Graphical abstract
Keywords: SARS-CoV-2, Hydroxychloroquine, Convalescent plasma, Anakinra, Corticosteroids, Vaccine
Abstract
Despite intense research there is currently no effective vaccine available against the new severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) emerged in the later 2019 and responsible for the COVID-19 pandemic. This infectious and communicable disease has become one of the major public health challenges in the world. The clinical management of COVID-19 has been limited to infection prevention and control measures associated with supportive care such as supplemental oxygen and mechanical ventilation. Meanwhile efforts to find an effective treatment to inhibit virus replication, mitigate the symptoms, increase survival and decrease mortality rate are ongoing. Several classes of drugs, many of them already in use for other diseases, are being evaluated based on the body of clinical knowledge obtained from infected patients regarding to the natural history and evolution of the infection. Herein we will provide an updated overview of the natural history and current knowledge on drugs and therapeutic agents being tested for the prevention and treatment of COVID-19. These include different classes of drugs such as antiviral agents (chloroquine, ivermectin, nitazoxanide, hydroxychloroquine, lopinavir, remdesivir, tocilizumab), supporting agents (Vitamin C, Vitamin D, azithromycin, corticosteroids) and promising investigational vaccines. Considering the controversies and excessive number of compounds being tested and reported in the literature we hope that this review can provide useful and updated consolidated information on potential drugs used to prevent, control and treat COVID-19 patients worldwide.
1. Introduction
Coronavirus Disease 2019 (COVID-19) was declared as pandemic by the World Health Organization on March 11th, 2020 mainly due to the speed and scale of the transmission of the disease [1]. Before that, it started as an epidemic in mainland China with the focus being firstly reported in the city of Wuhan, Hubei province in February 26th [[2], [3], [4]]. The etiologic agent of COVID-19 was isolated and identified as a novel coronavirus, initially designated as 2019-nCoV [5]. Later, the virus genome was sequenced [6] and because it was genetically related to the coronavirus outbreak responsible for the SARS outbreak of 2003, the virus was named as severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) by the International Committee for Taxonomy of Viruses [7,8].
The origin and source of the SARS-CoV-2 remains unknown, although the initial cases have been associated with the Huanan South China Seafood Market where snakes, birds and other animals such as bats were sold. Considering that many of the early patients worked in or visited the market in contrast to the exported cases, it was suggested either a human to human transmission or a more widespread animal source [9]. A suspected bat origin was suggested after 96 % genome sequence identity was demonstrated between SARS-CoV-2 and another coronavirus named Bat-CoV-RaTG13 isolated from bat species which colonized a province nearly 2000 km away from Wuhan [6,10]. Pangolins were also suggested as natural host of coronaviruses [11,12]. However, evidence of human to human transmission became strongly supported on January 22nd, 2020 after a visit conducted by a WHO delegation to the city of Wuhan [13]. Since the first outbreak recognized in February 2020, the disease spread rapidly around the world. According to the European Centre for Disease Prevention and Control, as of 17th of June 2020; 8,142,129 cases of COVID-19 and 443,488 deaths have been reported worldwide since 31st December 2019. American continent was among the ones with highest number of cases (3,987,543) with United States and Brazil the leading countries (2,137,731 and 923,189 respectively).
Several SARS-CoV-2 samples have been isolated from different people and genomic sequences have been available aiming to better understand the virus and to provide information for the development of diagnostic tools and a potential vaccine. To date more than 42,000 SARS-CoV-2 RNA genomes have been uploaded in the Global Initiative on Sharing All Influenza Data, known as GISAID [14].
SARS-CoV-2 belongs to the beta subgrouping of the Coronaviridae family and are enveloped virus containing a positive-sense, single- stranded RNA with 29,891 bases of size [15,16]. The genome encodes for 29 proteins involved in the infection, replication and virion assembly process. Like other coronaviruses they are characterized by the presence of crown-like spikes on their surface [17]. The spike S protein from SARS-CoV-2 contains a receptor binding domain (RBD) that binds the human angiotensin-converting enzyme 2 (ACE2) and thereby, promotes membrane fusion and uptake of the virus into human cells by endocytosis [18,19]. The RBD present in the spike protein is the most variable region of the coronavirus genome [6,20]. Structural and biochemical studies have suggested that RBD from SARS-CoV-2 binds with high affinity to ACE2 compared to other SARS-CoV viruses [[21], [22], [23]]. However, the human ACE2 protein variability may also be a factor for the high binding affinity [21].
2. SARS-CoV-2 infection, replication and clinical implications
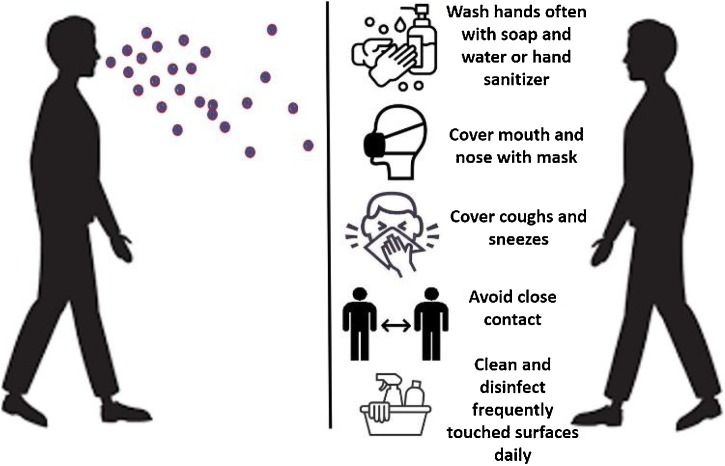
SARS-CoV-2 can be transmitted human to human by respiratory droplets, close contact with diseased patients, and possibly by fecal-oral and aerosol contact [[24], [25], [26]]. It was recently shown that airborne transmission is highly virulent and represents the dominant route to spread the disease [27]. This finding was obtained based on the analysis of the trend and mitigation measures in three different cities considered epicenters of COVID-19: Wuhan, China, Italy, and New York City, in the period from January 23 to May 9, 2020. Importantly, this result reveals that among the adopted mitigation measures such as social distancing and wearing of masks, the difference with and without mandated face covering represents the determinant in shaping the trends of the pandemic and spread of the disease. Majority of SARS-CoV-2 infected individuals (80 %) are asymptomatic or present mild symptoms most likely due to a good immune response able to control the advance of the disease [28,29]. There is evidence that these asymptomatic people can infect others with SARS-CoV-2 [30,31]. In the other hand, symptomatic individuals may evolve to more severe symptoms and eventual death. The best way to prevent transmission and illness is to avoid being exposed to the virus. Therefore, some recommendations include wash hands often, avoid close contact, cover mouth and nose with a mask, cover coughs and sneezes, and clean and disinfect frequently touched surfaces daily [32]. In this regard, wearing of face masks in public corresponds to the most effective means to prevent interhuman transmission [27] (Fig. 1 ).
Fig. 1.
Preventive measures to avoid the spread of SARS-CoV-2. The virus spread mainly from person-to-person between people who are in close contact with one another and through respiratory droplets produced when an infected person cough, sneezes or talk. The best way to prevent is to avoid being exposed to the virus.
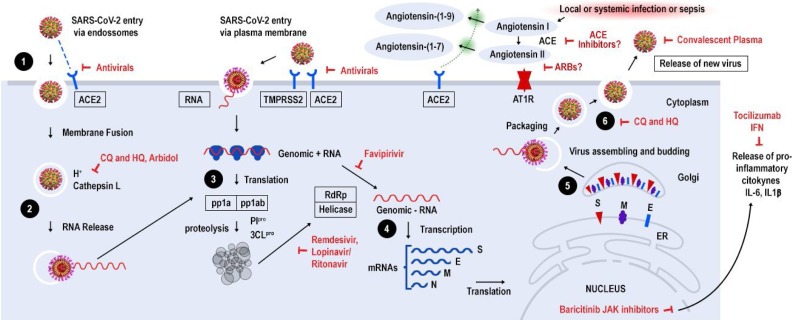
Upon cell contact, the virus can enter the cells in two ways, either via endosomes or plasma membrane fusion. In both ways spike proteins (S1 e S2) from SARS-CoV-2 mediate attachment to the cell membrane by binding to the ACE2 as the entry receptor [33]. On the other hand, virions are taken up into endosomes, spike proteins are activated by cathepsin L or alternatively by transmembrane protease serine 2 (TMPRSS2) in close proximity to ACE2 receptor, which initiates fusion of the viral membrane with the plasma membrane. The latter mechanism is less likely to trigger an antiviral immune response and is more efficient for viral replication [34].
Once inside the cell, viral RNA is released, and polyproteins are translated. Coronavirus genomic RNA encodes nonstructural proteins (NS), that play a critical role in viral RNA synthesis, and structural proteins which are important for new virion assembly. First NS proteins 1a and 1ab are translated and cleaved by the papain-like protease (PIpro) and 3C-like protease (3CLpro) to form functional NS proteins such as helicase or RNA-dependent RNA polymerase complex (RdRp). Structural proteins S1, S2, envelope (E), membrane (M) are translated by ribosomes bound to the endoplasmic reticulum (ER) and presented on its surface as a preparation of virion assembly. The nucleocapsids (N) remain in the cytoplasm and are assembled together with the genomic RNA. The virion precursor is then transported from the ER through the Golgi apparatus to the cell surface via vesicles. Finally, virions are released from the infected cell through exocytosis and a new replication cycle begins [15,35]. (Fig. 2 ).
Fig. 2.
Life cycle of SARS-CoV-2 and potential drug targets. 1) SARS-CoV-2 enters target cells via two ways, either via endosomes or plasma membrane fusion. In both ways spike proteins (S1 e S2) mediate attachment to the cell membrane by binding to the ACE2 receptor, 2) In the endosomal via, spike proteins are activated by cathepsin L or alternatively by transmembrane protease serine 2 (TMPRSS2) in close proximity to ACE2 receptor, which initiates fusion of the viral membrane with the plasma membrane, 3) viral RNA is released and part is translated to produce polyproteins pp1a and ppab, which are cleaved by proteases PIpro and 3CLpro to yield 16 non-structural proteins that form the RNA replicase-transcriptase complex, 4) This complex drives the production of negative-sense RNAs through both replication and transcription. A subset of around 9 subgenomic RNAs including those encoding all structural proteins (S-spike, M-membrane, N-nucleocapsid and E-envelope) are translated, 5) Viral nucleocapsids are assembled from genomic RNA and N protein in the cytoplasm, followed by budding into the lumen of endoplasmic reticulum (ER)- Golgi complex, 6) Virions are then released through exocytosis. Potential SARS-CoV-2 targets and drugs are shown in red. The drugs and treatment strategies investigated aim to inhibit viral entry/replication into human cells, avoid cytokine storm or decrease hyperinflammation and lung injury. ACE - Angiotensin-Converting Enzyme, ARB – Angiotensin Receptor Blocker, CQ - Chloroquine, HQ - Hydroxychloroquine, TMPRSS2–Transmembrane serine protease 2, IL-interleukin, JAK- Janus kinase.
Symptoms and signs associated with viral pneumonia such as fever, cough, sore throat, headache, fatigue, myalgia and dyspnea are frequently shown by patients during the onset of COVID-19 [[36], [37], [38], [39], [40], [41]]. Additionally, loss of taste or smell and gastrointestinal symptoms like nausea, vomiting or diarrhea has also been reported by infected patients [[42], [43], [44]]. Nevertheless, disease severity seems to be strongly associated with underlying host conditions including age, sex and overall health. The latter seems to play a critical role in susceptibility and contribute to the risk of infection. When severe and non severe patients are compared, conditions such as hypertension, diabetes, cardiovascular and kidney diseases increase the risk of infection two to three-fold [45].
3. Current therapeutic treatment for COVID-19 related to the onset and physiopathology of the disease
Better understanding of the mode of transmission, incubation period, molecular mechanisms underlying the virus infectivity and replication, as well as the pathophysiology and genetic factors associated to host, are crucial for the development of treatment strategies for COVID-19. Almost all patients with COVID-19 have lung involvement, as demonstrated by chest radiography, whereas severe complications are only observed in a small group of patients. Although observational studies reported older age and the presence of comorbidities as risk factors for increased disease severity in patients with COVID-19, it rapidly became clear that severe disease can also occur in younger patients with no pre-existing medical conditions [46]. Severe COVID-19 is strongly associated with hyperinflammation as evidenced by higher levels of C-reactive protein, ferritin and D-dimers in blood as well as increased neutrophil-to-lymphocyte ratio and serum levels of several inflammatory cytokines and chemokines [39,[47], [48], [49]].
Among hospitalized patients with COVID-19 complications such as pneumonia, sepsis, respiratory failure, and acute respiratory distress syndrome (ARD) are frequently found [50]. SARS-CoV-2 -induced ARDs exhibit similarities to that observed in other viruses and bacteria infections [51,52]. Overproduction of pro-inflammatory cytokines in response to SARS-CoV-2, known as cytokine storm, leads to increased risk of vascular permeability, organ failure and consequently death if uncontrolled [53,54]. It has been demonstrated that genes encoding for interleukins such as IL-1α, IL-1β, IL-6, IL-10, chemokines (CCl2, CCl3, CCL5, CCL10), and interferon (IFN-α2, IFN-β1, IFN-2) are highly expressed in patients, after 24 h post infection with SARS-CoV-2, and this is associated with increased infiltration of T cells, NK cells and monocytes [55,56]. This observation is similar to what was reported for other coronaviruses infections such as Middle East respiratory syndrome (MERS) caused by MERS-CoV, where interleukins (IL-6, IL-23α, IL-10, IL-7, IL-1α, IL1β) and interferon (IFN-α2, IFN2, IFN-γ) have increased dramatically in a period of 24 h post infection [57]. Furthermore, subsequent follow up after 24 h of 463 severely infected COVID-19 patients showed decreased number of total lymphocytes, CD3+, CD4+, and CD8 + T lymphocytes, which could be the factor resulting in lethal pneumonia [55]. Increased concentrations of IL-15, IL-17 and TNF-α also has been reported for MERS-CoV infections [58].
Interleukin 17A (IL-17A) is a member of a multifunctional cytokine family that has shown to be a key driver in chronic tissue inflammation, specially in joint and skin such as psoriasis, psoriatic arthritis and ankylosing spondylitis [59,60]. The role of IL-17A can be protective, in defense from both extracellular bacteria and viruses that infect airway mucous membranes or can lead to hyper-inflammation. Furthermore, its role seems to be dependent on which tissue it is expressed (gut, lung or skin) [61]. IL-17A is mainly produced by Th17 cells, but also by innate and other adaptive immune cell components such as natural killer T cells, macrophages, neutrophils, CD8 + T cells, γδ T cells and innate lymphoid cells [62]. IL-17A is known to stimulate the production of IL-8, monocyte chemoattractant protein-1 (MCP-1) and growth-regulated oncogene-α (Gro-α), which increase the recruitment of neutrophils and monocytes; it also stimulates the production of IL-6 in response to extracellular microorganisms; and also, granulocyte-colony stimulating factor (G-CSF) and granulocyte-macrophage (GM)-CSF, which in turn, stimulate the expansion of myeloid lineages and the production of other mediators such as IL-1, TNF-α and Prostaglandin E2 (PGE2) [63]. It has been recently shown that peripheral mononuclear blood cells from patients with severe COVID-19 infection presented strikingly high numbers of circulating Th17 cells, parallel with increased levels of cytokines including IL-1β, IL-2, IL-7, IL-10, IL-17, G-CSF, interferon γ-induced protein 10 (IP-10), MCP-1, macrophage inflammatory proteins (MIPs) and TNF-α. Due to the role of IL-17A in tissue inflammation and its putative protective function, IL-17A has been considered as new therapeutic target for the treatment and/or management of COVID-19 [[64], [65], [66]]. Furthermore, to face this typical cytokine storm, it was suggested that the drug fedratinib, a Janus kinase 2 (JAK2) small molecule inhibitor could be a potential therapeutic agent used for COVID-19 patients with this increased Th17 profile [67]. Therefore, one of the treatment strategies for COVID-19 includes anticytokine therapies or immunomodulators to target overactive cytokine response [68].
Besides IFN type 1 and IFN type 2, a third type of interferon family, termed lambda (IFN-λ), was identified. In fact, this family consists of four members in humans: IFN-λ1/IL-29, IFN-λ2/IL-28A, IFN-λ3/IL-28B, and IFN-λ4. They share low homology with type I IFNs and IL-10 and exhibit potent antiviral activity [69]. IFN-λ act by binding to a heterodimeric IFN-λ receptor (IFNLR) complex, activating a STAT phosphorylation-dependent signaling cascade and thereby inducing several genes that modulate immunity through a complex forward and feedback loops [70]. It has been shown that IFN-λ are induced at lower viral burden in influenza virus infections and before type I IFNs. This is considered a mechanism to limit the initial infection by inducing viral resistance to cells and helping them deal with the virus load [71]. Also, IFN-λ seems to lack the strong pro-inflammatory effects of type I IFNs and are rather tissue-protective and anti-inflammatory and therefore has been proposed as a potential strategy for the treatment of COVID-19 patients to help with two main clinical problems: persistent virus presence in the lung and induction of a “cytokine storm” [72].
Cytokine storm is also, the result of activation of coagulation pathways during the immune response to infection, which leads to an unbalance between -pro and -anticoagulant factors and results in micro thrombosis, disseminated intra vascular coagulation, and multiorgan failure evident in COVID-19 pneumonia [68]. Thus, prophylactic dose heparin has been recommended for hospitalized patients [73].
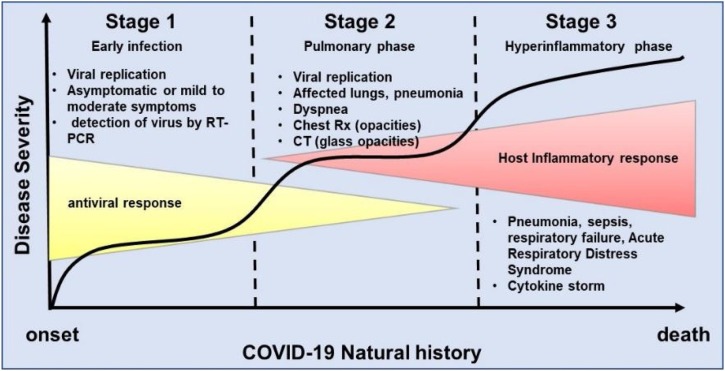
Considering the current knowledge acquired with research data from other coronavirus infections such as Middle East Respiratory Syndrome (MERS) and Acute Respiratory Syndrome (SARS), associated with the clinical features observed in patients infected with SARS-CoV-2, it is possible to identify basically three stages or phases in the natural history of COVID-19, regarding disease severity. The first phase is related to the onset of the disease and generally characterized by the development of influenza-like symptoms from mild to moderate [36,74]. In this phase virus can be detected by molecular analysis via reverse transcriptase-polymerase chain reaction (RT-PCR). Most patients in this initial phase may be asymptomatic and even transmit the disease to other people, however, depending on yet unknown factors they may progress to a second stage known as pulmonary phase. In this phase, it is possible to detect pneumonia-like symptoms evidenced as lung opacities as seen in chest radiography or as glass opacities in computed tomography (CT) [75,76]. COVID-19 pneumonia presents particularly distinctive features such as severe hypoxemia often associated with near normal respiratory system compliance with variable degrees of severity [77]. Depending on the severity of phase 2 patients can improve or worsen with the necessity of intubation and ventilation. These patients are typical examples of the phase 3 which is characterized by hyperinflammation and sepsis of lungs and patient often requires intensive care unit (ICU) and most of them unfortunately can not overcome the infection (Fig. 3 ). These preliminary observations based on medical experiences since the outbreak of COVID-19 have been driving the search of novel or repurposed drugs to treat this disease.
Fig. 3.
Schematic representation of the natural history of COVID-19 from the onset to recovery or death. There are basically three stages or phases in the natural history of COVID-19, regarding disease severity. The first phase is related to the onset of the disease and is generally characterized by the development of influenza-like symptoms from mild to moderate. Some individuals recover and some progress to the second phase. In this phase, it is possible to detect pneumonia-like symptoms evidenced as lung opacities as seen in chest radiography or as glass opacities in computed tomography (CT). Depending on the severity of phase 2 patients can improve or worsen with the necessity of intubation and ventilation. These patients are typical examples of the phase 3 which is characterized by hyperinflammation and sepsis of lungs and patient often requires intensive care unit (ICU) and most of them unfortunately can not overcome the infection and eventually die.
Proteins involved in the SARS-CoV-2 entry and replication mechanism into host cell have been the main targets for drug testing and development. As mentioned before coronaviruses are composed by nonstructural proteins (NS) and structural proteins (S). In fact, the SARS-CoV-2 RNA genome consists of eleven open reading frames (ORFs), disposed in the following order: ORF1ab, ORF2 (Spike protein), ORF3a, ORF4 (Envelope protein), ORF5 (Membrane protein), ORF6, ORF7a, ORF7b, ORF8, ORF9 (Nucleocapsid protein), and ORF10 in the 5' to 3' direction. The ORF1a/b codes for a polyprotein (PP1a and PP1ab), which comprises 16 nonstructural proteins (NSPs) [81]. NSP1, known as Leader protein, binds to 40S ribosome from host cell to inactivate host mRNA translation by degradation, while keeps viral RNA intact [79,82]. NSP2, a conserved protein in SARS-CoV-1, was shown to bind two host proteins: prohibitin 1 and prohibitin 2 (PHB1 and PHB2) involved in cell cycle progression, cell migration, cellular differentiation, apoptosis, and mitochondrial biogenesis [80,83]. NSP3 is a large papain-like proteinase with approximately 200 kDa in size, whose sequence contains several conserved domains including ssRNA binding, ADPr binding, G-quadruplex binding, protease (papain-like protease), and NSP4 binding domains besides a transmembrane domain [81]. The papain like protease domain is responsible for the release of NSP1, NSP2, and NSP3 from the N-terminal region of polyproteins 1a and 1ab from coronaviruses and therefore is considered an important target for antiviral agents [82]. NSP4 is a protein that interacts with NSP3 which is essential for viral replication. It contains a transmembrane domain and possibly interacts with host proteins with a function of membrane rearrangement in SARS-CoV-1 [83]. NSP5 is a 3C-like proteinase (3CLpro) with homology to the Middle East Respiratory Syndrome (MERS) coronavirus protease. This protease is able to cleave at eleven different sites to yield mature and intermediate nonstructural proteins [84]. NSP6 is thought to be involved in the generation of autophagosomes from the endoplasmic reticulum based on studies with avian coronavirus NSP6, which facilitate assembly of replicase proteins and avoid degradation of viral components [85]. NSP7 form a complex with NSP8 and NSP12 proteins to produce an active RNA polymerase [86]. Based on studies with porcine reproductive and respiratory syndrome virus (PRRSV), NSP9 has been shown to interact with the DEAD-box RNA helicase 5 (DDX5) cellular protein, an important association for viral replication [87]. NSP10 interacts with NSP14 resulting in the stimulation of the activity of this latter protein, which function as an S-adenosylmethionine (SAM)-dependent (guanine-N7) methyl transferase (N7-MTase) [88]. NSP10 also interacts with NSP16, a 2′-O-methyltransferase, whose activity is stimulated as result of this interaction [89]. NSP11 is a small protein with 13 amino acids that has an unknown function. Its first nine amino acids are identical to the first nine amino acids of NSP12 protein. This latter protein is an RNA-dependent RNA polymerase (RdRp) that makes copies of the viral RNA. NSP12 forms a complex with NSP7-NSP8 essential for its activity [90]. NSP13 works as a helicase that seems to interact with NSP12 and have 5′-triphosphatase activity as well. This activity is important to introduce 5′-terminal cap of the viral mRNA during its processing [91] together with NSP14, which has 3′-5′ exoribonuclease activity and N7-methyltransferase activity [92]. NSP15 has been characterized as an endoribonuclease that cleaves RNA at specific regions [93]. NSP15 proteins prevent the host immune sensing system from detecting the virus by degrading viral polyuridine sequences [94]. NSP16 is a 2′‑O‑Ribose‑Methyltransferase that methylates the 2′-hydroxy group of adenines during viral RNA processing by using S-adenosylmethionine as the methyl source [95].
The spike glycoprotein S is a main target of strategies using neutralizing antibodies since SARS-CoV-2 uses this protein to bind to its receptor to mediate membrane fusion and virus entry. Protein S has a trimeric structure with each monomer consisting of two subunits, named S1 and S2, that together account for a molecular weight of approximately 180 kDa [96]. It was shown that SARS-CoV-2 S protein is less stable than SARS-CoV-1, another coronavirus responsible for SARS, and antibodies anti-SARS-CoV-1 S1 protein is able to inhibit SARS-CoV-1 entry but not SARS-CoV-2. Also, sera from recovered SARS and COVID-19 patients showed limited cross- neutralization suggesting that a possible recovery from one infection may not protect against other [96]. Interestingly, the S protein of SARS-COV-2 was shown to have a furin cleavage site which is lacking in the S protein of SARS-COV-1 [22]. This could be one of the explanations for the difference in pathogenicity of these two viruses [78]. Besides spike (S) protein, nucleocapsid (N), envelope (E) and membrane (M) proteins, as well as 3CL protease (3CLpro), papain like protease and RNA-dependent RNA polymerase complex (RdRp) proteins which include the helicase protein have been suggested to be antiviral targets [97].
Recently Gordon et al. [98] reported an interesting approach to try to find new druggable targets for treatment of COVID-19. The authors cloned, tagged, and expressed 26 of the 29 SARS-Cov-2 proteins in human cells and by using affinity-purification mass spectrometry identified the human proteins physically associated with each one. Sixty six out of 332 human proteins identified were shown to be targeted by 69 compounds, some of them FDA-approved or in clinical and preclinical trials. A similar approach based on virtual screening was used to identify potential drugs that bind specifically to SARS-CoV-2 3-C like protease (3CLpro), essential for virus replication. Three-dimensional model of the protease using crystal structure of the high similar protease ortholog SARS-CoV was prepared and revealed 16 candidates for evaluation including two repurposed drugs such as velpatasvir and ledispavir [99].
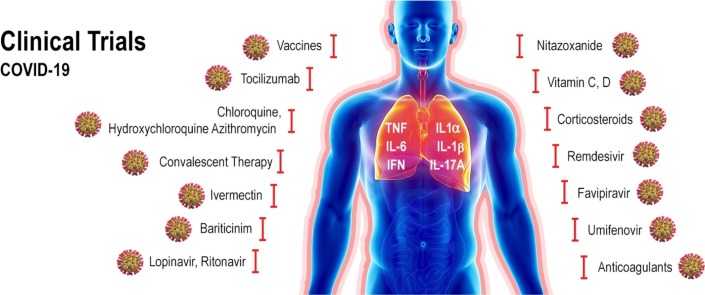
One of the approaches used to find a potential effective drug against SARS-CoV-2 has been the testing of repurposed drugs. Among these drugs are antiviral agents, anti-inflammatory agents and other pharmacological drugs aiming to modulate the immune response or to inhibit the overproduction of cytokines. Another alternative has been the use of strategies adopted in other virus epidemic such as convalescent plasma therapy. The structure and target of some of the repurposed drugs evaluated for treatment of COVID-19 is shown in Table 1 . Updated results on current knowledge regarding the use of these drugs and other therapeutic approaches are presented and discussed below.
Table 1.
Chemical structure of repurposed drugs discussed in the present review that are being tested as potential use for treatment of COVID-19.
| Drug | Structure | Action | Reference |
|---|---|---|---|
| Anakinra | 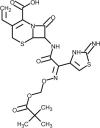 |
Recombinant IL-1 receptor antagonist, block cytokine storm. | [184] |
| Azithromycin | 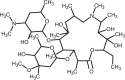 |
Antiviral, anti-inflammatory. | [241,248] |
| Baricitinib | 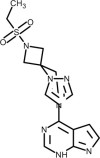 |
Anti-inflammatory Janus Kinase (JAK) inhibitor, block cytokine storm. | [187] |
| Chloroquine | 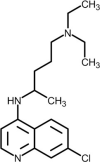 |
Increases endosomal pH and alters glycosylation of ACE-2, interfering with virus/receptor interaction. | [249] |
| Dexamethasone | 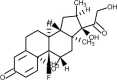 |
Steroid, anti-inflammatory, suppression of cytokine storm | [170] |
| Favipiravir | 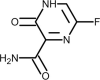 |
Antiviral, RNA-dependent RNA polymerase inhibitor. | [156] |
| Heparin | 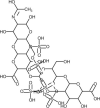 |
It can reverse the hypercoagulation in severe cases of COVID-19. | [173] |
| Hydroxychloroquine | 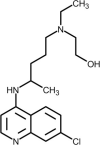 |
Increases endosomal pH and alters glycosylation of ACE-2, interfering with virus/receptor interaction. | [241] |
| Interferons (IFN) | – | Inhibits viral RNA transcription, protein translation and post translational modification. | [191] [198], |
| Ivermectin | 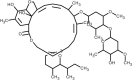 |
Antiviral, inhibits viral replication and assembly of new virions. | [136] [137], |
| Lopinavir | 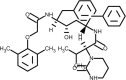 |
Inhibits 3CL protease activity, Blockage of protein processing. | [144] |
| Losartan | 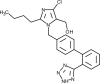 |
Angiotensin receptor blocker, interferes with renin-angiotensin system and blood vessels constriction/vasodilation. | [210] |
| Methylprednisolone | 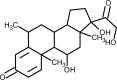 |
Anti-inflammatory, suppress cytokine- related lung injury, block cytokine storm | [168] |
| Nitazoxanide | 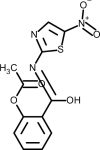 |
Antiviral, interferes with 3CL protease activity | [147] |
| Remdesivir | 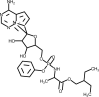 |
Inhibits RdRp polymerase inhibiting RNA synthesis. | [120,152] |
| Ritonavir | 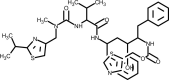 |
Inhibits 3CL protease activity, blockage of protein processing. | [144] |
| Tocilizumab | – | Interleukin-6 inhibitor, Humanized mAb targeting IL-6, Immunosuppressive, blockage of cytokine storm. | [189,190] |
| Umifenovir (Arbidol) | 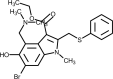 |
Interacts with spike glycoprotein and impedes its trimerization, which is key for host cell adhesion and hijacking. | [250] |
| Vitamin C | 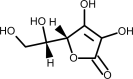 |
Boosts immunity by stimulating IFN production, stimulating lymphocyte proliferation and enhancing neutrophil phagocytic capability. | [231] |
| Vitamin D | 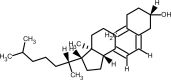 |
immunomodulatory property. | [234] |
Structures from National Center for Biotechnology Information. PubChem Database. https://pubchem.ncbi.nlm.nih.gov (accessed on June 16, 2020).
3.1. Azithromycin
The macrocyclic lactone antibiotic azithromycin has been shown to have in vitro activity against Zika and Ebola viruses [100,101]. Azithromycin has also shown to exhibit anti-inflammatory activity [102,103]. Additionally, it has shown to induce interferon type I (IFN-α, IFN-β) and type III (IFN-λ) in cells from chronic obstructive pulmonary disease patients and decrease rhinovirus-16 viral load in bronchial epithelial cells [104]. Most of the cytokines induced by azithromycin is related to viral infection response and in turn induce resistance to viral replication in target cells [105,106]. The in vitro concentration EC50 (50 % effective concentration) for azithromycin against SARS-CoV-2 was determined as 2.12 μM following 72 h incubation period post infection [107]. Several studies in COVID-19 patients have been showing limitations considering that they focused mainly on viral load as end point and detailed clinical outcomes are generally lacking. Nevertheless, collectively they present preliminary evidence that inclusion of azithromycin in various treatment regimens can influence the course of viral infection and potentially influence clinical outcomes [108,109]. Yet there are still controversies regarding efficacy of azithromycin in combination with hydroxychloroquine in COVID-19. While a French study have shown 100 % reduction of viral load in six patients treated with azithromycin and hydroxychloroquine in contrast with 57.1 % of patients treated with only hydroxychloroquine another study showed no efficacy at all in a case series with eleven patients treated with the same combination [109,110]. Thus, confirmation and validation are still needed before a conclusion on the efficacy of azithromycin can be reached. The contradictory results reported so far is one of the reasons suggesting that additional randomized controlled trials is essential to confirm these preliminary data and help to understand the benefits and the role of azithromycin in treatment combinations used to treat COVID-19 infection. Clinical trials are currently ongoing to fill up these gaps.
3.2. Chloroquine and hydroxychloroquine
Chloroquine is a drug that has been used worldwide as anti-malarial as well as for the treatment of immune disorders such as rheumatoid arthritis and Lupus [111,112]. The first indication of a potential effect of chloroquine on SARS-CoV-2 infection came from a report during China outbreak. In this study, results from more than 100 patients demonstrated that chloroquine inhibited the exacerbation of pneumonia, improved lung imaging findings, promoted a virus negative conversion and shortening of the disease course [113]. Before the COVID-19 outbreak, previous studies with chloroquine had shown its ability to inhibit in vitro the viral replication of another coronavirus (SARS-CoV), responsible for the Severe Acute Respiratory syndrome [114,115].
Hydroxychloroquine is a less toxic derivative of chloroquine that has shown lower half maximal inhibitory concentration (IC50) compared to chloroquine in inhibiting SARS-CoV-2 in vitro [116]. Additional studies confirmed this finding [[117], [118], [119], [120]]. A small study open-label non-randomized clinical trial with 36 confirmed COVID-19 patients was conducted in France. The end point was the presence or absence of virus after six days from the inclusion in the protocol. The results showed a significant association between hydroxychloroquine treatment and viral load reduction/disappearance and this effect was reinforced by azithromycin [109]. Important to note that in this study, clinical severity of the patients ranged from asymptomatic to pneumonia, i.e, none of them was critically ill. Also, the results of this study was questioned by the International Society of Antimicrobial Chemotherapy (ISAC), which declared that due to the lack of clear explanation of the inclusion criteria and the triage of patients to ensure patient safety, it did not meet the expected standard of that Organization. Nonetheless, an analysis of the data reported in Gautret work, using a pharmacokinetic model claim to confirm that co-treatment of COVID-19 with hydroxychloroquine and azithromycin increases the probability of negative-PCR in patients. The analysis also showed that clinical status affects the treatment outcome. Since the analysis was based on limited published data the results need to be interpreted with caution [121].
Basically, three mechanisms have been proposed for the antiviral activity of hydroxychloroquine/chloroquine. First the drugs interfere with the terminal glycosylation of the cellular receptor ACE2, thus preventing virus-receptor binding; Secondly, the drugs increase the pH of acidic cellular organelles, hampering endocytosis at intermediate stages with negative effects on virion transport and potentially altering post-translational modification of newly synthesized viral proteins; and third the drugs may disturb the process of virion assembly and viral protein synthesis [119,122].
Despite the lack of strong and reliable evidence of efficacy, due to the pressure that COVID-19 has imposed worldwide, many health authorities have implemented official guidelines on the use hydroxychloroquine and chloroquine for the treatment of patients with COVID-19 [[123], [124], [125]]. Regarding the side effects of hydroxychloroquine and chloroquine, both have been in clinical use for several years, thus their safety profile is well established [126]. Gastrointestinal upset has been reported with hydroxychloroquine and retinal toxicity has been described with long term use of both drugs [[127], [128], [129]]. Retinal toxicity may also be related to over-dosage [130]. Additionally, cardiomyopathy and heart arrhythmia have been reported [131,132]. To address the effect of chloroquine, hydroxychloroquine in combination or not with azithromycin on the QT interval (a measure of delayed ventricular repolarization), a prospective, observational study was performed in hospitalized patients with SARS-CoV-2. The primary outcome evaluated in this study was QT prolongation resulting in Torsade de pointes (TdP) and cardiac death. The results showed that for the two hundred one patients treated for COVID-19 with chloroquine/hydroxychloroquine, baseline QT intervals did not differ between patients treated with chloroquine/hydroxychloroquine and patients treated with these drugs and azithromycin. However, during treatment the maximum QT was significantly longer in the combination group compared to the monotherapy group and seven patients required discontinuation of the medications, but no arrhythmogenic deaths were reported. The authors concluded that although clinicians seldomly needed to discontinue therapy further studies are needed before final recommendations can be made [133]. A multinational registry evaluation of the use of chloroquine or hydroxychloroquine with or without a macrolide for the treatment of COVID-19, using data from 671 hospitals in six continents was recently published [134]. In this study was included patients hospitalized between Dec 20, 2019, and April 14, 2020, with a positive laboratory finding for SARS-CoV-2. Patients were included in one of four treatments (chloroquine alone, chloroquine with a macrolide, hydroxychloroquine alone, or hydroxychloroquine with a macrolide), and patients who received none of these treatments formed the control group. From a total of 96,032 patients analyzed, 14,888 patients were in the treatment groups (1,868 received chloroquine, 3,783 received chloroquine with a macrolide, 3,016 received hydroxychloroquine, and 6,221 received hydroxychloroquine with a macrolide) and 81,144 patients were in the control group. The study concluded that, after controlling for multiple confounding factors (age, sex, race or ethnicity, body-mass index, underlying cardiovascular disease and its risk factors, diabetes, underlying lung disease, smoking, immunosuppressed condition, and baseline disease severity), it was not possible to confirm a benefit of hydroxychloroquine or chloroquine, when used alone or with a macrolide, on in-hospital outcomes for COVID-19. Each of these drug regimens was associated with decreased in-hospital survival and an increased frequency of ventricular arrhythmias when used for treatment of COVID-19 [134]. Of note, this study was performed with hospitalized patients, most of them with associated comorbidities. Regrettably, this study was recently withdrawn under suspect of flawed data and ethical concerns about the methodology used in the analysis. Before this announcement, clinical trials designed to address the safety and efficacy of chloroquine and hydroxychloroquine alone or in combination with azithromycin were suspended temporarily by the World Health Organization authorities based on the published results. However, they stepped back and authorized the continuation of the ongoing clinical trials. Therefore, a great debate persists whether individuals with confirmed COVID-19 with mild symptoms or asymptomatic could benefit from treatment with these drugs.
3.3. Ivermectin
Ivermectin is best known as an antiparasitic agent sold with the commercial name STROMECTOL® but it has also demonstrated antiviral activities toward human immunodeficiency virus (HIV) and dengue virus [135]. Ivermectin targets the host nuclear transport importin α/β1 heterodimer which the virus relies for the replication and assembly of new virions [136]. This drug was shown to inhibit SARS-CoV-2 in vitro replication for up to 48 h at a concentration of 5 μM. The 50 % inhibition concentration (IC50) was determined as 2 μM which was much higher than the maximum plasma concentration [137]. A study was performed aiming to predict total (bound and unbound) and unbound plasma concentration-time profiles after the administration of FDA approved dose (200 mg/kg), 60 mg/kg and 120 mg/kg. The results of this study showed that plasma concentrations did not reach the IC50 even at doses higher than the approved. Furthermore, the authors concluded that after oral administration with the approved dose it is unlikely that ivermectin reaches the IC50 in the lungs and therefore the likelihood of a successful clinical trial would be low. Despite these results, combination therapy with other agents may be beneficial and it has been suggested [138]. Moreover, studies with animal models have shown up to 3-fold higher levels in pulmonary tissue than in plasma one week after oral dosing demonstrating the need for further research to better evaluate the effectivity of ivermectin for the treatment of respiratory viruses such as SARS-CoV-2 [139].
3.4. Lopinavir-Ritonavir
Lopinavir is an aspartate protease inhibitor that has demonstrated high specificity against human immunodeficiency virus (HIV) type 1. Lopinavir was developed by the pharmaceutical company Abbot and is sold by the brand name Kaletra®. This drug is generally administered in combination with ritonavir due to its poor oral bioavailability, rapid biotransformation and to increase its half-life through the inhibition of cytochrome P450 [140]. In the HIV-1 infection context, the lopinavir hydroxy ethylene bond mimics the normal peptide cleaved by the virus HIV-1 protease [141]. During the severe acute respiratory syndrome (SARS) epidemic in 2003, lopinavir was tested in vitro and showed inhibitory activity against SARS-CoV [140,142,143]. A multicenter retrospective study matched cohort study was performed by Chan et al. [144] to investigate possible benefits and adverse effects of the addition of lopinavir/ritonavir to a standard treatment protocol for the treatment of SARS. They showed that as an initial treatment, the protocol used was associated with a significant reduction in the overall death rate and intubation rate, when compared with a matched cohort who received standard treatment. Another study reported in 2004 suggested that the addition of lopinavir/ritonavir compared to a control group who received the antiviral ribavirin, reduced the risk of adverse clinical outcomes and viral load among patients with SARS [140]. One study reported a case of a single patient in the initial phase of SARS-CoV-2 outbreak in Korea who was identified as an index patient that caused secondary and tertiary transmission. The administration of lopinavir/ritonavir to this patient significantly decreased the viral load and no or little coronavirus titers were observed during follow-up [145].
Nevertheless, in another study reporting a randomized, controlled, open-label trial involving 199 hospitalized adult patients with confirmed SARS-Cov-2 infection, the use of lopinavir/ritonavir showed no benefit beyond standard care. This study had the time to clinical improvement as the primary end point [146].
3.5. Nitazoxanide
Nitazoxanide belongs to the class of drugs known as thiazolides. It has a broad spectrum antiparasitic and it has also shown to have broad spectrum antiviral activity. Nitazoxanide has been previously shown to exhibit in vitro activity against MERS-CoV and other coronaviruses [147]. Haffizulla et al. showed that when nitazoxanide was given in a regimen of 600 mg twice daily for 5 days, it reduced the duration of symptoms in patients with acute uncomplicated influenza with minor adverse effects [148]. Several recommendations have been made suggesting the potential use of nitazoxanide to treat SARS-CoV-2 infection, but they have been often ignored [149]. A recent review evaluated nine nitazoxanide research clinical trials to assess the safety, cost and potential use of this drug for COVID-19 [150]. The authors concluded that this drug demonstrate good safety profile at approved doses, although further evidence is required regarding hepatorenal and cardiovascular effects, as well as teratogenicity. If efficacy of nitazoxanide is clinically proven against COVID-19 it may represent an affordable and safe treatment. To date, the 15th of June 2020, twelve nitazoxanide clinical trials undergoing worldwide have been registered on the ClinicalTrial.gov website, five of them recruiting volunteers (Table 2 ).
Table 2.
Status of registered ongoing clinical trials testing different drugs and potential treatments for COVID-19.
| Drug/Treatment | aNumber of registered clinical trials | Not yet recruiting | Recruiting | Active not recruiting | Terminated | Enrolling by invitation | Withdrawn | Suspended | completed | Unknown status |
|---|---|---|---|---|---|---|---|---|---|---|
| Anakinra | 18 | 8 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Anticoagulants | 41 | 19 | 17 | 0 | 0 | 2 | 1 | 0 | 2 | 0 |
| ARB (Angiotensin Receptor Blockers) | 42 | 14 | 22 | 3 | 0 | 2 | 0 | 0 | 1 | 0 |
| Azithromycin | 93 | 35 | 44 | 5 | 1 | 1 | 1 | 5 | 1 | 0 |
| Baricitinib | 15 | 5 | 9 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Chloroquine | 73 | 28 | 35 | 2 | 0 | 3 | 1 | 2 | 2 | 0 |
| Convalescent plasma | 102 | 27 | 60 | 2 | 0 | 3 | 1 | 0 | 2 | 7 |
| Dexamethasone | 12 | 2 | 9 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Favipiravir | 24 | 13 | 8 | 2 | 0 | 1 | 0 | 0 | 0 | 0 |
| Heparin | 35 | 13 | 19 | 0 | 0 | 1 | 1 | 0 | 1 | 0 |
| Hydroxychloroquine | 218 | 71 | 102 | 16 | 2 | 8 | 3 | 8 | 8 | 0 |
| Interferon alpha | 17 | 7 | 8 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| Interferon beta | 14 | 3 | 6 | 1 | 0 | 2 | 0 | 0 | 2 | 0 |
| Interleukin 17A (IL-17A | 3 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ivermectin | 23 | 10 | 11 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| Lopinavir /Ritonavir | 75 | 25 | 37 | 3 | 1 | 3 | 0 | 0 | 6 | 0 |
| Losartan | 14 | 5 | 8 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Methylprednisolone | 25 | 8 | 12 | 0 | 0 | 1 | 0 | 0 | 4 | 0 |
| Nitazoxanide | 12 | 7 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Remdesivir | 33 | 10 | 15 | 2 | 1 | 1 | 0 | 1 | 1 | 2 |
| Tocilizumab | 55 | 12 | 36 | 5 | 0 | 1 | 0 | 0 | 1 | 0 |
| Umifenovir (Arbidol) | 8 | 3 | 2 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
| Vaccine | 119 | 44 | 63 | 9 | 0 | 1 | 0 | 0 | 2 | 0 |
| Vitamin C | 25 | 12 | 11 | 0 | 0 | 1 | 0 | 1 | 0 | 0 |
| Vitamin D | 26 | 14 | 9 | 2 | 0 | 1 | 0 | 0 | 0 | 0 |
| Zinc | 15 | 10 | 3 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
Source: ClinicalTrials.gov, aacessed 13 June, 2020.
3.6. Remdesivir
Remdesivir is a phosphoramidate nucleoside analogue prodrug and a broad-spectrum antiviral agent commercialized by the pharmaceutical company Gilead [151]. It has demonstrated in vitro and in vivo activity in animal models against coronaviruses causing MERS and SARS, which are structurally similar do SARS-CoV-2 [152]. Remdesivir demonstrated to potent block SARS-CoV-2 infection at low concentrations with a half-maximal effective concentration (EC50) of 0.77 μM [120]. A report on a single case patient described clinical improvement after use of remdesivir intravenously to treat the first United States case of COVID-19 [153]. Another study reported a compassionate use of remdesivir in hospitalized patients with confirmed SARS-CoV-2 infection who had an oxygen saturation of 94 % or less, or who were receiving oxygen support [154]. The authors observed clinical improvement in 36 of 53 patients (68 %) treated with remdesivir consisting of a loading dose of 200 mg intravenously on day 1, plus 100 mg daily for the following 9 days, however the authors suggested that efficacy will require ongoing randomized, placebo controlled trials.
Notwithstanding these results, a randomized, double-blind, placebo-controlled study with 237 patients enrolled showed that use of remdesivir was not associated with statistically significant clinical benefits. Patients receiving redemsivir had a numerical reduction in time to clinical improvement compared to placebo, although theses results were not statistically significant [155]. This study had to be terminated early due to the adverse events observed in the patients.
3.7. Favipiravir
Favipiravir, commercially sold as Avigan, is a pyrazinecarboxamide derivative and guanine analogue that has shown to selectively inhibit the RNA-dependent RNA polymerase (RdRP) of RNA viruses interfering with the assembly of viable virus [156,157]. This drug has shown activity against many RNA viruses such as influenza A virus, alphavirus, filovirus, arena and norovirus as well as Ebola virus [158]. A study designed to evaluate the potential antiviral activity against SARS-CoV-2 included favipiravir in a panel of drugs tested. Favipiravir showed efficacy in vitro in infected Vero E6 cells demonstrating a half-maximal effective concentration (EC50) of 61.88 μM and half cytotoxic concentration over 400 μM [120]. In contrast, Choy et al. showed that concentrations lower than 100 μM has no effect against SARS-CoV-2 in vitro [159]. On the other hand, it has been shown that COVID-19 patients treated with favipiravir show superior recovery rate (71.43 %) than patients treated with umifenovir (55.86 %) [160]. An open label nonrandomized controlled study examined the effects of favipiravir and lopinavir/ritonavir in 80 patients with confirmed COVID-19. The study comprised two arms, with one containing 35 patients treated with favipiravir (Day 1: 1600 mg twice daily; Days 2–14: 600 mg twice daily plus 5 kU twice daily IFN-α) and the other containing 45 patients treated with lopinavir/ritonavir (Days 1–14: 400 mg/100 mg twice daily plus 5 kU twice daily IFN-α) as a control group. Favipiravir treatment was associated with faster viral clearance, significantly higher improvement rate in chest imaging and fewer adverse effects compared to the lopinavir/ritonavir arm [161]. Non-randomized and randomized controlled clinical trials aiming to investigate efficacy and safety of favipiravir alone or in combination with tocilizumab or chloroquine phosphate are currently ongoing.
3.8. Umifenovir (Arbidol)
Umifenovir is an indole carboxylic acid derivative used for treating prophylaxis and infections associated with H1N1A and B arbovirus [162]. Also known as arbidol, umifenovir acts by blocking the virus-cell membrane fusion and virus-endosome fusion, through incorporation into cell membranes and interference with the phospholipids network organization [163]. This drug showed in vitro activity against SARS-CoV-1 virus and was speculated to have activity against SARS-CoV-2 [164]. Combination of umifenovir and lopinavir/ritonavir showed increased negative conversion rate of SARS-CoV-2 and improved chest CT scans results in a retrospective cohort study [165]. However, it was shown that patients treated with umifenovir demonstrated inferior outcome in clinical recovery rate and less improvement in symptoms like fever and cough, when compared to favipiravir [160]. In contrast, another study evaluated the antiviral effects and safety of lopinavir/ritonavir and arbidol in fifty patients with laboratory-confirmed COVID-19. They were divided into two groups: Lopinavir/ritonavir group consisting of 34 cases and arbidol group consisting of 16 cases. The first group received 400 mg/100 mg of Lopinavir/ritonavir, twice a day for a week, while the second group was given 0.2 g arbidol, three times a day. RT-PCR assay was used to detect SARS-CoV-2 virus during antiviral therapy. Patients in the arbidol group had a shorter duration of positive RNA test compared to those in the lopinavir/ritonavir group, suggesting that arbidol monotherapy may be superior to lopinavir/ritonavir in treating COVID-19 [166]. Clinical trials aiming to investigate efficacy and safety of umifenovir alone or in combination are currently ongoing.
3.9. Corticosteroids
Methylprednisolone is one of the classical immunosuppressive drugs used to stop or delay the progress of pneumonia and have been proved to effective for the treatment of acute respiratory distress syndrome (ARDs). Patients with COVID-19 pneumonia and severe cases develop rapidly to acute respiratory failure [39]. Therefore, it is not a surprise that methylprednisolone became a drug of choice to use in severe cases. In a retrospective cohort study with 201 patients with COVID-19 who developed ARD, administration of 1–2 mg/kg/daily IV for 5–7days apparently reduced the risk of death, although 42 % of patients who received this drug died compared to 61,8% who did not received the drug [167]. Early, low-dose and short application of methylprednisolone was associated to improvement in clinical symptoms and a shortened disease course in another study with 46 patients with severe COVID-19 [168]. A limited study with only 15 patients and no control group corroborated the benefits of low dose corticosteroids treatment in a subset of critically ill COVID-19 pneumonia patients [169]. Another steroid that has been used in the treatment of acute respiratory syndromes before is the fluorinated steroid dexamethasone, a synthetic member of the class of glucocorticoids. At the closing of this review on June 17th, the results of a randomized clinical trial established to examine the potential benefit of treatment with dexamethasone in COVID-19 patients were announced by the university of Oxford. This study was part of the proposal called RECOVERY (Randomized Evaluation of COVid-19 thERapY) and consisted of 2104 patients randomized to receive 6 mg once per day of dexamethasone (either orally or by intravenous injection) for ten days and 4321 patients randomized to receive standard care. As mentioned before, patients with severe COVID-19 infection requires ventilation and oxygen support and unfortunately, they are among the patients with high mortality rate. Dexamethasone reduced deaths by one-third among ventilated patients and by one fifth among those receiving oxygen. This benefit was not observed in patients who did not require respiratory support. The announced yet unpublished study is encouraging and give support to the hypothesis that corticosteroids can be useful in the treatment of severe COVID-19 patients reducing mortality [170].
3.10. Anticoagulants
Some studies have been reporting that severe COVID-19 patients present coagulopathy complications [39,171,172]. Disseminated intravascular coagulation is frequently observed in most of deaths caused by SARS-CoV-2 [39]. This observation led to the active application of anticoagulants such as heparin for patients with severe COVID-19 in China notwithstanding the lack of efficacy validation [173]. Nevertheless, Tang et al. [174] performed a retrospective study where they compared the 28-day mortality between a group of severe COVID-19 patients who received heparin and a group who did not receive this anticoagulant. No statistically difference in 28-day mortality was observed between groups (30.3 % vs 29.7 %, p = 0.910). However, in a group of patients presenting sepsis-induced coagulopathy with score greater than 4, those who received heparin showed a lower 28-day mortality than those who did not receive heparin (40.0 % vs 64.2 %, P = 0.029). On the other hand, Negri et al. [175] reported in a not yet peer-reviewed study, a series of 27 consecutive COVID-19 patients admitted at a hospital in São Paulo-Brazil and treated with heparin in therapeutic doses used in clinical severity. The treatment with heparin improved the ratio of arterial oxygen partial pressure to fractional inspired oxygen (PaO2/FiO2) and the mean time of discharge of 81 % of theses patients were 11.4 days [175]. Cardiac arrhytmias are often the immediate cause of death in severe patients [176], therefore, it has been suggested that the use of heparin in COVID-19 severe patients would be useful not only due to its anticoagulant activity but also its antiarrhythmic property [[177], [178], [179]].
3.11. Anti-inflammatory therapy approaches
Beyond the virus itself, another factor considered to be important as primary cause of mortality of patients infected with SARS-CoV-2 is the pro-inflammatory response which can promote a cytokine storm and secondary bacterial infection. Therefore, drugs acting as anti-inflammatory are thought to help decrease the severity of COVID-19 patients. Interleukins 1 (IL-1α and IL-1β) are cytokines associated with innate immunity and damaging inflammation [180]. IL-1β was shown to reach peak levels at the time of disease onset in a study of ARDS assessing serial bronchoalveolar lavage fluids for cytokine content [181]. Anakinra, sold by the brand name Kineret, is a recombinant IL-1 receptor antagonist used to treat autoinflammatory disorders [182]. A retrospective cohort study conducted in Italy with patients with COVID-19, moderate-to-severe ARDs and hyperinflammation demonstrated that high-dose intravenous anakinra was safe and associated with clinical improvement in 72 % of the patients [183]. Despite the recognized limitations of this study the results are promising and a randomized phase two clinical trial (NCT04324021) is ongoing. Meanwhile, a cohort prospective study was performed in France with participants who were admitted to Groupe Hospitalier Paris Saint-Joseph with severe COVID-19-related bilateral pneumonia on chest x-ray or lung CT scan from March 24 to April 6, 2020. The anakinra group consisted of 52 consecutive patients that received subcutaneous anakinra (100 mg twice a day for 72 h, then 100 mg daily for 7 days) as well as the standard treatments at the institution at the time. The control group consisted of 44 patients that received standard treatments and supportive care. Intensive Care Unit (ICU) admission or death occurred in 13 (25 %) patients in the anakinra group compared to 32 (73 %) patients in the control group [184]. These results suggest the need of more clinical trials to confirm the efficacy of anakinra to treat patients with severe COVID-19 infection.
Baricitinib is a Janus kinase (JAK) inhibitor licensed primarily for the treatment of rheumatoid arthritis and with good efficacy and safety records [185]. Interestingly, this anti-inflammatory drug has been considered an option for treatment of COVID-19 and its antiviral effect is believed to be related to its affinity for AP2 associated protein AAK1, reducing SARS-CoV-2 endocytosis [186]. Cantini et al. [187] reported a pilot study to evaluate the safety and clinical impact in COVID-19. Baricitinib was given to 12 patients with moderate COVID-19 at a dose of 4 mg/day/orally. No adverse events were recorded after two weeks and clinical and respiratory parameters significantly improved at two weeks with no patients required admission to ICU [187]. Caveats in this study is that no proper control group was included.
Interleukin 6 (IL-6), tumor necrosis factor (TNF) and interferon gama (IFN-γ) are other targetable cytokines that play important role in the pathogenesis of lung seen in COVID-19. Excessive IL-6 signaling induces several biological pathways including Janus kinase (JNK), which contribute to organ damage [188]. Tocilizumab is a recombinant humanized monoclonal anti-IL-6 antibody that binds both soluble and membrane bound IL-6 receptor [189]. A Chinese study aiming to assess the efficacy of tocilizumab in 21 patients with COVID-19 showed preliminary results demonstrating that this drug improved clinical outcome immediately in severe and critical patients and was an effective treatment to reduce mortality. All 21 patients in the study were discharged on average 15.1d after giving tocilizumab [190].
Interferons (IFNs) are the first line of immune defense against viral infections. Its antiviral activity occurs by blocking viral replication and eliminating the virus-infected cells [191]. Several cellular proteins, located in different compartments, contain Pattern Recognition Receptors (PRRs) such as Toll Like Receptors (TLRs), RIG-I Like Receptors (RLRs) and cyclic AMP-GMP synthase (cGAS)/stimulator of IFN genes (STING), that act as sensors for specific viral components [[192], [193], [194]]. Upon virus infection, transcription factors such as Interferon Regulatory Factors (IRFs) and Nuclear Factor-κB (NF-κB) are activated by signaling pathways triggered by Ligand-PRRs interaction. These transcription factors act cooperatively to induce the synthesis of Type I interferons, such as IFN-β, which in turn can induce the expression of several genes involved in blocking viral infection, via Janus Kinase (JAK)/Signal Transducer of Transcription (STAT) signaling pathway [195]. Interferon-β1b treatment have shown to improve outcome of MERS-CoV infection in a nonhuman primate model of common marmoset [196]. IFN-α and IFN-β have been tested in vitro against SARS-CoV-2 and shown potent antiviral activity. Infected Vero cells treated with IFN-α or IFN-β at a concentration of 50 international units (IU) per milliliter reduced viral titers by 3.4 log or over 4 log, respectively, demonstrating the potential therapeutic use of these biologicals in COVID-1 infections [197]. Addition of IFN-1β in the treatment of COVID-19 patients receiving the antivirals lopinavir/ritonavir was shown to be better than the treatment with the two antivirals alone as demonstrated by a open label, randomized, phase II clinical trial where patients in the triple combination treatment group had a significantly shorter median time from start of study treatment to negative nasopharyngeal swab (7 days) compared to the lopinavir/ritonavir control group (12 days) [198].
3.12. ACE inhibitors and angiotensin receptor 1 (AT1R) blockers
As mentioned before Angiotensin-Converting Enzyme 2 (ACE2) was identified as a functional receptor in SARS-CoV infection [199]. Structural and functional analysis of SARS-CoV-2 binding receptor showed that the spike glycoprotein of this virus also binds to ACE2 receptor [[200], [201], [202]]. It has been observed that patients with comorbidities such as hypertension and diabetes are frequently under medication with angiotensin-converting enzyme (ACE) inhibitors or angiotensin receptor blockers (ARB) which could result in overexpression of ACE2 and potentially increase availability of target molecules for SARS-CoV-2 entry [45,[203], [204], [205]]. ACE inhibitors and ARBs are used to treat patients with high blood pressure, heart and kidney problems and other conditions. It is believed that disbalance in ACE2, as caused by ACE inhibitors or nonsteroidal anti-inflammatory drug (NSAID) such as ibuprofen, an activator of ACE2 receptors expression, may predispose to severe disease [206]. On the other hand, there are some reports showing no association between these drugs and severe Covid-19 [207]. However, the renin-angiotensin-aldosterone system is very complex and it has been suggested that this complexity needs to be taken into consideration when treating COVID-19 pneumonia patients with comorbidities, due to the opposing physiological actions of ACE and ACE2 receptors [208]. ACE cleaves angiotensin I to generate angiotensin II, the peptide which binds to and activates angiotensin receptor 1 (AT1R) to constrict blood vessels, thereby elevating blood pressure. In contrast, ACE2 inactivates angiotensin II while generating angiotensin 1–7, a heptapeptide having a potent vasodilator function via activation of its Mas receptor [209]. In this context, it has been proposed that AT1R blockers, such as losartan and olmesartan, could reduce the aggressiveness and mortality from SARS‐CoV‐2 virus infections. The rationale appointed for this proposal is that when SARS-CoV-2 binds to ACE2 receptor, it causes a downregulation of ACE2 activity, which in turn results in excessive production of angiotensin-II by the related enzyme ACE, while less ACE2 is capable of converting it to the vasodilator heptapeptide angiotensin 1–7, which contributes to lung injury due to increased pulmonary vascular permeability [210]. Furthermore, observational studies on the use of ACE inhibitors and ARBs during the COVID-19 pandemic and in other acute respiratory syndromes do not support the hypothesis that these drugs increase the risk of infection with SARS-CoV-2 or have a negative impact on the clinical outcomes of COVID-19 patients. Some demonstrate the opposite of the initially raised concerns [211,212]. Although in preclinical animal model, treatment with losartan has been shown to upregulate ACE2 mRNA expression and activity, human studies showed no significant difference in ACE2 plasma levels in patients treated with ARBs or ACE inhibitors, although local tissue ACE2 levels have not been measured [[213], [214], [215], [216]]. ACE inhibitors and ARBs could mitigate the deleterious effects AT1R pathway activation, which in turn decreases inflammation and lung injury [217]. However, the real impact of ACE inhibitors or angiotensin receptor blockers on severe acute respiratory illness due to SARS CoV-2 is not well established and contradictory results have been published in the literature so far. Only controlled randomized trials will be able to definitively answer the question of whether ACE inhibitors or ARBs pose a harm to patients with Covid-19 or if they could even be beneficial. Despite controversies it has been suggested the use of ACE inhibitors and ARBs should be continued in patients that are in stable condition, who are at risk for infection, are being evaluated for infection, or have Covid-19 [218].
3.13. Convalescent plasma therapy
Convalescent plasma is a type of therapy where plasma is collected from individuals, following resolution of infection and development of antibodies. Transfusion of convalescent plasma may prevent infection or decrease clinical severity in individuals with recent exposure to the virus [219]. This type of therapy has been in use for over a century [220]. Human plasma from recovered COVID-19 patients is being considered a safe and potentially effective alternative for treatment and post-exposure prophylaxis [221,222]. A study with 173 SARS-CoV-2 infected patients detected the presence of antibodies in serial plasma samples and demonstrated seroconversion rates for antibody (Ab), IgM and IgG of 93.1 %, 82.7 % and 64.7 %, respectively. The presence of antibodies was <40 % among patients within 1-week since onset, and rapidly increased to 100.0 % (Ab), 94.3 % (IgM) and 79.8 % (IgG) since day-15 after onset demonstrating a strong empirical support for the routine application of serological testing in the diagnosis and management of COVID-19 patients [223]. There are still few data on the use of convalescent plasma therapy in COVID-19, however, reports from China has shown preliminary clinical benefits regarding improvement of symptoms like fever, cough, chest pain and no serious side effect [224,225]. Again, the need of randomized clinical trials is urgent to determine the true clinical effect of this treatment or whether the patients might have recovered without this therapy. According to the ClinicalTrials.gov database there are currently 102 studies evaluating this therapy for COVID-19.
3.14. Vitamin C
Vitamin C (Ascorbic acid) is an essential vitamin known for its antioxidant properties, important to boost the immune system [226]. Some studies have shown that various high-dose intravenous vitamin C infusions (e.g., 200 mg/kg body weight/day, divided into 4 doses) shortened the intensive care unit (ICU) stay by 7.8 % [227], and was accompanied by a significant reduction in the mortality rate in the treatment of severe sepsis and septic shock [228]. Moreover, vitamin C has also demonstrated antiviral activity which was recently reviewed [229]. Coronaviruses increase oxidative stress that promotes cellular malfunction and ultimately results in organ failure [230]. It is believed that high intravenous dose of vitamin C could be particularly effective by inhibiting the production of cytokines storm due to COVID-19 and many physicians in China have identified promising results using this approach against COVID-19 [231,232]. It was reported that high-dose intravenous vitamin C was successfully used in the treatment of 50 moderate to severe COVID-19 patients in China. The doses used varied between 10 g and 20 g per day, given over a period of 8–10 h. The oxygenation index was improving in real time and all the patients eventually cured and were discharged [231].
Recently a new clinical trial to test high-dose vitamin C in patients with COVID-19 (Identifier: NCT04264533) has begun in Wuhan, China. In this trial, the investigators will treat 140 patients with a placebo control or intravenous vitamin C at a dose of 24 g/day for 7 days. They will assess requirements for mechanical ventilation and vasopressor drugs, organ failure scores, ICU length of stay and 28-day mortality [232].
3.15. Vitamin D
It is known that vitamin D can stimulate innate immunity and modulate acquired immunity [233]. However, although there are several contradictory results reported in the literature regarding the effect of vitamin D on acute respiratory infections, considering that pneumonia, sepsis, respiratory failure, and acute respiratory distress are frequently found in SARS-CoV-2 infected patients, it has been suggested that this vitamin could be beneficial in patients with COVID-19. A meta-analysis study has shown evidence that vitamin D supplementation could protected against acute respiratory infections [234]. This benefit was observed in individuals who received frequent (e.g., daily) doses of vitamin D, but not in those who received bolus doses, and this effect was more evident in individuals with vitamin D deficiency. A study performed with 449 individuals recruited from a UK BioBank with confirmed COVID-19 aimed to establish whether 25-hydroxyvitamin D (25(OH)D) concentration was associated with COVID-19 risk [235]. This study showed no potential link between vitamin D concentrations and COVID-19 infection nor relation between this vitamin and the ethnic differences in COVID-19 infection. Despite this result, there are several clinical trials undergoing to better evaluate the role of vitamin D in COVID-19 infection (Table2).
3.16. Zinc
It has been shown that zinc inhibits coronavirus and retrovirus RNA polymerase activity and zinc ionophores exhibit the potential to block virus replication in vitro [236]. Moreover, zinc has also shown to play a role as anti-inflammatory agent in pneumonia in animal models, thus limiting tissue damage and systemic effects [237,238]. Furthermore, previous work performed before the COVID-19 pandemic demonstrated that chloroquine is a zinc ionophore increasing Zn2+ flux into the cell [239]. Therefore, zinc has been considered as supportive treatment in the therapy of COVID-19 infection. Recently it was reported a series of four patient cases (one 63 years old man and three women with 57, 41 and 26 years old) with clinical symptoms and/or laboratory confirmed COVID-19 that used high dose zinc salt oral lozenges to alleviate the symptoms of the disease. The treatment resulted in significant improvement in objective and symptomatic disease measures such as fever and PaO2, after one day of this high dose therapy [240]. Currently there is no standard dosage for zinc supplementation established in the treatment of COVID-19, however several clinical trials are underway to evaluate the effects of supplementation with zinc alone or in combination with other supplements or drugs such as vitamin C, chloroquine, Hydroxychloroquine and azithromycin on COVID-19 (Table 2).
3.17. Combination therapies
Hydroxychloroquine/azithromycin combination therapy has shown promising results in COVID-19 treatment although no clear mechanism of action has been established. Nevertheless, it was shown by molecular dynamics that both drugs act in synergy to prevent contact between the virus and the plasma membrane cells [241]. Azithromycin display similarity with the sugar moiety of ganglioside GM1, a lipid that act as important host attachment cofactor. This antibiotic interacts with the ganglioside-binding domain of SARS-CoV-2 spike protein and thereby prevents virus attachment to the host cell receptor. On the other hand, hydroxychloroquine saturates sites in the vicinity of the primary coronavirus receptor ACE2 [241].
The results from an open label, randomized, phase II clinical trial designed to assess the efficacy and safety of triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin for treating patients with COVID-19 were recently published. The study comprised 127 patients, from which 86 were randomly assigned to the combination group (14-day combination of lopinavir 400 mg and ritonavir 100 mg every 12 h, ribavirin 400 mg every 12 h, and three doses of 8 million international units of interferon beta-1b on alternate days) and 41patients were assigned to the control group (14 days of lopinavir 400 mg and ritonavir 100 mg every 12 h). The primary endpoint was the time to providing a nasopharyngeal swab negative for SARS-CoV-2 RT-PCR. The combination group had a significantly shorter median time from start of study treatment to negative nasopharyngeal swab (7 days) than the control group (12 days), p = 0·0010. The authors concluded that early triple antiviral therapy was safe and superior to lopinavir–ritonavir alone in alleviating symptoms and shortening the duration of viral shedding and hospital stay in patients with mild to moderate COVID-19 [198]. Several treatment combinations protocols with the drugs mentioned earlier are being tested on ongoing clinical trials (ClinicalTrials.gov 2020). A summary of the status of these clinical trials accessed in June 13th on the ClinicalTrials.gov databank is shown in Table 2.
3.18. First investigational vaccines
A vaccine named ChAdOx1 nCoV-19 was the first vaccine to enter phase I clinical trial aiming to assess whether health people can be protected from COVID-19. This investigational vaccine, developed at the University of Oxford Jenner Institute, protected six rhesus macaques from pneumonia caused by SARS-CoV-2 [242]. It started to recruit volunteers in Oxford on April 23rd, 2020. Around 1100 people is expected to take part in the trial with half receiving the vaccine and the other half control group. The vaccine is based on an adenovirus vaccine vector and the SARS-CoV-2 spike protein and has been produced in Oxford [243].
Another candidate vaccine for COVID-19 named mRNA-1273 was developed by the pharmaceutical Company Moderna and phase 1 data was announced on May 18th, 2020. mRNA-1273 is an mRNA vaccine against SARS-CoV-2 encoding for a prefusion stabilized form of the Spike (S) protein. The vaccine has shown to generate an immune response similar to the response seen in people infected by the virus and recovered. Despite the limited data, in phase I trial, eight patients who received low and middle doses of 25 and 100 μg of the vaccine respectively, developed neutralizing antibodies against SARS-CoV-2. One of the people who received 100 μg developed redness at the injection site, which is considered criteria for grade 3 adverse event. Three volunteers subjected to the highest dose tested of 250 μg presented these adverse events after receiving the second of two doses [244].
Zhu et al. [237] reported on May 22nd the results of a clinical trial (NCT04313127) where they tested the safety, tolerability and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine. Healthy adults aged between 18 and 60 years were sequentially enrolled and allocated to one of three dose groups (5 × 10¹⁰, 1 × 10¹¹, and 1·5 × 10¹¹ viral particles) to receive an intramuscular injection of vaccine. 108 participants (51 % male, 49 % female; mean age 36·3 years) were recruited and received the low dose (n = 36), middle dose (n = 36), or high dose (n = 36) of the vaccine. ELISA antibodies and neutralizing antibodies increased significantly at day 14 and peaked 28 days post-vaccination. Specific T-cell response peaked at day 14 post-vaccination. The authors concluded that this vaccine was tolerable, immunogenic and warrants further investigation [245]. Notwithstanding preliminary, these results are encouraging and shed hope for an effective treatment in the near future. Besides these vaccines there are more than 140 clinical trials registered in the ClinicalTrials.Gov databank.
4. Concluding remarks
Despite great efforts worldwide to try to find an effective drug against SARS-CoV-2 yet there is not a consensus about a definite therapy for COVID-19. The use of repurposed medications has shown to be a good alternative with promising results while an effective vaccine is still not available. In this review we incorporated only key drugs and treatments that have been tested and demonstrated potential for use against SARS-CoV-2 virus as of June 13th, 2020. However, the results obtained so far with the use of repurposed drugs must be faced with caution. Unfortunately, the pressure imposed by the increasingly death toll and the great attention of the media and politics on the pandemic have stimulated the publication of some small or incomplete studies in humans with questionable empirical clinical data, many of them without desirable scientific rigor. This fact has been the subject of broad debate. As a result, misinformation and confusion have occurred, hindering important actions to combat COVID-19 advance and leading to a needless discredit in science. In fact, some published research papers from recognized medical journals have been withdrawn recently after several concerns raised with respect to the veracity of the data and analyses conducted by the authors revealing the use of dubious methods of analysis and ethical problems [246]. This fact had a direct impact on ongoing clinical trials causing temporary suspension and compromising the long-awaited results. On the other hand, as stated by Zhang et al. [27]. “sound science should be effectively communicated to policy makers and should constitute the prime foundation in decision-making amid this pandemic”. Furthermore, only well designed, and rigorous randomized controlled trial can provide reliable and generalized data for safety and effective use of drugs to treat infected patients [247]. Despite the great challenge posed by the COVID-19 pandemic and the surfaced ethical issues, the ongoing efforts to fight the virus and the excellence and hardworking of many research groups across the world offer hope that in a near future we can win this battle.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
There is no conflict of interest
References
- 1.World Health Organization. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-mission-briefing-on-covid-19---12-march-2020. (accessed May 16, 2020).
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhan M., Qin Y., Xue X., Zhu S. Death from Covid-19 of 23 health care workers in China. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2005696. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verity R., Okell L.C., Dorigatti I., Winskill P., Whittaker C., Imai N., Cuomo-Dannenburg G., Thompson H., Walker P.G.T., Fu H., Dighe A., Griffin J.T., Baguelin M., Bhatia S., Boonyasiri A., Cori A., Cucunubá Z., FitzJohn R., Gaythorpe K., Green W., Hamlet A., Hinsley W., Laydon D., Nedjati-Gilani G., Riley S., van Elsland S., Volz E., Wang H., Wang Y., Xi X., Donnelly C.A., Ghani A.C., Ferguson N.M. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30243-7. Mar 30:S1473-3099(20)30243-7. doi: 10.1016/S1473-3099(20)30243-7. Epub ahead of print. Erratum in: Lancet Infect Dis. (2020) Apr 15;: Erratum in: Lancet Infect Dis. 2020 May 4;: PMID: 32240634; PMCID: PMC7158570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gralinski L.E., Menachery V.D. Return of the coronavirus: 2019-nCoV. Viruses. 2020;12(2):135. doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.World Health Organization . 2020. Naming the Coronavirus Disease (COVID-19) and the Virus That Causes It.https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance [Google Scholar]
- 8.Virological.org. Novel 2019 Coronavirus Genome. http://virological.org/t/issues-with-sars-cov-2-sequencing-data/473 (accessed 16 May, 2020).
- 9.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu P., Jiang J.Z., Wan X., Hua Y., Li L., Zhou J., Wang X., Hou F., Chen J., Zou J., Chen J. Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLoS Pathog. 2020;16(5) doi: 10.1371/journal.ppat.1008421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang T., Wu Q., Zhang Z. 2020. Pangolin Homology Associated With 2019-nCoV bioRxiv. [DOI] [Google Scholar]
- 13.World Health Organization . 2020. Mission summary: WHO Field Visit to Wuhan, China.https://www.who.int/china/news/detail/22-01-2020-field-visit-wuhan-china-jan-2020 20-21 January. (accessed May 16, 2020) [Google Scholar]
- 14.Global Initiative on Sharing All Influenza Data . 2020. 27,000 Viral Genomic Sequences of hCoV-19 Shared With Unprecedented Speed Via GISAID.https://www.gisaid.org/ [Google Scholar]
- 15.Chen Y., Liu Q., Guo D. Coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92(4):418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paraskevis D., Kostaki E.G., Magiorkinis G., Panayiotakopoulos G., Sourvinos G., Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect. Genet. Evol. 2020;79:104212. doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16(69):1–22. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11(1):1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94(7) doi: 10.1128/JVI.00127-20. e00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walls A.C., Park Y.-P., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;180:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hindson J. COVID-19: faecal–oral transmission? Nat. Rev. Gastroenterol. Hepatol. 2020;17:259. doi: 10.1038/s41575-020-0295-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I., Lloyd-Smith J.O., de Wit E., Munster V.J. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. New Engl. J. Med. 2020;382(16):1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y., Li X., Zhu B., Liang H., Fang C., Gong Y., Guo Q., Sun X., Zhao D., Shen J., Zhang H., Liu H., Xia H., Tang J., Zhang K., Gong S. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020;26(4):502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang R., Li Y., Zhang A.L., Wang Y., Molina M.J. Identifying airborne transmission as the dominant route for the spread of COVID-19. Proc. Natl. Acad. Sci. U. S. A. 2020:202009637. doi: 10.1073/pnas.2009637117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Channappanavar R., Zhao J., Perlman S. T cell-mediated immune response to respiratory coronaviruses. Immunol. Res. 2014;59(1–3):118–128. doi: 10.1007/s12026-014-8534-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelemans T., Kikkert M. Viral innate immune evasion and the pathogenesis of emerging rna virus infections. Viruses. 2019;11(10):961. doi: 10.3390/v11100961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aguirre-Duarte N. Can people with asymptomatic or pre-symptomatic COVID-19 infect others: a systematic review of primary data. medRxiv. 2020 doi: 10.1101/2020.04.08.20054023. [DOI] [Google Scholar]
- 31.Hu Z., Song C., Xu C., Jin G., Chen Y., Xu X., Ma H., Chen W., Lin Y., Zheng Y., Wang J., Hu Z., Yi Y., Shen H. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci. China Life Sci. 2020;63(5):706–711. doi: 10.1007/s11427-020-1661-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Center for Disease Control . 2020. Protect Youself.https://www.cdc.gov/coronavirus/2019-ncov/prevent-getting-sick/prevention.html [Google Scholar]
- 33.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shirato K., Kawase M., Matsuyama S. Wild-type human coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry. Virology. 2018;517:9–15. doi: 10.1016/j.virol.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14(8):523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan J.F.W., Yuan S., Kok K.H., To K.K.W., Chu H., Yang J., Xing F., Liu J., Yip C.C.-Y., Poon R.W.-S., Tsoi H.-W., Lo S.K.-F., Chan K.-H., Poon V.K.-M., Chan W.-M., Ip J.-D., Cai J.-P., Cheng V.C.-C., Chen H., Hui C.K.-M., Yuen K.-Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., Du B., Li L.J., Zeng G., Yuen K.Y., Chen R.C., Tang C.L., Wang T., Chen P.Y., Xiang J., Li S.Y., Wang J.L., Liang Z.J., Peng Y.X., Wei L., Liu Y., Hu Y.H., Peng P., Wang J.M., Liu J.Y., Chen Z., Li G., Zheng Z.J., Qiu S.Q., Luo J., Ye C.J., Zhu S.Y., Zhong N.S. China medical treatment expert group for Covid-19. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian S., Hu N., Lou J., Chen K., Kang X., Xiang Z., Chen H., Wang D., Liu N., Liu D., Chen G., Zhang Y., Li D., Li J., Lian H., Niu S., Zhang L., Zhang J. Characteristics of COVID-19 infection in Beijing. J. Infect. 2020;80(4):401–406. doi: 10.1016/j.jinf.2020.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paderno A., Schreiber A., Grammatica A., Raffetti E., Tomasoni M., Gualtieri T., Taboni S., Zorzi S., Lombardi D., Deganello A., Redaelli De Zinis L.O., Maroldi R., Mattavelli D. Smell and taste alterations in Covid-19: a cross-sectional analysis of different cohorts. Int. Forum Allergy Rhinol. 2020 doi: 10.1002/alr.22610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sedaghat A.R., Gengler I., Speth M.M. Olfactory dysfunction: a highly prevalent symptom of COVID-19 with public health significance. Otolaryngol. Head. Neck Surg. 2020 doi: 10.1177/0194599820926464. 194599820926464. [DOI] [PubMed] [Google Scholar]
- 44.Yan C.H., Faraji F., Prajapati D.P., Boone C.E., DeConde A.S. Association of chemosensory dysfunction and Covid-19 in patients presenting with influenza-like symptoms. Int. Forum Allergy Rhinol. 2020 doi: 10.1002/alr.22579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J., Zheng Y., Gou X., Pu K., Chen Z., Guo Q., Ji R., Wang H., Wang Y., Zhou Y. Prevalence of comorbidities and its effects in patients infected with SARS-CoV-2: a systematic review and meta-analysis. Int. J. Infect. Dis. 2020;94:91–95. doi: 10.1016/j.ijid.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020 doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang X., Yu Y., Xu J., Shu H., Xia J.A., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T., Wang Y., Pan S., Zou X., Yuan S., Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., Zhang X., Zhang M., Wu S., Song J., Chen T., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130(5):2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.S. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perrone L.A., Plowden J.K., García-Sastre A., Katz J.M., Tumpey T.M. H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.D’Elia R.V., Harrison K., Oyston P.C., Lukaszewski R.A., Clark G.C. Targeting the ‘cytokine storm’ for therapeutic benefit. Clin. Vaccine Immunol. 2013;20(3):319–327. doi: 10.1128/CVI.00636-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meduri G.U., Kohler G., Headley S., Tolley E., Stentz F., Postlethwaite A. Inflammatory cytokines in the BAL of patients with ARDS. Persistent elevation over time predicts poor outcome. Chest. 1995;108:1303–1314. doi: 10.1378/chest.108.5.1303. [DOI] [PubMed] [Google Scholar]
- 54.Bouros D., Alexandrakis M.G., Antoniou K.M., Agouridakis P., Pneumatikos I., Anevlavis S., Pataka A., Patlakas G., Karkavitsas N., Kyriakou D. The clinical significance of serum and bronchoalveolar lavage inflammatory cytokines in patients at risk for Acute Respiratory Distress Syndrome. BMC Pulm. Med. 2004;4:6. doi: 10.1186/1471-2466-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yao Z., Zheng Z., Wu K., Junhua Z. Immune environment modulation in pneumonia patients caused by coronavirus: SARS-CoV, MERS-CoV and SARS-CoV-2. Aging (Albany NY) 2020;12(9):7639–7651. doi: 10.18632/aging.103101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 2020;9(1):761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wong C.K., Lam C.W., Wu A.K., Ip W.K., Lee N.L., Chan I.H., Lit L.C., Hui D.S., Chan M.H., Chung S.S., Sung J.J. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. Exp. Immunol. 2004;136:95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mahallawi W.H., Khabour O.F., Zhang Q., Makhdoum H.M., Suliman B.A. MERS-CoV infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine. 2018;104:8–13. doi: 10.1016/j.cyto.2018.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Durham L.E., Kirkham B.W., Taams L.S. Contribution of the IL-17 pathway to psoriasis and psoriatic arthritis. Curr. Rheumatol. Rep. 2015;17:55. doi: 10.1007/s11926-015-0529-9. [DOI] [PubMed] [Google Scholar]
- 60.Raychaudhuri S.P., Raychaudhuri S.K. Mechanistic rationales for targeting interleukin17A in spondyloarthritis. Arthritis Res. Ther. 2017;19:51. doi: 10.1186/s13075-017-1249-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pacha O., Sallman M.A., Evans S.E. COVID-19: a case for inhibiting IL-17? Nat. Rev. Immunol. 2020;20:345–346. doi: 10.1038/s41577-020-0328-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gao Y., Li T., Han M., Li X., Wu D., Xu Y., Zhu Y., Liu Y., Wang X., Wang L. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID-19. J. Med. Virol. 2020 doi: 10.1002/jmv.25770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.D’Acquisto F., Maione F., Pederzoli-Ribeil M. From IL-15 to IL-33: the never-ending list of new players in inflammation. Is it time to forget the humble aspirin and move ahead? Biochem. Pharmacol. 2010;79:525–534. doi: 10.1016/j.bcp.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 64.Casillo G.M., Mansour A.A., Raucci F., Saviano A., Mascolo N., Iqbal A.J., Maione F. Could IL-17 represent a new therapeutic target for the treatment and/or management of COVID-19-related respiratory syndrome? Pharmacol. Res. 2020;156:104791. doi: 10.1016/j.phrs.2020.104791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Raucci F., Mansour A.A., Casillo G.M., Saviano A., Caso F., Scarpa R., Mascolo N., Iqbal A.J., Maione F. Interleukin-17A (IL-17A), a key molecule of innate and adaptive immunity, and its potential involvement in COVID-19-related thrombotic and vascular mechanisms. Autoimmun. Rev. 2020;19(7):102572. doi: 10.1016/j.autrev.2020.102572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ceccarelli G., Nardi K., Marchesani F. Letter to the Editor in response to the article “Could IL-17 represent a new therapeutic target for the treatment and/or management of COVID-19-related respiratory syndrome?”. Pharmacol. Res. 2020;158:104933. doi: 10.1016/j.phrs.2020.104933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu D., Yang X.O. TH17 responses in cytokine storm of COVID-19: an emerging target of JAK2 inhibitor fedratinib. J. Microbiol. Immunol. Infect. 2020 doi: 10.1016/j.jmii.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jose R.J., Manuel A. COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir. Med. 2020;20 doi: 10.1016/S2213-2600(20)30216-2. S2213-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Andreakos E., Zanoni I., Galani I.E. Lambda interferons come to light: dual function cytokines mediating antiviral immunity and damage control. Curr. Opin. Immunol. 2019;56:67–75. doi: 10.1016/j.coi.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Syedbasha M., Egli A. Interferon lambda: modulating immunity in infectious diseases. Front. Immunol. 2017;8:119. doi: 10.3389/fimmu.2017.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Galani I.E., Triantafyllia V., Eleminiadou E.E., Koltsida O., Stavropoulos A., Manioudaki M., Thanos D., Doyle S.E., Kotenko S.V., Thanopoulou K., et al. Interferon-lambda mediates nonredundant front-line antiviral protection against influenza virus infection without compromising host fitness. Immunity. 2017;46:875–890. doi: 10.1016/j.immuni.2017.04.025. e6. [DOI] [PubMed] [Google Scholar]
- 72.Andreakos E., Tsiodras S. COVID-19: lambda interferon against viral load and hyperinflammation. EMBO Mol. Med. 2020;12(6):e12465. doi: 10.15252/emmm.202012465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tang N., Bai H., Chen X., Gong J., Li D., Sun Z. Anticoagulant treatment is associated with decreased mortality in severe coronavirus disease 2019 patients with coagulopathy. J. Thromb. Haemost. 2020:1–6. doi: 10.1111/jth.14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kong W., Li Y., Peng M., Kong D., Yang X., Wang L., Liu M. SARS-CoV-2 detection in patients with influenza-like illness. Nat. Microbiol. 2020;5:675–678. doi: 10.1038/s41564-020-0713-1. [DOI] [PubMed] [Google Scholar]
- 75.Kanne J.P. Chest CT findings in 2019 novel coronavirus (2019-nCoV) infections from Wuhan, China: key points for the radiologist. Radiology. 2020 doi: 10.1148/radiol.2020200241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou S., Wang Y., Zhu T., Xia L. CT features of coronavirus disease 2019 (COVID-19) pneumonia in 62 patients in Wuhan, China. AJR Am. J. Roentgenol. 2020;5:1–8. doi: 10.2214/AJR.20.22975. [DOI] [PubMed] [Google Scholar]
- 77.Gattinoni L., Chiumello D., Caironi P., Busana M., Romitti F., Brazzi L., Camporota L. COVID-19 pneumonia: different respiratory treatments for different phenotypes? Intensive Care Med. 2020 doi: 10.1007/s00134-020-06033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoshimoto F.K. The Proteins of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS CoV-2 or n-COV19), the Cause of COVID-19. Protein J. 2020;39:198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang C., Lokugamage K.G., Rozovics J.M., Narayanan K., Semler B.L., Makino S. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cornillez-Ty C.T., Liao L., Yates J.R., Kuhn P., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. J. Virol. 2009;83:10314–10318. doi: 10.1128/JVI.00842-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lei J., Kusov Y., Hilgenfeld R. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antiviral Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baez-Santos Y.M., St. John S.E., Mesecar A.D. The SARS coronavirus papain-like protease: structure, function, and inhibition by designed antiviral compounds. Antiviral Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sakai Y., Kawachi K., Terada Y., Omori H., Matsura Y., Kamitani W. Two-amino acids change in the nsp4 of SARS coronavirus abolishes viral replication. Virology. 2017;510:165–174. doi: 10.1016/j.virol.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tomar S., Johnston M.L., St. John S.E., Osswald H.L., Nyalapatla P.R., Paul L.N., Ghosh A.K., Denison M.R., Mesecar A.D. Ligand induced dimerization of middle east respiratory syndrome (MERS) coronavirus nsp5 protease (3CLpro) implications for nsp5 Regulation and the Development of Antivirals. J. Biol. Chem. 2015;290:19403–19422. doi: 10.1074/jbc.M115.651463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cottam E.M., Whelband M.C., Wileman T. Coronavirus NSP6 restricts autophagosome expansion. Autophagy. 2014;10:1426–1441. doi: 10.4161/auto.29309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Te Velthuis A.J., van de Worm S.H., Snijder E. The SARScoronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012;40:1737–1747. doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhao S., Ge X., Wang X., Liu A., Guo X., Zhou L., Yu K., Yang H. The DEAD-box RNA helicase 5 positively regulates the replication of porcine reproductive and respiratory syndrome virus by interacting with viral Nsp9 in vitro. Virus Res. 2015;195:217–224. doi: 10.1016/j.virusres.2014.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ma Y., Wu L., Shaw N., Gao Y., Wang J., Sun Y., Lou Z., Yan L., Zhang R., Rao Z. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc. Natl. Acad. Sci. U. S. A. 2015;112:9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang Y., Sun Y., Wu A., Xu S., Pan R., Zeng C., Jin X., Ge X., Shi Z., Ahola T., Guo D. Coronavirus nsp10/nsp16 methyltransferase can be targeted by nsp10-derived peptide in vitro and in vivo to reduce replication and pathogenesis. J. Virol. 2015;89:8416–8427. doi: 10.1128/JVI.00948-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Case J.B., Ashbrook A.W., Dermody T.S., Denison M.R. Mutagenesis of S-adenosyl-l-methionine-binding residues in coronavirus nsp14 N7-methyltransferase demonstrates differing requirements for genome translation and resistance to innate immunity. J. Virol. 2016;90:7248–7256. doi: 10.1128/JVI.00542-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bhardwaj K., Sun J., Holzenburg A., Guarino L.A., Kao C.C. RNA recognition and cleavage by the SARS coronavirus endoribonuclease. J. Mol. Biol. 2006;361:243–256. doi: 10.1016/j.jmb.2006.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hackbart M., Deng X., Baker S. C Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc. Natl. Acad. Sci. U. S. A. 2020;117:8094–8103. doi: 10.1073/pnas.1921485117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Decroly E., Debarnot C., Ferron F., Bouvet M., Coutard B., Imbert I., Gluais L., Papageorgiou N., Sharff A., Bricogne G., Ortiz-Lombardia M., Lescar J., Canard B. Crystal structure and functional analysis of the sars-coronavirus RNA cap 2’-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11(1):1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Calligari P., Bobone S., Ricci G., Bocedi A. Molecular investigation of SARS-CoV-2 proteins and their interactions with antiviral drugs. Viruses. 2020;12:445. doi: 10.3390/v12040445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gordon D.E., Jang G.M., Bouhaddou M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020 doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen Y.W., Yiu C.B., Wong K.Y. Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL pro) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Res. 2020;9:129. doi: 10.12688/f1000research.22457.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Retallack H., Di Lullo E., Arias C., Knopp K.A., Laurie M.T., Sandoval-Espinosa C., Mancia Leon W.R., Krencik R., Ullian E.M., Spatazza J., Pollen A.A., Mandel-Brehm C., Nowakowski T.J., Kriegstein A.R., DeRisi J.L. Zika virus cell tropism in the developing human brain and inhibition by azithromycin. Proc. Natl. Acad. Sci. U. S. A. 2016;113(50):14408–14413. doi: 10.1073/pnas.1618029113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Madrid P.B., Panchal R.G., Warren T.K., Shurtleff A.C., Endsley A.N., Green C.E., Kolokoltsov A., Davey R., Manger I.D., Gilfillan L., Bavari S., Tanga M.J. Evaluation of Ebola virus inhibitors for drug repurposing. ACS Infect. Dis. 2015;1(7):317–326. doi: 10.1021/acsinfecdis.5b00030. [DOI] [PubMed] [Google Scholar]
- 102.Jaffé A., Bush A. Anti‐inflammatory effects of macrolides in lung disease. Pediatr. Pulmonol. 2001;31:464–473. doi: 10.1002/ppul.1076. [DOI] [PubMed] [Google Scholar]
- 103.Gibson P.G., Yang I.A., Upham J.W., Reynolds P.N., Hodge S., James A.L., Jenkins C., Peters M.J., Marks G.B., Baraket M., Powell H., Taylor S.L., Leong L.E.X., Rogers G.B., Simpson J.L. Effect of azithromycin on asthma exacerbations and quality of life in adults with persistent uncontrolled asthma (AMAZES): a randomised, double-blind, placebo-controlled trial. Lancet. 2017;390(10095):659–668. doi: 10.1016/S0140-6736(17)31281-3. [DOI] [PubMed] [Google Scholar]
- 104.Menzel M., Akbarshahi H., Bjermer L., Uller L. Azithromycin induces anti-viral effects in cultured bronchial epithelial cells from COPD patients. Sci. Rep. 2016;6:28698–28709. doi: 10.1038/srep28698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Iversen M.B., Paludan S.R. Mechanisms of type III interferon expression. J. Interferon Cytokine Res. 2010;30:573–578. doi: 10.1089/jir.2010.0063. [DOI] [PubMed] [Google Scholar]
- 106.Levy D.E., Marié I.J., Durbin J.E. Induction and function of type I and III interferon in response to viral infection. Curr. Opin. Virol. 2011;6:476–486. doi: 10.1016/j.coviro.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Touret F., Gilles M., Barral K., Nougairède A., Decroly E., Lamballerie X., Coutard B. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS‐CoV‐2 replication. bioRxiv. 2020 doi: 10.1101/2020.04.03.023846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Damle B., Vourvahis M., Wang E., Leaney J., Corrigan B. Clinical pharmacology perspectives on the antiviral activity of azithromycin and use in COVID‐19. Clin. Pharm. Therap. 2020 doi: 10.1002/cpt.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gautret P., Lagier J.C., Parola P., Hoang V.T., Meddeb L., Mailhe, Doudier B., Courjon J., Giordanengo V., Vieira V.E., Dupont H.T., Honoré S., Colson P., Chabrière E., La Scola B., Rolain J.M., Brouqui P., Raoult D. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020:105949. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Molina J.M., Delaugerre C., LeGoff J., Mela-Lima B., Ponscarme D., Goldwirt L., et al. No evidence of rapid antiviral clearance or clinical benefit with the combination of hydroxychloroquine and azithromycin in patients with severe COVID-19 infection. Med. Mal. Infect. 2020 doi: 10.1016/j.medmal.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rynes R. Antimalarial drugs in the treatment of rheumatological diseases. Rheumatology. 1997;36(7):799–805. doi: 10.1093/rheumatology/36.7.799. [DOI] [PubMed] [Google Scholar]
- 112.Ben-Zvi I., Kivity S., Langevitz P., Shoenfeld Y. Hydroxychloroquine: from malaria to autoimmunity. Clin. Rev. Allergy Immunol. 2012;42:145–153. doi: 10.1007/s12016-010-8243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gao J., Tian Z., Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends. 2020;14:72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 114.Keyaerts E., Vijgen L., Maes P., Neyts J., Van Ranst M. In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem. Biophys. Res. Commun. 2004;323:264–268. doi: 10.1016/j.bbrc.2004.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vincent M.J., Bergeron E., Benjannet S., Erickson B.R., Rollin P.E., Ksiazek T.G., Seidah N.G., Nichol S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005;2:69. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yao X., Ye F., Zhang M., Cui C., Huang B., Niu P., Liu D., Zhao L., Dong E., Song C., Zhan S., Lu R., Li H., Tan W., Liu D. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu J., Can R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gendrot M., Javelle E., Clerc A., Savini H., Pradines B. Chloroquine as a prophylactic agent against COVID-19? Int. J. Antimicrob. Agents. 2020:105980. doi: 10.1016/j.ijantimicag.2020.105980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cortegiani A., Ingoglia G., Ippolito M., Giarratano A., Einav S. A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19. J. Crit. Care. 2020;57:279–283. doi: 10.1016/j.jcrc.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Okour M., Al-Kofahi M., Austin D. Hydroxychloroquine and azithromycin as potential treatments for COVID-19; clinical status impacts the outcome. J. Pharmacokinet. Pharmacodyn. 2020 doi: 10.1007/s10928-020-09689-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Savarino A., Boelaert J.R., Cassone A., Majori G., Cauda R. Effects of chloroquine on viral infections: an old drug against today’s diseases? Lancet Infect. Dis. 2003;3:722–727. doi: 10.1016/S1473-3099(03)00806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.U.S. Food and Drug Administration . 2020. EUA Hydroxychloroquine Sulfate Health Care Provider: Fact Sheet for Patients and parent/caregivers Emergency Use Authorization (EUA) of Hydroxychloroquine Sulfate for Treatment of COVID-19 in Certain Hospitalized Patients.www.fda.gov [Google Scholar]
- 124.Multicenter collaboration group of Department of Science and Technology of Guangdong Province and Health Commission of Guangdong Province for chloroquine in the treatment of novelcoronavirus pneumonia. Zhonghua Jie He He Hu Xi Za Zhi. 2020;43(0):E019. doi: 10.3760/cma.j.issn.1001-0939.2020.0019. [DOI] [PubMed] [Google Scholar]
- 125.Agencia Española de Medicamentos y Productos Sanitarios Tratamientos disponibles para el manejo de la infección respiratoria por SARS-CoV-2. https://www.aemps.gob.es/laAEMPS/docs/medicamentos-disponibles-SARS-CoV-2-28-3-2020.pdf (Accessed 19 May, 2020)
- 126.Devaux C.A., Rolain J.M., Colson P., Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int. J. Antimicrob. Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Srinivasa A., Tosounidou S., Gordon C. Increased incidence of gastrointestinal side effects in patients taking hydroxychloroquine: a brand-related issue? J. Rheumatol. 2017;44(3) doi: 10.3899/jrheum.161063. 398-398. [DOI] [PubMed] [Google Scholar]
- 128.Mavrikakis M., Papazoglou S., Sfikakis P.P., Vaiopoulos G., Rougas K. Retinal toxicity in long term hydroxychloroquine treatment. Ann. Rheum. Dis. 1996;55(3):187–189. doi: 10.1136/ard.55.3.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Easterbrook M. The ocular safety of hydroxychloroquine. Seminars in Arthritis and Rheumatism; WB Saunders Eds.; 1993. pp. 62–67. Vol. 23, 2. [DOI] [PubMed] [Google Scholar]
- 130.Browning D.J. Hydroxychloroquine and chloroquine retinopathy: screening for drug toxicity. Am. J. Ophthalmol. Case Rep. 2002;133(5):649–656. doi: 10.1016/s0002-9394(02)01392-2. [DOI] [PubMed] [Google Scholar]
- 131.Cubero G.I., Reguero J.R., Ortega J.R. Restrictive cardiomyopathy caused by chloroquine. Heart. 1993;69(5):451–452. doi: 10.1136/hrt.69.5.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Costedoat-Chalumeau N., Hulot J.S., Amoura Z., Leroux G., Lechat P., Funck-Brentano C., Piette J.C. Heart conduction disorders related to antimalarials toxicity: an analysis of electrocardiograms in 85 patients treated with hydroxychloroquine for connective tissue diseases. Rheumatology. 2007;46(5):808–810. doi: 10.1093/rheumatology/kel402. [DOI] [PubMed] [Google Scholar]
- 133.Saleh M., Gabriels J., Chang D., Kim B.S., Mansoor A., Mahmood E., Makker P., Ismail H., Goldner B., Willner J., Beldner S., Mitra R., John R., Chinitz J., Skipitaris N., Mountantonakis S., Epstein L.M. The Effect of Chloroquine, Hydroxychloroquine and Azithromycin on the Corrected QT Interval in Patients with SARS-CoV-2 Infection. Circ. Arrhythm. Electrophysiol. 2020 doi: 10.1161/CIRCEP.120.008662. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mehra M.R., Desai S.S., Ruschitzka F., Patel A.N. Hydroxychloroquine or chloroquine with or without a macrolide for treatment of COVID-19: a multinational registry analysis. Lancet. 2020 doi: 10.1016/S0140-6736(20)31180-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 135.Wagstaff K.M., Sivakumaran H., Heaton S.M., Harrich D., Jans D.A. Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 2012;443(3):851–856. doi: 10.1042/BJ20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yang S.N.Y., Atkinson S.C., Wang C., Lee A., Bogoyevitch M.A., Borg N.A., Jans D.A. The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer. Antiviral Res. 2020;177 doi: 10.1016/j.antiviral.2020.104760. [DOI] [PubMed] [Google Scholar]
- 137.Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020;178 doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schmith V.D., Zhou J., Lohmer L.R.L. The approved dose of ivermectin alone is not the ideal dose for the treatment of COVID-19. medRxiv. 2020 doi: 10.1101/2020.04.21.20073262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lespine A., Alvinerie M., Sutra J.F., Pors I., Chartier C. Influence of the route of administration on efficacy and tissue distribution of ivermectin in goat. Vet. Parasitol. 2005;128:251–260. doi: 10.1016/j.vetpar.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 140.Chu C.M., Cheng V.C., Hung I.F., Wong M.M., Chan K.H., Chan K.S., Kao R.Y., Poon L.L., Wong C.L., Guan Y., Peiris J.S., Yuen K.Y. HKU/UCH SARS Study Group. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59:252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.De Clercq E. Anti-HIV drugs:25 compounds approved within 25 years after the discovery of HIV. Int. J. Antimicrob. Agents. 2009;33(4):307–320. doi: 10.1016/j.ijantimicag.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 142.Chen F., Chan K.H., Jiang Y., Kao R.Y., Lu H.T., Fan K.W., Cheng V.C., Tsui W.H., Hung I.F., Lee T.S., Guan Y., Peiris J.S., Yuen K.Y. In vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds. J. Clin. Virol. 2004;31:69–75. doi: 10.1016/j.jcv.2004.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wu C.-Y., Jan J.-T., Ma S.-H., Kuo C.J., Juan H.F., Cheng Y.S., Hsu H.H., Huang H.C., Wu D., Brik A., Liang F.S., Liu R.S., Fang J.M., Chen S.T., Liang P.H., Wong C.H. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Natl. Acad. Sci. U. S. A. 2004;101:10012–10017. doi: 10.1073/pnas.0403596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chan K.S., Lai S.T., Chu C.M., Tsui E., Tam C.Y., Wong M.M.L., Tse M.W., Que T.L., Peiris J.S.M., Sung J., Wong V.C.W., Yuen K.Y. Treatment of severe acute respiratory syndrome with lopinavir/ritonavir: A multicentre retrospective matched cohort study. Hong Kong Med. J. 2003;9:399–406. [PubMed] [Google Scholar]
- 145.Lim J., Jeon S., Shin H.Y., Kim M.J., Seong Y.M., Lee W.J., Choe K.W., Kang Y.M., Lee B., Park S.J. Case of the index patient who caused tertiary transmission of COVID19 infection in Korea: the application of lopinavir/ritonavir for the treatment of COVID-19 infected pneumonia monitored by quantitative RT-PCR. J. Korean Med. Sci. 2020;35(6):e79. doi: 10.3346/jkms.2020.35.e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M., Li X., Xia J., Chen N., Xiang J., Yu T., Bai T., Xie X., Zhang L., Li C., Yuan Y., Chen H., Li H., Huang H., Tu S., Gong F., Liu Y., Wei Y., Dong C., Zhou F., Gu X., Xu J., Liu Z., Zhang Y., Li H., Shang L., Wang K., Li K., Zhou X., Dong X., Qu Z., Lu S., Hu X., Ruan S., Luo S., Wu J., Peng L., Cheng F., Pan L., Zou J., Jia C., Wang J., Liu X., Wang S., Wu X., Ge Q., He J., Zhan H., Qiu F., Guo L., Huang C., Jaki T., Hayden F.G., Horby P.W., Zhang D., Wang C.A. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rossignol J.-F. Nitazoxanide, a new drug candidate for the treatment of Middle East respiratory syndrome coronavirus. J. Infection Public Health. 2016;9(3):227–230. doi: 10.1016/j.jiph.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Haffizulla J., Hartman A., Hoppers M. Effect of nitazoxanide in adults and adolescents with acute uncomplicated influenza: a double-blind, randomised, placebo-controlled, phase 2b/3 trial. Lancet Infect. Dis. 2014;14(7):609–618. doi: 10.1016/s1473-3099(14)70717-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kelleni M.T. Nitazoxanide/azithromycin combination for COVID-19: A suggested new protocol for early management [published online ahead of print, 2020 Apr 30] Pharmacol. Res. 2020;157:104874. doi: 10.1016/j.phrs.2020.104874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Pepperrell T., Pilkington V., Owen A., Wang J., Hill A.M. Review of safety and minimum pricing of nitazoxanide for potential treatment of COVID-19. J. Virus Erad. 2020;6(2):52–60. doi: 10.1016/S2055-6640(20)30017-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Siegel D., Hui H.C., Doerffler E., Clarke M.O., Chun K., Zhang L., Neville S., Carra E., Lew W., Ross B., Wang Q., Wolfe L., Jordan R., Soloveva V., Knox J., Perry J., Perron M., Stray K.M., Barauskas O., Feng J.Y., Xu Y., Lee G., Rheingold A.L., Ray A.S., Bannister R., Strickley R., Swaminathan S., Lee W.A., Bavari S., Cihlar T., Lo M.K., ;Warren T.K., Mackman R.L. Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f][triazin-4-amino] adenine C-nucleoside (GS-5734) for the treatment of Ebola and emerging viruses. J. Med. Chem. 2017;60(5):1648–1661. doi: 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- 152.Sheahan T.P., Sims A.C., Leist S.R., Schafer A., Won J., Brown A.J., Montgomery S.A., Hogg A., Babusis D., Clarke M.O., Spahn J.E., Bauer L., Sellers S., Porter D., Feng J.Y., Cihlar T., Jordan R., Denison M.R., Baric R.S. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11(1):222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Weldon W.C., Biggs H.M., Uyeki T.M., Pillai S.K. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., Flanigan T. Compassionate use of remdesivir for patients with severe Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Wang C. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395(10236):1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jin Z., Smith L.K., Rajwanshi V.K., Kim B., Deval J. The ambiguous base-pairing and high substrate efficiency of T-705 (favipiravir) ribofuranosyl 5’-triphosphate towards influenza A virus polymerase. PLoS One. 2014;8(7) doi: 10.1371/journal.pone.0068347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Furuta Y., Komeno T., Nakamura T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2017;93(7):449–463. doi: 10.2183/pjab.93.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Delang L., Abdelnabi R., Neyts J. Favipiravir as a potential countermeasure against neglected and emerging RNA viruses. Antiviral Res. 2018;153:85–94. doi: 10.1016/j.antiviral.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 159.Choy K.T., Wong A.Y.L., Kaewpreedee P., Sia S.F., Chen D. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res. 2020;178 doi: 10.1016/j.antiviral.2020.104786. 104786–104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chen C., Zhang Y., Huang J., Yin P., Cheng Z., Wu J., Wang X. Favipiravir versus arbidol for COVID-19: a randomized clinical trial. medRxiv. 2020 doi: 10.1101/2020.03.17.20037432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Cai Q., Yang M., Liu D., Chen J., Shu D. Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering. 2020 doi: 10.1016/j.eng.2020.03.007. S2095-8099(20)30063-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Boriskin Y.S., Leneva I.A., Pecheur E.I., Polyak S.J. Arbidol: a broad-spectrum antiviral compound that blocks viral fusion. Curr. Med. Chem. 2008;15(10):997–1005. doi: 10.2174/092986708784049658. [DOI] [PubMed] [Google Scholar]
- 163.Villalaín J. Membranotropic effects of arbidol, a broad anti-viral molecule, on phospholipid model membranes. J. Phys. Chem. B. 2010;114(25):8544–8554. doi: 10.1021/jp102619w. [DOI] [PubMed] [Google Scholar]
- 164.Dong L., Hu S., Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19) Drug Discov. Ther. 2020;14(1):58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- 165.Dong L., Li C., Zeng Q., Liu X., Li X., Zhang H. Arbidol combined with LPV/r versus LPV/r alone against corona virus disease 2019: a retrospective cohort study. J. Infect. 2020 doi: 10.1016/j.jinf.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhu Z., Lu Z., Xu T., Chen C., Yang G., Zha T., Lu J., Xue Y. Arbidol monotherapy is superior to lopinavir/ritonavir in treating COVID-19. J. Infect. 2020;81(1):e21–23. doi: 10.1016/j.jinf.2020.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wu C., Chen X., Cai Y., Song Y. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020 doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wang Y., Jiang W., He Q. A retrospective cohort study of methylprednisolone therapy in severe patients with COVID-19 pneumonia. Signal Transduct. Target. Ther. 2020;5(1):57. doi: 10.1038/s41392-020-0158-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhou W., Liu Y., Tian D. Potential benefits of precise corticosteroids therapy for severe 2019-nCoV pneumonia. Signal Transduct. Target. Ther. 2020;5(1):18. doi: 10.1038/s41392-020-0127-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Oxford . 2020. Dexamethasone Reduces Death in Hospitalised Patients With Severe Respiratory Complications of COVID-19.http://www.ox.ac.uk/news/2020-06-16-dexamethasone-reduces-death-hospitalised-patients-severe-respiratory-complications [Google Scholar]
- 171.Chen N., Zhou M., Dong X. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tang N., Li D., Wang X., Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemost. 2020 doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Shanghai Clinical Treatment Expert Group for COVID-19 Comprehensive treatment and management of coronavirus disease 2019: expert consensus statement from Shanghai (in Chinese) Chin. J. Infect. Dis. 2020;38 doi: 10.3760/cma.j.issn.1000-6680.2020.0016. Epub ahead of print. [DOI] [Google Scholar]
- 174.Tang N., Bai H., Chen X., Gong J., Li D., Sun Z. Anticoagulant treatment is associated with decreased mortality in severe coronavirus disease 2019 patients with coagulopathy. J. Thromb. Haemost. 2020;18(5):1094–1099. doi: 10.1111/jth.14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Negri E.M., Piloto B.M., Morinaga L.K., Jardim C.V.P., Lamy S.A.E.-D., D’Amico E.A., Deheinzelin D. Heparin therapy improving hypoxia in COVID-19 patients - a case series. São Paulo. MedRxiv. 2020 doi: 10.1101/2020.04.15.20067017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kochi A., Tagliari A.P., Forleo G.B., Fassini G.M., Tondo C. Cardiac and arrhythmic complications in patients with COVID-19. J. Cardiovasc. Electrophysiol. 2020:1–6. doi: 10.1111/jce.14479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Godoy C.M.G., Vasques Ê.R., Caricati-Neto A., Tavares J.G.P., Alves B.J., Duarte J., Miranda-Ferreira R., Lima M.A., Nader H.B., Tersariol I.L.S. Heparin oligosaccharides have antiarrhythmic effect by accelerating the sodium-calcium exchanger. Front. Cardiovasc. Med. 2018;5:67. doi: 10.3389/fcvm.2018.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Fender A.C., Wakili R., Dobrev D. Straight to the heart: pleiotropic antiarrhythmic actions of oral anticoagulants. Pharmacol. Res. Elsevier. 2019;145 doi: 10.1016/j.phrs.2019.104257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Menezes-Rodrigues F.S., Padrão Tavares J.G., Pires de Oliveira M. Anticoagulant and antiarrhythmic effects of heparin in the treatment of COVID-19 patients [published online ahead of print, 2020 May 14] J. Thromb. Haemost. 2020 doi: 10.1111/jth.14902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Dinarello C.A. Overview of the IL-1family ininnate inflammation and acquired immunity. Immunol. Rev. 2018;281(1):8–27. doi: 10.1111/imr.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Park W.Y., Goodman R.B., Steinberg K.P. Cytokine balance in the lungs of patients with acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 2001;164:1896–1903. doi: 10.1164/ajrccm.164.10.2104013. [DOI] [PubMed] [Google Scholar]
- 182.Cavalli G., Dinarello C.A. Treating rheumatological diseases and co-morbidities with interleukin-1 blocking therapies. Rheumatology. 2015;54:2134–2144. doi: 10.1093/rheumatology/kev269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Cavalli G., De Luca G., Campochiaro C., Della-Torre E., Ripa M., Canetti D., Oltolini C., Castiglioni B., Din C.T., Boffini N., Tomelleri A., Farina N., Ruggeri A., Rovere-Querini P., Di Lucca G., Martinenghi, Scotti R., Tresoldi M., Ciceri F., Landoni G., Zangrillo A., Scarpellini P., Dagna L. Interleukin-1 blockade with high-dose anakinra in patients with COVID-19, acute respiratory distress syndrome, and hyperinflammation: a retrospective cohort study. Lancet. 2020 doi: 10.1016/S2665-9913(20)30127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Huet T., Beaussier H., Voisin O., Jouveshomme S., Dauriat G., Lazareth I., Sacco E., Naccache J.-M., Bézie Y., Laplanche S., Berre A.L., Pavec J.L., Salmeron S., Emmerich J., Mourad J.J., Chatellier G., Hayem G. Anakinra for severe forms of COVID-19: a cohort study. Lancet Rheumath. 2020 doi: 10.1016/S2665-9913(20)30164-8. Published online May 29, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bechman K., Subesinghe S., Norton S. A systematic review and meta-analysis of infection risk with small molecule JAK inhibitors in rheumatoid arthritis. Rheumatology. 2019;58:1755–1766. doi: 10.1093/rheumatology/kez087. [DOI] [PubMed] [Google Scholar]
- 186.Richardson P., Griffin I., Tucker C. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020;395:e30–e31. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Cantini F., Niccoli L., Matarrese D., Nicastri E., Stobbione P., Goletti D. Baricitinib therapy in COVID-19: a pilot study on safety and clinical impact [published online ahead of print, 2020 Apr 23] J. Infect. 2020 doi: 10.1016/j.jinf.2020.04.017. S0163-4453(20)30228-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Liu B., Li M., Zhou Z., Guan X., Xiang Y. Can we use interleukin-6 (IL-6) blockade for coronavirus disease 2019 (COVID-19)-induced cytokine release syndrome (CRS)? J. Autoimmunity. 2020 doi: 10.1016/j.jaut.2020.102452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kaly L., Rosner I. Tocilizumab—a novel therapy for non-organ-specific autoimmune diseases. Best Pract. Res. Clin. Rheumatol. 2012;26:157–165. doi: 10.1016/j.berh.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 190.Xu X., Han M., Li T., Sun W., Wang D., Fu B., Zhou Y., Zheng X., Yang Y., Li X., Zhang X., Pan A., Wei H. Effective treatment of severe COVID-19 patients with tocilizumab. PNAS. 2020;117(20):10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Fenster V., Chattopadhyay S., Sen G.C. No love lost between viruses and interferons. Annu. Rev. Virol. 2015;2(1):549–572. doi: 10.1146/annurev-virology-100114-055249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Chattopadhyay S., Sen G.C. Tyrosine phosphorylation in Toll-like receptor signaling. Cytokine Growth Factor Rev. 2014;25(5):533–541. doi: 10.1016/j.cytogfr.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Barber G.N. STING-dependent cytosolic DNA sensing pathways. Trends Immunol. 2014;35(2):88–93. doi: 10.1016/j.it.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 194.Cai X., Chiu Y.H., Chen Z.J. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol. Cell. 2014;54(2):289–296. doi: 10.1016/j.molcel.2014.03.040. [DOI] [PubMed] [Google Scholar]
- 195.Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Chan J.F., Yao Y., Yeung M.L., et al. Treatment with lopinavir/ritonavir or interferon-β1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J. Infect. Dis. 2015;212:1904–1913. doi: 10.1093/infdis/jiv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Mantlo E., Bukreyeva N., Maruyama J., Paessler S., Huang C. Potent antiviral activities of type I interferons to SARS-CoV-2 infection. Antiviral Res. 2020;179 doi: 10.1016/j.antiviral.2020.104811. Apr 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Hung I.F., Lung K.C., Tso E.Y., Liu R., Chung T.W., Chu M.Y., Ng Y.Y., Lo J., Chan J., Tam A.R., Shum H.P., Chan V., Wu A.K., Sin K.M., Leung W.S., Law W.L., Lung D.C., Sin S., Yeung P., Yip C.C., Zhang R.R., Fung A.Y., Yan E.Y., Leung K.H., Ip J.D., Chu A.W., Chan W.M., Ng A.C., Lee R., Fung K., Yeung A.C., Wu T.C., Chan J.W., Yan W.W., Chan W.M., Chan J.F., Lie A.K., Tsang O.T., Cheng V.C., Que T.L., Lau C.S., Chan K.H., To K.K., Yuen K.Y. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395(10238):1695–1704. doi: 10.1016/S0140-6736(20)31042-4. May 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Li W., Moore M.J., Vasilieva N. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chen Y., Guo Y., Pan Y., Zhao Z.J. Structure analysis of the receptor binding of 2019-nCoV. Biochem. Biophys. Res. Com. 2020;525(1):135–140. doi: 10.1016/j.bbrc.2020.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and Antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5(4):562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Guan W.-J., Ni Z.-Y., Hu Y., Liang W.-H., Ou C.-Q., He J.-X. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020 doi: 10.1056/nejmoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zhang J.-J., Dong X., Cao Y.-Y., Yuan Y.-D., Yang Y.-B., Yan Y.-Q. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. 2020:14238. doi: 10.1111/all. [DOI] [PubMed] [Google Scholar]
- 205.Fang L., Karakiulakis G., Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir. Med. 2020;8(4) doi: 10.1016/s22132600(20)30116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Misra D.P., Agarwal V., Gasparyan A.Y., Zimba O. Rheumatologists’ perspective on coronavirus disease 19 (COVID-19) and potential therapeutic targets. Clin. Rheumatol. 2020 doi: 10.1007/s10067-020-05073-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Jarcho J.A., Ingelfinger J.R., Hamel M.B., D’Agostino R.B., Harrington D.P. Inhibitors of the renin-angiotensin-Aldosterone system and Covid-19. N. Engl. J. Med. 2020;382(25):2462–2464. doi: 10.1056/NEJMe2012924. Epub 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Smyth L.J., Cañadas-Garre M., Cappa R.C. Genetic associations between genes in the renin-angiotensin-aldosterone system and renal disease: a systematic review and meta-analysis. BMJ Open. 2019;9 doi: 10.1136/bmjopen-2018-026777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Santos R.A., Simoes e Silva A.C., Maric C., Silva D.M., Machado R.P., de Buhr I. Angiotensin‐(1‐7) is an endogenous ligand for the G protein‐coupled receptor Mas. PNAS. 2003;100(14):8258–8263. doi: 10.1073/pnas.1432869100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Gurwitz D. Angiotensin receptor blockers as tentative SARSCoV-2 therapeutics. Drug Dev. Res. 2020 doi: 10.1002/ddr.21656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Lai C.C., Wang Y.H., Wang C.Y., et al. Comparative effects of angiotensin-converting enzyme inhibitors and angiotensin II receptor blockers on the risk of pneumonia and severe exacerbations in patients with COPD. Int. J. Chron. Obstruct. Pulmon. Dis. 2018;13:867–874. doi: 10.2147/COPD.S158634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Mortensen E.M., Nakashima B., Cornell J., Copeland L.A., Pugh M.J., Anzueto A., Good C., Restrepo M.I., Downs J.R., Frei C.R., Fine M.J. Population-based study of statins, angiotensin II receptor blockers, and angiotensin-converting enzyme inhibitors on pneumonia-related outcomes. Clin. Infect. Dis. 2012;55(11):1466–1473. doi: 10.1093/cid/cis733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Bean D.M., Kraljevic Z., Searle T., Bendayan R., Kevin O.G., Pickles A., Folarin A., Roguski L., Noor K., Shek A., Zakeri R., Shah A.M., Teo J.T., Dobson R.J. ACE-inhibitors and Angiotensin-2 Receptor Blockers are not associated with severe SARS-COVID19 infection in a multi-site UK acute Hospital Trust. Eur. J. Heart Fail. 2020 doi: 10.1002/ejhf.1924. Jun 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Gao C., Cai Y., Zhang K., Zhou L., Zhang Y., Zhang X., Li Q., Li W., Yang S., Zhao X., Zhao Y., Wang H., Liu Y., Yin Z., Zhang R., Wang R., Yang M., Hui C., Wijns W., McEvoy J.W., Soliman O., Onuma Y., Serruys P.W., Tao L., Li F. Association of hypertension and antihypertensive treatment with COVID-19 mortality: a retrospective observational study. Eur. Heart J. 2020;41(22):2058–2066. doi: 10.1093/eurheartj/ehaa433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Gnavi R., Demaria M., Picariello R., Dalmasso M., Ricceri F., Costa G. Therapy with agents acting on the renin-angiotensin system and risk of SARS-CoV-2 infection. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa634. May 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.de Abajo F.J., Rodríguez-Martín S., Lerma V., Mejía-Abril G., Aguilar M., García-Luque A., Laredo L., Laosa O., Centeno-Soto G.A., Ángeles Gálvez M., Puerro M., González-Rojano E., Pedraza L., de Pablo I., Abad-Santos F., Rodríguez-Mañas L., Gil M., Tobías A., Rodríguez-Miguel A., Rodríguez-Puyol D. MED-ACE2-COVID19 study group. Use of renin-angiotensin-aldosterone system inhibitors and risk of COVID-19 requiring admission to hospital: a case-population study. Lancet. 2020;395(10238):1705–1714. doi: 10.1016/S0140-6736(20)31030-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ingraham N.E., Barakat A.G., Reilkoff R., Bezdicek T., Schacker T., Chipman J.G., Tignanelli C.J., Puskarich M.A. Understanding the Renin-Angiotensin-Aldosterone-SARS-CoV-Axis: a comprehensive review. Eur. Respir. J. 2020 doi: 10.1183/13993003.00912-2020. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Vaduganathan M., Vardeny O., Michel T., McMurray J.J.V., Pfeffer M.A., Solomon S.D. Renin-angiotensin-Aldosterone system inhibitors in patients with Covid-19. N. Engl. J. Med. 2020;382(17):1653–1659. doi: 10.1056/NEJMsr2005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Casadevall A., Scharff M.D. Serum therapy revisited: animal models of infection and development of passive antibody therapy. Antimicrob. Agents Chemother. 1994;38(8):1695–1702. doi: 10.1128/aac.38.8.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Luke T.C., Kilbane E.M., Jackson J.L., Hoffman S.L. Meta-analysis: convalescent blood products for Spanish influenza pneumonia: a future H5N1 treatment? Ann. Intern. Med. 2006;145(8):599–609. doi: 10.7326/0003-4819-145-8-200610170-00139. [DOI] [PubMed] [Google Scholar]
- 221.Bloch E.M., Shoham S., Casadevall A., Sachais B.S., Shaz B. Deployment of convalescent plasma for the prevention and treatment of COVID-19. J. Clin. Invest. 2020 doi: 10.1172/JCI138745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Casadevall A., Pirofski L.A. The convalescent sera option for containing COVID-19. J. Clin. Invest. 2020;130(4):1545–1548. doi: 10.1172/JCI138003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., Wang X., Yuan J., Li T., Li J., Qian S., Hong C., Wang F., Liu Y., Wang Z., He Q., Li Z., He B., Zhang T., Fu Y., Ge S., Liu L., Zhang J., Xia N., Zhang Z. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020:ciaa344. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020 doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Duan K., Liu B., Li C., Zhang H., Yu T., Qu J. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. U. S. A. 2020:202004168. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Carr A.C., Maggini S. Vitamin C and immune function. Nutrients. 2017;9(11) doi: 10.3390/nu9111211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Hemilä H., Chalker E. Vitamin C can shorten the length of stay in the ICU: a metaanalysis. Nutrients. 2019;11:708. doi: 10.3390/nu11040708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Marik P.E., Khangoora V., Rivera R., Hooper M.H., Catravas J. Hydrocortisone, vitamin C, and thiamine for the treatment of severe sepsis and septic shock: a retrospective before-after study. Chest. 2017;151:1229–1238. doi: 10.1016/j.chest.2016.11.036. [DOI] [PubMed] [Google Scholar]
- 229.Colunga-Biancatelli R.M.L., Berrill M., Marik P.E. The antiviral properties of vitamin C. Expert Rev. Anti. Ther. 2020;18(2):99–101. doi: 10.1080/14787210.2020.1706483. [DOI] [PubMed] [Google Scholar]
- 230.Boretti A., Banik B.K. Intravenous Vitamin C for reduction of cytokines storm in Acute Respiratory Distress Syndrome [published online ahead of print, 2020 Apr 21] PharmaNutrition. 2020;12:100190. doi: 10.1016/j.phanu.2020.100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Cheng R.Z. Can early and high intravenous dose of vitamin C prevent and treat coronavirus disease 2019 (COVID-19)? Med Drug Discov. 2020;5:100028. doi: 10.1016/j.medidd.2020.100028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Carr A.C. A new clinical trial to test high-dose vitamin C in patients with COVID-19. Crit. Care. 2020;24(1):133. doi: 10.1186/s13054-020-02851-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Brighenti S., Bergman P., Martineau A.R. Vitamin D and tuberculosis: where next? J. Intern. Med. 2018 doi: 10.1111/joim.12777. [DOI] [PubMed] [Google Scholar]
- 234.Martineau A.R., Jolliffe D.A., Hooper R.L. Vitamin D supplementation to prevent acute respiratory tract infections: systematic review and meta-analysis of individual participant data. BMJ. 2017;356:i6583. doi: 10.1136/bmj.i6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Hastie C.E., Mackay D., Ho F. Vitamin d concentrations and COVID-19 infection in UK Biobank. Diabetes Metab. Syndr. 2020;14(4):561–565. doi: 10.1016/j.dsx.2020.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Te Velthuis A.J., vanden Worm S.H., Sims A.C., Baric R.S., Snijder E.J., van Hemert M.J. Zn(2+) inhibits coronavirus and arterivirus RNA polymerase activity in vitro and zinc ionophores block the replication of these viruses in cell culture. PLoS Pathog. 2010;6(11) doi: 10.1371/journal.ppat.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Biaggio V.S., Pérez Chaca M.V., Valdéz S.R., Gómez N.N., Gimenez M.S. Alteration in the expression of inflammatory parameters as a result of oxidative stress produced by moderate zinc deficiency in rat lung. Exp. Lung Res. 2010;36:31–44. doi: 10.3109/01902140903061787. [DOI] [PubMed] [Google Scholar]
- 238.Bao S., Knoell D.L. Zinc modulates cytokine-induced lung epithelial cell barrier permeability. Am. J. Physiol. Lung Cell Mol. Physiol. 2006;291:L1132–L1141. doi: 10.1152/ajplung.00207.2006. [DOI] [PubMed] [Google Scholar]
- 239.Xue J., Moyer A., Peng B., Wu J., Hannafon B.N., WQ ding. Chloroquine is a zinc ionophore. PLoS One. 2014;9 doi: 10.1371/journal.pone.0109180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Finzi E. Treatment of SARS-CoV-2 with high dose oral zinc salts: a report on four patients [published online ahead of print, 2020 Jun 6] Int. J. Infect. Dis. 2020 doi: 10.1016/j.ijid.2020.06.006. S1201-9712(20)30441-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Fantini J., Chahinian H., Yahi N. Synergistic antiviral effect of hydroxychloroquine and azithromycin in combination against SARS-CoV-2: what molecular dynamics studies of virus-host interactions reveal. Int. J. Antimicrob. Agents. 2020 doi: 10.1016/j.ijantimicag.2020.106020. [DOI] [PubMed] [Google Scholar]
- 242.Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R., Avanzato V., Bushmaker T., Flaxman A., Ulaszewska M., Feldmann F., Allen E.R., Sharpe H., Schulz J., Holbrook M., Okumura A., Meade-White K., Pérez-Pérez L., Bissett C., Gilbride C., Williamson B.N., Rosenke R., Long D., Ishwarbhai A., Kailath R., Rose L., Morris S., Powers C., Lovaglio J., Hanley P.W., Dana Scott D., Saturday G., de Wit E., Gilbert S.C., Munster V.J. ChAdOx1 nCoV-19 vaccination prevents SARS-CoV-2 pneumonia in rhesus macaques. bioRxiv. 2020 doi: 10.1101/2020.05.13.093195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Oxford . 2020. Oxford COVID-19 Vaccine Begins Human Trial Stage.http://www.ox.ac.uk/news/2020-04-23-oxford-covid-19-vaccine-begins-human-trial-stage [Google Scholar]
- 244.Moderna . 2020. Moderna Announces Positive Interim Phase 1 Data for Its mRNA Vaccine (mRNA-1273) Against Novel Coronavirus.https://www.modernatx.com/modernas-work-potential-vaccine-against-covid-19 [Google Scholar]
- 245.Zhu F.-C., Li Y.-H., Guan X.-H., Hou L.-H., Wang W.-J., Li J.-X., Wu S.-P., Wang B.-S., Wang Z., Wang L., Jia S.-Y., Jiang H.-D., Wang L., Jiang T., Hu Y., Gou J.-B., Xu S.-B., Xu J.-J., Wang X.-W., Wang W., Chen W. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. 2020 doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Ledford H., Van Noorden R. High-profile coronavirus retractions raise concerns about data oversight. Nature. 2020;582:160. doi: 10.1038/d41586-020-01695-w. [DOI] [PubMed] [Google Scholar]
- 247.Kalil A.C. Treating COVID-19—off-label drug use, compassionate use, and randomized clinical trials during pandemics. JAMA. 2020 doi: 10.1001/jama.2020.4742. [DOI] [PubMed] [Google Scholar]
- 248.Kanoh S., Rubin B.K. Mechanisms of action and clinical application of macrolides as immunomodulatory medications. Clin. Microbiol. Rev. 2010;23(3):590–615. doi: 10.1128/CMR.00078-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Vincent M.J., Bergeron E., Benjannet S., Erickson B.R., Rollin P.E., Ksiazek T.G., et al. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005;2:69. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Vankadari N. Arbidol: a potential antiviral drug for the treatment of SARS-CoV-2 by blocking trimerization of the spike glycoprotein. Int. J. Antimicrob. Agents. 2020:105998. doi: 10.1016/j.ijantimicag.2020.105998. [DOI] [PMC free article] [PubMed] [Google Scholar]