Abstract
Background
ISR remains the major adverse outcome after percutaneous coronary intervention (PCI). MicroRNAs have been demonstrated to be associated with coronary plaque and stable in the blood and can be used as biomarkers/predictors. This study aimed to investigate whether circulating microRNAs could predict in-stent restenosis (ISR).
Methods
MicroRNA array was used to detect differently expressed microRNAs between 30 ISR patients and 30 non-ISR patients in the derivation cohort. Quantitative real-time polymerase chain reaction (qRT-PCR) was used to validate the microRNA array results and to detect levels of target microRNAs in the validation cohort. All patients were followed up for at least 1 year, and major adverse cardiac events (MACEs) were recorded. Univariate and multivariate logistic regression analysis were applied to find factors associated with ISR. Receiver operating characteristics (ROC) and Kaplan-Meier survival curves were used to analyze the predictive ability of the microRNA score for ISR.
Results
MicroRNA array and qRT-PCR showed that miR-143, 145, 425, 208, and let-7g were differently expressed between ISR patients and non-ISR patients. Multivariate analysis demonstrated that lower levels of mir-143 (OR =2.36, 95% CI: 1.43–3.67) and mir-145 (OR =2.12, 95% CI: 1.56–3.48) were associated with ISR. MicroRNA scores differed statistically between ISR patients and non-ISR patients (49.18±2.05 vs. 52.10±2.41, P<0.01) and has predictive ability for ISR with an area under the curve (AUC) of 0.8206 (95% CI: 0.7155–0.9256, P<0.01). In the validation cohort, Kaplan-Meier survival curves demonstrated that patients with higher microRNA scores have better prognosis in 1 year of follow-up.
Conclusions
A lower plasma level of mir-143/145 predicts a higher risk of ISR and a worse outcome.
Keywords: In-stent restenosis (ISR), microRNA, drug-eluting stent (DES), percutaneous coronary intervention (PCI), coronary heart disease
Introduction
Stent implantation is a milestone treatment for patients with coronary heart diseases (1,2), but 17–32% of patients treated with bare metal stents (BMS) will develop in-stent restenosis (ISR) after the procedure (3). With the introduction of drug-eluting stents (DES), the incidence of ISR has decreased to about 3.4% to 12% (4-6). However, the absolute number of stenting cases have increased dramatically in recent years. As a result, the cases of ISR have concurrently risen. Considering this problem, researchers have identified several risk factors of stent restenosis, including diabetes, age, minimum stent diameter, total stent length, etc. Progress has also been made in the treatment of ISR, such as the use of drug-eluting balloon, stent-in-stent technology. Recent studies have demonstrated that some microRNAs could serve as predictive or diagnostic biomarkers for diseases or adverse events (7). For example, miR-126 could predict myocardial infarction (8), while a combination of miR-375 and miR-142-5p could predict the recurrence of gastric cancer (9). Based on this information, we conducted a study aimed at identifying the circulating microRNAs in blood samples extracted before percutaneous coronary intervention (PCI) as predictors for ISR.
We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-4227).
Methods
Study population
In this study, to find those circulating microRNAs with predictive value for ISR, we enrolled patients who had received drug-eluting stent (DES) implantation for symptomatic coronary heart disease in our center from January 2009 to June 2012 (derivation cohort). Furthermore, to validate this result and investigate the relationship between circulating microRNAs and major adverse cardiac events (MACEs), we prospectively enrolled patients who had undergone DES implantation in another two centers from January 2011 to June 2012 (validation cohort). Data including demographic, clinical, and treatment variables were collected from in-hospital and follow-up medical records. The inclusion criteria for this study were the following: received DES implantation, received regular follow-up for at least 1 year (11 to 13 months), a blood sample taken before stent implantation, and a detailed and traceable medical record. The exclusion criteria were the following: repeated stent implantation before restenosis identification, tumor, heart failure, dialysis, and connective tissue disease. For clinical relevance, ISR was identified as target lesion revascularization (TLR). In this study, TLR was defined as repeat PCI or bypass graft placement for a stenosis in the DES placed at index PCI, or occurring within 5 mm of the stent (“edge effect”) as determined by the interventional cardiologists in each center (4). Patients with missing follow-up data or those who died during follow-up (without having TLR before death) were included in the analysis but were counted as having no TLR events. The study was conducted in accordance with the Declaration of Helsinki and the protocol of this study was approved by the ethics committee of each center, and all patients gave their written informed consent before blood extraction.
Plasma isolation and storage
Blood samples were taken via direct venous puncture into 10 mL sodium citrate–containing tubes. Plasma was isolated within 4 hours after collection of whole blood by centrifugation at 1,800 g for 10 min at room temperature and was aliquoted into RNAse/DNAse-free Eppendorf tubes and stored at −80 °C before RNA isolation. To minimize the effect of repeat freeze-thawing on circulating microRNAs, we only used plasma samples which had not been previously thawed, and results from this method have proven to be reliable (7).
RNA extraction
RNA was isolated as described previously (7). In brief, plasma was thawed on ice, and RNA was isolated using the miRNeasy RNA isolation kit (Qiagen) according to the manufacturer’s instruction for serum and plasma. For microRNA array, six pools were created by mixing plasma samples from 30 TLR patients (10 plasma samples per pool) and 30 sex- and age-matched controls (10 plasma samples per pool) for triple biological repeat (one microRNA array chip for one pooled sample). After binding to the membrane of the RNeasy Mini spin column and subsequent washing, RNA was eluted with 40 µL of RNase-free water. For all quantitative real-time polymerase chain reaction (qRT-PCR) experiments, RNA extracted from 200 µL of plasma was eluted with 14 µL of RNase-free water. To normalize the sample-to-sample variation in RNA isolation, synthetic Caenorhabditis elegans microRNAs, cel-miR-39, and cel-miR-54 (Qiagen) were added as controls before acid phenol–chloroform extraction.
MicroRNA array
Pooled total RNA was dephosphorylated and labeled with pCp-Cy3 (Agilent Technologies) and T4 RNA ligase (GE Healthcare). Then, the labeled sample was purified using Micro Bio-Spin 6 columns (Bio-Rad) and hybridized to Human miRNA Microarray V3 kit (Agilent Technologies) platform containing 866 human and 89 human viral microRNAs documented in the Sanger miRbase (version 12.0). After 20 hours of hybridizations carried out at 55 °C, microarrays were washed and scanned using an Agilent scanner controlled by Agilent Scan Control software (version 7.0) and then analyzed with Agilent Feature Extraction software (version 9.5.3.1). The raw microRNA expression data were normalized using quantile normalization and analyzed with GeneSpring GX (Agilent Technologies, version 11.5) according to recommendations from Agilent Technologies.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Plasma levels of target microRNAs were further tested by quantitative reverse transcription-polymerase chain reaction using the TaqMan method [TaqMan miRNA Reverse Transcription Kit and microRNA-specific stem-loop primers (Applied Bio Systems)] in a small-scale RT reaction for primers sequence, see Supplementary file. The RT reaction consists of a fixed volume of 1.67 µL total RNA solution of each sample as input, 1.387 µL of H2O, 0.5 µL of 10× reverse-transcription buffer, 0.063 µL of RNase-inhibitor (20 U/µL), 0.05 µL of 100 mM deoxynucleotide triphosphates (dNTPs) with deoxythymidine triphosphate (dTTP), and 0.33 µL of MultiScribe Reverse-Transcriptase (50 U/µL). The 5 µL reactions were incubated in an Applied Biosystems 7500 Real Time PCR System in a 96-well plate at 16 °C for 30 min, 42 °C for 30 min, and 85 °C for 5 min, and held at 4 °C. All RT reactions were run in duplicate. The levels of the synthetic spiked-in RNAs were measured in each of the samples and showed no signs of abnormal RNA loss during the extraction process of any of the samples.
Data processing and statistical analysis
Continuous variables were expressed as mean ± standard deviation (SD) and compared using the Student’s t-test. Categorical variables were expressed as numbers and percentages and were compared using the χ2 test.
In microarray analysis, the independent-samples t-test was performed to determine the expression differences between the ISR group and the non-ISR (NISR) group. An adjusted P value was obtained using the Benjamini-Hochberg false discover rate (FDR) for multiple comparisons.
Quantitative RT-PCR data were normalized by scaling with the mean Ct of the samples: for each sample the average Ct of all miRNAs measured in the sample was subtracted from the Ct of each miRNA. In order to return the signals to a scale which is easier to interpret, a constant (the average Ct over the entire sample set) was added back. Normalized signals were compared between the groups in order to find those miRNAs which were differently expressed between the groups. The significance of differences was assessed by a two-sided unpaired t-test. The Benjamini-Hochberg FDR method was used to control for multiple hypothesis testing, using an FDR of 0.05. Fold change was calculated as 2−ΔΔCt, where ΔCt is the absolute difference in the mean normalized Ct of a given miRNA and the mean of cel-miR-39 and cel-miR-54 in the ISR group or NISR group, yielding ΔCtISR and ΔCtNISR. ΔΔCt is the difference of ΔCtISR and ΔCtNISR.
Inverted-normalized signals were calculated for each miRNA by subtracting the normalized Ct from 50, so that high values represented high levels. The sum of inverted-normalized Ct of validated microRNAs were used as a microRNA score for analysis of the relationship of microRNAs with ISR. Univariate and multivariate logistic regression analysis were used to identify factors associated with ISR. This method has been proved to be solid and reliable. P values <0.05 were considered statistically significant.
To determine the predictive value and optimal cut point of a microRNA score for ISR, receiver operating characteristic (ROC) curves were generated. When the optimal cut point was determined, two groups in the validation cohort were categorized according to microRNA score: patients with a higher microRNA score and patients with a lower microRNA score. Then, the Kaplan-Meier survival curves were generated to further analyze the predictive value of this microRNA score for TLR and MACEs after PCI.
Statistical analysis was carried out using SPSS17.0 software (SPSS, Inc., IL, USA). P values <0.05 were considered statistically significant.
Results
Baseline clinical and procedural characteristics of patients with or without ISR
From January 2009 to June 2012, 3,485 patients received PCI for coronary heart diseases in our center. Among these patients, 3,271 underwent DES implantation. According to the inclusion and exclusion criteria, 3,046 patients were included into this study (derivation cohort) while 225 patients were excluded. In the final study population, 163 patients received TLR (ISR Group, 5.4%) within 1 year after initial DES implantation. Baseline characteristics of patients in the ISR Group and NISR Group (NISR Group, n=2,883, 94.6%) are shown in Table 1. Meanwhile, in another two centers, we included 1,034 patients (1,137 patients of which 103 were excluded) into the validation cohort for final analysis. In this validation cohort, 63 patients (6.1%) received TLR within 1 year of follow-up. Baseline characteristics of this validation cohort are displayed in Table 2. There was no statistical difference in the rate of TLR between these two cohorts (5.4% vs. 6.1%, P=0.3677).
Table 1. Baseline characteristics of patients in the ISR group and NISR group.
| Items | ISR group (n=163) | NISR group (n=2,883) | P value |
|---|---|---|---|
| Age (years old) | 59.3±10.2 | 61.7±12.4 | 0.1573 |
| Gender (male, %) | 127 (77.9%) | 2,343 (81.3%) | 0.2872 |
| History, n (%) | |||
| Hyperlipidaemia | 69 (42.3) | 1,147 (39.8) | 0.5184 |
| Hypertension | 77 (47.2) | 1,421 (49.3) | 0.6106 |
| Current smoking | 45 (27.6) | 868 (30.1) | 0.4979 |
| Current alcohol | 62 (38.0) | 1,049 (36.4) | 0.6701 |
| Diabetes | 57 (35.0) | 1,075 (37.3) | 0.5513 |
| PE findings on admission | |||
| BMI (kg/m2) | 27.2±6.7 | 26.5±5.4 | 0.1125 |
| HR (bpm) | 73.8±12.6 | 72.3±14.7 | 0.2019 |
| SBP (mmHg) | 133.6±26.2 | 131.4±30.5 | 0.3670 |
| DBP (mmHg) | 75.2±10.9 | 73.8±9.6 | 0.0723 |
| On admission tests | |||
| Blood glucose (mmol/L) | 6.2±4.1 | 5.1±3.6 | 0.0002 |
| LDL-C (mmol/L) | 2.8±0.9 | 2.3±0.8 | 0.0000 |
| HDL-C (mmol/L) | 0.9±0.4 | 1.0±0.6 | 0.0357 |
| TG (mmol/L) | 2.3±2.1 | 2.1±1.9 | 0.1938 |
| Medication at follow-up, n (%) | |||
| ACEI | 68 (41.7) | 1,254 (48.5) | 0.6558 |
| ARB | 34 (20.9) | 528 (18.3) | 0.4152 |
| BB | 49 (30.1) | 1,043 (36.2) | 0.1132 |
| Statins | 163 (100.0) | 2,883 (100.0) | 1.0000 |
| ASA | 163 (100.0) | 2,883 (100.0) | 1.0000 |
| CLOP | 163 (100.0) | 2,883 (100.0) | 1.0000 |
| CCB | 56 (34.4) | 908 (31.5) | 0.4449 |
| Procedural parameters | |||
| Total stent length (mm) | 37.5±8.6 | 31.6±7.9 | 0.0000 |
| Minimum stents diameters (mm) | 2.7±0.6 | 2.9±0.8 | 0.0017 |
BMI, body mass index; HR, heart rate; SBP, systolic blood pressure; DBP, diastolic blood pressure; LDL-C, low-density lipoprotein cholesterol; HDL-C, high-density lipoprotein cholesterol; TG, triglyceride; PE, physical examination; ACEI, angiotensin convert enzyme inhibitor; ARB, angiotensin receptor blocker; BB, β-receptor blocker; ASA, aspirin; CLOP, clopidogrel; CCB, calcium channel blocker.
Table 2. Baseline characteristics of patients in validation cohort.
| Items | ISR group (n=63) | NISR group (n=971) | P value |
|---|---|---|---|
| Age (years old) | 60.6±11.8 | 61.8±12.8 | 0.4690 |
| Gender (male, %) | 51 (81.0%) | 763 (78.6%) | 0.6555 |
| History, n (%) | |||
| Hyperlipidaemia | 25 (39.7) | 401 (41.3) | 0.8007 |
| Hypertension | 29 (46.0) | 458 (4.2) | 0.8610 |
| Current smoking | 19 (30.2) | 270 (27.8) | 0.6868 |
| Current alcohol | 23 (36.5) | 341 (35.1) | 0.8229 |
| Diabetes | 21 (33.3) | 303 (31.2) | 0.7421 |
| PE findings on admission | |||
| BMI (kg/m2) | 26.1±6.9 | 26.7±7.3 | 0.5261 |
| HR (bpm) | 71.6±13.1 | 70.8±12.9 | 0.6338 |
| SBP (mmHg) | 135.3±27.6 | 134.5±29.1 | 0.8321 |
| DBP (mmHg) | 75.2±10.9 | 73.8±9.6 | 0.0083 |
| On admission tests | |||
| Blood glucose (mmol/L) | 5.8±3.9 | 5.3±3.5 | 0.2756 |
| LDL-C (mmol/L) | 2.6±0.9 | 2.1±0.8 | 0.0000 |
| HDL-C (mmol/L) | 0.9±0.5 | 1.0±0.6 | 0.1960 |
| TG (mmol/L) | 2.2±2.1 | 2.0±2.0 | 0.4434 |
| Medication at follow-up, n (%) | |||
| ACEI | 30 (47.6) | 448 (46.1) | 0.8193 |
| ARB | 12 (19.0) | 180 (18.5) | 0.9196 |
| BB | 17 (27.0) | 251 (25.8) | 0.8422 |
| Statins | 63 (100.0) | 971 (100.0) | 1.0000 |
| ASA | 63 (100.0) | 971 (100.0) | 1.0000 |
| CLOP | 63 (100.0) | 971 (100.0) | 1.0000 |
| CCB | 20 (31.7) | 292 (30.1) | 0.7791 |
| Procedural parameters | |||
| Stent length (mm) | 36.7±9.1 | 30.4±8.2 | 0.0000 |
| Stent diameters (mm) | 2.6±0.7 | 2.9±0.9 | 0.0096 |
BMI, body mass index; HR, heart rate; SBP, systolic blood pressure; DBP, diastolic blood pressure; LDL-C, low-density lipoprotein cholesterol; HDL-C, high-density lipoprotein cholesterol; TG, triglyceride; PE, physical examination; ACEI, angiotensin convert enzyme inhibitor; ARB, angiotensin receptor blocker; BB, β-receptor blocker; ASA, aspirin; CLOP, clopidogrel; CCB, calcium channel blocker.
MicroRNA array and plasma level of selected microRNAs
The characteristics of 60 patients (30 ISR patients vs. 30 NISR patients) in the derivation cohort are summarized in Table 3. MicroRNA array revealed that a total of 5 miRNAs were significantly differentially expressed between the ISR group and NISR group (Table 4). This result was further validated by qRT-PCR. The fold change of these 5 microRNAs in qRT-PCR is also displayed in Table 4.
Table 3. Baseline characteristics of 30 ISR patients and 30 NISR patients.
| Items | ISR group (n=30) | NISR group (n=30) | P value |
|---|---|---|---|
| Age (years old) | 60.1±10.4 | 62.8±11.2 | 0.3373 |
| Gender (male, %) | 22 (73.3%) | 24 (80.0%) | 0.5416 |
| History, n (%) | |||
| Hyperlipidaemia | 11 (36.7) | 9 (30.0) | 0.5839 |
| Hypertension | 14 (46.7) | 15 (50.0) | 0.7961 |
| Current smoking | 5 (16.7) | 4 (13.3) | 0.7177 |
| Current alcohol, | 10 (33.3) | 8 (26.7) | 0.5731 |
| Diabetes | 9 (30.0) | 2 (6.7) | 0.0453 |
| PE findings on admission | |||
| BMI (kg/m2) | 27.2±5.2 | 25.3±4.7 | 0.1430 |
| HR (bpm) | 73.8±11.4 | 70.6±11.1 | 0.2752 |
| SBP (mmHg) | 135.5±21.9 | 131.2±20.5 | 0.4356 |
| DBP (mmHg) | 77.4±11.6 | 72.4±10.1 | 0.0802 |
| On admission tests | |||
| Blood glucose (mmol/L) | 6.8±2.9 | 5.1±2.5 | 0.0181 |
| LDL-C (mmol/L) | 2.6±0.8 | 2.1±0.8 | 0.0186 |
| HDL-C (mmol/L) | 0.9±0.5 | 1.1±0.6 | 0.1661 |
| TG (mmol/L) | 2.4±2.1 | 2.1±2.0 | 0.5732 |
| Medication at follow-up, n (%) | |||
| ACEI | 9 (30.0) | 10 (33.3) | 0.7814 |
| ARB | 4 (13.3) | 5 (16.7) | 0.7177 |
| BB | 7 (21.2) | 9 (30.0) | 0.5593 |
| Statins | 30 (100.0) | 30 (100.0) | 1.0000 |
| ASA | 30 (100.0) | 30 (100.0) | 1.0000 |
| CLOP | 30 (100.0) | 30 (100.0) | 1.0000 |
| CCB | 11 (36.7) | 12 (40.0) | 0.7906 |
| Procedural parameters | |||
| Stent length (mm) | 36.9±8.6 | 29.8±6.2 | 0.0005 |
| Stent diameters (mm) | 2.6±0.7 | 3.0±0.8 | 0.0438 |
BMI, body mass index; HR, heart rate; SBP, systolic blood pressure; DBP, diastolic blood pressure; LDL-C, low-density lipoprotein cholesterol; HDL-C, high-density lipoprotein cholesterol; TG, triglyceride; PE, physical examination; ACEI, angiotensin convert enzyme inhibitor; ARB, angiotensin receptor blocker; BB, β-receptor blocker; ASA, aspirin; CLOP, clopidogrel; CCB, calcium channel blocker.
Table 4. Fold change (ISR vs. NISR) of selected microRNAs in microRNA array and qRT-PCR.
| MicroRNA | MicroRNA array | qRT-PCR | |||
|---|---|---|---|---|---|
| Fold change | Adjusted P value | Fold change | P value | ||
| miR-143 | 0.28 | 0.0037 | 0.37 | 0.0076 | |
| miR-145 | 0.32 | 0.0042 | 0.39 | 0.0052 | |
| miR-425 | 1.63 | 0.0081 | 1.71 | 0.0138 | |
| miR-208 | 1.89 | 0.0163 | 1.84 | 0.0185 | |
| let-7g | 2.01 | 0.0209 | 2.13 | 0.0318 | |
ISR, in-stent restenosis; qRT-PCR, quantitative real-time polymerase chain reaction.
Relationship of microRNA score with ISR
Univariate and multivariate logistic regression analysis revealed that along with six clinical factors, plasma levels of miR-143 (OR =2.36, 95% CI: 1.43–3.67) and mir-145 (OR =2.12, 95% CI: 1.56–3.48) were statistically associated with ISR. MicroRNA scores were calculated to represent the cumulative plasma level of microRNA-143 and microRNA-145. The mean microRNA score in the ISR Group was significantly lower than that in the NISR Group (49.18±2.05 vs. 52.10±2.41, P<0.01), as shown in Figure 1 and Table 5. The relationship of microRNA score with in-future ISR is shown in Figure 2. The ability of this microRNA score to discriminate those who would develop ISR from those who would not is suggested by an area under the curve (AUC) of 0.8206 (95% CI: 0.7155–0.9256, P<0.01).
Figure 1.
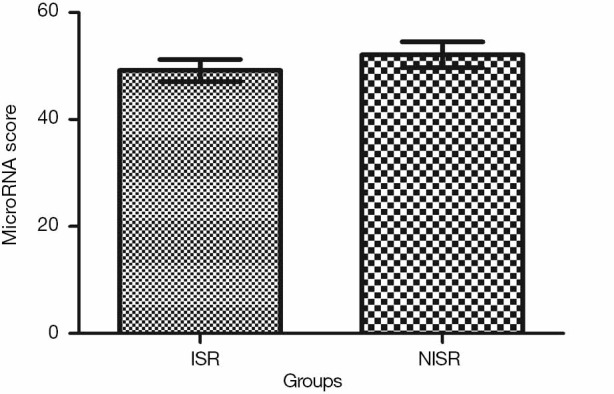
MicroRNA score in ISR group and NISR group in the derivation cohort. ISR, in-stent restenosis; NISR, non-ISR.
Table 5. Correlation of microRNAs with ISR.
| microRNA | OR value | 95% CI | P value |
|---|---|---|---|
| miR-143 | 2.36 | 1.43–3.67 | 0.0068 |
| miR-145 | 2.12 | 1.56–3.48 | 0.0079 |
| miR-425 | 1.29 | 0.86–1.57 | 0.1035 |
| miR-208 | 1.37 | 0.94–1.76 | 0.0824 |
| let-7g | 1.13 | 0.81–1.62 | 0.1352 |
ISR, in-stent restenosis.
Figure 2.
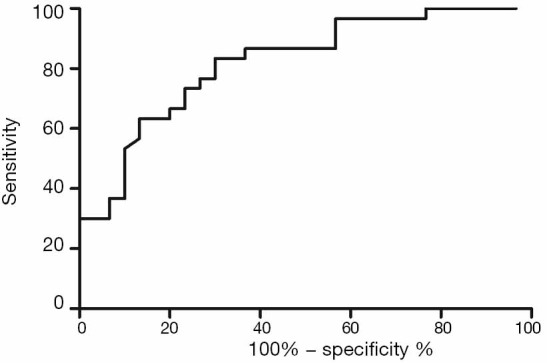
ROC of the microRNA score with ISR in the derivation cohort. ROC, receiver operating characteristic; ISR, in-stent restenosis.
Validation of the predictive value of microRNA score in a new cohort and its relationship with MACEs. To validate whether the microRNA score was fitted to patients from other centers or not, we enrolled 1,034 patients, including 63 patients who had received TLR, from another two hospitals. Plasma levels of microRNA-143 and microRNA-145 were also detected by qRT-PCR in the validation cohort, and the microRNA score of each patient was calculated according to the methods described above. From this validation cohort, we also established two groups: the ISR group (n=63) and the NISR group (n=63). These two groups were also age- and sex-matched. The results revealed that in validation cohort, there was also a significant difference of microRNA score between the ISR group and the NISR group (48.78±1.97 vs. 51.51±2.06, P<0.01) as shown in Figure 3. The ROC also gave us an AUC of 0.8396 (95% CI: 0.7669–0.9122, P<0.01; Figure 4). The ROC also showed that the optimal cutoff point of microRNA to identify patients with a higher risk of in-future ISR was 50.16 with a sensitivity of 80.33% and a specificity of 78.69% (P<0.01). According to this cutoff value of microRNA score, we divided all patients in the validation cohort into two groups: the microRNA score ≥50.16 group and the microRNA score <50.16 group. Kaplan-Meier survival curves (Figure 5) demonstrated that patients with a higher microRNA score (≥50.16, n=663, MACEs =43, 6.5%) had better outcomes with a 35.0% reduction in total MACEs compared with those patients with a lower microRNA score (<50.16, n=371, MACEs =37, 10.0%).
Figure 3.
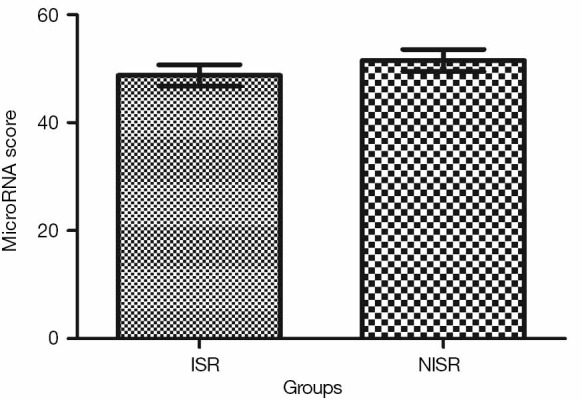
MicroRNA score in ISR Group and NISR Group in the validation cohort. ISR, in-stent restenosis; NISR, non-ISR.
Figure 4.
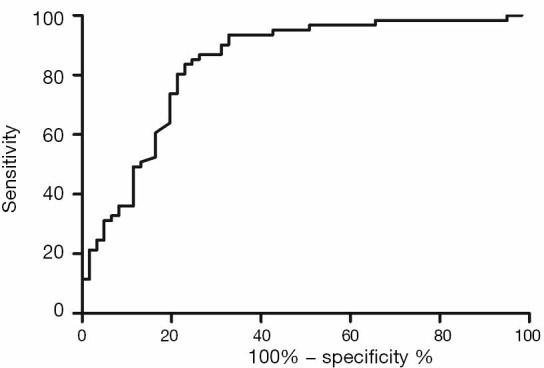
ROC of the microRNA score with ISR in the validation cohort. ROC, receiver operating characteristic; ISR, in-stent restenosis.
Figure 5.
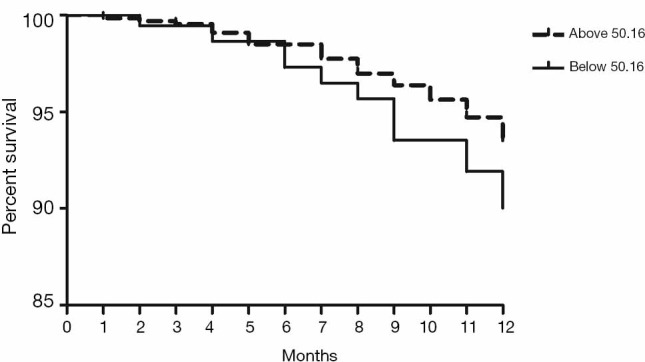
Kaplan-Meier survival curves of free from MACEs within 1 year of follow-up in the validation cohort according to microRNA score. P=0.0441. MACEs, major adverse cardiac events.
Discussion
Clinical and basic research on ISR has been conducted, and significant progress in risk prediction, prevention, and therapy has been subsequently achieved in recent years. Despite these advancements, ISR is still the main adverse outcome after PCI. Intensive antiplatelet and statin therapy and strict post-procedural management appears to fail to effectively prevent ISR in some patients. The rate of recurrent ISR after DES or drug-eluting balloon (DEB) for first ISR is relatively high. All these facts suggest that, for some patients, the genetic background needs to be considered for ISR. MicroRNA provides a convenient method to detect genetic background difference because of its stability in circulating blood and relative cell/tissue specificity. For the first time, the present study demonstrated that patients with a lower plasma level of mir-143/145 are at higher risk for ISR after DES implantation, independent of other factors.
The basic mechanism of ISR involves aggregation of platelets, leucocytes, and macrophages leading to medial smooth muscle cell migration and proliferation (5,6,10). Lately, researchers have found that about one-third of ISR incidents were due to in-stent neoatherosclerosis (ISNA) which is obviously different from the classic mechanism (11). However, there does exist a common phenomenon across these mechanisms which is the inflammation of ISR and ISNA. In previous studies, mir-143/145 has been confirmed to be highly and specifically expressed in vascular smooth muscle cells (VSMCs) and to play an important role in the process of proliferation and migration of VSMCs and in the function of arteries (12-14). Further studies demonstrated a direct association of mir-143/145 with cardiovascular disease in human and experimental animal models (15-17). Mir-143/145 was shown to be downregulated during neointimal formation in the rat carotid artery (17). In animal models of injured or atherosclerotic vessels, such as atherosclerotic aortas of ApoE knockout mice, mouse ligation-induced carotid artery, and rat carotid balloon-injured carotid artery, researchers found a significant decrease in the expression of miR-143/145 (15,16). All these findings suggest that mir-143/145 is involved in the process of atherosclerosis and ISR caused by VSMC migration and proliferation. Besides this, a few recent studies also found that mir-143/145 may possibly play an important role in some inflammatory diseases (18,19). In patients with allergic rhinitis, the authors found that the mir-143 level was downregulated compared to that in non-allergic patients (18). In another study, the authors showed that mir-143/145 was also downregulated in ulcerative colitis (19). These findings give us reason to believe that mir-143/145 is probably critical in ISR/ISNA-related inflammation. A pathology study also exists showing that, during restenosis progression, the quantity of mir-143/145 decreases in the media and intima of the coronary artery (20).
Our study, for the first time, demonstrated that in some patients with confirmed coronary heart disease, plasma levels of mir-143/145 are relatively lower than those in other coronary heart disease patients before PCI. This result suggests that plasma levels of mir-143/145 are part of the innate genetic background of some coronary heart disease patients. Furthermore, our results indicate that patients with lower plasma levels of mir-143/145 are prone to develop ISR after DES implantation and that this is independent of other factors. Actually, in some ISR patients with a lower plasma level of mir-143/145, we found that there were no reported risk factors for ISR (4-6,11,21,22). This finding supports the notion that a lower plasma level of mir-143/145 is an independent risk factor for ISR and that the detection of mir-143/145 before PCI may help us to identify patients with a high risk of ISR.
In this study, we defined ISR as restenosis needing TLR. In our clinical practice, the incidence of follow-up angiographic restenosis is about 10% and is relatively higher than the rate of TLR due to restenosis. This phenomenon suggests that some angiographic restenosis is stable and of little clinical importance. Further Kaplan–Meier survival analysis demonstrated that patients with a higher microRNA score have a better prognosis after PCI with a 35.0% reduction in total MACEs. In clinical practice, we should take these risk factors, along with the levels of miR-143 and miR-145, into consideration when deciding whether to implant DES. Accordingly, we should pay more attention to patients with these risk factors, and intensive therapy and follow-up should be initiated immediately after PCI procedure.
There are certain limitations to our study. First, we could not establish a direct, molecular relationship of mir-143/145 with ISR. For now, we only know that patients with lower plasma levels of miR-143 or miR-145 are at higher risk of ISR, while the exact and precise mechanism for this remains unclear, and needs to be elucidated in the future. Second, we could not determine the influence of drug therapy on the levels of these microRNAs, whether certain therapies could upregulate or downregulate these microRNAs, and their relationship to outcomes. Further study should aim to reveal the relationship of certain drug therapies with the change of plasma levels of microRNAs and the related prognosis.
Conclusions
A lower plasma level of mir-143/145 predicts a higher risk of ISR and a worse outcome.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: None.
Supplementary
Specific primers
miR-143 Forward primer 5'-CCTGGCCTGAGATGAAGCAC-3'
Reverse primer 5'-CAGTGCTGGGTCCGAGTGA-3'
miR-145 Forward primer 5'-GGCGTCCAGTTTTCCCAG-3'
Reverse primer 5'-CAGTGCTGGGTCCGAGTGA-3'
miR-425 Forward primer 5'-GCTACTGCGCAGGAAGTAAC-3'
Reverse primer 5'-CGCTTGAGTCGAGCGAGGT-3'
miR-208 Forward primer 5'-CGGCTCCGATGATAGAGACC-3'
Reverse primer 5'-CCTGTGGCGGCAGTGATGA-3'
let-7g Forward primer 5'-CCTGCTGAGCTGAGAGCAAC-3'
Reverse primer 5'-CTGGTGCGGATCAGTCGGA-3'
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and the protocol of this study was approved by the ethics committee of each center, and all patients gave their written informed consent before blood extraction.
Footnotes
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-4227
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-4227
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-4227). The authors have no conflicts of interest to declare.
References
- 1.Jabbar AY, Baydoun H, Janbain M, et al. Current concepts in the management of stable ischemic heart disease and acute coronary syndrome in patients with hemophilia. Ann Transl Med 2018;6:299. 10.21037/atm.2018.05.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmerini T, Biondi-Zoccai G, Della Riva D, et al. Clinical outcomes with drug-eluting and bare-metal stents in patients with ST-segment elevation myocardial infarction: evidence from a comprehensive network meta-analysis. J Am Coll Cardiol 2013;62:496-504. 10.1016/j.jacc.2013.05.022 [DOI] [PubMed] [Google Scholar]
- 3.Tsigkas GG, Karantalis V, Hahalis G, et al. Stent restenosis, pathophysiology and treatment options: a 2010 update. Hellenic J Cardiol 2011;52:149-57. [PubMed] [Google Scholar]
- 4.Stolker JM, Kennedy KF, Lindsey JB, et al. Predicting restenosis of drug-eluting stents placed in real-world clinical practice: derivation and validation of a risk model from the EVENT registry. Circ Cardiovasc Interv 2010;3:327-34. 10.1161/CIRCINTERVENTIONS.110.946939 [DOI] [PubMed] [Google Scholar]
- 5.Zhou H, Zhang S, Sun X, et al. Lipid management for coronary heart disease patients: an appraisal of updated international guidelines applying Appraisal of Guidelines for Research and Evaluation II—clinical practice guideline appraisal for lipid management in coronary heart disease. J Thorac Dis 2019;11:3534-46. 10.21037/jtd.2019.07.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Minha S, Pichard AD, Waksman R. In-stent restenosis of drug-eluting stents. Future Cardiol 2013;9:721-31. 10.2217/fca.13.45 [DOI] [PubMed] [Google Scholar]
- 7.Kroh EM, Parkin RK, Mitchell PS, et al. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods 2010;50:298-301. 10.1016/j.ymeth.2010.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zampetaki A, Willeit P, Tilling L, et al. Prospective study on circulating MicroRNAs and risk of myocardial infarction. J Am Coll Cardiol 2012;60:290-9. 10.1016/j.jacc.2012.03.056 [DOI] [PubMed] [Google Scholar]
- 9.Zhang X, Yan Z, Zhang J, et al. Combination of hsa-miR-375 and hsa-miR-142-5p as a predictor for recurrence risk in gastric cancer patients following surgical resection. Ann Oncol 2011;22:2257-66. 10.1093/annonc/mdq758 [DOI] [PubMed] [Google Scholar]
- 10.Carter AJ, Laird JR, Farb A, et al. Morphologic characteristics of lesion formation and time course of smooth muscle cell proliferation in a porcine proliferative restenosis model. J Am Coll Cardiol 1994;24:1398-405. 10.1016/0735-1097(94)90126-0 [DOI] [PubMed] [Google Scholar]
- 11.Park SJ, Kang SJ, Virmani R, et al. In-stent neoatherosclerosis: a final common pathway of late stent failure. J Am Coll Cardiol 2012;59:2051-7. 10.1016/j.jacc.2011.10.909 [DOI] [PubMed] [Google Scholar]
- 12.Rangrez AY, Massy ZA, Metzinger-Le Meuth V, et al. miR-143 and miR-145: molecular keys to switch the phenotype of vascular smooth muscle cells. Circ Cardiovasc Genet 2011;4:197-205. 10.1161/CIRCGENETICS.110.958702 [DOI] [PubMed] [Google Scholar]
- 13.Norata GD, Pinna C, Zappella F, et al. MicroRNA 143-145 deficiency impairs vascular function. Int J Immunopathol Pharmacol 2012;25:467-74. 10.1177/039463201202500216 [DOI] [PubMed] [Google Scholar]
- 14.Kohlstedt K, Trouvain C, Boettger T, S, et al. AMP-activated protein kinase regulates endothelial cell angiotensin-converting enzyme expression via p53 and the post-transcriptional regulation of microRNA-143/145. Circ Res 2013;112:1150-8. 10.1161/CIRCRESAHA.113.301282 [DOI] [PubMed] [Google Scholar]
- 15.Ji R, Cheng Y, Yue J, et al. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res 2007;100:1579-88. 10.1161/CIRCRESAHA.106.141986 [DOI] [PubMed] [Google Scholar]
- 16.Cheng Y, Liu X, Yang J, et al. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ Res 2009;105:158-66. 10.1161/CIRCRESAHA.109.197517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cordes KR, Sheehy NT, White MP, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 2009;460:705-10. 10.1038/nature08195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaoqing Y, Ruxin Z, Guojun L, et al. Microarray analysis of differentially expressed microRNAs in allergic rhinitis. Am J Rhinol Allergy 2011;25:e242-6. 10.2500/ajra.2011.25.3682 [DOI] [PubMed] [Google Scholar]
- 19.Pekow JR, Dougherty U, Mustafi R, et al. miR-143 and miR-145 are downregulated in ulcerative colitis: putative regulators of inflammation and protooncogenes. Inflamm Bowel Dis 2012;18:94-100. 10.1002/ibd.21742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Popovich IM. The role of micro-RNA/143/145 in evolution of intra-stent restenosis. Kardiologiia 2011;51:17-21. [PubMed] [Google Scholar]
- 21.Yonetsu T, Kato K, Kim SJ, et al. Predictors for neoatherosclerosis: a retrospective observational study from the optical coherence tomography registry. Circ Cardiovasc Imaging 2012;5:660-6. 10.1161/CIRCIMAGING.112.976167 [DOI] [PubMed] [Google Scholar]
- 22.Rathore S, Terashima M, Katoh O, et al. Predictors of angiographic restenosis after drug eluting stents in the coronary arteries: contemporary practice in real world patients. EuroIntervention 2009;5:349-54. 10.4244/V5I3A55 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as


