Abstract
Background
The abnormal expression of genes is an essential factor affecting the prognosis of cancer. RNA modification is a way of regulating post-transcriptional levels, including m6A, m5C, m1A RNA methylation. Studies have found that RNA methylation regulates tumorigenesis development and stem cell regeneration. However, there are few studies on lung adenocarcinoma. This study aims to explore the clinical value of RNA methylation for lung adenocarcinoma.
Methods
We summarized thirty-one RNA methylation regulators. The training set was obtained from The Cancer Genome Atlas (TCGA) database, and the test set was obtained from the Gene Expression Omnibus (GEO) database. The Wilcoxon test was used to analyze the expression of RNA methylation regulators. We constructed tumor subgroup models and risk models based on the expression of those regulators. Principal component analysis (PCA) and the receiver operating characteristic (ROC) confirmed the accuracy of the models. Real-time polymerase chain reaction (PCR) validates the results in vitro.
Results
Most RNA methylation regulators had distinct expressions in tumor tissues and adjacent tissues (P<0.05). All the models showed high predictive performance (AUC: 0.65–0.82), and the five-year survival of patients in each group was statistically different (P<0.05). The patients in the high-risk group were more likely to have a higher stage, more lymph node metastases, and distant metastases, showing a poor clinical outcome. Patients with high expression of NOP2 or HNRNP were more likely to have a poorly differentiated in vitro experiment.
Conclusions
With our study, we found that the expressions of most RNA methylation regulators were significantly different in cancer and para-cancerous tissues. Different molecular phenotypes constructed by RNA methylation regulators can be independent risk factors for the prognosis of lung adenocarcinoma. Our study demonstrates the critical role of RNA methylation in lung adenocarcinoma, and it is expected to supply a reference for the prognostic stratification and treatment strategy development of lung adenocarcinoma.
Keywords: RNA methylation, prognostic signature, lung adenocarcinoma, The Cancer Genome Atlas (TCGA), epigenetic modification
Introduction
Lung cancer is one of the most common malignant tumors in the world. It is estimated that lung cancer-related deaths account for 23% of the total cancer-related deaths in 2020 (1). Non-small cell lung cancer accounts for 75% to 80% of all lung cancers, including adenocarcinoma and squamous cell carcinoma (2). Surgery is the primary treatment of non-small cell lung cancer. Tumor volume and lymph node metastasis are critical indicators to assess whether patients can undergo surgery and tumor staging (3). Traditional tumor staging is often used to predict the prognosis of lung cancer patients. However, patients with the same tumor stage sometimes have different prognoses due to individual differences (4). Recent studies have found that molecular expression is a key factor influencing lung cancer prognosis. For example, PD-1 expression reflects the response to immunotherapy (5), which suggests the vital value of molecular level studies in lung cancer prognosis.
RNA modification is a way of regulating post-transcriptional levels (6,7). More than 100 types of chemical RNA modifications, which are widely distributed in messenger RNA (mRNA), transfer RNA (tRNA), non-coding small RNA, and long non-coding RNA (lncRNA), have been identified at present (6,8,9). N6-methyladenosine (m6A) methylation is the most common type of RNA modification on mRNA (10). Limited by the low sensitivity of early detection techniques, it was not until 2011 that the biological function of the first m6A methylation regulators on mass and obesity (FTO) was clarified (11). 5-methylcytosine (m5C) and N1-methyladenosine (m1A) are new types of RNA modifications that have attracted full attention in recent years. Bisulfite-based transcriptome sequencing study finds thousands of m5C modification sites on mRNAs in HeLa cells, and research on RIP sequencing technology found that m1A is a novel transcriptome control with evolutionary conservation (12,13). However, the characteristics and functions of m5C and m1A modification in mRNA are not entirely apparent.
RNA methylation regulates transcriptome expression under the dynamic regulation of methyltransferases (“writers”), binding proteins (“readers”), and demethylases (“erasers”) (14). Through the study of RNA methylation related proteins, thirty-one RNA methylation regulators were found (Table 1) (15-17). Recent studies have demonstrated that RNA methylation dynamically and reversibly regulates critical biological functions such as RNA metabolism and processing, as well as directional differentiation of stem cells (18). In the field of cancer research, it has been found that RNA methylation exists in multiple processes of tumorigenesis, development, and metastasis (19). For example, a study based on The Cancer Genome Atlas (TCGA) database found that m6A methylation regulators are involved in malignant progression and can predict the prognosis of liver cancer patients (20). The m6A demethylase FTO promotes the growth of lung cancer cells by regulating the expression of USP7 mRNA, and m6A methylation, which is also associated with afatinib resistance in lung cancer cells (21,22). However, m5C and m1A methylation are rarely studied in lung cancer, and the overall level of the predictive value of m6A methylation in lung cancer is insufficient.
Table 1. RNA methylation regulators of m6A, m5C, and m1A methylation.
| RNA methylation | Writer | Reader | Eraser |
|---|---|---|---|
| m6A | METTL3, METTL14, RBM1, WTAP, ZC3H13, KIAA1429 | HNRNPC, YTHDC1, YTHDC2, YTHDF1, YTHDF2 | ALKBH5, FTO |
| m5C | NOP2, NSUN2, NSUN3, NSUN4, NSUN5, NSUN7, DNMT1, TRDMT1, DNMT3A, DNMT3B | ALYREF | TET2 |
| m1A | TRMT6, TRMT10C, BMT2, RRP8 | ALKBH1, ALKBH3 |
In this study, we systematically analyzed the expression of m6A, m5C, and m1A methylation regulators in lung adenocarcinoma, as well as the associations between the clinicopathological characteristics. We constructed different tumor subgroup models and risk models to show the predictive value of RNA methylation for lung adenocarcinoma.
Methods
Datasets acquisition
The RNA-seq transcriptome data and corresponding clinical information of LUAD were downloaded from TCGA (https://cancergenome.nih.gov/) database. RNA-seq transcription of LUAD data included 59 cases of para-cancerous tissues and 535 cases of tumor tissues. Two hundred and ninety-four clinical cases were obtained after removing invalid data. Clinical information included age, gender, stage, T, N, M, overall survival (OS) time, and survival status (Table 2).
Table 2. Clinicopathological features of patients included in TCGA database.
| Variable | Number | Percentage |
|---|---|---|
| Total cases | 294 | 100.00 |
| Age | ||
| <65 | 124 | 42.18 |
| ≥65 | 170 | 57.82 |
| Gender | ||
| Male | 145 | 49.32 |
| Female | 149 | 50.68 |
| Stage | ||
| Stage I | 152 | 51.70 |
| Stage II | 71 | 24.15 |
| Stage III | 51 | 17.35 |
| Stage IV | 20 | 6.80 |
| T | ||
| T1 | 88 | 29.94 |
| T2 | 156 | 53.06 |
| T3 | 25 | 8.50 |
| T4 | 25 | 8.50 |
| N | ||
| N0 | 186 | 63.27 |
| N1 | 64 | 21.77 |
| N2 | 44 | 14.96 |
| M | ||
| M0 | 274 | 93.20 |
| M1 | 20 | 6.80 |
TCGA, The Cancer Genome Atlas.
Also, the test set data was downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) under the accession number (GSE37745). It is used to test the reliability of the Cox risk models constructed from TCGA. Sixty-seven tumor cases from lung adenocarcinoma and clinical information include age, gender, stage, OS, and survival status (Table 3).
Table 3. Clinicopathological features of patients included in GEO database.
| Variable | Number | Percentage |
|---|---|---|
| Total cases | 67 | 100.00% |
| Age | ||
| <65 | 38 | 56.72% |
| ≥65 | 29 | 43.28% |
| Gender | ||
| Male | 27 | 40.30% |
| Female | 40 | 59.70% |
| Stage | ||
| Stage i | 46 | 68.66% |
| Stage ii | 13 | 19.40% |
| Stage iii | 5 | 7.46% |
| Stage iv | 3 | 4.48% |
GEO, Gene Expression Omnibus.
Lung adenocarcinoma and adjacent tissues from 11 patients (average age =48.0±13.4 years; 8 male patients and 3 female patients) were collected from The First Affiliated Hospital of China Medical University. The surgery and samples collection period is October 2019 to present. The study was conducted in accordance with the Declaration of Helsinki. The study was approved by ethics board of The First Affiliated Hospital of China Medical University (No. YB M-05-02) and informed consent was taken from all the patients.
Bioinformatic analysis
After correcting for transcriptome expression on the training and test sets, we separately extracted expression data of the thirteen m6A RNA methylation regulators, twelve m5C RNA methylation regulators, six m1A RNA methylation regulators, and all the regulators. According to the classification of tumor tissues and adjacent tissues, we analyze the expression of those RNA methylation regulators. Violin maps and heatmaps were drawn to observe the expression distribution of RNA methylation regulators.
To investigate the role of different RNA methylation regulators in lung adenocarcinoma, we removed adjacent samples and grouped tumor samples using the “Consensus Cluster Plus” package. Principal component analysis (PCA) analysis was used to test the clustering effects. Combined clinical information, we draw survival curves to analyze the survival differences of different subgroups and draw heatmaps to explore the correlation between subgroups and clinical characteristics.
To clarify the prognosis risk of the RNA methylation regulators, we use the “survival” package to constructed Cox models to screen out risk molecules and divide patients into the high-risk group or low-risk group. We calculated the risk score using the following formula, where Coefi is the coefficient, and xi is the expression value of each selected molecule. Receiver operating characteristic (ROC) curve was used to test model effectiveness. Multivariate Cox regression was used to analyze the independent prognostic role of the risk model.
Lastly, gene set enrichment analysis (GSEA) is used to find the possible mechanism of RNA methylation affecting the prognosis of lung adenocarcinoma. The whole gene expression data of patients in the various risk groups were uploaded to the GSEA v4.0.3 (www.gsea.com) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) software for analysis, which was performed with 1,000 iterations. The pathways that may be enriched in the different risk groups are scored. Screen the KEGG pathway based on the enrichment score.
In vitro experiment
Real-time polymerase chain reaction (PCR) was validated in vitro after screening RNA methylation regulators associated with clinical pathology by bioinformatic analysis. RNA extraction from lung adenocarcinoma and adjacent tissues was performed using TRIzol reagent (TIANGEN Biotech Company, China). RNA was reverse-transcribed into cDNA with the FastKing RT Kit (TIANGEN Biotech Company, China). Real-time PCR was on TL988 Real-Time PCR Detection System (TIANLONG, China), and the levels were normalized to the level of β - actin. The primers were as follows: HNRNPC: forward: 5'-TCCTCCTCCTATTGCTCGGG-3' and reverse: 5'-GTGTTTCCTGATACACGCTGA-3'. NOP2: forward: 5'-AAGGGTGCCGAGACAGAACT-3' and reverse: 5'-GAGCACGACTAGACAGCCTC-3'. β-actin: forward: 5'-ATAGCACAGCCTGGATAGCAACGTAC-3' and reverse: 5'-CACCTTCTACAATGAGCTGCGTGTG-3'.
Statistical analysis
Statistical analysis of all RNA-seq transcriptome data was conducted using R v3.4.1 (https://www.r-project.org/). Wilcoxon test was used to compare the differences in the expression of RNA methylation regulators between different tumor stage or between tumor tissues and adjacent tissues. OS is defined as the interval from the date of diagnosis to the date of death. The Kaplan-Meier method was used to compare the OS of the patients in distinct groups. A chi-square test was used to analyze the correlation between the separate groups and clinical characteristics. P<0.05 was considered statistically significant.
Results
Expression of different RNA methylation regulators in lung adenocarcinoma
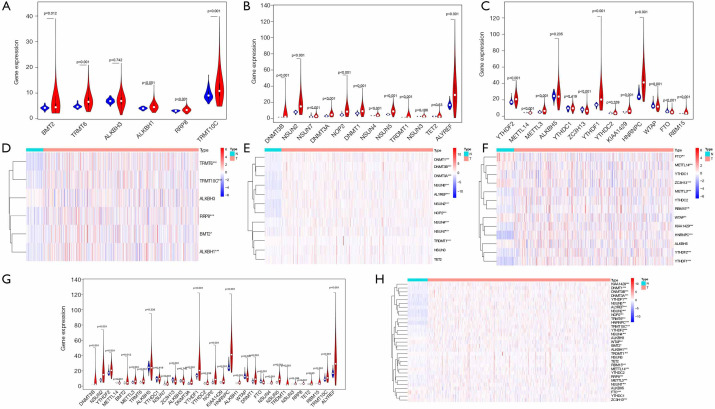
According to the classification of cancer tissues and adjacent tissues, we respectively analyzed the expression levels of the distinct types of RNA methylation regulators. The expressions of five m1A molecules, nine m5A molecules, and six m6A molecules were higher in tumor tissues than that in the adjacent tissues. One m5C molecule and four m6A molecules were lower in cancer tissues than in the adjacent tissues (P<0.05) (Figure 1A,B,C,D,E,F). Overall, the expression of most RNA methylation regulators in lung adenocarcinoma tissues is specific (P<0.05), which are shown in Figure 1G,H. These results suggest that RNA methylation may play an essential role in the malignant progression of lung adenocarcinoma.
Figure 1.
Expression of RNA methylation regulators in TCGA database. (A,D) The expression levels of each m1A RNA methylation regulators. (B,E) The expression levels of each m5C RNA methylation regulators. (C,F) The expression levels of each m6A RNA methylation regulators. (G,H) The expression levels of all RNA methylation regulators. *, P<0.05; **, P<0.01 and ***, P<0.001. TCGA, The Cancer Genome Atlas.
Consensus clustering of RNA methylation regulators identified cluster subgroups of lung adenocarcinoma with different clinical features
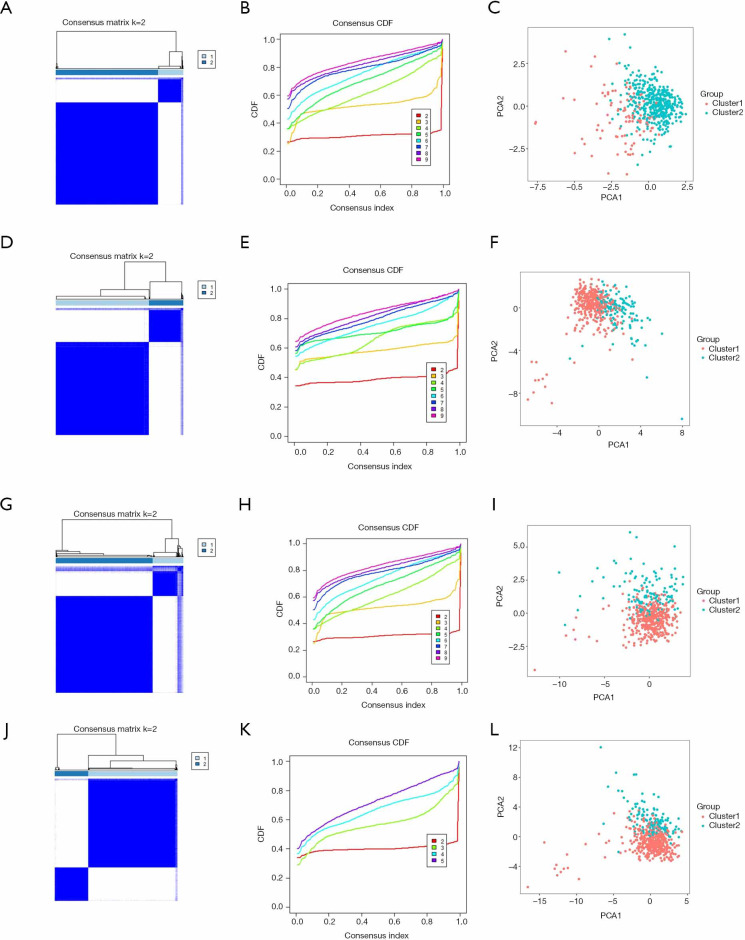
After removed adjacent samples, we grouped cancer samples using the “Consensus Cluster Plus” package. Considering the clustering stability and the number of each group, we divided the patients into two subgroups clustered by k=2. In the cluster models of lung adenocarcinoma, we can see that the distribution of the two subgroups of the m1A model overlaps (Figure 2A,B,C), while the other models show good dispersion by PCA (Figure 2D,E,F,G,H,I,J,K,L). These results show that our classification by RNA methylation regulators are correct.
Figure 2.
Identification of consensus clusters by RNA methylation regulators in lung adenocarcinoma. (A,B,C) Cluster model and PCA based on m1A RNA methylation regulators. (D,E,F) Cluster model and PCA based on m5C RNA methylation regulators. (G,H,I) Cluster model and PCA based on m6A RNA methylation regulators. (J,K,L) Cluster model and PCA based on all RNA methylation regulators. PCA, principal component analysis.
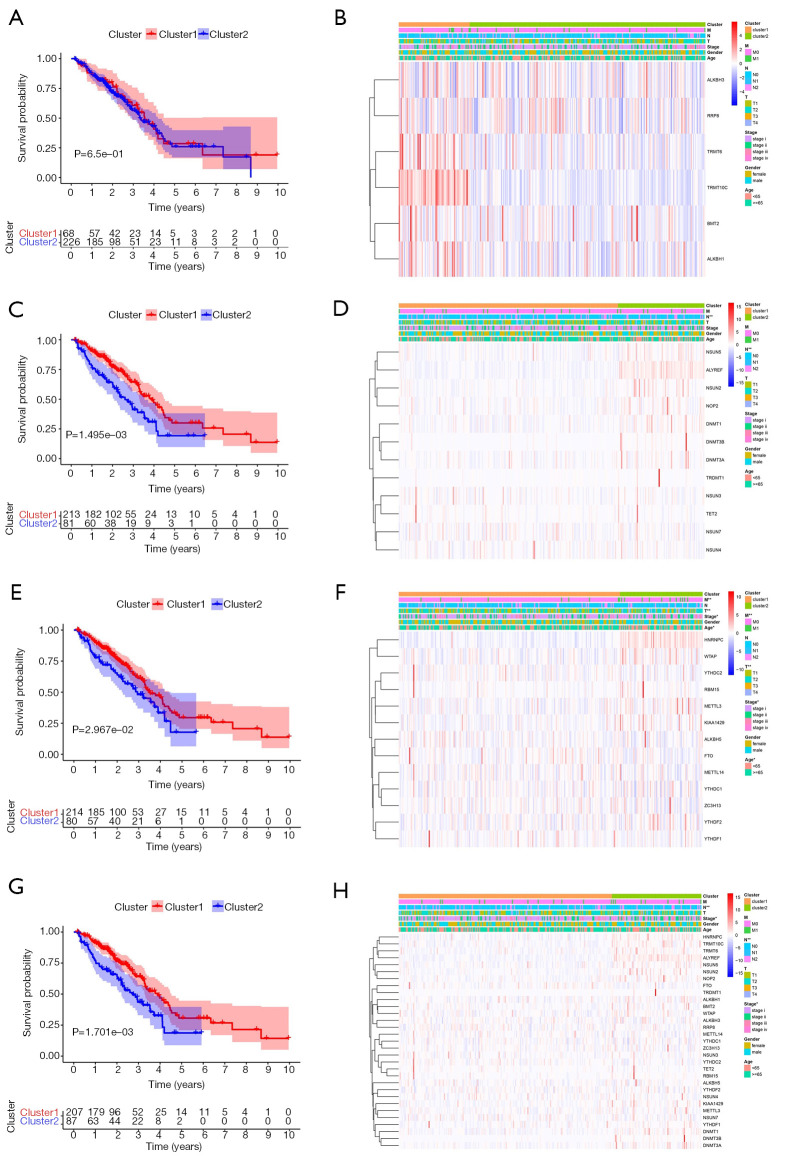
To better understand the correlation between clustering results and clinicopathological features, we performed a survival analysis with the OS. The model constructed by m1A methylation regulators does not show differences in survival or correlation with clinicopathological features (P>0.05) (Figure 3A,B). However, there are significant differences in survival between tumor subgroups of m5C and m6A models, which is also related to the clinical characteristics of stage, T, N, and M classification (P<0.05) (Figure 3C,D,E,F). The analysis results for all methylation regulators model are consistent (Figure 3G,H). Overall, these results suggest are consistent. Further analysis of the models suggests that cluster 2 patients had a lower five-year survival rate and this population is more likely to have a higher stage, lymph node metastases, and distant metastasis, indicating a poor clinical prognosis.
Figure 3.
Correlation of different subgroups with OS and clinical characteristics in TCGA database. (A,B) Survival curve and clinical characteristics of the clustering model based on m1A RNA methylation regulators. (C,D) Survival curve and clinical characteristics of the clustering model based on m5C RNA methylation regulators. (E,F) Survival curve and clinical characteristics of the clustering model based on m6A RNA methylation regulators. (G,H) Survival curve and clinical characteristics of the clustering model based on all RNA methylation regulators. *, P<0.05; **, P<0.01. OS, overall survival; TCGA, The Cancer Genome Atlas.
Prognostic value of RNA methylation regulators, and risk models built using selected RNA methylation regulators
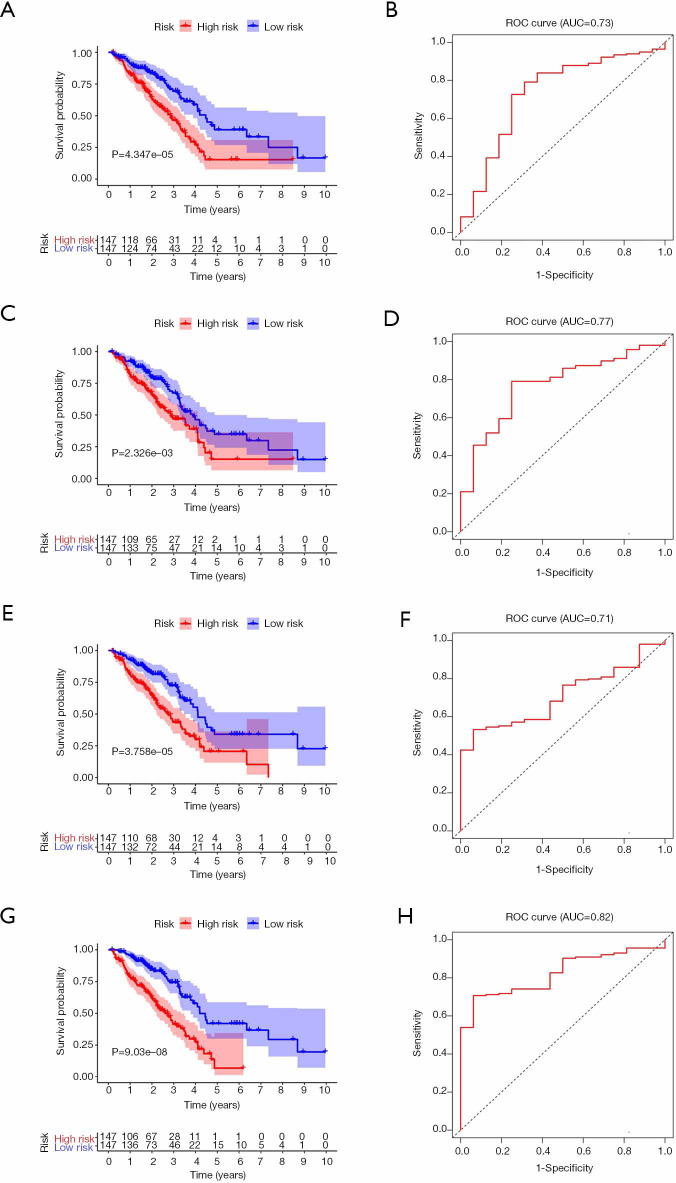
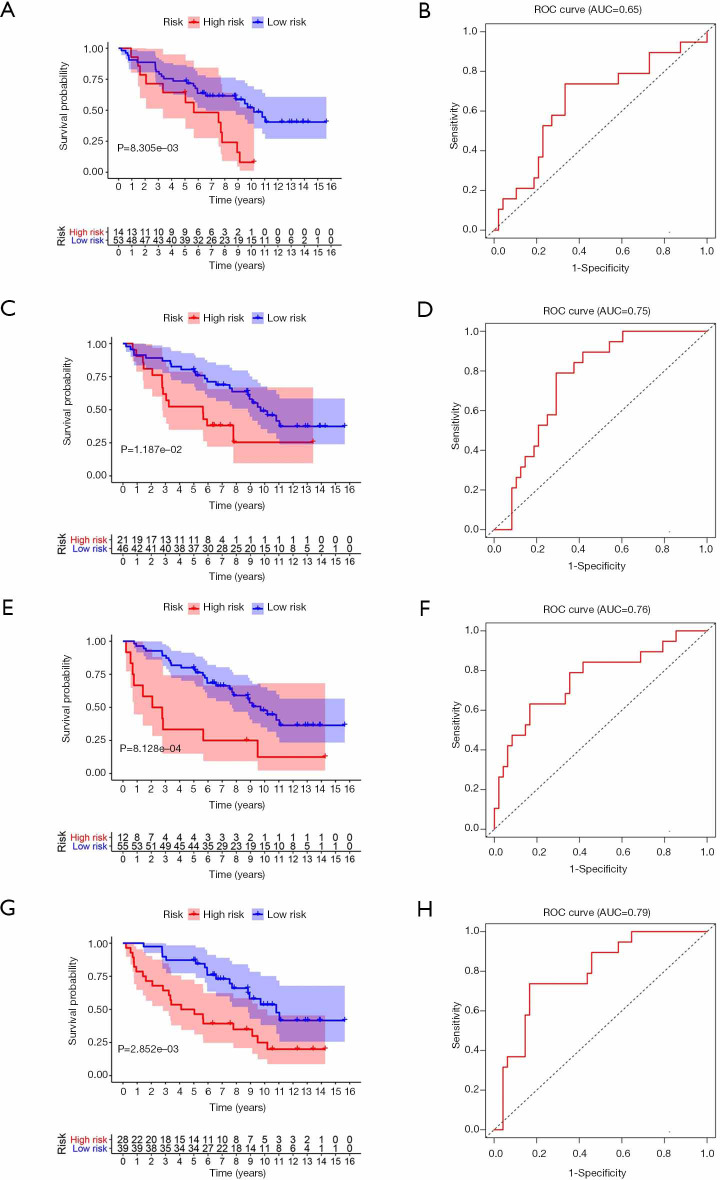
To better understand the prognostic role of RNA methylation regulators in lung adenocarcinoma, we constructed Cox models combining with the expression of RNA methylation regulators and OS from the TCGA database. Based on the risk score, patients were divided into high-risk and low-risk groups. The risk score coefficients of different risk models are shown in Table 4. The survival rate of the high-risk group in m1A prognostic signature is lower than that of the low-risk group (P<0.05), and the prediction effect is high (AUC=0.73) (Figure 4A,B). The prognostic signatures of m5C and m6A also show significant differences in survival (P<0.05), of which m5C signature has a higher predictive effect (AUC: 0.77) and m6A signature has the lowest predictive effect (AUC: 0.71) (Figure 4C,D,E,F). The prognostic signature constructed by all RNA methylation regulators has the highest predictive effect (AUC=0.82) (Figure 4G,H).
Table 4. Risk model constructed by m1A, m5C, m6A, or all methylation regulators.
| Risk model | Gene | Coefficient | HR | HR.95L | HR.95H | P |
|---|---|---|---|---|---|---|
| m1A RNA methylation regulators | BMT2 | −0.062 | 0.940 | 0.866 | 1.021 | 0.141 |
| TRMT6 | 0.052 | 1.053 | 1.003 | 1.106 | 0.037 | |
| RRP8 | −0.316 | 0.729 | 0.563 | 0.945 | 0.017 | |
| m5C RNA methylation regulators | NSUN2 | 0.012 | 1.012 | 0.998 | 1.026 | 0.095 |
| NSUN4 | −0.319 | 0.727 | 0.598 | 0.883 | 0.001 | |
| TET2 | 0.164 | 1.178 | 1.039 | 1.335 | 0.010 | |
| ALYREF | 0.013 | 1.013 | 1.004 | 1.022 | 0.003 | |
| m6A RNA methylation regulators | METTL14 | −0.255 | 0.775 | 0.590 | 1.018 | 0.067 |
| METTL3 | −0.070 | 0.932 | 0.867 | 1.002 | 0.057 | |
| ZC3H13 | 0.115 | 1.122 | 1.050 | 1.198 | 0.001 | |
| YTHDF1 | −0.017 | 0.983 | 0.967 | 1.001 | 0.060 | |
| HNRNPC | 0.018 | 1.018 | 1.005 | 1.032 | 0.006 | |
| RBM15 | 0.144 | 1.155 | 1.040 | 1.283 | 0.007 | |
| All RNA methylation regulators | YTHDF2 | 0.026 | 1.026 | 0.992 | 1.062 | 0.131 |
| BMT2 | −0.086 | 0.918 | 0.840 | 1.002 | 0.055 | |
| YTHDC1 | −0.071 | 0.931 | 0.846 | 1.024 | 0.141 | |
| ZC3H13 | 0.093 | 1.098 | 1.018 | 1.184 | 0.016 | |
| YTHDF1 | −0.028 | 0.973 | 0.953 | 0.993 | 0.009 | |
| NOP2 | 0.034 | 1.035 | 1.006 | 1.065 | 0.020 | |
| HNRNPC | 0.023 | 1.024 | 1.008 | 1.040 | 0.003 | |
| NSUN4 | −0.413 | 0.662 | 0.540 | 0.812 | <0.001 | |
| RBM15 | 0.219 | 1.245 | 1.101 | 1.407 | <0.001 | |
| TRMT10C | −0.039 | 0.962 | 0.921 | 1.004 | 0.074 | |
| ALYREF | 0.012 | 1.012 | 1.003 | 1.022 | 0.012 |
Figure 4.
Survival and ROC curves for different risk models from TCGA database. (A,B) The risk model constructed by m1A methylation regulators. (C,D) The risk model constructed by m5C methylation regulators. (E,F) The risk model constructed by m6A methylation regulators. (G,H) The risk model constructed by all methylation regulators. ROC, receiver operating characteristic; TCGA, The Cancer Genome Atlas.
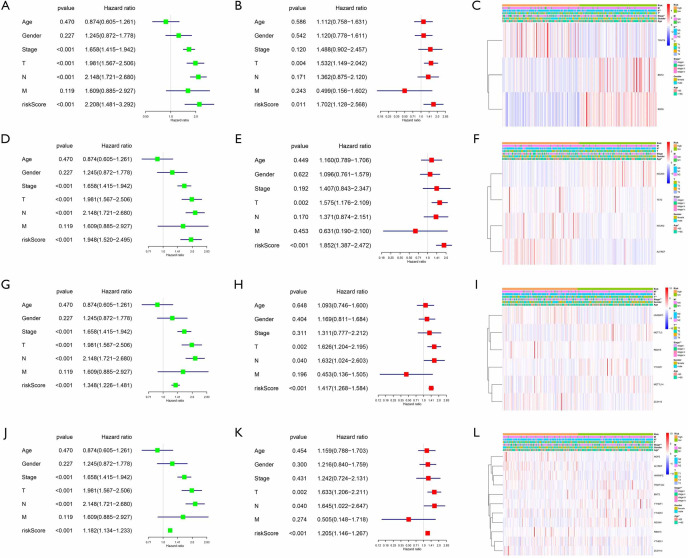
We further used univariate and multivariate Cox regression to explore the predictive value of the signatures, as well as the associations between the clinicopathological characteristics. The analysis results of all prognostic signatures are consistent. Risk score in those signatures can be independent risk factors affecting the prognosis of lung adenocarcinoma (HR>1, P<0.05), and the high-risk patients are more likely to have a higher stage, more lymph node metastases, and distant metastases, indicating a poor clinical prognosis (Figure 5).
Figure 5.
Clinical characteristics of different risk groups from TCGA database. (A,B,C) The risk model constructed by m1A methylation regulators. (D,E,F) The risk model constructed by m5C methylation regulators. (G,H,I) The risk model constructed by m6A methylation regulators. (J,K,L) The risk model constructed by all methylation regulators. *, P<0.05; **, P<0.01. TCGA, The Cancer Genome Atlas.
Verify the reliability of the risk model constructed by the TCGA database through the GEO database
To test the reliability of the risk models based on TCGA data, we use the GSE37745 dataset from the GEO for validation. By calculating the patient’s risk score, all the patients are divided into a high-risk group or low-risk group. The survival rate of the high-risk group in m1A prognostic signature is lower than that of the low-risk group (P<0.05), which is consistent with the training set, but the prediction effect is general (AUC =0.65) (Figure 6A,B). The prognostic signatures of m5C and m6A also show significant differences in survival (P<0.05), with higher predictive effects (AUC: 0.75–0.76) (Figure 6C,D,E,F). The prognostic signature constructed by all RNA methylation regulators has the highest predictive effect (AUC =0.79) (Figure 6G,H).
Figure 6.
Survival and ROC curves for different risk models in the test set. (A,B) Verification of the risk model constructed by m1A methylation regulators. (C,D) Verification of the risk model constructed by m5C methylation regulators. (E,F) Verification of the risk model constructed by m6A methylation regulators. (G,H) Verification of the risk model constructed by all methylation regulators. ROC, receiver operating characteristic.
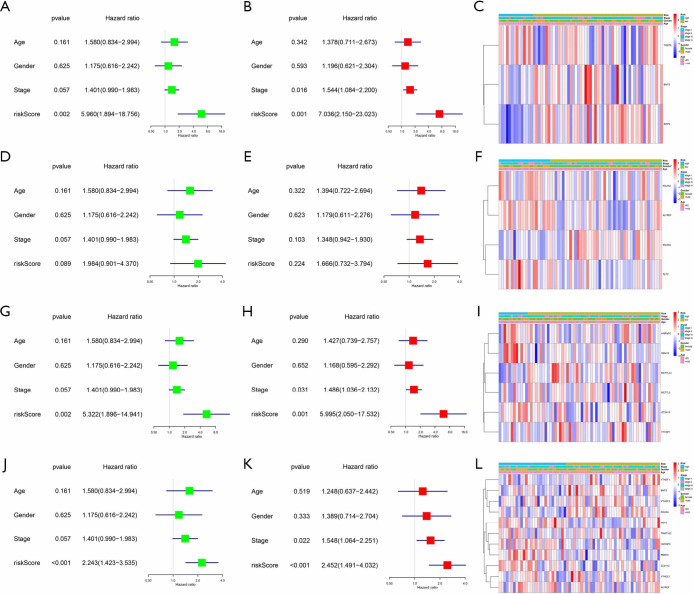
The test set only followed the patient’s gender, age, and stages, and we also performed univariate and multivariate Cox regression analysis. These results are a bit different. The risk score of the m1A signature can be a risk factor that affects the prognosis of lung adenocarcinoma (HR >1, P<0.05), but it has no correlation with clinical features (Figure 7A,B,C). The analysis of the m5C signature is not statistically significant (P>0.05) (Figure 7D,E,F). The risk score of other signatures also can be independent risk factors for the prognosis of lung adenocarcinoma (HR >1, P<0.05) (Figure 7G,H,I,J,K,L). Generally, the test set successfully verifies the results of the training set, which suggests that RNA methylation is a valid marker for predicting patients with lung adenocarcinoma.
Figure 7.
Clinical characteristics of different risk groups f in the test set. (A,B,C) Verification of the risk model constructed by m1A methylation regulators. (D,E,F) Verification of the risk model constructed by m5C methylation regulators. (G,H,I) Verification of the risk model constructed by m6A methylation regulators. (J,K,L) Verification of the risk model constructed by all methylation regulators. *, P<0.05.
Gene set enrichment analysis
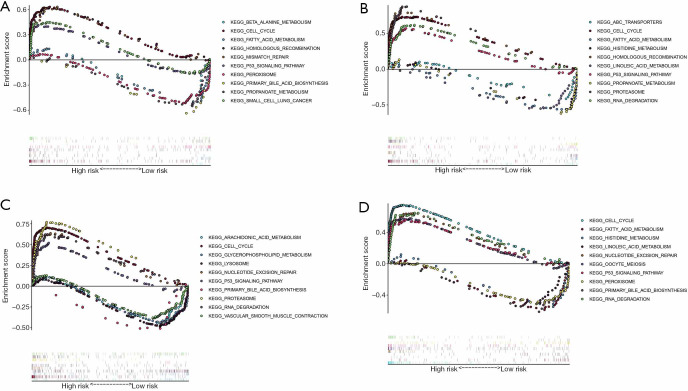
We also performed KEGG analysis of gene expression in patients in the different risk groups. The results of different risk models were consistent. Overall, cell cycle, RNA degradation, P53 signaling pathway, homologous recombination, and mismatch repair were significantly enriched in the high-risk group. Fatty acid metabolism, histidine metabolism, and primary bile acid biosynthesis were significantly enriched in the low-risk group (Figure 8). These results supply a reference for further functions and mechanisms of RNA methylation in lung adenocarcinoma.
Figure 8.
KEGG analysis of different risk groups in lung adenocarcinoma. (A) The risk model constructed by m1A methylation regulators. (B) The risk model constructed by m5C methylation regulators. (C) The risk model constructed by m6A methylation regulators. (D) The risk model constructed by all methylation regulators. KEGG, Kyoto Encyclopedia of Genes and Genome.
Validation of RNA methylation regulators expression and clinical pathological correlations in lung adenocarcinoma by real-time PCR
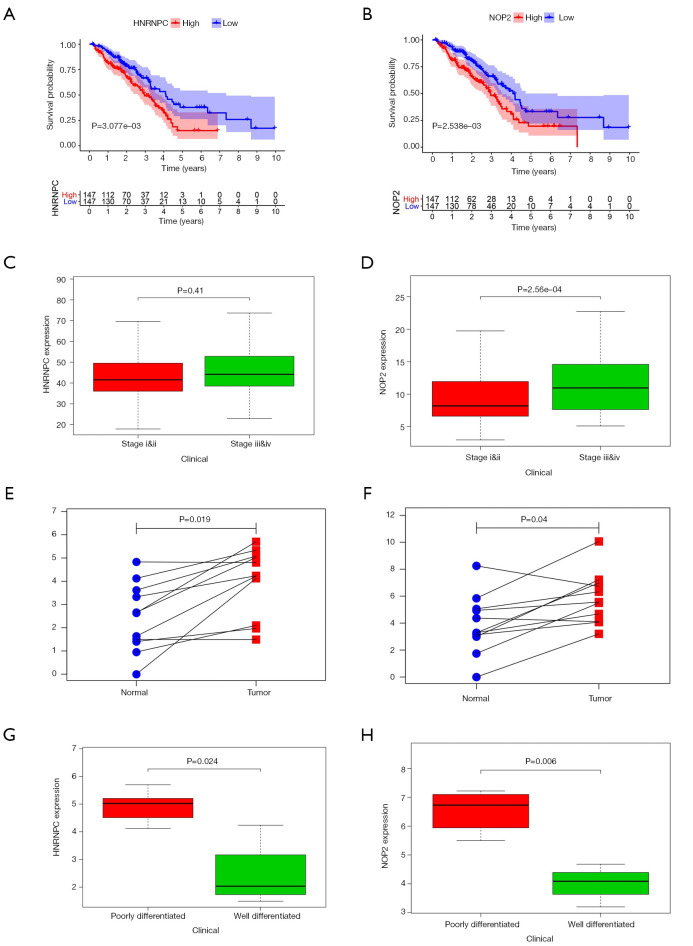
Survival analysis of each molecule identified nine RNA methylation regulators (RRP8, HNRNPC, NOP2, NSUN4, DNMT3B, RBM15, TRDMT1, DNMT1, YTHDF2) that were associated with patient survival (P<0.05) (Figure 9A,B). Among these RNA methylation regulators, patients with high expression of HNRNPC or NOP2 were more likely to be in advanced tumor stage (P<0.05) (Figure 9C,D). By performing real-time PCR on 11 lung adenocarcinoma patient tissues, we found that the expression of HNRNPC and NOP2 was higher in tumor tissues than that in the adjacent tissues, which is consistent with the results of bioinformatic analysis (P<0.05) (Figure 9E,F). Furthermore, patients with high expression of HNRNPC or NOP2 were more likely to have a poorly differentiated (P<0.05) (Figure 9G,H), indicating a poor clinical prognosis. These results suggest the impact of RNA methylation regulators in the clinical pathology of different tumors.
Figure 9.
Validation of RNA methylation regulators expression and clinical pathological correlations in lung adenocarcinoma by real-time PCR. (A,B) Correlation of HNRNPC and NOP2 expression with patient survival from TCGA. (C,D) Correlation of HNRNPC and NOP2 expression with tumor staging of the patient from TCGA. (E,F) Expression of HNRNPC and NOP2 in lung adenocarcinoma and adjacent tissues by real-time PCR. (G,H) Correlation of HNRNPC and NOP2 expression with tumor differentiation of the patient from by real-time PCR. PCR, polymerase chain reaction.
Discussion
Over the past decade, people recognized that the occurrence and development of lung cancer is a complex process involving many factors, and different molecular expressions have variable drug responses and clinical prognosis to patients. Traditional tumor staging only depends on tumor volume and metastasis, which cannot take into account the patient’s gene expression level (23). The mutation rate of functional driver genes in lung adenocarcinoma is about 60%, of which KRAS, EGFR mutation, and EML4-ALK fusion is the most common driver gene, accounting for about 35% to 40% (24). Kris’s study found that the median survival time of patients with such type of driver gene mutation receiving targeted therapy was prolonged by 2–4 years compared with other patients (25). The study of genetic changes in lung squamous cell carcinoma was later than lung adenocarcinoma. TP53, KRAS, GRM are the common gene mutations (26). Weiss’s research found that about 20% of lung squamous cell carcinomas have FGFR1 amplification. Inhibition of FGFR1 in cell lines and mouse models can inhibit cell growth and induce apoptosis (27). These studies suggest that the new molecular typing research is expected to predict the clinical prognosis of patients more from the molecular level.
RNA modification is a way of regulating post-transcriptional levels. For example, m6A modification refers to a modification in which one hydrogen atom (-H) attached to the sixth nitrogen atom (N6) on the adenine molecule is substituted with a methyl group (-CH4) (28). The m6A methyltransferase promotes mRNA methylation of adenine, and demethylase can eliminate it, and the binding proteins play a recognition role in this process (28,29). m5C and m1A RNA methylation also regulate gene expression by acting at different sites. RNA methylation is a dynamic and reversible modification method, and the entire process is regulated by methyltransferases, demethylase, and binding proteins. FTO is the first identified m6A demethylase, and its m6A-mediated modification can be a novel cis-element to regulate mRNA splicing and adipose precursor cell differentiation (18). Also, it was found that FTO can demethylate m1A, too (30). Yang discovered the distribution of m5C methylation, identified the main methyltransferase NSUN2 and the first binding protein ALYREF (17). David’s study found that Arabidopsis m5C modification regulates tissue development, and the methyltransferase is TRM4B (31). There is less research on m1A methylation. Li has developed a single-base-resolution m1A sequencing method and found that nuclear and mitochondrial coding transcripts have different types of m1A methylation (32).
RNA methylation also regulates tumorigenesis and development, drug response, and stem cell renewal. Zhou’s study found that FTO enhances the chemo-radiotherapy resistance both in vitro and in vivo through regulating the expression of β-catenin by reducing m6A levels in its mRNA transcripts (33). Cui’s research suggests that m6A mRNA modification is critical for glioblastoma stem cell self-renewal and tumorigenesis. Knockdown of METTL3 or METTL14, key components of the RNA methyltransferase complex, promotes human glioblastoma stem cell growth, self-renewal, and tumorigenesis (34). Chen’s research found that m5C RNA methylation promotes the pathogenesis of bladder cancer through stabilizing mRNAs, and high expression of NUSN2 predicts poor survival (35). In the study of TCGA, literature reported that m6A RNA methylation regulators contribute to malignant progression and have a clinical prognostic impact (15,36). These studies have suggested that RNA methylation not only plays a role in tumor progression but can also be a signature to predict clinical prognosis, but there are few studies on the effect of RNA methylation on lung cancer. We obtained a large number of patient cases and clinicopathological features by downloading lung adenocarcinoma data from the TCG database. We likewise mined the GEO database, but with a lower number of cases, which lends itself to doing studies that validate the TCGA results. The analysis and validation of these data provide the basis for a prognostic study of RNA methylation in lung adenocarcinoma.
Through the study of RNA methylation, we summarized thirty-one RNA methylation regulators. Further analysis showed that the expression of these regulators in tumor tissues differed from that in adjacent tissues, which suggested that RNA methylation plays a vital role in the development of lung adenocarcinoma. To explore the clinical value of RNA methylation, we constructed several subgroups and risk models, PCA and ROC curves show the excellent accuracy of the models. In different models, we found that the expression of RNA methylation regulators is related to the survival of patients, and different molecular phenotypes can be independent risk factors for the prognosis of lung adenocarcinoma. Real-time PCR validated the results of the bioinformatic analysis. Our study explains the critical role of RNA methylation in lung cancer, and it is expected to supply a reference for the prognostic stratification and treatment strategy development of lung adenocarcinoma.
Supplementary
The article’s supplementary files as
Acknowledgments
We thank our anonymous reviewers for their valuable comments on this manuscript, which have led to many improvements to the article.
Funding: This study was supported by the Program Funded by Liaoning Province Education Administration (grant No. QN2019003).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics board of The First Affiliated Hospital of China Medical University (No. YB M-05-02) and informed consent was taken from all the patients.
Footnotes
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-3744
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-3744). The authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 2.Kramer H, Groen HJ. Current concepts in the mediastinal lymph node staging of nonsmall cell lung cancer. Ann Surg 2003;238:180-8. 10.1097/01.SLA.0000081086.37779.1a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Schil PE, Rami-Porta R, Asamura H. The 8th TNM edition for lung cancer: a critical analysis. Ann Transl Med 2018;6:87. 10.21037/atm.2017.06.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu C, Li Y, Wei M, et al. Identification of a novel glycolysis-related gene signature that can predict the survival of patients with lung adenocarcinoma. Cell Cycle 2019;18:568-79. 10.1080/15384101.2019.1578146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Velez MA, Burns TF. Is the game over for PD-1 inhibitors in EGFR mutant non-small cell lung cancer? Transl Lung Cancer Res 2019;8:S339-42. 10.21037/tlcr.2019.04.09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roundtree IA, Evans ME, Pan T, et al. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017;169:1187-200. 10.1016/j.cell.2017.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taoka M, Nobe Y, Hori M, et al. A mass spectrometry-based method for comprehensive quantitative determination of post-transcriptional RNA modifications: the complete chemical structure of Schizosaccharomyces pombe ribosomal RNAs. Nucl Acids Res 2015;43:e115. 10.1093/nar/gkv560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agris PF, Narendran A, Sarachan K, et al. The Importance of Being Modified: The Role of RNA Modifications in Translational Fidelity. Enzymes 2017;41:1-50. 10.1016/bs.enz.2017.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keffer-Wilkes LC, Veerareddygari GR, Kothe U. RNA modification enzyme TruB is a tRNA chaperone. Proc Nat Aca Sci U S A 2016;113:14306-11. 10.1073/pnas.1607512113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hastings MH. m(6)A mRNA methylation: a new circadian pacesetter. Cell 2013;155:740-1. 10.1016/j.cell.2013.10.028 [DOI] [PubMed] [Google Scholar]
- 11.Ren W, Guo J, Jiang F, et al. CCAAT/enhancer-binding protein α is a crucial regulator of human fat mass and obesity associated gene transcription and expression. BioMed Res Int 2014;2014:406909. [DOI] [PMC free article] [PubMed]
- 12.Safra M, Sas-Chen A, Nir R, et al. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 2017;551:251-5. 10.1038/nature24456 [DOI] [PubMed] [Google Scholar]
- 13.Squires JE, Patel HR, Nousch M, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucl Acids Res 2012;40:5023-33. 10.1093/nar/gks144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, Hsu PJ, Chen YS, et al. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res 2018;28:616-24. 10.1038/s41422-018-0040-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chai RC, Wu F, Wang QX, et al. m6A RNA methylation regulators contribute to malignant progression and have clinical prognostic impact in gliomas. Aging 2019;11:1204-25. 10.18632/aging.101829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu F, Clark W, Luo G, et al. ALKBH1-Mediated tRNA Demethylation Regulates Translation. Cell 2016;167:1897. 10.1016/j.cell.2016.11.045 [DOI] [PubMed] [Google Scholar]
- 17.Yang X, Yang Y, Sun BF, et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res 2017;27:606-25. 10.1038/cr.2017.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X, Yang Y, Sun BF, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res 2014;24:1403-19. 10.1038/cr.2014.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixit D, Xie Q, Rich JN, et al. Messenger RNA Methylation Regulates Glioblastoma Tumorigenesis. Cancer Cell 2017;31:474-5. 10.1016/j.ccell.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, Li F, Peng Y, et al. Identification of three m6A-related mRNAs signature and risk score for the prognostication of hepatocellular carcinoma. Cancer Med 2020;9:1877-89. 10.1002/cam4.2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Han Y, Zhang H, et al. The m6A demethylase FTO promotes the growth of lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem Biophys Res Commun 2019;512:479-85. 10.1016/j.bbrc.2019.03.093 [DOI] [PubMed] [Google Scholar]
- 22.Meng Q, Wang S, Zhou S, et al. Dissecting the m6A methylation affection on afatinib resistance in non-small cell lung cancer. Pharmacogenomics J 2020;20:227-34. 10.1038/s41397-019-0110-4 [DOI] [PubMed] [Google Scholar]
- 23.Tian Z, Wen S, Zhang Y, et al. Identification of dysregulated long non-coding RNAs/microRNAs/mRNAs in TNM I stage lung adenocarcinoma. Oncotarget 2017;8:51703-18. 10.18632/oncotarget.18512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pikor LA, Ramnarine VR, Lam S, et al. Genetic alterations defining NSCLC subtypes and their therapeutic implications. Lung Cancer 2013;82:179-89. 10.1016/j.lungcan.2013.07.025 [DOI] [PubMed] [Google Scholar]
- 25.Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014;311:1998-2006. 10.1001/jama.2014.3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kan Z, Jaiswal BS, Stinson J, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 2010;466:869-73. 10.1038/nature09208 [DOI] [PubMed] [Google Scholar]
- 27.Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med 2010;2:62ra93. 10.1126/scitranslmed.3001451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu Y, Dominissini D, Rechavi G, et al. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet 2014;15:293-306. 10.1038/nrg3724 [DOI] [PubMed] [Google Scholar]
- 29.Wang CX, Cui GS, Liu X, et al. METTL3-mediated m6A modification is required for cerebellar development. PLoS Biol 2018;16:e2004880. 10.1371/journal.pbio.2004880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei J, Liu F, Lu Z, et al. Differential m6A, m6Am, and m1A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol Cell 2018;71:973-85.e5. 10.1016/j.molcel.2018.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.David R, Burgess A, Parker B, et al. Transcriptome-Wide Mapping of RNA 5-Methylcytosine in Arabidopsis mRNAs and Noncoding RNAs. Plant cell 2017;29:445-60. 10.1105/tpc.16.00751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Xiong X, Zhang M, et al. Base-Resolution Mapping Reveals Distinct m1A Methylome in Nuclear- and Mitochondrial-Encoded Transcripts. Mol Cell 2017;68:993-1005.e9. 10.1016/j.molcel.2017.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou S, Bai ZL, Xia D, et al. FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting β-catenin through mRNA demethylation. Mol Carcinog 2018;57:590-7. 10.1002/mc.22782 [DOI] [PubMed] [Google Scholar]
- 34.Cui Q, Shi H, Ye P, et al. m6A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep 2017;18:2622-34. 10.1016/j.celrep.2017.02.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen X, Li A, Sun BF, et al. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell Biol 2019;21:978-90. 10.1038/s41556-019-0361-y [DOI] [PubMed] [Google Scholar]
- 36.Su Y, Huang J, Hu J. m6A RNA Methylation Regulators Contribute to Malignant Progression and Have Clinical Prognostic Impact in Gastric Cancer. Front Oncol 2019;9:1038. 10.3389/fonc.2019.01038 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as