Abstract
The dynamics of viral load (VL) of the severe acute respiratory syndrome coronavirus 2 and its association with different clinical parameters remain poorly characterized in the US patient population. Herein, we investigate associations between VL and parameters, such as severity of symptoms, disposition (admission versus direct discharge), length of hospitalization, admission to the intensive care unit, length of oxygen support, and overall survival in 205 patients from a tertiary care center in New York City. VL was determined using quantitative PCR and log10 transformed for normalization. Associations were tested with univariate and multivariate regression models. Diagnostic VL was significantly lower in hospitalized patients than in patients not hospitalized (log10 VL = 3.3 versus 4.0; P = 0.018) after adjusting for age, sex, race, body mass index, and comorbidities. Higher VL was associated with shorter duration of the symptoms in all patients and hospitalized patients only and shorter hospital stay (coefficient = −2.02, −2.61, and −2.18; P < 0.001, P = 0.002, and P = 0.013, respectively). No significant association was noted between VL, admission to intensive care unit, length of oxygen support, and overall survival. Our findings suggest a higher shedding risk in less symptomatic patients, an important consideration for containment strategies. Furthermore, we identify a novel association between VL and history of cancer. Larger studies are warranted to validate our findings.
The 2019 novel coronavirus [severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)] was first documented in December 2019 in Wuhan, China, and has resulted thus far in significant global morbidity and mortality.1 SARS-CoV-2 is genetically related to SARS-associated coronavirus (SARS-CoV), which caused an epidemic in 2002 to 2003. Although not proved, it is believed that the epidemic of SARS-CoV was successfully contained through public health interventions; nonetheless, besides differences in policy timeliness and effectiveness between the two epidemics, SARS-CoV-2 has proved to be significantly more difficult to contain. Both viruses utilize the same receptor for cell entry; however, SARS-CoV-2 uses a modified spike protein, which has a higher avidity for the corresponding human receptor and likely amplifies viral infectivity.2,3 Moreover, the two viruses appear to have different shedding kinetics. SARS-CoV viral load (VL) peaks on average 10 days after symptom onset; in comparison, a recent study with 94 subjects from the Guangzhou Eighth People's Hospital in China showed that SARS-CoV-2 viral load peaks at 0.7 days before symptom onset, suggesting that transmission occurs early in the course of the infection.4, 5, 6, 7 As a result, disease containment may require strategies different from those utilized in the containment of SARS-CoV. In addition, authors compared viral load kinetics in mild and severe cases of coronavirus disease 2019 (COVID-19) and reported no differences between the two groups.7 In a similar manner, a study with 5000 subjects from Lombardy, Italy, showed no difference in viral loads between asymptomatic carriers and symptomatic COVID-19 patients.8 On the other hand, a study with 76 patients in Nanchang, China, showed that hospitalized patients with severe disease tend to have a high viral load and a longer virus-shedding period compared with milder cases.9
To date, there are no studies investigating association between viral load with different clinical parameters and outcome in the US patient population. Herein, we sought to identify potential associations between viral load obtained from SARS-CoV-2–positive patients at the emergency department in a tertiary care center in New York City and its association with different clinical parameters. These parameters included severity of symptoms, disposition (admission versus direct discharge), length of hospitalization, admission to the intensive care unit (ICU), length of need for oxygen support, and overall survival. We also sought to address whether certain comorbidities are associated with a higher viral load or not.
Materials and Methods
Patient Population and Data Collection
After review and approval by the institutional review board [New York University (NYU) Langone Health, study i20-00616], a retrospective observational study was conducted to include patients with confirmed COVID-19 that presented to our emergency department at NYU Langone Health in Manhattan, NY, between March 12 and March 18, 2020. Only adult patients (>18 years old) are included in this study. Clinical and demographic data were collected by the Center for Biospecimen Research and Development at New York University under an approved institutional review board protocol (study S16-00122) universal mechanism of human biospecimen collection and storage for research.10 Patient data were collected from the electronic medical record, and baseline demographics, severity of symptoms, therapeutic management, clinical course, laboratory data, and clinical outcomes were analyzed. Duration of symptoms was calculated from the day of onset to the collection of the nasopharyngeal swab. Analysis of certain comorbidities, such as hypertension, asthma, chronic obstructive pulmonary disease, diabetes, chronic kidney disease, and cancer history, was included. Follow-up duration was 8 weeks.
Molecular Testing for SARS-CoV-2
Nasopharyngeal swabs were collected from all patients presenting to our emergency department with suspected COVID-19 infection. Collection of the nasopharyngeal sample was performed by trained medical personnel. The nasopharyngeal samples were transferred to viral transport media immediately after collection and transported to microbiology and molecular pathology for further testing. A total of 205 sequential patients without any exclusions were tested using both qualitative RT-PCR and quantitative RT-PCR to obtain diagnostic SARS-CoV-2 viral load (CDC kit that was submitted for US Food and Drug Administration review under emergency authorization utilization on March 24, 2020) at initial presentation.
For qualitative RT-PCR, the cobas SARS-CoV-2 Qualitative Assay (Emergency Use Authorization; Roche Diagnostics, Basel, Switzerland) for use with cobas 6800/8800 systems (Roche) was used, following the manufacturer's instructions. The system is based on fully automated sample preparation (nucleic acid extraction and purification), followed by PCR amplification and detection of two target regions in the SARS-CoV-2 genome (Roche). The result was considered valid only when the CT value of the reference gene was <40. The result was considered positive when the CT values of both target genes were <40 and negative when they were both ≥40. For quantitative real-time RT-PCR, the CDC's SARS-CoV-2 assay test was used. This assay uses the US CDC design, targeting three regions of the virus nucleocapsid N gene. An additional primer/probe targeting the human RNAse P gene is included, serving as the internal control. Viral RNA was extracted within using the Promega (Madison, WI) Maxwell RSC Instrument and Swab kit, according to the manufacturer's instructions. All quantitative RT-PCR assays were performed using Thermo Fisher Scientific (Waltham, MA) TaqPath 1-Step RT-qPCR Master Mix, CG (catalog number A15299) on the Applied Biosystems (Foster City, CA) 7500 Fast Dx RealTime PCR Instrument. Similarly, the result was considered valid only when the CT value of the reference gene was <40. The result was considered positive when the CT values of all three targets were <40 and negative when they were both ≥40. Using the positive controls provided by the CDC, which are normalized to 1000 copies/μL, we converted the different CT positive in copies/μL. This was done using the ΔΔCT method, applying the formula: Power (2, (CT (sample, N1 gene) -CT (PC, N1 gene))-(CT (sample, RP gene) -CT (PC, RP gene)))∗1000.
Statistical Analysis
Descriptive analyses of the baseline characteristics and clinical parameters in frequencies and proportions for categorical variables and medians and interquartile ranges for continuous variables were performed. Wilcoxon rank-sum test (two sided) and Pearson's χ2 test were used to assess the difference between hospitalized and nonhospitalized patients for continuous and categorical baseline characteristics, respectively. The Fisher exact test was used instead for analyzing categorical variables when one or more of the observed cell counts were less than five. Viral load was log10 transformed for normalization before analysis. The association between VL on admission and clinical parameters was assessed among all patients and patients who were hospitalized. Specifically, simple linear regression of continuous clinical parameters against the viral load was fitted to the data to assess the association between VL and clinical parameters. Logistic regression for dichotomous clinical parameters against the viral load was fitted to the data to assess the association between VL and clinical parameters. The associations were further assessed with regression models adjusting for age, sex, race, body mass index (BMI), and comorbidities. The effect sizes of association were shown as regression coefficients and 95% CIs for continuous parameters and odds ratios and 95% CIs for dichotomous parameters. Analyses were performed using R version 3.6.1 (https://www.r-project.org).11 P < 0.05 was indicated as statistically significant, and P < 0.10 was indicated as marginally significant.
Results
Baseline Clinical Characteristics of the Cohort
A total of 205 patients were included in the study, including 165 discharged patients from the emergency department and 40 hospitalized patients. Nonhospitalized patients were overall younger (median age, 45 years) compared with hospitalized patients (median age, 60 years; P < 0.001). Other baseline characteristics, including sex, race, BMI, and comorbidities, showed no statistically significant difference in terms of distribution between the two groups (Table 1). The group of hospitalized patients was expectedly enriched in patients with severe or critical COVID-19 (P < 0.001) (Table 2). The median duration of symptoms from symptom onset to nasopharyngeal swab collection in that group was 5 days as opposed to 3 days in the nonhospitalized group (P = 0.017). Median hospitalization duration was 6.5 days (Table 2).
Table 1.
Baseline Characteristics of Enrolled Patients with Coronavirus Disease 2019 (COVID-19) by Hospitalization Status
| Characteristic | Not hospitalized (n = 165) | Hospitalized (n = 40) | P value |
|---|---|---|---|
| Median age, years (interquartile range) | 45 (33–58) | 60 (51–71.8) | <0.001 |
| Sex (male) | 84 (50.9) | 24 (60.0) | 0.392 |
| Race | |||
| White | 82 (49.7) | 23 (57.5) | 0.142 |
| African American | 16 (9.7) | 5 (12.5) | |
| Asian | 10 (6.1) | 5 (12.5) | |
| Other | 57 (34.6) | 7 (17.5) | |
| BMI, kg/m2 | |||
| Underweight (BMI < 18.5) | 2 (1.2) | 0 (0) | 0.301 |
| Normal weight (18.5 ≤ BMI < 24.9) | 32 (19.4) | 15 (37.5) | |
| Overweight (25 ≤ BMI < 29.9) | 40 (24.2) | 13 (32.5) | |
| Obese (BMI ≥ 30) | 38 (23.0) | 10 (25.0) | |
| Unknown | 0 (0) | 2 (5.0) | |
| Comorbidities (yes) | 74 (44.9) | 26 (65.0) | 0.221 |
Data are given as n (%) unless otherwise indicated. n = 205 enrolled patients.
BMI, body mass index.
Table 2.
Key Clinical Parameters and Viral Load Quantified by qPCR for Enrolled Patients by Hospitalization Status
| Variable | Not hospitalized (n = 165) | Hospitalized (n = 40) | P value |
|---|---|---|---|
| COVID-19 severity category | <0.001 | ||
| Tested for COVID-19 with no symptoms | 1 (0.6) | 0 (0) | |
| Tested for COVID-19 with mild symptoms (not admitted to hospital) | 139 (84.2) | 0 (0) | |
| Tested for COVID-19 with mild symptoms (admitted to hospital) | 0 (0) | 11 (27.5) | |
| Tested for COVID-19 with severe symptoms [dyspnea (RR > 30/min), hypoxia (<93% O2 saturation), or >50% lung involved on imaging in 24–48 hours] | 0 (0) | 3 (7.5) | |
| Tested for COVID-19 with critical symptoms (respiratory failure, septic shock, or multiorgan dysfunction) | 3 (1.8) | 26 (65.0) | |
| Unknown | 22 (13.3) | 0 (0) | |
| Duration of symptoms before testing, days | (n = 117) 3 (2–5) |
(n = 39) 5 (2.5–7) |
0.017 |
| Length of hospital stay, days | 6.5 (3–18) | ||
| Admission to ICU (yes) | 14 (35.0) | ||
| Oxygen support, days | (n = 2) 4 (2.5–5.5) |
(n = 26) 10 (4.3–16.8) |
0.210 |
| Deceased (yes) | 1 (0.6) | 9 (22.5) | <0.001 |
| Clinical outcome for alive patients | (n = 31) | ||
| Discharged | 31 (77.5) | ||
| Still in admission | 0 (0) | ||
| Log10 viral load∗ | 4.0 (2.6–4.9) | 3.3 (1.2–4.3) | 0.014 |
Data are given as n (%) or median (interquartile range). n = 205 enrolled patients.
COVID-19, coronavirus disease 2019; ICU, intensive care unit; qPCR, quantitative PCR; RR, respiratory rate.
The viral load is significantly lower in hospitalized patients than in patients not hospitalized after adjusting for age, sex, race, body mass index, and comorbidities (P = 0.018).
Viral Load in Hospitalized and Nonhospitalized Patients
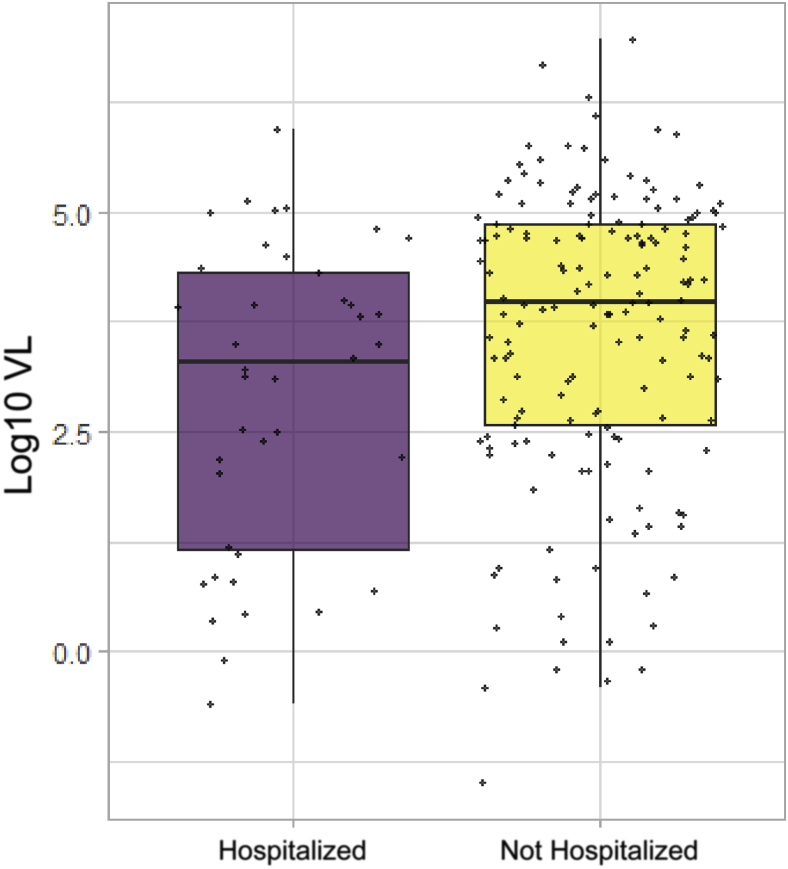
Diagnostic viral load titers were significantly lower in patients who ended up being admitted compared with patients who did not (log10 VL = 3.3 versus 4.0; P = 0.014) (Figure 1 and Table 2). Multivariate linear regression analysis corroborated these findings, showing that this association is independent of the effect of other confounders, including age, sex, race, comorbidities, and BMI (P = 0.018) (Table 2).
Figure 1.
Log10 viral load (VL) values in 205 (both hospitalized and nonhospitalized) patients (P = 0.014) with coronavirus disease 2019 (COVID-19). Bold horizontal lines represent medians.
Association of VL at Admission and Duration of Symptoms, Length of Hospital Stay, Length of Oxygen Support, and Disease Severity
Univariate analysis showed an inverse correlation between duration of symptoms and viral load (P < 0.001 and P = 0.005) in all enrolled patients and hospitalized patients, respectively (Table 3). A similar inverse correlation was noted between length of hospitalization and viral load in hospitalized patients (P = 0.013) (Table 3). Multivariate analysis, adjusting for age, sex, race, BMI, and comorbidities, corroborated these findings, showing a significant inverse correlation between viral load and the duration of symptoms (P < 0.001 and P = 0.008) and a trend with length of hospitalization (P = 0.078) (Table 4). As noted above, higher viral loads also showed an inverse correlation with COVID-19 severity across the cohort and in the subgroup of hospitalized patients, which marginally did not meet statistical significance in a univariate analysis (P = 0.056 and P = 0.097) (Table 3), but was statistically significant in the hospitalized group after adjusting for age, sex, race, BMI, and comorbidities (P = 0.045) (Table 4). A total of 14 patients (35% of the hospitalized patients and 7% of the whole cohort) were admitted to the ICU, and 8 of them died (22.5%) (Table 2). Viral load showed no statistically significant correlation with duration of oxygen support, admission to ICU, or mortality (Tables 3 and 4). Assessment of the relationship of the diagnostic viral load with separate comorbidities in all enrolled patients showed a significant positive correlation with a history of cancer (P = 0.033) (Table 5) and cardiovascular disorders (such as coronary artery disease, arrhythmias, and chronic heart failure), other than hypertension (P = 0.022) (Table 5). Both associations were still statistically significant when adjusted for age (P = 0.019 and P = 0.005, respectively) (Table 5).
Table 3.
Association of Viral Load on Admission with Key Clinical Parameters for all Enrolled Patients and Hospitalized Patients: Univariate Analysis
| Continuous clinical parameters | All patients (hospitalized versus not hospitalized) |
Hospitalized patients only |
||
|---|---|---|---|---|
| Coefficient (95% CI) | P value | Coefficient (95% CI) | P value | |
| Duration of symptoms before testing, days | −0.79 (−1.03 to −0.55) | <0.001 | −0.97 (−1.61 to −0.32) | 0.005 |
| Length of hospital stay, days | −2.18 (−3.86 to −0.50) | 0.013 | ||
| Oxygen support, days | −1.98 (−4.47 to 0.51) | 0.114 | −1.53 (−5.73 to 2.67) | 0.248 |
| Categorical clinical parameters | OR (95% CI) | P value | OR (95% CI) | P value |
| COVID-19 severity (severe/nonsevere) | 0.81 (0.65 to 1.01) | 0.056 | 0.67 (0.39 to 1.04) | 0.097 |
| Admission to ICU (yes/no) | 0.73 (0.48 to 1.07) | 0.111 | ||
| Deceased (yes/no) | 1.00 (0.69 to 1.55) | 0.987 | 1.07 (0.681 to 1.749) | 0.772 |
n = 205 enrolled patients; n = 40 hospitalized patients.
COVID-19, coronavirus disease 2019; ICU, intensive care unit; OR, odds ratio.
Table 4.
Association of Viral Load on Admission with Key Clinical Parameters for all Enrolled Patients and Hospitalized Patients: Multivariate Analysis with Adjustment for Age, Sex, Race, BMI, and Comorbidities
| Continuous clinical parameters | All patients (hospitalized versus not hospitalized) |
Hospitalized patients only |
||
|---|---|---|---|---|
| Coefficient (95% CI) | P value | Coefficient (95% CI) | P value | |
| Duration of symptoms before testing, days | −0.87 (−1.18 to −0.57) | <0.001 | −1.14 (−1.94 to −0.32) | 0.008 |
| Length of hospital stay, days | −1.96 (−4.14 to 0.23) | 0.078 | ||
| Oxygen support, days | −1.83 (−5.33 to 1.68) | 0.289 | −1.54 (−5.88 to 2.80) | 0.465 |
| Categorical clinical parameters | OR (95% CI) | OR (95% CI) | ||
| COVID-19 severity (severe/nonsevere) | 0.77 (0.58 to 1.01) | 0.062 | 0.08 (0.002 to 0.49) | 0.045 |
| Admission to ICU (yes/no) | 0.63 (0.35 to 1.04) | 0.093 | ||
| Deceased (yes/no) | 1.08 (0.66 to 1.82) | 0.772 | 2.30 (0.95 to 9.48) | 0.122 |
n = 205 enrolled patients; n = 40 hospitalized patients.
BMI, body mass index; COVID-19, coronavirus disease 2019; ICU, intensive care unit; OR, odds ratio.
Table 5.
Association between VL on Admission with Comorbidities for All Enrolled Patients with Coronavirus Disease 2019 (COVID-19)
| Variable | Log10 VL in patients with comorbidity |
Log10 VL in patients without comorbidity |
P value |
|---|---|---|---|
| n, mean (IQR) | n, median (IQR) | ||
| COPD | 6, 2.194 (0.6161–3.767) | 175, 3.858 (2.426–4.812) | 0.145 |
| HTN | 47, 3.978 (2.531–4.918) | 133, 3.823 (2.337–4.795) | 0.392 |
| DM | 25, 4.013 (3.135–4.947) | 156, 3.833 (2.34–4.798) | 0.360 |
| CKD | 13, 3.484 (0.8421–4.297) | 168, 3.871 (2.432–4.809) | 0.290 |
| Obesity | 16, 3.802 (2.535–4.82) | 165, 3.844 (2.395–4.795) | 0.936 |
| Asthma | 26, 4.012 (2.634–4.689) | 155, 3.842 (2.339–4.814) | 0.757 |
| Hx of cancer | 16, 4.601 (3.767–4.882) | 165, 3.797 (2.31–4.807) | 0.033∗ |
| Other lung disease | 7, 4.077 (2.653–4.325) | 174, 3.844 (2.399–4.814) | 0.874 |
| Other cardiovascular | 51, 4.323 (3.174–4.989) | 130, 3.566 (2.197–4.736) | 0.022∗ |
n = 205 enrolled patients.
CKD, chronic kidney disease; COPD, chronic obstructive pulmonary disease; DM, diabetes mellitus; HTN, hypertension; Hx, history; IQR, interquartile range; VL, viral load.
P < 0.05.
Discussion
Although qualitative SARS-CoV-2 RT-PCR assays have been widely implemented for the diagnosis of COVID-19 infection, the clinical utility of quantitative SARS-CoV-2 RT-PCR for the determination of viral copies remains undetermined. Studies thus far have shown conflicting evidence in regard to the viral shedding kinetics between mild and severe COVID-19, while there has been no link to clinical outcomes.7,9 The goal of this study was to examine whether the diagnostic viral load has any effect or association with known clinical parameters and outcomes.
We showed that diagnostic viral load is higher in nonhospitalized patients and has a significant inverse correlation with duration of symptoms. This inverse relationship of viral load and duration of symptoms was also observed in the hospitalized subgroup, assessed in a univariate and multivariate analysis. These results corroborate that viral loads peak shortly after symptom onset and support the results of initial Asian cohorts, which established that viral load peaks during the presymptomatic stage of the disease and slowly declines to become undetectable by day 18 to 21.7 Furthermore, we showed that viral loads are inversely correlated with disease severity. We believe that higher viral loads are seen in mild disease rather than severe disease, as they reflect the time from onset of infection. These findings are interesting when interpreted within our current knowledge of the natural history of COVID-19. Although many COVID-19 patients recover from their initial symptoms over the course of 14 to 21 days, a sizable proportion of patients show clinical deterioration to severe disease, which usually occurs a week after symptom onset.12 In the current study cohort, viral loads detected from nasopharyngeal sampling in patients requiring hospitalization (most of whom had severe or critical symptoms) were obtained at a later time point in the disease course. This fact is on par with prior studies demonstrating that longer disease duration is not necessarily associated with a higher viral load but rather a lower viral load.7 The results also show that patients with mild COVID-19 symptoms may represent the most important overlooked source of shedding given the higher viral load. We believe containment strategies can utilize quantitative viral loads to identify patients with high viral titers. In the hospital triage setting, the information from the viral load can be utilized to determine which patient would receive a negative pressure room. In the outpatient setting, clinicians can utilize viral load as a quantitative metric to reinforce the importance of self-isolation and face coverings to reduce the risk of spreading the infection.
Wölfel et al13 found that patients with COVID-19 infections had upper respiratory viral load peaks within the first week of symptom development. Despite 100% seroconversion of the study cohort by day 14 and cessation of symptoms, patients continued to have active viral replication in upper respiratory tract tissue detectable by PCR. Although this study does not provide a temporal characterization of viremia in COVID-19 patients, significantly prolonged symptomatic interval/duration in patients with severe disease was observed. Conversely, patients with a shorter symptomatic interval present mild symptoms, as witnessed in the nonhospitalized patients in our cohort. Chen et al14 described the temporal progression of COVID-19 in 249 patients from Shanghai; the authors identified that ICU patients remained PCR positive and have a prolonged symptom interval/duration compared with non-ICU patients. Moreover, Liu et al9 showed that mild cases have significantly lower viral loads compared with severe cases. Nonetheless, in both studies, cohorts were composed solely of hospitalized patients, as opposed to the current study, which included both nonhospitalized and hospitalized patients.
Moreover, studies thus far have not assessed the relationship between viral load and COVID-19 clinical outcomes. We showed that there was no significant association between viral load and clinical outcomes, including length of stay, oxygen support requirement, or survival. Interestingly, multiple publications have highlighted severe cases of COVID-19 with high morbidity occurring late in the disease process, suggesting that severe symptoms are likely unrelated to high viral titers.12 Instead, deterioration of patients seems to be related to acute respiratory distress syndrome and multisystem organ failure, related to immunologic hyperactivation, characterized by high levels of cytokines, like IL-6 and tumor necrosis factor-α, lymphocyte activation, T-helper 17 differentiation, and concomitant severe lymphopenia, most likely due to activation-induced cell death.15,16 High morbidity and mortality related to COVID-19 appear to be directly linked to this unregulated inflammatory response rather than being caused by high viral load titers per se.
This study is limited in that the analyzed cohort was predominantly composed of nonhospitalized patients and only a small subset of patients exhibited severe COVID-19 infection. Also, viral loads were only obtained from the upper respiratory tract using nasopharyngeal swabs and at a single time point. Although this is the recommended specimen by the CDC, additional synchronous and longitudinal sampling from other sources, such as lower respiratory tract and bronchoalveolar lavages (in intubated patients), would have been important comparator to have. Although dangerous because of technical challenges of specimen collection and increased risk of viral exposure, further exploration of quantitative viral loads from lower respiratory tract tissue and blood in patients exhibiting severe disease may prove to be a better predictor for clinical outcomes.
Although it seems that diagnostic viral load has no prognostic utility in terms of outcome prediction, it is an important surrogate epidemiologic marker of infectivity, notably in mildly symptomatic, nonhospitalized patients. Therefore, our analysis provides the rationale to implement diagnostic viral load in combination with qualitative methods as part of the initial diagnostic armamentarium for patients presenting to emergency department with suspicion of COVID-19. Nevertheless, although this implementation is most crucial in the asymptomatic or mildly symptomatic patient population, the manual nature of the quantitative assay could limit its large-scale integration in the diagnostic laboratory workflow. Automation of quantitative methods could overcome this issue in the future.
Footnotes
Supported by the New York University (NYU) Cancer Center NIH support grant P30CA016087 (G.J.) and the NYU Melanoma National Cancer Institute Specialized Programs of Research Excellence (SPORE) grant P50CA016087 (I.O., P.C.).
K.V.A. and A.S. contributed equally to this work.
Disclosures: None declared.
Contributor Information
Paolo Cotzia, Email: paolo.cotzia@nyulangone.org.
George Jour, Email: george.jour@nyulangone.org.
References
- 1.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peiris J.S., Chu C.M., Cheng V.C., Chan K.S., Hung I.F., Poon L.L., Law K.I., Tang B.S., Hon T.Y., Chan C.S., Chan K.H., Ng J.S., Zheng B.J., Ng W.L., Lai R.W., Guan Y., Yuen K.Y., Group H.U.S.S. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riley S., Fraser C., Donnelly C.A., Ghani A.C., Abu-Raddad L.J., Hedley A.J., Leung G.M., Ho L.M., Lam T.H., Thach T.Q., Chau P., Chan K.P., Lo S.V., Leung P.Y., Tsang T., Ho W., Lee K.H., Lau E.M., Ferguson N.M., Anderson R.M. Transmission dynamics of the etiological agent of SARS in Hong Kong: impact of public health interventions. Science. 2003;300:1961–1966. doi: 10.1126/science.1086478. [DOI] [PubMed] [Google Scholar]
- 6.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H.L., Peiris M., Wu J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X., Mo X., Chen Y., Liao B., Chen W., Hu F., Zhang Q., Zhong M., Wu Y., Zhao L., Zhang F., Cowling B.J., Li F., Leung G.M. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 8.Cereda D.T.M., Rovida F., Demicheli V., Ajelli M., Poletti P., Trentini F., Guzzetta G., Marziano V., Barone A., Magoni M., Deandrea S., Diurno G., Lombardo M., Faccini M., Pan A., Bruno R., Pariani E., Grasselli G., Piatti A., Gramegna M., Baldanti F., Melegaro A., Merler S. The early phase of the COVID-19 outbreak in Lombardy, Italy. arXiv. 2020 2003.09320. [Google Scholar]
- 9.Liu Y., Yan L.M., Wan L., Xiang T.X., Le A., Liu J.M., Peiris M., Poon L.L.M., Zhang W. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis. 2020;20:656–657. doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osman I., Cotzia P., Moran U., Donnelly D., Arguelles-Grande C., Mendoza S., Moreira A. The urgency of utilizing COVID-19 biospecimens for research in the heart of the global pandemic. J Transl Med. 2020;18:219. doi: 10.1186/s12967-020-02388-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.R Core Team: R . R Foundation for Statistical Computing; Vienna, Austria: 2013. a language and environment for statistical computing. [Google Scholar]
- 12.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brunink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 14.Chen J., Qi T., Liu L., Ling Y., Qian Z., Li T., Li F., Xu Q., Zhang Y., Xu S., Song Z., Zeng Y., Shen Y., Shi Y., Zhu T., Lu H. Clinical progression of patients with COVID-19 in Shanghai, China. J Infect. 2020;80:e1–e6. doi: 10.1016/j.jinf.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye Q., Wang B., Mao J. The pathogenesis and treatment of the “cytokine storm” in COVID-19. J Infect. 2020;80:607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felsenstein S., Herbert J.A., McNamara P.S., Hedrich C.M. COVID-19: immunology and treatment options. Clin Immunol. 2020;215:108448. doi: 10.1016/j.clim.2020.108448. [DOI] [PMC free article] [PubMed] [Google Scholar]