Abstract
Nutrient transporters, being polytopic membrane proteins, are believed, but not formally shown, to traffic from their site of synthesis, the ER, to the plasma membrane through Golgi‐dependent vesicular trafficking. Here, we develop a novel genetic system to investigate the trafficking of a neosynthesized model transporter, the well‐studied UapA purine transporter of Aspergillus nidulans. We show that sorting of neosynthesized UapA to the plasma membrane (PM) bypasses the Golgi and does not necessitate key Rab GTPases, AP adaptors, microtubules or endosomes. UapA PM localization is found to be dependent on functional COPII vesicles, actin polymerization, clathrin heavy chain and the PM t‐SNARE SsoA. Actin polymerization proved to primarily affect COPII vesicle formation, whereas the essential role of ClaH seems indirect and less clear. We provide evidence that other evolutionary and functionally distinct transporters of A. nidulans also follow the herein identified Golgi‐independent trafficking route of UapA. Importantly, our findings suggest that specific membrane cargoes drive the formation of distinct COPII subpopulations that bypass the Golgi to be sorted non‐polarly to the PM, and thus serving house‐keeping cell functions.
Keywords: Aspergillus nidulans, polarity, secretion, traffic, translocation
Subject Categories: Membrane & Intracellular Transport
Nutrient transporter translocation to the cell membrane operates via a novel trafficking route that does not involve functioning of the Golgi in fungi.
Introduction
Plasma membrane (PM) transporters mediating the selective cellular uptake or efflux of solutes and drugs are essential proteins in all organisms. The first step in their biogenesis, being polytopic transmembrane proteins, is their co‐translational translocation into the membrane of the endoplasmic reticulum (ER). The current belief is that after translocation into the ER, transporters are sorted into nascent ER‐exit sites, pack into budding COPII vesicles that fuse to the cis‐Golgi and then reach the trans‐Golgi network (TGN) via Golgi maturation 1, 2, 3. From the TGN, transporters are thought to be secreted towards the PM, similar to other membrane cargoes, either directly or indirectly via the endosomal compartment, in AP‐1/clathrin coated vesicles, the trafficking of which is controlled by multiple Rab GTPases and the microtubule cytoskeleton 4, 5. However, some lines of evidence support that specific transporters might not follow known conventional Golgi and post‐Golgi dependent routes. For example, genetic knock‐out of proteins involved in TGN‐dependent membrane cargo sorting (e.g. Arfrp1, golgin‐160 or AP‐1) leads to accumulation of the insulin‐regulated GLUT4 glucose carrier in the PM, rather than retention in the Golgi or other intracellular compartments, suggesting the presence of alternative routes out of the TGN, or even Golgi‐independent mechanisms 6. In addition, kinesin motor proteins or microtubule disruption has a moderate or no effect on GLUT4 accumulation at the PM 7, 8. Additionally, it has been shown recently that neosynthesized GLUT4 is sorted to the PM from an early secretory compartment, bypassing the TGN 9. Noticeably also, a specific form of the CFTR transmembrane protein (ΔF508‐CFTR), an ATP‐binding cassette (ABC) transporter that functions as a low conductance Cl− selective channel associated with cystic fibrosis, has been formally shown to translocate to the PM via Golgi bypass under specific stress conditions 10, 11. Finally, the mammalian potassium channel Kv2.1 is known to be sorted to the PM of the initial segment (AIS) of neurons via a mechanism that bypasses the Golgi 12. In fact, no formal evidence exists on whether neosynthesized transporters traffic through the Golgi/TGN compartment in any type of cell. A possible explanation for this might be that transporter passage and exit from the Golgi is very rapid, never leading to accumulation of sufficient steady‐state levels for detection with standard fluorescence microscopy. However, evidence against this explanation is also the fact that no mutation or specific condition has been shown to block PM transporters in the Golgi. In contrast, several mutations affecting the proper folding or altering specific motifs in transporters are well‐known to lead to retention in the ER, which is often associated with ubiquitination‐dependent turnover by proteasome degradation and/or selective autophagy 13, 14. Unconventional trafficking routes that bypass the Golgi have also been described for a handful of PM transmembrane proteins other than transporters and are collectively classified as type IV unconventional protein secretion (UPS) 10, 11, 15, 16, 17.
In the course of experiments addressing cargo trafficking in the model fungus Aspergillus nidulans, we noticed that sorting to the PM of the well‐studied UapA transporter 18 is not affected by repression of transcription of the AP‐1 adaptor complex, a key effector of conventional secretion of most membrane cargoes 19. This result contrasted to those obtained with polarly localized membrane cargoes (e.g. Chitin synthase ChsB, Synaptobrevin SynA, or lipid flippases DfnA or DfnB), which all needed AP‐1 for their apical localization 20, 21. Interestingly, endocytosis of UapA and other nutrient transporters has been shown to involve a mechanism distinct from that of apical membrane cargoes. In particular, transporters are internalized all along the hyphal membrane via a clathrin‐dependent, but AP‐2‐independent mechanism, destined for vacuolar degradation in response to physiological or stress signals 19. In contrast, polar cargoes are constitutively recycled in the apical region of hyphae via clathrin‐independent, but AP‐2‐dependent, endocytosis, a process essential for polarity maintenance and filamentous polar growth 19, 20. Thus, current evidence suggests that subcellular trafficking routes of nutrient transporters, which are localized homogenously along the hyphal membrane, might be mechanistically distinct from that of polarly localized apical membrane cargoes. Several additional observations prompted us to dissect the mechanism of biogenesis of transporters. First, we have never obtained any genetic or microscopic evidence that A. nidulans transporters pass from Golgi‐like structures. Second, all A. nidulans transporters studied in our lab (more then 30, all relative to uptake of nutrients such as purines, pyrimidines, amino acids and carboxylic acids) are not glycosylated. A similar observation holds true for most nutrient transporters studied in Saccharomyces cerevisiae. Finally, in cases where specific transporters in other systems have been reported to localize in the trans‐Golgi network (TGN), the experiments described do not distinguish whether this is the result of transporter recycling from the PM or secretion of de novo made molecules from the ER.
In the present work, we used UapA as a model cargo to investigate its dynamic trafficking to the PM. For doing so, we developed a controllable genetic system to study the trafficking of de novo made UapA when different steps of the conventional secretion pathway are tightly repressed. This system, combined with relative co‐localization studies of UapA and molecular markers of the secretory pathway, showed that UapA localization to the PM occurs without the need of Golgi‐ or post‐Golgi‐dependent cargo sorting mechanisms. We further showed that UapA localization to the PM is dependent on the formation of COPII vesicles, clathrin, actin polymerization and the PM t‐SNARE SsoA. To extend our findings, we also performed key experiments with additional transporters of A. nidulans and found that in all cases, their trafficking mechanism is similar to that identified with UapA. We finally discuss our findings within the general context of trafficking of nutrient transporter in polarized eukaryotic cells.
Results
In vivo trafficking of neosynthesized UapA
We established conditions to follow ab initio the traffic and subcellular localization of de novo made transporters in single hyphae of A. nidulans. As a prototype transporter‐cargo, we used the well‐studied UapA purine transporter, functionally tagged with GFP 18. Conidiospores of an otherwise wild‐type A. nidulans strain, containing an in‐locus targeted uapA‐gfp allele, were allowed to germinate overnight (14–16 h, 25°C) under conditions that repress uapA‐gfp transcription. This is achieved using either the native uapA promoter or the regulatable alcA p promoter (for details, see Materials and Methods). After this period, uapA‐gfp transcription was induced in germlings (i.e. very young hyphae) via a shift to derepressing conditions (0–8 h). Derepressed protein levels of UapA‐GFP driven by either the native or the alcA p promoter are very similar, at all‐time points. The strategy for repression–derepression of UapA synthesis is depicted in Fig 1A.
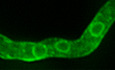
Figure 1. Subcellular localization of neosynthesized UapA.
-
ACartoon depicting the strategy for following the trafficking of neosynthesized UapA. The subcellular localization of UapA is shown in green. Red circles indicate positioning of nuclei. (for more details, see main text).
-
BIn vivo epifluorescence microscopy following de novo expressed UapA‐GFP in a single growing germling at 70, 80, 100, 120, 140, 160 and 170 min after derepression of transcription via its native uapA promoter (for details, see text and Materials and Methods). Notice that in the growing germling tip UapA localizes in a membranous mesh and some cytosolic puncta, whereas in more tip‐distal parts of the germlings, UapA is in PM‐localized puncta (see white arrows), which progressively become more abundant so that at more posterior areas the PM is homogeneously labelled.
-
CIn vivo epifluorescence microscopy following de novo expressed UapA‐GFP in a germling maturing to young hypha at 180, 240, 300, 360 and 400 min after derepression of transcription via its native uapA promoter. During this developmental transition, UapA apical localization is gradually diminished. As the cell tip acquires a faster growth rate, UapA is completely retracted from the hyphal apex (see zoom‐in panels on the right and white arrows).
-
DIn a mature hyphal cell, UapA is no longer found in the PM of the extreme apical compartment, but in a cytoplasmic membrane network resembling the ER. As the distance from the apex increases, UapA progressively populates the PM as cortical puncta (see white arrows).
-
ECartoon depicting the rearrangement of actin and UapA at the tip of A. nidulans during germling to hyphal maturation. In germlings, dense arrays of actin cables are present in the apex, known as apical actin array (AAA). Actin patches are concentrated, but not restricted, to the apex. In mature hyphae, a core of actin appears in the position of the Spitzenkorper and actin patches are restricted to the subapical endocytic collar (≈2 μm behind the apex). Actin cables are retracted from the apex and shifted to a more distal region of the hyphal tip, the subapical actin web (SAW). In the apex, actin cables are now found associated mostly as sparse arrays across the cell cortex. The localization of UapA during hyphal maturation seems to follow the pattern of actin cables 26, 27.
-
FIn vivo epifluorescence microscopy following neosynthesized alcA p‐UapA‐GFP in a single hypha at 120, 130, 140, 150, 160 and 170 min, under overexpressing (derepression/ethanol‐induction) conditions, as described in Materials and Methods. Notice that UapA labels the perinuclear ER rings (white arrows).
-
GIn vivo epifluorescence microscopy following de novo expressed alcA p‐GFP‐SynA in a single young hyphal cell, at 60, 70, 80, 90 and 100 min after transcriptional derepression (for strain details, see Materials and Methods). SynA is a standard polar membrane cargo that traffics to the hyphal tip via Golgi‐dependent secretion. Notice the very distinct GFP fluorescent signals obtained using UapA (non‐polar membrane network and PM) versus SynA (Golgi‐like puncta and polar depositioning at the tip; see also alter).
-
HSimilar experiment as in (B) performed in an isogenic strain where ArtA, the arrestin required for UapA endocytosis, is genetically depleted.
Figure 1B and C shows examples of images obtained following UapA‐GFP localization, when expressed from its native promoter, in single germlings (young hyphae). Similar results were obtained in several experiments. UapA‐GFP appears as cytoplasmic weak fluorescence at 70 min after the onset of transcription. Subsequently, UapA‐GFP labels a membranous cytoplasmic network (80–120 min) resembling A. nidulans ER membranes 14, 22 and progressively migrates to the cell periphery, where it appears as dispersed cortical puncta (120–160 min). With time, cortical puncta increase in number to eventually label the entire PM of hyphae in a rather homogeneous manner (170 min). The timing of appearance of UapA molecules in the periphery of cells is compatible with previous studies which show that UapA transport activity reaches its steady state 3 h after transcriptional derepression 23. Importantly, we did not detect fluorescence‐labelled cytoplasmic structures resembling early or late Golgi compartments (see later). Relatively increased accumulation of UapA in the PM of the growing apical region (or in the apex of secondary germ tubes emerging from the conidiospore head) was observed in young germlings (see 140‐ to 170‐min samples in Fig 1B). Noticeably, as germlings grow longer, the relatively increased apical cortical localization of UapA is progressively diminished (Fig 1C, see 240–400 min), and in longer more mature hyphae is lost (Fig 1D). Thus, in long hyphae, UapA is imaged to clearly label an ER‐like membranous network at the apical region, but in subapical regions, it progressively populates the PM at distinct puncta that are rather unevenly distributed. Interestingly, the change in the localization of UapA at the apical region is co‐incident to important cellular changes that underlie the developmental transition from slow‐growing germlings to fast‐growing mature hyphae observed in A. nidulans and other filamentous ascomycetes 24. Transition to 5‐ to 10‐fold faster apical growth is related to the formation of an apical secretory vesicular organizing centre, known as the Spitzenkörper, and re‐organization of apical actin cables from a dense network (in germlings) to less dense and cortical localization (in mature hyphae) (25, 26; also depicted in Fig 1E). During this transition, an apical actin array also retracts to a subapical actin web, compatible with the change in the apical localization of UapA in germlings versus mature hyphae, respectively 26, 27. These observations will become more apparent later, when actin polymerization is shown to be essential for UapA trafficking. Overall, results highlighted in Fig 1B–D suggested that neosynthesized UapA labels the ER, as probably expected, but it then appears in the PM with no indication of passing from other recognizable cytoplasmic structures, such as the Golgi or motile endosomes 28.
To acquire additional evidence that UapA labels mostly the ER and not Golgi‐like structures, we also followed the localization of de novo made UapA‐GFP under conditions that the transporter is significantly overexpressed. For this, we made use of the alcA p promoter expressed under conditions of derepression plus ethanol‐induction (for details, see Materials and Methods). Results shown in Fig 1F revealed that UapA overexpression labels, in addition to a diffuse membrane network, the characteristic perinuclear ER rings that are often seen in fungi (see 130‐ to 170‐min samples). Notably, however, UapA‐GFP overexpression did not label Golgi‐like or other punctuate structures.
In addition to UapA, we also examined the dynamic localization of a standard de novo made apical marker, such as Synaptobrevin A tagged with GFP (GFP‐SynA; 25). In this case, unlike UapA, we were able to detect SynA in several Golgi‐like and other punctuate cytoplasmic structures (Fig 1G). Some of the punctuate structures at later time points, when SynA has already reached its apical localization, showed endosome‐like motility, as expected due to apical recycling 25. Thus, the picture of UapA versus SynA secretion was strikingly different, the former marking mostly the ER network and eventually the PM, the latter marking the ER, Golgi‐like or other punctuate structures, and the tip of the apical region. Notably, UapA appearance in the PM does not seem to occur via lateral diffusion from the tip area. This is concluded based on two observations. First, there is no apparent continuous gradient of UapA from the apical area towards subapical parts, but instead isolated UapA cortical puncta (see arrows in Fig 1B, at 140 and 160 min). Second, the tip region in mature hyphae possesses practically no UapA, while the subapical compartments show several cortical and unevenly distributed UapA puncta (Fig 1D). These observations are also compatible with previous reports showing that membrane later diffusion of large transmembrane proteins is extremely slow or short distance 29, 30. Thus, the simplest scenario is that UapA localization to the PM takes place by short‐range lateral sorting from the ER network.
Previous studies have shown that UapA can be endocytosed and degraded in the vacuole in response to its activity or due physiological signals 31, 32. The latter study has also strongly supported that internalized UapA is not recycled back to the PM after endocytosis. Also, later in this work, we further show that UapA localization is independent of the function of recycling endosomes (see Appendix Fig S3). Thus, the subcellular localization of UapA shown in Fig 1B–D reflects strictly secretion of neosynthesized UapA. To exclude any doubt on the role of endocytosis and recycling in the images obtained, we repeated here the microscopic analysis in a strain that is genetically blocked in UapA ubiquitination and endocytosis, and thus to possible recycling, due to a null mutation in the specific arrestin adaptor ArtA 32. The result, shown in Fig 1H, was practically identical to the one obtained in the wild‐type strain.
Neosynthesized UapA does not co‐localize with Golgi markers
To obtain stronger evidence that de novo made UapA does not pass from the Golgi compartments as suggested by results highlighted in Fig 1, we performed co‐localization studies of UapA‐GFP with Golgi‐specific molecular markers tagged with mRFP or mCherry. In A. nidulans, fluorescent‐tagged protein markers distinguishing “early” (corresponding to cis) and “late” (corresponding to trans) Golgi have been very rigorously established 21, 33. Early Golgi is commonly marked with SedVSed5 or GeaAGea1 and late Golgi/TGN with HypBSec7 or PHOSBP. Both compartments appear as numerous, rather immotile, cytoplasmic puncta, all along the length of hyphae (see cartoon in lower left panel of Fig 2). Both compartments are transient, consistent with the cisternae maturation model, where late Golgi has an average lifetime of approximately 2‐3 min 34. Figure 2A and B shows representative time course experiments of trafficking of neosynthesized UapA‐GFP, driven by the alcA p promoter, in strains co‐expressing either SedV (early Golgi) or PHOSBP (late Golgi/TGN). In both cases, we did not detect significant co‐localization of UapA with the Golgi markers used. The result depicted in Fig 2A and B was statistically supported as Pearson correlation coefficients for co‐localization were very low in both cases (PCC < 0.35, P < 0.0001). Notice that Pearson correlation coefficients ≤ 0.35 are often obtained in co‐localization studies of markers of distinct cytoplasmic compartments that are topologically close (e.g. ER and Golgi). Thus, our findings are taken as a significant indication that UapA might not pass from the Golgi.
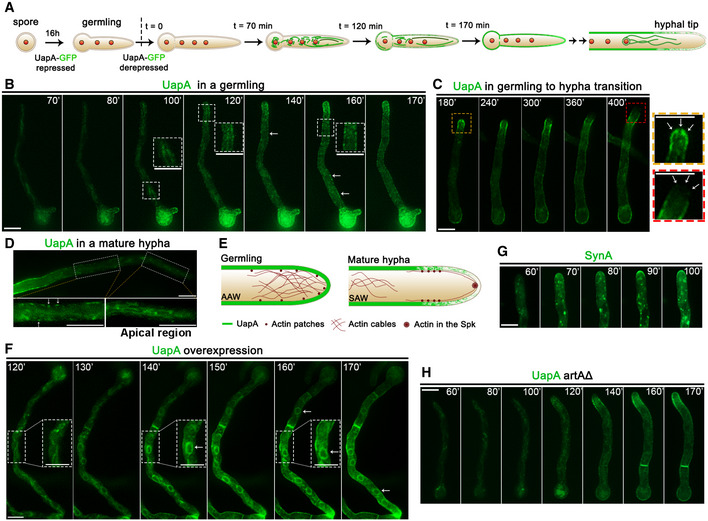
Figure 2. Neosynthesized UapA does not co‐localize with Golgi markers.
-
A, BCo‐localization analysis of neosynthesized alcA p‐UapA‐GFP with early (SedV) or late (PHOSBP) markers, respectively, tagged with mCherry or mRFP. Images show single hyphal cells. For conditions of transcriptional derepression, see Materials and Methods. Quantification of co‐localization was performed by calculating Pearson's correlation coefficient (PCC). One sample t‐test was performed to test the significance of differences in PCCs. Biological/technical replicates:2/9 for alcA p‐UapA‐GFP mCherry‐SedV and 2/8 for alcA p‐UapA‐GFP mRFP‐PHOSBP. For the definition of the two categories of replicates, see Materials and Methods. Results of quantification, shown on the middle, suggest that there is no significant overlapping fluorescent signal of UapA with SedV (PCC = 0.25 ± 0.09, P < 0.0001) or with PHOSBP (PCC = 0.34 ± 0.06, P < 0.0001), as PCC values close to 0.2–0.3 are also commonly obtained when distinct compartments (e.g. ER and early Golgi or early and late Golgi) are followed with different fluorophores. Scale bars: 2 μm.
-
C, DCo‐localization analysis of neosynthesized alcA p‐GFP‐SynA, used as a conventional cargo that traffics through the Golgi, with early (SedV) or late (PHOSBP) markers tagged with mCherry or mRFP, respectively. Images show single hyphal cells. For conditions of transcriptional derepression, see Materials and Methods. Quantification of co‐localization and statistical analysis was performed as in (A, B). Biological/technical replicates: 2/10 for each strain. Statistical analysis showed significant co‐localization of SynA with the late Golgi marker (PCC = 0.67 ± 0.04, P < 0.0001), but not with early Golgi marker (PCC = 0.37 ± 0.06, P < 0.0001). An explanation for lack of co‐localization of SynA with SedV is given in the text. Scale bars: 2 μm. The cartoon at the bottom depicts the localization of early or late markers as established here in several previous studies (see 20, 21).
Source data are available online for this figure.
To obtain more concrete evidence that we have not missed the time window for passage of UapA from the Golgi, we repeated essentially the same co‐localization experiment using a standard secreted cargo, namely SynA (synaptobrevin A), which is localized polarly in the apical region of hyphae via the conventional Golgi‐dependent pathway 21. Figure 2C and D represents experiments showing that de novo made SynA, expressed from alcA p, is significantly co‐localized with the late Golgi/TGN PHOSBP marker (PCC = 0.67, P < 0.0001), but only little with the early Golgi SedV marker (PCC = 0.37, P < 0.0001). These results suggest that SynA passes from the late Golgi/TGN, as might be expected (but never shown before in A. nidulans). On the other hand, we did not detect significant co‐localization of cargoes (SynA or UapA) at the level of the early Golgi. Given that the presence of SynA at the TGN would, in principle, necessitate sorting from the early Golgi, this means that our methodology is not sensitive enough to detect transient passage from early secretory compartments. Thus, at this point, this technical limitation forced us to consider that a fraction of UapA might still be sorted at least to the early Golgi. Thus, we subsequently looked for more rigorous evidence supporting the suspected Golgi bypass of UapA trafficking.
Localization of de novo made UapA in the PM in the absence of conventional secretion
To obtain more compelling evidence for the apparent bypass of Golgi by UapA, we followed the trafficking of neosynthesized UapA‐GFP in genetic backgrounds where distinct steps of the Golgi‐dependent conventional secretory pathway were blocked for a defined period of time, via tight transcriptional repression of specific genes. For this, we constructed strains where the promoters of endogenous genes encoding SedVSed5 or GeaAGea1 (early/medial‐Golgi), HypBSec7 (late‐Golgi/TGN), RabERab11 (TGN/post‐Golgi), AP‐1σ (post‐Golgi secretion) and ClaH (clathrin heavy chain/post‐Golgi secretion) were replaced, through targeted homologous recombination, by the thiA promoter (thiA p). This promoter is tightly repressible upon addition of thiamine in the growth medium (Fig 3A). Western blot analysis showed that addition of thiamine to a culture of conidiospores of A. nidulans at the onset of incubation (ab initio) leads to dramatic reduction or absence of the relative protein after 12 h of germination (Fig 3B). At 12‐14 h, when secretory proteins are not or hardly detected in Western blots, growth of germlings stops (Fig 3C) and the tip of A. nidulans swells in mutants lacking SedV, GeaA, RabE or ClaH, but less so in the case of HypB or AP‐1 (Fig 3D). This dramatic morphological change in the apical part of growing hyphae is strong evidence that Golgi‐ and post‐Golgi‐dependent cargo trafficking is inhibited because cargo secretion in filamentous fungi is essential for maintaining polarity and growth. During establishment of repression of secretion, the transcription of UapA from its endogenous promoter is also kept fully repressed, using ammonium ions as sole N source 35. After establishment of repression and blocking of secretion (12‐ to 14‐h germination), as evidenced by Western blot analysis, swelling of the tip and arrested growth, the transcription of UapA is derepressed essentially as previously described, via its native promoter. This system allowed us to follow UapA localization, while the conventional pathway is dramatically blocked at distinct Golgi‐dependent steps.
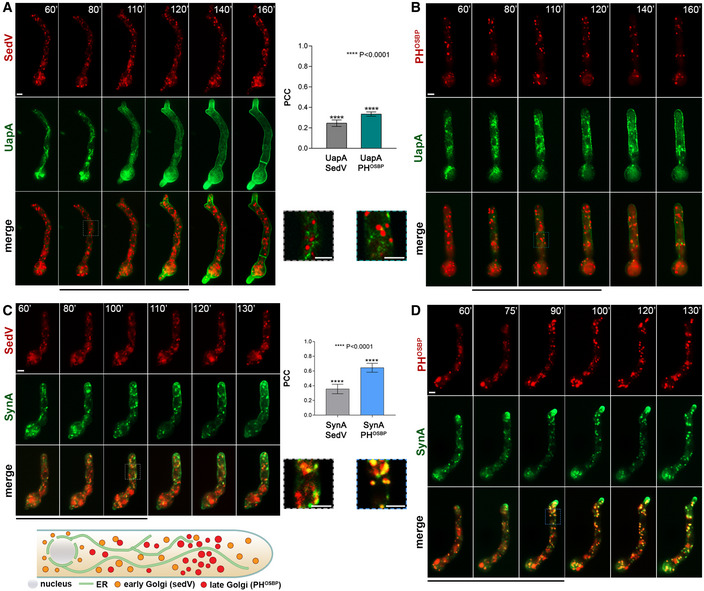
Figure 3. Localization of neosynthesized UapA in the PM when conventional secretion is blocked.
-
ASchema depicting the strategy for blocking conventional secretion.
-
BKey endogenous genes controlling Golgi (sedV, geaA, hypB) or post‐Golgi trafficking (rabE, ap1 σ , claH) were genetically replaced by versions transcribed under the highly repressible thiA p promoter via targeted homologous recombination. In the absence of thiamine from the growth medium (derepressed conditions), the relative proteins are expressed, while upon addition of thiamine at the onset of conidiospore germination (ab initio repression), the expression of these proteins is tightly repressed. Proteins are detected by a standard Western blot analysis using either anti‐FLAG or anti‐GFP antibodies for Golgi and Post‐Golgi proteins. Equal loading and protein steady‐state levels are normalized against the amount of actin, detected with a specific antibody.
-
CIn the absence of thiamine from the growth medium (derepressed conditions), the corresponding strains grow nearly as an isogenic wild‐type control, although a delay in growth is observed for thiA p ‐sedV and less so for thiAp‐ap1 σ (upper row). In the presence of thiamine to the growth medium, most cells do not form colonies, except for thiA p ‐hypB which forms a compact slow‐growing colony (lower row).
-
DMicroscopic examination of the corresponding strains under thiamine (repressing conditions) shows that, in most cases, the apical region of germlings is enlarged and growth is arrested. This morphological phenotype, taken as a strong indication of blocked secretion, is more evident in thiA p ‐sedV and thiA p ‐rabE but concerns all strains, except for thiAp‐hypB. Scale bar: 5 μm.
-
ESubcellular localization of UapA, after 6–8 h of initiation of transcription, via its native uapA promoter, while conventional sedV, geaA or hypB transcription is repressed by thiamine ab initio. Scale bars: 2 μm. Notice that UapA‐GFP translocates in the PM of germlings in all cases. Results shown are confirmed by quantification (right panel) of UapA‐GFP PM/cytosolic intensity ratios for the four strains (for details, see Materials and Methods). Mean PM/cytosolic intensity ratios for wild‐type, thiA p ‐sedV, thiA p ‐geaA and thiA p ‐hypB are 0.53 ± 0.02, 0.52 ± 0.02, 0.52 ± 0.02 and 0.52 ± 0.02, respectively. For the statistical analysis, Tukey's multiple comparison test was performed (one‐way ANOVA). No statistical significance was found between the wild type and each of the mutant strains. Biological/technical replicates: 3/15 for wild type, 3/18 for thiA p ‐sedV, 3/15 for thiA p ‐geaA and 3/18 for thiA p ‐hypB.
-
FalcA p‐UapA‐GFP, under derepressing conditions, translocates to the PM of a strain carrying a thermosensitive mutation in SedVts both at the permissive (25°C) and restrictive temperature (42°C). A degree of increased degradation of UapA, caused by exposure to high temperature (42°C), is apparent as cytosolic puncta, which correspond to membrane aggregates and sorting to vacuoles. Scale bar: 5 μm.
-
GUnlike UapA, de novo made apical markers, such as ChsB or SynA, lose their polar localization, when the expression of sedV, geaA or hypB is repressed. Scale bars: 2 μm. Quantification: GFP‐ChsB PM/cytosolic and alcA p ‐GFP‐SynA PM/cytosolic intensity ratios are plotted to the right upper and lower panel, respectively. For GFP‐ChsB, mean PM/cytosolic intensity ratios are 0.60 ± 0.02 (wild type), 0.11 ± 0.01 (thiA p ‐sedV), 0.26 ± 0.02 (thiA p ‐geaA) and 0.34 ± 0.01 (thiA p ‐hypB). For alcA p ‐GFP‐SynA, mean PM/cytosolic intensity ratios for wild type, thiA p ‐sedV, thiA p ‐geaA and thiA p ‐hypB are 0.61 ± 0.03, 0.14 ± 0.02, 0.26 ± 0.03 and 0.39 ± 0.03, respectively. The statistical analysis was performed as in (E). A significant difference (****P < 0.0001) of GFP‐ChsB fluorescence in the apical PM membrane was found between the wild‐type and the three strains lacking the key‐Golgi proteins. This is also the case for SynA, as seen in the scatter plot on the lower right panel, where the fluorescence intensity of SynA is diminished (****P < 0.0001) when sedV, geaA and hypB are repressed. Biological/technical replicates: 3/15 for each strain.
Source data are available online for this figure.
Images shown in Fig 3E show that de novo made UapA is principally localized to the PM, even when essential for secretion and growth Golgi markers, such as SedV, GeaA and HypB, are absent due to transcriptional repression. This result is confirmed via relative quantification of translocation of UapA to the PM, after 6–8 h of derepression shown in the right panel of Fig 3E. While the timing of localization of UapA to the PM in the absence of HypB is similar to that obtained in a wild‐type background (i.e. steady‐state maximum localization at 4 h), in the absence of GeaA and mostly SedV, UapA secretion is delayed, requiring 6–8 h of transcriptional derepression before reaching massively the PM (Appendix Fig S1). This might be due to a more critical role for growth of early Golgi compared to late Golgi in Aspergillus. This assumption is in line with the observed more pronounced reduction in growth and swelling of the tip when SedV or GeaA is prepressed, compared to repression of HypB (see Fig 3C and D).
To further demonstrate the redundancy of SedV for UapA localization to the PM, we also made use a standard thermosensitive mutant of SedV 36. Figure 3F shows UapA localization to the PM takes place even at the restrictive temperature (42°C) where SedV activity is dramatically lost. The concurrent appearance of several cytoplasmic UapA‐GFP aggregates in this case is normal as elevated temperature is known to lead to UapA endocytosis and vacuolar turnover.
To formally show that our repression system is proper for detecting the necessity of Golgi in secretion, we also followed the secretion of conventional polar cargoes, such as SynA or ChsB (chitin synthase) in the absence of either SedV or HypB. SynA trafficking was driven by the regulatable alcA promoter, whereas that of ChsB by its native constitutive promoter. Thus, in the former case we could follow the localization of newly made SynA at expression periods similar to the ones we followed UapA (6–8 h), whereas in the latter case, we followed the steady‐state biogenesis of ChsB expressed from the onset of conidiospores incubation. Images and relative quantification of results depicted in Fig 3G showed that sorting of both polar cargoes to the apical region of hyphae is impaired when Golgi functioning is blocked. This validated our system as efficient in repressing Golgi‐dependent secretion, and thus confirmed that UapA localization to the PM bypasses sorting to the Golgi.
UapA translocation to the PM is COPII‐dependent
Given that UapA seems to traffic to the PM via a Golgi‐independent route, we asked whether ER exit is COPII‐dependent or COPII‐independent. To test trafficking dependence on COPII, we tested whether de novo made UapA secretion is blocked upon repression of Sec24, the major component of COPII‐coated vesicles that mediates cargo binding and vesicular exit from the ER. Figure 4A shows that repression of sec24 leads to growth arrest, reflected to inability of this strain to form colonies in the presence of thiamine. However, initial germination and germling formation are not arrested, which allowed us to examine the effect of Sec24 repression on de novo UapA localization. Figure 4B shows that after Sec24 repression is established (12 h), neosynthesized UapA‐GFP never reaches PM. Instead, UapA molecules remain in the ER membrane network and in cytosolic puncta. A similar Sec24‐dependent total block in secretion was observed, as expected, when we followed, as a control, the trafficking of an apical membrane cargo ChsB (Fig 4C). Quantification of PM localization of UapA and ChsB from relevant experiments (±thiamine) confirmed the essential role of Sec24 in the secretion of both cargoes (see right panels of Fig 1B and C).
Figure 4. COPII vesicle formation is required for ER‐exit and PM localization of UapA.
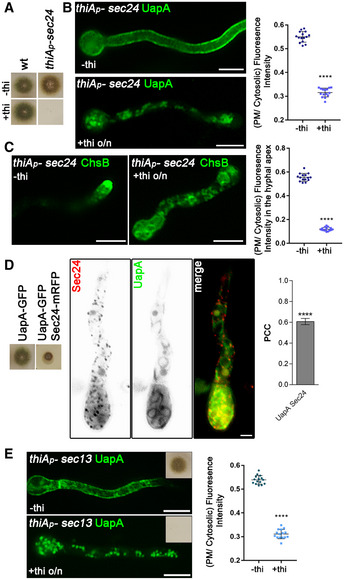
-
AGrowth test showing that Sec24 expression is essential for growth as its transcriptional repression by thiamine (+thi) leads to the absence of colony formation, despite initial germination (images at panels B and C). The strains shown are isogenic except for the sec24 locus. thiA p ‐sec24 signifies the strain where the endogenous sec24 promoter was replaced by the thiA p promoter.
-
BEpifluorescence microscopy analysis of the subcellular localization of UapA‐GFP under conditions where sec24 transcription is ab initio derepressed (upper panel) or repressed by thiamine (lower panel). o/n (overnight) means addition of thiamine from the onset of germination. Germination of conidiospores takes place until full repression of Sec24 is achieved (10‐12 h). Notice the total lack of PM‐associated signal of UapA under conditions of Sec24 repression. Scale bar: 5 μm. Quantification: In the scatter plot on the right, UapA‐GFP PM/cytosolic intensity ratios are quantified when sec24 is derepressed (−thi) or repressed (+thi). Mean PM/cytosolic intensity ratios are 0.55 ± 0.02 and 0.32 ± 0.02, respectively. To test the significance of differences, an unpaired t‐test was performed, which verified the significant difference (****P < 0.0001) in the presence of thiamine. Biological/technical replicates: 2/15 for each condition.
-
CEpifluorescence microscopy analysis of the subcellular localization of the apical marker chitin synthase (GFP‐ChsB) under sec24 derepressed or repressed conditions. Notice the loss of apical depositioning and the concurrent labelling of cytoplasmic foci and membranous structures under repression conditions. Scale bar: 5 μm. Quantification: GFP‐ChsB PM/cytosolic intensity ratios are plotted in cases where sec24 is derepressed (‐thi) or repressed (+thi). Mean values are 0.56 ± 0.03 and 0.12 ± 0.02, respectively. The fluorescence intensity of GFP‐ChsB is statistically lower (****P < 0.0001) when the expression of sec24 is repressed (statistical analysis as in B). Biological/technical replicates: 2/15 for each condition.
-
DCo‐localization analysis by epifluorescence microscopy of Sec24‐mRFP and de novo made UapA‐GFP (8 h derepressed transcription). Quantification of co‐localization was made by calculating Pearson's correlation coefficient. For the statistical analysis, one sample t‐test was used. Notice the significant co‐localization (PCC = 0.61 ± 0.07, ****P < 0.0001). Due to the partially functional mRFP‐tagged version of Sec24 (evident in the growth test shown on the left panel), UapA trafficking is significantly delayed, reflected in the appearance of prominent labelling of uniform perinuclear ER. Scale bar: 5 μm. Biological/technical replicates: 3/5.
-
EEpifluorescence microscopy analysis of the subcellular localization of UapA‐GFP under conditions where sec13 transcription is ab initio derepressed (left panel) or repressed by thiamine (right panel). The inserts in the upper right corner in both panels reflect growth tests showing that Sec13 repression leads to arrest in growth and absence of colony formation. Scale bars: 5 μm. All images reflect practically identical results obtained in several experiments. Quantification: UapA‐GFP PM/cytosolic intensity ratios are quantified when sec13 is derepressed (‐thi) or repressed (+thi), with mean values being 0.54 ± 0.02 and 0.31 ± 0.02, respectively. There is a significant difference on UapA PM fluorescence intensity (****P < 0.0001) in the absence of the COPII outer coat protein (statistical analysis as in B). Biological/technical replicates: 2/15 for each condition.
Source data are available online for this figure.
We also tested whether UapA and Sec24 interact, using relative co‐localization. Despite the partial functioning of Sec24‐mRFP, the appearance of Sec24‐specific puncta was typical of ERes 33 and co‐localized significantly (PCC = 0.6, P < 0.0001) with UapA‐labelled ER membranes (Fig 4D – Movie EV1). Unexpectedly, the co‐localization experiment also showed that UapA secretion is dramatically delayed in the strain expressing mRFP‐tagged Sec24. This delay is reflected in the prominent appearance of UapA‐labelled perinuclear ER rings, not seen in an untagged Sec24 background. Thus, the partial functioning Sec24‐mRFP fortuitously further confirmed that Sec24 is essential for proper UapA secretion.
To further confirm that conventional COPII formation is necessary for UapA secretion, we also tested the role of Sec13, an essential constituent of the outer coat of COPII. Images in Fig 4E and associated relative quantification (±thiamine) show that UapA sorting to the PM is absolutely dependent on the presence of Sec13, as repression of Sec13 synthesis led to the appearance of numerous cytosolic fluorescent puncta that resemble COPII‐prebudding structures. Thus, within the limits of our genetic and microscopic approaches, UapA‐specific COPII vesicles seem to contain the basic elements of standard COPII vesicles. However, unlike standard COPII vesicles, which are sorted to the cis‐Golgi, UapA‐specific COPII vesicles seem to bypass the Golgi, suggesting the existence of structurally and mechanistically distinct subpopulations of COPII vesicles. What differentiates these COPII subpopulations remains unknown, but some hypotheses are discussed later.
UapA translocation to the PM is RabERab11 and AP‐1 independent but requires clathrin
We have previously reported that UapA sorting to the PM is independent of AP‐1 and AP‐3 complexes, or clathrin light chain ClaL, which are essential for post‐Golgi traffic of several other cargoes, but depends on clathrin heavy chain (ClaH) 19. Here, we examined whether ab initio established repression of AP‐1, RabERab11 and ClaH has an effect on the localization of neosynthesized synthesized UapA. Images in Fig 5A and associated relative quantification of results show that repression of AP‐1 and RabE did not block the translocation of de novo made UapA to the PM. In contrast, ClaH repression led to dramatic impairment of translocation to the PM, as de novo made UapA remained trapped in a cytosolic mesh and never reached the periphery of cells. This result confirms that while ClaH is critical for the trafficking of de novo made UapA to the PM, two key TGN‐localized upstream effectors of ClaH function, AP‐1 and RabE, proved redundant for UapA secretion. In contrast to the redundancy of RabE and AP‐1, the polar localization of a standard apical cargo (e.g. SynA) was abolished in the absence of all three TGN‐related proteins (Fig 5B), in line with previous reports 20.
Figure 5. UapA translocation to the PM takes place without involvement of conventional post‐Golgi machinery.
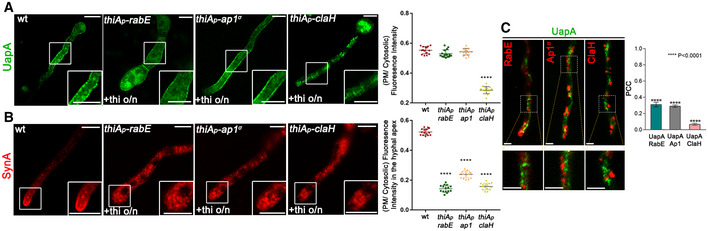
-
AEpifluorescence microscopy analysis examining the subcellular localization of de novo made UapA‐GFP, after 6–8 h of initiation of transcription in strains where the expression of RabE, AP‐1σ or ClaH (thiA p ‐rabE, thiA p ‐ap1 σ or thiA p ‐claH) has been repressed ab initio (o/n) by addition of thiamine. Notice that when rabE or ap1 σ is repressed UapA‐GFP still reaches the PM. In contrast, repression of claH abolishes labelling of the PM and leads to the cytosolic puncta and a membranous network. Scale bars: 5 μm. Quantification: UapA‐GFP PM/cytosolic intensity ratios are plotted on the right. Mean ratios for wild type, thiA p ‐rabE, thiA p ‐ap1 σ and thiA p ‐claH are 0.55 ± 0.02, 0.53 ± 0.03, 0.54 ± 0.02 and 0.29 ± 0.02, respectively. For the statistical analysis, Tukey's multiple comparison test was performed (one‐way ANOVA). After 6‐8 h of transcriptional derepression, UapA‐GFP fluorescence to the PM does not change when rabE or ap1 σ are repressed. On the other hand, in the absence of ClaH, UapA does not reach the PM, as its fluorescence intensity there is statistically lower (****P < 0.0001) in comparison with that of the wild‐type strain. Biological/technical replicates: 2/15 for each strain.
-
BIn a similar experiment, the apical polarized localization of de novo made SynA, used as a control of conventional secretion, is shown to be abolished in all three strains when RabE, AP‐1σ or ClaH is repressed. Scale bars: 5 μm. mCherry‐SynA PM/cytosolic intensity ratios are plotted with values being 0.52 ± 0.02 for wild type, 0.14 ± 0.02 for thiA p ‐rabE, 0.24 ± 0.02 for thiA p ‐ap1 σ and 0.16 ± 0.02 for thiA p ‐claH. The statistical evaluation was performed as in (A). There is a significant difference (****P < 0.0001) of mCherry‐SynA fluorescence in the apical PM membrane between the wild‐type and the three strains lacking the post‐Golgi proteins. Biological/technical replicates: 2/15 for each strain.
-
CCo‐localization analysis and relevant quantification of strains co‐expressing de novo made UapA‐GFP (120 min after initiation of transcription) with RabE‐mRFP, AP‐1‐mRFP or ClaH‐mRFP. Scale bars: 2 μm. Quantification by calculating Pearson's correlation coefficient (PCC) shows clear non‐co‐localization of UapA with all the Post‐Golgi markers tested, as confirmed by one sample t‐test (PCC = 0.31 ± 0.08, 0.29 ± 0.04 or 0.07 ± 0.01, with ****P < 0.0001, for RabE, AP‐1 or ClaH, respectively). Biological/technical replicates: 3/7 for each strain.
Source data are available online for this figure.
To better understand the role of ClaH in UapA secretion, we performed co‐localization studies of de novo made UapA with ClaH, but also with RabE and AP‐1 (Fig 5C). Statistical evaluation of the images obtained showed that there is no significant co‐localization of UapA with any of the AP‐1, RabE or ClaH proteins (PCC < 0.1–0.3, P < 0.0001). While the non‐co‐localization of UapA with AP‐1 or RabE might have been expected given the redundancy of these Golgi proteins for UapA secretion, the clear non‐co‐localization of UapA and ClaH (PCC < 0.1), despite the critical role of the latter in UapA secretion, strongly suggested that ClaH plays an indirect role in secretion, other than cargo vesicle coat packaging at the TGN (discussed later).
UapA sorting to the PM is microtubule independent but actin‐dependent
Conventional cargo secretion in A. nidulans and other eukaryotes is known to be microtubule and actin network dependent 27, 37, 38, 39. To investigate whether UapA secretion is dependent on distinct components of the cytoskeleton, we used drugs that block microtubule (benomyl) or actin (latrunculin B) organization. Figure 6A shows that addition of latrunculin B led to severe block in the proper secretion of UapA. On the other hand, addition of benomyl (Fig 6B), at conditions that had destructing effect on tubulin polymerization (lower panel), had absolutely no effect on de novo made UapA translocation to the PM, after 190 min of derepression (upper panel). The apical localization of a standard apical cargo, such as ChsB, was inhibited by the addition of both latrunculin and benomyl (Fig 6C). Thus, proper actin network organization, but not microtubule organization, is required for UapA trafficking to the PM.
Figure 6. Actin organization, but not microtubules, is essential for UapA translocation to the PM.
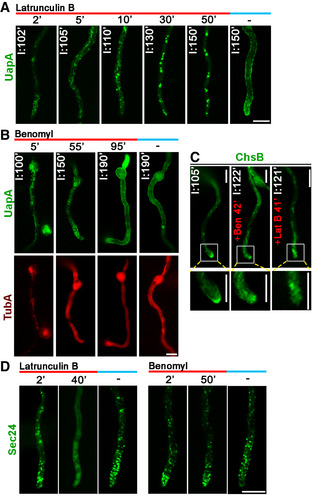
-
ATime course of treatment with the actin polymerization drug latrunculin B for 2, 5, 10, 30 or 50 min of a strain expressing neosynthesized UapA‐GFP under conditions of derepression compared to an untreated strain included as control (150 min). In all cases, latrunculin B was added at 100 min of UapA derepression, so that total time of UapA‐GFP expression was 102, 105, 110, 130 or 150 min in the different samples. Notice the abolishment of sorting of UapA to the PM after 130 or 150 min of expression when latrunculin B was present for the last 30 or 50 min, respectively. Scale bar: 5 μm.
-
BTime course of treatment of strains expressing neosynthesized alcA p‐UapA‐GFP and mCherry‐TubA with the anti‐microtubule drug benomyl for 5, 55 or 95 min. In all cases, benomyl was added at 95 min of UapA derepression, so that total time of UapA‐GFP expression was 100, 150 or 190 min. Benomyl abolished the thread‐like appearance of microtubules in all samples added, evident by the diffuse cytoplasmic signal of mCherry‐TubA. Notice that UapA reaches normally the PM (at 190 min of derepression), similarly to the untreated control (right panel). Scale bar: 5 μm.
-
CSubcellular localization of neosynthesized alcA p ‐GFP‐ChsB in the absence (left panel) or presence (≈40 min) of benomyl (middle panel) or latrunculin B (right panel), after ≈2 h of derepression. Notice the abolishment of proper polar localization of ChsB at the apical tip in both cases. Scale bars: 5 μm.
-
DEffect of latrunculin B or benomyl on COPII vesicle formation, followed through the subcellular localization Sec24‐GFP. Notice that 40 min of latrunculin B led to loss of the wild‐type punctuate appearance of Sec24 cytoplasmic puncta and the appearance of a diffuse fluorescent haze in the cytoplasm. In contrast, benomyl treatment did not affect the wild‐type punctuate localization of Sec24. Scale bar: 5 μm.
Source data are available online for this figure.
Given that UapA trafficking was shown to be COPII dependent, we examined whether the negative effect of actin is somehow related to COPII functioning. Figure 6D shows that inhibition of actin polymerization by latrunculin B led to loss of COPII punctuate organization on the ER membrane, as evidenced by Sec24‐GFP. The apparent demolishing effect of latrunculin B on ERes formation easily accounts for the loss of UapA sorting to the PM, also coincident with the appearance of large UapA aggregates (see samples after 30–50 min with latrunculin B). Our results do not exclude that actin might also be needed for UapA trafficking events occurring downstream from COPII formation. In contrast to latrunculin B, benomyl had no effect on COPII appearance. This result showed that actin is essential, whereas microtubules are redundant, for COPII vesicle formation in A. nidulans. This contrasts the reported need for microtubules in COPII secretion in mammalian cells 40, 41, 42.
We also examined whether sorting endosomes, which are key carriers of apical cargo sorting and recycling, play any role in the trafficking of de novo made UapA. As shown in Appendix Fig S2, trafficking of newly made UapA is independent of early/recycling endosome functioning.
The SsoA t‐SNARE is necessary for UapA localization to the PM
We also addressed whether UapA‐specific vesicles necessitate the PM t‐SNARE SsoA, a protein that serves as a major membrane‐specific element in the docking of secretory vesicles to the PM 25. Previous studies have shown that SsoA labels the entire PM of Aspergillus 25, 43. Noticeably, SsoA co‐localizes significantly with UapA, as not only both proteins cover the entire PM, but also upon imposing a hypertonic shock SsoA and UapA appear in the same punctuate PM microdomains that correspond to localized plasma membrane invaginations 44. Here, we followed the subcellular dynamics of the de novo localization of SsoA before reaching the PM with an experimental setup analogous to that for UapA. SsoA is repressed overnight and derepressed to moderate levels in the next day, via expression from the regulatable alcAp. Results depicted in Fig 7 showed that SsoA labels the ER (notice also the perinuclear ER rings) and eventually reaches the PM, without however labelling a prominent number of cytoplasmic punctuate structures, as we observed with the apical marker SynA (see Fig 1G). The picture of SsoA seems to resemble that obtained with UapA, rather than that of apical markers, but whether SsoA trafficking is Golgi‐dependent or Golgi‐independent needs further investigation that falls beyond the scope of this work.
Figure 7. The t‐SNARE SsoA is essential for UapA translocation to the PM.
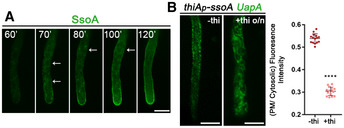
-
ASubcellular localization of de novo made alcA p ‐GFP‐SsoA in the PM in a single hypha. Notice the ER‐like membranous network (white arrows) and several cytosolic puncta labelled before SsoA reaches eventually the PM. Scale bar: 5 μm.
-
BSubcellular localization of neosynthesized UapA‐GFP (8 h after transcriptional depression) while ssoA transcription is ab initio repressed. Notice the localization of UapA in the ER‐membranous network, often at very close proximity with the PM. Scale bar: 5 μm. In the scatter plot on the right, UapA‐GFP PM/cytosolic intensity ratios are quantified when ssoA is derepressed (−thi) or repressed (+thi), with mean values being 0.54 ± 0.02 and 0.30 ± 0.02, respectively. There is statistically lower fluorescence intensity to the PM (****P < 0.0001) when SsoA is absent from the cell, as confirmed by an unpaired t‐test. Biological/technical replicates: 2/15 for each condition.
Source data are available online for this figure.
Most importantly, ab initio repression of SsoA fully blocked the translocation of UapA to the PM. Instead, UapA appears to label an extensive cytoplasmic membrane network which resembles the cortical and perinuclear ER (Fig 7B). This result is confirmed by relative quantification shown on the right panel of Fig 7B (±thiamine). Thus, UapA localization to the PM necessitates the SsoA t‐SNARE.
Distinct transporters follow the UapA trafficking route
To investigate whether the trafficking route of UapA reflects a general rather than a specific sorting pathway of nutrient transporters, we performed key experiments for following the sorting fate of two other well‐studied transporters, namely AzgA and FurA. AzgA is the major purine (adenine–guanine–hypoxanthine) transporter of A. nidulans and most fungi and defines a family structurally similar but evolutionary and functionally distinct from that of UapA 45. FurA is an allantoin transporter and member of the NCS1 superfamily, which is present in all fungi, some plants and bacteria, and is similar to numerous transporters of the LeuT fold present metazoan 46, 47.
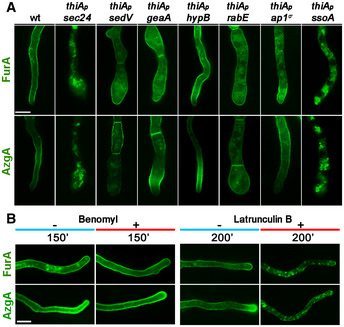
Using approaches already discussed for UapA, we tested whether neosynthesized AzgA or FurA localization to the PM bypasses the Golgi or requires microtubules or actin polymerization. In particular, we followed the localization of neosynthesized AzgA or FurA, tagged with GFP, in genetic backgrounds where Sec24, SedV, HypB, RabE, AP‐1 or SsoA could be repressed ab initio using the thiA p promoter (Fig 8A), or in the presence of benomyl or latrunculin B (Fig 8B). As shown in Fig 8, the de novo sorting of these two transporters to the PM was dependent on COPII formation, actin polymerization and SsoA, but independent of SedV, HypB, RabE, AP‐1 or microtubules, as is the case of UapA. These results strongly suggest that several nutrient transporters follow the same unconventional sorting route to be localized to the PM of A. nidulans.
Figure 8. Other transporters follow the trafficking route of UapA.
-
AEpifluorescence microscopy analysis examining the subcellular localization of de novo made alcA p‐AzgA‐GFP and alcA p‐FurA‐GFP, after 6–8 h of transcriptional derepression in strains where the expression of Sec24, SedV, GeaA, HypB, RabE, AP‐1σ and SsoA was repressed ab initio (o/n) by addition of thiamine. Images showing that sorting of AzgA or FurA to the PM do not require early (SedV, GeaA), late (HypB) or post‐Golgi (RabE, AP‐1σ) key proteins. On the other hand, COPII coat protein Sec24 and PM t‐SNARE SsoA are essential for proper localization of the two transporters to the PM. Scale bar: 5 μm.
-
BEpifluorescence microscopy analysis of the localization of de novo made AzgA and FurA transporters (see text for details) in the absence or presence of benomyl or latrunculin B after derepression of the relevant genes for the time indicated. Notice that addition of benomyl has no effect on translocation of AzgA and FurA to the PM, whereas latrunculin B abolishes their PM localization, as seen also for UapA. Scale bar: 5 μm.
Source data are available online for this figure.
Discussion
Golgi‐ and microtubule‐independent translocation of nutrient transporters to the PM of A. nidulans
Here, we uncover a novel trafficking route concerning the translocation of neosynthesized nutrient transporters to the PM of A. nidulans. This route is independent of (i) key Golgi proteins involved in secretion, (ii) acquisition of post‐Golgi secretory vesicle identity (i.e. independent of RabERab11 and AP‐1), (iii) microtubule organization, and (iv) endocytosis and sorting endosomes. The mechanism underlying this route is dependent on COPII formation, actin network organization, clathrin heavy chain and the PM t‐SNARE SsoA. These findings are in line with the observation that neosynthesized UapA is not detected, at least within the limits of fluorescent microscopy, in Golgi and post‐Golgi secretory vesicles in the course of “forward” subcellular traffic. Following this Golgi‐independent trafficking, route nutrient transporters are localized all along the hyphal cell PM. Unlike apical membrane cargoes, which show polar conventional secretion towards actively growing apical tips, localization of transporters to the PM is not polarized, occurring laterally at multiple distinct sites of the hyphal cell membrane. We are, however, aware that we cannot conclusively exclude that a subpool of transporters might transiently localize to the early Golgi, as the methodology used failed to detect passage of either UapA or SynA from this compartment.
The simple fact that UapA localization to the PM is Golgi‐independent but needs functional COPII vesicles (Sec24‐ and Sec13‐dependent) means that there must be a subpopulation of COPII vesicles carrying UapA (or other transporters) that does not migrate to the early Golgi, but is rather directed laterally towards the most proximal PM. The issue of distinct cargo‐dependent COPII subpopulations has been raised before in yeast 48, 49. Interestingly, one of the three distinct COPII populations identified in yeast is highly specific for a sugar transporter (Htx1), while the other two carry soluble cargoes or several GPI‐anchored proteins. The fact that during sorting from the ER to the PM we basically detect UapA in the ER membranous network and the PM, and very little in punctuate intermediate structures (see Fig 1), suggests that UapA‐specific COPII vesicles might directly fuse with the PM. In an alternative scenario, UapA‐specific COPII vesicles might undergo homotypic fusion to form a transporter‐specific ER‐to‐PM intermediate compartment, from where vesicles bud and fuse with the PM. Although the involvement of an ER‐to‐PM intermediate compartment is entirely speculative and remains to be shown, it offers the advantage of providing an extra step where the COPII coat is lost and a clathrin heavy chain coat is introduced. Interestingly, in mammals there are two distinct clathrin heavy chain genes, with the less prevalent one (CHC22) serving the trafficking of neosynthesized GLUT4, from an early secretory compartment to the PM, via a TGN‐independent route 9, 50. Although A. nidulans and other fungi have a single clathrin heavy chain, apparently serving both Golgi‐dependent and independent routes, we consider the possibility that the biogenesis of GLUT4, and possibly of other mammalian transporters, might present important mechanistic and physiological analogies with the trafficking mechanism discovered here through the study of fungal transporters.
The involvement of COPII and a PM t‐SNARE, SsoA, in addition to the fact that transporters are large transmembrane proteins, argues against a direct protein transport through ER‐PM direct contact sites, as those described for membrane lipid traffic and homeostasis 51. Thus, in the most probable scenario, UapA‐specific vesicles bud in bulk from the cortical ER, undergo short‐range local transport and fuse with the most proximal PM, via the tethering activity of SsoA. How SsoA recognizes secretory vesicles is not answered in the present work, as we have not identified a possible matching v‐SVARE needed for UapA or other cargo translocation to the PM. A rather provocative hypothesis is that the cargo itself contains the information to drive specific transporter vesicles directly to the PM. In this case, v‐SNAREs do not need to be involved, but instead, cytoplasmic domains of transporters might assist SsoA‐dependent tethering. In line with the importance of cis‐acting trafficking elements on the cargo itself, a conserved di‐hydrophobic motif in the cytoplasmic N‐terminal segment of UapA has been shown to be essential for ER‐exit and translocation to the PM 52. In fact, many transporters include cytoplasmic motifs affecting their specific sorting and PM translocation without affecting transport activity 53.
The transporter trafficking route discovered here is also related to proper actin polymerization and actin subcellular localization. However, the exact role of actin in transporter trafficking remains unknown. Surprisingly, actin polymerization, rather than microtubule polymerization 40, 41, 42, was found to be essential for COPII formation in A. nidulans. This observation is sufficient to explain why actin is needed for membrane cargo trafficking in general. Thus, we could not test whether actin has additional specific roles in downstream steps of transporter trafficking, as for example transporter vesicle short‐range movement and fusion to the PM. Finally, as actin scaffolding has been reported to require clathrin heavy chain in mammalian cells 54, 55, this might also provide an alternative reason why clathrin is needed for transporter trafficking, as described herein.
Based on the above, in Fig 9, we propose a speculative model on how nutrient transporters traffic to the PM through Golgi bypass in A. nidulans.
Figure 9. Speculative model of the subcellular biogenesis of neosynthesized UapA.
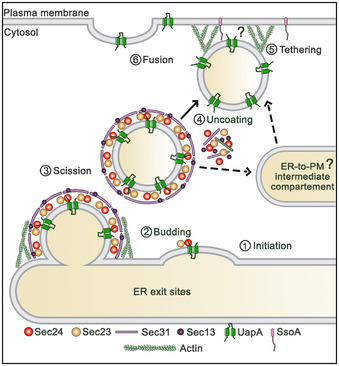
Evidence for the dependence of UapA translocation to the PM on Sec24, Sec13, actin and SsoA tSNARE, as well as, for the need for actin polymerization in COPII formation, is described in the text. The role of actin for the hypothesized close‐range transport and fusion of uncoated vesicles carrying UapA to the PM is highly speculative, as well as the existence of an ER‐to‐PM intermediate compartment. For details, see main text.
Trafficking of A. nidulans transporters to the PM of A. nidulans operates via a novel mechanism
Alternative pathways of unconventional protein secretion (UPS) have been described for a limited number of specific cargoes, but most are Golgi‐dependent. Additionally, most also depend on the Golgi‐associated GRASP protein 10, 11. Also, in most cases, cargoes of UPS are leaderless non‐vesicular or vesicular proteins that need to be secreted extracellularly (called type I and II or III UPS, respectively). UPS routes for the sorting of a handful of transmembrane cargoes to the PM by bypassing the Golgi have been described and termed type IV UPS 10, 11, 56. Distinct subtypes of UPS IV are classified based on associated proteins, such as GRASP (ΔF508‐CFTR, MpI), DNAJC14 (H723R‐Pendrin) or Rab8A (M2 mutant of Smoothened, Polycystin‐2) or proteins of the autophagy system, such as ATG5 or ATG7 10, 11, 57. Based on these observations, we also tested the possible involvement of A. nidulans homologues of GRASP and ATG5/7 in UapA trafficking and showed that none of these proteins is needed for translocation of UapA into the PM (Appendix Fig S3). Importantly, most cases of type IV UPS are COPII‐independent. The only COPII‐dependent case known concerned the translocation to the PM of peripherin, a 4‐TMS protein essential for the formation and maintenance of disk membrane of rods and cones 58. One of the best‐documented examples of biogenesis of a polytopic transmembrane protein that bypasses the Golgi is a mutant version of the CFTR transporter associated with cystic fibrosis. While wild‐type CFTR translocates to the PM via the conventional ER/Golgi‐dependent route, a partially misfolded version of CFTR (ΔF508‐CFTR) bypasses the Golgi and translocates to the PM under ER stress conditions 10, 11, 59, 60. The trafficking of ΔF508‐CFTR, similar to Aspergillus transporters, does not depend on Sed5, Arf1 or Rab, effectors that are essential for the conventional secretory pathway. However, unlike Aspergillus transporters, ΔF508‐CFTR sorting to the PM is COPII‐independent and GRASP‐ or ATG5/7‐dependent. Still another example of a transmembrane cargo that is secreted in Golgi‐ and COPII‐independent manner is the yeast Ist2 protein, which is essential for tethering cortical ER to the PM 10, 11. Interestingly, Ist2 seems to be required for efficient trafficking from the ER to the PM of newly synthesized leucine transporter Bap2 61. Finally, some signal‐peptide‐containing proteins also reach the cell surface in a COPII‐ and Golgi‐independent manner 10, 11, 56.
A cargo‐centric view of trafficking mechanisms has a strong physiological rationale
The term secretion is often used in a very broad sense to describe the “forward” trafficking of protein cargoes towards the PM. This definition, however, is limiting as it does not distinguish cargoes that are secreted extracellularly, usually hydrophilic proteins, from transmembrane cargoes that are integral PM constituents. Cargoes also differ in respect to whether they need to be directed to specific membranes (e.g. apical region of fungi, basolateral versus apical PM, axonal versus synaptic membranes in mammals), or just integrate homogeneously to the entire PM. In the first case, sorting to the PM is directional while in the second case is bulk and rather random. Cargoes in the first case usually serve polarized functions (e.g. fungal growth or neurotransmission), while in the second case are part of house‐keeping functions, such as cell nutrition or pH regulation. Also, some cargoes are constitutively expressed, while others are expressed in response to specific signal or stress. Thus, it is logical to assume that the biophysical properties and functions of cargoes should contain the information for their distinct trafficking and eventual membrane targeting. Although the conventional, Golgi‐ and microtubule‐dependent, secretory pathway is considered to be the principle route for the great majority of protein trafficking, this might prove not to be so. Current knowledge is principally based on hydrophilic cargoes that are secreted out of the cell, or membrane cargoes that are polarly delivered to specialized membrane domains. Importantly, very few studies have followed systematically the trafficking routes of transporters, which form the most abundant type of PM proteins in all cells. Moreover, to our knowledge, only a single other study, that on GLUT4, has addressed specifically the trafficking of neosynthesized transporters, showing that they also bypass the TGN during their biogenesis 9.
As most transporters serve cell nutrition, there is no obvious physiological need to drive transporters to a specialized domain of the PM, at least in free living microbial cells. This is well documented in fungi where transporters are localized non‐polarly and rather homogenously all over the PM of hyphae. In contrast to transporters, fungal membrane cargoes that serve membrane or cell wall synthesis, polarity maintenance and apical growth, are polarly localized at the hyphal tips via Golgi vesicular secretion on microtubules. An interesting observation, in line with the existence of mechanistic differences in the trafficking of apical cargoes and transporters, is also the change in UapA localization at the apical region of cells during transition from slow‐growing germlings to fast‐growing mature hyphae. While in germlings UapA localizes strongly in the PM of the apical region, in fast maturing hyphae UapA labels primarily the ER, in a way that suggests its secretion towards the apical PM is either slower than the rate of apical growth, or somehow actively excluded from the tip area. This is not seen for apical cargoes, which are equally efficiently localized at the tips of both germlings and mature hyphae. Slower secretion of transporters, compared to apical cargoes, might well be rationalized when we consider the non‐involvement of microtubules, unlike what is the case for apical cargo secretion. Along these lines, we have also showed previously that the endocytosis of transporters differs mechanistically and topologically from that of apical cargoes. The first operates all along the PM in a clathrin‐dependent, but AP‐2 independent, mechanism, whereas the second occurs locally at the subapical region and is clathrin‐independent, but AP‐2 dependent 19.
The direct lateral and homogenous sorting of transporters to the fungal PM seems to serve more efficiently cell nutrition in growing hyphae. Bypassing the Golgi might also serve the need to avoid glycosylation, which is in line with the observation that most fungal transporters studied are not glycosylated. In mammalian neurons, most cargoes involved in neurotransmission are polarly localized in the synaptic region by conventional Golgi‐dependent secretion, but specific cargoes serving dendrite, soma homeostasis or the initial segment (AIS), such as glutamate receptor GluA1, neuroligin or the potassium channel Kv2.1 are known to be sorted via distinct mechanisms that bypass the Golgi 12, 62, 63, 64. In epithelial cells, cargo‐dependent distinct subcellular sorting pathways have been reported in respect to targeting to basolateral or apical membrane 53, 65, 66, 67, 68. Last but not least, the recent discovery of distinct trafficking routes of mammalian glucose transporters serving different physiological needs might well reflect a more general necessity to generate, by distinct mechanisms, transporter‐specific vesicles, analogous to the GLUT4 insulin‐responsive vesicles 9.
A multiplicity of trafficking mechanisms seems to have evolved to better serve the need of regulating the targeting and topogenesis of specific cargoes, which in turn suggest that the cargoes themselves might contain intrinsic information to select the mechanism and route for driving their own proper subcellular targeting. It would thus not be surprising if the trafficking route discovered here by following the subcellular fate of fungal transporters proves to be a major mechanism of biogenesis of specific transporters, or other house‐keeping membrane cargoes, in other eukaryotic organisms.
Materials and Methods
Media, strains, growth conditions and transformation
Standard complete and minimal media for A. nidulans were used (FGSC, http://www.fgsc.net). Media and chemical reagents were obtained from Sigma‐Aldrich (Life Science Chemilab SA, Hellas) or AppliChem (Bioline Scientific SA, Hellas). Glucose 0.1–1% (w/v) or fructose 0.1% (w/v) was used as carbon sources. NH4 + (di‐ammonium tartrate) and NaNO3 were used as nitrogen sources at 10 mM. Thiamine hydrochloride was used at a final concentration of 10–20 μM as a repressor of the thiA promoter 69 in microscopic or Western blot analysis, respectively. Aspergillus nidulans transformation was performed by generating protoplasts from germinating conidiospores as described previously in Ref. 70 , using TNO2A7 as a recipient strain that allow selection of transformants via complementation of a pyrimidine autotrophy 71. Integrations of gene fusions with fluorescent tags, promoter replacement fusions or deletion cassettes were selected using the A. fumigatus markers orotidine‐5′‐phosphate‐decarboxylase (AFpyrG, Afu2g0836), GTP‐cyclohydrolase II (AFriboB, Afu1g13300) or a pyridoxine biosynthesis gene (AFpyroA, Afu5g08090), resulting in complementation of the relevant auxotrophies. Transformants were verified by PCR and Southern analysis. Combinations of mutations and fluorescent epitope‐tagged strains were generated by standard genetic crossing and progeny analysis. E. coli strains used were DΗ5α. A. nidulans strains used are listed in Table 1.
Table 1.
Strains used in this study. All strains carry the veA1 mutation affecting sporulation. pabaA1, pyroA4, riboB2, argB2, pyrG89, pantoB100, biA1, nicA2 and inoB2 are auxotrophic mutations for p‐aminobenzoic acid, pyridoxine, riboflavin, arginine, uracil/uridine, D‐pantothenic acid, biotin, nicotinic acid and inositol, respectively. yA2 and wA3 or wA4 are mutations resulting in yellow and white conidiospore colours, respectively
| Name | Genotype | Reference |
|---|---|---|
| TNO2A7 | nkuAΔ::argB pyrG89 pyroA4 riboB2 | 71 |
| Δ7 | uapAΔ uapCΔ::AFpyrG azgAΔ fcyBΔ::argB furDΔ::AFriboB furAΔ::AFriboB cntAΔ::AFriboB pantoB100 pabaA1 | 46 |
| mRFP‐PHOSBP | pyroA4[pyroA::gpdA m p ::mRFP‐PH OSBP ] inoB2 niiA4 wA4 | 33 |
| mCherry‐sedV | pyroA4[pyroA::gpdA m p ::mCherry‐sedV] nkuAΔ::bar, wA4, niiA4 inoB2 | 76 |
| mCherry‐synA | AFpyG‐mCherry‐synA yA::AFpyroA GFP‐tpmA fwA1 pyrG89 pyroA4 nicA2 nkuAΔ::argB | 25 |
| sedVts | sedVR238G::AFpyrG pyroA4 pyrG89 nkuAΔ::bar | 36 |
| mCherry‐tubA | alcA p ::mCherry‐tubA::pyroA nkuAΔ::argB pyrG89 pyroA4 | 77 |
| alcAp‐mRFP‐rabB | alcA p ‐mRFP‐rabB::pyroA nkuAΔ::bar inoB2 pyroA4 niiA4 wA4 | 28 |
| uapA‐GFP | uapAΔ::uapA‐GFP::AFriboB uapCΔ::AfpyrG nkuAΔ::argB pabaA1 pyroA4 riboB2 | 14 |
| alcAp‐uapA‐GFP | uapAΔ::alcA p ::uapA‐GFP::AFriboB uapCΔ::AfpyrG nkuAΔ::argB pabaA1 pyroA4 riboB2 | 14 |
| alcAp‐GFP‐chsB | alcAp‐GFP‐chsB::Ncpyr4 nkuAΔ::argB pyrG89 pyroA4 | 77 |
| alcAp‐GFP‐synA | alcAp‐GFP‐synA::AFpyrG nkuAΔ::argB pyrG89 pyroA4 riboB2 | This study |
| alcAp‐uapA‐GFP artAΔ | uapAΔ::alcA p ‐uapA‐GFP::AFriboB artAΔ::AFriboB nkuAΔ::argB pyroA4 | This study |
| GFP‐chsB | GFP‐chsB::AFpyrG nkuAΔ::argB pyrG89 pyroA4 riboB2 | This study |
| alcAp‐uapA‐GFP sedVts | (pBS‐argB)‐alcAp‐uapA‐GFP sedVR238G::AFpyrG uapAΔ pabaA1 pyroA4 | This study |
| alcAp‐furA‐GFP | (pGEM‐alcA p ‐panB)alcA p ‐furA‐GFP uapAΔ uapCΔ::AFpyrG azgAΔ fcyBΔ::argB furDΔ::AFriboB furAΔ::AFriboB cntAΔ::AFriboB pantoB100 pabaA1 | This study |
| alcAp‐azgA‐GFP | (pGEM‐alcA p ‐panB)alcA p ‐azgA‐GFP uapAΔ uapCΔ::AFpyrG azgAΔ fcyBΔ::argB furDΔ::AFriboB furAΔ::AFriboB cntAΔ::AFriboB pantoB100 pabaA1 | This study |
| alcAp‐uapA‐GFP mRFP‐PHOSBP | pyroA:: gpdA m p‐mRFP OSBP uapAΔ::alcA p ‐uapA‐GFP::AFriboB pabaA1 inoB2 | This study |
| alcAp‐uapA‐GFP mCherry‐sedV | pyroA::gpdA m p‐mCherry‐sedV uapAΔ::alcA p ‐uapA‐GFP::AFriboB pabaA1 inoB2 | This study |
| alcAp‐GFP‐synA mRFP‐PHOSBP | pyroA::gpdA m p‐mRFP OSBP alcA p ‐GFP‐synA::AFpyrG wA4 inoB2 | This study |
| alcAp‐GFP‐synA mCherry‐sedV | pyroA::gpdA m p‐mCherry‐sedV alcA p ‐GFP‐synA::AFpyrG wA4 inoB2 | This study |
| alcAp‐uapA‐GFP sec24‐mRFP | sec24‐ (5xGA) mRFP:: AFpyrG pyrG89 uapAΔ::alcA p ‐uapA‐GFP::AFriboB nkuAΔ::argB pabaA1 pyroA4 riboB2 | This study |
| alcAp‐uapA‐GFP ap1σ‐mRFP | ap1 σ ‐ (5xGA) mRFP::AFpyrG uapAΔ::alcA p ‐uapA‐GFP::AFriboB nkuAΔ::argB pyrG89 pyroA4 riboB2 | This study |
| alcAp‐uapA‐GFP mRFP‐rabE | mRFP‐rabE::AFpyrG uapAΔ::alcA p ‐uapA‐GFP::AFriboB nkuAΔ::argB pyrG89 pyroA4 riboB2 | This study |
| alcAp‐uapA‐GFP claH‐mRFP | claH‐ (5xGA) mRFP::AFpyrG uapAΔ::alcA p ‐uapA‐GFP::AFriboB nkuAΔ::argB pyrG89 pyroA4 riboB2 | This study |
| alcAp‐uapA‐GFP mCherry‐tubA | alcA p ‐mCherry‐tubA::pyroA uapAΔ::alcA p ‐uapA‐GFP::AFriboB nkuAΔ::argB pabaA1 | This study |
| alcAp‐uapA‐GFP alcAp‐mRFP‐rabB | alcA p‐mRFP‐rabB::pyroA uapAΔ::alcA p ‐uapA‐GFP::AFriboB wA3 pabaA1 | This study |
| thiAp‐sedV uapA‐GFP | thiA p ‐sedV::AFpyrG uapAΔ::uapA‐GFP nkuAΔ::argB pyrG89 pyroA4 | This study |
| thiAp‐hypB uapA‐GFP | thiA p ‐hypB::AFpyrG uapAΔ::uapA‐GFP nkuAΔ::argB pyrG89 pyroA4 | This study |
| thiAp‐geaA uapA‐GFP | thiA p ‐geaA::AFpyrG uapAΔ::uapA‐GFP nkuAΔ::argB pyrG89 pyroA4 | This study |
| thiAp‐sec24 alcAp‐uapA‐GFP | thiA p ‐sec24::AFpyrG uapAΔ::alcA p ‐uapA‐GFP nkuAΔ::argB pyrG89 pyroA4 | This study |
| thiAp‐sec13 alcAp‐uapA‐GFP | thiA p ‐sec13::AFpyrG uapAΔ::alcA p ‐uapA‐GFP::AFriboB nkuAΔ::argB pyrG89 pyroA4 riboB2 | This study |
| thiAp‐sec24 GFP‐chsB | thiA p ‐sec24::AFriboB GFP‐chsB::AFpyrG nkuAΔ::argB pyrG89 pyroA4 riboB2 | This study |
| thiAp‐sedV GFP‐chsB | thiA p ‐sedV::AFriboB GFP‐chsB::AFpyrG nkuAΔ::argB pyrG89 pyroA4 riboB2 | This study |
| thiAp‐hypB GFP‐chsB | thiA p ‐hypB::AFriboB GFP‐chsB::AFpyrG nkuAΔ::argB pyrG89 pyroA4 riboB2 | This study |
| thiAp‐geaA GFP‐chsB | thiA p ‐geaA::AFriboB GFP‐chsB::AFpyrG nkuAΔ::argB pyrG89 pyroA4 riboB2 | This study |
| thiAp‐ap1σalcAp‐uapA‐GFP | thiA p‐FLAG ap1 σ ::AFriboB (pBS‐argB)alcA p ‐uapA‐GFP pabaA1 | 19 |
| thiAp‐claH uapA‐GFP | uapAΔ::uapA‐GFP::AFriboB thiA p ‐claH::AFpyroA nkuAΔ::argB pyroA4 pabaA1 | 19 |
| thiAp‐rabE uapA‐GFP | thiA p ‐rabE::AFpyrG uapAΔ::uapA‐GFP nkuAΔ::argB pyrG89 pyroA4 | This study |
| thiAp‐sedV alcAp‐GFP‐synA | thiA p ‐sedV::AFriboB alcA p ‐GFP‐synA::AFpyrG nkuAΔ::argB pyrG89 pyroA4 riboB2 | This study |
| thiAp‐hypB alcAp‐GFP‐synA | thiA p ‐hypB::AFpyrG alcA p ‐GFP‐synA::AFpyrG nkuAΔ::argB pyrG89 wA4 pyroA4 | This study |
| thiAp‐geaA alcAp‐GFP‐synA | thiA p ‐geaA::AFriboB alcA p ‐GFP‐synA::AFpyrG nkuAΔ::argB pyrG89 pyroA4 | This study |
| thiAp‐ap1σ mCherry‐synA | thiA p ‐ap1σ::AFriboB yA::AFpyroA tpmA p ‐GFP‐tpmA AFpyrG::mCherry‐synA nkuAΔ::argB pyrG89 | This study |
| thiAp‐claH mCherry‐synA | ap2σ‐ (5xGA) GFP::AFpyrG claH::thiA p ‐claH::AFpyroA AFpyG::mCherry‐synA nkuAΔ:argB pyrG89 pyroA4 | This study |
| thiAp‐rabE mCherry‐synA | thiA p ‐rabE::AFriboB AFpyrG::mCherry‐synA nkuAΔ::argB pyrG89 pyroA4 | This study |
| alcAp‐uapA‐GFP rabBΔ thiAp‐rabA | thiA p ‐rabA::AFpyroA rabBΔ::AFpyrG uapAΔ::alcA p ‐uapA‐GFP::AFriboB nkuAΔ::argB pyrG89 pyroA4 riboB2 | This study |
| thiAp‐FLAG‐hypB | thiA p ‐ FLAG hypB::AFpyrG uapAΔ::uapA‐GFP nkuAΔ::argB pyrG89 pyroA4 | This study |
| thiAp‐FLAG‐geaA | thiA p ‐ FLAG geaA::AFpyrG uapAΔ::uapA‐GFP nkuAΔ::argB pyrG89 pyroA4 | This study |
| thiAp‐FLAG‐rabC | thiA p ‐ FLAG rabC::AFpyrG uapAΔ::uapA‐GFP nkuAΔ::argB pyrG89 pyroA4 | This study |
| thiAp‐FLAG‐rabE | thiA p ‐ FLAG rabE::AFpyrG uapAΔ::uapA‐GFP nkuAΔ::argB pyrG89 pyroA4 | This study |
| thiAp‐claH‐GFP | AFpyroA::thiA p ‐claΗ‐(5xGA)GFP::AFpyrG pyrG89 pyroA4 riboB2 | 19 |
| thiAp‐sec24 alcAp‐azgA‐GFP | thiA p ‐sec24::AFpyrG alcA p ‐azgA‐GFP uapAΔ uapCΔ::AFpyrG pyroA4 pabaA1 | This study |
| thiAp‐sedV alcAp‐azgA‐GFP | thiA p ‐sedV::AFriboB alcA p ‐azgA‐GFP pabaA1 pyroA4 | This study |
| thiAp‐hypB alcAp‐azgA‐GFP | thiA p ‐hypB::AFpyrG alcA p ‐azgA‐GFP pyroA4 | This study |
| thiAp‐geaA alcAp‐azgA‐GFP | thiA p ‐geaA::AFriboB alcA p ‐azgA‐GFP pabaA1 pyroA4 | This study |
| thiAp‐ap1σ alcAp‐azgA‐GFP | thiA p ‐FLAGap1 σ ::AFriboB alcA p‐ azgA‐GFP pabaA1 | This study |
| thiAp‐rabE alcAp‐azgA‐GFP | thiA p ‐rabE::AFpyrG alcA p ‐azgA‐GFP pabaA1 | This study |
| thiAp‐ssoA alcAp‐azgA‐GFP | thiA p ‐ssoA::AFpyrG alcA p ‐azgA‐GFP pyroA4 pabaA1 | This study |
| thiAp‐sec24 alcAp‐furA‐GFP | thiA p ‐sec24::AFpyrG alcA p ‐furA‐GFP uapAΔ pyroA4 pabaA1 | This study |
| thiAp‐sedV alcAp‐furA‐GFP | thiA p ‐sedV::AFriboB alcA p ‐furA‐GFP pyroA4 | This study |
| thiAp‐hypB alcAp‐furA‐GFP | thiA p ‐hypB::AFpyrG alcA p ‐furA‐GFP pabaA1 pyroA4 | This study |
| thiAp‐geaA alcAp‐furA‐GFP | thiA p ‐geaA::AFriboB alcA p ‐furA‐GFP pyroA4 | This study |
| thiAp‐ap1σ alcAp‐furA‐GFP | thiA p ‐FLAGap1 σ ::AFriboB alcA p ‐furA‐GFP pabaA1 pyroA4 | This study |
| thiAp‐rabE alcAp‐furA‐GFP | thiA p ‐rabE::AFriboB alcA p ‐furA‐GFP pyroA4 | This study |
| thiAp‐ssoA alcAp‐furA‐GFP | thiA p ‐ssoA::AFpyrG alcA p ‐furA‐GFP pabaA2 | This study |
| alcAp‐GFP‐ssoA | alcA p ‐GFP‐ssoA::AFpyrG nkuAΔ::argB pyrG89 pyroA4 riboB2 | This study |
| thiAp‐ssoA uapA‐GFP | thiA p ‐ssoA::AFpyrG uapAΔ::uapA‐GFP nkuAΔ::argB pyrG89 pyroA4 | This study |
| sec24‐GFP | sec24–(5xGA) GFP::AFpyrG nkuAΔ::argB pyrG89 pabaA1 | 14 |
| uapA‐GFP graspΔ | uapAΔ::uapA‐GFP graspΔ(grhA)::AFpyrG nkuAΔ::argB pyrG89 pyroA4 | This study |
| uapA‐GFP atg5Δ | uapAΔ::uapA‐GFP atg5Δ(AN5174)::AFpyrG nkuAΔ::argB pyrG89 pyroA4 | This study |
| uapA‐GFP atg7Δ | uapAΔ::uapA‐GFP atg7Δ(AN7428)::AFpyrG nkuAΔ::argB pyrG89 pyroA4 | This study |
Nucleic acid manipulations and plasmid constructions
Genomic DNA extraction was performed as described in FGSC (http://www.fgsc.net). All DNA fragments used in the various constructs were amplified from a TNO2A7 strain. Plasmid preparation and DNA gel extraction were performed using the Nucleospin Plasmid and the Nucleospin Extract II kits (Macherey‐Nagel, Lab Supplies Scientific SA, Hellas). Restriction enzymes were from Takara Bio or Minotech (Lab Supplies Scientific SA, Hellas). DNA sequences were determined by Eurofins‐Genomics (Vienna, Austria). Conventional PCRs and high‐fidelity amplifications were performed using KAPA Taq DNA and Kapa HiFi polymerases (Kapa Biosystems, Roche Diagnostics, Hellas). Gene cassettes were generated by sequential cloning of the relevant fragments in the pGEM‐T plasmid, which served as template to PCR‐amplify the relevant linear cassettes. For more details, see Supplementary information. For primers, see Appendix Table S1.
Description of promoters, conditions and strains used to express transporters and other cargoes
For following the subcellular trafficking and localization of de novo made UapA‐GFP, we used either the uapA native promoter or the regulatable alcA promoter 72. The uapA promoter can be tightly repressed in the presence of 10 mM ammonium tartrate supplied as a N source in the growth medium and depressed by a shift of the culture of germlings in a medium where ammonium is replaced by 10 mM sodium nitrate. This is a well‐studied transcriptional regulatory circuit 35. Conidiospores of a strain harbouring an in‐locus integrated copy of uapA‐gfp chimeric gene were grown overnight (o/n, 14–16 h) under repressed conditions to obtain germlings, and then, the culture was shifted in nitrate media for varying periods of time (1–8 h usually), before microscopic analysis was carried out. No UapA‐GFP signal was visible before the shift. UapA‐GFP started becoming visible after 40–60 min and progressively trafficked to the PM (see Results section). alcA p, used as an alternative of the native uapA p, is a promoter that can drive transcription at various levels depending on the medium of the A. nidulans culture used 72. In the presence of 1% (w/v) glucose as C source alcA p is tightly repressed. Upon replacement of glucose with 0.1% (w/v) fructose alcA p is derepressed, driving significant, but still moderate, levels of transcription. We have previously shown that upon derepression the strength of alcA p is similar to that of the uapA native promoter in nitrate media (preprint: 73). Finally, alcA p can also be used for obtaining significant overexpression of genes. This occurs when a culture is grown in derepressing medium (i.e. fructose), but in addition, inducers, such as 0.4% (v/v) ethanol, are added in the medium (derepressing‐inducing conditions). The levels of overexpression compared to derepressing, non‐inducing, conditions are approximately 10‐fold. For following the trafficking and localization of de novo made UapA‐GFP expressed from alcA p , we used a repression–derepression setup, analogous to the one described for the native uapA promoter. Conidiospores of a strain harbouring an in‐locus integrated copy of alcA p ‐uapA‐gfp gene were grown overnight (o/n, 14–16 h) under repressed conditions [1% (w/v) glucose) to obtain germlings, and then, the culture was shifted in 0.1% (w/v) fructose media for varying periods of time (1–8 h), before microscopic analysis was carried out. No UapA‐GFP signal was visible before the shift. UapA‐GFP became visible after 40–60 min and progressively trafficked to the membrane, similarly to the dynamics of appearance of UapA‐GFP when driven by its native promoter. The same setup was used for following the trafficking of the other two transporters studied here, namely AzgA and FurA, expressed from alcA p. We used the alcA p system to also overexpress UapA‐GFP in a single experiment shown in Fig 1F. For this experiment, the o/n repressed culture [1% (w/v) glucose] was shifted to derepressing‐inducing conditions [0.1% (w/v) fructose plus 0.4% (v/v) ethanol], before microscopic analysis was performed. In this case, UapA‐GFP fluorescence labelled more strongly the ER and allowed clear detection of perinuclear ER membranes (see text).
For following the trafficking of other control cargoes (ChsB, SynA), we used again either their native promoter or the alcA p promoter. In the former case, we could only follow the trafficking of constitutively expressed cargo, as chsB or synA promoter is not regulatable. In cases where we used the regulatable alcA p promoter, we could follow the trafficking of de novo polar cargo after a shift to derepressing conditions, as we did with transporters. All relevant strains used were the product of in‐locus gene replacement of the endogenous genes with the GFP‐tagged gene versions of the cargoes.
For most trafficking markers tagged with fluorescent epitopes (e.g. Sec24, SedV, AP‐1, RabE, ClaH, RabB, SsoA), except TubA and PHOSBP, we used native promoters for expression. For PHOSBP, which is an artificial marker of the TGN 33, we used the strong constitutive promoter gpdA p 74, whereas for TubA, we used the alcA p promoter expressed at moderate levels (derepressed, non‐induced). Again, all relevant strains carrying GFP‐ or RFP‐tagged versions of trafficking markers were the product of in‐locus gene replacements.
Protein extraction and Western blots
Total protein extraction was performed as previously described using dry mycelia from cultures grown in minimal media supplemented with NaNO3 at 25°C 47. Total proteins (50 μg, estimated by Bradford assays) were separated in a 10% (w/v) polyacrylamide gel and were transferred on PVDF membranes (GE Healthcare Life Sciences Amersham). Immunodetection was performed with an anti‐FLAG M2 monoclonal antibody (F3165, Sigma‐Aldrich), an anti‐GFP monoclonal antibody (11814460001, Roche Diagnostics), an anti‐actin monoclonal (C4) antibody (SKU0869100‐CF, MP Biomedicals Europe) and an HRP‐linked antibody (7076, Cell Signaling Technology Inc). Blots were developed using the LumiSensor Chemiluminescent HRP Substrate kit (Genscript USA) and SuperRX Fuji medical X‐Ray films (FujiFILM Europe).
Fluorescence microscopy and statistical analysis
Samples were prepared as previously described 19. Unless otherwise stated, conidiospores were incubated overnight in glass bottom 35‐mm μ‐dishes (ibidi, Lab Supplies Scientific SA, Hellas) in liquid minimal media, for 16–22 h at 25°C, under conditions of transcriptional repression of uapA [10 mM NH4 + when expressed from its native promoter or 1% (w/v) glucose when expressed from alcA p ‐uapA] and of genes involved in trafficking expressed under the thiA p promoter (10 μM thiamine). Transcriptional repression of uapA was followed by a derepression period, through a shift in media containing either NaNO3 as a nitrogen source for native uapA, or fructose as a carbon source for alcA p ‐uapA. Derepression periods ranged from to 30 min to 12 h, according to experiments. Benomyl (Sigma‐Aldrich) and latrunculin B (Sigma‐Aldrich) were used at 2.5 and 100 μg/ml final concentrations, respectively. Images were obtained using an inverted Zeiss Axio Observer Z1 equipped with an Axio Cam HR R3 camera. Contrast adjustment, area selection and colour combining were made using the Zen lite 2012 software. Scale bars were added using the FigureJ plugin of the ImageJ software. Images were further processed and annotated in Adobe Photoshop CS4 Extended version 11.0.2.
Technical replicates correspond to different hyphal cells observed within each sample, while biological replicates correspond to different samples 19. For quantifying co‐localization 75, Pearson's correlation coefficient (PCC) above thresholds, for a selected region of interest (ROI), was calculated using the ICY co‐localization studio plugin (pixel‐based method) (http://icy.bioimageanalysis.org/). One sample t‐test was performed to test the significance of differences in PCCs, using the GraphPad Prism software. Confidence interval was set to 95%. The fluorescence intensity of UapA‐GFP, GFP‐ChsB and GFP/mRFP‐SynA was quantified using the ICY software. For quantifying the fluorescence of UapA, two ROIs in the same region were drawn manually, using the Area Selection tool, one including both the PM and the cytoplasm and another identical one, excluding the PM. Concerning the fluorescence of the apical cargoes, two ROIs in the same region were drawn manually, one including the whole hyphal area between the hyphal apex and the endocytic collar and another identical one, excluding the PM. In the corresponding figures, PM/cytoplasmic mean fluorescence intensity ratios for each strain/condition are presented in box scatter plots, using the GraphPad Prism software. To test the significance of differences in PM/cytoplasmic fluorescence of measurements in Figs 3 and 5, Tukey's multiple comparison test was performed (one‐way ANOVA), while for Figs 4 and 7, an unpaired (two‐sample) t‐test was performed using the GraphPad Prism software. Confidence interval was set to 95%.
Author contributions
SD: Investigation, Validation, Formal Analysis, Visualization and Manuscript preparation. OM, MD, SA and VB: Investigation and Visualization. GD: Conceptualization, Writing—original draft preparation, Writing—review and editing, Visualization, Supervision, Project administration, Funding acquisition and Conceptualization.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Movie EV1
Source Data for Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Source Data for Figure 8
Acknowledgements
We thank Valentina D’ Autilia (Erasmus student) for help in initial experiments concerning transporters other than UapA. We are grateful to S. Efthmiopoulos for microscopy facilities. The current research was supported by the Fondation Santé to GD and SD and the Stavros Niarchos Foundation to GD and MD.
EMBO Reports (2020) 21: e49929
Data availability
No data were deposited in a public database.
References
- 1. Zanetti G, Prinz S, Daum S, Meister A, Schekman R, Bacia K, Briggs JA (2013) The structure of the COPII transport‐vesicle coat assembled on membranes. Elife 2: e00951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Feyder S, De Craene JO, Bar S, Bertazzi DL, Friant S (2015) Membrane trafficking in the yeast Saccharomyces cerevisiae model. Int J Mol Sci 16: 1509–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gomez‐Navarro N, Miller E (2016) Protein sorting at the ER‐Golgi interface. J Cell Biol 215: 769–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Robinson MS (2015) Forty years of clathrin‐coated vesicles. Traffic 16: 1210–1238 [DOI] [PubMed] [Google Scholar]
- 5. Zeng J, Feng S, Wu B, Guo W (2017) Polarized exocytosis. Cold Spring Harb Perspect Biol 9: a027870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hesse D, Hommel A, Jaschke A, Moser M, Bernhardt U, Zahn C, Kluge R, Wittschen P, Gruber AD, Al‐Hasani H et al (2010) Altered GLUT4 trafficking in adipocytes in the absence of the GTPase Arfrp1. Biochem Biophys Res Commun 394: 896–903 [DOI] [PubMed] [Google Scholar]
- 7. Takazawa K, Noguchi T, Hosooka T, Yoshioka T, Tobimatsu K, Kasuga M (2008) Insulin‐induced GLUT4 movements in C2C12 myoblasts: evidence against a role of conventional kinesin motor proteins. Kobe J Med Sci 54: E14–E22 [PubMed] [Google Scholar]
- 8. Dawicki‐McKenna JM, Goldman YE, Ostap EM (2012) Sites of glucose transporter‐4 vesicle fusion with the plasma membrane correlate spatially with microtubules. PLoS ONE 7: e43662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Camus SM, Camus MD, Figueras‐Novoa C, Boncompain G, Sadacca LA, Esk C, Bigot A, Gould GW, Kioumourtzoglou D, Perez F et al (2020) CHC22 clathrin mediates traffic from early secretory compartments for human GLUT4 pathway biogenesis. J Cell Biol 219: e201812135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rabouille C (2017) Pathways of unconventional protein secretion. Trends Cell Biol 27: 230–240 [DOI] [PubMed] [Google Scholar]
- 11. Gee HY, Kim J, Lee MG (2018) Unconventional secretion of transmembrane proteins. Semin Cell Dev Biol 83: 59–66 [DOI] [PubMed] [Google Scholar]
- 12. Jensen CS, Watanabe S, Stas JI, Klaphaak J, Yamane A, Schmitt N, Olesen SP, Trimmer JS, Rasmussen HB, Misonou H (2017) Trafficking of Kv2.1 channels to the axon initial segment by a novel nonconventional secretory pathway. J Neurosci 37: 11523–11536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Benyair R, Ron E, Lederkremer GZ (2011) Protein quality control, retention, and degradation at the endoplasmic reticulum. Int Rev Cell Mol Biol 292: 197–280 [DOI] [PubMed] [Google Scholar]
- 14. Evangelinos M, Martzoukou O, Chorozian K, Amillis S, Diallinas G (2016) BsdA(Bsd2) ‐dependent vacuolar turnover of a misfolded version of the UapA transporter along the secretory pathway: prominent role of selective autophagy. Mol Microbiol 100: 893–911 [DOI] [PubMed] [Google Scholar]
- 15. Malhotra V (2013) Unconventional protein secretion: an evolving mechanism. EMBO J 32: 1660–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Villeneuve J, Bassaganyas L, Lepreux S, Chiritoiu M, Costet P, Ripoche J, Malhotra V, Schekman R (2018) Unconventional secretion of FABP4 by endosomes and secretory lysosomes. J Cell Biol 217: 649–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cruz‐Garcia D, Malhotra V, Curwin AJ (2018) Unconventional protein secretion triggered by nutrient starvation. Semin Cell Dev Biol 83: 22–28 [DOI] [PubMed] [Google Scholar]
- 18. Diallinas G (2016) Dissection of transporter function: from genetics to structure. Trends Genet 32: 576–590 [DOI] [PubMed] [Google Scholar]
- 19. Martzoukou O, Amillis S, Zervakou A, Christoforidis S, Diallinas G (2017) The AP‐2 complex has a specialized clathrin‐independent role in apical endocytosis and polar growth in fungi. Elife 6: e20083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martzoukou O, Diallinas G, Amillis S (2018) Secretory vesicle polar sorting, endosome recycling and cytoskeleton organization require the AP‐1 complex in Aspergillus nidulans . Genetics 209: 1121–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pantazopoulou A (2016) The Golgi apparatus: insights from filamentous fungi. Mycologia 108: 603–622 [DOI] [PubMed] [Google Scholar]
- 22. Erpapazoglou Z, Kafasla P, Sophianopoulou V (2006) The product of the SHR3 orthologue of Aspergillus nidulans has restricted range of amino acid transporter targets. Fungal Genet Biol 43: 222–233 [DOI] [PubMed] [Google Scholar]
- 23. Pantazopoulou A, Lemuh ND, Hatzinikolaou DG, Drevet C, Cecchetto G, Scazzocchio C, Diallinas G (2007) Differential physiological and developmental expression of the UapA and AzgA purine transporters in Aspergillus nidulans . Fungal Genet Biol 44: 627–640 [DOI] [PubMed] [Google Scholar]
- 24. Horio T, Oakley BR (2005) The role of microtubules in rapid hyphal tip growth of Aspergillus nidulans . Mol Biol Cell 16: 918–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Taheri‐Talesh N, Horio T, Araujo‐Bazan L, Dou X, Espeso EA, Penalva MA, Osmani SA, Oakley BR (2008) The tip growth apparatus of Aspergillus nidulans . Mol Biol Cell 19: 1439–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schultzhaus Z, Quintanilla L, Hilton A, Shaw BD (2016) Live cell imaging of actin dynamics in the filamentous fungus Aspergillus nidulans . Microsc Microanal 22: 264–274 [DOI] [PubMed] [Google Scholar]
- 27. Berepiki A, Lichius A, Read ND (2011) Actin organization and dynamics in filamentous fungi. Nat Rev Microbiol 9: 876–887 [DOI] [PubMed] [Google Scholar]
- 28. Abenza JF, Pantazopoulou A, Rodriguez JM, Galindo A, Penalva MA (2009) Long‐distance movement of Aspergillus nidulans early endosomes on microtubule tracks. Traffic 10: 57–75 [DOI] [PubMed] [Google Scholar]
- 29. Valdez‐Taubas J, Pelham HR (2003) Slow diffusion of proteins in the yeast plasma membrane allows polarity to be maintained by endocytic cycling. Curr Biol 13: 1636–1640 [DOI] [PubMed] [Google Scholar]
- 30. Bianchi F, Syga L, Moiset G, Spakman D, Schavemaker PE, Punter CM, Seinen AB, van Oijen AM, Robinson A, Poolman B (2018) Steric exclusion and protein conformation determine the localization of plasma membrane transporters. Nat Commun 9: 501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gournas C, Amillis S, Vlanti A, Diallinas G (2010) Transport‐dependent endocytosis and turnover of a uric acid‐xanthine permease. Mol Microbiol 75: 246–260 [DOI] [PubMed] [Google Scholar]
- 32. Karachaliou M, Amillis S, Evangelinos M, Kokotos AC, Yalelis V, Diallinas G (2013) The arrestin‐like protein ArtA is essential for ubiquitination and endocytosis of the UapA transporter in response to both broad‐range and specific signals. Mol Microbiol 88: 301–317 [DOI] [PubMed] [Google Scholar]
- 33. Pantazopoulou A, Penalva MA (2009) Organization and dynamics of the Aspergillus nidulans Golgi during apical extension and mitosis. Mol Biol Cell 20: 4335–4347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pantazopoulou A, Pinar M, Xiang X, Penalva MA (2014) Maturation of late Golgi cisternae into RabE(RAB11) exocytic post‐Golgi carriers visualized in vivo . Mol Biol Cell 25: 2428–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gorfinkiel L, Diallinas G, Scazzocchio C (1993) Sequence and regulation of the uapA gene encoding a uric acid‐xanthine permease in the fungus Aspergillus nidulans . J Biol Chem 268: 23376–23381 [PubMed] [Google Scholar]
- 36. Pinar M, Pantazopoulou A, Arst HN Jr, Penalva MA (2013) Acute inactivation of the Aspergillus nidulans Golgi membrane fusion machinery: correlation of apical extension arrest and tip swelling with cisternal disorganization. Mol Microbiol 89: 228–248 [DOI] [PubMed] [Google Scholar]
- 37. Ross JL, Ali MY, Warshaw DM (2008) Cargo transport: molecular motors navigate a complex cytoskeleton. Curr Opin Cell Biol 20: 41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bergs A, Ishitsuka Y, Evangelinos M, Nienhaus GU, Takeshita N (2016) Dynamics of actin cables in polarized growth of the filamentous fungus Aspergillus nidulans . Front Microbiol 7: 682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schuh M (2011) An actin‐dependent mechanism for long‐range vesicle transport. Nat Cell Biol 13: 1431–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Watson P, Forster R, Palmer KJ, Pepperkok R, Stephens DJ (2005) Coupling of ER exit to microtubules through direct interaction of COPII with dynactin. Nat Cell Biol 7: 48–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fromme JC, Orci L, Schekman R (2008) Coordination of COPII vesicle trafficking by Sec23. Trends Cell Biol 18: 330–336 [DOI] [PubMed] [Google Scholar]
- 42. Verissimo F, Halavatyi A, Pepperkok R, Weiss M (2015) A microtubule‐independent role of p150glued in secretory cargo concentration at endoplasmic reticulum exit sites. J Cell Sci 128: 4160–4170 [DOI] [PubMed] [Google Scholar]
- 43. Kuratsu M, Taura A, Shoji JY, Kikuchi S, Arioka M, Kitamoto K (2007) Systematic analysis of SNARE localization in the filamentous fungus Aspergillus oryzae . Fungal Genet Biol 44: 1310–1323 [DOI] [PubMed] [Google Scholar]
- 44. Bitsikas V, Karachaliou M, Gournas C, Diallinas G (2011) Hypertonic conditions trigger transient plasmolysis, growth arrest and blockage of transporter endocytosis in Aspergillus nidulans and Saccharomyces cerevisiae . Mol Membr Biol 28: 54–68 [DOI] [PubMed] [Google Scholar]
- 45. Cecchetto G, Amillis S, Diallinas G, Scazzocchio C, Drevet C (2004) The AzgA purine transporter of Aspergillus nidulans. Characterization of a protein belonging to a new phylogenetic cluster. J Biol Chem 279: 3132–3141 [DOI] [PubMed] [Google Scholar]
- 46. Krypotou E, Evangelidis T, Bobonis J, Pittis AA, Gabaldon T, Scazzocchio C, Mikros E, Diallinas G (2015) Origin, diversification and substrate specificity in the family of NCS1/FUR transporters. Mol Microbiol 96: 927–950 [DOI] [PubMed] [Google Scholar]
- 47. Papadaki GF, Amillis S, Diallinas G (2017) Substrate specificity of the FurE transporter is determined by cytoplasmic terminal domain interactions. Genetics 207: 1387–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Castillon GA, Watanabe R, Taylor M, Schwabe TM, Riezman H (2009) Concentration of GPI‐anchored proteins upon ER exit in yeast. Traffic 10: 186–200 [DOI] [PubMed] [Google Scholar]
- 49. Manzano‐Lopez J, Perez‐Linero AM, Aguilera‐Romero A, Martin ME, Okano T, Silva DV, Seeberger PH, Riezman H, Funato K, Goder V et al (2015) COPII coat composition is actively regulated by luminal cargo maturation. Curr Biol 25: 152–162 [DOI] [PubMed] [Google Scholar]
- 50. Dannhauser PN, Camus SM, Sakamoto K, Sadacca LA, Torres JA, Camus MD, Briant K, Vassilopoulos S, Rothnie A, Smith CJ et al (2017) CHC22 and CHC17 clathrins have distinct biochemical properties and display differential regulation and function. J Biol Chem 292: 20834–20844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Saheki Y, De Camilli P (2017) Endoplasmic reticulum‐plasma membrane contact sites. Annu Rev Biochem 86: 659–684 [DOI] [PubMed] [Google Scholar]
- 52. Martzoukou O, Karachaliou M, Yalelis V, Leung J, Byrne B, Amillis S, Diallinas G (2015) Oligomerization of the UapA purine transporter is critical for ER‐exit, plasma membrane localization and turnover. J Mol Biol 427: 2679–2696 [DOI] [PubMed] [Google Scholar]
- 53. Mikros E, Diallinas G (2019) Tales of tails in transporters. Open Biol 9: 190083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brodsky FM (2012) Diversity of clathrin function: new tricks for an old protein. Annu Rev Cell Dev Biol 28: 309–336 [DOI] [PubMed] [Google Scholar]
- 55. Vassilopoulos S, Gentil C, Laine J, Buclez PO, Franck A, Ferry A, Precigout G, Roth R, Heuser JE, Brodsky FM et al (2014) Actin scaffolding by clathrin heavy chain is required for skeletal muscle sarcomere organization. J Cell Biol 205: 377–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kim J, Gee HY, Lee MG (2018) Unconventional protein secretion ‐ new insights into the pathogenesis and therapeutic targets of human diseases. J Cell Sci 131: jcs213686 [DOI] [PubMed] [Google Scholar]
- 57. New J, Thomas SM (2019) Autophagy‐dependent secretion: mechanism, factors secreted, and disease implications. Autophagy 15: 1682–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tian G, Ropelewski P, Nemet I, Lee R, Lodowski KH, Imanishi Y (2014) An unconventional secretory pathway mediates the cilia targeting of peripherin/rds. J Neurosci 34: 992–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yoo JS, Moyer BD, Bannykh S, Yoo HM, Riordan JR, Balch WE (2002) Non‐conventional trafficking of the cystic fibrosis transmembrane conductance regulator through the early secretory pathway. J Biol Chem 277: 11401–11409 [DOI] [PubMed] [Google Scholar]
- 60. Wang X, Matteson J, An Y, Moyer B, Yoo JS, Bannykh S, Wilson IA, Riordan JR, Balch WE (2004) COPII‐dependent export of cystic fibrosis transmembrane conductance regulator from the ER uses a di‐acidic exit code. J Cell Biol 167: 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wolf W, Meese K, Seedorf M (2014) Ist2 in the yeast cortical endoplasmic reticulum promotes trafficking of the amino acid transporter Bap2 to the plasma membrane. PLoS ONE 9: e85418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Arnold DB, Gallo G (2014) Structure meets function: actin filaments and myosin motors in the axon. J Neurochem 129: 213–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bowen AB, Bourke AM, Hiester BG, Hanus C, Kennedy MJ (2017) Golgi‐independent secretory trafficking through recycling endosomes in neuronal dendrites and spines. Elife 6: e27362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gumy LF, Hoogenraad CC (2018) Local mechanisms regulating selective cargo entry and long‐range trafficking in axons. Curr Opin Neurobiol 51: 23–28 [DOI] [PubMed] [Google Scholar]
- 65. Gu HH, Wu X, Giros B, Caron MG, Caplan MJ, Rudnick G (2001) The NH(2)‐terminus of norepinephrine transporter contains a basolateral localization signal for epithelial cells. Mol Biol Cell 12: 3797–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Brown A, Muth T, Caplan M (2004) The COOH‐terminal tail of the GAT‐2 GABA transporter contains a novel motif that plays a role in basolateral targeting. Am J Physiol Cell Physiol 286: C1071–C1077 [DOI] [PubMed] [Google Scholar]
- 67. Varma S, Sobey K, Campbell CE, Kuo SM (2009) Hierarchal contribution of N‐ and C‐terminal sequences to the differential localization of homologous sodium‐dependent vitamin C transporters, SVCT1 and SVCT2, in epithelial cells. Biochemistry 48: 2969–2980 [DOI] [PubMed] [Google Scholar]
- 68. Kuo SM, Wang LY, Yu S, Campbell CE, Valiyaparambil SA, Rance M, Blumenthal KM (2013) The N‐terminal basolateral targeting signal unlikely acts alone in the differential trafficking of membrane transporters in MDCK cells. Biochemistry 52: 5103–5116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Apostolaki A, Harispe L, Calcagno‐Pizarelli AM, Vangelatos I, Sophianopoulou V, Arst HN Jr, Penalva MA, Amillis S, Scazzocchio C (2012) Aspergillus nidulans CkiA is an essential casein kinase I required for delivery of amino acid transporters to the plasma membrane. Mol Microbiol 84: 530–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Koukaki M, Giannoutsou E, Karagouni A, Diallinas G (2003) A novel improved method for Aspergillus nidulans transformation. J Microbiol Methods 55: 687–695 [DOI] [PubMed] [Google Scholar]
- 71. Nayak T, Szewczyk E, Oakley CE, Osmani A, Ukil L, Murray SL, Hynes MJ, Osmani SA, Oakley BR (2006) A versatile and efficient gene‐targeting system for Aspergillus nidulans . Genetics 172: 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Waring RB, May GS, Morris NR (1989) Characterization of an inducible expression system in Aspergillus nidulans using alcA and tubulin‐coding genes. Gene 79: 119–130 [DOI] [PubMed] [Google Scholar]
- 73. Bouris V, Martzoukou O, Amillis S, Diallinas G (2019) Nutrient transporter translocation to the plasma membrane via a Golgi‐independent unconventional route. bioRxiv 10.1101/540203 [PREPRINT] [DOI] [Google Scholar]
- 74. Punt PJ, Dingemanse MA, Kuyvenhoven A, Soede RD, Pouwels PH, van den Hondel CA (1990) Functional elements in the promoter region of the Aspergillus nidulans gpdA gene encoding glyceraldehyde‐3‐phosphate dehydrogenase. Gene 93: 101–109 [DOI] [PubMed] [Google Scholar]
- 75. Dunn KW, Kamocka MM, McDonald JH (2011) A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol 300: C723–C742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pantazopoulou A, Penalva MA (2011) Characterization of Aspergillus nidulans RabC/Rab6. Traffic 12: 386–406 [DOI] [PubMed] [Google Scholar]
- 77. Takeshita N, Mania D, Herrero S, Ishitsuka Y, Nienhaus GU, Podolski M, Howard J, Fischer R (2013) The cell‐end marker TeaA and the microtubule polymerase AlpA contribute to microtubule guidance at the hyphal tip cortex of Aspergillus nidulans to provide polarity maintenance. J Cell Sci 126: 5400–5411 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Movie EV1
Source Data for Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Source Data for Figure 8
Data Availability Statement
No data were deposited in a public database.


