. 10.1080/07391102.2020.1761881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A., & Azzam E. B. (2020). Novel guanosine derivatives against MERS CoV polymerase: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1758789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmezayen A. D., Al-Obaidi A., Şahin A. T., & Yelekçi K. (2020). Drug repurposing for coronavirus (COVID-19): in silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1758791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enayatkhani M., Hasaniazad M., Faezi S., Guklani H., Davoodian P., Ahmadi N., Einakian M. A., Karmostaji A., & Ahmadi K. (2020). Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: An in silico study. Journal of Biomolecular Structure and Dynamics, 1–19. 10.1080/07391102.2020.1756411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enmozhi S. K., Raja K., Sebastine I., & Joseph J. (2020). Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: An in silico approach. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1760136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahmy N. M., Al-Sayed E., Moghannem S., Azam F., El-Shazly M., & Singab A. N. (2020). Breaking down the barriers to a natural antiviral agent: Antiviral activity and molecular docking of erythrina speciosa extract, fractions, and the major compound. Chemistry & Biodiversity, 17(2), e1900511 10.1002/cbdv.201900511 [DOI] [PubMed] [Google Scholar]
- Gatbonton-Schwager T. N., Letterio J. J., & Tochtrop G. P. (2012). Bryonolic acid transcriptional control of anti-inflammatory and antioxidant genes in macrophages in vitro and in vivo. Journal of Natural Products, 75(4), 591–598. 10.1021/np200823p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M. K., Vemula S., Donde R., Gouda G., Behera L., & Vadde R. (2020). In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. Journal of Biomolecular Structure and Dynamics, 1–17. 10.1080/07391102.2020.1751300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyebi G. A., Ogunro O. B., Adegunloye A. P., Ogunyemi O. M., & Afolabi S. O. (2020). Potential Inhibitors of Coronavirus 3-Chymotrypsin-Like Protease (3CLpro): An in silico screening of Alkaloids and Terpenoids from African medicinal plants. Journal of Biomolecular Structure and Dynamics, 1–19. 10.1080/07391102.2020.1764868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibzadeh P., & Stoneman E. K. (2020). The novel coronavirus: A Bird’s eye view. The International Journal of Occupational and Environmental Medicine, 11(2), 65–1965. 10.15171/ijoem.2020.1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A., Paray B. A., Hussain A., Qadir F. A., Attar F., Aziz F. M., Sharifi M., Derakhshankhah H., Rasti B., Mehrabi M., & Shahpasand K. (2020). A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1754293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan S. T. S., Berchová-Bímová K., Petráš J., & Hassan K. T. S. (2017). Cucurbitacin B interacts synergistically with antibiotics against Staphylococcus aureus clinical isolates and exhibits antiviral activity against HSV-1. South African Journal of Botany, 108, 90–94. 10.1016/j.sajb.2016.10.001 [DOI] [Google Scholar]
- Hayashi K., Minoda K., Nagaoka Y., Hayashi T., & Uesato S. (2007). Antiviral activity of berberine and related compounds against human cytomegalovirus. Bioorganic & Medicinal Chemistry Letters, 17(6), 1562–1564. 10.1016/j.bmcl.2006.12.085 [DOI] [PubMed] [Google Scholar]
- He M., Min J. W., Kong W. L., He X. H., Li J. X., & Peng B. W. (2016). A review on the pharmacological effects of vitexin and isovitexin. Fitoterapia, 115, 74–85. 10.1016/j.fitote.2016.09.011 [DOI] [PubMed] [Google Scholar]
- Hsu C. H., Hwang K. C., Chao C. L., Chang S. G., Ho M. S., Lin J. G., Chang H. H., Kao S. T., Chen Y. M., & Chou P. (2008). An evaluation of the additive effect of natural herbal medicine on SARS or SARS-like infectious diseases in 2003: A randomized, double-blind, and controlled pilot study. Evidence-Based Complementary and Alternative Medicine: ECAM, 5(3), 355–362. 10.1093/ecam/nem035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T. C., Jassey A., Liu C. H., Lin C. J., Lin C. C., Wong S. H., Wang J. Y., Yen M. H., & Lin L. T. (2019). Berberine inhibits hepatitis C virus entry by targeting the viral E2 glycoprotein. Phytomedicine: International Journal of Phytotherapy and Phytopharmacology, 53, 62–69. 10.1016/j.phymed.2018.09.025 [DOI] [PubMed] [Google Scholar]
- Islam R., Parves R., Paul A. S., Uddin N., Rahman M. S., Mamun A. A., Hossain M. N., Ali M. A., & Halim M. A. (2020). A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. Journal of Biomolecular Structure and Dynamics, 1–20. 10.1080/07391102.2020.1761883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain J., Kumar A., Narayanan V., Ramaswamy R. S., Sathiyarajeswaran P., Devi M. S., Kannan M., & Sunil S. (2019). Antiviral activity of ethanolic extract of Nilavembu Kudineer against dengue and chikungunya virus through in vitro evaluation. Journal of Ayurveda and Integrative Medicine. S0975-9476(18)30073-1, 1-7. 10.1016/j.jaim.2018.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I. S., Baik Y. S., Bae S. J., Sun S. H., Lee J. S., & Han C. H. (2009). An overview of the herbal remedies for Severe Acute Respiratory Syndrome (SARS) in WHO official report (2004). The Korean Journal of Internal Medicine, 30(3), 571–581. [Google Scholar]
- Jiang Z. Y., Liu W. F., Zhang X. M., Luo J., Ma Y. B., & Chen J. J. (2013). Anti-HBV active constituents from Piper longum. Bioorganic & Medicinal Chemistry Letters, 23(7), 2123–2127. 10.1016/j.bmcl.2013.01.118 [DOI] [PubMed] [Google Scholar]
- Joshi R. S., Jagdale S. S., Bansode S. B., Shankar S. S., Tellis M. B., Pandya V. K., Chugh A., Giri A. P., & Kulkarni M. J. (2020). Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. Journal of Biomolecular Structure and Dynamics, 1–16. 10.1080/07391102.2020.1760137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamalarajan P., Muthuraman S., Ganesh M. R., & Valan M. F. (2019). Phytochemical investigation of nilavembu kudineer chooranam ethyl acetate extract and its ability to reduce intracellular antioxidant levels in THP-I cells. European Journal of Medicinal Plants, 30(4), 1–13. 10.9734/ejmp/2019/v30i430187 [DOI] [Google Scholar]
- Kaushik U., Aeri V., & Mir S. R. (2015). Cucurbitacins—An insight into medicinal leads from nature. Pharmacognosy Reviews, 9(17), 12–18. 10.4103/0973-7847.156314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavinilavan R., Mekala P., Raja M. J., Arthanari Eswaran M., & Thirumalaisamy G. (2017). Exploration of immunomodulatory effect of nilavembu kudineer chooranam against newcastle disease virus in backyard chicken. Journal of Pharmacognosy and Phytochemistry, 6, 749–751. [Google Scholar]
- Khallouki F., Owen R. W., Silvente-Poirot S., & Poirot M. (2018). Bryonolic acid blocks cancer cell clonogenicity and invasiveness through the inhibition of fatty acid: Cholesteryl ester formation. Biomedicines, 6(1), 21 10.3390/biomedicines6010021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan R. J., Jha R. K., Amera G., Jain M., Singh E., Pathak A., Singh R. P., Muthukumaran J., & Singh A. K. (2020. a). Targeting SARS-Cov-2: A systematic drug repurposing approach to identify promising inhibitors against 3C-like Proteinase and 2’-O-RiboseMethyltransferase. Journal of Biomolecular Structure and Dynamics, 1–40. 10.1080/07391102.2020.1753577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A., Zia K., Ashraf S., Uddin R., & Ul-Haq Z. (2020. b). Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1751298 [DOI] [PubMed] [Google Scholar]
- Kim H. Y., Shin H. S., Park H., Kim Y. C., Yun Y. G., Park S., Shin H. J., & Kim K. (2008). In vitro inhibition of coronavirus replications by the traditionally used medicinal herbal extracts, Cimicifuga rhizoma, Meliae cortex, Coptidis rhizoma, and Phellodendron cortex. Journal of Clinical Virology: The Official Publication of the Pan American Society for Clinical Virology, 41(2), 122–128. 10.1016/j.jcv.2007.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipping K., Garssen J., & van’t L. (2012). An evaluation of the inhibitory effects against rotavirus infection of edible plant extracts. Virology Journal, 9(1), 137 10.1186/1743-422X-9-137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D., Kumari K., Jayaraj A., Kumar V., Kumar R. V., Dass S. K., Chandra R., & Singh P. (2020). Understanding the binding affinity of noscapines with protease of SARS-CoV-2 for COVID-19 using MD simulations at different temperatures. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1752310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam K. Y., Ling A. P. K., Koh R. Y., Wong Y. P., & Say Y. H. (2016). A review on medicinal properties of orientin. Advances in Pharmacological Sciences, 2016, 4104595 10.1155/2016/4104595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau J. T., Leung P. C., Wong E. L. Y., Fong C., Cheng K. F., Zhang S. C., Lam C. W. K., Wong V., Choy K. M., & Ko W. M. (2005. b). The use of an herbal formula by hospital care workers during the severe acute respiratory syndrome epidemic in Hong Kong to prevent severe acute respiratory syndrome transmission, relieve influenza-related symptoms, and improve quality of life: A prospective cohort study. Journal of Alternative and Complementary Medicine, 11(1), 49–55. 10.1089/acm.2005.11.49 [DOI] [PubMed] [Google Scholar]
- Lau T. F., Leung P. C., Wong E. L. Y., Fong C., Cheng K. F., Zhang S. C., Lam C. W. K., Wong V., Choy K. M., & Ko W. M. (2005. a). Using herbal medicine as a means of prevention experience during the SARS crisis. The American Journal of Chinese Medicine, 33(3), 345–356. 10.1142/S0192415X05002965 [DOI] [PubMed] [Google Scholar]
- Li S.-Y., Chen C., Zhang H.-Q., Guo H.-Y., Wang H., Wang L., Zhang X., Hua S.-N., Yu J., Xiao P.-G., Li R.-S., & Tan X. (2005). Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antiviral Research, 67(1), 18–23. 10.1016/j.antiviral.2005.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. L., Ma S. C., Yang Y. T., Ye S. M., & But P. P. (2002). Antiviral activities of flavonoids and organic acid from Trollius chinensis Bunge. Journal of Ethnopharmacology, 79(3), 365–368. 10.1016/S0378-8741(01)00410-X [DOI] [PubMed] [Google Scholar]
- Lobo-Galo N., Terrazas-López M., Martínez-Martínez A., & Díaz-Sánchez Á. G. (2020). FDA-approved thiol-reacting drugs that potentially bind into the SARS-CoV-2 main protease, essential for viral replication. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1764393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattummal R., Gopi D. K., Parameswaran S. R., & Narayana S. K. K. (2018). Bioactive molecules in Siddha Polyherbal Nilavembu Kudineer alleviating symptoms of Dengue/Chikugunya. Traditional Medicine Research, 3(5), 215–229. 10.12032/TMR201813080 [DOI] [Google Scholar]
- Meng X. Y., Zhang H. X., Mezei M., & Cui M. (2011). Molecular docking: A powerful approach for structure-based drug discovery. Current Computer-Aided Drug Design, 7(2), 146–157. 10.2174/157340911795677602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed S. M., Hassan E. M., & Ibrahim N. A. (2010). Cytotoxic and antiviral activities of aporphine alkaloids of Magnolia grandiflora L. Natural Product Research, 24(15), 1395–1402. 10.1080/14786410902906959 [DOI] [PubMed] [Google Scholar]
- Muralidharan N., Sakthivel R., Velmurugan D., & Gromiha M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 Protease against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–7. 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- Nakkeeran C., Selvakumari P., Kasthury T., & Kumar R. T. (2016). FTIR analysis on Nilavembu Kudineer Churanam and acetominaphen. Journal of Chemical and Pharmaceutical Research, 8(3), 634–639. [Google Scholar]
- Neag M. A., Mocan A., Echeverría J., Pop R. M., Bocsan C. I., Crişan G., & Buzoianu A. D. (2018). Berberine: Botanical occurrence, traditional uses, extraction methods, and relevance in cardiovascular, metabolic, hepatic, and renal disorders. Frontiers in Pharmacology, 9, 557 10.3389/fphar.2018.00557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norgan A. P., Coffman P. K., Kocher J. P. A., Katzmann D. J., & Sosa C. P. (2011). Multilevel parallelization of AutoDock 4.2. Journal of Cheminformatics, 3(1), 12 10.1186/1758-2946-3-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira O. V. D., Gerd B. R., Andrew S P., & Luciano T C. (2020). Repurposing approved drugs as inhibitors of SARS-CoV-2 S-protein from molecular modeling and virtual screening. Journal of Biomolecular Structure and Dynamics, 1–22. 10.1080/07391102.2020.1773318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant S., Singh M., Ravichandiran V., Murty U. S. N., & Srivastava H. K. (2020). Peptide-like and small-molecule inhibitors against Covid-19. Journal of Biomolecular Structure and Dynamics, 1–15. 10.1080/07391102.2020.1757510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M. B., & Mishra S. M. (2012). Magnoflorine from Tinospora cordifolia stem inhibits α-glucosidase and is antiglycemic in rats. Journal of Functional Foods, 4(1), 79–86. 10.1016/j.jff.2011.08.002 [DOI] [Google Scholar]
- Peng L., Kang S., Yin Z., Jia R., Song X., Li L., Li Z., Zou Y., Liang X., Li L., He C., Ye G., Yin L., Shi F., Lv C., & Jing B. (2015). Antibacterial activity and mechanism of berberine against Streptococcus agalactiae. International Journal of Clinical and Experimental Pathology, 8(5), 5217–5223. [PMC free article] [PubMed] [Google Scholar]
- Ram A., Joseph D. A., Balachandar S., & Singh V. P. (2009). Medicinal plants from Siddha system of medicine useful for treating respiratory diseases. Journal of Pharmaceutical Analysis, 1(2), 20. [Google Scholar]
- Ramanathan M., Subramanian L., Poongodi T., Manish S., Muneeswari E., Pavithra P., & Pugalendran S. (2019). Formulation and evaluation of Nilavembu kudineer capsules. Asian Journal of Pharmaceutical Research and Development, 7(1), 41–45. 10.22270/ajprd.v7i1.468 [DOI] [Google Scholar]
- Reddy V. L., Reddy S. M., Ravikanth V., Krishnaiah P., Goud T. V., Rao T. P., Ram T. S., Gonnade R. G., Bhadbhade M., & Venkateswarlu Y. (2005). A new bis-andrographolide ether from Andrographis paniculata nees and evaluation of anti-HIV activity . Nat. Prod. Res, 19(3), 223–230. 10.1080/14786410410001709197 [DOI] [PubMed] [Google Scholar]
- Sarma P., Sekhar N., Prajapat M., Avti P., Kaur H., Kumar S., Singh S., Kumar H., Prakash A., Dhibar D. P., & Medhi B. (2020). In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1753580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P., Prakash O., Shukla A., Rajpurohit C. S., Vasudev P. G., Luqman S., Srivastava S. K., Pant A. B., & Khan F. (2016). Structure-activity relationship studies on holy basil (Ocimum sanctum L.) based flavonoid orientin and its analogue for cytotoxic activity in liver cancer cell line HepG2. Combinatorial Chemistry & High Throughput Screening, 19(8), 656–666. 10.2174/1386207319666160709192801 [DOI] [PubMed] [Google Scholar]
- Shukla P., Jain S. D., Agrawal A., & Gupta A. K. (2019). Indian herbal plants used as antipyretic: A review. International Journal of Pharmacy and Life Sciences, 10(11/12), 6406–6409. [Google Scholar]
- Sinha S. K., Shakya A., Prasad S. K., Singh S., Gurav N. S., Prasad R. S., & Gurav S. S. (2020). An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1762741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Systemes D. (2015). BIOVIA, discovery studio modeling environment. Release 4.5. Dassault Systemes. [Google Scholar]
- Tan J., Wang J., Yang C., Zhu C., Guo G., Tang J., & Shen H. (2019). Antimicrobial characteristics of Berberine against prosthetic joint infection-related Staphylococcus aureus of different multi-locus sequence types. BMC Complementary and Alternative Medicine, 19(1), 218 10.1186/s12906-019-2558-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Uno C., Akimoto M., Tabata M., Honda C., & Kamisako W. (1991). Anti-allergic effect of bryonolic acid from Luffa cylindrica cell suspension cultures. Planta Medica, 57(6), 527–530. 10.1055/s-2006-960199 [DOI] [PubMed] [Google Scholar]
- Umesh K., D., Selvaraj C., Singh S. K., & Dubey V. K. (2020). Identification of new anti-nCoV drug chemical compounds from Indian spices exploiting SARS-CoV-2 main protease as target. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1763202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese F. S., Thaa B., Amrun S. N., Simarmata D., Rausalu K., Nyman T. A., Merits A., McInerney G. M., Ng L., & Ahola T. (2016). The antiviral alkaloid berberine reduces chikungunya virus-induced mitogen-activated protein kinase signaling. Journal of Virology, 90(21), 9743–9757. 10.1128/JVI.01382-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeramachaneni G. K., Thunuguntla V. B. S. C., Janaki Ram B., & Bondili J. S. (2020). Structural and Simulation analysis of hot spot residues interactions of SARS-CoV 2 with human ACE2 receptor. Journal of Biomolecular Structure and Dynamics, 1–24. 10.1080/07391102.2020.1773318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahedi H. M., Ahmad S., & Abbasi S. W. (2020). Stilbene-based natural compounds as promising drug candidates against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–16. 10.1080/07391102.2020.1762743 [DOI] [PubMed] [Google Scholar]
- Wang H., Li K., Ma L., Wu S., Hu J., Yan H., Jiang J., & Li Y. (2017). Berberine inhibits enterovirus 71 replication by downregulating the MEK/ERK signaling pathway and autophagy. Virology Journal, 14(1), 2 10.1186/s12985-016-0674-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A., Ji W., Yan L., Zhu Y., Zhu C., Fang X., Yang X., Huang Y., Gao H., Liu F., Ge J., Sun Q., Yang X., Xu W., … Rao Z. (2020). Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell, 181(4), 894–904. 10.1016/j.cell.2020.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen C. C., Shyur L. F., Jan J. T., Liang P. H., Kuo C. J., Arulselvan P., Wu J. B., Kuo S. C., & Yang N. S. (2011). Traditional Chinese medicine herbal extracts of Cibotium barometz, Gentiana scabra, Dioscorea batatas, Cassia tora, and Taxillus chinensis inhibit SARS-CoV replication. Journal of Traditional and Complementary Medicine, 1(1), 41–50. 10.1016/S2225-4110(16)30055-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., & Sheng J. (2020). Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host & Microbe, 27(3), 325–328. 10.1016/j.chom.2020.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Li J-q., Kim Y-j., Wu J., Wang Q., & Hao Y. (2011). In vivo and in vitro antiviral effects of berberine on influenza virus. Chinese Journal of Integrative Medicine, 17(6), 444–452. 10.1007/s11655-011-0640-3 [DOI] [PubMed] [Google Scholar]
- Xie Y., Liu X., & Zhou P. (2020). In vitro antifungal effects of berberine against Candida spp. In planktonic and biofilm conditions. Drug Des Devel Ther, 14, 87–101. 10.2147/DDDT.S230857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., & Zhang Y. (2011). Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophysical Journal, 101(10), 2525–2534. 10.1016/j.bpj.2011.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav M., Song F., Huang J., Chakravarti A., & Jacob N. K. (2018). Ocimum flavone Orientin as a countermeasure for thrombocytopenia. Scientific Reports, 8(1), 5075 10.1038/s41598-018-23419-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. H., Kim K. J., Cha J. D., Kim H. K., Lee Y. E., Choi N. Y., & You Y. O. (2005). Antimicrobial activity of berberine alone and in combination with ampicillin or oxacillin against methicillin-resistant Staphylococcus aureus. J Med Food, 8(4), 454–461. 10.1089/jmf.2005.8.454 [DOI] [PubMed] [Google Scholar]
- Zhang L., Lin D., Kusov Y., Nian Y., Ma Q., Wang J., von Brunn A., Leyssen P., Lanko K., Neyts J., de Wilde A., Snijder E. J., Liu H., & Hilgenfeld R. (2020a). α-Ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: Structure-based design, synthesis, and activity assessment. Journal of Medicinal Chemistry, 63(9), 4562–4578. 10.1021/acs.jmedchem.9b01828 [DOI] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., & Hilgenfeld R. (2020b). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (New York, NY.), 368(6489), 409–412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M. F., & Shen Y. Q. (1989). Antidiarrheal and anti-inflammatory effects of berberine. Zhongguo Yao li Xue Bao = Acta Pharmacologica Sinica, 10(2), 174–176. 10.1038/aps.2016.125 [DOI] [PubMed] [Google Scholar]
- Zumla A., Chan J. F., Azhar E. I., Hui D. S., & Yuen K. Y. (2016). Coronaviruses—drug discovery and therapeutic options. Nature Reviews. Drug Discovery, 15(5), 327–347. 10.1038/nrd.2015.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are openly available in GenBank database.