Abstract
Our understanding of the genetics of acute myeloid leukemia (AML) development from myelodysplastic syndrome (MDS) has advanced significantly as a result of next-generation sequencing technology. Although differences in cell biology and maturation exist between MDS and AML secondary to MDS, these 2 diseases are genetically related. MDS and secondary AML cells harbor mutations in many of the same genes and functional categories, including chromatin modification, DNA methylation, RNA splicing, cohesin complex, transcription factors, cell signaling, and DNA damage, confirming that they are a disease continuum. Differences in the frequency of mutated genes in MDS and secondary AML indicate that the order of mutation acquisition is not random during progression. In almost every case, disease progression is associated with clonal evolution, typically defined by the expansion or emergence of a subclone with a unique set of mutations. Monitoring tumor burden and clonal evolution using sequencing provides advantages over using the blast count, which underestimates tumor burden, and could allow for early detection of disease progression prior to clinical deterioration. In this review, we outline advances in the study of MDS to secondary AML progression, with a focus on the genetics of progression, and discuss the advantages of incorporating molecular genetic data in the diagnosis, classification, and monitoring of MDS to secondary AML progression. Because sequencing is becoming routine in the clinic, ongoing research is needed to define the optimal assay to use in different clinical situations and how the data can be used to improve outcomes for patients with MDS and secondary AML.
Visual Abstract
Introduction
Myelodysplastic syndromes (MDSs) are a heterogenous group of clonal bone marrow disorders characterized by cytopenia, bone marrow dysplasia, ineffective hematopoiesis, and a high risk for transformation to acute myeloid leukemia (AML).1,2 MDS is typically diagnosed in older patients (median age, 70 years),3 and it is estimated that ≥45 000 new cases are diagnosed per year in the United States,4-7 making it one of the most common adult myeloid malignancies. By definition, MDS patients have a myeloblast count < 20%.
Approximately 30% of MDS patients eventually progress to AML, which is diagnosed by an increase in blast count to ≥20% of total nucleated cells in the bone marrow and is commonly termed “secondary AML to MDS.” Secondary AML accounts for up to 25% to 35% of total AML cases,8,9 with most (60-80%) arising from an antecedent MDS.10 The 2016 World Health Organization classification of hematologic malignancies classifies AML developing from MDS as a distinct clinicopathologic entity termed “AML with myelodysplasia-related changes” (AML-MRC).3 MDS patients who progress to secondary AML typically have inferior rates of complete remission, relapse-free survival, and overall survival compared with patients with de novo AML.8,9,11,12
Although MDS and secondary AML are classified as distinct entities, they represent a disease continuum that undergoes genetic clonal evolution. The discovery that similar genes are mutated in age-related clonal hematopoiesis (ARCH), also known as clonal hematopoiesis of indeterminate potential (CHIP), and myeloid malignancies suggests that progression from clonal hematopoiesis to MDS or AML is possible.13-22 Studies have shown that mutant cells in this spectrum of clonal myeloid diseases acquire additional mutations and clonally evolve during progression.13-15,17-21,23-27 Because of space constraints, this review will primarily focus on the genetic progression from MDS to secondary AML and address how monitoring for disease response and progression using sequencing could be implemented in the clinic.
Clinical and biologic aspects of MDS and secondary AML
MDS patients generally have a poor prognosis, with a median overall survival of only 5 years.28,29 This poor prognosis is not solely due to progression to secondary AML, indicating that MDS without progression is associated with mortality. In fact, as defined by the revised International Prognostic Scoring System,29 the overall survival of high-risk and very high-risk MDS (1.6 and 0.8 years, respectively)29 is similar to AML-MRC (10 months),12,30 indicating that higher-risk MDS and secondary AML are clinically similar. Making it clear that MDS and secondary AML represent cancers on a disease continuum can have a significant impact on a patient’s understanding of their disease.31,32
Cellular differences between MDS and secondary AML
Hematopoietic cell differentiation and maturation are abnormal in MDS and secondary AML. AML is characterized by a block in hematopoietic cell maturation, leading to an accumulation of myeloblasts in the blood or bone marrow (≥20% of nucleated cells). In contrast, MDS patients produce mature blood cells, although maturation and cell morphology are abnormal, and mature cells are reduced in number. Although the block in maturation is a continuum between MDS and secondary AML, higher blast percentage generally confers a worse prognosis in MDS.28,29 A major limitation of using blast count to differentiate MDS from secondary AML is that the blast count is a subjective measurement, relying on a morphological assessment sensitive to interobserver variability.33,34 In addition, MDS blasts can be difficult to distinguish from normal blasts by microscopy, making it difficult to monitor or detect clonal disease in the bone marrow in patients with a normal blast count (eg, <5%). In these cases, the presence of dysplasia (subjective as well) and/or genetic abnormalities is needed to diagnose MDS.
Cell death and proliferation also differ between MDS and secondary AML. Lower-grade MDS is often characterized by increased apoptosis, a noninflammatory cell death that contributes to ineffective myelopoiesis.35-40 Recent research has also linked MDS to increased inflammatory cell death processes, such as pyroptosis and necroptosis.41,42 Unlike apoptosis, these release inflammatory signals and provide a link between cell death and the proinflammatory bone marrow environment observed in MDS patients.41-47 In contrast, secondary AML, and AML in general, is associated with increased cell survival and proliferation. Collectively, the data suggest that the increased cell death phenotype observed in lower-grade MDS gives way to a prosurvival phenotype during progression to higher-grade MDS and secondary AML. Recent work has identified transcriptome, splicing, and DNA methylation alterations in MDS and secondary AML, including in genes associated with hematopoietic cell differentiation, signaling pathways, proliferation, and DNA damage repair. Some of these alterations have been associated with progression to secondary AML and/or clinical outcome.48-51 Future studies using newly developed single-cell techniques (eg, mass cytometer [cytometry by time of flight (CyTOF)], single-cell RNA sequencing) will likely provide additional differences in cell biology between MDS and secondary AML.
The blast count underestimates tumor burden in MDS
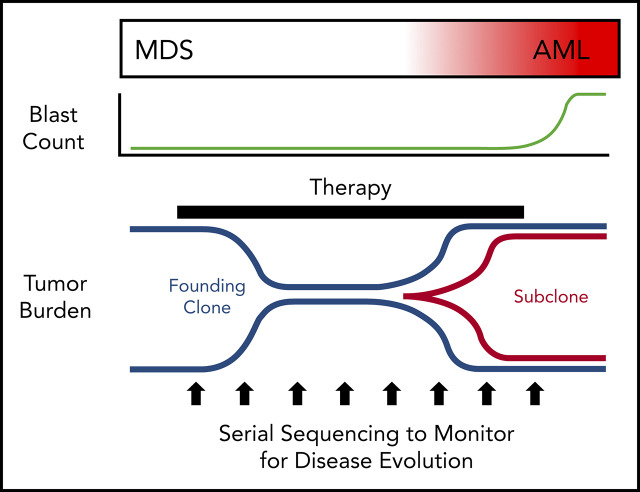
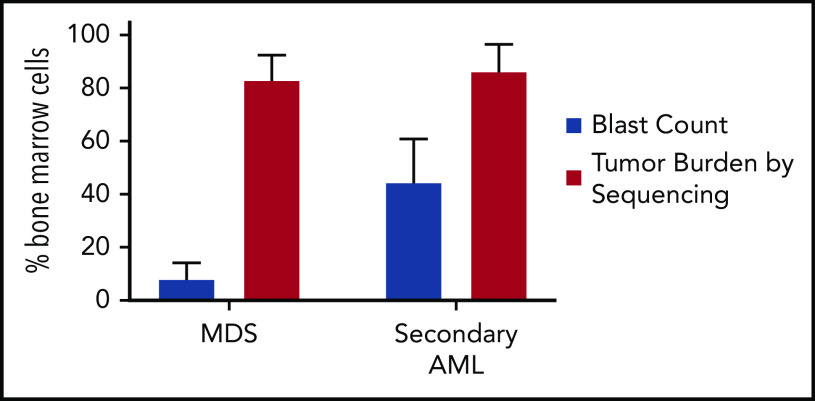
MDS was first demonstrated to be a clonal disease by studies of skewed X chromosome inactivation in women,52-54 followed by the identification of recurrent chromosomal abnormalities in up to 50% of MDS patients.55-63 By combining targeted gene panel sequencing with cytogenetics, ≥78% of MDS patients have a clonal genetic abnormality.18,64-67 Using whole-exome sequencing (WES) or whole-genome sequencing (WGS), we now know that nearly all MDS patients have a clonal mutation. Although these genetic studies can identify clonality in MDS, they also provide a measure of tumor burden that often greatly exceeds the bone marrow blast percentage. Indeed, MDS patients with a normal blast count (ie, <5%) can have an abnormal karyotype or mutations in nearly all bone marrow cells using WGS, regardless of the blast count (Figure 1).17,18,68 Although new mutations may not be acquired between high-risk MDS and leukemia, using the blast count to define a boundary between MDS and AML secondary to MDS has limitations, because disease progression is a continuum.69,70 In addition, the presence of specific genetic abnormalities can be diagnostic of leukemia, even with a blast count < 20% (eg, translocations involving RUNX1-RUNX1T1, CBFB-MYH11), providing further support that a blast count cutoff may not always be ideal. Moving forward, incorporating serial sequencing to monitor dynamic changes in MDS tumor burden over time may be useful along with current monitoring criteria, including blast count, to help identify progression earlier, including the emergence and/or expansion of high-risk genetic abnormalities (eg, TP53, RUNX1, and RAS genes).71,72
Figure 1.
Tumor burden is similar at MDS and secondary AML. Tumor burden at MDS and secondary AML (percentage of bone marrow cells) from 8 patients assessed at both time points.17,18 Tumor burden was measured by morphology using the blast count percentage and sequencing of total bone marrow cells (ie, percentage of clonal cells based on the mutations’ variant allele frequency). Although the blast count increases significantly from MDS to secondary AML, the percentage of clonal cells based on sequencing is similar at both time points. Data are mean ± standard deviation.
The genetics of progression from MDS to secondary AML
Genetic abnormalities in MDS and secondary AML
Chromosome abnormalities and structural variants (including copy number alterations [CNAs], inversions, and translocations) are common in MDS and secondary AML. Similar cytogenetic abnormalities are shared between MDS and secondary AML, such as del(5q), del(7q), del(20q), complex karyotype, and others. MDS-associated cytogenetic abnormalities often result in CNAs, as opposed to balanced rearrangements, which are more common in de novo AML [eg, t(15;17), t(8;21), inv(16), and KMT2A rearrangements].58,63,73,74 Additionally, smaller CNAs (<20 kb) and copy number neutral loss of heterozygosity (or uniparental disomy) occur in MDS and secondary AML and can be detected using single-nucleotide polymorphism arrays. CNA and uniparental disomy regions have been reported to harbor driver genes in MDS and secondary AML.75-80 In fact, a set of “MDS-associated” cytogenetic abnormalities is considered diagnostic of the World Health Organization–defined AML-MRC, even in the absence of a prior MDS diagnosis or morphologic dysplasia.3
Large-scale next-generation sequencing studies have consistently identified mutations in genes in ≥6 major cellular pathways that are shared between MDS and secondary AML, including spliceosome genes, epigenetic modifiers, transcription factors, activated signaling genes, cohesin factors, and TP53.17,18,64-67,81-83 In addition to the previously mentioned cytogenetic abnormalities that distinguish secondary AML from de novo AML, the presence of mutations in spliceosome genes (eg, SRSF2, SF3B1, U2AF1), EZH2, BCOR, and STAG2 is highly suggestive that an AML evolved from MDS, even without a known antecedent MDS diagnosis.65,81,84
Although there is a large degree of overlap in the genes mutated in MDS and secondary AML, the frequency of gene mutations is often different between MDS and secondary AML (Figure 2). Mutations in epigenetic modifiers (eg, TET2, DNMT3A) and TP53 are common across a variety of clonal hematopoietic diseases (eg, ARCH/CHIP, MDS, secondary AML, and de novo AML), and splicing factor gene mutations are most common in MDS and secondary AML.66,85,86 This suggests that these mutations occur early in MDS pathogenesis prior to secondary AML progression. In contrast, mutations in transcription factor (eg, RUNX1, GATA2, CEBPA) and activating signaling genes (eg, RAS family genes, FLT3) are more common in secondary AML, suggesting that these mutations are acquired later during disease progression in a subset of cells that expand.49,65,72,81,87-89 Although our understanding of what genes are mutated in MDS and secondary AML has advanced rapidly thanks to next-generation sequencing, a major advantage of this technology is that it can be used to impute tumor clonality and provide insight into clonal evolution.
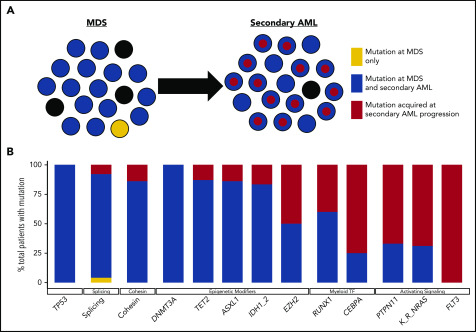
Figure 2.
Clonal evolution during progression from MDS to secondary AML. Using previously published paired MDS and secondary AML samples from the same patients (N = 60),17,18,23,65,66,88 the order of mutation acquisition was inferred by assessing the presence or absence of specific mutations at each sampling. (A) A model for sequential accumulation of mutations during progression from MDS to secondary AML. Black cells are normal at MDS and secondary AML. Mutations in blue are acquired early, define the founding clone, and expand to become the most abundant clone in the marrow at MDS diagnosis. These cells then acquire red mutations, form a subclone, and expand at the time of progression to secondary AML. (B) Percentage of patients with a mutation detectable at MDS only (yellow), secondary AML only (red), or at MDS and persisting during disease progression (blue). Mutations in specific functional categories are enriched in 1 of these patterns, with most TP53, epigenetic modifiers, and spliceosome gene mutations present at MDS (eg, 100% of DNMT3A mutations are blue), whereas mutations in transcription factors (eg, RUNX1, CEBPA) and activating signaling genes (RAS family, PTPN11, FLT3) typically expand or are acquired and emerge at progression (eg, 100% of FLT3 mutations are red). This suggests a typical order of mutation acquisition, with blue mutations being acquired early and present in the majority of marrow cells at MDS diagnosis, followed by the red mutations, which expand or are acquired and expand at progression. Only 1 mutation (shown in yellow) that was detected at MDS was not detected at secondary AML (SRSF2 mutation coding for the P95R substitution).88 Adapted from Lindsley et al.65
Tumor clonality heterogeneity in MDS and secondary AML
WGS of MDS and secondary AML bone marrow samples revealed that these tumors are made up of multiple genetically related, but distinct, clones.17,68 Imputing tumor clonality is possible because there are hundreds of mutations detected using WGS90 and dozens of mutations detected by WES.23,66,88,91 Individual mutations can be clustered based on their variant allele frequency (VAF), with mutations having similar VAFs being present in the same cell or clone.90 The validity of imputing clonality has been confirmed by sequencing single-cell colonies grown from MDS or AML patient bone marrow cells, which showed that sequencing of single-cell–derived colonies recapitulated the imputed clonal architecture in bulk cells.90,92,93 Multiple studies have shown that a patient’s bone marrow sample can be clonally complex and contain multiple genetically related, but unique, clones, including a founding clone and subclone(s) derived from the founding clone.17,18,26,66,68,88,93,94 Imputing accurate clonal architecture is limited when mutations are present at low VAFs, making it challenging to determine whether mutations co-occur in the same cell or exist in parallel subclones. In these cases, single-cell sequencing technologies may be needed; however, they can be limited by allele drop-out.
The combination of mutations that occurs within a patient or clone is not necessarily random, with some mutations co-occurring at frequencies greater (or less) than expected by chance. For example, the combination of ASXL1 and U2AF1 has been reported at a greater-than-expected rate, as has STAG2 with several mutations (including RUNX1, SRSF2, and EZH2).64,95,96 Combinations go beyond mutations alone (eg, SF3B1 mutations are associated with overexpression of EV11, including some with EVI1 rearrangements).49 In contrast, mutations in splicing factor genes are rarely comutated, and the same is seen with cohesin complex genes.19,67,83,97,98 The lack of co-occurrence of these mutations may be driven by functional complementarity/redundancy or synthetic lethality.99 Together, these studies demonstrate that MDS and secondary AML tumors are made up of multiple clones, each containing multiple mutations that likely cooperate to cause the disease phenotype.
The frequency of mutations in epigenetic modifier genes in MDS and secondary AML suggests that epigenetic alterations and heterogeneity may play a significant role in disease progression. Although outside the scope of this review, this topic has been covered in recent reviews.86,100,101 Further work in this area may provide new information regarding how genetic and nongenetic abnormalities affect cell phenotypes and could provide insight into effective treatments.
Order of mutation acquisition
Although the co-occurrence of gene mutations is important, the order of mutation acquisition is likely critical for disease pathogenesis and may also have an important impact on the design of clinical trials using targeted therapies. Deciphering the exact order of mutation acquisition in a patient can be difficult. We can gain insight into this question based on several observations. Recent studies have shown that up to 10% of people older than 70 years of age have clonal hematopoiesis, defined by somatic mutations in their blood cells, and are classified as having ARCH/CHIP.13,14,20,21,25,102,103 Many of these mutations (eg, DNMT3A, TET2, ASXL1, and spliceosome genes) are common, with high VAFs in MDS and secondary AML, suggesting that mutations in these genes occur early in the disease pathogenesis and are present in the majority of cells.17,18,64-67,81,82 Consistent with this, mutations in these genes occur in the founding clone of MDS and secondary AML patients, whereas transcription factor and signaling gene mutations typically occur in subclones derived from the founding clone or other subclones (Figure 2).17,23,24,65,66,68 By sequencing a large number of low- and high-risk MDS samples, as well as secondary AML patients, Makishima et al identified 2 classes of genes that were mutated: type 1 (enriched in secondary AML compared with high-risk MDS) and type 2 (enriched in high-risk compared with low-risk MDS).66 Mutations in these genes suggest a typical order of mutation acquisition during progression. However, our understanding of the order of mutation acquisition remains incomplete, because most data are based on the frequency of gene mutations in cohorts of unpaired MDS and secondary AML patients rather than serial samples from the same patient.
To address this limitation, we identified 60 patients in the literature with paired serial MDS/secondary AML samples (Figure 2).17,18,23,65,66,88 Mutations in common ARCH/CHIP-associated genes (eg, TP53, spliceosome, and epigenetic modifiers) detected at secondary AML were already present in the MDS sample (100%, 92%, and 84% of mutations were present at MDS, respectively) with high average VAFs (33%, 43%, and 31%, respectively), indicating that they are acquired early in the disease course. In contrast, mutations in signaling genes were less commonly detected at MDS (34% of mutations present at MDS), indicating that they are gained later in disease progression (Figure 2).17,18,23,65,66,88 The wide range of sequencing approaches used across these studies limited the ability to consistently detect low-level mutations and define clonality. However, these studies do provide important evidence that a typical order of mutation acquisition may exist during progression from MDS to secondary AML. Given that the order of mutation acquisition has been shown to be important for the hematopoietic phenotype in myeloproliferative neoplasms,104,105 the specific order of mutation acquisition may be important for progression from MDS to secondary AML. Future studies using WGS coupled with ultradeep error-corrected sequencing of somatic mutations from a larger number of paired MDS and secondary AML serial samples from the same patient could help to address these questions.
Subclone expansion defines disease progression
In addition to elucidating the clonality of MDS and secondary AML, WGS and other sequencing platforms have revealed that the expansion of a subclone is a common feature of progression from MDS to secondary AML (Figure 3). The expansion of subclones during progression can occur over a relatively short period of time (eg, weeks to months).17,18 However, some subclones that become the most abundant clone at secondary AML can be detected at low levels in MDS months to years prior to progression, suggesting that their presence may be prognostic for future progression.17,23,66,82,88,106 Indeed, subclonal driver mutations are an independent risk factor for disease progression in chronic lymphocytic leukemia.107 Which gene mutations preexist at MDS will also likely matter. Initial studies by Bejar et al identified mutations (eg, RUNX1, TP53, EZH2) that were independent predictors of poor outcome for MDS, although not necessarily disease progression.71 The presence of rare RAS family member gene mutations at MDS was shown to have the same poor outcome regardless of VAF, suggesting that the presence of these mutations is important.72 Consistent with this, several studies with paired MDS and secondary AML samples have shown that signaling gene mutations, along with new cytogenetic abnormalities, expand at the time of disease progression.17,23,88
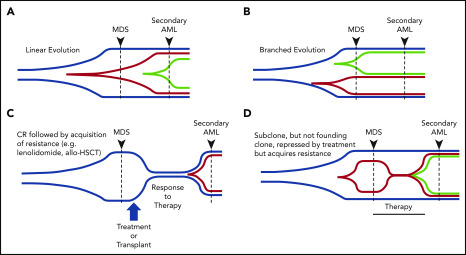
Figure 3.
Patterns of clonal evolution during the progression of MDS to secondary AML. Multiple patterns of subclone expansion are associated with progression from MDS to secondary AML. Subclones (red, green) can be acquired from the founding clone (blue) in a sequential (ie, linear) order (A) or in parallel (ie, branching) (B). Clonal evolution can also be influenced by treatment, including transplant and chemotherapy. (C) Although chemotherapy can suppress the founding clone [eg, lenalidomide in del(5q)-associated MDS], the acquisition of additional mutations (eg, TP53) or cytogenetic abnormalities can occur during disease progression and contribute to subclone expansion. Similar patterns of progression can occur following progression after a transplant. (D) Treatment may also cause subclone clearance while sparing the founding clone (eg, MEK inhibitor repressing a RAS-mutated subclone). Progression can occur when a new subclone emerges carrying additional mutations (green) that drive progression to secondary AML. allo-HSCT, allogeneic hematopoietic stem call transplant; CR, complete remission. Adapted from Nangalia et al.108
Sequencing serial samples is critical to define expanding subclones. Analysis of clonal evolution has shown that evolution can occur in 1 of 2 patterns: linear, in which each successive subclone occurs within the previous parental clone, or branched, in which a founding clone can generate several parallel subclones (Figure 3A-B).17,23,66,68,88,108 In other cases, it has been shown that a subclone can expand to occupy the entire tumor, forcing the collapse of other subclones, a process termed “clone sweeping.”66 WGS of additional paired MDS and secondary AML samples, capable of identifying hundreds of mutations in each sample, will help to define the patterns of clonal evolution (clone collapse and expansion) associated with disease progression. Identifying the genetic patterns of clonal expansion during disease progression may provide important information regarding genetic drivers of subclone expansion during progression to secondary AML.
Clonal diversity of hematopoietic stem cells in MDS and secondary AML
MDS and secondary AML are diseases of hematopoietic stem cells,109-114 with somatic mutations being present in myeloid and lymphoid cells from MDS patients.88,115,116 This suggests that the hematopoietic stem cell, myeloid biased or otherwise, is the cell of origin of disease. Recently, Chen et al sequenced a limited number of phenotypically defined stem cell populations in MDS/secondary AML patients using cell sorting, followed by whole-genome amplification and targeted sequencing.26 The results indicate that subclonal diversity may be higher in stem cells than in blasts at MDS and secondary AML. Furthermore, they show that the dominant clone in the stem cells at MDS (ie, the clone of stem cells giving rise to blasts) is not always the same as the dominant clone at AML, indicating a potential nonlinear path of clonal evolution at the stem cell level. These results indicate a possible greater level of clonal diversity than appreciated using bulk bone marrow samples. These data raise interesting questions regarding the clonal diversity of stem cells in MDS and secondary AML and warrant further study.
Effects of MDS treatment on the clonal evolution of secondary AML
As outlined above, clonal evolution during progression to secondary AML is characterized by the persistence of founding clone mutations and, typically, the expansion of a subclone with unique mutations. Although this pattern is similar for patients receiving treatment or supportive care (transfusions, erythropoietin, thrombopoietin receptor agonists, or granulocyte colony-stimulating factor),17,23,68 several studies have addressed whether specific therapies influence the pattern of clonal evolution.
Clonal evolution following lenalidomide treatment in patients with del(5q) MDS
One of the most common chromosomal abnormalities in MDS/secondary AML is del(5q).74 These patients can achieve a complete remission following lenalidomide treatment.117,118 Although lenalidomide is associated with clinical responses and a decrease in clonal del(5q) cells in the marrow, del(5q) was shown to persist in the quiescent stem cell population (CD34+/CD38−/CD90+).112 These patients eventually relapse, with an expansion of the del(5q) cells containing additional abnormalities (eg, TP53 mutations) (Figure 3C).23,112 Consistent with these results, TP53 mutations were present at low levels months to years prior to disease progression in del(5q)-associated MDS.119 These studies demonstrate that selective pressure from chemotherapy can impact clonal evolution during progression to secondary AML.
Effect of hypomethylating agents on disease progression
Using DNA-hypomethylating agents (HMAs) to treat higher-risk MDS or older AML patients is common.120,121 Although this represents a noncurative therapy, HMAs are associated with improved overall survival.121-124 Predicting response to HMAs based on a patient’s gene mutations remains challenging, and results are variable across studies. Various studies have identified that MDS patients with mutations in epigenetic modifiers (eg, TET2, DNMT3A) and MDS and AML patients with complex cytogenetics and/or TP53 mutations respond better to decitabine.82,125-129 Of note, HMA therapy has been shown to induce C>G transversions in vitro, and is associated with C>G transversions in MDS and AML cells from patients, indicating that HMAs may influence clonal evolution.82,106,130 These C>G transversion mutations can occur in genes implicated in disease pathogenesis, as well as in subclones that emerge at relapse, raising questions of whether HMAs may induce pathogenic mutations.106 Previous studies of decitabine-treated patients utilizing fluorescent in situ hybridization and flow-sorted hematopoietic stem and progenitor cells showed that genetic relapse can predate morphological relapse,113,131 providing the rationale to incorporate sequencing to monitor tumor burden in patients. Tracking all clones will be important, because some patients have a reduction in a subclone with no change in the size of the founding clone (Figure 3D).82 Future studies are needed to define how to incorporate serial sequencing results into traditional response criteria to monitor tumor burden and detect expanding clones.
Disease progression following bone marrow transplantation for MDS and secondary AML
The only curative therapy for MDS and secondary AML is allogeneic hematopoietic stem cell transplant (HSCT). For patients who qualify, most often those with intermediate-2 or higher risk by the International Prognostic Scoring System who are younger than 75 years and in good health with minimal comorbidities,132 HSCT can improve outcomes, although relapse is still common. The use of reduced-intensity conditioning regimens has increased the number of older MDS patients receiving an HSCT.133,134 The pattern of clonal evolution and disease progression following transplant shares some similarities with postchemotherapy progression (Figure 3C). A study of 9 MDS patients who progressed following transplant showed that subclones emerged in 8 of 9 patients at progression, often harboring a new structural variant.106 Furthermore, founding clone mutations were detectable at relapse in all cases, with many mutations being detectable as early as 30 and/or 100 days posttransplant.106 These results suggest that monitoring patients for molecular residual disease may identify a high-risk group of individuals who could be considered for an early intervention in a clinical trial.
Monitoring disease progression
What is the optimal approach to monitor MDS patients for disease progression? Flow cytometry is able to identify dyspoiesis in MDS patients that correlates with clinical outcomes and genetic abnormalities.135-139 The utility of flow cytometry in the clinical work-up of MDS patients has been outlined previously by the European LeukemiaNet Working Group.140,141 Thus, along with morphology, flow cytometry provides a tool for assessment and monitoring of MDS patients. Given the prevalence of genetic abnormalities in these patients, incorporating sequencing results into traditional response criteria may allow for better tumor burden monitoring, particularly in patients with a normal karyotype (Figure 1).17,18,82,91,106,142,143 However, this will have to be tested in prospective clinical trials that use International Working Group response criteria, serial sequencing (for mutation VAFs and clones), and outcomes including survival.144 The optimal sequencing approach to monitor tumor burden and disease MDS progression remains an open question. There are advantages and limitations for various sequencing platforms that could influence how they get incorporated in the clinic.
Whole-genome sequencing
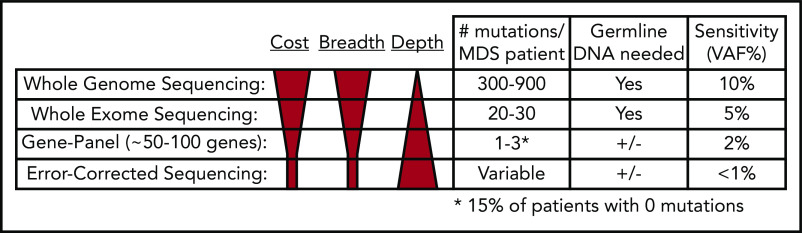
WGS can detect hundreds of mutations per sample, making it ideal for imputing tumor clonality17,18,68,92,93; however, sequencing depth is often limited by costs, making it difficult to detect mutations with VAFs < 10%. Therefore, low-level subclones can be missed. Additionally, sequencing of paired normal DNA is necessary to confidently identify somatic mutations (Figure 4). Despite these limitations, in situations in which the goal is to define the clonal architecture of a sample or discover structural variants and mutations in noncoding regions of the genome, WGS remains an ideal approach.
Figure 4.
Comparison of next-generation sequencing platforms. The optimal sequencing platform to use for clinical testing is dependent on several variables, include the cost, breadth of sequencing coverage (ie, the number of mutations that can be detected), and the typical sequencing depth obtained (ie, the sensitivity of variant detection). Although WGS provides the greatest breadth, the standard depth of coverage (30-45×) limits detection of variants to those with a VAF typically >10%. WES only provides coverage of coding bases in the genome, but the greater sequencing depth (typically 75-150×) allows for detection of mutations with VAFs as low as 5%. Gene panel sequencing is limited to the most commonly mutated genes but can detect mutations at lower VAFs. A typical 100-gene panel with 1000× coverage depth can detect mutations with VAFs as low as 2%, at a reasonable cost. Finally, error-corrected sequencing provides extremely high-depth coverage (10 000×) along with error correction via unique molecular indexes, together allowing for detection of VAFs < 1%. Error-corrected sequencing and gene panel sequencing approaches offer a large degree of flexibility, because they can be used to validate mutations (ie, those detected by WGS), track mutations, or discover mutations. In addition, these methods are often used without paired normal DNA to detect variants. However, germline DNA is necessary to definitively identify somatic mutations (+/−). Adapted from Jacoby et al.90
Whole-exome sequencing
In contrast to WGS, WES typically only detects dozens of mutations in MDS and secondary AML samples; however, greater sequencing depths can be achieved at a reasonable cost using WES, allowing detection of mutation VAFs as low as 5%.23,66,88,91 Germline DNA is still required to confidently identify somatic mutations (Figure 4). Although most patients have a detectable coding mutation, it remains challenging to accurately define clones; however, retrospective WES studies have shown that subclones that expand at progression can be detected prior to progression.66,82,105 If clinically relevant subclonal mutations could be identified in real time, it may be possible to implement a clinical intervention to prevent progression.
Gene-panel sequencing
Targeted gene-panel sequencing is widely available in the clinic.18,64,66,67,97 Gene panels of ∼100 genes or mutation hotspots are able to detect a mutation in most patients (Figure 4); however, gene panel sequencing is limited by the fact that a subset of patients will not have a detectable mutation. Two large sequencing studies interrogated 944 and 738 MDS patients for mutations in 104 and 111 genes, respectively.64 Mutations were identified in 845 (89.5%) and 549 (74%) patients, respectively.64,67 Similar findings were reported for secondary AML; a panel covering 40 genes identified a mutation in 90 of 93 (97%) patients.65 A major limitation of gene panel sequencing is that too few mutations are identified to confidently identify clones, with many clones being completely missed if they do not harbor a mutation in one of the sequenced genes.
Error-corrected sequencing
An error-corrected sequencing (ECS) approach is based on using unique molecular identifiers coupled with high-sequencing coverage depths.82,91,106,145,146 ECS allows for very low detection of a designated set of mutations (<1% VAF). It can be used in 2 ways: as a gene panel to discover mutations or to validate and track previously identified mutations (ie, by WGS) (Figure 4). ECS may provide significant utility in the setting of residual disease detection. In the transplant setting, residual disease detection has clinical relevance when detected by morphology, quantitative polymerase chain reaction, chimerism analysis, and flow cytometry147-151; however, these technologies have limitations. In an exploratory study of MDS patients who received allogeneic HSCT, Duncavage et al found that detection of persistent mutations at VAF ≥ 0.5% at day 30 posttransplant was associated with increased disease progression and lower progression-free survival.91 Patients had a median of 67 days between day-30 mutation detection and disease progression,91 a period that may allow for salvage therapy in some patients.152-156 Kim et al also observed that detecting mutations posttransplant for de novo AML impacts outcomes.157
Together, the benefits and limitations of these sequencing technologies indicate that the best approach is dependent on the specific clinical question being asked. The optimal sequencing technique will depend on whether the goal is diagnostic, the identification of clones, or the monitoring of residual disease. It will be critical to test the feasibility and utility of each application in well-controlled prospective clinical trials before implementation in the clinic becomes standard.
Concluding thoughts
The introduction of next-generation sequencing for MDS and secondary AML confirms that clonal evolution accompanies disease progression, as previously observed using cytogenetics. These studies show that MDS and secondary AML represent a disease continuum that genetically evolves, rather than a new disease. Sequencing results have consistently shown that the blast count assessment, although an important risk factor, often underestimates the tumor burden in MDS. Based on our current knowledge, we suggest that it is time to augment our clinical assessment of tumor burden based on morphology with next-generation sequencing data and develop an approach to incorporate molecular-based disease classification and tumor burden to better diagnose and monitor MDS. Future studies, especially ones involving paired MDS and secondary AML samples from the same patient, could provide insight into a critical conserved order of mutation acquisition that drives disease pathogenesis and is predictive of MDS progression, potentially allowing for early intervention.
Acknowledgments
The authors thank Meagan A. Jacoby and Eric J. Duncavage for helpful scientific discussions and Sridhar N. Srivatsan for assistance with figures.
This work was supported by National Institutes of Health, National Cancer Institute (grants P01 CA101937 and R33CA217700), the Edward P. Evans Foundation, and the Lottie Caroline Hardy Trust (all M.J.W.).
The authors apologize to investigators whose work could not be included because of space constraints.
Authorship
Contribution: A.J.M. and M.J.W. jointly wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Matthew J. Walter, Washington University School of Medicine, 660 S. Euclid Ave, Campus Box 8007, St. Louis, MO 63110; e-mail: mjwalter@wustl.edu.
REFERENCES
- 1.Nimer SD. Myelodysplastic syndromes. Blood. 2008;111(10):4841-4851. [DOI] [PubMed] [Google Scholar]
- 2.Tefferi A, Vardiman JW. Myelodysplastic syndromes. N Engl J Med. 2009;361(19):1872-1885. [DOI] [PubMed] [Google Scholar]
- 3.Swerdlow SH, Campo E, Harris NL, et al. . WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th ed Lyon, France: International Agency for Research on Cancer; 2008. [Google Scholar]
- 4.Cogle CR, Craig BM, Rollison DE, List AF. Incidence of the myelodysplastic syndromes using a novel claims-based algorithm: high number of uncaptured cases by cancer registries. Blood. 2011;117(26):7121-7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldberg SL, Chen E, Corral M, et al. . Incidence and clinical complications of myelodysplastic syndromes among United States Medicare beneficiaries. J Clin Oncol. 2010;28(17):2847-2852. [DOI] [PubMed] [Google Scholar]
- 6.Ma X. Epidemiology of myelodysplastic syndromes. Am J Med. 2012;125(7 suppl):S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeidan AM, Shallis RM, Wang R, Davidoff A, Ma X. Epidemiology of myelodysplastic syndromes: why characterizing the beast is a prerequisite to taming it. Blood Rev. 2019;34:1-15. [DOI] [PubMed] [Google Scholar]
- 8.Granfeldt Østgård LS, Medeiros BC, Sengeløv H, et al. . Epidemiology and clinical significance of secondary and therapy-related acute myeloid leukemia: a national population-based cohort study. J Clin Oncol. 2015;33(31):3641-3649. [DOI] [PubMed] [Google Scholar]
- 9.Hulegårdh E, Nilsson C, Lazarevic V, et al. . Characterization and prognostic features of secondary acute myeloid leukemia in a population-based setting: a report from the Swedish Acute Leukemia Registry. Am J Hematol. 2015;90(3):208-214. [DOI] [PubMed] [Google Scholar]
- 10.Leone G, Mele L, Pulsoni A, Equitani F, Pagano L. The incidence of secondary leukemias. Haematologica. 1999;84(10):937-945. [PubMed] [Google Scholar]
- 11.Borthakur G, Lin E, Jain N, et al. . Survival is poorer in patients with secondary core-binding factor acute myelogenous leukemia compared with de novo core-binding factor leukemia. Cancer. 2009;115(14):3217-3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu XQ, Wang JM, Gao L, et al. . Characteristics of acute myeloid leukemia with myelodysplasia-related changes: A retrospective analysis in a cohort of Chinese patients. Am J Hematol. 2014;89(9):874-881. [DOI] [PubMed] [Google Scholar]
- 13.Genovese G, Kähler AK, Handsaker RE, et al. . Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaiswal S, Fontanillas P, Flannick J, et al. . Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ley TJ, Miller C, Ding L, et al. ; Cancer Genome Atlas Research Network . Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Link DC, Walter MJ. “CHIP”ping away at clonal hematopoiesis. Leukemia. 2016;30(8):1633-1635. [DOI] [PubMed] [Google Scholar]
- 17.Walter MJ, Shen D, Ding L, et al. . Clonal architecture of secondary acute myeloid leukemia. N Engl J Med. 2012;366(12):1090-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walter MJ, Shen D, Shao J, et al. . Clonal diversity of recurrently mutated genes in myelodysplastic syndromes. Leukemia. 2013;27(6):1275-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welch JS, Ley TJ, Link DC, et al. . The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150(2):264-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie M, Lu C, Wang J, et al. . Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young AL, Challen GA, Birmann BM, Druley TE. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat Commun. 2016;7(1):12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young AL, Tong RS, Birmann BM, Druley TE. Clonal hematopoiesis and risk of acute myeloid leukemia. Haematologica. 2019;104(12):2410-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.da Silva-Coelho P, Kroeze LI, Yoshida K, et al. . Clonal evolution in myelodysplastic syndromes. Nat Commun. 2017;8(1):15099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mossner M, Jann JC, Wittig J, et al. . Mutational hierarchies in myelodysplastic syndromes dynamically adapt and evolve upon therapy response and failure. Blood. 2016;128(9):1246-1259. [DOI] [PubMed] [Google Scholar]
- 25.Zink F, Stacey SN, Norddahl GL, et al. . Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood. 2017;130(6):742-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J, Kao YR, Sun D, et al. . Myelodysplastic syndrome progression to acute myeloid leukemia at the stem cell level [published correction appears in Nat Med. 2019;25(3):529]. Nat Med. 2019;25(1):103-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chesnais V, Renneville A, Toma A, et al. ; Groupe Francophone des Myélodysplasies . Effect of lenalidomide treatment on clonal architecture of myelodysplastic syndromes without 5q deletion. Blood. 2016;127(6):749-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenberg P, Cox C, LeBeau MM, et al. . International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89(6):2079-2088. [PubMed] [Google Scholar]
- 29.Greenberg PL, Tuechler H, Schanz J, et al. . Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012;120(12):2454-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weinberg OK, Seetharam M, Ren L, et al. . Clinical characterization of acute myeloid leukemia with myelodysplasia-related changes as defined by the 2008 WHO classification system. Blood. 2009;113(9):1906-1908. [DOI] [PubMed] [Google Scholar]
- 31.Steensma DP. Are myelodysplastic syndromes “cancer”? Unexpected adverse consequences of linguistic ambiguity. Leuk Res. 2006;30(10):1227-1233. [DOI] [PubMed] [Google Scholar]
- 32.Lichtman MA. Language and the clonal myeloid diseases. Blood. 2002;99(2):725-726. [DOI] [PubMed] [Google Scholar]
- 33.Ramos F, Fernández-Ferrero S, Suárez D, et al. . Myelodysplastic syndrome: a search for minimal diagnostic criteria. Leuk Res. 1999;23(3):283-290. [DOI] [PubMed] [Google Scholar]
- 34.Senent L, Arenillas L, Luño E, Ruiz JC, Sanz G, Florensa L. Reproducibility of the World Health Organization 2008 criteria for myelodysplastic syndromes. Haematologica. 2013;98(4):568-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hellström-Lindberg E, Kanter-Lewensohn L, Ost A. Morphological changes and apoptosis in bone marrow from patients with myelodysplastic syndromes treated with granulocyte-CSF and erythropoietin. Leuk Res. 1997;21(5):415-425. [DOI] [PubMed] [Google Scholar]
- 36.Parker JE, Mufti GJ, Rasool F, Mijovic A, Devereux S, Pagliuca A. The role of apoptosis, proliferation, and the Bcl-2-related proteins in the myelodysplastic syndromes and acute myeloid leukemia secondary to MDS. Blood. 2000;96(12):3932-3938. [PubMed] [Google Scholar]
- 37.Raza A, Alvi S, Borok RZ, et al. . Excessive proliferation matched by excessive apoptosis in myelodysplastic syndromes: the cause-effect relationship. Leuk Lymphoma. 1997;27(1-2):111-118. [DOI] [PubMed] [Google Scholar]
- 38.Raza A, Gezer S, Mundle S, et al. . Apoptosis in bone marrow biopsy samples involving stromal and hematopoietic cells in 50 patients with myelodysplastic syndromes. Blood. 1995;86(1):268-276. [PubMed] [Google Scholar]
- 39.Span LF, Rutten E, Gemmink A, Boezeman JB, Raymakers RA, de Witte T. Bone marrow mononuclear cells of MDS patients are characterized by in vitro proliferation and increased apoptosis independently of stromal interactions. Leuk Res. 2007;31(12):1659-1667. [DOI] [PubMed] [Google Scholar]
- 40.Tehranchi R, Fadeel B, Forsblom AM, et al. . Granulocyte colony-stimulating factor inhibits spontaneous cytochrome c release and mitochondria-dependent apoptosis of myelodysplastic syndrome hematopoietic progenitors. Blood. 2003;101(3):1080-1086. [DOI] [PubMed] [Google Scholar]
- 41.Basiorka AA, McGraw KL, Eksioglu EA, et al. . The NLRP3 inflammasome functions as a driver of the myelodysplastic syndrome phenotype. Blood. 2016;128(25):2960-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagner PN, Shi Q, Salisbury-Ruf CT, et al. . Increased Ripk1-mediated bone marrow necroptosis leads to myelodysplasia and bone marrow failure in mice. Blood. 2019;133(2):107-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allampallam K, Shetty V, Mundle S, et al. . Biological significance of proliferation, apoptosis, cytokines, and monocyte/macrophage cells in bone marrow biopsies of 145 patients with myelodysplastic syndrome. Int J Hematol. 2002;75(3):289-297. [DOI] [PubMed] [Google Scholar]
- 44.Chen X, Eksioglu EA, Zhou J, et al. . Induction of myelodysplasia by myeloid-derived suppressor cells. J Clin Invest. 2013;123(11):4595-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mundle SD, Venugopal P, Cartlidge JD, et al. . Indication of an involvement of interleukin-1 beta converting enzyme-like protease in intramedullary apoptotic cell death in the bone marrow of patients with myelodysplastic syndromes. Blood. 1996;88(7):2640-2647. [PubMed] [Google Scholar]
- 46.Navas T, Zhou L, Estes M, et al. . Inhibition of p38alpha MAPK disrupts the pathological loop of proinflammatory factor production in the myelodysplastic syndrome bone marrow microenvironment [published correction appears in Leuk Lymphoma 2009;50(9):1554]. Leuk Lymphoma. 2008;49(10):1963-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takizawa H, Boettcher S, Manz MG. Demand-adapted regulation of early hematopoiesis in infection and inflammation. Blood. 2012;119(13):2991-3002. [DOI] [PubMed] [Google Scholar]
- 48.Taskesen E, Havermans M, van Lom K, et al. . Two splice-factor mutant leukemia subgroups uncovered at the boundaries of MDS and AML using combined gene expression and DNA-methylation profiling. Blood. 2014;123(21):3327-3335. [DOI] [PubMed] [Google Scholar]
- 49.Shiozawa Y, Malcovati L, Gallì A, et al. . Gene expression and risk of leukemic transformation in myelodysplasia [published correction appears in Blood 2018;132(8):869-875]. Blood. 2017;130(24):2642-2653. [DOI] [PubMed] [Google Scholar]
- 50.Im H, Rao V, Sridhar K, et al. . Distinct transcriptomic and exomic abnormalities within myelodysplastic syndrome marrow cells. Leuk Lymphoma. 2018;59(12):2952-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang YT, Chiu YC, Kao CJ, et al. . The prognostic significance of global aberrant alternative splicing in patients with myelodysplastic syndrome. Blood Cancer J. 2018;8(8):78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janssen JW, Buschle M, Layton M, et al. . Clonal analysis of myelodysplastic syndromes: evidence of multipotent stem cell origin. Blood. 1989;73(1):248-254. [PubMed] [Google Scholar]
- 53.Prchal JT, Throckmorton DW, Carroll AJ III, Fuson EW, Gams RA, Prchal JF. A common progenitor for human myeloid and lymphoid cells. Nature. 1978;274(5671):590-591. [DOI] [PubMed] [Google Scholar]
- 54.Raskind WH, Tirumali N, Jacobson R, Singer J, Fialkow PJ. Evidence for a multistep pathogenesis of a myelodysplastic syndrome. Blood. 1984;63(6):1318-1323. [PubMed] [Google Scholar]
- 55.Second International Workshop on Chromosomes in Leukemia. Cancer Res. 1980;40(12):4826-4827. [PubMed] [Google Scholar]
- 56.Jacobs RH, Cornbleet MA, Vardiman JW, Larson RA, Le Beau MM, Rowley JD. Prognostic implications of morphology and karyotype in primary myelodysplastic syndromes. Blood. 1986;67(6):1765-1772. [PubMed] [Google Scholar]
- 57.Le Beau MM, Albain KS, Larson RA, et al. . Clinical and cytogenetic correlations in 63 patients with therapy-related myelodysplastic syndromes and acute nonlymphocytic leukemia: further evidence for characteristic abnormalities of chromosomes no. 5 and 7. J Clin Oncol. 1986;4(3):325-345. [DOI] [PubMed] [Google Scholar]
- 58.Le Beau MM, Espinosa R III, Davis EM, Eisenbart JD, Larson RA, Green ED. Cytogenetic and molecular delineation of a region of chromosome 7 commonly deleted in malignant myeloid diseases. Blood. 1996;88(6):1930-1935. [PubMed] [Google Scholar]
- 59.Nowell P, Finan J. Chromosome studies in preleukemic states. IV. Myeloproliferative versus cytopenic disorders. Cancer. 1978;42(5):2254-2261. [DOI] [PubMed] [Google Scholar]
- 60.Sokal G, Michaux JL, van den Berghe H. The karyotype in refractory anaemia and pre-leukaemia. Clin Haematol. 1980;9(1):129-139. [PubMed] [Google Scholar]
- 61.Sokal G, Michaux JL, Van Den Berghe H, et al. . A new hematologic syndrome with a distinct karyotype: the 5 q--chromosome. Blood. 1975;46(4):519-533. [PubMed] [Google Scholar]
- 62.Streuli RA, Testa JR, Vardiman JW, Mintz U, Golomb HM, Rowley JD. Dysmyelopoietic syndrome: sequential clinical and cytogenetic studies. Blood. 1980;55(4):636-644. [PubMed] [Google Scholar]
- 63.Yunis JJ, Rydell RE, Oken MM, Arnesen MA, Mayer MG, Lobell M. Refined chromosome analysis as an independent prognostic indicator in de novo myelodysplastic syndromes. Blood. 1986;67(6):1721-1730. [PubMed] [Google Scholar]
- 64.Haferlach T, Nagata Y, Grossmann V, et al. . Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28(2):241-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lindsley RC, Mar BG, Mazzola E, et al. . Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood. 2015;125(9):1367-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Makishima H, Yoshizato T, Yoshida K, et al. . Dynamics of clonal evolution in myelodysplastic syndromes. Nat Genet. 2017;49(2):204-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Papaemmanuil E, Gerstung M, Malcovati L, et al. ; Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium . Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122(22):3616-3627, quiz 3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu L, Gu ZH, Li Y, et al. . Genomic landscape of CD34+ hematopoietic cells in myelodysplastic syndrome and gene mutation profiles as prognostic markers. Proc Natl Acad Sci USA. 2014;111(23):8589-8594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lichtman MA. Does a diagnosis of myelogenous leukemia require 20% marrow myeloblasts, and does <5% marrow myeloblasts represent a remission? The history and ambiguity of arbitrary diagnostic boundaries in the understanding of myelodysplasia. Oncologist. 2013;18(9):973-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lichtman MA. Distinguishing clonal evolution from so-called secondary acute myelogenous leukemia: Adhering to unifying concepts of the genetic basis of leukemogenesis. Blood Cells Mol Dis. 2015;55(1):1-2. [DOI] [PubMed] [Google Scholar]
- 71.Bejar R, Stevenson K, Abdel-Wahab O, et al. . Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364(26):2496-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murphy DM, Bejar R, Stevenson K, et al. . NRAS mutations with low allele burden have independent prognostic significance for patients with lower risk myelodysplastic syndromes. Leukemia. 2013;27(10):2077-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haase D. Cytogenetic features in myelodysplastic syndromes. Ann Hematol. 2008;87(7):515-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haase D, Germing U, Schanz J, et al. . New insights into the prognostic impact of the karyotype in MDS and correlation with subtypes: evidence from a core dataset of 2124 patients. Blood. 2007;110(13):4385-4395. [DOI] [PubMed] [Google Scholar]
- 75.Gondek LP, Dunbar AJ, Szpurka H, McDevitt MA, Maciejewski JP. SNP array karyotyping allows for the detection of uniparental disomy and cryptic chromosomal abnormalities in MDS/MPD-U and MPD. PLoS One. 2007;2(11):e1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gondek LP, Haddad AS, O’Keefe CL, et al. . Detection of cryptic chromosomal lesions including acquired segmental uniparental disomy in advanced and low-risk myelodysplastic syndromes. Exp Hematol. 2007;35(11):1728-1738. [DOI] [PubMed] [Google Scholar]
- 77.Gondek LP, Tiu R, O’Keefe CL, Sekeres MA, Theil KS, Maciejewski JP. Chromosomal lesions and uniparental disomy detected by SNP arrays in MDS, MDS/MPD, and MDS-derived AML. Blood. 2008;111(3):1534-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Heinrichs S, Kulkarni RV, Bueso-Ramos CE, et al. . Accurate detection of uniparental disomy and microdeletions by SNP array analysis in myelodysplastic syndromes with normal cytogenetics. Leukemia. 2009;23(9):1605-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mohamedali A, Gäken J, Twine NA, et al. . Prevalence and prognostic significance of allelic imbalance by single-nucleotide polymorphism analysis in low-risk myelodysplastic syndromes. Blood. 2007;110(9):3365-3373. [DOI] [PubMed] [Google Scholar]
- 80.Sato-Otsubo A, Sanada M, Ogawa S. Single-nucleotide polymorphism array karyotyping in clinical practice: where, when, and how? Semin Oncol. 2012;39(1):13-25. [DOI] [PubMed] [Google Scholar]
- 81.Papaemmanuil E, Gerstung M, Bullinger L, et al. . Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374(23):2209-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Uy GL, Duncavage EJ, Chang GS, et al. . Dynamic changes in the clonal structure of MDS and AML in response to epigenetic therapy. Leukemia. 2017;31(4):872-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yoshida K, Sanada M, Shiraishi Y, et al. . Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64-69. [DOI] [PubMed] [Google Scholar]
- 84.Yokoyama K, Shimizu E, Yokoyama N, et al. . Cell-lineage level-targeted sequencing to identify acute myeloid leukemia with myelodysplasia-related changes. Blood Adv. 2018;2(19):2513-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Saez B, Walter MJ, Graubert TA. Splicing factor gene mutations in hematologic malignancies. Blood. 2017;129(10):1260-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sperling AS, Gibson CJ, Ebert BL. The genetics of myelodysplastic syndrome: from clonal haematopoiesis to secondary leukaemia. Nat Rev Cancer. 2017;17(1):5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci USA. 2014;111(7):2548-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim T, Tyndel MS, Kim HJ, et al. . The clonal origins of leukemic progression of myelodysplasia. Leukemia. 2017;31(9):1928-1935. [DOI] [PubMed] [Google Scholar]
- 89.Takahashi K, Jabbour E, Wang X, et al. . Dynamic acquisition of FLT3 or RAS alterations drive a subset of patients with lower risk MDS to secondary AML. Leukemia. 2013;27(10):2081-2083. [DOI] [PubMed] [Google Scholar]
- 90.Jacoby MA, Duncavage EJ, Walter MJ. Implications of tumor clonal heterogeneity in the era of next-generation sequencing. Trends Cancer. 2015;1(4):231-241. [DOI] [PubMed] [Google Scholar]
- 91.Duncavage EJ, Jacoby MA, Chang GS, et al. . Mutation clearance after transplantation for myelodysplastic syndrome. N Engl J Med. 2018;379(11):1028-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hughes AE, Magrini V, Demeter R, et al. . Clonal architecture of secondary acute myeloid leukemia defined by single-cell sequencing. PLoS Genet. 2014;10(7):e1004462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Klco JM, Spencer DH, Miller CA, et al. . Functional heterogeneity of genetically defined subclones in acute myeloid leukemia. Cancer Cell. 2014;25(3):379-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ding L, Ley TJ, Larson DE, et al. . Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481(7382):506-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Alberti MO, Srivatsan SN, Shao J, et al. . Discriminating a common somatic ASXL1 mutation (c.1934dup; p.G646Wfs*12) from artifact in myeloid malignancies using NGS. Leukemia. 2018;32(8):1874-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ogawa S. Genetics of MDS. Blood. 2019;133(10):1049-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gerstung M, Pellagatti A, Malcovati L, et al. . Combining gene mutation with gene expression data improves outcome prediction in myelodysplastic syndromes. Nat Commun. 2015;6(1):5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kon A, Shih LY, Minamino M, et al. . Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat Genet. 2013;45(10):1232-1237. [DOI] [PubMed] [Google Scholar]
- 99.Lee SC, North K, Kim E, et al. . Synthetic lethal and convergent biological effects of cancer-associated spliceosomal gene mutations. Cancer Cell. 2018;34(2):225-241.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Heuser M, Yun H, Thol F. Epigenetics in myelodysplastic syndromes. Semin Cancer Biol. 2018;51:170-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Itzykson R, Fenaux P. Epigenetics of myelodysplastic syndromes. Leukemia. 2014;28(3):497-506. [DOI] [PubMed] [Google Scholar]
- 102.McKerrell T, Park N, Moreno T, et al. ; Understanding Society Scientific Group . Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep. 2015;10(8):1239-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Steensma DP, Bejar R, Jaiswal S, et al. . Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nangalia J, Nice FL, Wedge DC, et al. . DNMT3A mutations occur early or late in patients with myeloproliferative neoplasms and mutation order influences phenotype. Haematologica. 2015;100(11):e438-e442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ortmann CA, Kent DG, Nangalia J, et al. . Effect of mutation order on myeloproliferative neoplasms. N Engl J Med. 2015;372(7):601-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jacoby MA, Duncavage EJ, Chang GS, et al. . Subclones dominate at MDS progression following allogeneic hematopoietic cell transplant. JCI Insight. 2018;3(5):98962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Landau DA, Carter SL, Stojanov P, et al. . Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152(4):714-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nangalia J, Mitchell E, Green AR. Clonal approaches to understanding the impact of mutations on hematologic disease development. Blood. 2019;133(13):1436-1445. [DOI] [PubMed] [Google Scholar]
- 109.Fialkow PJ, Singer JW, Raskind WH, et al. . Clonal development, stem-cell differentiation, and clinical remissions in acute nonlymphocytic leukemia. N Engl J Med. 1987;317(8):468-473. [DOI] [PubMed] [Google Scholar]
- 110.Nilsson L, Astrand-Grundström I, Anderson K, et al. . Involvement and functional impairment of the CD34(+)CD38(−)Thy-1(+) hematopoietic stem cell pool in myelodysplastic syndromes with trisomy 8. Blood. 2002;100(1):259-267. [DOI] [PubMed] [Google Scholar]
- 111.Pang WW, Pluvinage JV, Price EA, et al. . Hematopoietic stem cell and progenitor cell mechanisms in myelodysplastic syndromes. Proc Natl Acad Sci USA. 2013;110(8):3011-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tehranchi R, Woll PS, Anderson K, et al. . Persistent malignant stem cells in del(5q) myelodysplasia in remission. N Engl J Med. 2010;363(11):1025-1037. [DOI] [PubMed] [Google Scholar]
- 113.Will B, Zhou L, Vogler TO, et al. . Stem and progenitor cells in myelodysplastic syndromes show aberrant stage-specific expansion and harbor genetic and epigenetic alterations. Blood. 2012;120(10):2076-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Woll PS, Kjällquist U, Chowdhury O, et al. . Myelodysplastic syndromes are propagated by rare and distinct human cancer stem cells in vivo [published corrections appear in Cancer Cell. 2014;25(6):861 and 2015;27(4):603-605]. Cancer Cell. 2014;25(6):794-808. [DOI] [PubMed] [Google Scholar]
- 115.Mortera-Blanco T, Dimitriou M, Woll PS, et al. . SF3B1-initiating mutations in MDS-RSs target lymphomyeloid hematopoietic stem cells. Blood. 2017;130(7):881-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nilsson L, Astrand-Grundström I, Arvidsson I, et al. . Isolation and characterization of hematopoietic progenitor/stem cells in 5q-deleted myelodysplastic syndromes: evidence for involvement at the hematopoietic stem cell level. Blood. 2000;96(6):2012-2021. [PubMed] [Google Scholar]
- 117.List A, Dewald G, Bennett J, et al. ; Myelodysplastic Syndrome-003 Study Investigators . Lenalidomide in the myelodysplastic syndrome with chromosome 5q deletion. N Engl J Med. 2006;355(14):1456-1465. [DOI] [PubMed] [Google Scholar]
- 118.List A, Kurtin S, Roe DJ, et al. . Efficacy of lenalidomide in myelodysplastic syndromes. N Engl J Med. 2005;352(6):549-557. [DOI] [PubMed] [Google Scholar]
- 119.Jädersten M, Saft L, Smith A, et al. . TP53 mutations in low-risk myelodysplastic syndromes with del(5q) predict disease progression. J Clin Oncol. 2011;29(15):1971-1979. [DOI] [PubMed] [Google Scholar]
- 120.Kantarjian H, Issa JP, Rosenfeld CS, et al. . Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer. 2006;106(8):1794-1803. [DOI] [PubMed] [Google Scholar]
- 121.Silverman LR, Demakos EP, Peterson BL, et al. . Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J Clin Oncol. 2002;20(10):2429-2440. [DOI] [PubMed] [Google Scholar]
- 122.Dombret H, Seymour JF, Butrym A, et al. . International phase 3 study of azacitidine vs conventional care regimens in older patients with newly diagnosed AML with >30% blasts. Blood. 2015;126(3):291-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fenaux P, Mufti GJ, Hellstrom-Lindberg E, et al. ; International Vidaza High-Risk MDS Survival Study Group . Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol. 2009;10(3):223-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fenaux P, Mufti GJ, Hellström-Lindberg E, et al. . Azacitidine prolongs overall survival compared with conventional care regimens in elderly patients with low bone marrow blast count acute myeloid leukemia. J Clin Oncol. 2010;28(4):562-569. [DOI] [PubMed] [Google Scholar]
- 125.Bejar R, Lord A, Stevenson K, et al. . TET2 mutations predict response to hypomethylating agents in myelodysplastic syndrome patients. Blood. 2014;124(17):2705-2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Itzykson R, Kosmider O, Cluzeau T, et al. ; Groupe Francophone des Myelodysplasies (GFM) . Impact of TET2 mutations on response rate to azacitidine in myelodysplastic syndromes and low blast count acute myeloid leukemias. Leukemia. 2011;25(7):1147-1152. [DOI] [PubMed] [Google Scholar]
- 127.Metzeler KH, Walker A, Geyer S, et al. . DNMT3A mutations and response to the hypomethylating agent decitabine in acute myeloid leukemia. Leukemia. 2012;26(5):1106-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Traina F, Visconte V, Elson P, et al. . Impact of molecular mutations on treatment response to DNMT inhibitors in myelodysplasia and related neoplasms. Leukemia. 2014;28(1):78-87. [DOI] [PubMed] [Google Scholar]
- 129.Welch JS, Petti AA, Miller CA, et al. . TP53 and decitabine in acute myeloid leukemia and myelodysplastic syndromes. N Engl J Med. 2016;375(21):2023-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jackson-Grusby L, Laird PW, Magge SN, Moeller BJ, Jaenisch R. Mutagenicity of 5-aza-2′-deoxycytidine is mediated by the mammalian DNA methyltransferase. Proc Natl Acad Sci USA. 1997;94(9):4681-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Craddock C, Quek L, Goardon N, et al. . Azacitidine fails to eradicate leukemic stem/progenitor cell populations in patients with acute myeloid leukemia and myelodysplasia. Leukemia. 2013;27(5):1028-1036. [DOI] [PubMed] [Google Scholar]
- 132.Cutler CS, Lee SJ, Greenberg P, et al. . A decision analysis of allogeneic bone marrow transplantation for the myelodysplastic syndromes: delayed transplantation for low-risk myelodysplasia is associated with improved outcome. Blood. 2004;104(2):579-585. [DOI] [PubMed] [Google Scholar]
- 133.Kröger N, Iacobelli S, Franke GN, et al. . Dose-reduced versus standard conditioning followed by allogeneic stem-cell transplantation for patients with myelodysplastic syndrome: a prospective randomized phase III study of the EBMT (RICMAC Trial). J Clin Oncol. 2017;35(19):2157-2164. [DOI] [PubMed] [Google Scholar]
- 134.Scott BL, Pasquini MC, Logan BR, et al. . Myeloablative versus reduced-intensity hematopoietic cell transplantation for acute myeloid leukemia and myelodysplastic syndromes. J Clin Oncol. 2017;35(11):1154-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wells DA, Benesch M, Loken MR, et al. . Myeloid and monocytic dyspoiesis as determined by flow cytometric scoring in myelodysplastic syndrome correlates with the IPSS and with outcome after hematopoietic stem cell transplantation. Blood. 2003;102(1):394-403. [DOI] [PubMed] [Google Scholar]
- 136.Kern W, Haferlach C, Schnittger S, Alpermann T, Haferlach T. Serial assessment of suspected myelodysplastic syndromes: significance of flow cytometric findings validated by cytomorphology, cytogenetics, and molecular genetics. Haematologica. 2013;98(2):201-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Scott BL, Wells DA, Loken MR, Myerson D, Leisenring WM, Deeg HJ. Validation of a flow cytometric scoring system as a prognostic indicator for posttransplantation outcome in patients with myelodysplastic syndrome. Blood. 2008;112(7):2681-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Della Porta MG, Picone C, Pascutto C, et al. . Multicenter validation of a reproducible flow cytometric score for the diagnosis of low-grade myelodysplastic syndromes: results of a European LeukemiaNET study. Haematologica. 2012;97(8):1209-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ogata K, Della Porta MG, Malcovati L, et al. . Diagnostic utility of flow cytometry in low-grade myelodysplastic syndromes: a prospective validation study. Haematologica. 2009;94(8):1066-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Porwit A, van de Loosdrecht AA, Bettelheim P, et al. . Revisiting guidelines for integration of flow cytometry results in the WHO classification of myelodysplastic syndromes-proposal from the International/European LeukemiaNet Working Group for Flow Cytometry in MDS. Leukemia. 2014;28(9):1793-1798. [DOI] [PubMed] [Google Scholar]
- 141.van de Loosdrecht AA, Ireland R, Kern W, et al. . Rationale for the clinical application of flow cytometry in patients with myelodysplastic syndromes: position paper of an International Consortium and the European LeukemiaNet Working Group. Leuk Lymphoma. 2013;54(3):472-475. [DOI] [PubMed] [Google Scholar]
- 142.Cheson BD, Bennett JM, Kopecky KJ, et al. ; International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia . Revised recommendations of the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia [published correction appears in J Clin Oncol. 2004;22(3):576]. J Clin Oncol. 2003;21(24):4642-4649. [DOI] [PubMed] [Google Scholar]
- 143.Cheson BD, Greenberg PL, Bennett JM, et al. . Clinical application and proposal for modification of the International Working Group (IWG) response criteria in myelodysplasia. Blood. 2006;108(2):419-425. [DOI] [PubMed] [Google Scholar]
- 144.Sekeres MA, Steensma DP. Rethinking clinical trial endpoints in myelodysplastic syndromes. Leukemia. 2019;33(3):570-575. [DOI] [PubMed] [Google Scholar]
- 145.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci USA. 2011;108(23):9530-9535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Schmitt MW, Kennedy SR, Salk JJ, Fox EJ, Hiatt JB, Loeb LA. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci USA. 2012;109(36):14508-14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bornhäuser M, Oelschlaegel U, Platzbecker U, et al. . Monitoring of donor chimerism in sorted CD34+ peripheral blood cells allows the sensitive detection of imminent relapse after allogeneic stem cell transplantation. Haematologica. 2009;94(11):1613-1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Christopeit M, Miersch K, Klyuchnikov E, et al. . Evaluation of BM cytomorphology after allo-SCT in patients with AML. Bone Marrow Transplant. 2012;47(12):1538-1544. [DOI] [PubMed] [Google Scholar]
- 149.Christopeit M, Ocheni S, Haferlach T, et al. . Evaluation of BM cytomorphology after allo-SCT in patients with MDS. Bone Marrow Transplant. 2013;48(3):465-466. [DOI] [PubMed] [Google Scholar]
- 150.Díez-Campelo M, Pérez-Simón JA, Pérez J, et al. . Minimal residual disease monitoring after allogeneic transplantation may help to individualize post-transplant therapeutic strategies in acute myeloid malignancies. Am J Hematol. 2009;84(3):149-152. [DOI] [PubMed] [Google Scholar]
- 151.Tobiasson M, Olsson R, Hellström-Lindberg E, Mattsson J. Early detection of relapse in patients with myelodysplastic syndrome after allo-SCT. Bone Marrow Transplant. 2011;46(5):719-726. [DOI] [PubMed] [Google Scholar]
- 152.Platzbecker U, Middeke JM, Sockel K, et al. . Measurable residual disease-guided treatment with azacitidine to prevent haematological relapse in patients with myelodysplastic syndrome and acute myeloid leukaemia (RELAZA2): an open-label, multicentre, phase 2 trial. Lancet Oncol. 2018;19(12):1668-1679. [DOI] [PubMed] [Google Scholar]
- 153.Platzbecker U, Wermke M, Radke J, et al. . Azacitidine for treatment of imminent relapse in MDS or AML patients after allogeneic HSCT: results of the RELAZA trial. Leukemia. 2012;26(3):381-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rosenow F, Berkemeier A, Krug U, et al. . CD34(+) lineage specific donor cell chimerism for the diagnosis and treatment of impending relapse of AML or myelodysplastic syndrome after allo-SCT. Bone Marrow Transplant. 2013;48(8):1070-1076. [DOI] [PubMed] [Google Scholar]
- 155.Schroeder T, Czibere A, Platzbecker U, et al. . Azacitidine and donor lymphocyte infusions as first salvage therapy for relapse of AML or MDS after allogeneic stem cell transplantation. Leukemia. 2013;27(6):1229-1235. [DOI] [PubMed] [Google Scholar]
- 156.Woo J, Howard NP, Storer BE, et al. . Mutational analysis in serial marrow samples during azacitidine treatment in patients with post-transplant relapse of acute myeloid leukemia or myelodysplastic syndromes. Haematologica. 2017;102(6):e216-e218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kim T, Moon JH, Ahn JS, et al. . Next-generation sequencing-based posttransplant monitoring of acute myeloid leukemia identifies patients at high risk of relapse. Blood. 2018;132(15):1604-1613. [DOI] [PubMed] [Google Scholar]