Abstract
Listeria monocytogenes (L. monocytogenes) is a ubiquitous foodborne pathogen that comprises 14 serotypes, of which serovar 4h is a novel serotype recently reported. Serovar 4h L. monocytogenes belonging to hybrid sub-lineage II exhibit hypervirulent features. Conventional biochemical tests and widely used PCR-based serogrouping schemes could not distinguish serovar 4h strains. In this study, we developed a new multiplex PCR assay for rapid detection of serotype 4h L. monocytogenes. Three primer pairs based on the target genes, LMxysn_1095, lmo1083, and smcL, were designed. The multiplex PCR results showed that serovar 4h strains could be specifically identified from all tested strains, including various L. monocytogenes serovars, Listeria spp., and other species. The detection limits of the multiplex PCR were 291 fg/μL for genomic DNA and 5.5 × 106 CFU/mL for bacterial suspension. Furthermore, pork meat artificially contaminated with serovar 4h L. monocytogenes in a concentration of 1.8 × 103–1.8 × 100 CFU/10 g were successfully detected within 10–16 h. These results demonstrate that the multiplex PCR with high specificity and sensitivity is applicable for the rapid detection of L. monocytogenes serotype 4h strains.
Keywords: Listeria monocytogenes, serovar 4h, multiplex PCR, identification, hypervirulent, somatic antigen, Listeria ivanovii
Introduction
Listeria monocytogenes (L. monocytogenes) is a Gram-positive, facultative intracellular foodborne pathogen that is widely distributed in the environment (Vazquez-Boland et al., 2001; Liu et al., 2005). The bacterium is capable of causing spontaneous abortion, sepsis, and meningoencephalitis with a mortality rate of 20–30% in human (Farber and Peterkin, 1991; Barbuddhe and Chakraborty, 2009). Immunocompromised patients, pregnant women, newborns, and the elderly are at a greater risk of listeriosis (Lomonaco et al., 2009; Radoshevich and Cossart, 2017). Although the minimum infection dose of L. monocytogenes is at least 100 colony forming units (CFU) per gram of food (Yang et al., 2007), considering the possible serious consequences, many countries, including the United States, Australia, and China, have issued a zero-tolerance policy for L. monocytogenes in ready-to-eat (RTE) food (Churchill et al., 2006; Tao et al., 2016).
Because of the genetically heterogeneous nature of L. monocytogenes, serotyping as the primary characterization in epidemiological surveillance has been widely accepted. Based on somatic (O) and flagellar (H) antigens, 13 serotypes of L. monocytogenes have been described and divided into four phylogenetic subdivisions, lineages I to IV (Seeliger and Höhne, 1979; Ward et al., 2008). Lineage I includes serotypes 1/2b, 3b, 4b, 4d, 4e, and 7; lineage II includes serotypes 1/2a, 1/2c, 3a, and 3c; lineage III and IV include serotypes 4a, 4ab, 4c, and atypical 4b (Roberts et al., 2006; Ward et al., 2008; Orsi et al., 2011). In recent years, many PCR-based methods have been developed and widely used for L. monocytogenes serotyping. Doumith et al. (2004) established a PCR-serogroup method to subdivide L. monocytogenes into four molecular groups: 1/2a-3a, 1/2b-3b-7, 1/2c-3c, and 4b-4d-4e. Chen and Knabel (2007) proposed a multiplex PCR, which could detect Listeria spp. L. monocytogenes serotypes 1/2a and 4b and epidemic clones I, II, and III simultaneously. Nho et al. (2015) distinguished L. monocytogenes high-risk serotypes 1/2a and 1/2c in lineage I from low-risk serotypes 3a and 3c using a multiplex PCR assay. Alía et al. (2019) developed a quadruplex real-time quantitative PCR (qPCR) method capable of differentiating the four most predominant L. monocytogenes serotypes (1/2a, 1/2b, 1/2c, and 4b).
Serovar 4h was reported by Yin et al. (2019), that is, the 14th serotype of L. monocytogenes belonging to a hybrid sublineage of the major lineage II (HSL-II). L. monocytogenes serovar 4h strains are atypical because they fail to ferment rhamnose and cannot be recognized by the commonly accepted PCR serotyping method of Doumith et al. (2004). Strikingly, L. monocytogenes serotype 4h isolates harbor pan-species virulence genes acquiring from truncated pathogenicity island 2 (LIPI-2) of Listeria ivanovii (L. ivanovii) and gene cluster associated with galactosylation of wall teichoic acid (WTA) from Lineage I serovar 4b strains, which leads to hypervirulent properties of the bacteria (Yin et al., 2019). Therefore, it is urgent to implement robust monitoring and develop a rapid method of detecting L. monocytogenes serotype 4h strains.
In this study, we establish a multiplex PCR to distinguish L. monocytogenes serotype 4h from other serogroups and evaluate the specificity, sensitivity, and stability of this method. Besides, natural-occurring L. monocytogenes isolates and artificially contaminated meat samples were tested by the PCR assay. The multiplex PCR will be a useful tool for detection of serotype 4h L. monocytogenes from different sources of samples.
Materials and Methods
Bacterial Strains and Culture Conditions
The strains used in this study were commercially available or isolated by our laboratory, including 143 strains of Listeria spp. and 14 strains of other species. All tested strains were grown in brain heart infusion (BHI) (BD) or Luria-Bertani medium (LB) (OXOID), respectively, at 37°C overnight under constant shaking.
Target Genes
To differentiate serotype 4h L. monocytogenes from other L. monocytogenes serogroups by multiplex PCR, we downloaded various L. monocytogenes serotypes and non-L. monocytogenes genome sequences from the GenBank database1. Three target genes, LMxysn_1095 (GenBank accession no. CP007583.1 segment 1113983-1115773), lmo1083 (Gene ID: 986257), and smcL (GenBank accession no. CP007583.1 segment 1245812-1246819), were selected by comparative genomics analysis. The corresponding primers were designed based on the conserved region of target genes by Primer Premier 5.0 software (PREMIER Biosoft International, Palo Alto, CA, United States) and determined by NCBI Primer-BLAST tool (Table 1).
TABLE 1.
Nucleotide sequence of the primers used in this study.
| Target gene | Primer sequence (5′-3′) | Product size | Protein encoded | Serotype specificity |
| LMxysn_1095 | F: AATACTTGGACAGACGGAACGC | 602 bp | Galactosyltransferase | Serotypes 4h, 4b, 4d, and 4e |
| R: TCATCTGGCTCTTTTAGAACCG | ||||
| lmo1083 | F: CACAAATGGTCTTGACGGGG | 390 bp | L-rhamnosyl transferase | Serotypes 1/2, 3, and 7 and Listeria seeligery (1/2b) |
| R: TTTGCGCGTGATTTTAGTGG | ||||
| smcL | F: CACAGACCATTGTGGTGACTTG | 889 bp | Sphingomyelinase C | Serotype 4h and L. ivanovii |
| R: CGGTGCTTTCATTTTTTTACTC |
F, forward primer; R, reverse primer.
Genomic DNA Extraction
The extraction of Listeria genomic DNA was conducted by an optimized boiling lysis method. Bacteria culture (0.5–1 × 109 CFU) was rinsed with phosphate-buffered saline and then centrifuged at 8000 rpm for 2 min. The pellet was mixed thoroughly with 88 μL of sterile distilled water and 10 μL of lysozyme (20 mg/mL) and incubated at 37°C for 10 min; after that, 1 μL of protease K (20 mg/mL) and 1 μL of RNase A (10 mg/mL) were added and incubated at 58°C for 10 min. Finally, the suspension was heated at 95°C for 5 min and centrifuged at 12,000 rpm for 2 min. The supernatant was used as the DNA template for multiplex PCR. The concentration of genomic DNA was measured by a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, United States).
Multiplex PCR
The multiplex PCR mixture contained 12.5 μL of 2× Taq Master mix (Vazyme Biotech Co., Ltd., China), 0.4 μM of each primer, bacterial genomic DNA, and ultrapure water to fill the final volume to 25 μL. The PCR procedure consisted of an initial denaturation step at 95°C for 5 min; 30 cycles of quantification at 95°C for 0.5 min, 55°C for 0.5 min, and 72°C for 1 min, and a final cycle at 72°C for 10 min. The amplicons were separated on 1% agarose gels (ethidium bromide dyed) in 1× TAE buffer and visualized using a Gel Doc XR system (Bio-Rad, Hercules, CA, United States).
Specificity Evaluation of Multiplex PCR
The specificity of multiplex PCR for detection of serovar 4h L. monocytogenes was evaluated. The genomic DNA was extracted from 14 strains of L. monocytogenes (including three serovar 4h strains) and 14 strains of other species (Table 2), and 1 μL of genomic DNA was used as the PCR template.
TABLE 2.
Listeria spp. and other strains used in this study for multiplex PCR specificity evaluation.
| Species (number of isolates) | Strain | Serotype | PCR results (LMxysn_1095/lmo1083/smcL) |
| L. monocytogenes (14) | XYSN | 4h | +/−/+ |
| L. monocytogenes (14) | 15LG | 4h | +/−/+ |
| L. monocytogenes (14) | 16E | 4h | +/−/+ |
| L. monocytogenes (14) | EGD-e | 1/2a | −/+/− |
| L. monocytogenes (14) | LmBJ113 | 1/2b | −/+/− |
| L. monocytogenes (14) | LmBJ114 | 1/2c | −/+/− |
| L. monocytogenes (14) | LmBJ115 | 3a | −/+/− |
| L. monocytogenes (14) | LmBJ117 | 3c | −/+/− |
| L. monocytogenes (14) | LmBJ118 | 4a | −/−/− |
| L. monocytogenes (14) | LmBJ119 | 4ab | −/−/− |
| L. monocytogenes (14) | NTSN | 4b | +/−/− |
| L. monocytogenes (14) | LmBJ121 | 4d | +/−/− |
| L. monocytogenes (14) | LmBJ122 | 4c | −/−/− |
| L. monocytogenes (14) | LmBJ123 | 7 | −/+/− |
| L. ivanovii (2) | YZU0805 | – | −/−/+ |
| LIV ZJU | – | −/−/+ | |
| Listeria innocua (1) | LBJ131 | – | −/−/− |
| Listeria grayi (1) | LBJ132 | – | −/−/− |
| Listeria seeligery (1) | LBJ133 | – | −/+/− |
| Listeria welshimeri (1) | LBJ137 | – | −/−/− |
| S. Enteritidis (1) | C50041 | – | −/−/− |
| Salmonella Pullorum (1) | S06004 | – | −/−/− |
| Salmonella Typhimurium (1) | – | – | −/−/− |
| Vibrio parahaemolyticus (1) | – | – | −/−/− |
| E. coli (1) | ATCC25922 | – | −/−/− |
| Staphylococcus aureus (1) | – | – | −/−/− |
| Campylobacter jejuni (1) | – | – | −/−/− |
| Campylobacter coli (1) | – | – | −/−/− |
Detection Limits Evaluation of Multiplex PCR
For detection limit evaluation, genomic DNA of serovar 4h L. monocytogenes strain XYSN was extracted and serially diluted 10-fold from 291 ng/μL to 291 ag/μL. Meanwhile, bacteria suspension of the strain XYSN was serially diluted 10-fold from 5.5 × 109 to 5.5 × 100 CFU/mL, and 1 μL of genomic DNA and 5 μL of bacteria suspension as the PCR template were added to the PCR mixture, respectively. The detection limits of the multiplex PCR were judged by electrophoresis.
Stability Evaluation of Multiplex PCR
Stability evaluation of multiplex PCR was conducted, referring to the methodology by Tao et al. (2016). Simply, serotype 4h L. monocytogenes strain XYSN, Escherichia coli (E. coli) strain ATCC25922, and Salmonella Enteritidis (S. Enteritidis) strain C50041 were cultured at 37°C overnight and serially 10-fold diluted. The concentration of serotype 4h L. monocytogenes was set at 104 CFU/mL and the other two strains were prepared from 104 to 109 CFU/mL. The bacteria culture of serotype 4h L. monocytogenes was mixed with E. coli and S. Enteritidis in ratios of 1:1, 1:10, 1:103, and 1:105. Genomic DNA of each mixture was extracted, and 3 μL of genomic DNA were used as the template for multiplex PCR.
Listeria monocytogenes Isolates Testing
Naturally contaminated samples between September 2018 to September 2019 were collected from goat farms, slaughterhouses, and markets in Jiangsu, China, according to the testing methodology for Listeria species or L. monocytogenes in environmental samples (Anonymous, 2015). Briefly, all samples were incubated in UVM broth (BD) at 30°C for 24 h. Then, the UVM enrichment (100 μL) was transferred to Fraser broth (BD) at 37°C for 24 h. Afterward, the culture was streaked onto CHROMagarTM plates and incubated at 37°C for 36 h. Colonies with a blue/green sheen and opaque halos were purified on BHI agar plates and confirmed by VITEK-2 system (bioMérieux, France). To evaluate the efficacy of the multiplex PCR for identifying serotype 4h strains, all L. monocytogenes isolates were further subjected to the developed multiplex PCR assay. Meanwhile, the serovar of isolates was determined by traditional slide agglutination test (Denka-Seiken Co., Ltd., Japan). The results obtained by multiplex PCR and the agglutination test were compared according to ISO 16140:2003 (Anonymous, 2003).
Artificial Contamination Experiment
The artificial contamination experiment was performed based on the procedure by Sheng et al. (2018). Briefly, L. monocytogenes serotype 4h strain XYSN was cultured overnight at 37°C and then prepared at different concentrations ranging from 101 to 105 CFU/mL. The raw pork was fully sterilized by UV for half an hour in advance. Each dilution of bacteria (100 μL) was sprayed separately on the upper surface of 10 g meat (around 10 cm2). When the bacteria suspension on the surface was completely dry, the artificially contaminated pork was put into sampling bags containing 90 mL BHI medium and incubated at 37°C for 4, 6, 8, 10, and 12 h. Genomic DNA were extracted from 1 mL of BHI medium and detected by multiplex PCR. The experiment was repeated three times.
Results
Designing of Primers
LMxysn_1095 and lmo1083 are somatic antigen-related genes involved in glycosylation modification of the WTA in L. monocytogenes (Carvalho et al., 2015; Yin et al., 2019). Comparative genomics analysis revealed that LMxysn_1095 presents in L. monocytogenes serovars 4h, 4b, 4d, and 4e, and lmo1083 presents in serovars 1/2, 3, and 7 as well as Listeria seeligery (L. seeligery, 1/2b). smcL is one of the virulence factors located on the LIPI-2 of L. ivanovii (Rodríguez-Lázaro et al., 2010; Wang et al., 2014). Genome of serotype 4h L. monocytogenes carry a partial sequence from LIPI-2 of L. ivanovii, which contains the smcL gene. Therefore, three primer pairs were designed to distinguish L. monocytogenes serotype 4h from other serovars, specifically targeting LMxysn_1095, lmo1083, and smcL (Table 1).
Specificity of Multiplex PCR
Twenty-eight bacteria strains were used to detect the specificity of multiplex PCR. The results showed that strains of L. monocytogenes serotype 4h generated two amplicons of 602 and 889 bp for the LMxysn_1095 and smcL, respectively. Strains of L. monocytogenes serovars 4b and 4d amplified the 602 bp band for LMxysn_1095. L. ivanovii generated the 889 bp band for smcL. Strains of L. monocytogenes serovars 1/2a, 1/2b, 1/2c, 3a, 3c, and 7 and L. seeligery generated the 390 bp band for lmo1083. The remaining strains presented no amplicon (Table 2). It was concluded that the multiplex PCR can specifically differentiate serovar 4h L. monocytogenes from other strains through the characteristic PCR band patterns.
Detection Limits of Multiplex PCR
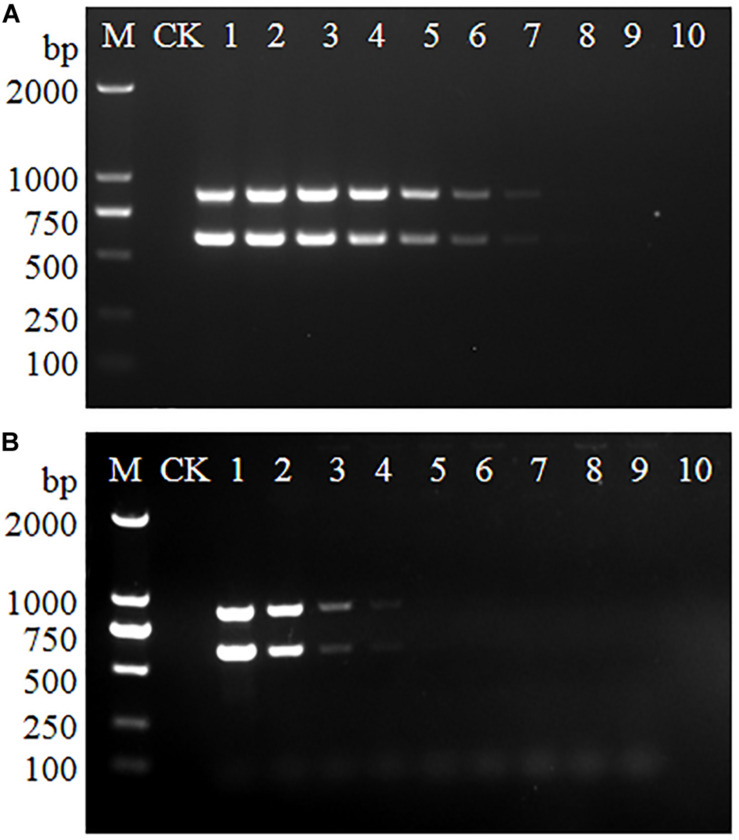
To determine the detection limits of the multiplex PCR, serially diluted genomic DNA and bacteria suspension of serotype 4h L. monocytogenes were used as the PCR template. As shown in Figures 1A,B, the 602 and 889 bp bands could be observed when the genomic DNA concentration ranged from 291 ng/μL to 291 fg/μL and the bacteria suspension concentration decreased from 5.5 × 109 to 5.5 × 106 CFU/mL. The results indicate that at least 291 fg/μL of genomic DNA or 5.5 × 106 CFU/mL of bacteria suspension are needed for multiplex PCR detection of serovar 4h L. monocytogenes.
FIGURE 1.
Detection limits of multiplex PCR for L. monocytogenes serotype 4h. The genomic DNA (A) and bacteria suspension (B) of serotype 4h L. monocytogenes were used as the PCR templates. Lane M: DL2000 DNA ladder; Lane CK: negative control. (A) Lanes 1–10: 291 ng/μL, 29.1 ng/μL, 2.91 ng/μL, 291 pg/μL, 29.1 pg/μL, 2.91 pg/μL, 291 fg/μL, 29.1 fg/μL, 2.91 fg/μL, 291 ag/μL. (B) Lanes 1–10: 5.5 × 109, 5.5 × 108, 5.5 × 107, 5.5 × 106, 5.5 × 105, 5.5 × 104, 5.5 × 103, 5.5 × 102, 5.5 × 101, 5.5 × 100 CFU/mL.
Stability of Multiplex PCR
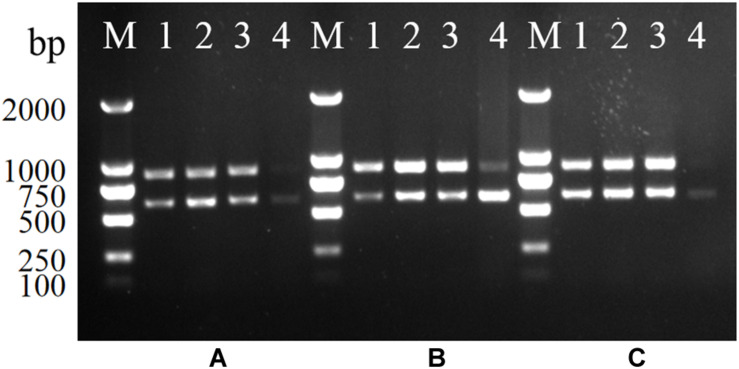
Serovar 4h L. monocytogenes was mixed with E. coli, S. Enteritidis, and two bacteria co-cultures at the ratios of 1:1, 1:10, 1:103, and 105. The genomic DNA of each mixture was extracted and used as PCR templates. As shown in Figure 2, two specific bands were generated for each mixture, indicating that the multiplex PCR was not affected by other interference strains present in a multicomponent sample.
FIGURE 2.
Stability of the multiplex PCR for L. monocytogenes serotype 4h. (A–C): serotype 4h L. monocytogenes were mixed with S. Enteritidis (A), E. coli (B), and two bacteria co-cultures (C). Lanes 1–4: bacteria suspension of serotype 4h mixing with other bacteria in ratios of 1:1, 1:10, 1:103, and 1:105; Lane M: DL2000 DNA ladder.
Identification of Listeria monocytogenes Isolates
All 129 L. monocytogenes isolates from food and environmental samples were identified by multiplex PCR and serotyped by slide agglutination in parallel (Table 3 and Supplementary Table S1). A traditional slide agglutination test showed that the 129 isolates belonged to six serovars (1/2a, 1/2b, 1/2c, 3c, 4b, and 4h), including two serotype 4h strains. The PCR results suggest that two isolates generated two specific bands for LMxysn_1095 and smcL, which were identified as serotype 4h L. monocytogenes. These results were consistent with that of the slide agglutination test. All serovar 4h isolates were identified using the multiplex PCR method, and no amplification was found for non-serovar 4h strains, indicating that the relative sensitivity and specificity of the multiplex PCR were both 100%.
TABLE 3.
L. monocytogenes serotype 4 strains isolated from diverse sources.
| Isolate no. | Serotype | PCR results (LMxysn_1095/lmo1083/smcL) | Sample types | Sources |
| P161020-B18 | 4h | +/−/+ | Environment | Goat farm |
| P161020-D14 | 4h | +/−/+ | Environment | Goat farm |
| Lm03032001 | 4b | +/−/− | Raw pork meat | Market |
| Lm03061801 | 4b | +/−/− | Raw pork meat | Market |
| Lm03060104 | 4b | +/−/− | Environment | Market |
| Lm03061102 | 4b | +/−/− | Environment | Market |
| Lm03061104 | 4b | +/−/− | RTE meat | Market |
| Lm03061807 | 4b | +/−/− | RTE meat | Market |
| Lm03061808 | 4b | +/−/− | RTE meat | Market |
Detection of Artificially Contaminated Meat
The multiplex PCR was applied to detect serotype 4h L. monocytogenes in artificially contaminated pork. The initial bacterial inoculation dose per 10 g of meat was 1.8 × 104, 1.8 × 103, 1.8 × 102, 1.8 × 101, and 1.8 × 100 CFU. Afterward, all samples were incubated at 37°C in BHI medium for 4, 6, 8, 10, and 12 h. As shown in Table 4 and Supplementary Figure S1, with the prolongation of enrichment time, the detection limit of the PCR assay gradually improved. After 6, 8, 10, and 12 h of enrichment, the detection limit of multiplex PCR was 1.8 × 103, 1.8 × 102, 1.8 × 101, and 1.8 × 100 CFU/10 g, respectively. Considering the time required for other operation procedures (genomic DNA extraction, PCR, and electrophoresis), it can be speculated that meat samples with the contamination dose less than 1.8 × 103 CFU/10 g can be detected within 10–16 h.
TABLE 4.
Detection of artificially contaminated pork by multiplex PCR.
| Initial inoculum | Multiplex PCR results of | ||||
| (CFU/10 g) | different enrichment time |
||||
| 4 h | 6 h | 8 h | 10 h | 12 h | |
| 1.8 × 104 | − | + | + | + | + |
| 1.8 × 103 | − | + | + | + | + |
| 1.8 × 102 | − | − | + | + | + |
| 1.8 × 101 | − | − | − | + | + |
| 1.8 × 100 | − | − | − | − | + |
| 0 | − | − | − | − | − |
+, positive PCR result; −, negative PCR result.
Discussion
Listeria monocytogenes is the main etiology of the fatal disease listeriosis, which poses a serious threat to human health. The vast majority of listeriosis clinical cases were caused by L. monocytogenes serotypes 1/2a, 1/2b, and 4b (Vines and Swaminathan, 1998). Hypervirulent serotype 4h L. monocytogenes were discovered and reported from goat listeriosis outbreaks and environments. Newly named serotype 4h can be identified by slide agglutination that reacts positively to O- antigens V/VI and VI and to H-antigens A and B (Yin et al., 2019), but this method is limited by the high cost of antisera and only if it is the pure colony. Thus, establishment of a rapid detection method for serotype 4h L. monocytogenes is of great significance for effective prevention and control of the pathogen.
In this study, a multiplex PCR method comprising three primer pairs, LMxysn_1095, lmo1083, and smcL, was developed for detection of L. monocytogenes serotype 4h. The sensitivity of the multiplex PCR was evaluated using both serovar 4h L. monocytogenes pure culture and their genomic DNA. It was found that the detection limit of multiplex PCR using genomic DNA as the template is close to previously reported PCR methods for L. monocytogenes subtyping (Tao et al., 2016; Razei et al., 2017; Sheng et al., 2018). However, the detection limit from bacteria culture was lower than genomic DNA because of the impact of the cell wall, nuclease, and metabolites in bacterial suspension (Sheng et al., 2018). Low contamination levels in samples make it difficult to detect target pathogens; hence, enrichment steps were applied in the process of L. monocytogenes detection (Garrido et al., 2013; Ding et al., 2017; Parichehr et al., 2019). Target bacteria and background flora, such as E. coli and Salmonella, coexist in enrichment medium, which may interfere with the stability of detection methods. Nevertheless, the multiplex PCR was not affected by existence of other strains even though the genomic DNA concentration of background flora was significantly higher than the target bacteria.
Contamination of meat products with L. monocytogenes is a public health concern because many human listeriosis outbreaks have been linked to contaminated meat products (Jacquet et al., 1995; Loncarevic et al., 1997; Jensen et al., 2016; Anna et al., 2018). Thus, rapid detection of L. monocytogenes from contaminated meat, etc., provides an effective guarantee for the food safety system. Conventional detection methods for L. monocytogenes are laborious and time-consuming, including procedures of primary isolation with enrichment and selective medium, biochemical analysis and serological tests, which take up to 5–10 days (Grady et al., 2008; Vanegas et al., 2009; Frece et al., 2010). In the artificial contamination experiment of this study, the multiplex PCR could detect serotype 4h L. monocytogenes in meat with a contaminated concentration of less than 1.8 × 103 CFU/10 g within 10–16 h, which dramatically shortened the detection time (10–16 h vs. 5–10 days). It is noteworthy that the optimized boiling lysis method was applied to extract genomic DNA directly from enrichment medium as the PCR template. The full process could be accomplished in 25–30 min, which is more proper for rapid extraction of genomic DNA from large-scale samples than the conventional extraction method by kit (25–30 min vs. 3 h).
In general, the multiplex PCR could detect serovar 4h L. monocytogenes in artificially contaminated meat rapidly and accurately, which is expected to be adopted for laboratory or clinical tests of naturally contaminated samples. Up to now, the collection of L. monocytogenes serovar 4h isolates was limited; five 4h strains have been isolated from goat listeriosis and environment. More L. monocytogenes serovar 4h strains are possible to be detected with the multiplex PCR assay established in this study.
Conclusion
In conclusion, a multiplex PCR method comprising three primer pairs, LMxysn_1095, lmo1083, and smcL, was proposed in this study. PCR results demonstrate that the multiplex PCR with high specificity, sensitivity, and stability is applicable to detect L. monocytogenes serotype 4h rapidly and effectively. Our research affords a useful detection and identification technique to enhance surveillance for the emergence of hypervirulent serovar 4h L. monocytogenes.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
YY and XJ designed the experiments. YF, HY, SC, and XS collected all strains, performed the PCR assays, and analyzed the results. YY, XJ, and YF wrote the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by the National Key R&D Program of China (2017YFC1601203, 2017YFC1601201, and 2018YFD0500502), the National Natural Science Foundation of China (No. 31472193), Key Research and Development Program (Modern Agriculture) Project of Jiangsu Province (BE2017341), Jiangsu Agricultural Science and Technology Independent Innovation Funds [CX(16)1028], the Science and Technology Support Program of Jiangsu Province (BE2012367), and the Priority Academic Development Program of Jiangsu Higher Education Institutions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01309/full#supplementary-material
Detection results of pork meat contaminated with serotype 4h L. monocytogenes by multiplex PCR. Lanes 1–6: the initial bacterial inoculation dose was 0, 1.8×100, 1.8×101, 1.8×102, 1.8×103, and 1.8×104 CFU, respectively; Lane M: DL2000 DNA ladder.
References
- Alía A., Andrade M. J., Córdoba J. J., Martín I., Rodríguez A. (2019). Development of a multiplex real-time PCR to differentiate the four major Listeria monocytogenes serotypes in isolates from meat processing plants. Food Microbiol. 87:103367. 10.1016/j.fm.2019.103367 [DOI] [PubMed] [Google Scholar]
- Anna D., Michela S., Giuliana B., Aniela A. V., Francesco P. (2018). A severe outbreak of listeriosis in central Italy with a rare pulsotype associated with processed pork products. J. Med. Microbiol. 67 1351–1360. 10.1099/jmm.0.000785 [DOI] [PubMed] [Google Scholar]
- Anonymous (2003). EN ISO 16140:2003: Microbiology of Food and Animal Feeding Stuffs- Protocol for the Validation of Alternative Methods. Geneva: International Organization for Standardization. [Google Scholar]
- Anonymous (2015). Testing Methodology for Listeria species or L. monocytogenes in Environmental Samples, Version 1. Maryland: U.S Food and Drug Administration Center for Food Safety and Applied Nutrition. [Google Scholar]
- Barbuddhe S. B., Chakraborty T. (2009). Listeria as an enteroinvasive gastrointestinal pathogen. Curr. Top Microbiol. Immunol. 337 173–195. 10.1007/978-3-642-01846-6_6 [DOI] [PubMed] [Google Scholar]
- Carvalho F., Atilano M. L., Pombinho R., Covas G., Gallo R. L., Filipe S. R., et al. (2015). L-Rhamnosylation of Listeria monocytogenes wall teichoic acids promotes resistance to antimicrobial peptides by delaying interaction with the membrane. PLoS Pathog. 11:e1004919. 10.1371/journal.ppat.1004919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Knabel S. J. (2007). Multiplex PCR for simultaneous detection of bacteria of the genus Listeria, Listeria monocytogenes, and major serotypes and epidemic clones of L. monocytogenes. Appl. Environ. Microbiol. 73 6299–6304. 10.1128/AEM.00961-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill R. L. T., Lee H., Hall J. C. (2006). Detection of Listeria monocytogenes and the toxin listeriolysin O in food. J. Microbiol. Meth. 64 141–170. 10.1016/j.mimet.2005.10.007 [DOI] [PubMed] [Google Scholar]
- Ding T., Suo Y., Zhang Z., Liu D., Ye X., Chen S., et al. (2017). A Multiplex RT-PCR Assay for S. aureus, L. monocytogenes, and Salmonella spp. Detection in raw milk with pre-enrichment. Front. Microbiol. 8:989. 10.3389/fmicb.2017.00989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumith M., Buchrieser C., Glaser P., Jacquet C., Martin P. (2004). Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 42 3819–3822. 10.1128/JCM.42.8.3819-3822.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber J. M., Peterkin P. I. (1991). Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55 476–511. 10.1128/mmbr.55.3.476-511.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frece J., Markov K., Čvek D., Kolarec K., Delaš F. (2010). Comparison of conventional and molecular methods for the routine confirmation of Listeria monocytogenes in milk products produced domestically in Croatia. J. Dairy Res. 77 112–116. 10.1017/s0022029909990379 [DOI] [PubMed] [Google Scholar]
- Garrido A., Chapela M. J., Román B., Fajardo P., Lago J., Vieites J. M., et al. (2013). A new multiplex real-time pcr developed method for Salmonella spp. and Listeria monocytogenes detection in food and environmental samples. Food Control 30 76–85. 10.1016/j.foodcont.2012.06.029 [DOI] [Google Scholar]
- Grady J. O., Sedano-Balbas S., Maher M., Smith T., Barry T. (2008). Rapid real-time PCR detection of Listeria monocytogenes in enriched food samples based on the ssrA gene, a novel diagnostic target. Food Microbiol. 25 75–84. 10.1016/j.fm.2007.07.007 [DOI] [PubMed] [Google Scholar]
- Jacquet C. B., Catimel R., Brosch R., Buchreiser C., Dehaumont P., Goulet V., et al. (1995). Investigations related to the epidemic strain involved in French listeriosis outbreak in 1992. Appl. Environ. Microbiol. 61 2242–2246. 10.1002/bit.260460514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen A. K., Nielsen E. M., Bjorkman J. T., Jensen T., Muller L., Persson S., et al. (2016). Whole-genome sequencing used to investigate a nationwide of listeriosis caused by Ready-to-eat delicatessen meat. Denmark, 2014. Clin. Infect. Dis. 63 64–70. 10.1093/cid/ciw192 [DOI] [PubMed] [Google Scholar]
- Liu D., Lawrence M. L., Ainsworth A. J., Austin F. W. (2005). Comparative assessment of acid, alkali and salt tolerance in Listeria monocytogenes virulent and avirulent strains. FEMS Microbiol. Lett. 243 373–378. 10.1016/j.femsle.2004.12.025 [DOI] [PubMed] [Google Scholar]
- Lomonaco S., Decastelli L., Nucera D., Gallina S., Manila Bianchi D., Civera T. (2009). Listeria monocytogenes in Gorgonzola: subtypes, diversity and persistence overtime. Int. J. Food Microbiol. 128 516–520. 10.1016/j.ijfoodmicro.2008.10.009 [DOI] [PubMed] [Google Scholar]
- Loncarevic S., Danielsson-Tham M. L., Martensson L., Ringner A., Runehagen A., Tham W. (1997). A case of listeriosis in Sweden. Lett. Appl. Microbiol. 24 65–68. 10.1046/j.1472-765X.1997.00353.x [DOI] [PubMed] [Google Scholar]
- Nho S. W., Abdelhamed H., Reddy S., Karsi A., Lawrence M. L. (2015). Identification of high-risk, Listeria monocytogenes, serotypes in lineage i (serotype 1/2a, 1/2c, 3a and 3c) using multiplex PCR. J. Appl. Microbiol. 119 845–852. 10.1111/jam.12876 [DOI] [PubMed] [Google Scholar]
- Orsi R. H., Bakker H. C. D., Wiedmann M. (2011). Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 301 79–96. 10.1016/j.ijmm.2010.05.002 [DOI] [PubMed] [Google Scholar]
- Parichehr M., Mohammad K., Abbas D., Mehdi K. (2019). Developing a multiplex real-time PCR with a new pre-enrichment to simultaneously detect four foodborne bacteria in milk. Future Microbiol. 14 885–898. 10.2217/fmb-2019-0044 [DOI] [PubMed] [Google Scholar]
- Radoshevich L., Cossart P. (2017). Listeria monocytogenes: towards a complete picture of its physiology and pathogenesis. Nat. Rev. Microbiol. 16 32–46. 10.1038/nrmicro.2017.126 [DOI] [PubMed] [Google Scholar]
- Razei A., Sorouri R., Mousavi S. L., Nazarian S., Aghamollaei H. (2017). Presenting a rapid method for detection of Bacillus cereus, Listeria monocytogenes and Campylobacter jejuni in food samples. Iran. J. Basic Med. Sci. 20 1050–1055. 10.22038/IJBMS.2017.9275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Nightingale K., Jeffers G., Fortes E., Kongo J. M., Wiedmann M. (2006). Genetic and phenotypic characterization of Listeria monocytogenes lineage III. Microbiology 152 685–693. 10.1099/mic.0.28503-0 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Lázaro D., López-Enríquez L., Hernández M. (2010). Smcl as a novel diagnostic marker for quantitative detection of Listeria ivanovii in biological samples. J. Appl. Microbiol. 109 863–872. 10.1111/j.1365-2672.2010.04712.x [DOI] [PubMed] [Google Scholar]
- Seeliger H. P. R., Höhne K. (1979). Chapter ii serotyping of Listeria monocytogenes and related species. Method Microbiol. 13 31–49. 10.1016/S0580-9517(08)70372-6 [DOI] [Google Scholar]
- Sheng J., Tao T., Zhu X., Bie X., Lv F., Zhao H., et al. (2018). A multiplex PCR detection method for milk based on novel primers specific for Listeria monocytogenes 1/2a serotype. Food Control 86 183–190. 10.1016/j.foodcont.2017.11.028 [DOI] [Google Scholar]
- Tao T., Chen Q., Bie X., Lu F., Lu Z. (2016). Investigation on prevalence of Listeria spp. and listeria monocytogenes in animal-derived foods by multiplex PCR assay targeting novel genes. Food Control 73 704–711. 10.1016/j.foodcont.2016.09.026 [DOI] [Google Scholar]
- Vanegas M. C., Vásquez E., Martinez A. J., Rueda A. M. (2009). Detection of Listeria monocytogenes in raw whole milk for human consumption in Colombia by real-time PCR. Food Control 20 430–432. 10.1016/j.foodcont.2008.07.007 [DOI] [Google Scholar]
- Vazquez-Boland J. A., Kuhn M., Berche P., Chakraborty T., Dominguez-Bernal G., Goebel W., et al. (2001). Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14 584–640. 10.1128/CMR.14.3.584-640.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vines A., Swaminathan B. (1998). Identification and characterization of nucleotide sequence differences in three virulence-associated genes of Listeria monocytogenes strains representing clinically important serotypes. Curr. Microbiol. 36 309–318. 10.1007/s002849900315 [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang Y., Xu H., Dai H., Meng S., Ye C. (2014). Rapid and sensitive detection of Listeria ivanovii by loop-mediated isothermal amplification of the smcl gene. PLoS One 9:e115868. 10.1371/journal.pone.0115868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward T. J., Ducey T. F., Usgaard T., Dunn K. A., Bielawski J. P. (2008). Multilocus genotyping assays for single nucleotide polymorphism-based subtyping of Listeria monocytogenes isolates. Appl. Environ. Microbiol. 74 7629–7642. 10.1128/AEM.01127-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Qu L., Wimbrow A. N., Jiang X., Sun Y. (2007). Rapid detection of Listeria monocytogenes by nanoparticle-based immunomagnetic separation and real-time pcr. Int. J. Food Microbiol. 118 132–138. 10.1016/j.ijfoodmicro.2007.06.019 [DOI] [PubMed] [Google Scholar]
- Yin Y., Yao H., Doijad S., Kong S., Shen Y., Cai X., et al. (2019). A hybrid sub-lineage of Listeria monocytogenes comprising hypervirulent isolates. Nat. Commun. 10 1–16. 10.1038/s41467-019-12072-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detection results of pork meat contaminated with serotype 4h L. monocytogenes by multiplex PCR. Lanes 1–6: the initial bacterial inoculation dose was 0, 1.8×100, 1.8×101, 1.8×102, 1.8×103, and 1.8×104 CFU, respectively; Lane M: DL2000 DNA ladder.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.