Abstract
Impediments to DNA replication threaten genome stability. The homologous recombination (HR) pathway has been involved in the restart of blocked replication forks. Here, we used a method to increase yeast cell permeability in order to study at the molecular level the fate of replication forks blocked by DNA topoisomerase I poisoning by camptothecin (CPT). Our results indicate that Rad52 and Rad51 HR factors are required to complete DNA replication in response to CPT. Recombination events occurring during S phase do not generally lead to the restart of DNA synthesis but rather protect blocked forks until they merge with convergent forks. This fusion generates structures requiring their resolution by the Mus81 endonuclease in G2/M. At the global genome level, the multiplicity of replication origins in eukaryotic genomes and the fork protection mechanism provided by HR appear therefore to be essential to complete DNA replication in response to fork blockage.
Keywords: BIR, fork restart, Mus81, recombination, replication
Subject Categories: DNA Replication, Repair & Recombination
Camptothecin induces replication fork blockage. Homologous recombination is required to protect blocked forks until they merge with convergent forks. This fusion generates structures requiring their resolution by the Mus81 endonuclease in G2/M.
Introduction
Chromosome duplication during the S phase is a crucial step of cell division. DNA replication in eukaryotes is initiated from multiple origins distributed on all chromosomes. Replication forks progress along chromosomes arms until merging with another incoming fork at termination regions. The progression of replication forks can be hindered by obstacles in the DNA template such as DNA lesions. Genome stability is particularly at risk when damaged DNA molecules are replicated. Failure in DNA damage repair can lead to the terminal arrest or breakage of replication forks and ultimately to the distribution of under‐replicated and/or broken chromosomes to the daughter cells after mitotic division. When a fork becomes dysfunctional, the completion of replication could be ensured by a converging functional fork, a process that can be favored by the firing of nearby dormant origins 1, 2. Alternatively, dysfunctional replication forks could also be restarted, a mechanism that requires the homologous recombination (HR) pathway 3, 4, 5. During HR, Rad52 and Rad51 are fundamental to coat the ssDNA generated by resection and carry out the strand invasion and exchange reactions 6. At dysfunctional forks, Rad51 loading by Rad52 also regulates nascent DNA strands degradation by exonucleases 7, 8, 9. Whether this protection mechanism is different from the restart mechanism has not been clearly determined 7, 10, 11, 12.
Recombination‐mediated replication restart has been mainly studied in the yeast cellular model using site‐specific tools to break or block replication forks at a unique location 3, 4, 13, 14, 15, 16, 17. Replication restart in Saccharomyces cerevisiae has been recently investigated using a Flp‐nick system, which produces a site‐specific DNA nick that is converted into a one‐ended DSB upon the passage of a replication fork 4, 14, 18. Recombination‐mediated restart seems to occur by break‐induced replication (BIR), an HR pathway mainly characterized outside of S phase 19, 20. BIR is favored when only one DSB end is able to invade a homologous template. In that case, DNA synthesis primed within the displacement loop (D‐loop) can proceed to the chromosome end by a conservative mechanism 21, 22, 23, 24. In particular, the non‐essential DNA polymerase δ subunit Pol32 is required for priming DNA synthesis inside the D‐loop and the Pif1 DNA helicase is required to promote its migration 24, 25, 26. Lastly, BIR synthesis is highly mutagenic 4, 27 and subjected to template switching, leading to complex chromosomal rearrangements 28, 29, 30, 31. Terminally blocked replication forks at the RTS1 barrier in Schizosaccharomyces pombe can be restarted by HR without DSB formation 3, 7, 32, 33, 34, 35. Similar to BIR in S. cerevisiae, replication restart at RTS1 requires the DNA polymerase δ and the Pfh1 DNA helicase, is highly mutagenic and prone to template switching 35, 36, 37, 38. In that context, HR is not thought to be primed from a DSB but from the tip of a reversed fork, which results from the displacement and reannealing of the nascent strands together 32, 39, 40.
It remains to be determined whether replication restart studied at locus‐specific barriers occurs in the same way at the genome‐wide level in response to natural replication impediments. For example, the DNA topoisomerase 1 (Top1) normally introduces a transient nick to relax supercoiled DNA during transcription and replication 41. During this reaction, Top1 remains covalently attached to the 3′ end of the break, forming a “cleavage complex” (Top1cc), before the relaxation of DNA and religation of the break. Cells are constantly challenged with blocked Top1ccs, which is a major driver of mutagenesis in highly transcribed genes 42, 43. Furthermore, cells devoid of both Tdp1 and Wss1, two factors involved in the removal of Top1ccs, have a severe Top1‐dependent growth defect 44, 45, which indicates that Top1ccs are natural threats to cell survival. Top1ccs can also be stabilized by the drug camptothecin (CPT) 46. CPT is thought to cause the formation of a one‐ended DSB, the typical substrate for BIR, upon the collision of the replication fork with the nick in the Top1cc, thus inducing a replication stress 47, 48, 49. However, CPT‐induced DSBs have not been observed in yeast cells 50, 51. More recently, it has been shown that CPT treatment of yeast cells induces fork reversal 50, 52, as a possible consequence of accumulation of positive supercoils ahead of replication forks due to Top1 inhibition 53. Hence, replication fork blockage by CPT‐stabilized Top1ccs appears as a relevant model to address whether replication restart occurs at the global genomic level.
Finally, it remains unclear which factors participate in the resolution of the recombination intermediates during HR‐mediated restart of DNA replication. The structure‐selective endonuclease Mus81 has been proposed to cleave either blocked or reversed forks to promote repair by HR 54, 55, 56. However, Mus81 is not required for replication restart at RTS1 in fission yeast, nor for the repair of a replication‐born DSB in budding yeast 3, 17. Mus81 is nevertheless involved in the processing of these recombination events, as they accumulate in its absence 3, 17, resulting in a decreased amount of final repair products 17, 57. By using the Flp‐nick system, it has been proposed that Mus81 limits replication restart by Pol32‐dependent BIR in S phase by processing the migrating D‐loop 4. Mus81 catalytic activity is normally very low in S phase and only increases at the G2/M transition, when the Mms4 regulating subunit of the complex is hyper‐phosphorylated by multiple kinases 58, 59, 60, 61. Hence, the role of Mus81 in replication restart by HR in S phase appears contradictory to the regulation of its activity. We took advantage of our study to clarify the role of Mus81 in recombination‐mediated restart of DNA replication.
Our data reveal that HR is primarily providing a replication fork protection mechanism rather than promoting the restart of DNA synthesis from blocked forks. Thanks to a method to increase CPT entry into yeast cells, we show that S phase completion in response to Top1 poisoning requires Rad52 and Rad51. We propose that HR promotes the formation of a D‐loop structure that protects blocked forks until they merge with converging forks. Completion of replication also requires Mus81 in G2/M to promote the termination of DNA replication at protected forks. We confirmed our results obtained in CPT‐treated cells by using an independent system in which a specific mutation of the Rad3 DNA helicase generates a similar replication stress. This allowed us to demonstrate that the mechanism we have characterized upon Top1 poisoning is independent of the accumulation of supercoiled DNA and DNA‐protein crosslinks. The engagement of HR to protect blocked forks and to ensure the completion of DNA replication appears therefore as a general mechanism to respond to replication stress.
Results
Top1 poisoning by CPT and the rad3‐102 allele exert a genetically similar replication stress
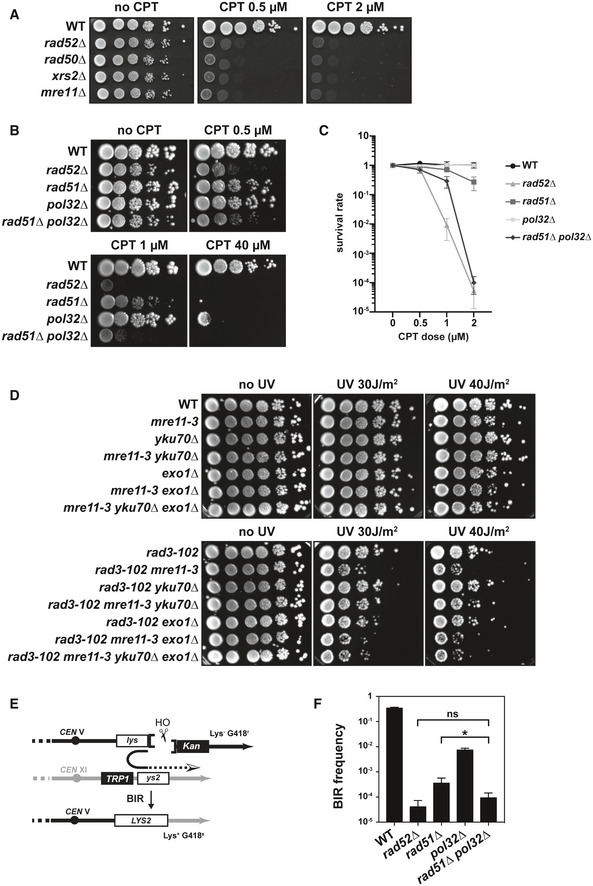
In this study, we have chosen to study the dynamics of DNA replication challenged by CPT treatment, which exacerbates at the genome‐wide level the presence of trapped Top1 on the DNA template. Resistance to CPT‐induced DNA damage absolutely requires the homologous recombination (HR) machinery, as null mutations of RAD52, MRE11, RAD50, and XRS2 lead to the highest sensitivities to low doses of CPT compared to wild‐type cells (Fig 1A) 62, 63. rad51∆ mutants are also highly sensitive to low CPT doses but to a lesser extent than its upstream regulator Rad52 (Fig 1B and C, Appendix Fig S1A) 4. Interestingly, Rad52 and the members of the MRX complex (Mre11‐Rad50‐Xrs2), but not Rad51, are essential for the survival of cells bearing the rad3‐102 mutation: This mutation impairs the nucleotide excision repair (NER) pathway by increasing the binding of the TFIIH complex to a single‐strand DNA gap intermediate, which prevents its subsequent filling 64, 65. Additionally, it has been described that the elevated CPT sensitivity of a nuclease‐deficient Mre11 mutant (mre11‐3) can be suppressed by the absence of the Ku complex and in an Exo1‐dependent manner (Appendix Fig S1B) 66. This suggests that Mre11 counteracts the action of Ku at CPT‐induced DSBs ends, which limits Exo1‐dependent DNA resection. Similarly, although the combination of mre11‐3 with rad3‐102 is not lethal, it sensitizes the cells to UV exposure, a context in which replication stress is enhanced in the rad3‐102 background 65 (Fig 1D). Strikingly, the increased UV sensitivity of rad3‐102 mre11‐3 compared to rad3‐102 can be partially suppressed by the absence of Ku in an Exo1‐dependent manner (Fig 1D and Appendix Fig S1C). These data show that Mre11, Ku, and Exo1 play similar roles in the repair of DNA damage induced by rad3‐102 or CPT. Finally, rad3‐102 has been found lethal in combination with both rad51∆ and pol32∆, leading to the proposal that DNA repair in the absence of Rad51 in these cells is backed up by the presence of the Pol32 non‐essential subunit of the DNA polymerase ∂ 65, 67. Remarkably, we found that the rad51∆ pol32∆ double mutant was more sensitive to CPT than the rad51∆ single mutant (Fig 1B and C), suggesting that, as in rad3‐102 cells, Pol32 also partially compensates the absence of Rad51 to cope with CPT‐induced DNA damage.
Figure 1. CPT‐treated or rad3‐102 cells share the same genetic requirements to cope with DNA damage.
-
A, BCPT sensitivity assayed by 10‐fold serial dilutions of different mutant combinations on YPAD plates. Four biological replicates have been performed.
-
CColony‐forming unit assays to increasing CPT concentrations of the same mutant combinations as in (B). Mean values ± SD are plotted. Four biological replicates have been performed.
-
DUV sensitivity assayed by 10‐fold serial dilutions of different mutant combinations with rad3‐102 allele on YPAD plates after exposure to the indicated UV‐C doses. Five biological replicates have been performed.
-
ESchematic representation of the BIR assay. Two fragments of the LYS2 gene sharing 2.1 kb of homology (lys and ys2) were integrated on chromosomes V and XI, respectively. Induction of HO endonuclease expression under control of the GAL1 promoter produces a DSB next to the lys fragment that can be repaired by BIR. BIR events can be scored by selecting survivor colonies, which harbor a functional LYS2 gene.
-
FBIR frequencies (Lys+ survivors among total cells) for the WT and indicated mutant strains are plotted on a logarithmic scale. Data represent the mean ± SD from 3 to 6 biological replicates. ns, non‐significant difference, *P = 0.0134, Mann–Whitney unpaired t‐test.
Thus, rad3‐102 cells suffer from a replication stress that mimics the effect of CPT. However, accumulation of topological stress is not expected in rad3‐102 cells, since they are not affected in DNA supercoils removal by Top1. Neither Tdp1 (tyrosyl‐DNA phosphodiesterase 1) nor the metalloprotease Wss1 are required for survival in rad3‐102 cells, consistent with the TFIIH complex not being covalently linked to DNA (Appendix Fig S1D). Overall, in view of the strikingly similar genetic requirements for the cell survival in response to CPT and in the rad3‐102 background, we decided to use these two systems to further investigate the repair mechanisms required to cope with this type of replication stress, independently from the accumulation of DNA supercoiling and DNA‐protein crosslinks.
Rad51 and Pol32 independently promote DSB repair by break‐induced replication
First, we asked what could be the specific contribution of Pol32 to DNA repair in the absence of Rad51. We previously proposed that Pol32 may stabilize strand invasion in the absence of Rad51 by promoting the priming of DNA synthesis 67, further supported by the essential role of Pol32 for viability of cells lacking Rad51 that are defective in histone H3K56 deacetylation 68. Among DSB repair pathways mediated by HR, Pol32 has only an essential role during BIR. In the absence of Pol32, the initiation of DNA synthesis during BIR is compromised 25. Moreover, the recovery of both Rad51‐dependent and Rad51‐independent survivors in cells lacking telomerase, thought to occur by BIR, also relies on Pol32 25. Finally, the BIR pathway has been proposed to be involved in the repair of broken replication forks 4. We thus wondered whether Pol32 compensatory function in the absence of Rad51 in cells exposed to CPT or in the rad3‐102 background could be related to its function in BIR. To assess the redundant role of Pol32 over Rad51 in BIR, we used a well‐described chromosomal system 23 in which a single DSB is induced by the HO endonuclease. In this system, only one of the two ends can undergo homology‐dependent strand invasion at an ectopic location. Subsequent priming and elongation of DNA synthesis reaching the chromosome end lead to the production of viable Lys2+ recombinants (Fig 1E). In this system, the absence of Rad52 decreased the BIR frequency by about three orders of magnitude compared to the wild type (Fig 1F). Deletion of RAD51 or POL32 also significantly decreased the BIR frequency compared to the wild type (Fig 1F) 23. However, only the combined absence of Rad51 and Pol32 did affect the BIR frequency as much as in the absence of Rad52 (Fig 1F). These results show that Pol32 can promote BIR in the absence of Rad51 and indicate that Pol32 functions in BIR to facilitate both strand invasion and priming of extensive DNA synthesis. The absence of Pol32 alone affects cell survival only in response to high CPT concentrations (Fig 1B) 69. Altogether, these results suggest Pol32‐dependent BIR‐mediated synthesis is not the main pathway required to cope with CPT‐induced DNA damage and that the role of Pol32 in the absence of Rad51 may be ascribed to its function in facilitating strand invasion during HR.
Mus81 is involved in HR‐mediated repair involving both Rad51 and Pol32
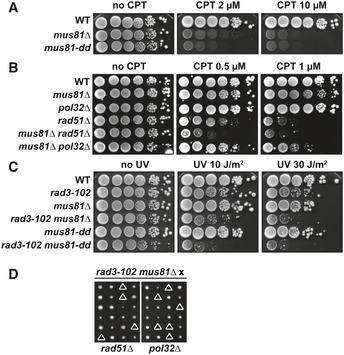
In human cells, the structure‐specific endonuclease (SSE) Mus81 has been shown to generate DSBs in response to CPT treatment, leading to the suggestion that Mus81 may cleave stalled or reversed replication forks upon Top1 poisoning to promote fork restart 54. In budding yeast, null and catalytically dead (mus81‐dd) mutations of MUS81 have been shown to sensitize cells to mild CPT doses (Fig 2A) 62, 63, 70, 71, 72. The absence of Mus81 is backed up by the Yen1 SSE, as shown by the higher CPT sensitivity of the double mus81∆ yen1∆ mutant compared to mus81∆ (Fig EV1A) 71, 72. Moreover, Mus81 and Yen1 have been proposed to limit BIR synthesis initiated from a replication‐born DSB 4.
Figure 2. Mus81 is required in the restart pathway involving both Rad51 and Pol32.
-
A, BCPT sensitivity assayed by 10‐fold serial dilutions of different mutant combinations on YPAD plates. Four biological replicates have been performed.
-
CUV sensitivity assayed by 10‐fold serial dilutions of different mutant combinations with rad3‐102 allele on YPAD plates after exposure to the indicated UV‐C doses. Four biological replicates have been performed.
-
DSynthetic combinations of rad3‐102 mus81∆ with rad51∆ and pol32∆. Tetrads dissected on YPAD medium are shown. Triangles indicate either a severe growth defect or lethality.
Figure EV1. Yen1 backs up Mus81 in CPT‐treated and rad3‐102 cells.
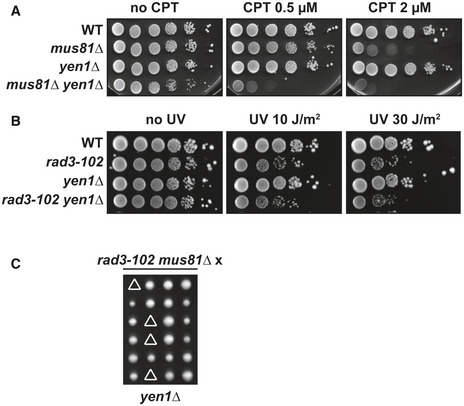
- CPT sensitivity assayed by 10‐fold serial dilutions of different mutant combinations between mus81∆ and yen1∆ on YPAD plates. Two biological replicates have been performed.
- UV sensitivity assayed by 10‐fold serial dilutions of yen1∆ in combination with rad3‐102 allele on YPAD plates after exposure to the indicated UV‐C doses. Two biological replicates have been performed.
- Synthetic combinations of rad3‐102 mus81∆ with yen1∆ and pol32∆. Tetrads dissected on YPAD medium are shown. Triangles indicate lethality. Three biological replicates have been performed.
We asked whether Mus81 could have a similar role in CPT‐induced DNA damage repair. First, we observed that the combination of mus81∆ with rad51∆ and pol32∆ mutations resulted in an increased CPT sensitivity than either single mutants (Fig 2B) 4, suggesting that Mus81 participates in Rad51‐ and Pol32‐dependent repair of DNA damage induced by CPT. As for, rad51∆, mus81∆, or mus81‐dd are not lethal in combination with rad3‐102 but the double mutant cells exhibited a much higher sensitivity to UV than single mutant cells (Fig 2C) 65. We analyzed the redundancy between Mus81 and Yen1 for survival in the rad3‐102 background. rad3‐102 yen1∆ cells were not more sensitive to UV than rad3‐102 cells (Fig EV1B), consistent with yen1∆ cells not being sensitive to CPT (Fig EV1A). The rad3‐102 mus81∆ yen1∆ triple mutants were unviable (Fig EV1C), indicating that Yen1 also backs up Mus81 in rad3‐102 cells. Finally, we found that rad3‐102 mus81∆ pol32∆ mutants were unviable and rad3‐102 mus81∆ rad51∆ cells had a severe growth defect (Fig 2D). These results indicate that Mus81 is required for repair mediated by both Rad51 and the polymerase δ subunit Pol32 in rad3‐102 cells.
Mus81 and Pol32 are not required for replication progression upon Top1 poisoning
Next, we reasoned that if an increased CPT sensitivity is associated with replication defects, we should observe an impaired progression through S phase. Opposite to other cellular models, it was reported that an acute exposure to CPT in liquid cultures of S. cerevisiae cells did not induce a delay in S phase progression but rather a prolonged arrest in G2/M 50, 51, 73. CPT is highly insoluble in culture media, and the water‐soluble derivatives of CPT (topotecan and irinotecan) affect neither yeast cell growth nor their survival 74, thus not being exploitable for our study. We suspected a permeability issue for CPT entry into the cells, as described for the proteasome inhibitor MG132 and the DNA polymerase inhibitor aphidicolin 75, 76. We therefore used a modified culture medium described to render cells more permeable to the antifungal agent brefeldin A 77 in order to increase the cell permeability to CPT. We took advantage of the natural blue fluorescence emitted by the CPT compound when excited by 350 nm UV light to quantify the relative amount of CPT inside cells. Incubation of an asynchronous cell culture with CPT for 30 min in MPD +SDS medium (minimal‐proline‐dextrose + 0.003% SDS) 77 allowed the detection of blue fluorescence inside cells, whose quantification was up to 10 times higher than in cells incubated with CPT for 30 min in standard MAD medium (minimal‐ammonium‐dextrose) (Fig EV2A and B). We then used these conditions to analyze the progression through a single S phase in the presence of CPT by flow cytometry. Wild‐type cells were synchronized in G1 with α‐factor, incubated with CPT for 1 h and then released from G1 into S phase, still in the presence of CPT. As observed by others, CPT did not alter S phase progression when cells were grown in normal MAD medium (Fig EV2C) 50, 51, 73, but when incubated with CPT in MPD +SDS medium, they progressed significantly slower through S phase than control wild‐type cells incubated with DMSO, thus reaching the G2/M phase later (Fig EV2C). Of note, the addition of SDS in the culture medium did not perturb the G1‐S transition, as previously reported with higher concentrations of SDS 78. Thus, these experimental conditions allowed us to perform the first characterization of the effect of CPT on DNA replication using S. cerevisiae cells as a model system.
Figure EV2. Increased cell permeability allows detection of CPT effects on a single S phase.
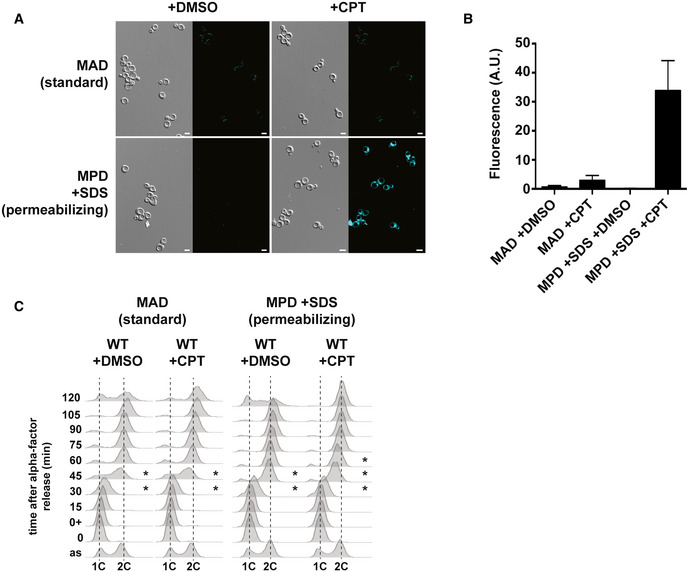
- Visualization of CPT‐containing yeast cells. Exponentially growing wild‐type cells in either MAD (minimal‐ammonium‐dextrose) or MPD +SDS (minimal‐proline‐dextrose + 0.003% SDS) medium were incubated with DMSO or 50 μM CPT for 30 min and visualized by fluorescence microscopy. Scale bar = 5 μm. Three biological replicates have been performed.
- Quantification of fluorescence emitted by CPT in cells grown as described in (A). Mean ± SEM is indicated for each experimental condition. A.U. arbitrary units.
- Analysis of DNA content by flow cytometry of G1 phase synchronized wild‐type cells and further released into S phase. Cells were synchronized in G1 with α‐factor in either MAD or MPD +SDS medium, treated with DMSO or 50 μM CPT, let in G1 for 30 min, and released into S phase. Three biological replicates have been performed. Asterisks indicate the progression of cells in S phase.
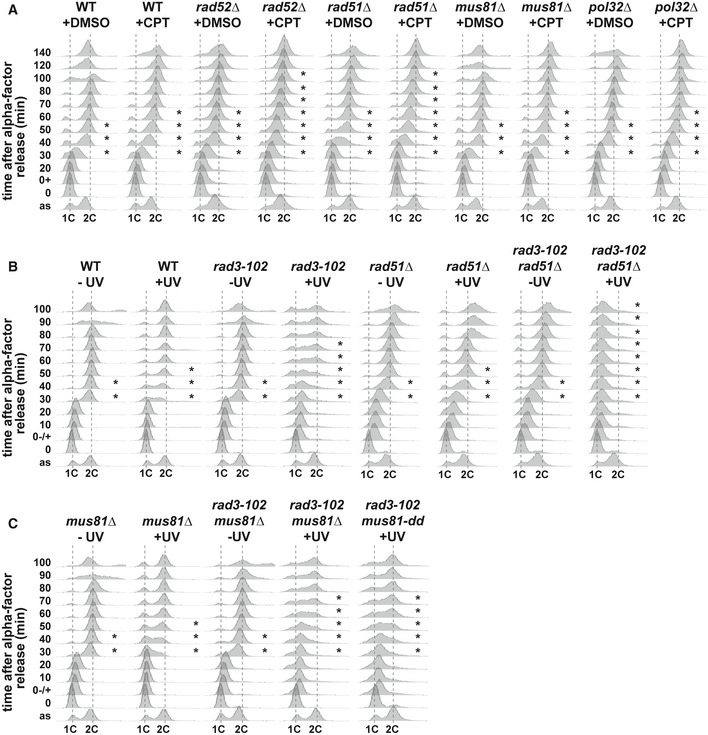
Using these conditions, we next asked how the HR factors Rad52 and Rad51, the Mus81 resolvase, and the polymerase δ subunit Pol32 contribute to the progression of replication forks encountering trapped Top1. At the global level, using flow cytometry, we could observe that the S phase delay was strikingly increased in CPT‐treated cells lacking one of the two main HR factors, Rad52 or Rad51, compared to wild‐type cells (Fig 3A). However, in the absence of Mus81 or Pol32, S phase progression was as affected by CPT as in wild‐type cells. We confirmed these results using the rad3‐102 mutation. We synchronized cells in G1 phase with α‐factor and irradiated them with UV‐C. Cells were kept for 2 h in G1 before release into S phase, in order to let compromised NER leave DNA gaps or nicks bound by the TFIIH complex in the rad3‐102 mutant, as described previously 65. As expected, UV irradiation only affected S phase progression in rad3‐102 cells and to a greater extent in rad3‐102 cells lacking the HR factor Rad51 (Fig 3B). However, rad3‐102 cells lacking Mus81 or Mus81 nuclease activity did not show an increased S phase delay compared with rad3‐102 single mutants (Fig 3C). All these results indicate that HR factors Rad52 and Rad51 are required to promote S phase progression when replication stress is induced by CPT. However, neither Mus81 nor Pol32 is required for bulk DNA synthesis in these conditions.
Figure 3. Mus81 is not required for S phase progression in CPT‐treated and rad3‐102 cells.
-
AAnalysis of DNA content by flow cytometry of G1 phase synchronized wild‐type, rad52∆, rad51∆, mus81∆, and pol32∆ cells and further released into S phase. Cells were synchronized in G1 with α‐factor, treated with DMSO or 100 μM CPT, let in G1 for 1 h, and released into S phase. Three biological replicates have been performed.
-
B, CAnalysis of DNA content by flow cytometry of G1 phase synchronized wild‐type, rad3‐102, rad3‐102, rad51∆, rad3‐102 mus81, and rad3‐102 mus81‐dd cells and further released into S phase. Cells were synchronized in G1 with α‐factor, untreated, or irradiated with 20 J/m2 UV‐C, let in G1 for 2 h, and released into S phase. Three biological replicates have been performed.
To confirm these data at the molecular level, we analyzed nascent DNA synthesis by DNA combing from asynchronous cell cultures treated with CPT. DNA combing allows monitoring replication fork progression genome‐wide though at the level of individual DNA molecules 79, 80. After incubation with CPT for 2 h, cells were pulse‐labeled with the thymidine analog EdU for 20 min before combing analysis (Fig 4A). We first compared the length of the EdU tracks in all strains under unchallenged conditions (+DMSO). Only mus81∆ cells showed a significant decrease of the EdU track length compared to wild‐type cells (Fig 4A; P = 0.0015). This result agrees with a recent report that proposed a role for human Mus81 in DNA replication in the absence of exogenous damage 81. Then, we focused our attention onto the effect of CPT on fork progression in each strain. As expected from cell cycle analyses (Fig 3A), CPT treatment significantly affected fork progression in all strains compared to the DMSO control (Fig 4A). The EdU track length of wild‐type cells was reduced by 47% and that of mutant cells was further reduced to 55% in rad51∆ cells and 75% in rad52∆ cells (Fig 4A). However, the reduction in fork progression caused by CPT in mus81∆ (45%) and in pol32∆ (37%) cells was more similar to that of wild‐type cells (Fig 4A). To accurately evaluate the effect of CPT in mutants compared to the wild type, we calculated a conversion factor to equal the median of each DMSO‐treated samples from mutants to that of the wild type. We then used this normalization factor for each mutant to convert all track length values of CPT‐treated samples. The statistical analysis of the normalized data shows that only rad52∆ and rad51∆ mutants are more affected by CPT than wild‐type cells (Fig EV3A). Our results thus argue against a role of Mus81 in replication restart in CPT‐treated cells, as was suggested in human cells 54. This was confirmed in rad3‐102 mus81∆ cells, in which replication fork progression assayed by DNA combing was not more affected than in single mutants (Fig EV3B). Overall, our results show that CPT‐mediated Top1 poisoning induces a global replication stress that requires HR factors for replication fork progression. However, Pol32 is not required to promote fork progression in response to CPT, suggesting that the strong reduction of nascent DNA tracks in rad52∆ and rad51∆ mutants is not due to the inability of blocked forks to perform BIR‐mediated DNA synthesis. These results indicate that Pol32‐dependent BIR synthesis is not the main pathway promoting the progression of replication forks blocked by Top1ccs. Even though we do not exclude that HR may promote the restart of blocked forks by BIR, we propose that HR is primarily required to protect blocked forks from degradation in response to Top1 poisoning.
Figure 4. Rad52 and Rad51 are required for HR‐mediated replication fork protection.
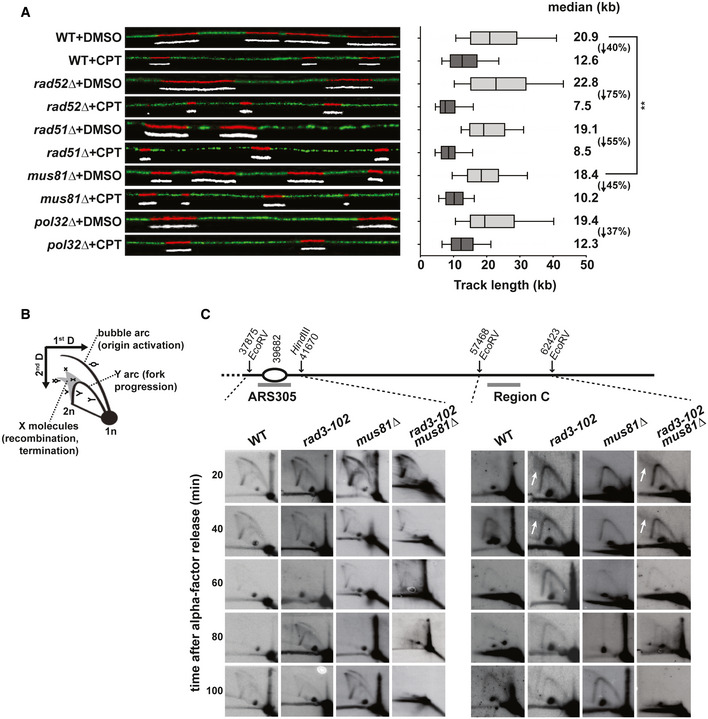
- Analysis of replicated DNA tracks length by single‐molecule DNA combing in WT, rad52∆, rad51∆, mus81∆, and pol32∆ cells exposed to CPT. Exponentially growing cells were treated with DMSO or 50 μM CPT for 2 h and then pulse‐labeled with 50 μM EdU for 20 min. DNA fibers were combed on silanized coverslips, and EdU‐labeled DNA was detected by Click chemistry. Graph depicts the distribution of EdU tracks length in kb. Box and whiskers indicate 25‐75 and 10‐90 percentiles, respectively. Median EdU tracks length is indicated in kb (values from two biological replicates were pooled). Asterisks indicate the P‐value of the Mann–Whitney unpaired t‐test, **P = 0.0015. The percentage of EdU track length decrease between the DMSO and CPT conditions is indicated between parentheses for each strain. Representative images of DNA fibers are shown. Red and white: EdU, green: DNA.
- Schematic representation of replication intermediates visualized by 2D gels.
- Analysis of replication intermediates by 2D‐gel electrophoresis. Replication intermediates were monitored at early origin ARS305 and region C in WT, rad3‐102, mus81∆, and rad3‐102 mus81∆ cells. Cells were synchronized in G1 with α‐factor and collected at the indicated time points after release into S phase. A scheme of the studied chromosomal region is shown (drawn to scale). Relevant probes are indicated by gray bars, and coordinates of ARS and restriction sites are indicated in bp. Two biological replicates have been performed.
Figure EV3. Analysis of replication fork progression in CPT‐treated and in rad3‐102 cells.
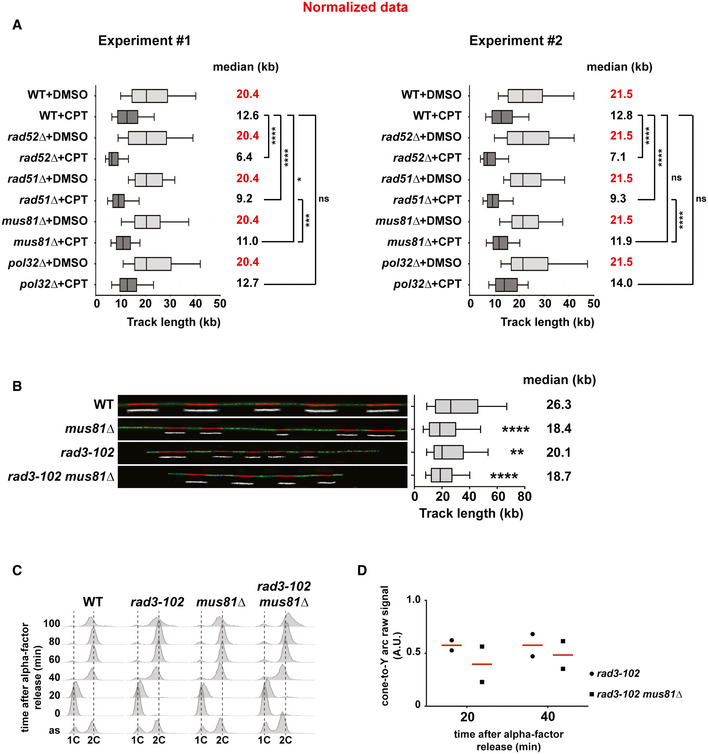
- Statistical analysis of the two biological replicates of DNA combing experiments, corresponding to the data presented in Fig 4A. For each replicate, track length values of DMSO‐treated samples have been normalized to the median of wild‐type values. In each mutant, track length values of CPT‐treated samples have been converted using the same normalization factor as for DMSO‐treated samples. Graphs depict the distribution of normalized EdU track length in kb. Box and whiskers indicate 25–75 and 10–90 percentiles, respectively. Normalized median EdU tracks length is indicated in kb. Asterisks indicate the P‐value of the Mann–Whitney unpaired t‐test, ****P < 0.0001, ***P < 0.001, *P = 0.238, ns non‐significant difference.
- Analysis of replicated DNA tracks length by single‐molecule DNA combing in wild‐type, mus81∆, rad3‐102 and rad3‐102 mus81∆ cells. Exponentially growing cells pulse‐labeled with 50 μM EdU for 20 min. DNA fibers were combed on silanized coverslips, and EdU‐labeled DNA was detected by Click chemistry. Graph depicts the distribution of EdU tracks length in kb. Box and whiskers indicate 25–75 and 10–90 percentiles, respectively. Median EdU tracks length is indicated in kb (values from two biological replicates were pooled). Asterisks indicate the P‐value of the Mann–Whitney unpaired t‐test, ****P < 0.0001, **P < 0.01. Representative images of DNA fibers are shown. Red and white: EdU, green: DNA.
- Analysis of DNA content by flow cytometry of cells collected for 2D‐gel analyses in Fig 4C. Cells were synchronized in G1 with α‐factor in YPAD medium, released into S phase, and collected at the indicated time points. Two biological replicates have been performed.
- Quantification of recombination intermediates (cone signal relative to Y arc signal) in two independent experiments illustrated in Fig 4C. Individual and mean values are plotted for each time point analyzed.
Source data are available online for this figure.
Mus81 resolves S phase‐induced recombination events in G2/M
Our data indicate that Mus81 is required to cope with the replication stress induced by Top1 poisoning but it does not promote replication progression in S phase. In human cells, Mus81 has been proposed to promote fork restart by cleaving stalled or reversed replication forks 54. To understand this apparent contradiction and define the precise window of Mus81 activity, we assessed replication progression by two‐dimensional (2D) neutral–neutral gel electrophoresis in synchronized cultures after release from G1 phase, which was previously used to characterize the replication defects in rad3‐102 cells. We studied the early replication origin ARS305 and the passively replicated region C besides it 82. Notably, rad3‐102 cells accumulated complex branched structures at region C (Fig 4B and C, see arrows), described as recombination or fork reversal events 65. This assay gave us the opportunity to study the role of Mus81 in replication fork progression and recombination intermediate resolution in the rad3‐102 background. As previously described, firing at ARS305 occurs slightly earlier in rad3‐102 cells compared to wild type 65. In the absence of Mus81 (mus81∆ and rad3‐102 mus81∆ mutants), slower replication was observed around the ARS305, as the Y arc signal was still clearly observable at 60 min, while it had already disappeared in wild‐type and rad3‐102 cells (Fig 4C, left panel). This is consistent with a slower replication fork progression in mus81∆ than in wild‐type cells observed by DNA combing in the absence of CPT‐induced DNA damage (Figs 4A and EV3B). Complex branched structures did not accumulate in region C in the absence of Mus81 in rad3‐102 mus81∆ cells (Fig 4C, see arrows and Fig EV3D), suggesting that Mus81 does not process these substrates, observed in rad3‐102 cells during S phase. More interestingly, we noted that, in a 100‐min time‐window after G1 release, ARS305 fired twice in wild‐type, mus81∆, and rad3‐102 cells, implying two rounds of replication. This was not observed in the rad3‐102 mus81∆ mutant (Fig 4C, left panel). As for ARS305 region, replication forks progressed only once through region C in rad3‐102 mus81∆ cells (Fig 4C, right panel). These results made us consider a cell cycle delay that could stem from a replication termination defect in the absence of Mus81 in rad3‐102 cells.
To explore this, we monitored the appearance of Rad52‐YFP foci in cells exposed to CPT (Fig 5A). Wild‐type cells were synchronized in G1 with α‐factor, incubated with CPT for 30 min, and then released from G1 into S phase in the presence of CPT. Wild‐type cells incubated with DMSO showed the appearance of a low amount of Rad52 foci in S phase 40 and 60 min after release from G1 (Fig 5C). These foci disappeared when cells reached G2/M (Fig 5B and C), consistent with the observation that spontaneous Rad52 foci predominantly form in S phase cells 83. When wild‐type cells were released from the G1 arrest in the presence of CPT, they accumulated six times more Rad52 foci than in control cells only when cells entered S phase at 40 min (Fig 5B and C). This accumulation continuously increased until cells reached the G2/M phase at 80 min and then started to decrease in later time points (Fig 5B and C). These results show that S phase entry is required for the initiation of recombination events induced by CPT, and these events are resolved after the completion of DNA replication in G2/M. When assessing the contribution of Mus81, we made two main observations. First, the increased accumulation of Rad52 foci during S phase caused by CPT exposure was not suppressed in the absence of Mus81 (Fig 5B and C), suggesting that Mus81 is not required to generate the substrates for recombination. Second, we observed that CPT‐induced Rad52 foci in mus81∆ cells accumulated over the entire time course experiment until 150 min, not showing the decrease observed in wild‐type cells during the G2/M phase (Fig 5B and C). This phenotype was again confirmed in rad3‐102 mus81∆ cells, which accumulated more Rad52 foci than either single mutant (Fig EV4A). These results suggest that Mus81 processes S phase‐induced recombination events in G2/M.
Figure 5. Mus81 is involved in the processing of recombination intermediates in G2/M after replication restart.
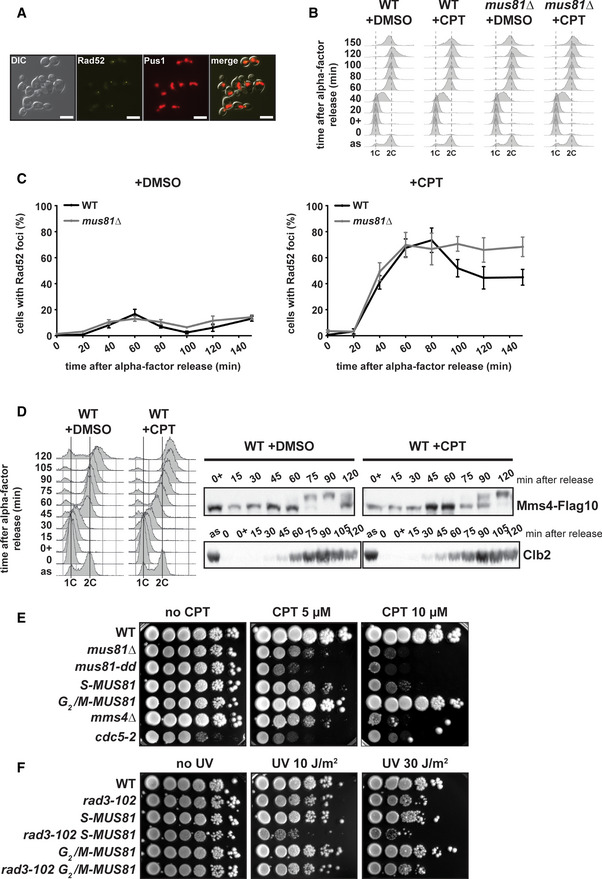
-
AAnalysis of nuclear Rad52‐YFP foci formation. An illustrative image of the experimental setup is shown. DIC, differential interference contrast; yellow, Rad52‐YFP; red, mCHERRY‐Pus1 (nuclear compartment marker). Scale bar = 5 μm.
-
B, CKinetic analysis of Rad52 foci formation. Wild‐type and mus81∆ cells containing Rad52‐YFP and mCHERRY‐Pus1 were synchronized in G1 with α‐factor, treated with DMSO or 100 μM CPT, let in G1 for 30 min, and released into S phase. Cells were collected at the indicated time points and visualized by fluorescence microscopy. Mean ± SEM of cells with Rad52 foci are shown for each time point. Flow cytometry profiles corresponding the experimental setup are shown. Four biological replicates have been performed.
-
DMms4‐Flag10 phosphorylation analyzed by immunoblot in wild‐type cells exposed to CPT. Wild‐type cells were synchronized in G1 with α‐factor, treated with DMSO or 50 μM CPT, let in G1 for 1 h, and released into S phase. Cells were collected at the indicated time points and Mms4 was immunodetected with anti‐Flag antibodies. Clb2 immunodetection serves as a marker for G2/M phase entry. FACS profiles corresponding the experimental setup are also shown. Two biological replicates have been performed.
-
ECPT sensitivity assayed by 10‐fold serial dilutions of S‐MUS81 and G 2/M‐MUS81 alleles compared to the wild‐type, mus81, mms4∆ and cdc5‐2 mutants. Three biological replicates have been performed.
-
FUV sensitivity assayed by 10‐fold serial dilutions of S‐MUS81 and G 2/M‐MUS81 alleles in combination with rad3‐102 on YPAD plates after exposure to the indicated UV‐C doses. Three biological replicates have been performed.
Source data are available online for this figure.
Figure EV4. Mms4 phosphorylation in G2/M is required for recombination intermediates processing after replication restart.
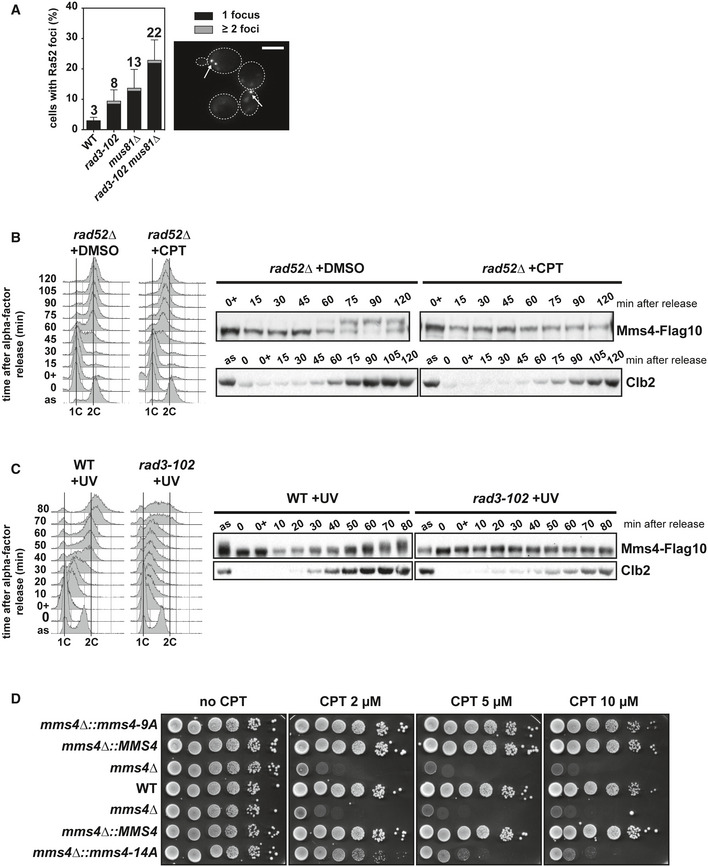
- Analysis of Rad52 foci formation. Cells with Rad52‐YFP foci were scored in exponentially growing wild‐type mus81∆, rad3‐102, and rad3‐102 mus81∆ cells. Mean and SEM of cells with Rad52 foci for three biological replicates are shown. An illustrative image is shown with Rad52 foci indicated by white arrows. Scale bar = 5 μm.
- Mms4‐Flag10 phosphorylation analyzed by immunoblot in rad52∆ cells exposed to CPT. rad52∆ cells were synchronized in G1 with α‐factor, treated with DMSO or 50 μM CPT, let in G1 for 1 h, and released into S phase. Cells were collected at the indicated time points and Mms4 was immunodetected with anti‐Flag antibodies. Clb2 immunodetection serves as a marker for G2/M phase entry. Flow cytometry profiles corresponding the experimental setup are shown. Two biological replicates have been performed.
- Mms4‐Flag10 phosphorylation analyzed by immunoblot in wild‐type and rad3‐102 cells. Cells were synchronized in G1 with α‐factor, untreated or irradiated with 30 J/m2 UV‐C, let in G1 for 2 h, and released into S phase. Cells were collected at the indicated time points, and Mms4 was immunodetected with anti‐Flag antibodies. Clb2 immunodetection serves as a marker for G2 phase entry. Flow cytometry profiles corresponding the experimental setup are shown. Three biological replicates have been performed.
- CPT sensitivity assayed by 10‐fold serial dilutions of wild‐type and mms4 mutants. Three biological replicates have been performed.
Source data are available online for this figure.
In an unperturbed cell cycle, the regulating subunit Mms4 of the Mus81‐Mms4 complex is hyper‐phosphorylated by multiple kinases at the G2/M transition, and this correlates with an enhanced activity of the complex on branched DNA structures in vitro 58, 59, 60, 61. To validate whether the temporal regulation of Mus81 activity fits with the timely requirement for Mus81 to process recombination intermediates generated during CPT‐induced damage repair (Fig 5C), we followed a synchronous culture going through a single S phase and analyzed Mms4 phosphorylation. Treatment of wild‐type cells with CPT caused a delay in S phase progression with respect to control cells, with a consequent delay in Mms4 phosphorylation, detected as an electrophoretic mobility shift by immunoblotting (Fig 5D). The highest degree of Mms4 phosphorylation was reached 90 and 120 min after G1 release in control cells and cells treated with CPT, respectively (Fig 5D). In rad52∆ cells exposed to CPT, cells arrested in late S phase and no phosphorylation of Mms4 was observed (Fig EV4B). We also analyzed Mms4 phosphorylation in wild‐type and rad3‐102 cells. First, it is worth noting that exposure to UV‐C in G1 phase did not induce any change in Mms4 mobility in both tested strains (Fig EV4C). In wild‐type cells, Mms4 underwent phosphorylation 60 to 70 min after release from G1, when most cells had reached the G2/M phase. However, no phosphorylation of Mms4 was observed in rad3‐102 cells, in agreement with their accumulation in S phase after G1 release (Fig EV4C). Thus, the nuclease activity of Mus81‐Mms4 is required to process S phase‐associated HR events once the cells reach G2/M (validated by the accumulation of the mitotic cyclin B2 (Clb2)) (Fig 5D). Indeed, hyper‐phosphorylation of Mms4 is required for the function of Mus81 in the repair of CPT‐induced DNA damage, as indicated by the increased sensitivity to CPT compared to wild‐type cells of the mms4‐14A mutant, which cannot undergo phosphorylation by Cdk1 and Cdc5, and of the cdc5‐2 kinase‐defective mutant (Figs 5E and EV4D) 59. However, we noticed that the CPT sensitivity of the mms4‐14A mutant was lower than in the complete absence of Mms4 (mms4∆), and the mms4‐9A mutant (mms4‐np, 58) was not found more CPT sensitive than the wild type (Fig EV4D). This discrepancy between mms4 mutants could be explained by the high number and redundancy of Mms4 phosphorylation sites 59, 61.
Last, to confirm the need of Mus81 only after the completion of genome duplication, we limited the availability of Mus81 to the S or G2/M phases by taking advantage of the regulatory elements of the cyclin B6 and B2, respectively 84, 85. Fused to the S‐tag 85, Mus81 was only present when cells entered the S phase and was targeted for degradation when reaching the G2/M phase (Appendix Fig S5A). Fused to the G2/M tag 84, Mus81 appeared once cells reached the G2/M phase and was targeted for degradation in the following G1 phase (Appendix Fig S5B). Cells bearing the S‐MUS81 allele were sensitive to CPT‐induced DNA damage, similar to cells lacking Mus81, Mms4, or the catalytic activity of Mus81 (mus81‐dd) (Fig 5E). On the contrary, cells bearing the G 2/M‐MUS81 allele were not sensitive to CPT‐induced DNA damage (Fig 5E). Analogously, rad3‐102 S‐MUS81 cells were more sensitive to UV than the rad3‐102 single mutant, whereas rad3‐102 G 2/M‐MUS81 was not (Fig 5F).
Overall, we conclude that HR factors are required for the restart of replication forks blocked by Top1 poisoning by mediating recombination events. Despite that these events occur in S phase, Mus81 nuclease is acting to process recombination events only when its activity is increased during the G2/M phase. Our results also show that Mus81 was not required for the assembly of Rad52 foci nor activated at the time of their appearance, indicating that Mus81 is unlikely to be involved in the generation of HR substrates by cleaving replication forks in our systems.
Mus81 processes recombination intermediates to complete replication
Our 2D‐gel analysis indicated that rad3‐102 mus81∆ cells synchronously released from a G1 block had a delay in starting the following cell cycle (Fig 4C). We confirmed this observation in cells exposed to Top1 poisoning by performing longer time course flow cytometry experiments. Indeed, mus81∆ cells treated with CPT started the following cell cycle 40 min later than wild‐type cells under the same treatment (Fig EV5A). This cell cycle delay could stem from the inability of cells to timely process recombination intermediates induced by replication fork restart. Since we proposed that HR may promote the restart of CPT‐induced blocked forks by BIR, this implies the formation of a D‐loop. Merging of the D‐loop with a converging replication fork would form a single Holliday junction, whose resolution would be mandatory for replication termination. We did not observe an accumulation of recombination or termination intermediates in rad3‐102 mus81∆ cells, yet they may have not accumulated in the region analyzed by 2D gels. Thus, we decided to look for the accumulation of termination intermediates at the genome level by pulsed‐field gel electrophoresis (PFGE). PFGE allows the separation of individual chromosomes according to their size in G1 and G2/M cells. However, when cells enter S phase, the chromosomes are trapped into the wells due to the presence of joint molecules (JMs), as replication bubbles and other replication intermediates. For instance, this could be observed for wild‐type or mus81∆ cells incubated with DMSO, for which chromosome bands disappeared from the gel 40 min after the release from G1 and could be detected back again at 60 min (Fig 6B, upper panel). After Southern blot analysis, we could observe that chromosome IV was retained in the gel well at S phase entry 40 min after release and then disappeared from the well when the cells reached G2/M (Fig 6B, lower panel). This is consistent with the FACS analysis, which shows that bulk DNA synthesis started at 40 min and ended 60 to 80 min after release (Fig 6A). Incubation of wild‐type cells with CPT induced a delay in S phase progression (Fig 6A), which was mirrored by the kinetics of accumulation of JMs in the gel wells (Fig 6B). As shown before (Fig 3A), CPT treatment induced the same S phase progression delay in mus81∆ and wild‐type cells (Fig 6A). Yet, opposite to the situation in wild‐type cells, the amount of JMs did not decrease 100 min after release from G1 in CPT‐treated mus81∆ cells and chromosomes hardly re‐entered the gel (Fig 6B). Since the bulk of DNA synthesis had already ended at that time, these JMs unlikely represents replication forks but rather single Holliday junctions that would form upon the merging of D‐loops with converging forks. The cdc5‐2 mutant fails to phosphorylate the Mus81‐Mms4 complex in G2/M in order to increase its nuclease activity 59. In CPT‐treated cdc5‐2 cells, we observed a similar accumulation of JMs after the bulk of DNA replication compared to the mus81∆ mutant, suggesting that Mus81‐Mms4 complex phosphorylation in G2/M is required for the nuclease to resolve CPT‐induced JMs (Fig 6B). In G 2/M‐MUS81 cells, in which Mus81 presence is strictly restricted to the G2/M phase (Appendix Fig S5B), CPT‐induced JMs did not accumulate over time, indicating that the restriction of Mus81 to G2/M is sufficient for JMs processing (Fig 6B). Altogether, these results indicate that Mus81 nuclease activity would be required in G2/M for the processing of termination intermediates that specifically form after the restart/protection of replication forks by HR during S phase.
Figure EV5. DSBs are not detected in wild‐type CPT‐treated and rad3‐102 cells.
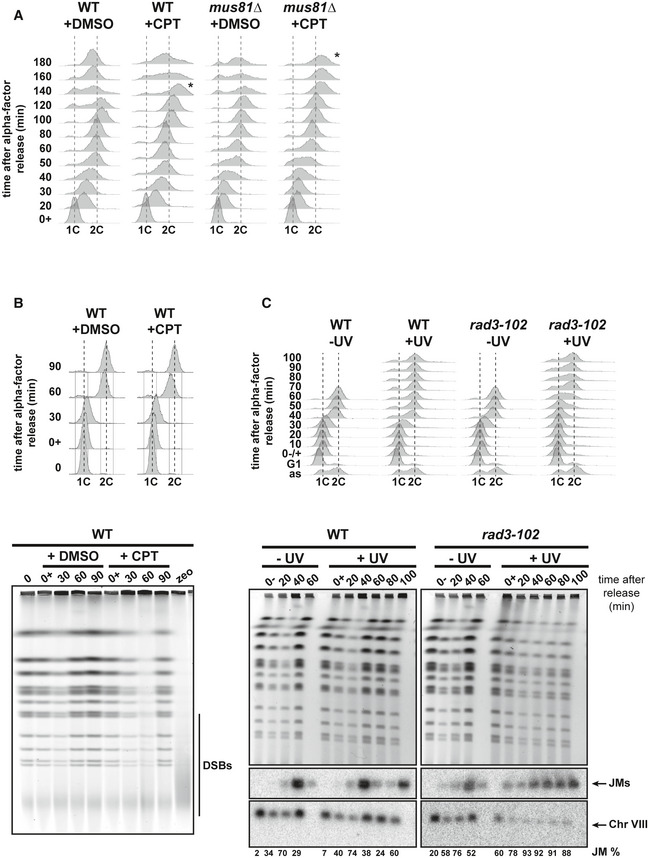
- Analysis of DNA content by flow cytometry of G1 phase synchronized wild‐type and mus81∆ cells and further released into S phase. Cells were synchronized in G1 with α‐factor, treated with DMSO or 100 μM CPT, let in G1 for 1 h, and released into S phase. Asterisks indicate when cells start the following cell cycle. Three biological replicates have been performed.
- Pulsed‐field gel electrophoresis (PFGE) analysis of wild‐type cells in response to CPT. Wild‐type cells were synchronized in G1 with α‐factor, treated with DMSO or 200 μM CPT, let in G1 for 1 h, and released into S phase. Cells were collected at the indicated time points. DNA contents were analyzed by flow cytometry, and the DNA extracted in agarose plugs was analyzed by PFGE. The agarose gel has been stained with ethidium bromide. zeo, wild‐type cells released from the G1 arrest in the presence of 1 mg/ml Zeocin for 1 h. Zeocin‐induced DSBs are indicated by a vertical bar. Three biological replicates have been performed.
- PFGE analysis of wild‐type and rad3‐102 cells. Wild‐type and rad3‐102 cells were synchronized in G1 with α‐factor, untreated or irradiated with 20 J/m2 UV‐C, let in G1 for 2 h, and released into S phase. Cells were collected at the indicated time points. DNA contents were analyzed by flow cytometry, and the DNA extracted in agarose plugs was analyzed by PFGE. Upper panel: agarose gel stained with ethidium bromide; lower panels: Southern blot using a chromosome VIII‐specific probe. JMs, joint molecules accumulated in the gel wells. The mean value of JMs relative to the total amount of DNA is indicated for each time point. Two biological replicates have been performed.
Source data are available online for this figure.
Figure 6. Recombination intermediates generated by replication restart accumulate in the absence of Mus81.
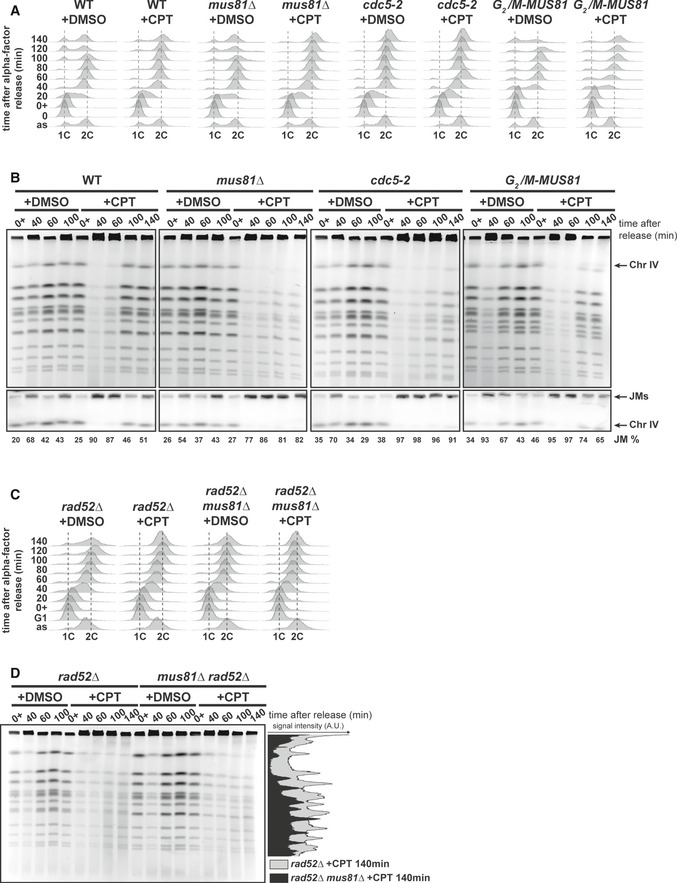
-
A, BPulsed‐field gel electrophoresis (PFGE) analysis of wild‐type, mus81∆, cdc5‐2, and G 2/M‐MUS81 cells in response to CPT. Cells were synchronized in G1 with α‐factor, treated with DMSO or 100 μM CPT, let in G1 for 1 h, and released into S phase. Cells were collected at the indicated time points. DNA contents were analyzed by flow cytometry, and the DNA extracted in agarose plugs was analyzed by PFGE. Upper panel: agarose gel stained with ethidium bromide; lower panel: Southern blot using a chromosome IV‐specific probe. JMs, joint molecules accumulated in the gel wells. The mean value of JMs relative to the total amount of DNA is indicated for each time point. Two to three biological replicates have been performed.
-
C, DPFGE analysis of rad52∆ and rad52∆ mus81∆ cells in response to CPT performed as in (A) (B). The gel has been stained with ethidium bromide and densitometry profiles corresponding to the +CPT 140 min time points in rad52∆ and rad52∆ mus81∆ cells are shown. Two biological replicates have been performed.
Source data are available online for this figure.
One last remaining question was whether this BIR‐mediated replication restart occurs from broken or reversed replication forks. The stabilization of a nicked DNA bound to Top1 by CPT has been initially proposed to induce a replication fork run‐off, leading to the formation of a one‐ended DSB 49. More recently, an alternative model proposed that Top1 inhibition by CPT causes the accumulation of topological constrains in DNA that may impede the progression of replication forks 50, 53. Since 20–25% of forks have been observed to be reversed by electron microscopy after in vivo psoralen crosslinking in CPT‐treated yeast cells, it was suggested that the accumulation of supercoils in front of replication forks promotes their reversal 50, 52. Reversed forks are formed upon the annealing of nascent strands together, exposing a single double‐strand DNA end that mimics a one‐ended DSB. In PFGE experiments, the occurrence of DSBs would result in the diminution of full‐length chromosome bands and the appearance of smeared signals. This could be readily observed in wild‐type cells released in S phase in the presence of the radiomimetic agent zeocin, which induces fragmentation of chromosomes (Fig EV5B). We could not observe such smears in wild‐type CPT‐treated cells when they progressed through S phase 30–60 min after release from the G1 block (Figs 6B and EV5B). Similarly, we could not observe any smeared signal in S phase rad3‐102 cells (Fig EV5C). In the absence of Rad52, which is strictly required for HR‐mediated repair of DSBs, smears could be detected only when CPT‐treated cells reached the G2/M phase 100 min after release (Fig 6C and D). This could be explained by the accumulation of unrepaired CPT‐induced DSBs that could not be detected in S phase. Alternatively, the appearance of broken chromosomes in rad52∆ cells could be the consequence of the cleavage of blocked or reversed forks that could not undergo restart through HR. The latter hypothesis is supported by densitometry analysis of PFGE, showing that the amount of smeared signals at 140 min postrelease in rad52∆ cells treated with CPT was reduced by 50% in the absence of the Mus81 nuclease (Fig 6C and D).
Altogether, our results favor a model in which Top1 poisoning by CPT in our experimental conditions mostly induces replication fork reversal rather than DSBs during S phase. Mus81 is required to process recombination intermediates that accumulate at termination sites following HR‐mediated replication forks restart and forks that could not be restarted because of an HR defect.
Discussion
In the present study, we have used Top1 poisoning by CPT to generate a replication stress genome‐wide and to study the restart of replication forks blocked by Top1ccs. Our results do not support a model involving Pol32‐dependent BIR to promote replication progression at the global level. However, HR factors Rad52 and Rad51 are required during S phase to protect blocked forks from degradation. We propose that HR mainly acts by generating a D‐loop structure protecting blocked forks until their merging with convergent forks. The Mus81 nuclease does not participate in the replication restart but appears to be essential to resolve recombination intermediates to promote the termination of protected/restarted forks. Thanks to a complementary, though independent, approach using the rad3‐102 allele, we conclude that this mechanism is independent of the accumulation of DNA supercoiling and DNA‐protein crosslinks naturally caused by CPT.
It is worth emphasizing that this work has been made possible because we set up conditions to improve CPT entry into yeast cells in liquid cultures. Using standard culture conditions, CPT treatment did not detectably delay S phase completion, even in rad52∆ mutants, whereas this phenotype was clearly observed in CPT‐treated human cells 50. The culture conditions we used allowed observing a CPT‐dependent S phase delay, which could be exacerbated by mutants defective in HR as rad52∆ and rad51∆. These S phase progression defects could be characterized at the molecular level by demonstrating replication defects by both DNA combing and pulsed‐field gel electrophoresis (PFGE) experiments. The latter experiments could also show physical evidence of chromosome breakage upon CPT treatment, which was not found in previous studies 50, 51. Nevertheless, DSBs only appeared in CPT‐treated rad52∆ cells and not during their progression through S phase, as expected from the conversions of nicked DNA into DSBs by the passage of replication forks 47, 48, 49. DSBs appeared when cells reached the G2/M phase and were partially dependent on the presence of the Mus81 nuclease. The remaining DSBs are expected to be dependent on the Yen1 nuclease because the survival of mus81∆ cells is sustained by the presence of Yen1 in response to CPT and in rad3‐102 cells 71, 72. Consistently, Yen1 catalytic activity is restricted to mitosis 59, 86, 87.
CPT‐induced DSBs were also detected in human cells and were partially Mus81‐dependent 54. In these cells, immunostaining of γ‐H2AX, a marker of DNA damage checkpoint activation, was co‐localizing with sites of neo‐synthesized DNA. This led the authors to propose that Mus81 cleaves stalled replication forks to promote their restart 54. However, these experiments were performed in HCT116 colorectal carcinoma cells, which are deficient for Mre11 88, 89. mre11∆ yeast cells are as sensitive to CPT as rad52∆ cells, and the combinations of mre11∆ or rad52∆ with rad3‐102 are lethal 65, suggesting that Mre11 is absolutely required for HR‐mediated replication restart. Thus, the results described in human HCT116 cells are reminiscent of the Mus81‐dependent DSBs we have observed in rad52∆ cells, which likely represent the cleavage of blocked or reversed forks that could not undergo restart through HR (Fig 7) 90.
Figure 7. Proposed model for HR‐mediated protection and restart of blocked replication forks.
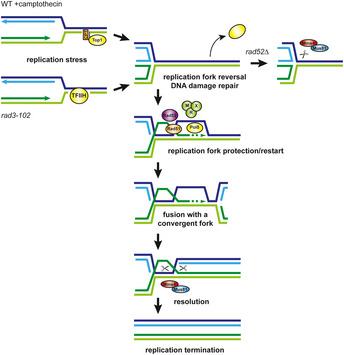
Replication fork block by CPT and in rad3‐102 cells would result in fork reversal, favoring the removal of the replication impediment. The tip of the reversed fork would serve as a substrate for the invasion of the parental duplex by HR, requiring Rad52, Rad51, the MRX complex, and the DNA polymerase δ (including Pol32). Replication could restart by BIR until the merging of the D‐loop with a convergent fork, producing a Holliday junction. Resolution by Mus81 would promote replication termination. Mus81 could also cleave replication forks that could not be restarted by HR (e.g. rad52∆ mutant).
Our results are consistent with a recent study on the survival to an inducible, replication‐born, one‐ended DSB, which primarily depends on Rad52 and Rad51 4. Since we did not detect DSBs in wild‐type CPT‐treated nor in rad3‐102 cells, we favor the hypothesis that replication restart in our systems would use the tip of reversed forks as substrates for HR, as proposed in fission yeast and in mammalian cells 32, 40. This would explain why the absence of the Ku complex, which is normally recruited to double‐stranded DNA ends, could suppress the resection defect caused by the lack of Mre11 nuclease activity in cells treated with CPT or containing the rad3‐102 allele 66. Additionally, replication intermediates corresponding to fork reversal were observed by 2D‐gel analyses in rad3‐102 cells 65. The occurrence of fork reversal in rad3‐102 cells would suggest that fork reversal upon Top1 poisoning would not be the consequence of the accumulation of topological stress ahead of replication forks 50, 53, 91. DNA end resection of reversed forks would involve the Mre11 and Exo1 nucleases 7, 92, 93, and Rad52 and Rad51 would promote the efficient invasion of the parental duplex (Fig 7).
We had previously reported that replication restart in the absence of Rad51 in rad3‐102 cells can still be compensated by the presence of Pol32, a non‐essential subunit of DNA polymerase δ 65. The repair of replication‐born DSBs by sister‐chromatid recombination in a plasmid‐based system confirmed the existence of a Rad51‐independent, Pol32‐dependent pathway 68. A similar pathway, termed MIDAS, has been described in mammalian cells and depends on RAD52 and POLD3, the homolog of Pol32, but not on RAD51 94, 95, 96. Here, we have shown that Pol32 also compensates the absence of Rad51 for CPT resistance and DSB repair by BIR. These results have led us to propose that Pol32 could facilitate strand invasion in the absence of Rad51. The absence of Pol32 alone only affects cell survival in response to high doses of CPT 69. Likewise, Pol32 is not primarily required for cell survival in response to broken replication forks in the Flp‐nick system 4. The Pif1 DNA helicase is required for extensive DNA synthesis during BIR 24, 26. However, the absence of nuclear Pif1 in pif1‐m2 mutants neither sensitizes cells to CPT 4 nor leads to lethality in rad3‐102 rad51∆ cells (unpublished observations). These results suggest that extensive BIR‐mediated synthesis is not essential in response to blocked forks. Because DNA replication is initiated from multiple origins along chromosomes, blocked replication forks could be restarted by BIR but rapidly stopped by a convergent fork (Fig 7) 4. Nevertheless, HR events are occurring in S phase in response to CPT and HR factors required for strand invasion, Rad52 and Rad51, are essential to complete replication in response to blocked forks. Track length of nascent DNA synthesis is reduced in the absence of these factors, suggesting that HR engagement could protect nascent strands from degradation until encountering a convergent fork (Fig 7). Consistently, it has been shown that nascent DNA strands at blocked or broken forks are extensively degraded by nucleolytic activities in the absence of Rad52 in fission yeast or BRCA2 in human cells 7, 8, 18, 90, preventing the merging with a convergent fork 7. This protection mechanism could happen through the formation of a D‐loop, a mechanism reminiscent of the protection of chromosome ends by the looping of telomeres (T‐loops) 97.
Mus81 is required for cell survival in response to CPT and in the rad3‐102 background. Our molecular analyses show that Mus81 is not required for HR‐mediated restart of blocked forks but rather for the processing of joint molecules after the bulk of DNA synthesis in G2/M phase. After the restart of a broken fork by BIR, it has been proposed that Mus81 could cleave the migrating D‐loop in order to limit the mutagenesis associated with BIR synthesis and re‐establish a stable fork structure 4. Our data argue against the possibility that Mus81 could fulfill this role during the S phase because Mus81 is not activated in S phase by the replication stress caused by CPT or rad3‐102 and the absence of Mus81 does neither affect, nor ameliorate, replication progression in these conditions. One possibility is that HR‐mediated replication restart is initiated in S phase but the priming of DNA synthesis is delayed until G2/M, when Mus81 catalytic activity is enhanced by the hyper‐phosphorylation of its regulatory partner Mms4. This hypothesis is consistent with the observed time delay between strand invasion and the initiation of DNA synthesis within the D‐loop when BIR initiated in G2/M 21. In the context of fork restart, this delay would give the opportunity to a convergent fork to reach the blocked forks protected by HR to ensure the completion of replication without challenging genome stability. The fusion of a D‐loop with a convergent replication fork would lead to the formation of a nicked Holliday junction, for which the Mus81 nuclease has a high affinity in vitro 98, 99. Indeed, we observed in cells treated with CPT and devoid of Mus81 activity, an accumulation of joint DNA molecules, which are consistent with the formation of Holliday junctions. Thus, in response to fork blockage, Mus81 could process Holliday junctions at termination sites (Fig 7), as suggested previously 4. We propose that this would be the essential function of Mus81 after the restart of blocked forks. During the preparation of this manuscript, and by using cell cycle‐restricted Mus81 constructs, the lab of Boris Pfander has come to the same conclusion that Mus81 acts post‐replicatively in response to various replication stresses 100.
In conclusion, our work describes how cells deal with blocked replication forks dispersed throughout the genome by Top1 poisoning. Although we do not exclude that HR factors Rad52 and Rad51 can orchestrate the restart of DNA synthesis by BIR, these factors primarily act to protect blocked forks until they merge with converging forks. The processing of intermediates formed during fork merging by Mus81 is then required for the completion of DNA replication. Because incomplete NER reactions as those observed in rad3‐102 cells also occur in wild‐type cells 101, and Top1ccs have been found to accumulate naturally 44, this mechanism is likely to be general in response to other replication blocks requiring HR to complete replication. Finally, the method we have described to use CPT in yeast cell cultures will allow performing a further characterization of Top1 poisoning by CPT at the molecular level. This will indubitably have important implications for understanding the effects of CPT as a chemotherapeutic agent.
Materials and Methods
Yeast strains
All Saccharomyces cerevisiae yeast strains used in this study are in W303‐1aR5 background (his3‐11, 15 leu2‐3, 112, trp1‐1 ura3‐1 ade2‐1 can1‐100 RAD5) unless indicated and are listed in Table EV1. Deletion mutants were either obtained by the PCR‐based gene replacement method (verified by PCR and drug sensitivity assays) or by genetic crosses (verified by tetrad analysis).
Sensitivity assays to camptothecin (CPT) and ultra‐violet irradiation (UV)
Cells from mid‐log cultures were counted using a CASY® (OLS system) and concentrated to 1 × 108 cells/ml, 10 μl of 10‐fold serial dilutions was spotted on rich YPAD plates +/− CPT ((S)‐(+)‐Camptothecin, Sigma #C9911), and plates were incubated for 3 days at 30°C. To assay UV sensitivity, 10‐fold serial dilutions were spotted on YPAD plates and plates were irradiated with UV‐C in a Bio‐Link™ BLX crosslinker and incubated for 3 days at 30°C in darkness. To quantify cell growth from drop tests, the box tool from Image J software was over‐imposed onto the five/six drops corresponding to each strain and the area under the curve of the derived plots was retrieved. Raw area values were directly exploited in GraphPad Prism for quantification and statistical analyses. Three to four independent biological replicates have been performed for each sensitivity assay.
Colony‐forming unit assays
Cells from mid‐log cultures were counted using a CASY® (OLS system) and concentrated to 1 × 107 cells/ml, and appropriate dilutions were plated on rich YPAD plates +/− CPT ((S)‐(+)‐Camptothecin, Sigma #C9911). Survival rates were determined by counting colonies after incubating the plates for 3–5 days at 30°C. Four independent biological replicates have been performed for each genotype assayed.
Cell cycle progression analyses
Overnight mid‐log cultures at 7 × 106 cells/ml were synchronized in G1 with α‐factor (0.5 μg/ml) in YPAD medium for 2–3 h at 30°C. For UV‐induced DNA damage, G1‐synchonized cells were resuspended in water onto Petri dishes as a 4 mm deep cell suspension, irradiated with UV‐C in a Bio‐Link™ BLX crosslinker, resuspended again in YPAD medium with α‐factor, incubated for 2 more h in darkness, and released into S phase by addition of pronase (50 μg/ml). For CPT‐induced DNA damage, G1‐synchonized cells were washed with MPD +SDS medium (0.17% yeast nitrogen base, 0.1% l‐proline, 2% glucose, and 0.003% SDS) 77, resuspended in MPD +SDS medium supplemented with α‐factor and DMSO or CPT, incubated for 1 more h, and released into S phase by addition of pronase (50 μg/ml). Samples (430 μl) were taken every 10 or 15 min and fixed with 100% ethanol for subsequent flow cytometry analysis. Cells were centrifuged for 1 min at 16,000 g, resuspended in 50 mM sodium citrate buffer containing 10 μl of RNase A (20 mg/ml, Qiagen 76254), and incubated for 2 h at 50°C. Then, 10 μl of proteinase K (Sigma, P6556) was added for further incubation 2 h at 50°C. Aggregates of cells were dissociated by sonication. Twenty μl of cell suspension was incubated with 200 μl of 50 mM sodium citrate buffer containing 4 μg/ml propidium iodide (Sigma) for at least 2 h in darkness. Data were acquired on a MACSQuant analyzer (Miltenyi Biotech) and analyzed with FlowJo software. Two to four independent biological replicates have been performed for each cell cycle progression analysis.
DNA combing
DNA combing was performed essentially as described 79. EdU incorporation and uptake by cells was performed using strains bearing 7 integrated copies of the Herpes simplex virus thymidine kinase (HSV‐TK) and the human equilibrative nucleoside transporter 1 (hENT1) on a plasmid. Overnight mid‐log cultures at 5x106 cells/ml in YPAD medium were washed with MPD +SDS medium and resuspended in MPD +SDS with DMSO or 50 μM CPT, incubated for 2 h, and pulse‐labeled with 50 μM EdU for 20 min. DNA fibers were combed on silanized coverslips. Total DNA was detected with YOYO®‐1 iodide (Molecular Probes Y3601), and EdU‐labeled DNA was detected by Click chemistry using 20 μl of the following mix per coverslip: 16.7 μl of H2O, 0.8 μl of 100 mM Copper sulfate, 0.5 μl of 0.5 mg/ml Alexa Fluor® 555 Azide (Molecular Probes A20012), and 2 μl of 100 mM sodium ascorbate. Coverslips were incubated for 1 h at 60°C in a humid chamber in obscurity. Images were recorded on a Leica DM6000 microscope equipped with a CoolSNAP HQ CCD camera (Roper Scientific) and processed as described 101. For comparing track length values of CPT‐treated samples, the median values of all DMSO‐treated mutant samples were normalized to the median value of the wild‐type strain as to obtain a normalization factor. For each mutant, we have used this normalization factor to convert all track length values of CPT‐treated samples. For statistical analysis, we used a Mann–Whitney test to compare, in each case, two unpaired groups with no Gaussian distribution. Two independent biological replicates were performed, summing at least 100 counted EdU tracks per condition.
Protein analyses
For time course experiments, proteins were extracted from cell pellets with acid‐washed glass beads in a denaturing cracking buffer (8 M Urea, 5% SDS, 40 mM Tris–HCl pH 6.8, 0.1 mM EDTA, 0.4 mg/ml bromophenol blue, 50 mM NaF, 150 mM β‐mercaptoethanol, 2 mM PMSF) supplemented with cOmplete™ (Roche) protease inhibitors, 20 min at 70°C under permanent agitation in a Thermomixer® (Eppendorf). Extracts were cleared by centrifugation, separated in Nupage® 3–8% (Invitrogen) polyacrylamide gels, and blotted on PVDF membranes using the Trans‐Blot® Turbo™ system (Bio‐Rad). Two to three independent biological replicates have been performed for each time course experiment.
For immunodetection, the following antibodies were used: anti‐FLAG® M2 (Sigma‐Aldrich, F1804), anti‐Clb2 (Santa Cruz Biotech, y‐180), anti‐HA (Abcam, ab9110), and anti‐tubulin YOL1/34 (Abcam, ab6161).
Pulsed‐field gel electrophoresis
Agarose plugs containing chromosomal DNA were made as described 80. Chromosomes were separated at 13°C in a 0.9% agarose gel in TBE 0.5× using a Rotaphor apparatus (Biometra) using the following parameters: interval from 100 to 10 s (logarithmic), angle from 120 to 110° (linear), and voltage from 200 to 150 V (logarithmic) during 24 h. The gel was subsequently stained with ethidium bromide and transferred to Hybond XL (GE Healthcare). Quantification of chromosome intensity was performed with ImageJ software after Southern blotting and hybridization using a radioactive probe specific for chromosome IV (ARS453) or VIII (RRM3 gene), using a PhosphorImager (Typhoon IP, GE). Two to three independent biological replicates were performed.
Microscopy analyses
For the visualization of CPT inside cells, images were recorded on a Leica DM6000 microscope after excitation of CPT compound with a 350 nm UV light. Fluorescence quantification was performed with Image J software for three independent biological replicates. For the analysis of spontaneous Rad52 foci, non‐fixed nuclei from exponentially growing cell cultures bearing plasmid pWJ1344 were stained with DAPI and Rad52‐YFP foci were counted in S‐G2 cells. For the kinetic analyses of CPT‐induced Rad52 foci, non‐fixed nuclei were visualized thanks to mCHERRY‐Pus1 and Rad52‐YFP foci were counted in all cells. Four independent biological replicates were performed.
2D gel electrophoresis
For DNA extraction, 50 ml of the desired cultures was collected in Falcon tubes containing 500 μl 10% Sodium Azide and kept in ice till processing. Cells were washed with 5 ml of chilled water, carefully resuspended in 1 ml of 1 M sorbitol‐10 mM EDTA pH 8‐0,1% β‐mercaptoethanol‐2 mg/ml Zymolyase 20T, and then incubated at 30°C during 1 h under soft agitation. After centrifugation, the spheroplast pellet was washed with 500 μl of cold water and then broken with 400 μl of cold water plus 500 μl of 1.4 M NaCl‐100 mM Tris–Cl pH 7.6‐25 mM EDTA pH 8–2% CTAB and incubated for 30 min at 50°C with 40 μl 10 mg/ml RNase. Next, 40 μl 20 mg/ml Proteinase K was added and incubation performed overnight under very soft agitation. After centrifugation, pellet and supernatant were treated separately. The supernatant was extracted with 500 μl (24:1) chloroform:isoamyl alcohol and DNA precipitated with two volumes of 50 mM Tris–Cl pH 7.6‐0 mM EDTA pH 8‐1% CTAB and further resuspended in 250 μl 1.4 M NaCl‐1 mM EDTA pH 8–10 mM Tris–Cl pH 7.6. The original pellet was resuspended in 400 μl 1.4 M NaCl‐1 mM EDTA pH 8‐10 mM Tris–Cl pH 7.6 and incubated during 1 h at 50°C, extracted with 200 μl (24:1) chloroform:isoamyl alcohol, and rejoined with the supernatant DNA. The whole sample was precipitated then with one volume room temperature isopropanol, centrifuged, washed with 70% ethanol, and resuspended in 100 μl 10 mM Tris–Cl pH 8. DNA was digested using 200 U of each EcoRV and HindIII during 5 h and NaCl‐precipitated. First dimension electrophoresis was carried out at room temperature in 0.4% agarose gels at 40 V during 20 h in 1× TBE and next stained with ethidium bromide, and a band comprised between 2 and 12 kb was cut and rotated 90° for the second dimension electrophoresis. Second dimension electrophoresis was carried out at 4°C in 1% agarose gels containing 0.34 μg/ml ethidium bromide at 130 V during 12 h in 1× TBE containing 0.34 μg/ml ethidium bromide. Gels were treated and transferred by standard procedures. For hybridization, coordinates of α32‐P PCR probes were 37883‐41883 for ARS305 and 57903‐61158 for region C on chromosome III. Signals were acquired using a Fujifilm FLA‐5100 PhosphorImager. Two independent biological replicates were performed.
BIR assay
Exponentially growing cells in YPAR (2% Raffinose) medium were plated on rich medium containing 2% glucose (YPAD) or 2% galactose (YPAG) and incubated 3 days at 30°C. Colonies on YPAG plates were replica‐plated onto synthetic complete (SC) medium lacking lysine. BIR frequencies were determined by dividing the number of Lys+ by the number of YPAD cfu. For statistical analysis, we used a Mann–Whitney test (two unpaired groups, non‐parametric). Three to six independent biological replicates were performed for each genotype assayed.
Statistical analysis
Statistical analyses for different experiments were done in GraphPad Prism 8. As indicated in the relevant figure legends, a Mann–Whitney non‐parametric test comparing unpaired groups of values was applied, for no Gaussian distributions were assumed. A P‐value of < 0.02 was deemed significant. Mean ± SEM are displayed for the concerned experiments, and sample size was not predetermined using any statistical method.
Author contributions
Conceived and designed the experiments: BP, MM‐C, AA and PP. Performed the experiments: BP, MM‐C and TV. Analyzed the data: BP, MM‐C, AA and PP. Wrote the paper: BP, MM‐C, AA and PP.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Source Data for Expanded View
Review Process File
Source Data for Figure 5
Source Data for Figure 6
Acknowledgements
We thank L.S. Symington, J.H. Petrini, S.C. West, W.D. Heyer, J.A. Tercero, D. Branzei, S Piatti, H. Hombauer, and R. Kolodner for yeast strains and reagents. We thank Etienne Schwob and the DNA combing facility of Montpellier for providing silanized coverslips. We acknowledge the imaging and flow cytometry facility MRI, a member of the national infrastructure France‐BioImaging supported by the “Agence Nationale pour la Recherche” (ANR‐10‐INBS‐04, Investissements d'avenir), France. This work was funded by grants from the “Agence Nationale pour la Recherche” (ANR, France), the “Institut National du Cancer” (INCa, France), the “Ligue contre le Cancer” (équipe labellisée, France), and the MSDAvenir Fund, France, to PP and from the Spanish Ministry of Economy and Competitiveness (BFU2013‐42918P, and Consolider Ingenio 2010 CSD2007‐015), the “Junta de Andalucia” (BIO‐1238 and CVI‐4567, Spain), and the European Union (FEDER) to AA. BP was supported by fellowships from the “Fondation Recherche Médicale” (SPF20121226243, France) and from the Spanish Ministry of Economy and Competitiveness (JCI 2009‐04101). MMC was recipient of a predoctoral FPU training grant from the Spanish Ministry of Economy and Competitiveness and a postdoctoral grant from the Labex EpiGenMed, an “Investissements d'Avenir” program (ANR‐10‐LABX‐12‐01), France.
EMBO Reports (2020) 21: e49367
Contributor Information
Benjamin Pardo, Email: benjamin.pardo@igh.cnrs.fr.
Andrés Aguilera, Email: aguilo@us.es.
Data availability
Raw data that support the findings of this study others than the source data associated to this article (e.g., microscopy images) have not been deposited in a public database and are available from the corresponding authors upon reasonable request.
References
- 1. Brambati A, Zardoni L, Achar YJ, Piccini D, Galanti L, Colosio A, Foiani M, Liberi G (2018) Dormant origins and fork protection mechanisms rescue sister forks arrested by transcription. Nucleic Acids Res 46: 1227–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yekezare M, Gomez‐Gonzalez B, Diffley JF (2013) Controlling DNA replication origins in response to DNA damage ‐ inhibit globally, activate locally. J Cell Sci 126: 1297–1306 [DOI] [PubMed] [Google Scholar]
- 3. Lambert S, Mizuno K, Blaisonneau J, Martineau S, Chanet R, Freon K, Murray JM, Carr AM, Baldacci G (2010) Homologous recombination restarts blocked replication forks at the expense of genome rearrangements by template exchange. Mol Cell 39: 346–359 [DOI] [PubMed] [Google Scholar]
- 4. Mayle R, Campbell IM, Beck CR, Yu Y, Wilson M, Shaw CA, Bjergbaek L, Lupski JR, Ira G (2015) DNA REPAIR. Mus81 and converging forks limit the mutagenicity of replication fork breakage. Science 349: 742–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ait Saada A, Lambert SAE, Carr AM (2018) Preserving replication fork integrity and competence via the homologous recombination pathway. DNA Repair 71: 135–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Krogh BO, Symington LS (2004) Recombination proteins in yeast. Annu Rev Genet 38: 233–271 [DOI] [PubMed] [Google Scholar]
- 7. Ait Saada A, Teixeira‐Silva A, Iraqui I, Costes A, Hardy J, Paoletti G, Freon K, Lambert SAE (2017) Unprotected replication forks are converted into mitotic sister chromatid bridges. Mol Cell 66: 398–410 e4 [DOI] [PubMed] [Google Scholar]
- 8. Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M (2011) Double‐strand break repair‐independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145: 529–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vallerga MB, Mansilla SF, Federico MB, Bertolin AP, Gottifredi V (2015) Rad51 recombinase prevents Mre11 nuclease‐dependent degradation and excessive PrimPol‐mediated elongation of nascent DNA after UV irradiation. Proc Natl Acad Sci USA 112: E6624–E6633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ameziane N, May P, Haitjema A, van de Vrugt HJ, van Rossum‐Fikkert SE, Ristic D, Williams GJ, Balk J, Rockx D, Li H et al (2015) A novel Fanconi anaemia subtype associated with a dominant‐negative mutation in RAD51. Nat Commun 6: 8829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang AT, Kim T, Wagner JE, Conti BA, Lach FP, Huang AL, Molina H, Sanborn EM, Zierhut H, Cornes BK et al (2015) A dominant mutation in human RAD51 reveals its function in DNA interstrand crosslink repair independent of homologous recombination. Mol Cell 59: 478–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zadorozhny K, Sannino V, Belan O, Mlcouskova J, Spirek M, Costanzo V, Krejci L (2017) Fanconi‐anemia‐associated mutations destabilize RAD51 filaments and impair replication fork protection. Cell Rep 21: 333–340 [DOI] [PubMed] [Google Scholar]
- 13. Mohebi S, Lambert SA, Carr AM (2015) Analyzing the response to dysfunction replication forks using the RTS1 barrier system in fission yeast. Methods Mol Biol 1300: 239–259 [DOI] [PubMed] [Google Scholar]
- 14. Nielsen I, Bentsen IB, Lisby M, Hansen S, Mundbjerg K, Andersen AH, Bjergbaek L (2009) A Flp‐nick system to study repair of a single protein‐bound nick in vivo . Nat Methods 6: 753–757 [DOI] [PubMed] [Google Scholar]
- 15. Cortes‐Ledesma F, Aguilera A (2006) Double‐strand breaks arising by replication through a nick are repaired by cohesin‐dependent sister‐chromatid exchange. EMBO Rep 7: 919–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gonzalez‐Barrera S, Cortes‐Ledesma F, Wellinger RE, Aguilera A (2003) Equal sister chromatid exchange is a major mechanism of double‐strand break repair in yeast. Mol Cell 11: 1661–1671 [DOI] [PubMed] [Google Scholar]
- 17. Roseaulin L, Yamada Y, Tsutsui Y, Russell P, Iwasaki H, Arcangioli B (2008) Mus81 is essential for sister chromatid recombination at broken replication forks. EMBO J 27: 1378–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jakobsen KP, Nielsen KO, Lovschal KV, Rodgaard M, Andersen AH, Bjergbaek L (2019) Minimal resection takes place during break‐induced replication repair of collapsed replication forks and is controlled by strand invasion. Cell Rep 26: 836–844 e3 [DOI] [PubMed] [Google Scholar]
- 19. Anand RP, Lovett ST, Haber JE (2013) Break‐induced DNA replication. Cold Spring Harb Perspect Biol 5: a010397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kramara J, Osia B, Malkova A (2018) Break‐induced replication: the where, the why, and the how. Trends Genet 34: 518–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Malkova A, Naylor ML, Yamaguchi M, Ira G, Haber JE (2005) RAD51‐dependent break‐induced replication differs in kinetics and checkpoint responses from RAD51‐mediated gene conversion. Mol Cell Biol 25: 933–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morrow DM, Connelly C, Hieter P (1997) “Break copy” duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae . Genetics 147: 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Donnianni RA, Symington LS (2013) Break‐induced replication occurs by conservative DNA synthesis. Proc Natl Acad Sci USA 110: 13475–13480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, Deem A, Ira G, Haber JE, Lobachev KS, Malkova A (2013) Migrating bubble during break‐induced replication drives conservative DNA synthesis. Nature 502: 389–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lydeard JR, Jain S, Yamaguchi M, Haber JE (2007) Break‐induced replication and telomerase‐independent telomere maintenance require Pol32. Nature 448: 820–823 [DOI] [PubMed] [Google Scholar]
- 26. Wilson MA, Kwon Y, Xu Y, Chung WH, Chi P, Niu H, Mayle R, Chen X, Malkova A, Sung P et al (2013) Pif1 helicase and Poldelta promote recombination‐coupled DNA synthesis via bubble migration. Nature 502: 393–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Deem A, Keszthelyi A, Blackgrove T, Vayl A, Coffey B, Mathur R, Chabes A, Malkova A (2011) Break‐induced replication is highly inaccurate. PLoS Biol 9: e1000594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Anand RP, Tsaponina O, Greenwell PW, Lee CS, Du W, Petes TD, Haber JE (2014) Chromosome rearrangements via template switching between diverged repeated sequences. Genes Dev 28: 2394–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pardo B, Aguilera A (2012) Complex chromosomal rearrangements generated by break‐induced replication and the role of structure‐selective endonucleases. PLoS Genet 8: e1002979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ruiz JF, Gomez‐Gonzalez B, Aguilera A (2009) Chromosomal translocations caused by either pol32‐dependent or pol32‐independent triparental break‐induced replication. Mol Cell Biol 29: 5441–5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smith CE, Llorente B, Symington LS (2007) Template switching during break‐induced replication. Nature 447: 102–105 [DOI] [PubMed] [Google Scholar]
- 32. Teixeira‐Silva A, Ait Saada A, Hardy J, Iraqui I, Nocente MC, Freon K, Lambert SAE (2017) The end‐joining factor Ku acts in the end‐resection of double strand break‐free arrested replication forks. Nat Commun 8: 1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ahn JS, Osman F, Whitby MC (2005) Replication fork blockage by RTS1 at an ectopic site promotes recombination in fission yeast. EMBO J 24: 2011–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lambert S, Watson A, Sheedy DM, Martin B, Carr AM (2005) Gross chromosomal rearrangements and elevated recombination at an inducible site‐specific replication fork barrier. Cell 121: 689–702 [DOI] [PubMed] [Google Scholar]
- 35. Nguyen MO, Jalan M, Morrow CA, Osman F, Whitby MC (2015) Recombination occurs within minutes of replication blockage by RTS1 producing restarted forks that are prone to collapse. Elife 4: e04539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Iraqui I, Chekkal Y, Jmari N, Pietrobon V, Freon K, Costes A, Lambert SA (2012) Recovery of arrested replication forks by homologous recombination is error‐prone. PLoS Genet 8: e1002976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jalan M, Oehler J, Morrow CA, Osman F, Whitby MC (2019) Factors affecting template switch recombination associated with restarted DNA replication. Elife 8: e41697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Miyabe I, Mizuno K, Keszthelyi A, Daigaku Y, Skouteri M, Mohebi S, Kunkel TA, Murray JM, Carr AM (2015) Polymerase delta replicates both strands after homologous recombination‐dependent fork restart. Nat Struct Mol Biol 22: 932–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Meng X, Zhao X (2017) Replication fork regression and its regulation. FEMS Yeast Res 17, fow110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zellweger R, Dalcher D, Mutreja K, Berti M, Schmid JA, Herrador R, Vindigni A, Lopes M (2015) Rad51‐mediated replication fork reversal is a global response to genotoxic treatments in human cells. J Cell Biol 208: 563–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pommier Y, Sun Y, Huang SN, Nitiss JL (2016) Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat Rev 17: 703–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lippert MJ, Kim N, Cho JE, Larson RP, Schoenly NE, O'Shea SH, Jinks‐Robertson S (2011) Role for topoisomerase 1 in transcription‐associated mutagenesis in yeast. Proc Natl Acad Sci USA 108: 698–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Takahashi T, Burguiere‐Slezak G, Van der Kemp PA, Boiteux S (2011) Topoisomerase 1 provokes the formation of short deletions in repeated sequences upon high transcription in Saccharomyces cerevisiae . Proc Natl Acad Sci USA 108: 692–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stingele J, Schwarz MS, Bloemeke N, Wolf PG, Jentsch S (2014) A DNA‐dependent protease involved in DNA‐protein crosslink repair. Cell 158: 327–338 [DOI] [PubMed] [Google Scholar]
- 45. Balakirev MY, Mullally JE, Favier A, Assard N, Sulpice E, Lindsey DF, Rulina AV, Gidrol X, Wilkinson KD (2015) Wss1 metalloprotease partners with Cdc48/Doa1 in processing genotoxic SUMO conjugates. Elife 4: e06763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pommier Y, Leo E, Zhang H, Marchand C (2010) DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol 17: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hsiang YH, Lihou MG, Liu LF (1989) Arrest of replication forks by drug‐stabilized topoisomerase I‐DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res 49: 5077–5082 [PubMed] [Google Scholar]
- 48. Nitiss J, Wang JC (1988) DNA topoisomerase‐targeting antitumor drugs can be studied in yeast. Proc Natl Acad Sci USA 85: 7501–7505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Strumberg D, Pilon AA, Smith M, Hickey R, Malkas L, Pommier Y (2000) Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5′‐phosphorylated DNA double‐strand breaks by replication runoff. Mol Cell Biol 20: 3977–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ray Chaudhuri A, Hashimoto Y, Herrador R, Neelsen KJ, Fachinetti D, Bermejo R, Cocito A, Costanzo V, Lopes M (2012) Topoisomerase I poisoning results in PARP‐mediated replication fork reversal. Nat Struct Mol Biol 19: 417–423 [DOI] [PubMed] [Google Scholar]
- 51. Redon C, Pilch DR, Rogakou EP, Orr AH, Lowndes NF, Bonner WM (2003) Yeast histone 2A serine 129 is essential for the efficient repair of checkpoint‐blind DNA damage. EMBO Rep 4: 678–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Menin L, Ursich S, Trovesi C, Zellweger R, Lopes M, Longhese MP, Clerici M (2018) Tel1/ATM prevents degradation of replication forks that reverse after topoisomerase poisoning. EMBO Rep 19: e45535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Koster DA, Palle K, Bot ES, Bjornsti MA, Dekker NH (2007) Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature 448: 213–217 [DOI] [PubMed] [Google Scholar]
- 54. Regairaz M, Zhang YW, Fu H, Agama KK, Tata N, Agrawal S, Aladjem MI, Pommier Y (2011) Mus81‐mediated DNA cleavage resolves replication forks stalled by topoisomerase I‐DNA complexes. J Cell Biol 195: 739–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hanada K, Budzowska M, Davies SL, van Drunen E, Onizawa H, Beverloo HB, Maas A, Essers J, Hickson ID, Kanaar R (2007) The structure‐specific endonuclease Mus81 contributes to replication restart by generating double‐strand DNA breaks. Nat Struct Mol Biol 14: 1096–1104 [DOI] [PubMed] [Google Scholar]
- 56. Pepe A, West SC (2014) MUS81‐EME2 promotes replication fork restart. Cell Rep 7: 1048–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Munoz‐Galvan S, Tous C, Blanco MG, Schwartz EK, Ehmsen KT, West SC, Heyer WD, Aguilera A (2012) Distinct roles of mus81, yen1, slx1‐slx4, and rad1 nucleases in the repair of replication‐born double‐strand breaks by sister chromatid exchange. Mol Cell Biol 32: 1592–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gallo‐Fernandez M, Saugar I, Ortiz‐Bazan MA, Vazquez MV, Tercero JA (2012) Cell cycle‐dependent regulation of the nuclease activity of Mus81‐Eme1/Mms4. Nucleic Acids Res 40: 8325–8335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Matos J, Blanco MG, Maslen S, Skehel JM, West SC (2011) Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell 147: 158–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Matos J, Blanco MG, West SC (2013) Cell‐cycle kinases coordinate the resolution of recombination intermediates with chromosome segregation. Cell Rep 4: 76–86 [DOI] [PubMed] [Google Scholar]
- 61. Princz LN, Wild P, Bittmann J, Aguado FJ, Blanco MG, Matos J, Pfander B (2017) Dbf4‐dependent kinase and the Rtt107 scaffold promote Mus81‐Mms4 resolvase activation during mitosis. EMBO J 36: 664–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liu C, Pouliot JJ, Nash HA (2002) Repair of topoisomerase I covalent complexes in the absence of the tyrosyl‐DNA phosphodiesterase Tdp1. Proc Natl Acad Sci USA 99: 14970–14975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vance JR, Wilson TE (2002) Yeast Tdp1 and Rad1‐Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc Natl Acad Sci USA 99: 13669–13674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Herrera‐Moyano E, Moriel‐Carretero M, Montelone BA, Aguilera A (2014) The rem mutations in the ATP‐binding groove of the Rad3/XPD helicase lead to Xeroderma pigmentosum‐Cockayne syndrome‐like phenotypes. PLoS Genet 10: e1004859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Moriel‐Carretero M, Aguilera A (2010) A postincision‐deficient TFIIH causes replication fork breakage and uncovers alternative Rad51‐ or Pol32‐mediated restart mechanisms. Mol Cell 37: 690–701 [DOI] [PubMed] [Google Scholar]
- 66. Foster SS, Balestrini A, Petrini JH (2011) Functional interplay of the Mre11 nuclease and Ku in the response to replication‐associated DNA damage. Mol Cell Biol 31: 4379–4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Moriel‐Carretero M, Aguilera A (2010) Replication fork breakage and re‐start: new insights into Rad3/XPD‐associated deficiencies. Cell Cycle 9: 2958–2962 [PubMed] [Google Scholar]
- 68. Munoz‐Galvan S, Jimeno S, Rothstein R, Aguilera A (2013) Histone H3K56 acetylation, Rad52, and non‐DNA repair factors control double‐strand break repair choice with the sister chromatid. PLoS Genet 9: e1003237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kapitzky L, Beltrao P, Berens TJ, Gassner N, Zhou C, Wuster A, Wu J, Babu MM, Elledge SJ, Toczyski D et al (2010) Cross‐species chemogenomic profiling reveals evolutionarily conserved drug mode of action. Mol Syst Biol 6: 451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bastin‐Shanower SA, Fricke WM, Mullen JR, Brill SJ (2003) The mechanism of Mus81‐Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1‐Rad10. Mol Cell Biol 23: 3487–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Blanco MG, Matos J, Rass U, Ip SC, West SC (2010) Functional overlap between the structure‐specific nucleases Yen1 and Mus81‐Mms4 for DNA‐damage repair in S. cerevisiae . DNA Repair 9: 394–402 [DOI] [PubMed] [Google Scholar]
- 72. Tay YD, Wu L (2010) Overlapping roles for Yen1 and Mus81 in cellular Holliday junction processing. J Biol Chem 285: 11427–11432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Puddu F, Salguero I, Herzog M, Geisler NJ, Costanzo V, Jackson SP (2017) Chromatin determinants impart camptothecin sensitivity. EMBO Rep 18: 1000–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Del Poeta M, Chen SF, Von Hoff D, Dykstra CC, Wani MC, Manikumar G, Heitman J, Wall ME, Perfect JR (1999) Comparison of in vitro activities of camptothecin and nitidine derivatives against fungal and cancer cells. Antimicrob Agents Chemother 43: 2862–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Liu C, Apodaca J, Davis LE, Rao H (2007) Proteasome inhibition in wild‐type yeast Saccharomyces cerevisiae cells. Biotechniques 42: 158, 160, 162 [DOI] [PubMed] [Google Scholar]
- 76. Plevani P, Badaracco G, Ginelli E, Sora S (1980) Effect and mechanism of action of aphidicolin on yeast deoxyribonucleic acid polymerases. Antimicrob Agents Chemother 18: 50–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Pannunzio VG, Burgos HI, Alonso M, Mattoon JR, Ramos EH, Stella CA (2004) A simple chemical method for rendering wild‐type yeast permeable to brefeldin A that does not require the presence of an erg6 mutation. J Biomed Biotechnol 2004: 150–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kono K, Al‐Zain A, Schroeder L, Nakanishi M, Ikui AE (2016) Plasma membrane/cell wall perturbation activates a novel cell cycle checkpoint during G1 in Saccharomyces cerevisiae . Proc Natl Acad Sci USA 113: 6910–6915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bianco JN, Poli J, Saksouk J, Bacal J, Silva MJ, Yoshida K, Lin YL, Tourriere H, Lengronne A, Pasero P (2012) Analysis of DNA replication profiles in budding yeast and mammalian cells using DNA combing. Methods 57: 149–157 [DOI] [PubMed] [Google Scholar]
- 80. Tourrière H, Saksouk J, Lengronne A, Pasero P (2017) Single‐molecule analysis of DNA replication dynamics in budding yeast and human cells by DNA combing. Bio‐Protocol 7: e2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Fu H, Martin MM, Regairaz M, Huang L, You Y, Lin CM, Ryan M, Kim R, Shimura T, Pommier Y et al (2015) The DNA repair endonuclease Mus81 facilitates fast DNA replication in the absence of exogenous damage. Nat Commun 6: 6746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lopes M, Cotta‐Ramusino C, Liberi G, Foiani M (2003) Branch migrating sister chromatid junctions form at replication origins through Rad51/Rad52‐independent mechanisms. Mol Cell 12: 1499–1510 [DOI] [PubMed] [Google Scholar]
- 83. Lisby M, Rothstein R, Mortensen UH (2001) Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci USA 98: 8276–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Karras GI, Jentsch S (2010) The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell 141: 255–267 [DOI] [PubMed] [Google Scholar]
- 85. Hombauer H, Srivatsan A, Putnam CD, Kolodner RD (2011) Mismatch repair, but not heteroduplex rejection, is temporally coupled to DNA replication. Science 334: 1713–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Eissler CL, Mazon G, Powers BL, Savinov SN, Symington LS, Hall MC (2014) The Cdk/cDc14 module controls activation of the Yen1 holliday junction resolvase to promote genome stability. Mol Cell 54: 80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Blanco MG, Matos J, West SC (2014) Dual control of Yen1 nuclease activity and cellular localization by Cdk and Cdc14 prevents genome instability. Mol Cell 54: 94–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Giannini G, Ristori E, Cerignoli F, Rinaldi C, Zani M, Viel A, Ottini L, Crescenzi M, Martinotti S, Bignami M et al (2002) Human MRE11 is inactivated in mismatch repair‐deficient cancers. EMBO Rep 3: 248–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Takemura H, Rao VA, Sordet O, Furuta T, Miao ZH, Meng L, Zhang H, Pommier Y (2006) Defective Mre11‐dependent activation of Chk2 by ataxia telangiectasia mutated in colorectal carcinoma cells in response to replication‐dependent DNA double strand breaks. J Biol Chem 281: 30814–30823 [DOI] [PubMed] [Google Scholar]
- 90. Lemacon D, Jackson J, Quinet A, Brickner JR, Li S, Yazinski S, You Z, Ira G, Zou L, Mosammaparast N et al (2017) MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81‐dependent fork rescue in BRCA2‐deficient cells. Nat Commun 8: 860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Jakobsen KP, Andersen AH, Bjergbaek L (2019) Abortive activity of topoisomerase I: a challenge for genome integrity? Curr Genet 65: 1141–1144 [DOI] [PubMed] [Google Scholar]
- 92. Kim HS, Williamson EA, Nickoloff JA, Hromas RA, Lee SH (2017) Metnase mediates loading of exonuclease 1 onto single strand overhang DNA for end resection at stalled replication forks. J Biol Chem 292: 1414–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Tsang E, Miyabe I, Iraqui I, Zheng J, Lambert SA, Carr AM (2014) The extent of error‐prone replication restart by homologous recombination is controlled by Exo1 and checkpoint proteins. J Cell Sci 127: 2983–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Bhowmick R, Minocherhomji S, Hickson ID (2016) RAD52 facilitates mitotic DNA synthesis following replication stress. Mol Cell 64: 1117–1126 [DOI] [PubMed] [Google Scholar]
- 95. Costantino L, Sotiriou SK, Rantala JK, Magin S, Mladenov E, Helleday T, Haber JE, Iliakis G, Kallioniemi OP, Halazonetis TD (2014) Break‐induced replication repair of damaged forks induces genomic duplications in human cells. Science 343: 88–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Sotiriou SK, Kamileri I, Lugli N, Evangelou K, Da‐Re C, Huber F, Padayachy L, Tardy S, Nicati NL, Barriot S et al (2016) Mammalian RAD52 functions in break‐induced replication repair of collapsed DNA replication forks. Mol Cell 64: 1127–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T (1999) Mammalian telomeres end in a large duplex loop. Cell 97: 503–514 [DOI] [PubMed] [Google Scholar]
- 98. Ehmsen KT, Heyer WD (2009) A junction branch point adjacent to a DNA backbone nick directs substrate cleavage by Saccharomyces cerevisiae Mus81‐Mms4. Nucleic Acids Res 37: 2026–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Schwartz EK, Wright WD, Ehmsen KT, Evans JE, Stahlberg H, Heyer WD (2012) Mus81‐Mms4 functions as a single heterodimer to cleave nicked intermediates in recombinational DNA repair. Mol Cell Biol 32: 3065–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Bittmann J, Grigaitis R, Galanti L, Amarell S, Wilfling F, Matos J, Pfander B (2020) An advanced cell cycle tag toolbox reveals principles underlying temporal control of structure‐selective nucleases. Elife 9: e52459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Andersen SL, Zhang A, Dominska M, Moriel‐Carretero M, Herrera‐Moyano E, Aguilera A, Petes TD (2016) High‐resolution mapping of homologous recombination events in rad3 hyper‐recombination mutants in yeast. PLoS Genet 12: e1005938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Pasero P, Bensimon A, Schwob E (2002) Single‐molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes Dev 16: 2479–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Source Data for Expanded View
Review Process File
Source Data for Figure 5
Source Data for Figure 6
Data Availability Statement
Raw data that support the findings of this study others than the source data associated to this article (e.g., microscopy images) have not been deposited in a public database and are available from the corresponding authors upon reasonable request.


