Abstract
The adrenal gland has traditionally been viewed as a source of “weak androgens”; however, emerging evidence indicates 11-oxy-androgens of adrenal origin are metabolized in peripheral tissues to potent androgens. Also emerging is the role of gut bacteria in the conversion of C21 glucocorticoids to 11-oxygenated C19 androgens. Clostridium scindens ATCC 35704 is a gut microbe capable of converting cortisol into 11-oxy-androgens by cleaving the side-chain. The desA and desB genes encode steroid-17,20-desmolase. Our prior study indicated that the urinary tract bacterium, Propionimicrobium lymphophilum ACS-093-V-SCH5 encodes desAB and converts cortisol to 11β-hydroxyandrostenedione. We wanted to determine how widespread this function occurs in the human microbiome. Phylogenetic and sequence similarity network analyses indicated that the steroid-17,20-desmolase pathway is taxonomically rare and located in gut and urogenital microbiomes. Two microbes from each of these niches, C. scindens and Propionimicrobium lymphophilum, respectively, were screened for activity against endogenous (cortisol, cortisone, and allotetrahydrocortisol) and exogenous (prednisone, prednisolone, dexamethasone, and 9-fluorocortisol) glucocorticoids. LC/MS analysis showed that both microbes were able to side-chain cleave all glucocorticoids, forming 11-oxy-androgens. Pure recombinant DesAB from C. scindens showed the highest activity against prednisone, a commonly prescribed glucocorticoid. In addition, 0.1 nM 1,4-androstadiene-3,11,17-trione, bacterial side-chain cleavage product of prednisone, showed significant proliferation relative to vehicle in androgen-dependent growth LNCaP prostate cancer cells after 24 hours (2.3 fold; P < 0.01) and 72 hours (1.6 fold; P < 0.01). Taken together, DesAB-expressing microbes may be an overlooked source of androgens in the body, potentially contributing to various disease states, such as prostate cancer.
Keywords: steroid desmolase, prostate cancer, microbiome drug metabolism, prednisone
Introduction
The adrenal gland has traditionally been viewed as a source of “weak androgens”; however, emerging evidence indicates that several 11-oxygenated C19 androgens of adrenal origin are clinically important androgens (1). The adrenal synthesis of 11β-hydroxyandrostenedione (11β-OHAD) has been known since the 1970’s, but its contribution to the human androgen pool was largely rejected (1). Recent evidence has renewed the interest in 11-oxygenated C19 androgens as potentially major contributors to diseases such as castration resistant prostate cancer (CRPC), congenital adrenal hyperplasia (CAH), and polcystic ovary syndrome (PCOS) (1–4). Two routes have been proposed for the formation of 11β-OHAD. The first is 11β-hydroxylation via cytochrome P450 11β-hydroxylase (P450 11B1) in the adrenal, and the second is through side-chain cleavage of cortisol by a combination of the gut microbiota and a yet to be identified host enzyme (5, 6). Indeed, our recent work (7–10) confirmed and extended previous studies (11–17) into a novel bacterial enzyme, steroid-17,20-desmolase which side-chain cleaves cortisol and may significantly contribute to host 11-oxygenated C19 androgen formation.
Early clinical studies by Nabarro (18) and Wade (19) provided compelling evidence that gut bacteria are capable of converting hydrocortisone to 11-keto-androgens. After several decades, it was shown that incubation of radiolabelled cortisol with human and rat fecal slurries resulted in the formation of numerous metabolites, including 11-keto-androgen side-chain cleavage products (11, 12). Subsequently, a fecal isolate designated “Clostridium strain 19” was shown to convert cortisol to 11β-hydroxyandrostenedione (13) and was named Clostridium scindens ATCC 35704 (14). Studies by Krafft and Hylemon (15, 16) determined that the steroid-17,20-desmolase enzyme was induced by addition of cortisol to the growth medium. Their studies determined co-factor requirements and purified an accessory NADH-dependent cortisol 20α-hydroxysteroid dehydrogenase (20α-HSDH). Bokkenheuser et al. (17) identified steroid-17,20-desmolase activity by Clostridium cadavaris and Butyricicoccus desmolans (formerly Eubacterium desmolans) with accessory 20β-HSDH activity. Ridlon et al (2013) identified a cortisol-inducible operon (desABCD) in C. scindens ATCC 35704 and characterized the desC gene product, which encodes NADH-dependent cortisol 20α-HSDH (7). Additionally, we identified and characterized the gene encoding NADH-dependent 20β-HSDH (desE) from B. desmolans (9) and performed detailed biochemical characterization of recombinant 20β-HSDH from Bifidobacterium adolescentis and solved the apo- and holoenzyme crystal structures (10).
Recently, we developed an enzyme-coupled continuous spectrophotometric assay to characterize recombinant desAB gene product, which encodes steroid-17,20-desmolase (8). In that study, we determined substrate-specificity with endogenous derivatives of cortisol, including tetrahydrocortisol (5β-reduced), which was not found to be a substrate (8). Subsequent to that study, we identified allotetrahydrocortisol (5α-reduced epimer) as a substrate for side-chain cleavage by C. scindens ATCC 35704 and began testing synthetic glucocorticoid drugs with whole cells of C. scindens and recombinant DesAB (20). Recently, it was shown in a large drug screen that C. scindens is capable of metabolizing synthetic glucocorticoids in vitro and in vivo (21). Here, we report metabolism of endogenous cortisone and allotetrahydrocortisol, and glucocorticoid drugs and demonstrate that the DesAB is responsible for this drug metabolism. We also performed evolutionary and network-based functional analysis of the steroid transketolase (DesAB) in order to determine the diversity of this enzyme in the human microbiome.
Materials and Methods
Bacterial strains, enzymes, reagents, and materials
Clostridium scindens ATCC 35704 was purchased from the American Type Culture Collection (ATCC, Manassas, VA). Propionimicrobium lymphophilum ACS-093-V-SCH5 was obtained from the Culture Collection, University of Götesborg, Sweden. Turbo Competent Escherichia coli DH5α (High Efficiency) cells were purchased from New England Biolabs (NEB, Ipswich, MA, USA). BL21-CodonPlus (DE3)-RIPL Competent E. coli cells were purchased from Agilent Technologies (Santa Clara, CA). Phusion High-Fidelity DNA Polymerase, Restriction Endonucleases, and T4 DNA Ligase were purchased from NEB. Isopropyl β-D-1-thiogalactopyranoside (IPTG) was purchased from Gold Biotechnology. TALON His-Tag Purification Resin was purchased from Takara Bio USA, Inc. Strep-Tactin® resin was purchased from IBA GmbH (Goettingen, Germany). Steroids were purchased from Steraloids (Newport, RI) and Sigma-Aldrich (St. Louis, MO). All other materials were purchased from Fisher Scientific and MilliporeSigma. LNCaP cells were obtained from American Type Culture Collection (ATCC).
Cloning of desA and desB from C. scindens and 17β-HSDH from C. lunatus
The desA and desB genes were cloned into pETDuet-1 and 17β-HSDH gene from C. lunatus was cloned into pET-51b(+) previously (14). Recombinant plasmids were transformed into chemically competent E. coli DH5a cells via the heat-shock method, plated, and grown for 16 h at 37°C on lysogeny broth (LB) agar plates supplemented with ampicillin (100 μg/ml). A single colony from each transformation was inoculated into LB medium (5 ml) containing ampicillin (100 μg/ml) and grown to saturation. The cells were subsequently centrifuged (3,220 g, 15 min, 4°C) and plasmids were extracted from the resulting cell pellet using the QIAprep Spin Miniprep kit (Qiagen, Valencia, CA). The sequences of the inserts were determined via Sanger sequencing (W. M. Keck Center for Comparative and Functional Genomics at the University of Illinois at Urbana-Champaign). Only plasmids with correct inserts were used for gene expression and purification.
Gene expression and purification
For protein expression, chemically competent E. coli BL-21 CodonPlus (DE3) RIPL cells were transformed as described above and plated LB agar supplemented with ampicillin (100 μg/ml) and chloramphenicol (50 μg/ml). After 16 h, five isolated colonies were used to inoculate 10 ml of fresh LB medium supplemented with antibiotics and grown at 37°C for 6 h with vigorous aeration. The pre-cultures were then added to fresh LB medium (1 L) supplemented with the same antibiotics and grown with vigorous aeration at 37°C. Once an OD600nm of 0.3 was reached, IPTG was added to each culture at a final concentration of 0.1 mM and the temperature was decreased to 16°C for 16 hours. Cells were pelleted by centrifugation (4,000 g, 30 min, 4°C) and resuspended in 30 ml of binding buffer (20 mM Tris-HCl and 150 mM NaCl at pH 7.9). The cell suspension was subjected to four passages through an EmulsiFlex C-3 cell homogenizer (Avestin, Ottawa, Canada), and the cell lysate was clarified by centrifugation at 20,000 g for 30 min at 4°C.
The recombinant DesAB (rDesAB) was then purified using TALON® Metal Affinity Resin (Clontech Laboratories, Mountain View, CA) per the manufacturer’s protocol. The recombinant protein was eluted using an elution buffer composed of 20 mM Tris-HCl, 150 mM NaCl, and 250 mM imidazole at pH 7.9. The recombinant 17β-HSDH was then purified using Strep-Tactin® resin (IBA GmbH, Goettingen, Germany) per manufacturer’s protocol. The recombinant 17β-HSDH was eluted using an elution buffer composed of 20 mM Tris-HCl, 150 mM NaCl, and 2.5 mM desthiobiotin at pH 7.0. The purity of the proteins was assessed by SDS-PAGE, and the protein bands were visualized by staining with Coomassie Brilliant Blue G-250 Dye. The protein concentrations were calculated based on the computed molecular mass and extinction coefficient. The observed subunit mass for each protein was calculated by migration distance of purified protein to standard proteins using ImageJ (https://imagej.nih.gov/ij/docs/faqs.html).
Continuous enzyme-coupled spectrophotometric assay
Steroid-17,20-desmolase activity was measured aerobically at 25°C by monitoring the conversion of NADPH to NADP+ 340 nM (e=6,220 M−1·cm−1) reflecting DesAB-catalyzed conversion of the glucocorticoid substrate to the side-chain cleaved product followed by NADPH-dependent 17β-HSDH conversion of the C-17 carbonyl to a β-hydroxyl group. Control reactions were run leaving out one component at a time. The standard reaction mixture contained 50 mM MOPS buffer (pH 7), 1 μM MnCl2, 10 μM TPP, 200 μM NADPH, 2.5 μM DesAB, 1.0 μM 17β-HSDH, and buffer to a final volume of 0.5 ml (8). The reaction was initiated by the addition of DesAB. The initial velocity of enzyme catalyzed reactions was plotted against the substrate concentrations, and the kinetic parameters were estimated by fitting the data to the Michaelis-Menten equation by nonlinear regression method using the enzyme kinetics module in GraphPad Prism (GraphPad Software, La Jolla, CA).
Whole-cell bacterial steroid conversion assay
For whole cell metabolism analysis, C. scindens ATCC 35704 and P. lymphophilum ACS-093-VSCH5 were grown for 48 hours in anaerobic blood heart infusion (BHI) and peptone yeast glucose (PYG) supplemented with 0.1% Tween 80, respectively, at 37°C until a dense optical density was reached. For each steroid analyzed, a 20% inoculum was added to 2 mL of BHI supplemented with 100 μM of the corresponding glucocorticoid. After incubation at 37°C for 48 hours, the medium was extracted with 2 volumes of ethyl acetate and recovering the organic phase. This extract was combined and evaporated under a stream of nitrogen gas and the residue was dissolved in 200 μL methanol. Total ion count (TIC) and single ion monitoring (SIM) mode were utilized to qualitatively assess if conversion occurred by detecting the expected mass of side-chain cleavage products (m/z = 60.02 less than the substrate).
We have previously validated that the molecular structure of the side-chain cleavage product of cortisol produced by C. scindens is 11β-OHAD using high-resolution mass spectrometry, 1H and 13C NMR spectroscopy, and X-ray crystallography (7). We have also shown that purified rDesAB from C. scindens forms 11β-OHAD, which were consistent with the standards, producing a product peak that was 60.02 amu less than the substrate, cortisol (8). For those reaction DesAB reaction products that are not commercially available, we determined that side-chain cleavage occurred if a LC/MS peak was 60.02 amu less than the substrate with analogous substrate/product separation pattern as observed with cortisol and 11β-OHAD. Reaction substrate peaks, and product peaks when possible, were validated with commercial standards by LC/MS.
DesAB substrate specificity
Recombinant steroid-17,20-desmolase (DesAB) substrate specificity was determined by incubating 2 μM of pure rDesAB enzyme with 100 μM of substrate in the presence of 1 μM MnCl2 and 10 μM TPP in 50 mM MOPS buffer (pH 7.0) at room temperature. After 2 minutes, while the reactions were still in the linear range, the reaction was stopped with the addition of 1/10 volume of 1 M HCl and vortexing to drop the pH below 2. A final concentration of 50 μM of internal standard, 20β-dihydrocortisol, was spiked into the reaction. The steroids were extracted twice by vortexing the reaction with ethyl acetate and prepared for LC/MS as described above. The residue was further diluted to bring the substrate concentration within the linear detection range of the LC/MS for accurate quantification. Standard curves were generated for each substrate and the internal standard in order to determine the linear range of the MS detector. Each substrate was tested in triplicate reactions, and values reported as ± standard error of the mean (SEM).
Relative response factor (RRF) was calculated for each substrate in relation to the internal standard in order to calculate the remaining concentration of substrate after 2 minutes. Product formation was calculated based on stoichiometric ratios of the DesAB reaction. Using the final product concentrations from each reaction, relative substrate-specificity of rDesAB was calculated as a ratio from the most converted substrate.
Liquid Chromatography/Mass Spectrometry (LC/MS) analysis
LC/MS was performed on a Waters Synapt G2Si Q-TOF mass spectrometer coupled with H Class Acquity UPLC system (Waters, Millford, MA). Samples were separated on a Waters Cortecs UPLC C18 column (1.6 μm particle size) (2.1 mm × 50 mm) with a column temperature of 25°C. The injection volume was 1.0 μl for each sample. The mobile phase consisted of Solvent A: 95% H2O, 5% ACN and 0.1% formic acid; and Solvent B: 95% ACN, 5% H2O and 0.1% formic acid. The initial mobile phase was 95% Solvent A. Over the next 7 min Solvent B was increased linearly until reaching 50% and was increased linearly again to 100% over the next 3 min period. At 10 min, the mobile phase was 100% Solvent B before returning to the initial mobile phase over the next 2 min. The flow rate was 0.5 ml min−1 with a total run time of 12 min. The Q-TOF MS utilized ESI in positive ion mode. The ion source and desolvation temperatures were 120°C and 400°C, respectively. The nebulizer gas pressure was set at 6.0 bar and the desolvation gas flow at 800 L/min. The detector voltage was 3000kV. High-purity Argon was used as collision cell gas. MassLynx 4.1 software (Waters) was used for the data acquisition and processing.
Phylogenetic analysis
Phylogenetic analyses were performed on protein sequences and followed a two-step protocol, due to the similarity of DesA and DesB with sugar transketolases. For the first analysis, a BLASTP (22) search using DesA (WP_004606448.1) or DesB (WP_004606449.1) from C. scindens ATCC 35704 as a query against the non-redundant NCBI protein database retrieved the top 40,000 matches for each protein (with a maximum E-value of 1E-6). To minimize the presence of fragmented sequences and false positives, proteins shorter than 200 or longer than 800 amino acids were then excluded. Each protein set was aligned by Clustal Omega v. 1.2.4 (23) with default parameters and, for maximum likelihood tree inference, alignments were analyzed by FastTree v. 2.1.10 (24) using the LG amino acid substitution model and gamma likelihoods of rate heterogeneity.
Each resulting first-phase tree was analyzed to identify which of the two large clusters contained the query sequence (short sequence cluster) and which contained the sugar transketolases (long sequence cluster). The sequences present in the desmolase subtree were aligned and analyzed again in the second phase, which used the same methods described above for alignment and tree inference.
In order to color branches according to organismal classification obtained from the taxonomic information file from NCBI, resulting trees were processed manually and by in-house Perl scripts and displayed by Dendroscope v. 3.5.10 (25) with further cosmetic adjustments performed in Inkscape (https://inkscape.org/).
Sequence similarity network analysis
Protein sequences were obtained from NCBI by collecting the top 500 results from BLAST searches from the amino acid sequence of DesA (WP_004606448.1) and DesB (WP_004606449.1) from Clostridium scindens ATCC 35704, resulting in 1,000 curated sequences on May 31, 2019. FASTA files from the different BLAST results were then combined, and duplicates were removed based on their annotation (identical sequences annotated as different entries were not eliminated). The EFI-EST (http://efi.igb.illinois.edu/efi-est/) web tool was utilized to generate sequence similarity networks (SSNs), using Option C of EFI-EST; sequence-function space in the SSNs was analyzed with Cytoscape (version 3.7.1), a desktop platform for visualizing complex networks (http://www.cytoscape.org/), using node attributes from the UniProtKB and NCBI to assist segregating the SSN into isofunctional clusters. Experimentally verified sequences were used to identify the members of the “Desmolase” family that has not been curated by Pfam. The alignment scores used for filtering the SSNs are given in the Figure legends.
Genomic context for the “Desmolase” family members was confirmed by performing whole genome alignments on the organisms identified from the SSN. Full genome information, contained in the FASTA and GFF3 files, was downloaded from NCBI for each organism. Alignment was performed using the Progressive MAUVE algorithm on Geneious Prime (version 2019.2.1). Poor alignments were manually curated based on the genomic annotation data extracted from the SSN.
Prostate cancer cell line and cell proliferation assay
LNCaP cells were purchased from the American Type Culture Collection (ATCC) (Manassas, VA). Cells were cultured in RPMI-1640 media supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (Corning, Corning, NY). Cells were maintained in a 37°C, 5% CO2 humidified chamber. LNCaP cells (passage 28) were seeded (2.5 × 104 cells) in 96-well plate. After 96 hours, the RPMI-1640 medium was removed. Cells were washed with phosphate-buffered saline (PBS) and resuspended in RPMI 1640 media containing 10% charcoal: dextran stripped FBS. 24 hours later, steroids were added to the media as a DMSO solution to a final concentration of 0.1 nM or 10 nM. The final DMSO content was 1% v/v of the medium. The MTS cell proliferation assay colorimetric kit (Abcam, Cambridge, England) was performed according to the manufacturer’s instructions 24 and 72 hours after steroids were added. Briefly, 10 μL of MTS reagent was added to each well, and incubated for three hours (either 21 or 69 hours after adding the steroids). The plate was briefly shaken and the absorbance was measured at 490 nm by a μQuant Microplate Spectrophotometer (BioTek, Winooski, VT). The experiments were performed in triplicate with four independent experiments. Cell proliferation was calculated as a percentage of cells treated with steroids compared to the cells that were treated with the vehicle (1% DMSO).
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 8.2 (GraphPad Software, San Diego, CA). Values are represented as mean ± SEM. Normality and homogeneity of variance were evaluated before further testing. For cellular proliferation/viability data, effects were tested by a one-way ANOVA followed by Dunnett’s multiple comparisons. All steroid treatments were compared to the vehicle control. Unless stated otherwise, all tests were conducted with α equal to 0.05.
RESULTS
The desA and desB gene products, encoding a steroid transketolase, are phylogenetically and functionally distinct from sugar transketolases.
We first wanted to determine the evolutionary relationships between desA and desB amino acid sequences and sugar transketolases and related proteins, and determine the extent of desAB-encoding bacteria in the human microbiome. After querying, we collected 40,000 DesA and 40,000 DesB amino acid sequences against the non-redundant NCBI database, and reduced the numbers to 37,894 and 38,186, respectively, after discarding sequences shorter than 200 or longer than 800 residues.
The overall phylogenies separated sequences in two clear clusters (Figure 1A) which, upon closer inspection, contained either short (~310 residues) or long (~640 residues) protein sequences; these were identified to be, respectively, the clades where the desmolases or sugar transketolases were placed. About half of the sequences were in each sub-tree: in the initial tree using DesA as a query (Figure 1A), 19,907 sequences were in the short-sequence clade and 17,988 were in the long-sequence one; for DesB, numbers were 16,910 and 21,276, respectively. The sequences from the short-sequence sub-trees were then realigned and reanalyzed by maximum-likelihood to better identify the placement of DesA (Figure 1B).
Figure 1. Large scale phylogenetic analysis of DesA.
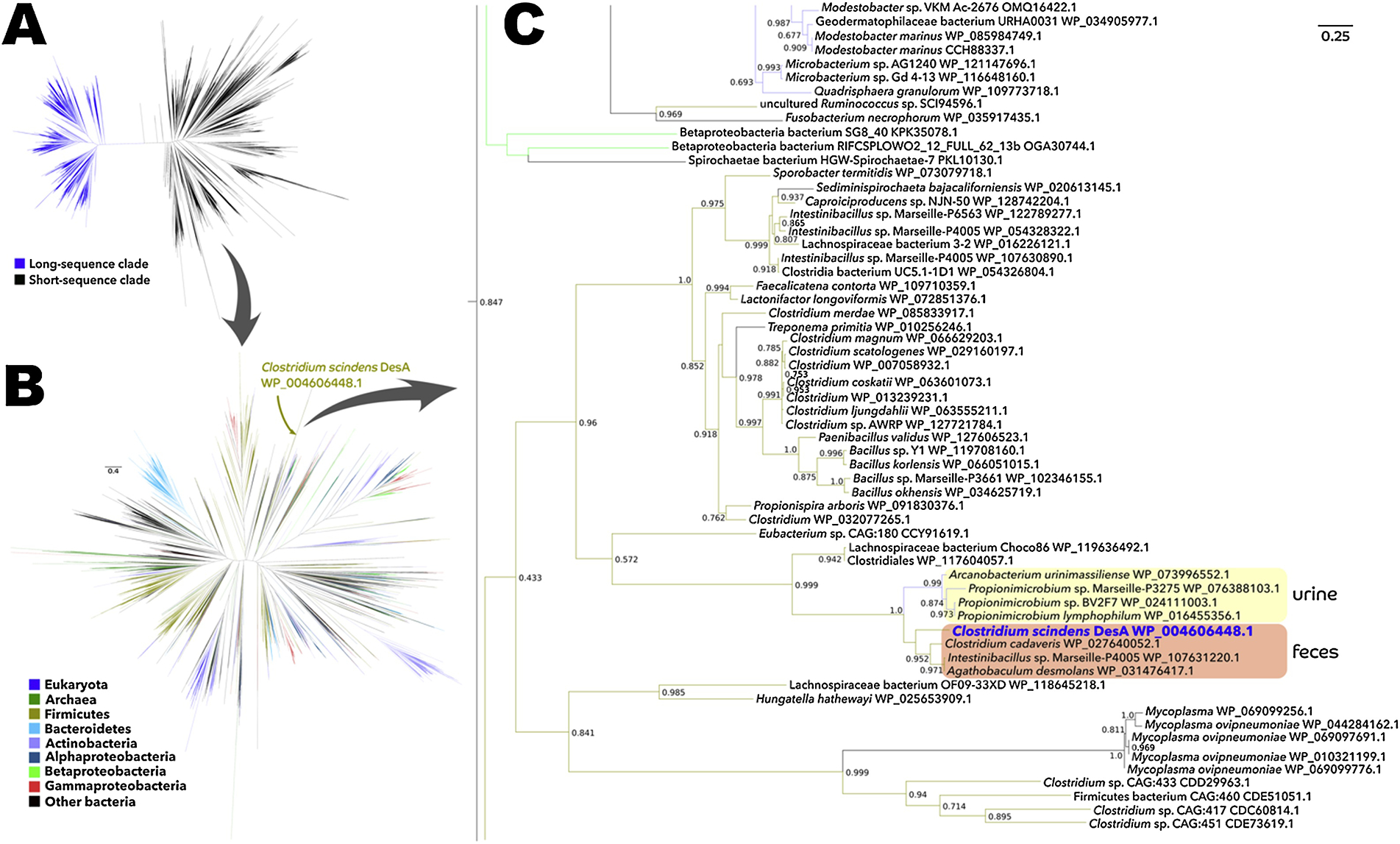
A: Maximum-likelihood phylogenetic tree of 37,894 protein sequences from NCBI’s non-redundant database that were similar to C. scindens DesA and were between 200 and 800 amino acids in length. B: Phylogenetic analysis of solely the sequences present in the short-sequence clade (i.e., the subtree where the DesA protein is placed). C: Zoom into the part of the tree where C. scindens DesA is located; numbers on nodes are FastTree-calculated support values, with only values above 0.5 being shown.
A sub-tree was identified containing both verified DesA sequences from C. scindens ATCC 35704, B. desmolans and Propionibacterium lymphophylum, as well as organisms such as Arcanobacterium urinimassilliense and Intestinibacillus sp. Marseille-P4005 which have not been characterized with respect to steroid metabolism (Figure 1C). Interestingly, the sub-tree bifurcates with high support value (1.0) based on colonization niche with Propionimicrobium and Arcanobacterium inhabiting the urogenital tract, and Clostridium cadaveris, Intestinibacillus, C. scindens, and B. desmolans inhabiting the GI tract. DesB was also associated solely with the taxa described above (Figure S1), and the gene appears to have diverged based on anatomical site of colonization.
Clostridium magnum DSM 2767, isolated from freshwater mud, harbors a gene cluster that appears to encode a low sequence identity desAB gene along with both desC and desE genes. The gene organization, desABCED indicates that this organism may encode steroid-17,20-desmolase (desAB), 20α-HSDH (desC), 20β-HSDH (desE), as well as a putative cortisol transporter (desD). Further study is needed to determine whether this bacterium expresses steroid-17,20-desmolase activity, which may function in the degradation of environmental steroids. Fusobacterium necrophorum has a desABC-like gene cluster whose DesC amino acid sequence is again only 37% identical to DesC from C. scindens ATCC 35704, but nonetheless shares conserved domains indicative of zinc-dependent dehydrogenase in the medium chain dehydrogenase/reductase (MDR) family. This may indicate possible side-chain cleavage of glucocorticoids, or alternatively an analogous oxidoreduction and lyase activity against another class of substrate.
Functional network-based analysis reveal distinct sub-clustering of DesA and DesB.
We then examined the similarity relationships implied by a network composed of proteins annotated as members of the transketolase superfamily at different stringency thresholds (Fig. 2). At lower stringencies, the sequences are roughly divided into two large groups, mainly based on their sequence lengths, but a less-significant separation tendency between different Pfam domains could be observed (not shown). With the increase in the stringency threshold, smaller groups are expected to become independent from the main clusters. The first small group to exhibit this behavior, at a stringency threshold of −log1075, was the cluster containing members of the DesA family (Fig. 2, red circle). This is evidence that the relationship between members of the DesA family and other transketolase representatives is the weakest amongst analyzed sequences. At very high thresholds (e.g. 150 or higher, Fig. 2D), most sequences break up into disconnected groups. The optimal separation of clusters for our purposes was observed at a stringency threshold of −log10100, and further analyses will be based on this network.
Figure 2. Thresholded sequence similarity networks represent sequences as nodes (circles) and pairwise sequence relationships (alignments) better than a set threshold as edges (lines).
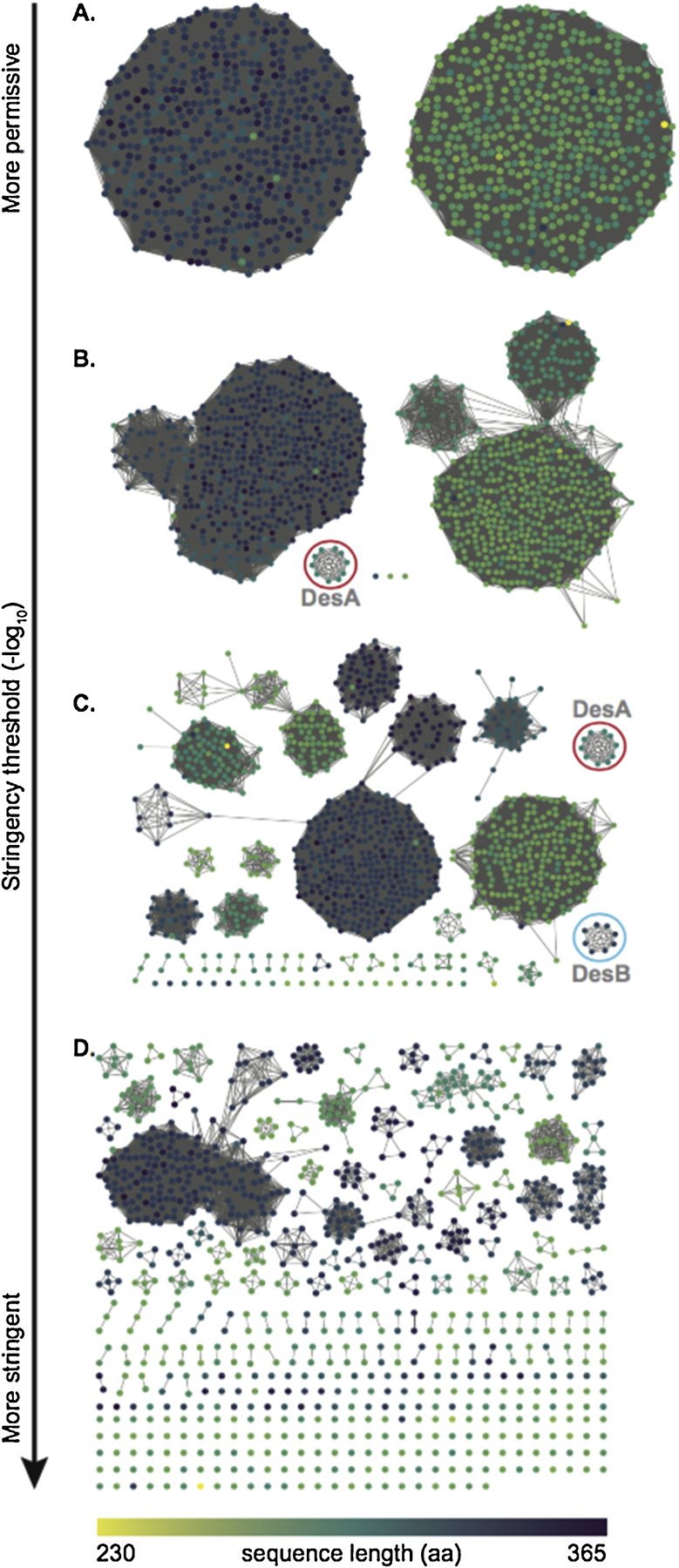
As the threshold becomes more stringent, sequences break up from the main cluster. Nodes have been colored according to sequence length in amino acids (scale at the bottom). A: At threshold values below 50, proteins sequence are divided in two main clusters. B: At a threshold value of 75, the first group of sequences becomes independent from the main clusters. This group (red circle) contains proteins from the DesA family. C: At a threshold value of 100, there is a clear separation into multiple groups. Proteins from the DesA family remain clustered (red circle), and proteins from the DesB family are separated into their own cluster (cyan circle). D: At threshold values above 150, the majority of sequences break up into disconnected groups.
When looking at the taxonomic composition of the smaller clusters containing members from the “Desmolase” families, a pattern can be noticed: between the 10 members of DesA and the 8 members of DesB, 7 are overlapping (Fig. 3A). Analysis of the genomic organization of these individuals revealed that both DesA and DesB are not only found in the same gene cluster, but were observed in tandem in all cases (Fig. 3B). Thus, the results based on phylogeny and functional network analysis agree.
Figure 3. In-depth view of the DesA and DesB groups and their genomic context.
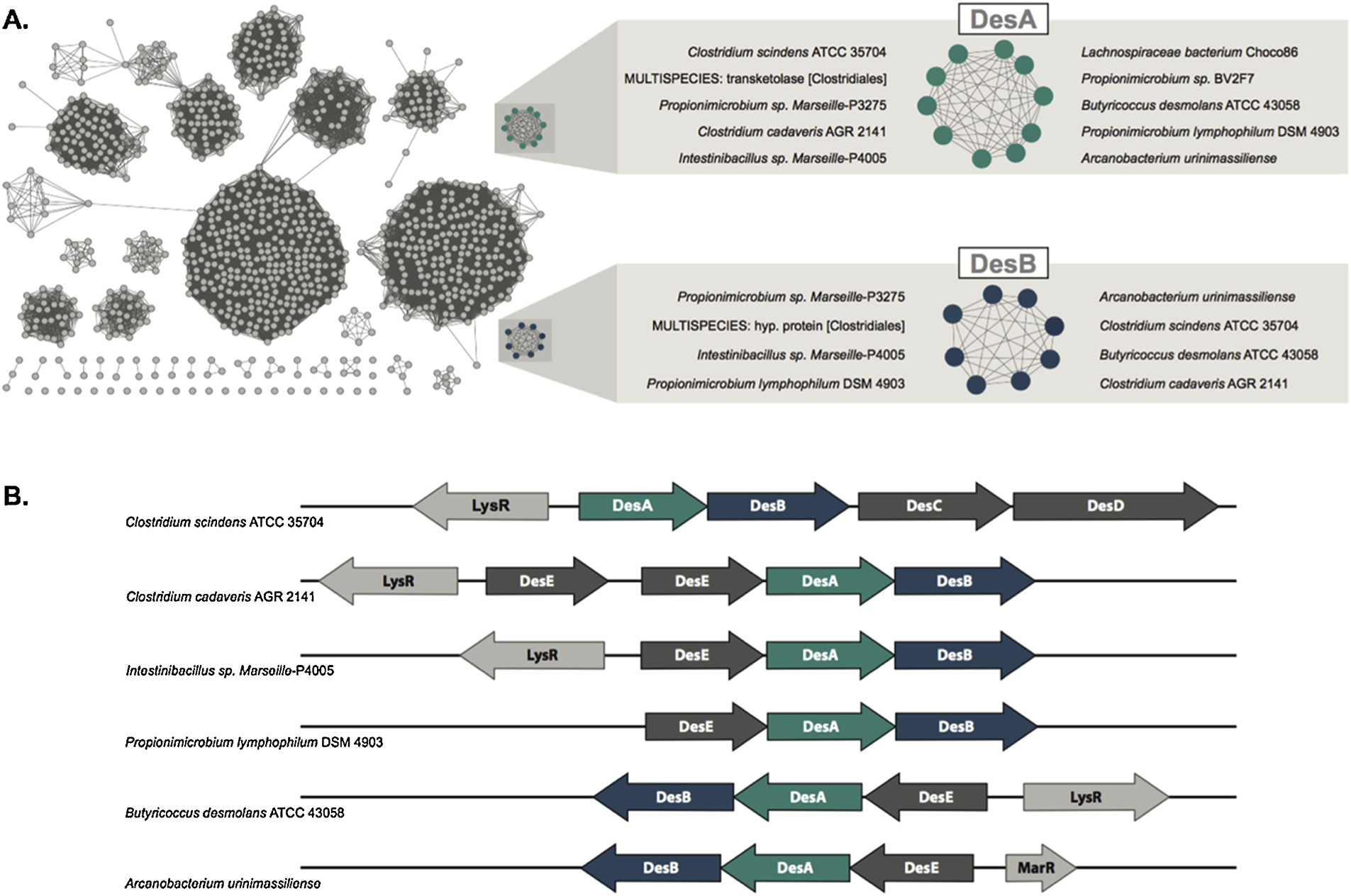
A: Species of organisms identified within the functional clusters of family DesA and DesB. Proteins labeled as “multispecies” are derived from metagenomics data and, thus, don’t have a single species associated with them. B: Genomic context of proteins identified within the DesA and DesB functional clusters.
The gut microbe, Clostridium scindens ATCC 35704, and urinary microbe, Propionimicrobium lymphophilum ACS-093-V-SCH5, are capable of side-chain cleavage of cortisol, allotetrahydrocortisol and pharmaceutical analogs of cortisol.
Our prior studies of cortisol metabolism by C. scindens ATCC 35704 (8) and P. lymphophilum ACS-093-V-SCH5 (9) led us to determine the extent to which “allo” (5α-reduced) derivatives of cortisol as well as glucocorticoid drugs were metabolized by whole cells. After 48 hour incubation of C. scindens ATCC 35704 or P. lymphophylum ACS-093-V-SCH5 with 100 μM substrates, the reaction products were separated by LC/MS. Cortisol (363.2165 m/z) and cortisone (361.20 m/z) were side-chain cleaved by both strains to 11β-hydroxyandrostenedione (303.1949 m/z) and 11-ketoandrostenedione (301.1790 m/z), respectively (Fig. 4). We previously reported that tetrahydrocortisol (5β-reduced) is not a substrate for steroid-17,20-desmolase (8). However, when allotetrahydrocortisol was tested as a substrate with C. scindens ATCC 35704, we detect both allotetrahydrocortisol (367.25 m/z) as well as allotetrahydro-11β-hydroxyandrostenedione (307.23 m/z). This is significant because the liver reduces cortisol to either 5β- or 5α-reduced derivatives (11). Only the 5α-reduced derivatives are androgenic (2, 26), and potentially prohypertensive through inhibition of host 11β-HSD1 & 2 isoforms in aortic smooth muscle and renal tubes similar to that caused by ingestion of licorice (26).
Figure 4. Side-chain cleavage of endogenous and glucocorticoid drugs by whole cells of gut bacterium Clostridium scindens ATCC 35704 and urinary tract isolate Propionimicrobium lymphophilum ACS-093-V-SCH5.
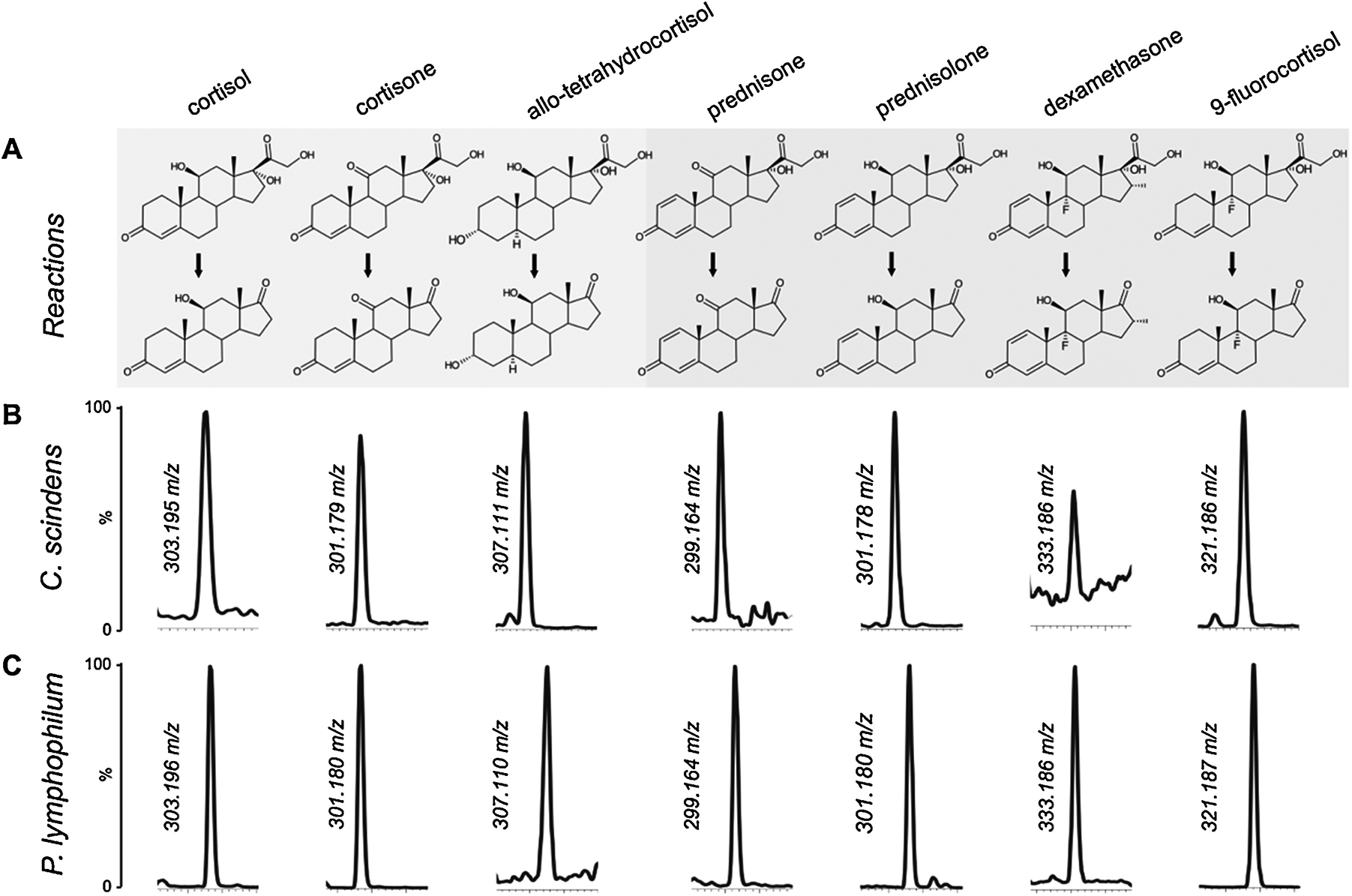
A: Steroid-17,20-desmolase reaction showing endogenous (highlighted in gray) and exogenous (highlighted in blue) compounds as substrates. Anticipated product (loss of C2O2H4) of side chain cleavage is 60.02 m/z smaller than substrate. B: LC/MS in single ion monitoring (SIM) mode of extracts from C. scindens incubated with 50 μM substrate. C: LC/MS in single ion monitoring (SIM) mode of extracts from P. lymphophilum incubated with 50 μM substrate. Chromatographs representative of three biological replicates.
All glucocorticoid drugs that we screened were metabolized by whole cells of C. scindens ATCC 35704 and P. lymphophilum ACS-093-V-SCH5 (Fig. 4; Figs. S2–S5). Anticipated side-chain cleavage products (loss of C2O2H4) detected by LC/MS are 60.02 m/z less than the substrates. Dexamethasone was converted to a product consistent with 9α-fluoro-11β-hydroxy-16α-methylandrosta-1,4-diene-3,17-dione (333.18 m/z), prednisolone was converted to a product consistent with 11β-hydroxy-1,4-androstene-3,17-dione (301.18 m/z), 9-fluorocortisol was converted to a product consistent with 9α-fluoro-11β-hydroxyandrosta-4-ene-3,17-dione (363.20 m/z), and prednisone was converted to a product consistent with 11-keto-1,4-androstene-3,17-dione (299.16 m/z). Formation of another significant product after incubation with prednisone was evident, (3.75 min) eluting before prednisone (4.00 min) that is 2 amu greater than prednisone. This is consistent with NADH-dependent 20α-HSDH activity reported previously (7). These results indicate that gut and urinary bacteria encoding the steroid-17,20-desmolase pathway metabolize endogenous glucocorticoids, including derivatives of cortisol including allotetrahydrocortisol, as well as synthetic pharmaceutical derivatives of cortisol.
Purified recombinant DesAB is sufficient to catalyze side-chain cleavage of pharmaceutical cortisol derivatives.
We previously reported the cloning and heterologous expression of desA and desB genes from C. scindens ATCC 35704 in pETDuet, and separate cloning and overexpression of a gene encoding NADP(H)-dependent 17β-hydroxysteroid dehydrogenase (17β-HSDH) from Cochliobolus lunatus in pET-51b(+) (8). Coupling 17β-HSDH with recombinant DesAB (rDesAB) provides a continuous spectrophotometric assay to quantify side-chain cleavage products of steroid-17,20-desmolase. The rDesA and rDesB were co-expressed and after affinity-purification, equal proportions of pure rDesA (32.0 kD ± 0.8) and pure rDesB (38.5 kD ± 0.5) were observed (Fig. 5). Recombinant 17β-HSDH eluted from streptavidin affinity chromatography was detected as a single-band on SDS-PAGE (31.9 kDa ± 0.3) (Fig. 5). When cortisol was substrate, and reaction conditions identical to our previous report (8), kinetic constants were consistent (Km 7.08 ± 0.42 μM; Vmax 13.19 ± 2.08 nmol min−1 mg−1). The assumptions behind a coupled-continuous assay are that 1) the rate of the coupled enzyme (r17β-HSDH) is far greater than the rate of the enzyme being coupled to (rDesAB), 2) the substrates for the coupled enzymes show similar affinity. To test this, we compared the reaction velocity of NADPH-dependent r17β-HSDH with and without rDesAB using 11β-OHAD or cortisol as substrates, respectively, and compared this to reaction velocities with a similar setup when prednisone or 11β-hydroxy-androsta-1,4-diene-3,17-dione (1,4–11β-OHAD) were substrates. When cortisol was substrate, the coupled reaction was ~10 fold slower than when 11β-OHAD was measured alone with 17β-HSDH, indicating that DesAB is rate-limiting (data not shown). When this experiment was performed with prednisone or 1,4–11β-OHAD, the coupled reaction with prednisone and 17β-HSDH with 1,4–11β-OHAD had the same rate, indicating that 17β-HSDH became rate-limiting. Indeed, it was determined that when 1,4–11β-OHAD was a substrate, the relative 17β-HSDH activity was only 9.67% relative to 11β-OHAD (data not shown). These results indicate current limitations for the coupled assay.
Figure 5. SDS-PAGE of purified recombinant DesA and DesB from Clostridium scindens ATCC 35704 and NADPH-dependent 17β-HSDH from Cochliobolus lunatus.
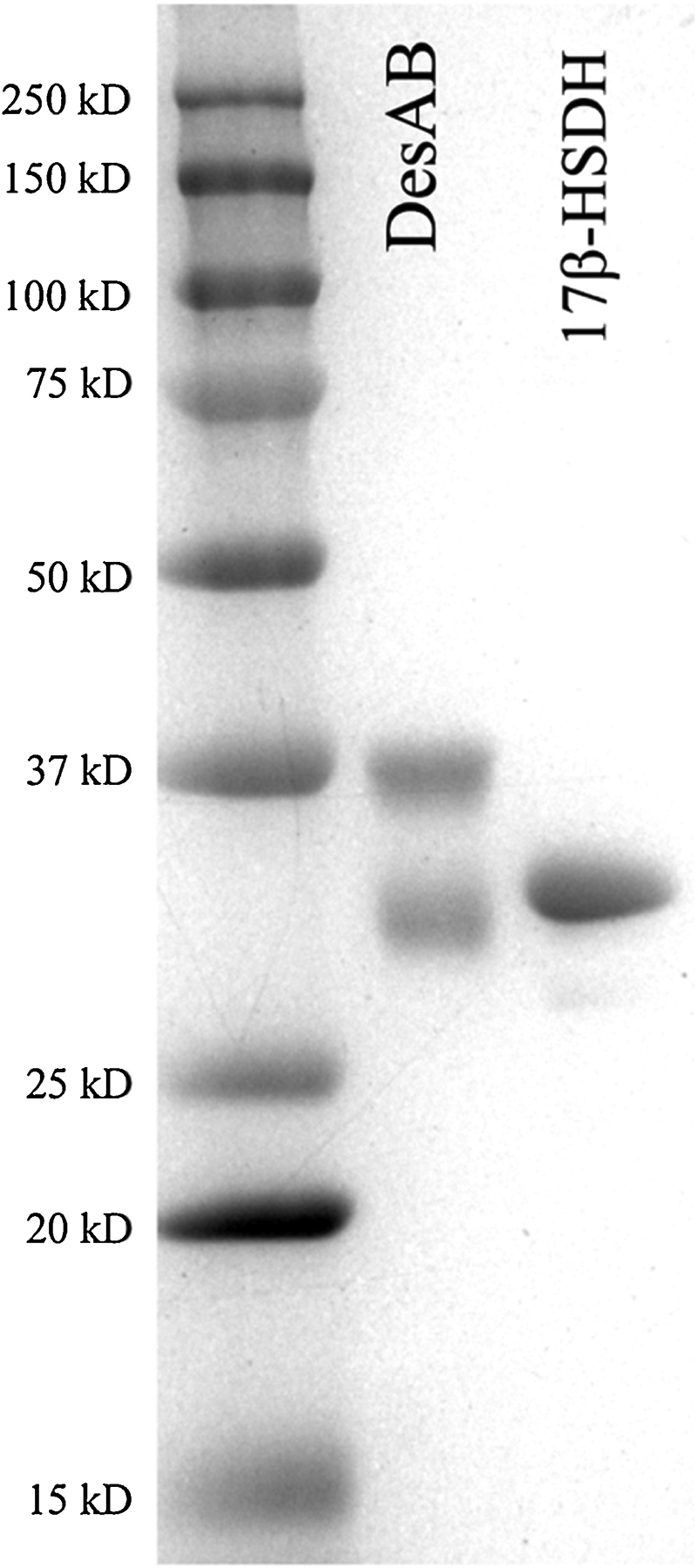
Affinity purified recombinant steroid-17,20-desmolase (rDesA and rDesB) co-expressed in pETDuet (Lane DesAB), affinity purified recombinant 17β-hydroxysteroid dehydrogenase expressed in pET21 (Lane 17β-HSDH), and molecular mass marker (Lane M).
We then directly measured rDesAB activity, comparing substrate conversion by integration of peak area of substrate relative to internal standard, 20β-dihydrocortisol, which is not a substrate for DesAB (8). In our previous report, tetrahydrocortisol (5β-reduced) was not a substrate; however, and importantly, rDesAB activity towards 5α-dihydrocortisol (5α-reduced) was comparable to that of cortisone and cortisol (Table 1). Of note, prednisone had the highest relative activity followed by prednisolone (71%) and 9-fluorocortisol (43%). Dexamethasone displayed 23% activity relative to prednisone, indicating that the 16α-methyl group is inhibitory. Mass spectra are reported in Fig. S6. Overall, these results indicate that rDesAB is sufficient to explain metabolism of synthetic glucocorticoids, which in the case of prednisone/predisolone are better substrates than endogenous host glucocorticoids.
Table 1.
Substrate-specificity of pure rDesAB from Clostridium scindens for endogenous and pharmaceutical glucocorticoids.
| Steroid | Trivial name | Relative activity %a |
|---|---|---|
| 1,4-pregnadien-17,21-diol-3,11,20-trione | prednisone | 100.00 ± 0.12 |
| 1,4-pregnadien-11β, 17, 21-triol-3,20-dione | prednisolone | 71.47 ± 0.43 |
| 4-pregnen-9α-fluoro-11β, 17, 21, triol-3, 20-dione | 9-fluorocortisol | 43.29 ± 0.35 |
| 5α-pregnan-3α, 11β, 17, 21-tetrol-20-one | allotetrahydrocortisol | 38.87 ± 9.19 |
| 4-pregnen-17,21-diol-3,11,20-trione | cortisone | 37.80 ± 1.82 |
| 4-pregnen-11β,17,21-triol-3,20-dione | cortisol | 34.57 ± 2.24 |
| 1,4-pregnadien-9α-fluoro-16α-methyl-11β,17,21-triol-3,20-dione | dexamethasone | 23.42 ± 3.33 |
Values represent mean ± SEM from three technical replicates.
The side-chain cleavage product of prednisone causes significant proliferation of LNCaP cells and expression of androgen-receptor downstream targets.
We then wanted to determine whether the side-chain cleavage product of prednisone stimulated androgen-dependent growth of androgen-responsive prostate cancer cells (LNCaP). Experiments included 4 biological replicates, each with 3 technical replicates. The product of cortisone side-chain cleavage, 11-keto-androstenedione (11-KA), at 0.1 nM caused significant growth of LNCaP cells relative to vehicle control (1.8 fold; P < 0.05) at 24 hrs but not 72 hrs. 5α-dihydrotestosterone (DHT) at 0.1 nM was significant after 24 hrs (1.7 fold; P = 0.052), and trending at 72 hrs (P = 0.071). 11β-hydroxyandrostenedione and 11-ketotestosterone (11KT) did not cause significant proliferation at 0.1 nM at either time point. By contrast, 1,4-androstadiene-3,11,17-trione (AT), the product of bacterial side-chain cleavage of prednisone, caused significant proliferation of LNCaP cells relative to vehicle at 24 hrs (2.3 fold; P < 0.01) and 72 hrs (1.6 fold; P < 0.01). Significant changes in proliferation were not observed at 10 nM for any of the compounds tested (Figure 7).
Figure 7. Measurement of LNCaP cell proliferation in the presence of steroid-17,20-desmolase reaction products.
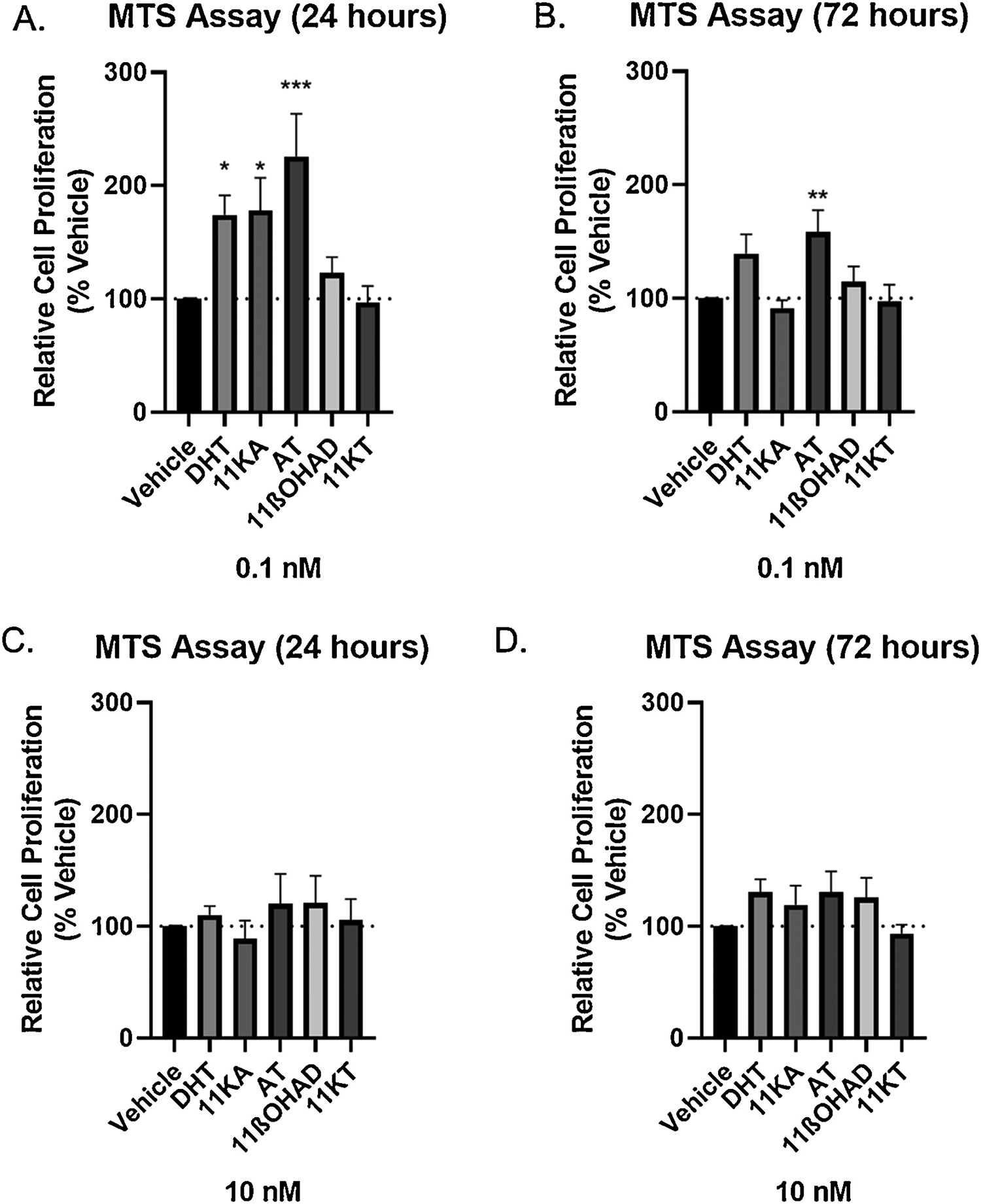
Induction of cell proliferation in androgen-dependent LNCaP cells by DHT (dihydrotestosterone), 11KA (11-keto-androstenedione), and AT (1,4-androstadiene-3,11,17-trione). Cells (2.5 × 104 cells) were seeded in a 96-well plate in RPMI 1640 media containing 10% charcoal: dextran stripped FBS for 24 hours before adding 0.1 and 1 nM steroid. MTS assay was carried out 24 and 72 hours after. Results are shown as mean ± SEM of four independent experiments with three replicates each. Differences between the compound of interest and Vehicle were conducted by ANOVA, followed by Dunnett’s test (* = p<0.05, ** = p<0.01, and *** = p<0.001 relative to the vehicle control).
Discussion
The purpose of the current study was to determine the diversity of taxa encoding desAB, the metabolism of endogenous and synthetic glucocorticoids by urinary tract and gut bacterial isolates, and the effect of the side-chain cleaved product of prednisone on androgen-dependent prostate cancer cell proliferation. The results of the current study address two areas that are gaining increased attention in the literature: drug metabolism by the gut microbiome (21, 27) and resurgent interest in the formation of 11-oxy-androgens (1–5). Clostridium scindens ATCC 35704 was originally isolated (and named) based on its ability to side-chain cleave cortisol forming 11β-OHAD (14); in addition, it is widely recognized as one of a few species of gut clostridia capable of converting the host bile acids cholic acid and chenodeoxycholic acid to deoxycholic acid and lithocholic acid, respectively, through a complex bile acid 7α-dehydroxylation pathway (28). While all strains of C. scindens so far characterized encode the bile acid inducible (bai) operon facilitating 7α-dehydroxylation pathway, only a few strains of C. scindens encode the steroid-17,20-desmolase pathway genes (7). Our phylogenetic and network-based analysis indicate that C. scindens ATCC 35704 may also be a rarity among bacterial strains inhabiting the human body in its ability to metabolize cortisol and cortisone as well as pharmaceutical derivatives (Figures 1–3).
C. scindens (29–31), C. cadaveris (17, 32), and B. desmolans (17, 33) are normal inhabitants of the human gastrointestinal tract, and P. lymphophilum is a normal inhabitant of the urogenital tract (34, 35). Since strains of C. scindens and P. lymphophilum often vary with respect to expression of steroid-17,20-desmolase (7, 9), there may be important inter-individual differences with respect to cortisol side-chain cleavage in the gut and urinary tract that may impact host physiology and disease risk. In addition, we identified other bacteria, including pathogenic bacteria, bacteria inhabiting the environment, and intestinal bacteria that have conserved gene organization with the steroid-17,20-desmolase but low amino acid sequence identity. It remains to be determined whether these organisms are capable of metabolizing cortisol. In addition, we cannot exclude the possibility that other bacterial or fungal enzymes inhabiting the vertebrate host have evolved to side-chain cleave 17-hydroxycorticosteroids. Indeed, Mycobacterium tuberculosis expresses a thiolase-derived enzyme involved in side-chain cleavage of cholesterol (36). Fungi, such as the plant pathogen, Nectria radicicola, express a mono-oxygenase capable of oxygen and NADPH-dependent oxidative side-chain cleavage of progesterone (37, 38).
An interesting commonality between the bile acid 7α-dehydroxylation pathway and cortisol steroid-17,20-desmolase pathway is the utilization of NAD(P)(H)-dependent hydroxysteroid dehydrogenase (HSDH). C. scindens constitutively expresses NADP(H)-dependent 7α-HSDH which reversibly converts cholic acid to 7-keto-deoxycholic acid, where the latter is not converted to deoxycholic acid (28). Likewise, the desC gene encodes NAD(H)-dependent 20α-HSDH involved in reversible conversion of cortisol to 20α-dihydrocortisol, which is not a substrate for DesAB (7,8). In this way, both bile acid 7α-dehydroxylation and steroid-17,20-desmolase appear to be regulated by the redox and energy status of the bacterial cell, but utilize different pyridine nucleotide pools. The use of HSDH enzymes as “switches” to regulate flux through steroid metabolizing pathways by altering their structure and catabolic enzyme specificity provides a rapid means to conserve steroid substrate based on energy demand and to regulate the ratio of NAD(P):NAD(P)H.
We also determined limitations in the coupled assay that we recently developed to continuously monitor steroid-17,20-desmolase (8). Because the 17β-HSDH from C. lunatis showed greater activity for endogenous steroids, it may be possible to engineer the 17β-HSDH from C. lunatis to broaden substrate-specificity. The presence of Δ1,4 bonds in the pharmaceutical drugs increased the activity of rDesAB relative to cortisol; however, a 16α-methyl was inhibitory. These experiments are thus providing information that will be useful in future mechanistic work on the DesAB.
Androgen receptor (AR) agonism has importance in development and function in both male and female reproductive systems. Indeed, premature puberty (adrenarche) in children is androgen-sensitive and 11β-OHAD is a biomarker for adrenarche (39). Beyond testosterone and DHT, 11β-OHAD and 11-oxy-androgens are now viewed as important in the pathology of CRPC in men (1–5). Levels of 11β-OHAD and 11-oxy-androgens are significantly elevated in women with PCOS (40). PCOS is a common endocrine disorder, affecting 10% of females of reproductive age (41). While both the ovary and adrenal contribute to androgen excess in this condition, the gut microbiome is also implicated although causal associations are still lacking (40, 41). Interestingly, prostate cancer (35, 42) also appears to have microbiome components that are only beginning to be understood. Of note, we recently reported that urinary tract isolate Propionibacterium lymphophylum encodes desABE genes and converts cortisol to 11β-OHAD (7). A recent study that compared urinary microbiome abundances between prostate cancer patients relative to biopsy negative controls showed significant elevation of P. lymphophilum in patients with prostate cancer (35). However, it is unclear what proportion of the strains of P. lymphophilum identified in patient urinary 16S rDNA sequences harbor the desAB genes, and to what extent the desAB genes are important in tumor androgen concentration. Another study detected 11-oxy-androgens in prostate tissue and serum from patients with benign prostatic hyperplasia (BPH) and characterized C19 11-oxy androgen in BPH-1 cells (43). The link between urinary tract microbial metabolism of glucocorticoids, BPH, as well as prostate cancer risk and disease progression remains to be established. However, LC/MS/MS methods now exist and are being further developed that will allow measurement of 11-oxy-androgens with increasing sensitivity and the ability to separate structurally related metabolites with greater precision (44).
Additionally, the formation of 11-oxy-androgens by the human microbiome may affect gastrointestinal immune function. The expression of AR has been detected in numerous immune cell lineages, including neutrophils, macrophages, mast cells, B cells, and T cells (45). In this way, members of the gut microbiota may be capable of modulating immune function via conversion of cortisol to 11-oxy-androgens. Colonocytes also express not only nuclear AR (nAR) but also membrane AR (mAR) (46, 47). It is therefore possible that formation of 11-oxy androgens results in altered colonic physiology including transit time, contractility, and enteric hormone secretion (41). While the role of sex hormones in immune and nervous system have received significant attention, very little is understood about the role of AR activation in the gastrointestinal system (41).
These studies also raise other important implications concerning the role of steroid-17,20-desmolase activity by gut microbiota on both endogenous glucocorticoids and exogenous glucocorticoids: Firstly, consideration must now be given to the effects of gut steroid 17,20-desmolase activity on the pharmacokinetics and bioavailability of the commonly prescribed synthetic glucocorticoids, prednisone and prednisolone. This may also apply to dexamethasone and 9a-fluorocortisol. Not only may the prescribed dosage be (effectively lowered) altered but the physiological consequences of the previously unrecognized steroid-17,20-desmolase transformed products have now to be taken into account. Timing and dosages whether in the morning, during the day, or at midnight will now need to be re-examined since subsets of patients will likely vary with respect to colonization of host-associated bacteria expressing steroid-17,20-desmolase and may metabolize these pharmaceuticals to different extents.
Secondly, the formation of 11-oxy-androgens from both endogenous and exogenous glucocorticoids must now be considered in what has become known as the “GALF hypothesis” (26). Many of these metabolites, particularly the (allo)-Ring A 5 reduced products of cortisol and cortisone will possess significant inhibitory properties towards both 11β-HSD1 & 2 isoforms in aortic smooth muscle and renal tubules as well as many other important glucocorticoid target tissues (26). These may lead to hypertension, similar to a form of hypertension known as apparent mineralocorticoid excess where pseudohyperaldosteronism is evident. The work of Honour indicates that gut bacteria play a substantial role in steroid hypertension (26, 48). Their effects may also reduce the risk of colorectal cancer by a mechanism similar to licorice (glycyrrhetinic acid) (49). Therefore, gut bacterial formation of 11-oxy androgens is potentially significant in the health and treatment of disease common to both male and females, as well as sex-dependent diseases such as PCOS and CRPC.
Our cell culture experiments provide initial indications that bacterial metabolism of replacement glucocorticoids such as prednisone may promote prostate cancer cell growth. Prednisone is commonly prescribed with abiraterone as a treatment for advanced or metastatic prostate cancer (50). Abiraterone inhibits tumoral androgen biosynthesis through inhibition of cytochrome P450 enzyme 17-hydroxylase-17,20-lyase (CYP17A1) with the goal of reducing accessible androgens (51). Interestingly, 11-keto-androstenedione (product of cortisone side-chain cleavage by steroid-17,20-desmolase) and AT (the product of bacterial side-chain cleavage of prednisone by steroid-17,20-desmolase) were capable of stimulating proliferation of LNCaP cells at a similar or greater level than DHT relative to the vehicle control. Dihydrotestosterone is considered the most potent known endogenous ligand for androgen receptor. Dihydrotestosterone has hormetic effects on proliferation with a maximal effect at 0.1 nM and inhibitory effects above 1 nM (52). Importantly, compounds cleaved by steroid-17,20-desmolase are capable of stimulating proliferation in LNCaP cells at similar rates to DHT, and steroid-17,20-desmolases should not be affected by abiraterone, which is the only FDA approved inhibitor of adrenal androgen synthesis. Based on the current data in this manuscript, both 11-ketoandrostenedione and AT appear to stimulate proliferation at similar physiological concentration to DHT within a similar period of time. Other novel androgens, such as 11KT are metabolized slower than DHT and might take longer to elicit functions (2). Because of this, we believe that our incubation period was possibly too short to observe changes in proliferation with some metabolites. Moreover, LNCaP cells express a mutant ligand-binding domain, allowing for potential receptor promiscuity (53). Future experiments will be needed to determine whether AT causes proliferation in cell lines with wild-type AR, which is most common among prostate cancer patients. More research is needed to elucidate the mechanism through which these compounds stimulation proliferation.
In this regard, a recently published large drug screen also identified metabolism of prednisolone and dexamethasone by C. scindens ATCC 35704, but also provided evidence that the host metabolizes dexamethasone (21). In that study, mono-association with C. scindens ATCC 35704 resulted in significant detection of the side-chain cleavage product of dexamethasone in cecum, colon, and feces, but not small intestine (21). This is consistent with known colonization of C. scindens in the large bowel (54). In addition, significant increase in the side-chain cleavage product was detected in serum and liver indicating that the metabolite is being absorbed from the GI tract (21). Intriguingly, germ-free mice in those experiments also had measurable levels of the side-chain cleavage product in serum, bile, liver, and GI tract (21). This may indicate that host CYP17A1 is capable of side-chain cleavage of dexamethasone, or the action of a yet unknown host enzyme suggested by work in CYP17A1-deficient patients (6). Prednisolone and other synthetic glucocorticoids are prescribed for diseases such as PCOS and CRPC and have significant side-effects. Thus, understanding bacterial side-chain cleavage may be important for modulating both endogenous as well as pharmaceutical glucocorticoid metabolism in host physiology and pathophysiology.
The current study builds from previous work from our group on the steroid-17,20-desmolase pathway (7–10), expanding the scope of substrate specificity of the DesAB from C. scindens ATCC 35704 to include pharmaceutical steroids. Some limitations with respect to our current phylogeny and network-based analysis is the relative paucity of urinary metagenomic datasets. Future metagenomic sequencing or culturomics approaches coupled with bioconversion assays may further expand the list of desAB harboring taxa. Additional work will also be necessary to fully characterize the androgen-dependent proliferation of LNCaP and other cell lines, as well as the steroid profile after incubation in cell culture. In conclusion, steroid-17,20-desmolase is a human microbiome function that contributes novel 11-oxy-androgens from synthetic glucocorticoids that may play a potential clinically significant role in the development and treatment of prostate cancer.
Supplementary Material
Figure 6. Total ion count chromatographs of rDesAB reaction products.
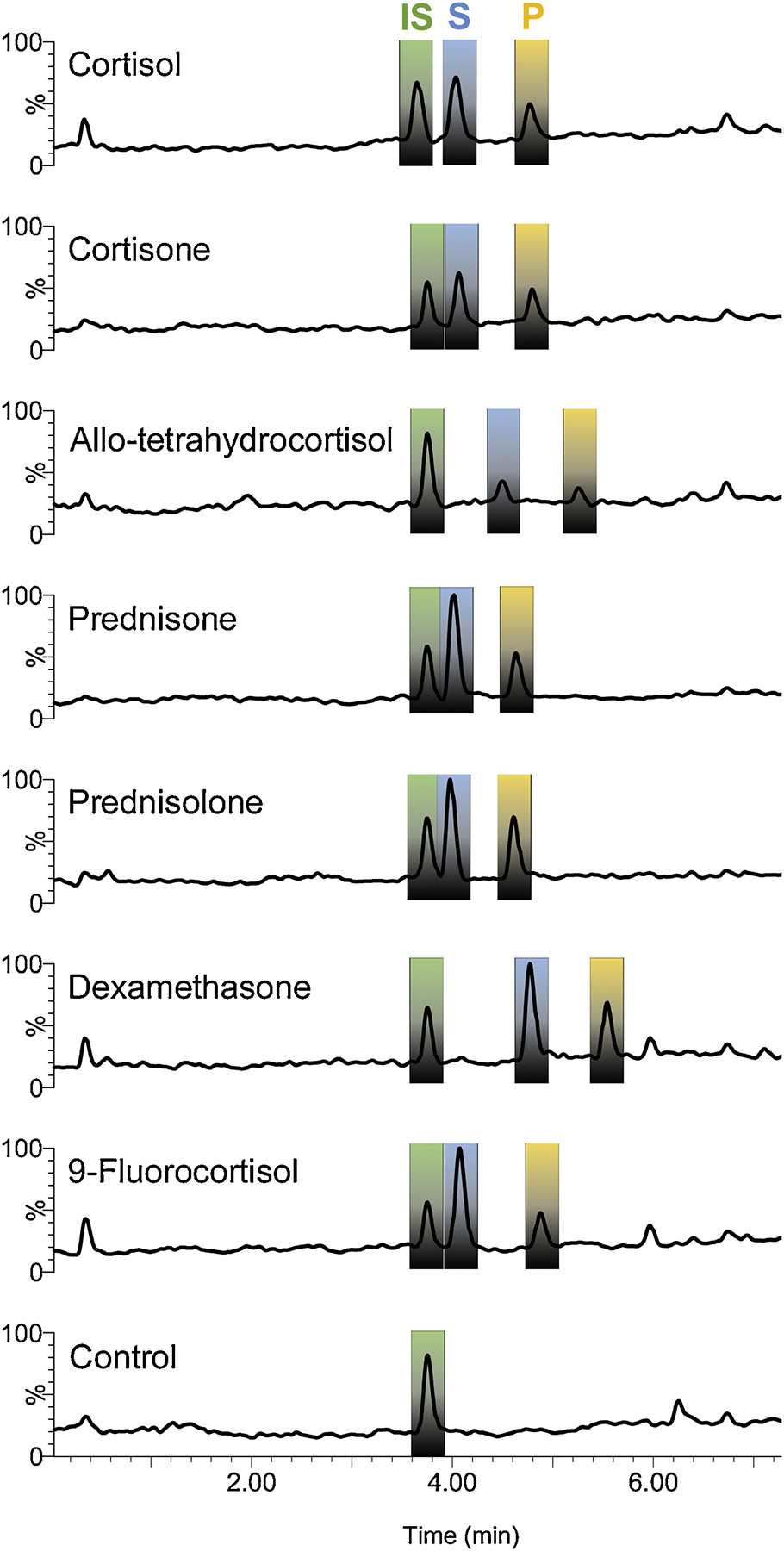
Reaction was initiated with 2 μM of pure rDesAB enzyme to 50 mM MOPS buffer (pH 7.0), 100 μM of substrate (blue) in the presence of 1 μM MnCl2 and 10 μM TPP at room temperature. After 2 minutes, while the reactions were stopped with the addition of 1/10 volume of 1 M HCl and vortexing to drop the pH below 2. A final concentration of 50 uM of internal standard, 20β-dihydrocortisol (green), was spiked into the reaction. Products formed were identical with respect to retention time and loss of 60 amu consistent with side-chain cleavage.
Figure 8. Hypothetical model of physiological relevance of steroid-17,20-desmolase activity in urinary microbes in the context of prostate cancer.
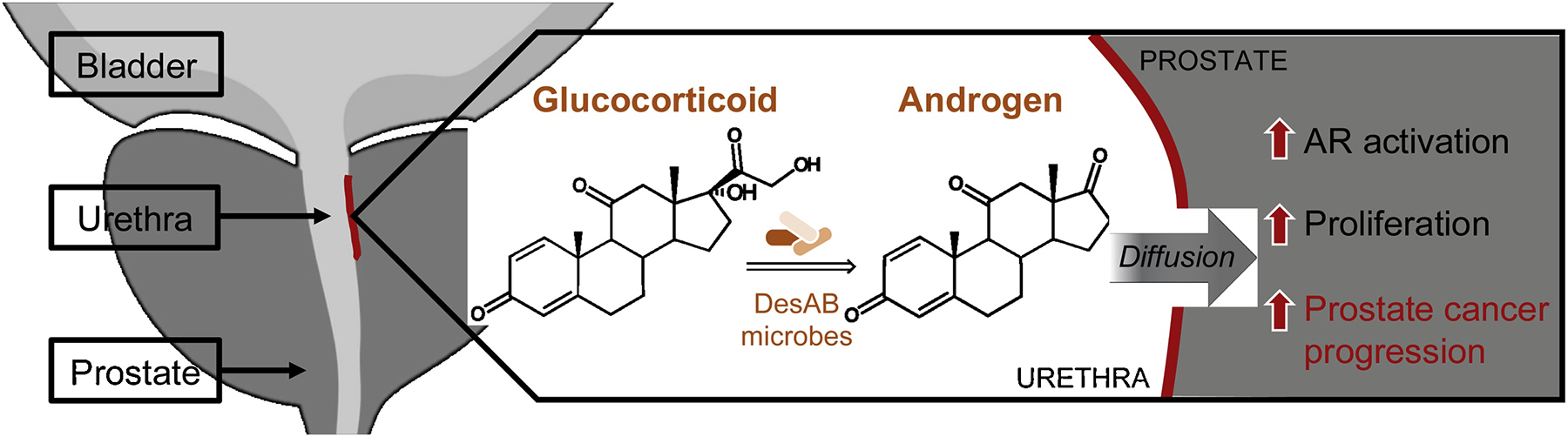
Urinary microbes inhabiting a minimal nutrient environment metabolize glucocorticoids for energy, generating androgens that diffuse into surrounding prostate tissue. Microbe-derived androgens activate androgen receptor (AR), leading to increased proliferation of androgen-dependent prostate cancer cells and ultimately resulting in prostate cancer progression.
Highlights:
Host-associated bacteria are capable of converting cortisol to 11-oxy-androgens via an enzyme known as steroid-17,20-desmolase encoded by the desAB genes
Phylogenetics and sequence similarity network analysis of bacterial DesA and DesB
Isolated bacteria from the urinary and GI tracts and purified recombinant DesAB are capable of metabolizing both endogenous glucocorticoids and glucocorticoid drugs including prednisone and dexamethasone
The side-chain cleavage product of prednisone was found to cause significant proliferation of prostate cancer cells (LNCaP), to a greater extent than dihydrotestosterone
Acknowledgements
We gratefully acknowledge the financial support provided to J.M.R. for new faculty startup through the Department of Animal Sciences at the University of Illinois at Urbana-Champaign (grant Hatch ILLU-538-916) as well as Illinois Campus Research Board RB18068. L.L. is supported by a Graduate Research Fellowship through the National Science Foundation. P.G.W is supported by the UIC Cancer Education and Career Development Training Program Administered by the Institute for Health Research and Policy at the University of Illinois at Chicago with funding by the National Cancer Institute (Grant No. T32CA057699).
Abbreviations:
- 11β-OHAD
11β-hydroxyandrostenedione
- CRPC
castration resistant prostate cancer
- PCOS
polcystic ovary syndrome
- CYP17 A1
cytochrome P450 17-hydroxylase/17,20-lyase
- 17β-HSDH
17β-hydroxysteroid dehydrogenase
- DesAB
steroid-17,20-desmolase
- rDesAB
recombinant DesAB
- SIM
Single ion monitoring mode
- RRF
Relative response factor
- LC/MS
Liquid Chromatography/Mass Spectrometry
- AT
1,4-androstadiene-3,11,17-trione
- 1,4–11β-OHAD
1,4-androstadiene-11β-ol,3,17-dione
- 11-KA
11-keto-androstenedione
- DHT
5α-dihydrotestosterone
- 11KT
11-ketotestosterone
- AR
androgen receptor
- CAH
congenital adrenal hyperplasia
- P450 11B1
11β-hydroxylation via cytochrome P450 11β-hydroxylase
- 20α-HSDH
20α-hydroxysteroid dehydrogenase
- BPH
benign prostatic hyperplasia
- FDA
Food and Drug Administration
- CYP17A1
cytochrome P450 enzyme 17-hydroxylase-17,20-lyase
- GI
gastrointestinal tract
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Turcu AF, Auchus RJ. Clinical significance of 11-oxygenated androgens. Curr Opin Endocrinol Diabetes Obes 24(3) (2017) pp. 252–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Pretorius E, Arlt W, Storbeck KH. 2017. A new dawn for androgens: Novel lessons from 11-oxygenated C19 steroids. Mol Cell Endocrinol 441 (2017) pp. 76–85. [DOI] [PubMed] [Google Scholar]
- [3].Pretorius E E, Africander DJ, Vlok M, Perkins MS, Quanson J, Storbeck K. 11-Ketotestosterone and 11-ketodihydrotesterone in castration resistant prostate cancer: potent androgens which can no longer be ignored. PLoS ONE 11(7) (2016) e0159867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mostaghel EA. Beyond T and DHT-novel steroid derivatives capable of wild type androgen receptor activation. Int J Biol Sci 10(6) (2014) pp. 602–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Swart AC, Storbeck K. 11β-hydroxyandrostenedione: Downstream metabolism by 11βHSD, 17βHSD and SRD5A produces novel substrates in familiar pathways. Mol Cell Endocrinol 408 (2015) pp. 114–123. [DOI] [PubMed] [Google Scholar]
- [6].Shackleton CH, Neres MS, Hughes BA, Stewart PM, Kater CE. 17-Hydroxylase/C17,20-lyase (CYP17) is not the enzyme responsible for side-chain cleavage of cortisol and its metabolites. Steroids. 73(6) (2008) pp. 652–656. [DOI] [PubMed] [Google Scholar]
- [7].Ridlon JM, Ikegawa S, Alves JMP, Zhou B, Kobayashi A, Iida T, Mitamura K, Tanabe G, Serrano M, De Guzman A, Cooper P, Buck GA, Hylemon PB. Clostridium scindens: a human gut microbe with a high potential to convert glucocorticoids into androgens. J Lipid Res 54(9) (2013) pp. 2437–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Devendran S, Mythen SM, Ridlon JM. The desA and desB genes from Clostridium scindens ATCC 35704 encode steroid-17,20-desmolase. J Lipid Res 59(6) (2018) pp. 1005–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Devendran S, Méndez-García C, Ridlon JM. Identification and characterization of a 20β-HSDH from the anaerobic gut bacterium Butyricicoccus desmolans ATCC 43058. J. Lipid Res 58(5) (2017) pp. 916–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Doden HL, Pollet RM, Mythen SM, Wawrzak Z, Devendran S, Cann I, Koropatkin NM, Ridlon JM. Structural and biochemical characterization of 20β-hydroxysteroid dehydrogenase from Bifidobacterium adolescentis strain L2–32. J Biol Chem 294(32) (2019) pp. 12040–12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Erikkson H, Gustafsson J. Excretion of steroid hormones in adults: steroids in faeces from adults. Eur J Biochem 18(1) (1971) pp. 146–150. [DOI] [PubMed] [Google Scholar]
- [12].Cerone-McLernon AM, Winter EH, Mosbach J, Bokkenheuser VD. Side-chain cleavage of cortisol by fecal flora. Biochim Biophys Acta 666(3) (1981) pp. 341–347. [DOI] [PubMed] [Google Scholar]
- [13].Bokkenheuser VD, Morris GN, Ritchie AE, Holdeman LV, Winter J. Biosynthesis of androgen from cortisol by a species of Clostridium recovered from human fecal flora. J Infect Dis. 149(4) (1984) pp. 489–494. [DOI] [PubMed] [Google Scholar]
- [14].Winter J, Morris GN, O’Rourke-Locascio S, Bokkenheuser VD, Mosbach EH, Cohen BI, Hylemon PB. Mode of action of steroid desmolase and reductases synthesized by Clostridium“scindens” (formerly Clostridium strain 19). J Lipid Res 25(10) (1984) pp. 1124–1131. [PubMed] [Google Scholar]
- [15].Krafft AE, Winter J, Bokkenheuser VD, Hylemon PB. Cofactor requirements of steroid-17,20-desmolase and 20alpha-hydroxysteroid dehydrogenase activities in cell extracts of Clostridium scindens. J Steroid Biochem 28(1) (1987) pp. 49–54. [DOI] [PubMed] [Google Scholar]
- [16].Krafft AE, Winter J, Hylemon PB. Purification and characterization of 20alpha-hydroxysteroid dehydrogenase from Clostridium scindens. J Bacteriol 171(6) (1989) pp. 2925–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bokkenheuser VD, Winter J, Morris GN, Locascio S. 1986. Steroid desmolase synthesis by Eubacterium desmolans and Clostridium cadavaris. Appl Environ Microbiol 52(5) (1986) pp. 1153–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Nabarro JDN, Moxham A, Walker G, Slater JDH. Rectal hydrocortisone. Br Med J. 2(5039) (1957) pp. 272–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wade AP, Slater JDM, Kellie AE, Holliday ME. Urinary excretion of 17-ketosteroids following rectal infusion of cortisol. J Clin Endocrinol Metab 19(4) (1959) pp. 444–453. [DOI] [PubMed] [Google Scholar]
- [20].Ly LK, Rowles JL, Devendran S, Erdman JW Jr., Ridlon JM. Desmolase (DesAB) activity from gut and urinary microbes forms 11-oxy-androgens from glucocorticoids in vitro. Congress on Gastrointestinal Function, Chicago Illinois, April (2019). [Google Scholar]
- [21].Zimmermann M, Zimmermann-Kogadeeva M. Wegmann R, Goodman AL. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 570(7762) (2019) pp. 462–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: Architecture and Applications. BMC Bioinformatics 10 (2009) 421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliams H, Remmert M, Söding J, Thompson JD, Higgins DG. Fast, Scalable Generation of High-Quality Protein Multiple Sequence Alignments Using Clustal Omega. Molecular Systems Biology 7 (2011) pp. 539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Price MN, Dehal PS, Arkin AP. 2010. FastTree 2 – Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 5(3) (2010) e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Huson DH, Scornavacca C. Dendroscope 3: An Interactive Tool for Rooted Phylogenetic Trees and Networks. Systematic Biology 61(6) (2012) pp. 1061–1067. [DOI] [PubMed] [Google Scholar]
- [26].Morris DJ, Ridlon JM. 2017. Glucocorticoids and gut bacteria: “The GALF Hypothesis” in the metagenomic era. Steroids 125 (2017) 1–13. [DOI] [PubMed] [Google Scholar]
- [27].Spanogiannopoulos P, Bess EN, Carmody RN, Turnbaugh PJ. The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat Rev Microbiol 14(5) (2016) pp. 273–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 47(2) (2006) pp. 241–59. [DOI] [PubMed] [Google Scholar]
- [29].Wells JE, Berr F, Thomas LA, Dowling RH, Hylemon PB. Isolation and characterization of cholic acid 7alpha-dehydroxylating fecal bacteria from cholesterol gallstone patients. J Hepatol 32(1) (2000) pp. 4–10. [DOI] [PubMed] [Google Scholar]
- [30].Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H H, Kinnebrew M, Viale A, Littmann E, van den Brink MR, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 517(7533) (2015) pp. 205–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331(6015) (2011) pp.337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, Le A, Cowan TM, Nolan GP, Fischbach MA, Sonnenburg JL. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551(7682) (2017) pp. 648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Jeraldo P, Hernandez A, Nielsen HB, Chen X, White BA, Goldenfeld N, Nelson H, Alhquist D, Boardman L, Chia N. Capturing One of the Human Gut Microbiome’s Most Wanted: Reconstructing the Genome of a Novel Butyrate-Producing, Clostridial Scavenger from Metagenomic Sequence Data. Front Microbiol 7 (2016) pp. 783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lewis DA, Brown R, Williams J, White P, Jacobson SK, Marchesi JR, Drake MJ. The human urinary microbiome; bacterial DNA in voided urine of asymptomatic adults. Front Cell Infect Microbiol 3 (2013) pp. 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Shrestha E, White JR, Yu SH, Kulac I, Ertunc O, De Marzo AM, Yegnasubramanian S, Mangold LA, Partin AW, Sfanos KS. Profiling the Urinary Microbiome in Men with Positive versus Negative Biopsies for Prostate Cancer. J Urol. 199(1) (2018) pp. 161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gilbert S, Hood L, Seah SYK. Characterization of an Aldolase Involved in Cholesterol Side Chain Degradation in Mycobacterium tuberculosis. J Bacteriol 200(2) (2017) pii: e00512–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Rahm MA, Sih CJ. Mechanisms of steroid oxidation by microorganisms. XI. Enzymatic cleavage of the pregnane side chain. J Biol Chem. 241(15) (1966) pp. 3615–3623. [PubMed] [Google Scholar]
- [38].Itagaki E. Studies on steroid monooxygenase from Cylindrocarpon radicicola ATCC 11011. Purification and characterization. J Biochem 99(3) (1986) pp. 815–824. [DOI] [PubMed] [Google Scholar]
- [39].Rege J, Rainey WE. 2012. The steroid metabolome of adrenarche. J Endrocrinol 214(2) (2012) pp. 133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].O’Reilly MW, Kempegowda P, Jenkinson C, Taylor AE, Quanson JL, Storbeck KH, Arlt W. 2017. 11-Oxygenated C19 Steroids Are the Predominant Androgens in Polycystic Ovary Syndrome. J Clin Endocrinol Metab 102(3) (2017) pp. 840–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Thackray VG. Sex, microbes, and polycystic ovary syndrome. Trends Endocrin Metab 30(1) (2019) pp. 54–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Golombos DM, Ayangbesan A, O’Malley P, Lewicki P, Barlow L, Barbieri CE, Chan C, DuLong C, Abu-Ali G, Huttenhower C, Scherr DS. The Role of Gut Microbiome in the Pathogenesis of Prostate Cancer: A Prospective, Pilot Study. Urology 111 (2018) pp. 122–128. [DOI] [PubMed] [Google Scholar]
- [43].du Toit T, Swart AC. The 11β-hydroxyandrostenedione pathway and C11-oxy C21 backdoor pathway are active in benign prostatic hyperplasia yielding 11keto-testosterone and 11keto-progesterone. J Steroid Biochem Mol Biol 196 (2019) pp.105497–105517. [DOI] [PubMed] [Google Scholar]
- [44].Bloem LM, Storbeck KH, Swart P, du Toit T, Schloms L, Swart AC. Advances in the analytical methodologies: Profiling steroids in familiar pathways-challenging dogmas. J Steroid Biochem Mol Biol 153 (2015) pp. 80–92. [DOI] [PubMed] [Google Scholar]
- [45].Trigunaite A, Dimo J, Jørgensen TN. Suppressive effects of androgens on the immune system. Cell Immunol 294(2) (2015) pp. 87–94. [DOI] [PubMed] [Google Scholar]
- [46].Gu S, Papadopoulou N, Gehring EM, Nasir O, Dimas K, Bhavsar SK, Föller M, Alevizopoulos K, Lang F, Stournaras C. Functional membrane androgen receptors in colon tumors trigger pro-apoptotic responses in vitro and reduce drastically tumor incidence in vivo. Mol Cancer 8 (2009) pp. 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Catalano MG, Pfeffer U, Raineri M, Ferro P, Curto A, Capuzzi P, Corno F, Berta L, Fortunati N. Altered expression of androgen-receptor isoforms in human colon-cancer tissues. Int J Cancer 86(3) (2000) pp. 325–30. [DOI] [PubMed] [Google Scholar]
- [48].Honour JW. Historical perspective: gut dysbiosis and hypertension. Physiol Genomics. 47(10) (2015) pp. 443–446. [DOI] [PubMed] [Google Scholar]
- [49].Jiang L, Yang S, Yin H, Fan X, Wang S, Yao B, Pozzi A, Chen X, Harris RC, Zhang MZ. Epithelial-specific deletion of 11β-HSD2 hinders Apcmin/+ mouse tumorigenesis. Mol Cancer Res 11(9) (2013) pp. 1040–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Rehman Y, Rosenberg JE. Abiraterone acetate: oral androgen biosynthesis inhibitor for treatment of castration-resistant prostate cancer. Drug Des Devel Ther. 6 (2012) pp. 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Rice MA, Malhotra SV, Stoyanova T. 2019. Second-Generation Antiandrogens: From Discovery to Standard of Care in Castration Resistant Prostate Cancer. Frontiers in Oncology 9 (2019) pp. 801. doi: 10.3389/fonc.2019.00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].de Launoit Y, Veilleux R, Dufour M, Simard J, Labrie F. Characteristics of the Biphasic Action of Androgens and of the Potent Antiproliferative Effects of the New Pure Antiestrogen EM-139 on Cell Cycle Kinetic Parameters in LNCaP Human Prostatic Cancer Cells. Cancer Research 51(19) (1991) pp. 5165–5170. [PubMed] [Google Scholar]
- [53].Veldscholte J, Berrevoets CA, Ris-Stalpers C, Kuiper GG, Jenster G, Trapman J, Brinkmann AO, Mulder E. The androgen receptor in LNCaP cells contains a mutation in the ligand binding domain which affects steroid binding characteristics and response to antiandrogens. J Steroid Biochem Mol Biol 41(3–8) (1992) pp. 665–9. [DOI] [PubMed] [Google Scholar]
- [54].Marion S, Studer N, Desharnais L, Menin L, Escrig S, Meibom A, Hapfelmeier S, Bernier-Latmani R. In vitro and in vivo characterization of Clostridium scindens bile acid transformations. Gut Microbes 10(4) (2019) pp. 481–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


