Abstract
Leaf senescence is a highly complex developmental process that is tightly controlled by multiple layers of regulation. Abscisic acid (ABA) and reactive oxygen species (ROS) are two well‐known factors that promote leaf senescence. We show here that the transcription factor CDF4 positively regulates leaf senescence. Constitutive and inducible overexpression of CDF4 accelerates leaf senescence, while knockdown of CDF4 delays it. CDF4 increases endogenous ABA levels by upregulating the transcription of the ABA biosynthesis genes 9‐cis‐epoxycarotenoid dioxygenase 2, 3 (NCED2, 3) and suppresses H2O2 scavenging by repressing expression of the catalase2 (CAT2) gene. NCED2, 3 knockout and CAT2 overexpression partially rescue premature leaf senescence caused by CDF4 overexpression. We also show that CDF4 promotes floral organ abscission by activating the polygalacturonase PGAZAT gene. Based on these results, we propose that the levels of CDF4, ABA, and ROS undergo a gradual increase driven by their interlinking positive feedback loops during the leaf senescence and floral organ abscission processes.
Keywords: ABA biosynthesis, DOF transcription factor, floral organ abscission, leaf senescence, ROS scavenging
Subject Categories: Plant Biology
This study discovers an amplification loop, involving CDF4, ABA, and ROS, and provides insights into the molecular regulatory network of leaf senescence and floral organ abscission in Arabidopsis.

Introduction
Leaf senescence is a very complicated and orderly developmental process that is regulated by age. It involves the degradation of macromolecules and the mobilization and translocation of metabolic products to the younger tissues of the plant 1. The progression of leaf senescence is regulated not only by many internal factors, such as the reproductive status of the plant, metabolism, and transcriptional regulators, but also by various stresses 2, 3. Its initiation, progression, and completion are tightly regulated by multiple layers of regulatory factors. Plant hormones play a key role in controlling the progression of leaf senescence. Cytokinins (CK) 4, 5 and gibberellic acid (GA) 4, 6, together with auxins 7, are thought to be senescence‐suppressing hormones, while ethylene (ETH), jasmonic acid (JA), salicylic acid (SA), and abscisic acid (ABA) are thought to be senescence‐promoting factors. ETH is a well‐known hormonal regulator of leaf senescence, and mutations that affect ETH biosynthesis or signaling alter the progression of leaf senescence 8, 9. In Arabidopsis thaliana, senescing leaves have higher concentrations of ABA and SA than non‐senescing leaves 10. In senescing Arabidopsis leaves, many ABA and SA biosynthesis‐related genes are upregulated 11.
ABA is a sesquiterpenoid, and its biosynthetic genes have been cloned 12. The NCEDs (9‐cis‐epoxycarotenoid dioxygenases) function in the cleavage of xanthophylls and are rate‐limiting enzymes that regulate ABA biosynthesis 13. ABA DEFICIENT 3 (ABA3) is involved in the synthesis of the molybdenum co‐factor that is needed for NCED3 activity 14. ABA plays important roles not only in the response to abiotic stress but also in seed dormancy, abscission, and senescence 12. Previous reports have shown that exogenous ABA treatment induces senescence and strongly upregulates senescence‐associated genes (SAGs), but the molecular mechanisms underlying this effect are not well characterized. One of the ABA regulators during senescence is the NAC transcription factor, NAC‐like, activated by AP3/P1 (NAP), which was shown to control expression of ABA biosynthesis genes 15. ATAF1 was shown to regulate NCED3 16. In addition, many ABA metabolism‐ and signaling‐related genes have roles in controlling senescence 17. The ABA receptor PYL9 positively regulates leaf senescence and improves plant drought tolerance 18.
Drought, salinity, high light levels, high temperature, and some important developmental changes can promote endogenous H2O2 accumulation, which is responsible for an accelerated leaf senescence process. Increased protein and lipid oxidation caused by upregulated H2O2 levels are characteristics of senescence‐associated syndrome 19. The Arabidopsis jub1 (JUNGBRUNNEN 1) and cpr5 (CONSTITUTIVE EXPRESSION OF PR GENES) mutants exhibit an early‐leaf‐senescence phenotype because of the accumulation of excessive amounts of H2O2 20, 21. However, the ntl4 (NAC WITH TRANSMEMBRANE MOTIF 1‐LIKE4) mutant, with downregulated H2O2 accumulation, shows delayed drought‐related leaf senescence 22. In addition, the functional relationship between ABA and ROS in leaf senescence has previously been reported in many studies. For example, ROS has been proposed to function as a second messenger in ABA signaling in guard cells 23. Mutations in the NADPH oxidase catalytic subunit genes AtRBOHD and AtRBOHF impair ABA‐induced stomatal closure and ABA‐induced cytosolic Ca2+ increases in guard cells 24. ABA is capable of increasing H2O2 levels in maize embryos and seedlings, further supporting the roles of ROS in ABA signaling 25.
Although regulation of leaf senescence in plants is important, the underlying molecular mechanisms are not fully understood. Many methods have been used to isolate a series of SAGs with altered transcript levels in senescing leaves 26. Transcriptome analyses have shown that hundreds of genes are specifically induced during age‐related leaf senescence 27. Furthermore, many plant‐specific transcription factors can regulate these SAGs 28. However, knockdown mutants of relatively few of these transcription factors produce altered leaf senescence phenotypes. For instance, AtWRKY53 is strongly upregulated early in leaf senescence, and its knockdown mutant is defective with respect to age‐related senescence 29. AtWRKY6 expression is induced by leaf senescence 30. Mutations of the genes encoding NAC transcription factors NAP or ORS1 (Oresara 1 sister 1) delay leaf senescence 31, 32. EIN3 accelerates leaf senescence by repressing transcription of the microRNA miR164 33. ORE1 (Oresara 1) regulates the leaf senescence process in the trifurcate feed‐forward pathway 34. VNI2 integrates abscisic acid signals with leaf senescence via the COR/RD genes 35. JUB1 negatively regulates leaf senescence 36, and NAC016 overexpression also induces leaf senescence 37. Environmental stimuli induce the transcriptional regulators PIF4 and PIF5, which also promote dark‐induced senescence by inducing ORE1 expression 38.
Organ abscission is a typical cell separation process that occurs throughout the plant's growth cycle 39. At the predetermined abscission position, an abscission zone (AZ) develops. The AZ cells recognize abscission signals that activate cell wall‐loosening proteins. Endoglucanases, polygalacturonases, and expansins have been well documented as the cell wall‐loosening proteins 40, 41. Many genes have been found to be involved in the regulation of organ abscission. The ida mutant has a delayed abscission phenotype 42, and the HAESA and HAESA‐LIKE2 receptor‐like kinases are activated by interactions with INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) 43. Moreover, the transcription factors AGL15, DOF4.7, and ZFP2 are involved in the regulation of the floral organ abscission process 44, 45, 46.
The DNA‐binding with one finger (DOF) family proteins have been reported to participate in many biological processes since the first DOF transcription factor, ZmDOF1, was found to be a light‐response factor in maize 47. Arabidopsis DOF proteins have been shown to function in many developmental processes, such as flowering time, leaf prioritization, light response, cell cycle progression, secondary metabolism, nitrate response, inter‐fascicular cambium formation, and root development 46, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58. By screening the DOF protein family, we identified the leaf senescence‐related transcription factor CDF4. Induced or constitutive overexpression of CDF4 promoted leaf senescence, while delayed leaf senescence was observed in transgenic plants in which CDF4 expression was downregulated. Furthermore, CDF4 controlled the endogenous ABA level and H2O2 scavenging by positively regulating the transcription of genes involved in ABA biosynthesis and by repressing catalase activity. Genetic analysis showed that CDF4‐induced ABA biosynthesis and H2O2 accumulation are critical for accelerating the progression of leaf senescence. We also found that CDF4 promotes floral organ abscission by regulating expression of the PGAZAT polygalacturonase gene. This study discovers a novel tripartite amplification loop, involving CDF4, ABA, and ROS, which are mutually promoted by distinct mechanisms, and provides insights into the molecular regulatory network of leaf senescence and floral organ abscission in Arabidopsis.
Results
The CDF4 gene encodes a leaf senescence‐related transcription factor
To uncover the genetic network behind the transcriptional regulation of leaf senescence, we systematically screened genes belonging to the plant‐specific DOF transcription factor family in Arabidopsis. We used real‐time quantitative PCR (qPCR) to quantify transcript abundance of the subfamily of DOF A, B, D, E family genes in Arabidopsis rosette leaves at different developmental stages (Appendix Fig S1A). Interestingly, the cycling DOF family gene CDF1‐5 expression patterns were age‐dependent (Appendix Fig S1B and C). The cycling DOF family genes were previously implicated in photomorphogenesis 59. Among the family members, CDF4 (AT2G34140) is of particular interest because changing its expression level, but not that of any other cycling DOF family gene, can affect leaf senescence 58, 60, 61, 62. CDF4 exhibited similarities with CDF1, CDF2, CDF3, and CDF5 in terms of amino acid (aa) sequence (Appendix Fig S2). CDF4 was previously reported to be a nucleus‐localized transcription factor 63. As with the other nucleus‐localized DOF transcription factors, the 171‐aa CDF4 protein had a DNA‐binding domain at the N‐terminus (Fig 1A).
Figure 1. Molecular characterization of the CDF4 gene and effects of ABA, SA, H2O2, abiotic stresses and darkness on its expression in Arabidopsis leaves.
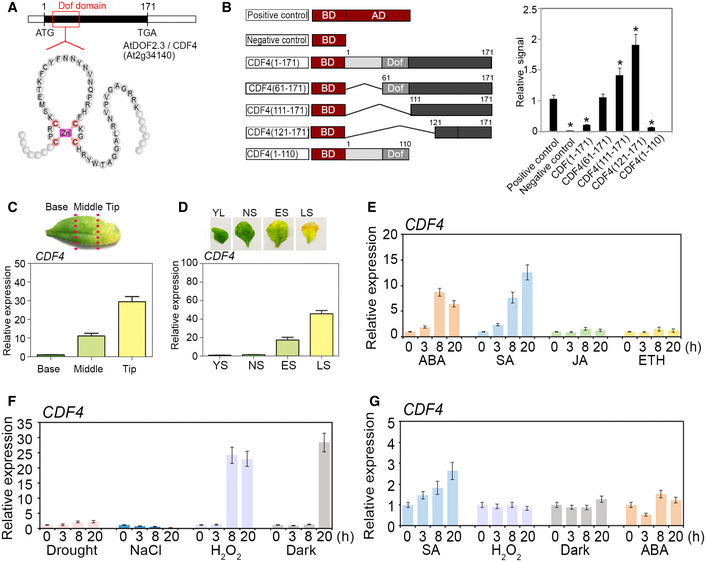
- The gene and protein structures of the DOF domain of CDF4. Cysteine residues conserved in the DOF domain are indicated in red, and the conserved DOF Zn‐figure DNA‐binding domain is found in CDF4.
- Trans‐activation activity assays of CDF4 in Arabidopsis protoplasts. The full‐length ORF of CDF4 and its four deletion constructs are described in the Methods section. DOF (gray boxes) represents the DNA‐binding domains of CDF4. Negative control was the effector vector without gene inserts. The effector vector used is shown in the left panel. The GAL4 transient expression assays were performed using Arabidopsis protoplasts, shown in the right panel. Deletion of the CDF4 N‐terminal domain compromised the transcriptional repression by CDF4 in our assay. Three independent experiments were conducted. Values are given as mean ± SD, n = 3. *P < 0.05 by Student's t‐test.
- Localized expression of CDF4 in a senescing rosette leaf. The ˜ 50% senesced rosette leaf was cut into three parts as indicated by the red lines. The level of CDF4 mRNA was determined in the three sections of the senescing leaf: base; middle; and tip. Three independent experiments were conducted. Values are given as mean ± SD, n = 3.
- CDF4 expression level at various developmental stages of rosette leaves: YL, young leaf; NS, fully expanded, non‐senescent leaf; ES, early‐senescent leaf, with 1/4 leaf area turned yellow; and LS, late‐senescent leaf, with > 50% leaf area turned yellow. Three independent experiments were conducted. Values are given as mean ± SD, n = 3.
- Effect of plant growth hormones (1 mM SA, 1 μM ABA, 10 μM JA, and 10 μM 1‐aminocyclopropane ‐1‐carboxylic acid (ACC)) on CDF4 expression. Three independent experiments were conducted. Values are given as mean ± SD, n = 3.
- Effects of abiotic stress treatments on CDF4 expression. Rosette leaves were treated with salt (100 mM NaCl), drought, H2O2 (2 mM), or darkness. Three independent experiments were conducted. Values are given as mean ± SD, n = 3.
- Effects of SA, H2O2, and dark treatment on CDF4 expression in the aba2‐1 mutant background. AtACT2 was used as an internal control. Three independent experiments were conducted. Values are given as mean ± SD, n = 3.
We wanted to determine whether the DOF family protein CDF4 has transcriptional activity. The DNA‐binding domain was at the N‐terminus of CDF4, which suggested that the C‐terminal sequence of CDF4 was critical for its transcriptional activity. Compared with expression of the vector control, expression of the CDF4 sequence repressed luciferase activity by approximately 74%, indicating that CDF4 was a transcriptional repressor. To examine its transcriptional regulatory domain in detail, we obtained a series of CDF4 coding sequence deletions, fused them to the GAL4 DNA‐binding domain (Fig 1B, left panel), and co‐transformed them into Arabidopsis protoplasts with a reporter vector. The negative control had no luciferase activity. The sequence consisting of residues 61–171, without the N‐terminal region compromised transcriptional repression by CDF4 in our assay. However, transcriptional activation activity was detected in the C‐terminal region consisting of aa residues 111–171 or 121–171, and expression of this region upregulated luciferase activity by 1.3‐ to 1.8‐fold. But the sequence consisting of aa residues 1–110 repressed luciferase activity by 88%. The N‐terminal region, including the DOF domain, somehow inhibited the trans‐activation activity of CDF4 (Fig 1B, right panel). These results showed that CDF4 was a senescence‐related functional transcription factor. Similarly, maize DOF2 and barley PBF could act as either activators or repressors, depending on the downstream promoter or interaction partners 47.
CDF4 transcript levels increase as leaves age and are regulated by dark and H2O2 treatments
To identify the role played by CDF4 in developmental processes, we used qPCR to check the CDF4 mRNA level in leaves at different developmental stages. When Arabidopsis rosette leaves displaying ~ 50% senescence were cut into three parts along the leaf axis (basal, middle, and tip; Fig 1C, top panel), the CDF4 transcript was detected at a low level in the basal part of the leaf, but was highly abundant in the leaf tip (Fig 1C). CDF4 mRNA levels were low in younger leaves but were more abundant at later stages when leaves were senescing (Fig 1D). Analysis of rosette leaves of proCDF4::GUS plants at the early‐senescence stage detected GUS signals in the leaf tip (Appendix Fig S3D). These results indicated that CDF4 expression is developmentally associated with leaf senescence. Therefore, we characterized the temporal and spatial expression patterns of CDF4. RNA samples obtained from various organs indicated that CDF4 was expressed at a relatively high level in the root, silique, and flower (Appendix Fig S4A).
To assess whether environmental cues affect CDF4 expression, we determined the effect of plant hormones and various environmental treatments on the CDF4 mRNA level in rosette leaves. The results showed that CDF4 expression was upregulated by ABA and SA treatments but was not affected by JA or ETH (Fig 1E). Twenty hours of dark treatment upregulated the CDF4 gene expression approximately 20‐fold, but salt and drought treatments did not affect CDF4 expression (Fig 1F). H2O2 has key roles in Arabidopsis stress responses, and abiotic stress results in the accumulation of endogenous H2O2. We confirmed that CDF4 expression responded positively to H2O2 treatment in Arabidopsis seedlings (Fig 1F). The effects of SA, H2O2 and dark treatments on CDF4 expression in the ABA‐deficient aba2‐1 mutant were analyzed to determine whether the induction of CDF4 relies on ABA 64 (Fig 1G). The results indicated that CDF4 expression was reduced significantly in the aba2‐1 mutant background. Therefore, the CDF4 gene is regulated by SA, H2O2, and dark treatments in a partly ABA‐dependent manner.
Leaf senescence is accelerated in CDF4‐overexpressing transgenic lines
To identify the potential role of CDF4 in the control of leaf senescence, we generated transgenic lines that constitutively overexpressed CDF4 under the control of the CaMV 35S promoter (Fig 2A). CDF4 overexpression in 35S::CDF4 transgenic lines was verified by qPCR (Appendix Fig S5A and B). Analysis of leaf longevity showed that the transgenic plants constitutively expressing CDF4 had an accelerated leaf senescence phenotype compared with the vector control, but the leaf senescence process was not different between wild‐type Col‐0 and the vector control plants (Appendix Fig S6). We propose that the phenotype of the 35S::CDF4 plants results from CDF4 promotion of the senescence program. Many genes, including SAG12, which encodes a cysteine protease, and SAG13, which encodes a short‐chain alcohol dehydrogenase, are known to be induced during developmentally related senescence. Therefore, we assayed SAG12 and SAG13 expression in the 35S::CDF4 transgenic plants. SAG12 was expressed at more than a 900‐fold higher level, whereas SAG13 expression was increased 50‐fold in 35S::CDF4 plants (Fig 2B). The upregulation of SAG12 and SAG13 verified that the phenotype of 35S::CDF4 was due to activation of the senescence program. The 35S::CDF4 transgenic plants were smaller than the vector control plants (Appendix Fig S5A and D), potentially due to reduced cell size (Appendix Fig S5C and F). It was interesting that constitutive overexpression of many DOF family transcription factor genes, such as OBP1, OBP2, OBP3, OBP4, AtDOF5.1, HCA2/AtDOF5.6, SCAP1/AtDOF5.7, and including CDF4, can cause plant dwarfing, although the other biological functions of these genes in plant growth and development are diverse 48, 50, 51, 53, 54, 55. We determined the expression levels of cell expansion‐related factor genes, and the results showed that many of them were downregulated in 35S::CDF4 transgenic plants (Appendix Fig S5G). The possible explanation is that these DOF transcription factors conservatively inhibited the expression of cell wall elongation factors and thus inhibited cell expansion. Taken together, we showed that the senescence‐related pathways were promoted in plants in which CDF4 was constitutively overexpressed, and cell expansion was inhibited.
Figure 2. Overexpression or knockdown of CDF4 in transgenic plants affects leaf senescence.
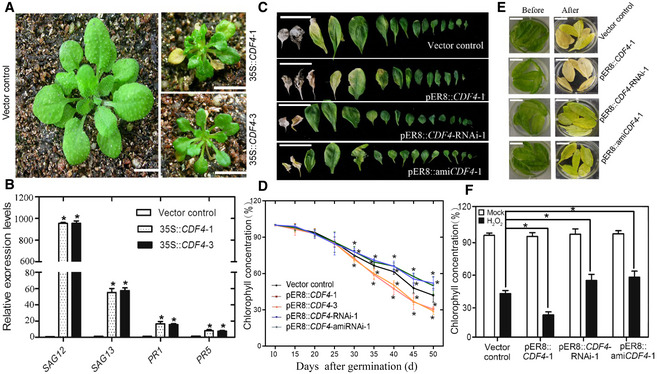
- The age‐dependent leaf senescence symptoms in the vector control and 35S::CDF4 lines grown under long‐day conditions were shown at 21 days old. The 35S::CDF4 plants had a small leaf size and extremely early‐leaf‐senescent phenotype, while the control vector leaves were fully green. Scale bars indicate 1 cm.
- The increase in SAG12, SAG13, PR1, and PR5 gene expression levels was investigated in 35S::CDF4 plants. Three independent experiments were conducted. Values are given as mean ± SD, n = 3. *P < 0.05 in comparison with the vector control by Student's t‐test.
- Phenotypes of vector control, pER8::CDF4, pER8::CDF4‐RNAi, and pER8::amiCDF4 plants, after treatment with 20 μM estradiol for 7 days at 18 days old. Scale bars indicate 1.5 cm.
- The leaf senescence phenotypes of detached rosette leaves in vector control, pER8::CDF4, pER8::CDF4‐RNAi, and pER8::amiCDF4 lines before and after treatment with 2 mM H2O2 in the dark for 4 days. Scale bars indicate 0.7 cm.
- Analysis of the relative chlorophyll concentration in the sixth and seventh rosette leaves from the vector control, pER8::CDF4, pER8::CDF4‐RNAi, and pER8::amiCDF4 lines shown at various development stages. Four independent experiments were conducted. Values are given as mean ± SD, n = 4. *P < 0.05 by Student's t‐test.
- The chlorophyll concentration in leaves from (D) was analyzed. The percentage indicates the chlorophyll content relative to vector control before treatment. Four independent experiments were conducted. Values are given as mean ± SD, n = 4. *P < 0.05 by Student's t‐test.
Source data are available online for this figure.
Perturbation of plant homeostasis could result in an early‐senescence phenotype. Thus, the early senescence observed in 35S::CDF4 transgenic plants could indirectly result from general physiological and/or metabolic disturbances. To rule out general metabolic perturbation due to constitutive overexpression of CDF4, transgenic plants of CDF4 gene under the control of an estradiol‐inducible promoter were obtained. RT–qPCR indicated that CDF4 expression in the transgenic plant was clearly induced by estradiol treatment (Appendix Fig S7). Whole plants were sprayed daily with estradiol for 1 week at 18 days after planting, and the effect of inducing CDF4 overexpression on the leaf senescence process was investigated. Estradiol treatment caused early leaf yellowing and reduced chlorophyll concentration in the transgenic plants with inducible CDF4 (Fig 2C and D). Furthermore, inducible overexpression of CDF4 in the young seedlings led to a stunted growth phenotype and obvious downregulation of the lateral root number (Appendix Fig S8). Thus, we concluded that temporary activation of overexpression of CDF4 promoted leaf senescence. H2O2 treatment is thought to cause leaf senescence. The accelerated leaf senescence phenotype of the inducible CDF4‐overexpressing plants was also assessed in response to H2O2 treatment under darkness. Leaves of the transgenic estradiol ‐induced CDF4‐overexpressing plants became more yellowish than the leaves of the vector control plant after 3 days in the dark (Fig 2E). Measurements of chlorophyll concentration indicated that leaves of the estradiol‐treated inducible CDF4 transgenic plants had less chlorophyll than did those of the vector control plants (Fig 2F). These data demonstrated that CDF4‐overexpression was sufficient to induce early leaf senescence.
Knockdown of CDF4 delays natural and H2O2‐induced leaf senescence
No T‐DNA insertion mutants for CDF4 were available in the public seed collections. Therefore, in order to continue to investigate CDF4 molecular functions in controlling senescence, we generated CDF4 RNA interference (RNAi) knockdown transgenic lines. RNAi transgenic lines with a high CDF4 repression level were utilized in the subsequent assays (Appendix Fig S7A). Due to the sequence similarity of DOF transcription factors, the specificity of RNAi in our assay was checked as well; the knockdown transgenic lines did not exhibit any significant change in the gene expression of the COG1 genes (Appendix Fig S7B). Compared with the vector control, leaf senescence was delayed in the pER8::CDF4‐RNAi transgenic lines following estradiol treatment (Fig 2C). When the vector control plants had brownish dead leaves, most of the leaves on the pER8::CDF4‐RNAi plants remained green. Consistent with this phenotype, we found that chlorophyll concentration declined to 60% of that in the vector control lines at 40 days after planting, and similar chlorophyll losses occurred at 44 days after planting in the pER8::CDF4‐RNAi plants (Fig 2D). In addition, inducible inhibition of CDF4 expression in 7‐day‐old seedlings promoted plant development, with an increased lateral root number (Appendix Fig S8A and B). We also used the artificial microRNA (amiRNA) gene silencing method to obtain pER8::amiCDF4 transgenic lines with a high CDF4 repression level. Compared with the wild‐type control, leaf senescence was delayed in pER8::amiCDF4 plants in response to estradiol treatment (Fig 2C). Previous research has shown that natural and dark‐induced leaf senescence overlap considerably in terms of cellular processes and molecular mechanisms 28. We found that H2O2, ABA, and dark‐induced leaf senescence were delayed in both pER8::CDF4‐RNAi and pER8::amiCDF4 plants (Fig 2E and Appendix Fig S8C). Consistent with this delayed senescence phenotype; chlorophyll loss was also delayed (Fig 2F). Furthermore, we found that ABA treatment partially restored the leaf senescence phenotype of CDF4 knockdown plants (Appendix Fig S9). And exogenous fluridone (ABA biosynthesis inhibitor) or ROS generation inhibitor DPI (diphenyleneiodonium) treatments delayed the leaf senescence phenotype of CDF4‐overexpressing plants (Appendix Fig S9). In addition, we obtained two mutants, CS91480 and CS87649, from the Nottingham Arabidopsis Stock Centre (NASC) mutant repository and found that each of them had a single amino acid substitution, at the 90 and 105aa positions, respectively, of the conserved DOF domain (Appendix Fig S10A). Compared with the wild‐type control, leaf senescence was delayed in both mutants (Appendix Fig S10B). Furthermore, phenotypic complementary test showed that the wild‐type CDF4 genomic DNA can complement the leaf senescence phenotype of the mutants. These results confirm the function of CDF4 in promoting leaf senescence.
CDF4 promotes ABA biosynthesis by inducing the transcription of NCED2 and NCED3
A previous research demonstrated that an atypical bipartite NLS with a 17 amino acid long linker between its flanking basic regions directs Arabidopsis thaliana DOF proteins to the cell nucleus 65. As expected, CDF4 proved to be a nuclear protein (Appendix Fig S4B). Next, we sought to explore how CDF4 promotes leaf senescence. To test whether endogenous hormone levels were altered in CDF4 transgenic plants, we first determined the ABA concentration in the vector control, CDF4‐overexpressing and CDF4‐repressing plants. In pER8::CDF4 leaves, the ABA concentration increased from 0.342 nmol/g fresh weight (FW) to 0.612 nmol/g FW and 0.576 nmol/g FW (fresh weight) (FW) over 3 days of treatment with estradiol, but the ABA levels in the pER8::CDF4‐RNAi plants decreased by approximately 60% after 4 days of estradiol treatment (Fig 3A). These results indicated that the CDF4 gene regulated endogenous ABA levels in Arabidopsis. To investigate the mechanisms underlying the function of CDF4 in ABA biosynthesis and leaf longevity, we searched for ABA biosynthesis‐ and signaling‐related genes which exhibited altered mRNA levels in CDF4‐overexpressing or knockdown transgenic lines; among these genes, we found that NCED2, NCED3, NCED6, AAO2, AAO3, and CYP707A1 were highly upregulated in the 35S::CDF4 lines (Fig 3B and C). However, the selected SA biosynthesis‐ and signaling‐related genes were not significantly altered in 35S::CDF4 lines (Appendix Fig S11A). Additionally, only NCED2 and NCED3 were clearly downregulated in the CDF4 knockdown transgenic plants (Appendix Fig S11B). The expression of the NCED family genes has been reported to increase in senescing leaves 26. Another Arabidopsis aldehyde oxidase (AAO) family gene, AAO3, is targeted by NAP, which promotes ABA accumulation and leaf senescence in Arabidopsis 59. By using the glucocorticoid‐mediated transcriptional induction system in transgenic plants, we wanted to determine whether NCED2 and NCED3 were immediately downstream of CDF4. And their expressions were induced in the CDF4GR transgenic plants after dexamethasone (DEX) induction in the absent or presence of cycloheximide (CHX) (Fig 3D–G). The DEX induction assays suggested that NCED2 and/or NCED3 were likely downstream targets of CDF4, which contributed to the altered ABA level in CDF4 transgenic plants.
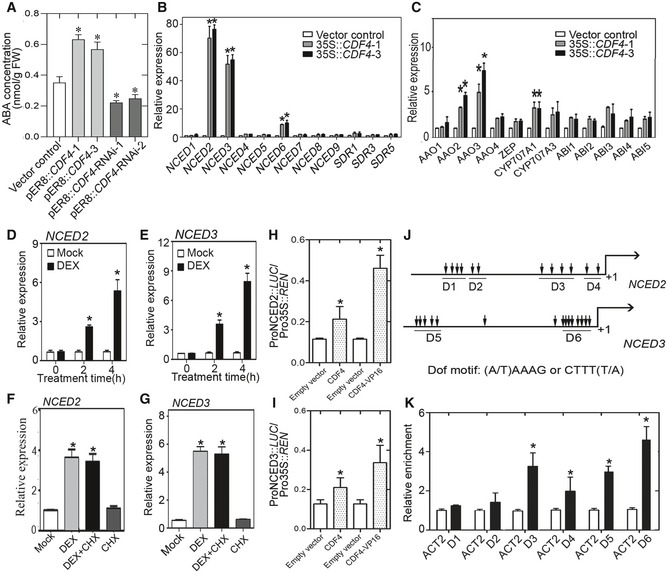
Figure 3. CDF4 is involved in ABA biosynthesis by directly activating the transcription of NCED2 and NCED3 .
-
AMeasurement of free ABA levels in the third and fourth rosette leaves from 21‐day‐old transgenic lines with altered CDF4 expression after estradiol induction for 3 days (once every day). Three independent experiments were conducted. Values are given as mean ± SD, n = 3. *P < 0.05 by Student's t‐test.
-
B, CqPCR analysis of ABA synthesis and signaling‐related genes in Col‐0 and 35S::CDF4 plants. Three independent experiments were conducted. Values are given as mean ± SD, n = 3. *P < 0.05 by Student's t‐test.
-
D, ERelative expression of NCED2 and NCED3 in 14‐day‐old CDF4GR transgenic plants treated with 20 μM β‐estradiol or mock treatment for 0, 2, or 4 h. The expression of the corresponding genes in mock‐treated plants was set to 1.0. Three independent experiments were conducted. Values are given as mean ± SD, n = 3. *P < 0.05 by Student's t‐test.
-
F, GRelative expression level of NCED2 and NCED3 in 14‐day‐old CDF4GR transgenic plants treated with 20 μM DEX, 100 μM CHX, DEX plus 100 μM CHX, or mock. The gene expression in mock‐treated plants was set to 1.0. Three independent experiments were conducted. Values are given as mean ± SD, n = 3. *P < 0.05 by Student's t‐test.
-
HSchematic diagram indicating the locations of six DOF motif clusters (D1–D6) in the NCED2 and NCED3 gene promoters.
-
IChIP‐qPCR analysis of the ability of CDF4 to bind to the promoters of NCED2 and NCED3. An anti‐HA monoclonal antibody was used for DNA immunoprecipitation from 3‐week‐old pER8::CDF4‐HA transgenic plants. Black bars indicate the enrichment fold changes normalized to that of ACT2. Three independent experiments were conducted. Values are given as mean ± SD, n = 3. *P < 0.05 by Student's t‐test.
-
J, KTransient dual‐luciferase reporter assay. The pGreenII‐0800 LUC construct containing the CDF4 promoter and the p62‐SK construct with or without the CDF4 coding region and CDF4‐VP16 were transiently co‐transformed into Col‐0 protoplasts. Firefly luciferase (LUC) and Renilla luciferase (REN) activities were measured after culturing the protoplasts under low light conditions for 16 h. The ProNCED2/3:LUC/Pro35S:REN ratio represents the relative activities of NCED2 and NCED3 transcription. Three independent experiments were conducted. Values are given as mean ± SD, n = 3. *P < 0.05 by Student's t‐test.
To further test whether CDF4 is a direct transcriptional regulator of NCED2 and NCED3, we used a LUC (luciferase)‐based trans‐activation experiment. We co‐expressed CDF4 with both NCED2 and NCED3 promoter‐LUC fusion reporters in mesophyll protoplasts of Arabidopsis, which resulted in an increase in luciferase activity, although they showed activities only 2.0‐ and 1.7‐fold higher than in the control. To increase transcriptional activity, we used the 35S::CDF4‐VP16 effector plasmid carrying the VP16 activation domain and observed much greater increases in luciferase activity (Fig 3H and I). Hence, we suggested that CDF4 probably acted in concert with co‐activators that are expressed in mature or senescing rosette leaves. Because young leaves were used for the earlier protoplast isolation in our study, they may not have contained any co‐activators. These results implied that CDF4 might bind to the promoters of NCED2 and NCED3 to induce their transcription. The (A/T) AAAG/CTTT (T/A) core sequence was reported to be crucial for DOF transcription factor binding. There were 12 and 15 DOF‐binding core sequences in the approximately 2.0‐kb NCED2 and NCED3 promoter sequences, respectively, that were assessed in the trans‐activation experiment (Fig 3J).
To provide further evidence for the interaction, chromatin immunoprecipitation (ChIP) assays were conducted to identify whether the CDF4 protein could bind to the gene promoters. pER8::CDF4::HA transgenic plants were used, which contained an HA‐coding sequence fused in‐frame to the 3′ end of the CDF4 gene. Quantitative real‐time ChIP‐qPCR assays, using an anti‐HA antibody, showed that CDF4 could bind to the conserved sequence motifs in the NCED2 and NCED3 gene promoters (Fig 3K). We also performed in vitro DNA–protein interaction ELISA (DPI‐ELISA) experiments with an epitope‐tagged CDF4 protein. A native‐sequence oligonucleotide, containing the functional DOF‐binding sites derived from the NCED2 and NCED3 promoter sequences and the mutated version, was used (Appendix Fig S12A–C). The result shows a positive correlation between the amount of glutathione S‐transferase (GST)‐CDF4 detected with the epitope‐specific antibody, indicating that GST‐CDF4 bound to the wild‐type oligonucleotide in a concentration‐dependent manner. As shown in Appendix Fig S12, the wild‐type, but not the mutated, oligonucleotide was able to reduce the amount of GST bound to the plate, thus confirming the specificity of the binding. We further performed the EMSA to verify the binding of CDF4 to the selected D3 and D4 domains of NCED2 gene promoter and D5 and D6 domains of NCED3 gene promoter (Appendix Fig S13A and B). These results demonstrated that CDF4 could target the NCED2 and NCED3 gene promoters and induce their transcription.
Mutation of NCED2 and NCED3 suppresses ABA accumulation and leaf senescence in pER8::CDF4
Our data indicated that CDF4 was involved in the control of leaf senescence by targeting the NCED2 and NCED3 promoters. Moreover, we wanted to determine whether the expression of NCED2 and NCED3 genes was related to leaf senescence control. To address this question, we measured the expression of NCED2 and NCED3 in senescing leaves. As with CDF4 expression, they exhibited high levels of expression in the leaf tip (Fig 1C). The temporal expression patterns of the NCED2 and NCED3 genes were similar to that of CDF4 (Fig 4A and C). Expression of NCED2 and NCED3 was upregulated during the leaf senescence process, but their expression was significantly reduced in the CDF4::RNAi plants, implying that NCED2 and NCED3 played an important role in CDF4‐mediated control of leaf senescence (Fig 4B and D). Interestingly, we also detected an increase in CDF4 expression in the plants that constitutively overexpressed NCED2 and NCED3, which is consistent with the finding that CDF4 expression is induced by ABA (Appendix Fig S14A–D).
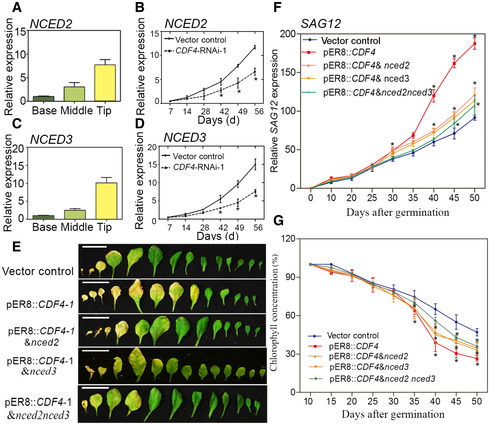
Figure 4. Involvement of NCED2 and NCED3 in the progression of leaf senescence in the vector control, pER8::CDF4, pER8::CDF4 & nced2, pER8::CDF4 & nced3, and pER8::CDF4 & nced2nced3 plants.
-
A–DTranscript levels were determined by qPCR and normalized to AtACT2. (A, C) Localized NCED2 and NCED3 expression in the wild‐type senescing leaf. Half‐senesced leaves were split into three parts, as shown in Fig 1A and C. (B, D) NCED2 and NCED3 expression during leaf senescence in the vector control and CDF4‐RNAi‐1 plants. Transgenic plant leaves were analyzed from 7 to 56 d (d, days after planting). Four independent experiments were conducted. Values are given as mean ± SD, n = 4. *P < 0.05 by Student's t‐test.
-
EObservation of rosette leaf longevity from the 26‐day‐old vector control, pER8::CDF4, pER8::CDF4 & nced2, pER8::CDF4 & nced3, and pER8::CDF4 & nced2nced3 transgenic plants after 20 μM estradiol induction for 2 weeks. Phenotype of detached rosette leaves arranged from oldest to youngest. Scale bars indicate 1.5 cm.
-
F, G(F) SAG12 expression level and (G) chlorophyll concentration in rosette leaves in (E) at various development stages. Three independent experiments were conducted. Values are given as mean ± SD, n = 4. *P < 0.05 by Student's t‐test.
Source data are available online for this figure.
To determine whether ABA accumulation in CDF4‐overexpressing plants depends on NCED2 and NCED3, we obtained NCED2 and NCED3 T‐DNA insertion knockout mutants (Appendix Fig S15A–C). A decrease in CDF4 transcript levels was observed in these nced2 and nced3 mutants (Appendix Fig S15D), which supported the hypothesis that NCED2/3‐mediated ABA biosynthesis and CDF4 expression mutually promoted one another. Then, a pER8::CDF4 transgenic plant was crossed with the nced2, nced3, and nced2nced3 double mutants. pER8::CDF4 & nced2, pER8::CDF4 & nced3, and pER8::CDF4 & nced2nced3 plants had higher CDF4 expression after induction with estradiol (Appendix Fig S16). We analyzed the progression of senescence in the vector control line and in various transgenic lines. Under normal growth conditions, pER8::CDF4 & nced2 and pER8::CDF4 & nced3 rosette leaves exhibited delayed senescence compared with pER8::CDF4 after induction with estradiol (Fig 4E). In addition, the pER8::CDF4 & nced2 and pER8::CDF4 & nced3 rosette leaves showed decreased SAG12 expression (Fig 4F), increased chlorophyll content (Fig 4G), and suppressed CDF4‐induced leaf senescence compared with the pER8::CDF4 plants. Moreover, pER8::CDF4 overexpression in the nced2nced3 mutant background further delayed the leaf senescence phenotype to the vector control level (Fig 4E–G). We also found that the nced2nced3 double mutant, rather than the nced2 and nced3 single mutants, delayed the aging process of leaves (Appendix Fig S14E). Taken together, we concluded that the loss of NCED2 and NCED3 function suppressed ABA production and leaf senescence progression in plants overexpressing CDF4.
CDF4‐RNAi plants exhibit enhanced tolerance to oxidative stress
The endogenous levels of H2O2 gradually increased during the leaf senescence process, and the accumulated H2O2 played an important role in the development of characteristics associated with senescence. Based on our findings showing the induction of CDF4 expression by H2O2 (Fig 1F), we next wanted to determine whether CDF4 participated in the regulation of oxidation‐related leaf senescence. We examined the H2O2‐induced senescence phenotype in the vector control and the transgenic plant leaves under dark conditions. When subjected to H2O2 treatment for 3 days, all pER8::CDF4 detached leaves turned yellow, while detached leaves of the vector control plants were pale green, but the detached CDF4‐RNAi leaves largely remained fully green (Fig 5A). DAB (3,3′‐diaminobenzidine) staining and quantitative measurements of H2O2 also revealed that CDF4‐RNAi leaves accumulated less endogenous H2O2 than did either the vector control or the pER8::CDF4 leaves in response to exogenous H2O2 application (Fig 5B and C). Measurement of chlorophyll concentration in dark/H2O2‐treated vector control and transgenic plant leaves supported the observed phenotype (Fig 5D). The DAB staining showed that, following exogenous H2O2 treatment, CDF4‐RNAi leaves displayed less H2O2 accumulation than did the vector control leaves, implying that CDF4‐RNAi leaves had greater H2O2 scavenging capacity (Fig 5B). Furthermore, similar results were obtained from comparisons of vector control and pER8::amiCDF4 transgenic plants (Fig 2E). Taken together, downregulation of CDF4 expression enhanced tolerance to oxidative stress and delays leaf senescence.
Figure 5. CDF4 suppresses H2O2 scavenging.
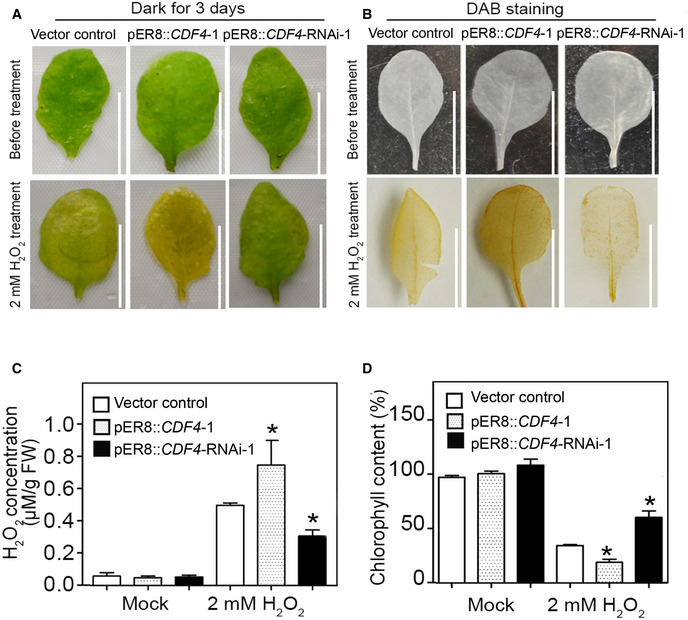
-
AThe senescence phenotype of the vector control, pER8::CDF4, and pER8::CDF4::RNAi detached leaves treated with H2O2 and incubated in the dark for 3 days. The third leaf in rosettes of 3‐week‐old plants was detached and incubated in MES buffer (2 mM MES, pH 5.8) treated with 2 mM H2O2 and 20 μM β‐estradiol under dark conditions for 3 days. Picture shows the leaves before and after treatment. Scale bars indicate 1 cm.
-
B, C(B) DAB staining and (C) measurement of H2O2 before and after 2 mM H2O2 plus 20 μM β‐estradiol treatment under dark conditions for 2 days. The brown color represents H2O2 accumulation. Three independent experiments were conducted. Values are given as mean ± SD, n = 3. *P < 0.05 by Student's t‐test. Scale bars indicate 1 cm.
-
DChlorophyll concentrations in the leaves shown in (A). Three independent experiments were conducted. Values are given as mean ± SD, n = 3. *P < 0.05 by Student's t‐test.
CDF4 suppresses catalase (CAT) activity by repressing CAT2 transcription
Given that CDF4‐RNAi plants showed reduced H2O2 accumulation and enhanced tolerance to exogenous H2O2 treatment, we investigated the relationship between CDF4 expression and the activity of H2O2 scavengers. Plants contain several antioxidant enzymes or proteins. Catalases (CATs) are primarily responsible for H2O2 scavenging. We first measured CAT activity, and found that CDF4‐RNAi plants exhibited higher CAT activities in both young and old leaves compared with the vector control (Fig 6A), indicating that CDF4 suppresses CAT activities. There are three catalase genes in the Arabidopsis genome. We found that the expression of CAT2 and CAT3, but not CAT1, was induced in CDF4‐RNAi plants (Fig 6B). Because CAT2 is the major catalase isoform and expression of only CAT2 declined in 35S::CDF4 transgenic plants (Appendix Fig S17A), we focused our analysis on CAT2. Interestingly, many DOF‐binding motif clusters were found in the promoter of the CAT2 gene (Fig 6C). Furthermore, we tested whether CDF4 could directly regulate CAT2 at the transcriptional level. ChIP assays were conducted using pER8::CDF4::HA transgenic plants to identify whether the CDF4 protein binds to the CAT2 promoter. ChIP‐qPCR assays, using an anti‐HA antibody, indicated that CDF4 could bind to the selected regions in the CAT2 promoter (Fig 6D). In addition, dual‐LUC reporter assays revealed that CDF4 repressed CAT2 transcription in Arabidopsis protoplasts (Fig 6E). In vitro DPI‐ELISA experiments, performed with an epitope‐tagged CDF4 protein, also verified that CDF4 binds selected DOF‐binding motif clusters in the CAT2 promoter (Appendix Fig S12D). We further performed the EMSA to verify the binding of CDF4 to the D2, D3 and D5 domains of CAT2 gene promoter (Appendix Fig S13C). All together, these results demonstrated that CDF4 repressed catalase activity by inhibiting CAT2 transcription.
Figure 6. CDF4 suppresses catalase activity by repressing CAT2 transcription.
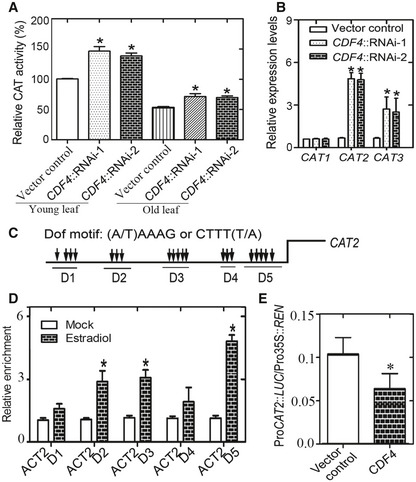
- Measurement of catalase activity in vector control and CDF4RNAi plants. Ten‐day‐old green seedlings (“Young”) and the third and fourth rosette leaves from 36‐day‐old plants (“Old”) were used, respectively. Three independent experiments were conducted. Data are represented as means ± SD, n = 3. Student's t‐test, *P < 0.05.
- qPCR analysis of CAT genes expression in the third and fourth rosette leaves from 36‐day‐old plants. The expression of CAT genes in the wild‐type plant is given as 1. Three independent experiments were conducted. Data are represented as means ± SD, n = 3. Student's t‐test, *P < 0.05.
- Schematic diagram indicating the locations of the putative CDF4‐binding motif clusters (D1–D5) in the promoter of CAT2.
- ChIP‐qPCR analysis of the relative binding of CDF4 to the promoter of CAT2. An anti‐HA monoclonal antibody was used for DNA immunoprecipitation from 32‐day‐old pER8::CDF4‐HA transgenic plants. Black columns indicate the enrichment fold changes normalized to that of ACT2. Three independent experiments were conducted. Data are represented as means ± SD, n = 3. Student's t‐test, *P < 0.05.
- Transient dual‐luciferase reporter assay. The pGreenII‐0800 LUC construct containing the CAT2 promoter and the p62‐SK construct with or without CDF4 were transiently co‐transformed into Col‐0 protoplasts. Firefly luciferase (LUC) and Renilla luciferase (REN) activity were measured after culturing the protoplasts under low light conditions for 16 h. Six independent experiments were conducted. Values are given as mean ± SD, n = 6. *P < 0.05 by Student's t‐test.
Moreover, we also determined the expression of other genes encoding H2O2‐scavenging enzymes, such as ascorbate peroxidases (APXs) and superoxide dismutases (SODs). Eight ascorbate peroxidase genes have been annotated on the Arabidopsis genome. We found that the expression of APX4‐6, sAPX, and tAPX, but not APX1‐3, was changed in the CDF4 transgenic plants (Appendix Fig S17B and C). Among them, APX5 expression was obviously affected in CDF4 transgenic plants (Appendix Fig S17B and C). In addition, the lower abundance levels of several SOD transcripts in 35S::CDF4 leaves indicated that CDF4 also repressed their expression (Appendix Fig S17D).
Increased CAT activity suppresses the early‐senescence phenotype of CDF4 overexpression
Given our findings that CDF4 suppressed CAT2 transcription, we investigated whether the increase in catalase activity in proSAG12::CDF4 plants could suppress their early‐leaf‐senescence phenotype. To address this question, we generated proSAG12::CDF4 & 35S::CAT2 transgenic plants by crossing proSAG12::CDF4 into the 35S constitutive CAT2‐overexpressing plant background. We found that the CAT2 gene overexpression partly rescued the early‐senescence phenotype of proSAG12::CDF4, with a reduced number of yellow leaves (Fig 7A), and CDF4 gene expression levels were high in the transgenic plants (Fig 7B). Consistent with the visible phenotype, the increase in H2O2 level and reduction in chlorophyll concentration observed in proSAG12::CDF4 plants were suppressed by CAT2 gene overexpression (Fig 7C–E). Interestingly, consistent with previous results, the cat2 knockout mutant showed an early‐senescence phenotype (Appendix Figs S15A, E, F and S18A). However, 35S::CAT2 plants in the Col‐0 background displayed a leaf senescence phenotype similar to that of Col‐0, implying that the H2O2‐scavenging activities are probably controlled at an adequate level during normal development (Appendix Fig S18B and C). These results suggested that the elevated H2O2 concentration in proSAG12::CDF4 plants served as another mechanism underlying their accelerated leaf senescence phenotype.
Figure 7. Overexpression of CAT2 represses proSAG12::CDF4 early‐senescence phenotype.
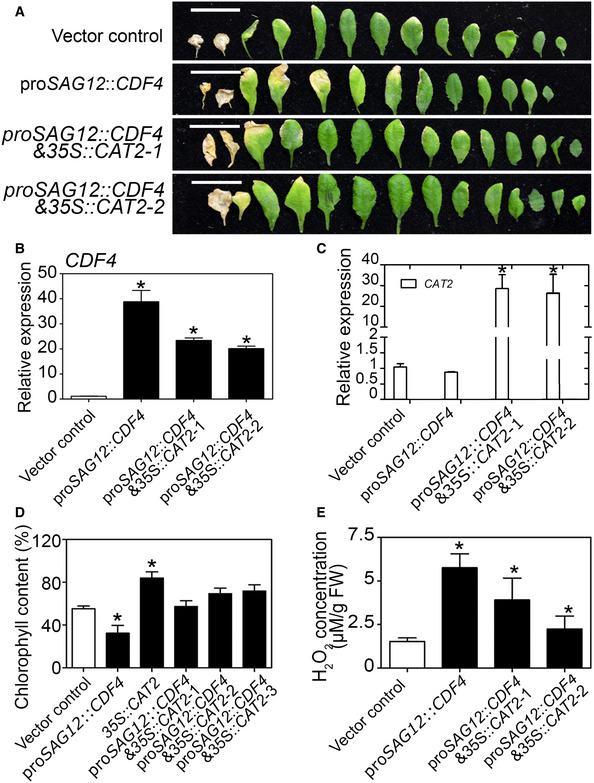
-
AThe senescence phenotypes of 5‐week‐old vector control, proSAG12::CDF4, and proSAG12::CDF4 & 35S::CAT2 plants, from old to young. Scale bars indicate 1.5 cm.
-
BqPCR analysis of the expression of CDF4 and CAT2 in the leaves shown in (A). Four independent experiments were conducted. Values are given as mean ± SD, n = 4. *P < 0.05 by Student's t‐test.
-
CqPCR analysis of the expression of CAT2 in the leaves shown in (A). Four independent experiments were conducted. Values are given as mean ± SD, n = 4. *P < 0.05 by Student's t‐test.
-
D, E(D) Measurement of chlorophyll and (E) H2O2 concentrations in the leaves shown in (A). The third leaves in the rosette were used. Four independent experiments were conducted. Data are presented as the mean ± SD, n = 4. *P < 0.05 by Student's t‐test.
CDF4 promotes floral organ abscission in Arabidopsis
Senescence usually occurs synchronously with the abscission process in Arabidopsis. For example, in the abscission‐delayed etr1‐1 mutant, the leaf senescence process is slowed 66. Here, we found a similar situation. Our study shows that CDF4 may play a role in the floral organ abscission regulation, because CDF4 was specifically expressed in the floral organ abscission zone (Appendix Fig S3B and C) and the clearly altered abscission process was observed in the plants overexpressing or downregulating expression of the CDF4 gene under the control of the floral organ abscission‐related peptide ligand INFLORESCENCE DEFICIENT IN ABSCISSION (AtIDA) promoter (Fig 8A and Appendix Fig S21). Given that CDF4 belongs to the DOF transcription factor family, we were interested in determining whether it regulates the expression of genes involved in abscission. Several components, with putative functional enzymatic activities or signaling roles in abscission, were selected for analysis of their expression levels. Combining transcriptome data with our qPCR analysis, the transcript levels for most of the selected genes, namely ADGP1, PGAZAT/ADPG2, QTR2, AtDOF4.7, HWS, HAESA, IDA, AGL15, AtEXP10, ARP4, and AT3G14380, were checked in positions 5–6 of young siliques of the control and the CDF4‐overexpression lines (Fig 8B). There were no significant changes in expression, except for the three polygalacturonase (PG) genes, ADGP1, PGAZAT, and QTR2. The level of expression of PGAZAT gene showed the greatest change. ADPG1, PGAZAT, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis 67. The relative PGAZAT expression ratios between the overexpression lines and the wild type were 28 in line 1 and 26 in line 3, with the PGAZAT gene expression level being obviously inhibited in the CDF4 knockdown lines (Fig 8C). These results showed that PGAZAT gene expression was obviously upregulated by CDF4 overexpression. In addition, when proIDA::CDF4 was crossed with the proPGAZAT::GUS line, the proIDA::CDF4 & proPGZAT::GUS plants significantly enhanced GUS staining activity compared with the proPGAZAT::GUS plants (Fig 8E). We also found that pER8:CDF4 and pER8:CDF4‐RNAi showed the altered floral abscission phenotype after a period of estradiol induction (Appendix Fig S19).
Figure 8. Altered floral organ abscission process in the CDF4 transgenic plants.
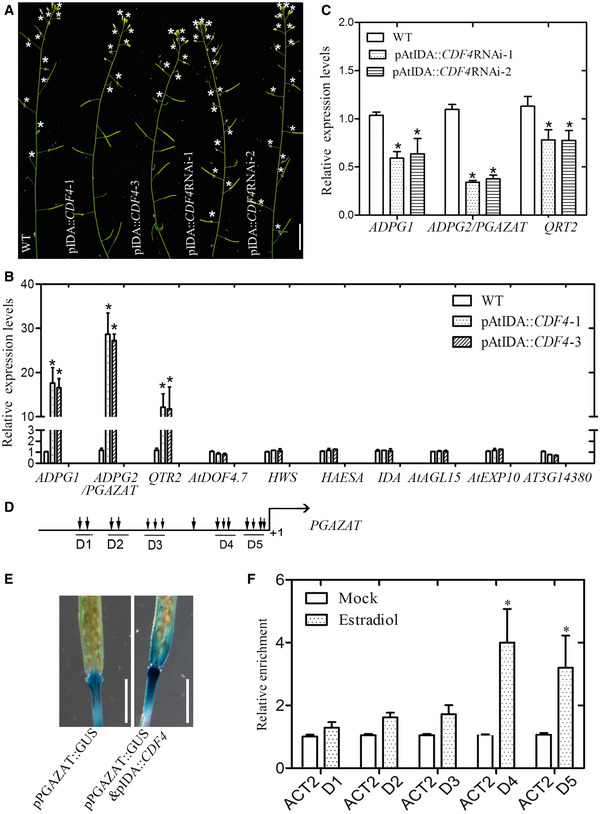
-
A–C(A) Observation of inflorescences of 7‐week‐old wild‐type (WT) Arabidopsis and the CDF4 transgenic plants. The asterisk represents the flower organ attached along the inflorescence. Scale bar indicates 1.5 cm. Selected abscission‐related gene expression levels in (B) WT and proIDA::CDF4 lines or (C) WT and proIDA::CDF4RNAi lines by using qPCR analysis. The expression of these selected genes in the wild‐type plant is given as 1. The relative expression level represents only the level of expression of the gene relative to the wild type. ACTIN2 was used as the internal control. Three independent experiments were conducted. Values are given as mean ± SD, n = 3. *P < 0.05 by Student's t‐test.
-
DSchematic diagram indicating the locations of the putative CDF4‐binding motif clusters (D1–D5) in the approximately 1.8‐kb promoter of PGAZAT.
-
EObservation of GUS staining activity in the proPGAZAT::GUS and proPGAZAT::GUS & proIDA::CDF4. Scale bar indicates 0.5 cm.
-
FChIP‐qPCR analysis of the relative binding of CDF4 to the promoter regions of PGAZAT. An anti‐HA monoclonal antibody was used for DNA immunoprecipitation from 6‐week‐old pER8::CDF4‐HA transgenic plants after estradiol induction. Black columns indicate the enrichment fold changes normalized to ACT2. Three independent experiments were conducted. Values are given as mean ± SD, n = 3. *P < 0.05 by Student's t‐test.
Furthermore, to examine whether the CDF4 protein regulates PGAZAT expression by binding to the promoter region, five regions with many DOF protein‐binding cis‐acting elements were selected for analysis (Fig 8D) 47. For ChIP analysis, formaldehyde cross‐linked chromatin from transgenic plants was fragmented and immunoprecipitated with the HA antibody. The PCR product of the PGAZAT promoter was enriched in pER8::CDF4‐HA (Fig 8F). We further performed the EMSA to verify the binding of CDF4 to the D4 and D5 domains of PGAZAT gene promoter (Appendix Fig S13D). The conclusion was that CDF4 targeted the PGAZAT gene and induced its expression. The expression of some endoglucanases, that are related to abscission, is also regulated by abscission signals, such as ABA and hydrogen peroxide 67, 68. Further investigation of the role of the two in promoting floral organ abscission is to be studied in the next step of our research. Taken together, our results from the CDF4 gene studies provide novel insights into the regulation of the activation of the abscission enzymes. These results showed that the CDF4 transcription factor integrated both ABA biosynthesis and endogenous H2O2 concentration to modulate leaf longevity and floral organ abscission. Overall, a proposed feedback loop model, involving CDF4, ABA and ROS, is shown in Fig 9.
Figure 9. The proposed model illustrating the amplification loop involving CDF4, ABA, and ROS.
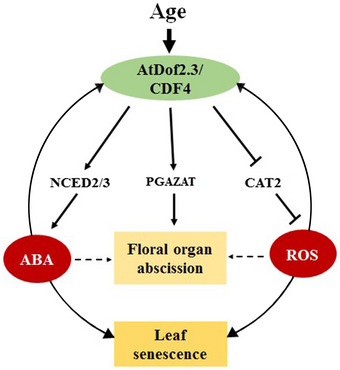
CDF4 is a senescence‐associated gene with expression induced by ABA and ROS, two well‐known leaf senescence inducers. CDF4 further promotes ABA biosynthesis by activating the transcription of NCED2 and NCED3 and repressing H2O2 scavenging by inhibiting CAT2 transcription. At the same time, CDF4 can bind to the PGAZAT promoter region, thus promoting the floral organ abscission process. Given that ABA and ROS also mutually promote each other's accumulation, CDF4, ABA, and ROS form a tripartite amplification loop to accelerate leaf senescence and floral organ abscission. As a result, the levels of CDF4, ABA, and ROS undergo a gradual increase driven by their interlinking positive feedback loops during the leaf senescence and floral organ abscission process. In the legend, the solid and dotted lines represent the direct interaction obtained in this research and indirect interaction, respectively. And the arrow‐ and T‐ending lines represent positive and negative regulatory pathways, respectively.
Discussion
Plant senescence is a well‐controlled developmental process that is largely regulated genetically. It ultimately leads to cell disintegration. Senescence clearly cannot occur in young leaves. It is possible that senescence‐associated inhibitors block the senescence program when leaves are young, while senescence‐related activators are turned on as the leaf ages. From molecular and genetic perspectives, many methods have been used to identify genes with changes in expression during leaf senescence. In such a genetic pathway, the activated transcription factors will turn on expression of a large number of leaf senescence‐related genes. Thus, the isolation and molecular characterization of leaf senescence‐related transcription factors will aid in understanding their roles in regulating gene expression. CDF4 belongs to the subfamily A or type II DOF family proteins 47, 59. CDF4 had previously been identified as a nucleus‐localized transcription factor; WOX5 represses CDF4 transcription by recruiting TPL/TPR and HDA19 at the CDF4 regulatory region. CDF4 promotes columella cell differentiation, and its repression by WOX5 is essential to maintain CSCs in an undifferentiated state 63. However, the mechanism of CDF4 involvement in leaf senescence regulation was unknown. Here, the CDF4 gene was isolated as a new leaf senescence‐related transcription factor gene. Expression analyses revealed that the CDF4 mRNA level increased at the late stage of leaf senescence (Fig 1C and D). This profile indicated that senescence could be controlled via the transcriptional regulation of genes involved in leaf senescence. To verify the function of CDF4, leaf senescence phenotypes in CDF4 transgenic lines were investigated. The leaf phenotype observed in 35S::CDF4 mainly proceeded via a senescence‐related pathway because the reliable senescence marker SAG12 was easily detected in these transgenic lines (Fig 2B). In addition, we observed inhibited cell expansion in 35S::CDF4 plants as a result of downregulation of cell elongation factors. Interestingly, the ability to inhibit cell expansion has been observed in many DOF family genes, implying that this function is conserved during evolution 48, 50, 51, 53, 54, 55. However, the phenotype of premature senescence in the rosette leaves was not found in other DOF transgenic plants, indicating that the promotion of plant dwarfing by overexpression of DOF family genes was not the direct cause of early leaf senescence. Our data supported the conclusion that CDF4 played an important role in the control of leaf senescence. The first evidence was derived from plants in which the constitutive and inducible overexpression of CDF4 promoted early leaf senescence (Fig 2A and B). Additional evidence was obtained from assays in which CDF4 was downregulated by RNAi‐ and amiRNA‐mediated gene silencing; these transgenic lines exhibited delayed developmental and H2O2‐inducible leaf senescence (Fig 2C and D).
Leaves start from leaf primordia and develop into photosynthetic organs through vegetative growth and maturation, which is completed through the coordination of cell division, expansion, and differentiation, and finally enter the senescence stage 1. Previous study on components of cytokinin and auxin signaling has demonstrated the relevance between leaf growth and senescence. For example, triple mutations of Arabidopsis HISTIDINE KINASE 2 (AHK2), AHK3, and AHK4 result in a smaller leaf size, as a result of reduced cell proliferation and early leaf senescence 69. Cytokinin response factors (CRFs) have been implicated in the control of leaf growth and senescence in Arabidopsis 70. In addition, the AUXIN RESPONSE FACTOR 2 (ARF2) mutation enhances leaf growth and retards leaf senescence 71. Our knowledge of the interrelationship between early leaf development and senescence is still limited. Here, we identified that CDF4 mediates cell expansion and senescence during leaf development. As mentioned here, in addition to the positive leaf senescence regulator CDF4, JUB1 is another TF related to leaf senescence. Both of them have the same effect of inhibiting cell extension and regulating the aging phenotype of leaves. However, the difference is that CDF4 suppresses H2O2 scavenging production by inhibiting the expression of CAT2, meanwhile promoting the aging process of leaves by upregulating endogenous ABA levels. But JUB1 overexpression strongly delays senescence, dampens intracellular H2O2 levels, and enhances stress tolerance and the induction of DREB2A expression. The growth of plants is accompanied by the increase of cell volume and the change of cell wall rigidity and the cell wall collapses during the late senescence phase of leaf development. Therefore, the regulation of cell wall plasticity and cell size is closely related to leaf senescence process. Thus, we believe that CDF4 might provide us with a good opportunity of investigating the mechanisms involved in mediating leaf development and senescence. This result also implies the relationship between plant regulation of leaf senescence and cell size regulation.
Leaf senescence occurs slowly and is associated with the efficient transfer of nutrients from the senescing leaves to the developing parts of the plant. Promotion of leaf senescence by ABA is a long‐term response that allows survival under extreme conditions. The DOF domain protein family is involved in various plant‐specific physiological processes. By selecting a probable senescence‐related DOF transcription factor, our assay revealed that CDF4 promotes leaf senescence, in part, through upregulated ABA biosynthesis. Because the CDF4 mRNA abundance was increased when leaf senescence was accelerated by the senescence‐accelerating hormone ABA, we also found that dark‐induced senescence in detached leaves was accelerated in CDF4‐overexpressing plants (Fig 2E). Further studies have indicated that CDF4 regulates leaf senescence by increasing ABA biosynthesis as a result of targeting the key ABA biosynthesis genes, NCED2 and NCED3. The nced2 and nced3 knockout mutants delayed CDF4‐induced leaf senescence (Fig 4E). In order to provide a more in‐depth insight into the role of CDF4 for ABA‐mediated regulation of leaf senescence, we measured the stomatal aperture of vector control and CDF4 transgenic plants. The results showed that stomatal aperture obviously decreased in CDF4 overexpression plant leaves and increased in CDF4 knockdown plants (Appendix Fig S20). The change of stomatal aperture may be due to the effect of CDF4 regulating ABA content in plants. These results indicated that the change of stomatal aperture was involved in CDF4‐induced leaf senescence. Taken together, we conclude that the CDF4 protein plays a role in integrating developmental age and environmental stimuli to initiate the leaf senescence process. A recent report has indicated that the mitochondrial protease FtSH4 promotes leaf senescence by inducing the expression of WRKY transcription factor genes as well as promoting ABA accumulation 72. Furthermore, the wrky70wrky54 double mutant exhibits increased ABA levels and exhibited an early‐senescence phenotype 73. These results have verified the important role of ABA in controlling leaf senescence.
In addition to promoting ABA biosynthesis, we discovered that CDF4 also functions in plant tolerance to oxidative signals such as H2O2. Exogenous application of H2O2 can dramatically accelerate the leaf senescence process, particularly in the dark. Downregulation of CDF4 led to enhanced tolerance to the effects of exogenous H2O2 application, as less endogenous H2O2 accumulated and higher chlorophyll concentration occurred in rosette leaves (Fig 5A–D). One of the reasons behind this effect was the increased catalase activity in CDF4 RNAi plants, which was due mainly due to the upregulation of expression of CAT2. Like NCED2 and NCED3, CAT2 is a target gene of CDF4, although the regulatory mechanisms of these two classes of gene were different. We identified various DOF‐binding motifs in the promoter of NCED2 and NCED3 that mediated their induction by CDF4, whereas the DOF‐binding motifs in the promoter of CAT2 mediated their repression by CDF4. Currently, it is not known how these seemingly identical DOF‐binding sequences play opposite roles in mediating CDF4‐directed effects on transcription. Exogenous application of H2O2 can dramatically accelerate the leaf senescence process. Knockdown of CDF4 led to enhanced tolerance to exogenous H2O2 application, as less endogenous H2O2 accumulated in plant rosette leaves (Fig 5C). Given that the downstream regulatory networks dictated by CDF4 in these processes are still unclear, our findings about the regulatory role of CDF4 in ABA biosynthesis and ROS scavenging in leaf senescence offer a potential mechanism for these developmental processes as well. It is conceivable that, upon leaf senescence, age‐dependent CDF4 expression progressively leads to concomitant increases in ABA and H2O2 levels, two well‐defined inducers of leaf senescence. We also found that CDF4 expression was induced by ABA and H2O2 treatment. Interestingly, ABA and ROS levels are highly correlated, and these compounds mutually induce each other's accumulation in many biological processes 74, 75, 76, 77, 78.
Floral organ abscission has been used as a model system for studying genetic regulation of the leaf abscission process in Arabidopsis 79. However, little is known about the molecular mechanisms behind signal transduction and cell wall dissolution in abscission. Here, by studying the position of floral organ withering in the wild‐type and the CDF4 transgenic plants, we found that CDF4 promoted not only leaf senescence but also floral organ abscission. Combining the expression pattern, transcriptome data and quantitative PCR results, we infer that CDF4 plays important roles in the transcriptional regulation of floral organ abscission. Furthermore, the in vivo ChIP experiment confirmed that the effect is mainly realized by regulating the cell wall hydrolase gene, PGAZAT. Because CDF4 can promote the synthesis of ABA and ROS accumulation, and ABA and ROS are two well‐known abscission inducing factors, it is possible to propose that, with the increase in leaf age, the increase in CDF4 gene expression promotes endogenous ABA synthesis and ROS accumulation, thus promoting the floral organ abscission process.
Based on the current findings and previous results, we propose a positive feedback loop model, showing how CDF4, ABA, and ROS are responsible for promoting leaf senescence and floral organ abscission (Fig 9). In this model, each of these three players promotes the accumulation of the other two via independent mechanisms. Thus, the levels of CDF4, ABA, and ROS undergo a gradual increase driven by their interlinking positive feedback loops, along with the leaf senescence process. It is straightforward to imagine that suppression of one of them would lead to a slowdown of senescence progression. Although our study established a signaling network involving CDF4, ABA, and ROS, other components could also be associated with this loop. Another positive regulator of the amplification loop might be the bZIP transcription factor GBF1, which induces H2O2 accumulation by downregulating CAT2 expression during leaf senescence 80. A recent report indicated that the WRKY family transcription factor WRKY75 promoted SA accumulation and H2O2 accumulation, with WRKY75, SA, and ROS forming an amplification loop to promote the progression of leaf senescence 81. Taken together, we have found that our positive feedback loop model provides a molecular framework connecting upstream signals, such as developmental age, ABA, and other environmental stresses, with the downstream regulatory network during leaf senescence and floral organ abscission progression.
Materials and Methods
Plant materials and growth conditions
The Col‐0 A. thaliana ecotype was used. Plants were grown in a climate‐controlled growth room at 22°C (± 2°C) with a relative humidity of 60% under 16‐h/8‐h light/dark lighting conditions and 120 mmol photons/m2/s of white light illumination. The nced2 (SALK‐004231), nced3 (CS331021), cat2‐1 (SALK _057998), and cat2‐2 (SALK_097354) mutants were obtained from the ABRC T‐DNA mutant repository. The mutants CS91480 and CS87649 were obtained from the NASC mutant repository. The pER8::CDF4 transgenic plant was crossed with the nced2, nced3 mutants, and 35S::CAT2 lines to obtain pER8::CDF4 & nced2, pER8::CDF4 & nced3, pER8::CDF4 & nced2nced3, and proSAG12::CDF4 & 35S::CAT2 transgenic plants. T2 transgenic plants were used in our assays. Sequence data for the genes described in this article can be found in the Arabidopsis TAIR database (https://www.arabidopsis.org) under the following accession numbers: At2g34140 for CDF4, At1g29160 for COG1, At4g18350 for NCED2, At3g14440 for NCED3, At2g14610 for PR1, At1g75040 for PR5, At3g20770 for SAG12, At1g20630 for CAT1, At1g58030 for CAT2, At1g20620 for CAT3, At1g07890 for APX1, At3g09640 for APX2, At4g35000 for APX3, At4g09010 for APX4, At4g35970 for APX5, At4g32320 for APX6, AT4G38000 for AtDOF4.7, At3g57510 for ADPG1, At2g41850 for PGAZAT/ADPG2, At3g07970 for QRT2, and At3g18780 for ACTIN2. Germplasm used included nced2 (SALK_004231), nced3 (GABI_129B08), cat2‐1 (SALK_057998), and cat2‐2 (SALK_097354) mutants.
Construction of plasmids and generation of transgenic plants
To generate transgenic plants overexpressing CDF4, NCED2, NCED3, and CAT2 genes, cDNAs were subcloned into a CaMV 35S‐controlled pHB vector. For inducible CDF4 overexpression, the CDF4 open reading frame (ORF) was cloned into the binary vector pER8. For the inducible knockdown of CDF4, the approximately 300‐bp target sequence was cloned from the genome. This sequence was introduced into the pCAMBIA1301 vector in the sense and antisense orientation. Next, the entire fragment was removed by digesting the pCAMBIA1301‐based construct, and the fragment was inserted into pER8 and pHB vectors to produce pER8::CDF4‐RNAi and pHB::CDF4‐RNAi. We obtained eleven inducible CDF4 knockdown transgenic lines, and the highly downregulated lines pER8::CDF4::RNAi‐1 and pER8::CDF4::RNAi‐2 were used for our experiments (Appendix Fig S7). For the proSAG12::CDF4 and proIDA::CDF4 constructs, the CDF4 coding sequence (CDS) was fused with the SAG12 and IDA gene promoters, and then assembled into the pCAMBIA1300 vector. For the proIDA::CDF4‐RNAi construct, the target sequence was introduced into the pCAMBIA1300 vector in the sense and antisense orientation under the control of the IDA gene promoter. For artificial microRNA (amiRNA)‐mediated gene silencing of CDF4, we used the WMD3 online tool (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi) to achieve the design of an amiRNA specific to the CDF4 coding region, with the artificial miRNA vector pRS300 being obtained from the Weigel Lab. The final fragment was inserted into pHB vector to produce pER8::amiCDF4 by using SpeI and XhoI restriction sites. The highly CDF4 downregulated lines pER8::amiCDF4‐1 and pER8::amiCDF4‐2 were used for our experiments (Appendix Fig S7). To generate 4Enhp CDF4‐CDF4GR, the CaMV 35S enhancer tetrad was amplified using pSKI015 as the template and cloned into pQDL4R1 to generate pQDL4R1‐4Enh. The GR domain was cloned from pTA7002, and the coding region of CDF4 was cloned from the cDNA. Both fragments were fused together and cloned into pQDR2L3 vector to generate pQDR2L3‐CDF4GR. The floral‐dip method was used to transform Arabidopsis. The primers used for vector construction are shown in Appendix Table S1.
qPCR analysis
Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen, USA) and stored at −80°C. The RNA samples were treated with DNase to eliminate genomic DNA. Then, 1 mg of total RNA was used for cDNA synthesis using the Novo Script® 1st Strand cDNA Synthesis SuperMix E041 (Novoprotein, China). qRT–PCRs were set up using Hieff® qPCR SYBR Green Master Mix (Yeasen, China). The data were analyzed with LightCycler 96 analysis software 1.1 (ΔΔC T method). The AtACT2 gene was used as an internal control. The qRT–PCR primers are shown in Appendix Table S2. All assays were performed in triplicate.
Subcellular localization study
The green fluorescent protein (GFP) signal was observed in the roots of 6‐day‐old transgenic Arabidopsis seedlings, which were constitutively transformed with the35S::GFP::CDF4 construct and analyzed by confocal microscopy (Zeiss LSM510; Jena, Germany).
Estradiol induction assay
The inducible overexpression or inhibition of CDF4 was induced by spraying whole plants with 20 μM estradiol at specific stages.
DEX induction assay
The rosette leaves of CDF4GR transgenic plants were treated with 20 μM DEX in DMSO, 20 μM DEX plus 100 μM CHX, or DMSO (mock). Leaves were collected at the indicated time and placed into liquid nitrogen immediately.
Hormone and abiotic stress treatments
For the dark treatment, leaves were detached and floated on 3 mM 2‐(N‐morpholino)ethanesulfonic acid (MES) buffer at pH 5.8 for a specific number of days. For the hormone treatment, mature rosette leaves were transferred to Murashige and Skoog (MS) liquid cultures containing 1 μM ABA, 1 mM SA, or 10 μM JA growth hormones. For ETH treatment, 10 μM 1‐aminocyclopropane‐1‐carboxylic acid (ACC) was used. To determine the effects of drought, salinity, or H2O2 on gene expression, leaves were transferred to dry 3 M paper or to a liquid culture containing 100 mM NaCl or 2 mM H2O2 for a period of time.
Natural and H2O2‐induced leaf senescence assays
Natural leaf senescence analysis was performed as described previously 82. The third and fourth rosette leaves of plants at different development stages were used for measurement of the SAG12 expression level and chlorophyll concentration. For H2O2‐induced leaf senescence, the third and fourth rosette leaves were detached and incubated in 2 mM EMS buffer (pH 5.8) with 2 mM H2O2 under darkness for a period of time.
Measurements of chlorophyll concentration
Chlorophyll was extracted from rosette leaves with N,N‐dimethyl formamide and measured as described previously 35.
Stomatal aperture assay
The assay was conducted as previously described with slight modifications 83. Epidermal peels from plant leaves were incubated for 4 h in a solution containing 10 mM KCl, 0.2 mM CaCl2, and 10 mM MES·KOH (pH 6.15) under white light (300 μmol/m2/s). The peeled strips were subsequently incubated in a solution containing the same buffer. Guard cells were photographed by using a light microscope equipped with a digital camera. More than 50 stomata apertures were measured for each mutant and WT plant.
Determination of endogenous ABA concentration
Leaves were collected and immediately frozen in liquid nitrogen. Frozen leaves were pulverized, and ABA was extracted as described previously 15. Quantitative determination of endogenous ABA was performed by the competitive ELISA method using a Phytodetek ABA test kit (Agdia).
DAB staining and H2O2 measurements
For DAB staining, tissues were washed three times with phosphate‐buffered saline (PBS) buffer, incubated in DAB staining solution (1 mg/mL DAB, 10 mM Na2HPO4, and 0.05% Tween 20, pH 3.8) in the dark for 4–8 h, and then decolorized in 95% ethanol. The intensity of brown coloration reflects the H2O2 content. Quantitative H2O2 measurements were performed using an Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit (Molecular Probes). Briefly, the samples were frozen in liquid nitrogen and ground into a fine powder, and 30 mg of each sample was fully suspended in 200 μl of H2O2 extraction buffer (25 mM sodium phosphate buffer, pH 6.5). The extract was centrifuged at 13,500 g for 15 min at 4°C, and the supernatant was prepared for the quantitative assay. The absorbance at 560 nm was measured with a TeCAN Infinite F200/M200 (TECAN) spectrophotometer. The H2O2 concentrations are reported in μM/g FW.
Protoplast transfection assays
For transient expression assays, several reporter and effector plasmids were constructed. The reporter plasmid contained the firefly luciferase (LUC) gene, which was controlled by a 35S promoter. CDF4 sequences were fused to the GAL4 DNA‐binding domain (BD) coding sequence and cloned into the effector plasmid. Expression of the fusion genes was driven by the 35S promoter. The vector containing the BD sequence represented the negative control. The reporter and effector plasmids were co‐transformed into Arabidopsis protoplasts. The luciferase assay was performed using the Luciferase Assay System Kit (Promega, Madison, WI, USA).
Transient dual‐luciferase reporter system
The approximately 1.8‐kb promoter sequences from NCED2 and NCED3 and approximately 1.6‐kb promoter sequence from CAT2 were amplified and inserted into the reporter plasmid pGreen II 0800‐LUC. The coding sequence of CDF4 was amplified by PCR and inserted into the effector plasmid pGreen II 62‐SK. Arabidopsis protoplasts were prepared as described previously 63 and co‐transfected with the constructs according to the manufacturer's instructions for the Dual‐Luciferase Reporter Assay System (Promega). The ratio of LUC to REN (Renilla luciferase) was determined for the Dual‐Luciferase Reporter Assay System (Promega, Madison, WI, USA) on a GLO‐MAX 20/20 luminometer (Promega) after culturing the protoplasts under low light conditions for 16 h. The LUC/REN ratio is an indicator of transcriptional activity.
ChIP assays
An HA‐coding sequence was fused in‐frame to the 3′ end of the CDF4 gene, and the gene fusion was subcloned into the pER8 vector. The expression construct was then transformed into Col‐0 plants. Two‐ or five‐week‐old pER8::CDF4::HA transgenic plants were grown on MS agar plates and those that expressed CDF4::HA via induction by estradiol for a period were used to conduct ChIP experiments according to a previously described method using anti‐HA polyclonal antibodies (Roche:118674231) 48, 84. The qRT–PCR primers used are listed in Appendix Table S3.
Recombinant protein purification
The full‐length CDF4 CDS was cloned into the pET30a and pET41b vectors and transformed into Escherichia coli. The recombinant protein was induced at 16°C and purified in its native form using Qiagen Ni‐NTA agarose (Limburg, The Netherlands) following the manufacturer's protocol.
EMSA
Construction of plasmid for the expression of recombinant CDF4 protein in E. coli and purification of CDF4 protein were conducted. A standard binding reaction was performed in a total volume of 10 μl by incubation of an appropriate amount of purified CDF4 protein with 10 fm of biotin‐labeled probe DNA and 1 μg poly (dI‐dC) in buffer (25 mM HEPES‐potassium hydroxide, pH 7.5, 100 mM KCl, 0.1 mM EDTA, 10% [v/v] glycerol, 1 mM DTT) at room temperature for 30 min. The binding reaction products were resolved on the 6% polyacrylamide gel run in 0.5 × TBE. The probes for EMSA are described in Appendix Table S3.
DNA–protein interaction ELISA
DNA–protein interaction enzyme‐linked immunosorbent assay (DPI‐ELISA) was performed as described in Brand et al 85. Full‐length glutathione S‐transferase (GST)‐CDF4 protein was produced in the BL21 strain and purified using Glutathione Superflow Resin (Qiagen). An antibody against GST conjugated with horseradish peroxidase (HRP) was used to detect the bound proteins.
Scanning electron microscopy
The leaves were cut and sputter‐coated with gold and visualized using a Hitachi JEOL JSM‐6360LV SEM (scanning electron microscope).
Analysis of catalase enzymatic activity
The catalase activity assay was performed using a catalase assay kit (Beyotime Biotechnology). Briefly, samples were frozen in liquid nitrogen and fully ground into a powder; each sample was suspended in 100 μl extraction buffer (10 mM Tris–HCl, pH 7.6, 150 mM NaCl, and 1% Nonidet P‐40) and centrifuged at 12,000 rpm for 12 min at 4°C. The supernatant was used to analyze catalase activity. Catalase activity is presented as units/mg protein. One unit of catalase activity represents the amount of enzyme that catalyzes the decomposition of 1 μM H2O2 per minute at 25°C. Protein concentration was measured using a BCA protein assay kit (Beyotime Biotechnology).
Statistical analysis and multiple alignments
Student's t‐test or ANOVA was carried out, and differences were considered significant when P < 0.05. Multiple alignments of the predicted amino acid sequences and phylogenetic analysis were performed using DNA MAN 6.0 and MEGA 4.1 software.
Author contributions
PX and WC conceived and designed the research. PX wrote the article with help from WC. PX conducted the experiments and contributed to the study design with help from HY. All authors approved the final draft of the manuscript and agreed to its submission.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Source Data for Appendix
Review Process File
Source Data for Figure 2
Source Data for Figure 4
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31500236, U1738107, 31600684, 31971172, and 31570859), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant Nos. XDA04020202‐15 and XDA04020415), and the China Manned Space Flight Technology Project.
EMBO Reports (2020) 21: e48967
Contributor Information
Peipei Xu, Email: ppxu@sibs.ac.cn.
Weiming Cai, Email: wmcai@sibs.ac.cn.
References
- 1. Nam HG (1997) The molecular genetic analysis of leaf senescence. Curr Opin Biotechnol 8: 200–207 [DOI] [PubMed] [Google Scholar]
- 2. Quirino BF, Noh YS, Himelblau E, Amasino RM (2000) Molecular aspects of leaf senescence. Trends Plant Sci 5: 278–282 [DOI] [PubMed] [Google Scholar]
- 3. Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58: 115–136 [DOI] [PubMed] [Google Scholar]
- 4. Aharoni N, Back A, Benyehoshua S, Richmond AE (1975) Exogenous gibberellic‐acid and cytokinin isopentenyladenine retardants of senescence in romaine lettuce. J Am Soc Hortic Sci 100: 4–6 [Google Scholar]
- 5. Gan S, Amasino RM (1995) Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270: 1986–1988 [DOI] [PubMed] [Google Scholar]
- 6. Yu K, Wei J, Ma Q, Yu D, Li J (2009) Senescence of aerial parts is impeded by exogenous gibberellic acid in herbaceous perennial Paris polyphylla . J Plant Physiol 166: 819–830 [DOI] [PubMed] [Google Scholar]
- 7. Kim JI, Murphy AS, Baek D, Lee SW, Yun DJ, Bressan RA, Narasimhan ML (2011) YUCCA6 over‐expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana . J Exp Bot 62: 3981–3992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zacarias L, Reid MS (1990) Role of growth‐regulators in the senescence of Arabidopsis‐Thaliana leaves. Physiol Plantarum 80: 549–554 [Google Scholar]
- 9. Reid MS, Wu MJ (1992) Ethylene and flower senescence. Plant Growth Regul 11: 37–43 [Google Scholar]
- 10. Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim YS, Penfold CA, Jenkins D et al (2011) High‐resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Van der Graaff E, Schwacke R, Schneider A, Desimone M, Flugge UI, Kunze R (2006) Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol 141: 776–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nambara E, Marion‐Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56: 165–185 [DOI] [PubMed] [Google Scholar]
- 13. Schwartz SH, Tan BC, McCarty DR, Welch W, Zeevaart JA (2003) Substrate specificity and kinetics for VP14, a carotenoid cleavage dioxygenase in the ABA biosynthetic pathway. Biochim Biophys Acta 1619: 9–14 [DOI] [PubMed] [Google Scholar]
- 14. Bittner F, Oreb M, Mendel RR (2001) ABA3 is a molybdenum cofactor sulfurase required for activation of aldehyde oxidase and xanthine dehydrogenase in Arabidopsis thaliana . J Biol Chem 276: 40381–40384 [DOI] [PubMed] [Google Scholar]
- 15. Yang J, Worley E, Udvardi M (2014a) A NAP‐AAO3 regulatory module promotes chlorophyll degradation via ABA biosynthesis in Arabidopsis leaves. Plant Cell 26: 4862–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jensen MK, Lindemose S, De Masi F, Reimer JJ, Nielsen ML, Perera V, Chris T, Franziska T, Murray R, Mundy J et al (2013) ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thaliana . FEBS Open Bio 3: 321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Weiner JJ, Peterson FC, Volkman BF, Cutler SR (2010) Structural and functional insights into core ABA signaling. Curr Opin Plant Biol 13: 495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Melcher K, Xu Y, Ng LM, Zhou XE, Soon FF, Chinnusamy V, Suino‐Powell KM, Kovach A, Tham FS, Cutler SR et al (2010) Identification and mechanism of ABA receptor antagonism. Nat Struct Mol Biol 17: 1102–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jing HC, Dijkwel PP (2008) CPR5: a Jack of all trades in plants. Plant Signal Behav 3: 562–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jing HC, Hebeler R, Oeljeklaus S, Sitek B, Stuhler K, Meyer HE, Sturre MJ, Hille J, Warscheid B, Dijkwel PP (2008) Early leaf senescence is associated with an altered cellular redox balance in Arabidopsis cpr5/old1 mutants. Plant Biol (Stuttg) 10(Suppl 1): 85–98 [DOI] [PubMed] [Google Scholar]
- 21. Shahnejatbushehri S, Tarkowska D, Sakuraba Y, Balazadeh S (2016) Arabidopsis NAC transcription factor JUB1 regulates GA/BR metabolism and signalling. Nat Plants 2: 16013 [DOI] [PubMed] [Google Scholar]
- 22. Lee S, Seo PJ, Lee HJ, Park CM (2012) A NAC transcription factor NTL4 promotes reactive oxygen species production during drought‐induced leaf senescence in Arabidopsis . Plant J 70: 831–844 [DOI] [PubMed] [Google Scholar]
- 23. Shi WL, Jia WS, Liu X, Zhang SQ (2004) Protein tyrosine phosphatases involved in signaling of the ABA‐induced H2O2 generation in guard cells of Vicia faba L. Chinese Sci Bull 49: 1841–1846 [Google Scholar]
- 24. Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JD, Schroeder JI (2003) NADPH oxidase AtrbohD and AtrbohF genes function in ROS‐dependent ABA signaling in Arabidopsis . EMBO J 22: 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu YL, Zhen SM, Wang S, Wang YP, Cao H, Zhang YZ, Li JR, Yan YM (2016) Comparative transcriptome analysis of wheat embryo and endosperm responses to ABA and H2O2 stresses during seed germination. BMC Genom 17: 97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zentgraf U, Jobst J, Kolb D, Rentsch D (2004) Senescence‐related gene expression profiles of rosette leaves of Arabidopsis thaliana: leaf age versus plant age. Plant Biol (Stuttg) 6: 178–183 [DOI] [PubMed] [Google Scholar]
- 27. Balazadeh S, Riano‐Pachon DM, Mueller‐Roeber B (2008a) Transcription factors regulating leaf senescence in Arabidopsis thaliana . Plant Biol (Stuttg) 10(Suppl 1): 63–75 [DOI] [PubMed] [Google Scholar]
- 28. Balazadeh S, Parlitz S, Mueller‐Roeber B, Meyer RC (2008b) Natural developmental variations in leaf and plant senescence in Arabidopsis thaliana . Plant Biol (Stuttg) 10(Suppl 1): 136–147 [DOI] [PubMed] [Google Scholar]
- 29. Miao Y, Zentgraf U (2007) The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell 19: 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang Y, Feng CZ, Ye Q, Wu WH, Chen YF (2016) Arabidopsis WRKY6 transcription factor acts as a positive regulator of abscisic acid signaling during seed germination and early seedling development. PLoS Genet 12: e1005833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Balazadeh S, Kwasniewski M, Caldana C, Mehrnia M, Zanor MI, Xue GP, Mueller‐Roeber B (2011) ORS1, an H(2)O(2)‐responsive NAC transcription factor, controls senescence in Arabidopsis thaliana . Mol Plant 4: 346–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kou X, Watkins CB, Gan SS (2012) Arabidopsis AtNAP regulates fruit senescence. J Exp Bot 63: 6139–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li Z, Peng J, Wen X, Guo H (2013) Ethylene‐insensitive3 is a senescence‐associated gene that accelerates age‐dependent leaf senescence by directly repressing mir164 transcription in Arabidopsis . Plant Cell 25: 3311–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Balazadeh S, Siddiqui H, Allu AD, Matallana‐Ramirez LP, Caldana C, Mehrnia M, Zanor MI, Kohler B, Mueller‐Roeber B (2010) A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt‐promoted senescence. Plant J 62: 250–264 [DOI] [PubMed] [Google Scholar]
- 35. Yang SD, Seo PJ, Yoon HK, Park CM (2011) The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23: 2155–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shahnejat‐Bushehri S, Nobmann B, Devi Allu A, Balazadeh S (2016) JUB1 suppresses Pseudomonas syringae‐induced defense responses through accumulation of DELLA proteins. Plant Signal Behav 11: e1181245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim YS, Sakuraba Y, Han SH, Yoo SC, Paek NC (2013) Mutation of the Arabidopsis NAC016 transcription factor delays leaf senescence. Plant Cell Physiol 54: 1660–1672 [DOI] [PubMed] [Google Scholar]
- 38. Sakuraba Y, Jeong J, Kang MY, Kim J, Paek NC, Choi G (2014) Phytochrome‐interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis . Nat Commun 5: 4636 [DOI] [PubMed] [Google Scholar]
- 39. Roberts JA, Elliott KA, Gonzalez‐Carranza ZH (2002) Abscission, dehiscence, and other cell separation processes. Annu Rev Plant Biol 53: 131–158 [DOI] [PubMed] [Google Scholar]
- 40. Del Campillo E (1999) Multiple endo‐1,4‐beta‐D‐glucanase (cellulase) genes in Arabidopsis . Curr Top Dev Biol 46: 39–61 [DOI] [PubMed] [Google Scholar]
- 41. Cho HT, Cosgrove DJ (2000) Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana . Proc Natl Acad Sci USA 97: 9783–9788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Butenko MA, Patterson SE, Grini PE, Stenvik GE, Amundsen SS, Mandal A, Aalen RB (2003) Inflorescence deficient in abscission controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell 15: 2296–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stenvik GE, Tandstad NM, Guo Y, Shi CL, Kristiansen W, Holmgren A, Clark SE, Aalen RB, Butenko MA (2008) The EPIP peptide of inflorescence deficient in abscission is sufficient to induce abscission in Arabidopsis through the receptor‐like kinases haesa and haesa‐like2. Plant Cell 20: 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fang SC, Fernandez DE (2002) Effect of regulated overexpression of the MADS domain factor AGL15 on flower senescence and fruit maturation. Plant Physiol 130: 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cai S, Lashbrook CC (2008) Stamen abscission zone transcriptome profiling reveals new candidates for abscission control: enhanced retention of floral organs in transgenic plants overexpressing Arabidopsis zinc finger protein2. Plant Physiol 146: 1305–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wei PC, Tan F, Gao XQ, Zhang XQ, Wang GQ, Xu H, Li LJ, Chen J, Wang XC (2010) Overexpression of AtDOF4.7, an Arabidopsis DOF family transcription factor, induces floral organ abscission deficiency in Arabidopsis . Plant Physiol 153: 1031–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yanagisawa S (2002) The Dof family of plant transcription factors. Trends Plant Sci 7: 555–560 [DOI] [PubMed] [Google Scholar]
- 48. Xu P, Chen H, Ying L, Cai W (2016) AtDOF5.4/OBP4, a DOF transcription factor gene that negatively regulates cell cycle progression and cell expansion in Arabidopsis thaliana . Sci Rep 6: 27705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Papi M, Sabatini S, Altamura MM, Hennig L, Schafer E, Costantino P, Vittorioso P (2002) Inactivation of the phloem‐specific Dof zinc finger gene DAG1 affects response to light and integrity of the testa of Arabidopsis seeds. Plant Physiol 128: 411–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ward JM, Cufr CA, Denzel MA, Neff MM (2005) The Dof transcription factor OBP3 modulates phytochrome and cryptochrome signaling in Arabidopsis . Plant Cell 17: 475–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Skirycz A, Reichelt M, Burow M, Birkemeyer C, Rolcik J, Kopka J, Zanor MI, Gershenzon J, Strnad M, Szopa J et al (2006) DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis . Plant J 47: 10–24 [DOI] [PubMed] [Google Scholar]
- 52. Park DH, Lim PO, Kim JS, Cho DS, Hong SH, Nam HG (2003) The Arabidopsis COG1 gene encodes a Dof domain transcription factor and negatively regulates phytochrome signaling. Plant J 34: 161–171 [DOI] [PubMed] [Google Scholar]
- 53. Negi J, Moriwaki K, Konishi M, Yokoyama R, Nakano T, Kusumi K, Hashimoto‐Sugimoto M, Schroeder JI, Nishitani K, Yanagisawa S et al (2013) A Dof transcription factor, SCAP1, is essential for the development of functional stomata in Arabidopsis . Curr Biol 23: 479–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Guo Y, Qin G, Gu H, Qu LJ (2009) Dof5.6/HCA2, a Dof transcription factor gene, regulates interfascicular cambium formation and vascular tissue development in Arabidopsis . Plant Cell 21: 3518–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim HS, Kim SJ, Abbasi N, Bressan RA, Yun DJ, Yoo SD, Kwon SY, Choi SB (2010) The DOF transcription factor Dof5.1 influences leaf axial patterning by promoting Revoluta transcription in Arabidopsis . Plant J 64: 524–535 [DOI] [PubMed] [Google Scholar]
- 56. Gardiner J, Sherr I, Scarpella E (2010) Expression of DOF genes identifies early stages of vascular development in Arabidopsis leaves. Int J Dev Biol 54: 1389–1396 [DOI] [PubMed] [Google Scholar]
- 57. Goralogia GS, Liu TK, Zhao L, Panipinto PM, Groover ED, Bains YS, Imaizumi T (2017) CYCLING DOF FACTOR 1 represses transcription through the topless co‐repressor to control photoperiodic flowering in Arabidopsis . Plant J 92: 244–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Corrales AR, Carrillo L, Lasierra P, Nebauer SG, Dominguez‐Figueroa J, Renau‐Morata B, Pollmann S, Granell A, Molina RV, Vicente‐Carbajosa J et al (2017) Multifaceted role of cycling DOF factor 3 (CDF3) in the regulation of flowering time and abiotic stress responses in Arabidopsis . Plant Cell Environ 40: 748–764 [DOI] [PubMed] [Google Scholar]
- 59. Fornara F, Panigrahi KC, Gissot L, Sauerbrunn N, Ruhl M, Jarillo JA, Coupland G (2009) Arabidopsis DOF transcription factors act redundantly to reduce constans expression and are essential for a photoperiodic flowering response. Dev Cell 17: 75–86 [DOI] [PubMed] [Google Scholar]
- 60. Henriques R, Wang H, Liu J, Boix M, Huang LF, Chua NH (2017) The antiphasic regulatory module comprising CDF5 and its antisense RNA FLORE links the circadian clock to photoperiodic flowering. New Phytol 216: 854–867 [DOI] [PubMed] [Google Scholar]
- 61. Sun Z, Guo T, Liu Y, Liu Q, Fang Y (2015) The roles of Arabidopsis CDF2 in transcriptional and posttranscriptional regulation of primary microRNAs. PLoS Genet 11: e1005598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Imaizumi T, Schultz TF, Harmon F, Ho LA, Kay SA (2005) FKF1 F‐box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis . Science 309: 293–297 [DOI] [PubMed] [Google Scholar]
- 63. Pi L, Aichinger E, van der Graaff E, Llavata‐Peris CI, Weijers D, Hennig L, Groot E, Laux T (2015) Organizer‐derived WOX5 signal maintains root columella stem cells through chromatin‐mediated repression of CDF4 expression. Dev Cell 33: 576–588 [DOI] [PubMed] [Google Scholar]
- 64. Schwartz SH, Leon‐Kloosterziel KM, Koornneef M, Zeevaart JA (1997) Biochemical characterization of the aba2 and aba3 mutants in Arabidopsis thaliana . Plant Physiol 114: 161–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Krebs J, Mueller‐Roeber B, Ruzicic S (2010) A novel bipartite nuclear localization signal with an atypically long linker in DOF transcription factors. J Plant Physiol 167: 583–586 [DOI] [PubMed] [Google Scholar]
- 66. Lashbrook CC, Klee HJ (1999) Ethylene regulation of abscission competence In Biology and biotechnology of the plant hormone ethylene II, Kanellis AK, Chang C, Klee H, Bleecker AB, Pech JC, Grierson D. (eds), pp 227–233. Dordrecht: Springer; [Google Scholar]
- 67. Ogawa M, Kay P, Wilson S, Swain SM (2009) Arabidopsis dehiscence zone polygalacturonase1 (adpg1), adpg2, and quartet2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis . Plant Cell 21: 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sawicki M, Ait Barka E, Clement C, Vaillant‐Gaveau N, Jacquard C (2015) Cross‐talk between environmental stresses and plant metabolism during reproductive organ abscission. J Exp Bot 66: 1707–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Riefler M, Novak O, Strnad M, Schmu T (2006) Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18: 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Raines T, Shanks C, Cheng CY, McPherson D, Argueso CT, Kim HJ, Franco‐Zorrilla JM, López‐Vidriero I, Solano R, Vaňková R et al (2016) The cytokinin response factors modulate root and shoot growth and promote leaf senescence in Arabidopsis . Plant Journal 85: 134–147 [DOI] [PubMed] [Google Scholar]
- 71. Lim PO, Lee IC, Kim J, Kim HJ, Ryu JS, Woo HR, Nam HG (2010) Auxin response factor 2 (arf2) plays a major role in regulating auxin‐mediated leaf longevity. J Exp Bot 61: 1419–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang S, Li C, Wang R, Chen Y, Shu S, Huang R, Zhang D, Li J, Xiao S, Yao N et al (2017) The Arabidopsis mitochondrial protease FtSH4 is involved in leaf senescence via regulation of WRKY‐dependent salicylic acid accumulation and signaling. Plant Physiol 173: 2294–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Besseau S, Li J, Palva ET (2012) WRKY54 and WRKY70 co‐operate as negative regulators of leaf senescence in Arabidopsis thaliana . J Exp Bot 63: 2667–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yang L, Zhang J, He J, Qin Y, Hua D, Duan Y, Chen Z, Gong Z (2014b) ABA‐mediated ROS in mitochondria regulate root meristem activity by controlling PLETHORA expression in Arabidopsis . PLoS Genet 10: e1004791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mittler R, Blumwald E (2015) The roles of ROS and ABA in systemic acquired acclimation. Plant Cell 27: 64–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wu A, Allu AD, Garapati P, Siddiqui H, Dortay H, Zanor M‐I, Asensi‐Fabado M, Munné‐Bosch S, Antonio A, Tohge T et al (2012) JUNGBRUNNEN1, a reactive oxygen species‐responsive NAC transcription factor, regulates longevity in Arabidopsis . Plant Cell 24: 482–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Qi W, Zhang L, Feng W, Xu H, Wang L, Jiao Z (2015) ROS and ABA signaling are involved in the growth stimulation induced by low‐dose gamma irradiation in Arabidopsis seedling. Appl Biochem Biotechnol 175: 1490–1506 [DOI] [PubMed] [Google Scholar]
- 78. Watkins JM, Chapman JM, Muday GK (2017) Abscisic acid‐induced reactive oxygen species are modulated by flavonols to control stomata aperture. Plant Physiol 175: 1807–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bleecker AB, Patterson SE (1997) Last exit: senescence, abscission, and meristem arrest in Arabidopsis . Plant Cell 9: 1169–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Smykowski A, Zimmermann P, Zentgraf U (2010) G‐Box binding factor1 reduces CATALASE2 expression and regulates the onset of leaf senescence in Arabidopsis . Plant Physiol 153: 1321–1331 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 81. Guo P, Li Z, Huang P, Li B, Fang S, Chu J, Guo H (2017) A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell 29: 2854–2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Seo M, Peeters AJ, Koiwai H, Oritani T, Marion‐Poll A, Zeevaart JA, Koornneef M, Kamiya Y, Koshiba T (2000) The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc Natl Acad Sci USA 97: 12908–12913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J (2002) Arabidopsis ost1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14: 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Xu P, Cai W (2019) Nitrate‐responsive OBP4‐XTH9 regulatory module controls lateral root development in Arabidopsis thaliana . PLoS Genet 15: e1008465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Brand LH, Kirchler T, Hummel S, Chaban C, Wanke D (2010) DPI‐ELISA: a fast and versatile method to specify the binding of plant transcription factors to DNA in vitro. Plant Methods 61: 25–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Source Data for Appendix
Review Process File
Source Data for Figure 2
Source Data for Figure 4


