Abstract
Background
Some multifunctional cellular proteins, as the monocyte chemotactic protein-induced protein 1 (ZC3H12A/MCPIP1) and the cyclin-dependent kinase inhibitor CDKN1A/p21, are able to modulate the cellular susceptibility to the human immunodeficiency virus type 1 (HIV-1). Several studies showed that CDKN1A/p21 is expressed at high levels ex vivo in cells from individuals who naturally control HIV-1 replication (HIC) and a recent study supports a coordinate regulation of ZC3H12A/MCPIP1 and CDKN1A/p21 transcripts in a model of renal carcinoma cells. Here, we explored the potential associations between mRNA expression of ZC3H12A/MCPIP1 and CDKN1A/p21 in HIC sustaining undetectable (elite controllers–EC) or low (viremic controllers–VC) viral loads.
Results
We found a selective upregulation of ZC3H12A/MCPIP1 and CDKN1A/p21 mRNA levels in PBMC from HIC compared with both ART–suppressed and HIV–negative control groups (P≤ 0.02) and higher MCPIP1 and p21 proteins levels in HIC than in HIV-1 negative subjects. There was a moderate positive correlation (r ≥ 0.57; P ≤ 0.014) between expressions of both transcripts in HIC and in HIC combined with control groups. We found positive correlations between the mRNA level of CDKN1A/p21 with activated CD4+ T cells levels in HIC (r ≥ 0.53; P ≤ 0.017) and between the mRNA levels of both CDKN1A/p21 (r = 0.74; P = 0.005) and ZC3H12A/MCPIP1 (r = 0.58; P = 0.040) with plasmatic levels of sCD14 in EC. Reanalysis of published transcriptomic data confirmed the positive association between ZC3H12A/MCPIP1 and CDKN1A/p21 mRNA levels in CD4+ T cells and monocytes from disparate cohorts of HIC and other HIV-positive control groups.
Conclusions
These data show for the first time the simultaneous upregulation of ZC3H12A/MCPIP1 and CDKN1A/p21 transcripts in the setting of natural suppression of HIV-1 replication in vivo and the positive correlation of the expression of these cellular factors in disparate cohorts of HIV-positive individuals. The existence of a common regulatory pathway connecting ZC3H12A/MCPIP1 and CDKN1A/p21 could have a synergistic effect on HIV-1 replication control and pharmacological manipulation of these multifunctional host factors may open novel therapeutic perspectives to prevent HIV-1 replication and disease progression.
Keyword: HIV-1 controllers, Restriction factors, MCPIP1, p21, Immune activation
Background
Among the individuals infected by the human immunodeficiency virus type 1 (HIV-1), a rare group called HIV controllers (HIC) suppress viral replication in absence of antiretroviral therapy, maintaining RNA viral loads (VL) below the limit of detection (LOD; elite controllers, EC) or at low levels (> LOD and < 2000 copies/ml; viremic controllers, VC). Natural control of HIV-1 replication is probably a multifactorial feature that involves different combinations of host and viral factors [1].
Some intrinsic host proteins, termed restriction factors (RF), are components of the innate immune response [2, 3] that have the ability to cause a significant reduction in viral infectivity by interacting directly with the pathogen. RF are, generally, interferon (IFN)-stimulated genes (ISGs) [4] and several has been shown to tackle different stages of HIV life cycle [3], including some classical RF such the Apolipoprotein B mRNA-Editing enzyme, Catalytic polypeptide-like (APOBEC3G), the Bone Stromal Tumor protein 2 (BST2)/Tetherin, the Sterile Alpha Motif domain and HD domain-containing protein 1 (SAMHD1) [2], and others more recently characterized like the Myxovirus resistance protein 2 (Mx2), the Interferon-inducible transmembrane family proteins (IFITM1-3 members), and Schlafen 11 (SLFN11) [3]. The mRNA levels of some RF including SAMHD1, Theterin, IFITM1, Mx2 and SLFN11 were described to be elevated in peripheral blood mononuclear cells (PBMC) or CD4+ T cells of HIC compared to antiretroviral (ART)-suppressed and/or HIV-uninfected individuals [5–9], although with contrasting findings across different HIC cohorts.
Others host multifunctional proteins, not recognized as classical RF, are also able to modulate the cellular susceptibility to HIV-1 infection. The cyclin-dependent kinase (CDK) inhibitor p21, encoded by the CDKN1A gene, modulates multiple relevant processes of the immune system, including proliferation of activated/memory T cells, macrophage activation and inflammation [10–17]. This protein also indirectly limits the HIV-1 replication in vitro in various cellular systems by blocking the biosynthesis of dNTPs required for viral reverse transcription and by inhibiting the CDK9 activity required for HIV-1 mRNA transcription [18–23]. Several studies described that CDKN1A/p21 is expressed at high levels ex vivo in CD4+ T cells from HICs [21, 24–26] and that p21 mRNA levels are correlated with CD4+ T cell activation in EC, but not in other HIV-infected groups [5]. These pieces of evidence suggest that the inducibility of CDKN1A/p21 to immune activation is a singular characteristic of EC and may contribute to the natural control of HIV-1 replication in vivo.
The monocyte chemotactic protein–induced protein 1 (MCPIP1), encoded by the Zc3h12a gene, is another newly discovered host multifunctional modulator of immune response with antiviral activity [27]. MCPIP1 plays a critical role in the regulation of the inflammatory response and immune homeostasis and also blocks HIV-1 replication in vitro by promoting the viral mRNA degradation through its RNase activity, particularly in quiescent CD4+ T cells [27, 28]. In activated CD4+ T cells, ZC3H12A/MCPIP1 is rapidly degraded [28] after its cleavage by the mucosa-associated lymphoid-tissue lymphoma-translocation 1 (MALT1) protein [29, 30]. In activated macrophage cells, by contrast, MCPIP1 transcripts are induced by TLR ligands and pro-inflammatory cytokines (mainly, TNF-α, IL-1β, and CCL2/MCP-1), and its expression stimulates a negative feedback loop that attenuates the inflammatory state by decreasing its fundamental mediators [27, 31]. The expression level of ZC3H12A/MCPIP1 in HIC was never described before.
Interestingly, a recent study of human renal carcinoma cell line (Caki-1 cells) revealed that ZC3H12A/MCPIP1 overexpression reduces the cellular growth by increasing the levels of CDKN1A/p21 transcripts, along with other proteins involved in cell cycle progression/arrest, supporting a coordinate regulation of ZC3H12A/MCPIP1 and CDKN1A/p21 in that cell-line [32]. This evidence prompted us to ask whether the expression of ZC3H12A/MCPIP1 could be elevated and positively correlated with CDKN1A/p21 in the setting of natural control of HIV-1 infection. To test this hypothesis, we quantified the ex vivo expression of ZC3H12A/MCPIP1 and CDKN1A/p21 mRNAs in PBMC from HIC, ART-suppressed, and HIV-uninfected individuals of disparate cohorts.
Results
The ZC3H12A/MCPIP1 and CDKN1A/p21 mRNA and protein expression levels are upregulated in PBMC from HIC
Twenty-nine HIV-1 positive (21 HIC and 8 ART-suppressed) and 10 HIV-negative individuals were included in this cross-sectional study. Most HIV-positive (59%) and HIV-negative (60%) individuals were females and all individuals displayed CD4+ T cells counts above 500 cells/μl (Table 1). Although the EC subgroup showed a higher proportion of females (77%) than other groups, the difference was not statistically significant (Additional file 1: Table S1).
Table 1.
Main clinical and epidemiologic characteristics of individuals of this study
| Characteristics | HIC (n = 21) | ART-suppressed (n = 10) | HIV-1 negative (n = 8) | |
|---|---|---|---|---|
| EC (n = 13) | VC (n = 8) | |||
| Sex, no. (%) | ||||
| Female | 10 (77) | 3 (38) | 4 (50) | 6 (60) |
| Male | 3 (23) | 5 (62) | 4 (50) | 4 (40) |
| Age (years)ab | 45 (39–60) | 44 (39–47) | 47 (38–53) | 47 (36–51) |
| Study point | ||||
| Time since HIV-1 diagnosis (years)b | 9 (5.5–15) | 12.5 (7–16) | NA | – |
| CD4+ T cell (cells/μl)b | 1027 (834–1255) | 664 (563–1228) | 889 (678–1097) | 1043 (784–1581) |
| Plasma HIV RNA (copies/ml)b | < 50 | 641 (327–915) | < 40 | – |
| CD4/CD8 ratiob | 1.33 (1.24–1.61) | 0.91 (0.67–1.23) | 1.06 (0.73–1.5) | 1.69 (1.62–2.00) |
HIC HIV controllers, ART antiretroviral therapy, EC elite controllers, VC viremic controllers, NA not available
aAge at study point; bInterquartile ranges are shown in parenthesis
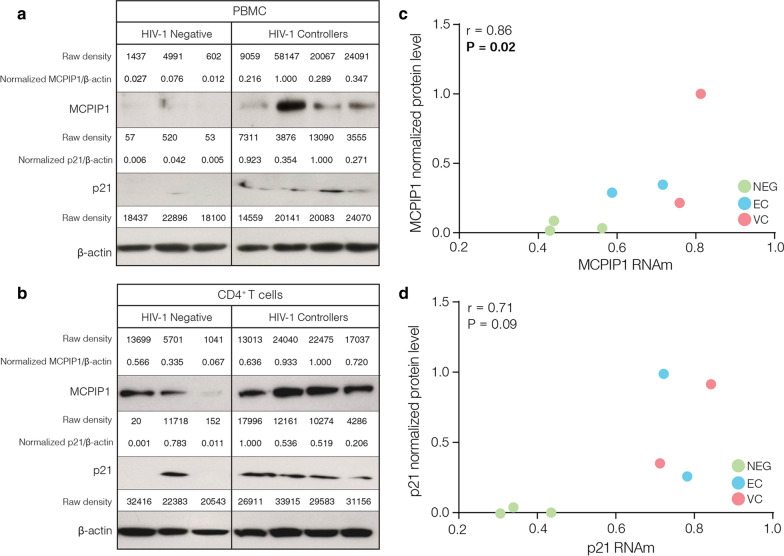
Analysis of the expression of multifunctional genes revealed a significant (P < 0.05) upregulation of both ZC3H12A/MCPIP1 (Fig. 1a) and CDKN1A/p21 (Fig. 1b) transcripts in PBMC from HIC compared to cells from ART-suppressed (1.68-fold increase and 1.63-fold increase, respectively) and HIV-negative (1.37-fold increase and 1.55-fold increase, respectively) individuals (Fig. 1). Analysis of the mRNA levels of several antiretroviral RF, by contrast, failed to reveal significant differences between the HIC and control groups, with the only exception of significant upregulation of IFITM1 in HIC respect to the HIV-negative group (Additional file 1: Figure S1). We also measured the expression level of MCPIP1 and CDKN1A proteins in PBMC and CD4+ T cells from a subset of four HIC (two EC and two VC) and three HIV-1 negative subjects (Additional file 1: Figure S2). Western blot analyses demonstrated that the median expression levels of MCPIP1 and p21 proteins in PBMC and CD4+ T cells from HIC were higher than in HIV-negative individuals (Fig. 2a, b). Moreover, we observed strong positive correlation between the mRNA and protein expression of ZC3H12A/MCPIP1 (r = 0.86, P = 0.024) and CDKN1A/p21 (r = 0.71, P = 0.09) in PBMC (Fig. 2c, d). These results support a selective upregulation of both ZC3H12A/MCPIP1 and CDKN1A/p21 mRNA levels in PBMC from our HIC cohort and further suggest that transcripts levels are correlated with levels of protein expression.
Fig. 1.
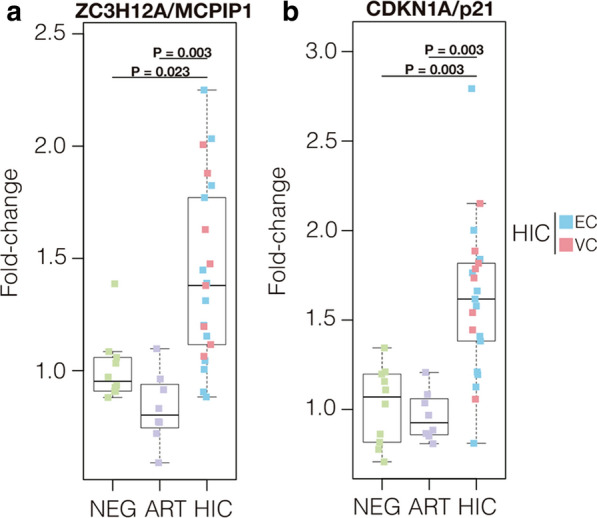
ZC3H12A/MCPIP1 and CDKN1A/p21 mRNA levels are upregulated in PBMC from HIC. Boxplots represent the interquartile and sample median (central solid black line) of the relative changes (fold-change values relative to the mean of HIV-1-uninfected (NEG) subjects) of ZC3H12A/MCPIP1 a and CDKN1A/p21 b expression comparing NEG and ART-suppressed subjects (ART) with HIV controllers (HIC). P-values < 0.05 were considered statistically significant
Fig. 2.
MCPIP1 and p21 protein levels are increased in HIC. The MCPIP1 and p21 proteins expression in PBMC a and CD4+ T cells b from HIV-negative and HIV controllers, was determined by western blot. The quantified MCPIP1 and p21 protein expression levels were normalized with β-actin protein levels and immunoblotting bands were quantified with ImageJ64 software. Correlation between normalized proteins and mRNA levels is shown to MCPIP1 c and p21 d. Correlation coefficients (Spearman’s ρ) are shown in the upper right corner of each graph. P-values < 0.05 were considered statistically significant
Positive correlation of ZC3H12A/MCPIP1 and CDKN1A/p21 mRNA expression in PBMC
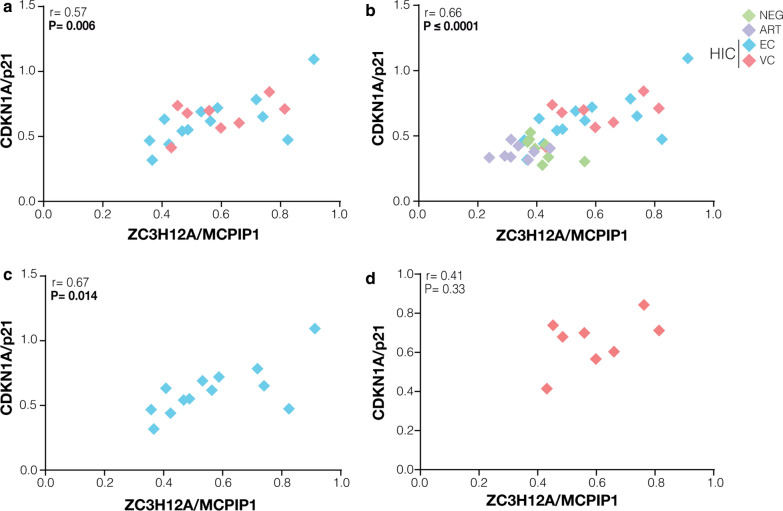
A recent study supports a coordinate regulation of ZC3H12A/MCPIP1 and CDKN1A/p21 in a human renal carcinoma cell line [32]. Consistent with this finding, we observed a significant positive correlation between the mRNA expression of ZC3H12A/MCPIP1 and CDKN1A/p21 in PBMC from HIC (r = 0.57; P = 0.006) (Fig. 3a). Positive correlations trends were also maintained when HIC were combined with control groups (r = 0.66; P ≤ 0.0001) (Fig. 3b), when EC (r = 0.67; P = 0.014) and VC (r = 0.41; P = 0.33) sub-groups were analyzed separately (Fig. 3c, d), and when individuals were subdivided by sex (Additional file 1: Figure S3). By contrast, no significant correlations were observed between the mRNA expression of ZC3H12A/MCPIP1 or CDKN1A/p21 and the RF analyzed, with the only exception of a significant negative correlation between expression of those multifunctional genes and APOBEC3G in HIC (Additional file 1: Figure S4). These results demonstrate a coordinated mRNA expression of ZC3H12A/MCPIP1 and CDKN1A/p21 in PBMC from HIC that was not positive correlated with expression of classical RF. These results further suggest that such association is probably not an exclusive characteristic of HIC, but can also be detected in PBMC from others groups of HIV-infected and uninfected individuals.
Fig. 3.
CDKN1A/p21 and ZC3H12A/MCPIP1 mRNA levels in PBMC from HIC are positively correlated. The CDKN1A/p21 and ZC3H12A/MCPIP1 normalized expression correlations were calculated considering HIC a, all groups b, elite controllers c, and viremic controllers d. The points’ colors indicate the patient group, accordingly to the legend. Correlation coefficients (Spearman’s ρ) are shown in the upper right corner of each graph. P-values < 0.05 were considered statistically significant
MCPIP1 and p21 mRNA expression is positive correlated with immune activation in HIC
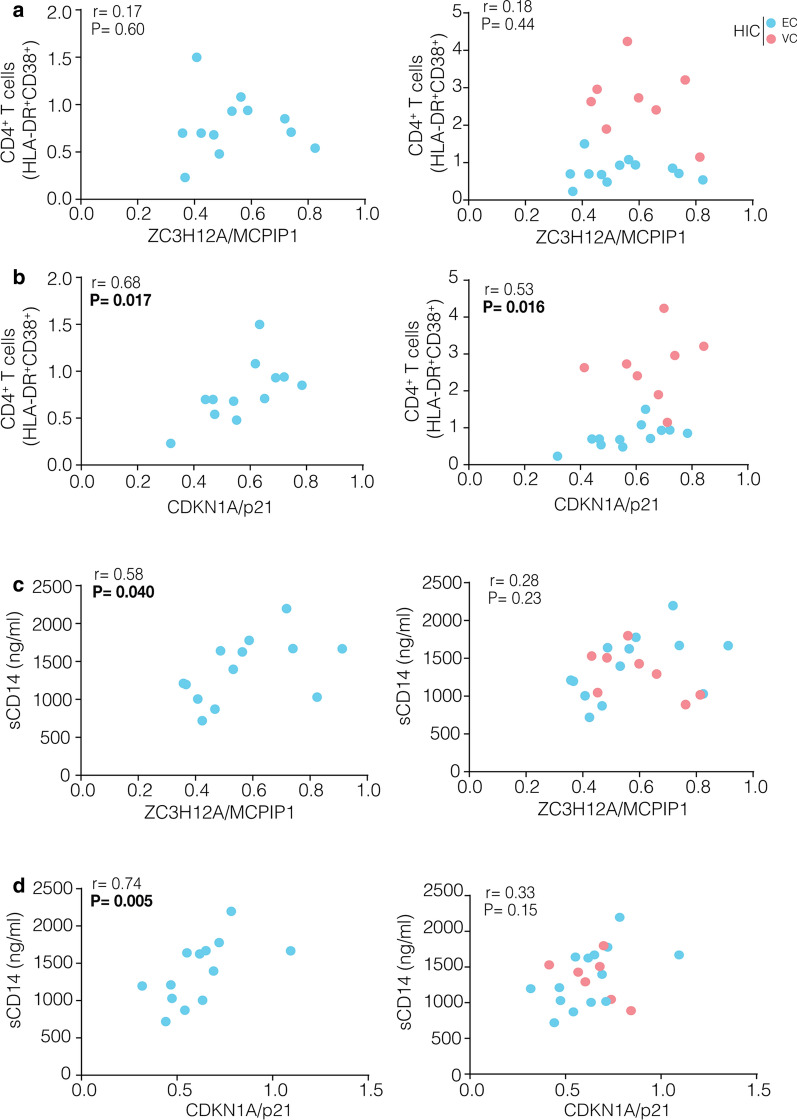
A previous study showed that CDKN1A/p21 mRNA levels are positive correlated with CD4+ T cell activation in HIC [5, 21]. To explore the potential relationship of CDKN1A/p21 or ZC3H12A/MCPIP1 expression with immune activation, we measured the frequency of HLA-DR+CD38+ phenotype on CD4+ and CD8+ T cells (T cell activation) and plasma levels of sCD14 (monocyte activation) in our cohort. Frequencies of activated CD4+ T cell populations in VC and ART-suppressed subjects were higher than in EC (P < 0.0001) and HIV-negative (P = 0.0002) individuals (Additional file 1: Figure S5A). The VC subgroup also had significantly higher frequencies of activated CD8+ T cell than EC (P = 0.0007) and control groups (P ≤ 0.006) (Additional file 1: Figure S5B). The median concentration of sCD14 in plasma was not significantly different across the groups (Additional file 1: Figure S5C). No significant correlations between mRNA levels of ZC3H12A/MCPIP1 and CD4+ T cell (Fig. 4a) or CD8+ T cell (Additional file 1: Figure S6A) activation were observed for HIC. The mRNA levels of CDKN1A/p21 were positively associated with activated CD4+ T cells levels in HIC (r = 0.53; P = 0.016) (Fig. 4b); but not with activated CD8+ T cell levels (Additional file 1: Figure S6B). Levels of sCD14 were positively correlated with both ZC3H12A/MCPIP1 (r = 0.58; P = 0.040) and CDKN1A/p21 (r = 0.74; P = 0.005) mRNA levels only in the EC subset (Fig. 4c, d). No significant correlations between mRNA levels of MCPIP1/p21 and CD4+/CD8+ T cell activation or sCD14 levels were observed when ART-suppressed and HIV-negative individuals were included (Additional file 1: Figure S7). Multivariate analysis showed that the upregulation of ZC3H12A/MCPIP1 was positively associated with the increase of CDKN1A/p21 expression in HIC (1.44-fold increase; P = 0.0035) (Additional file 1: Figure S8A). The frequency of activated CD4+ T cells also was positively associated with the increase of CDKN1A/p21 expression in both EC and VC (1.48-fold increase; P = 0.0116), although this increase of the CDKN1A/p21 expression was down-regulated by the increase of activated CD4+ T cells in VC when compared to EC (1.30-fold decrease by an increase of 1% CD4+HLA-DR+CD38+ T cells; P = 0.0284) (Additional file 1: Figure S8B). Overall, the model was highly significant (P = 0.003) and could explain as much as 70% (R2 = 0.492) of CDKN1A/p21 expression. These results suggest that expression of CDKN1A/p21 in PBMC from HIC may be modulated by both ZC3H12A/MCPIP1 expression and CD4+ T cell activation.
Fig. 4.
CDKN1A/p21 transcripts are positively correlated with CD4+ T cell and monocyte activation while ZC3H12A/MCPIP1transcripts are positively correlated only with monocyte activation in EC. The correlations were made evaluating the relationship between activated CD4+ T cells a, b or sCD14 levels c, d with the normalized expression of p21 and MCPIP1 for EC and HIC groups. The points’ colors present in each graph indicate the groups present according to the legend. Correlations coefficient (Spearman’s ρ) is shown in the upper left corner of each graph
Positive correlation between ZC3H12A/MCPIP1 and CDKN1A/p21 mRNA expression in CD4+ T cells and primary monocytes from disparate cohorts of HIV-infected and uninfected subjects
To further validate our observations, we analyzed the expression profiles of ZC3H12A/MCPIP1 and CDKN1A/p21 in HLA-DR− CD4+ T cells [33], total CD4+ T cells [34, 35], HIV-specific CD4+ T cells [36], total CD8+ T cells [35] and primary monocytes [37, 38] previously reported for other cohorts (Additional file 1: Table S2). All selected studies include data of HIC, with the only exception of the studies of monocytes, and different control groups including: ART-suppressed, ART-non-suppressed, untreated non-controllers typical progressors (NC-TP), untreated non-controllers long-term nonprogressors (NC-LTNP), and/or HIV-negative individuals [33–38]. Pairwise comparisons revealed modest (logFC ≤ 2.5), but significant (P < 0.05), upregulation of ZC3H12A/MCPIP1 and CDKN1A/p21 in HIC compared to HIV-negative individuals from all cohorts (Additional file 1: Table S2). We also detect significant upregulation of CDKN1A/p21 isoform in HLA-DR− CD4+ T cells from EC respect to ART-suppressed individuals and significant upregulation of both CDKN1A/p21 and ZC3H12A/MCPIP1 in HIV-specific CD4+ T cells from HIC respect to NC-TP and ART-suppressed individuals (Additional file 1: Table S2). These results suggest that upregulation of CDKN1A/p21 and ZC3H12A/MCPIP1 in HIC respect to other HIV-infected groups is a characteristic common to several, but not all, cohorts.
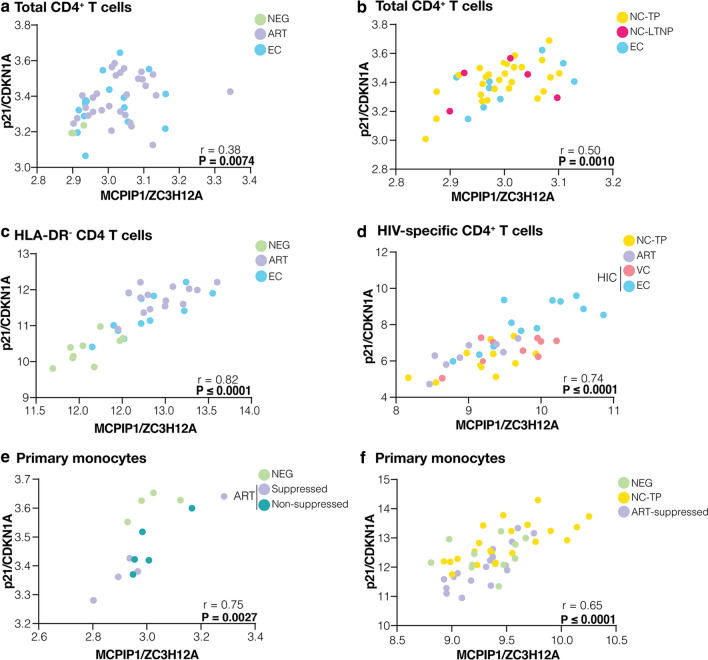
Analyses of published PBMC transcriptomic data confirm a significant positive correlation between the mRNA expression of ZC3H12A/MCPIP1 and CDKN1A/p21 (r ≥ 0.38; P ≤ 0.0074) in all cohorts analyzed for CD4+ T cells and primary monocytes cell types (Fig. 5), but not for CD8+ T cells (Additional file 1: Figure S9). Overall, the most abundant CDKN1A isoform demonstrated a much better significant positive correlation with ZC3H12A/MCPIP1 mRNA expression (Fig. 5) than the less abundant CDKN1A.1 isoform (r ≥ 0.30; P ≤ 0.45) (Additional file 1: Figure S10). Correlations coefficients between expression of ZC3H12A/MCPIP1 and CDKN1A/p21 in non-activated HLA-DR− CD4 T cells (r = 0.82), HIV-specific CD4+ T cells (r = 0.74) and monocytes (r ≥ 0.65) were higher than in total CD4+ T cells (0.27 ≥ r ≥ 0.50). Significant positive correlations between the mRNA expression of ZC3H12A/MCPIP1 and CDKN1A/p21 in CD4+ T cells and primary monocytes were observed across studies combining different groups including: HIC, ART-suppressed and HIV-negative individuals (Fig. 5a, c); EC and non-controllers individuals (Fig. 5b); HIC, ART-suppressed individuals and non-controllers individuals (Fig. 5d); ART-suppressed, ART-non-suppressed and HIV-negative individuals (Fig. 5e); and ART-suppressed, non-controllers and HIV-negative individuals (Fig. 5f). Positive correlations were also observed when different HIV-positive groups were analyzed separately (Additional file 1: Figure S11). These data clearly support that coordinated regulation of mRNA expression of the ZC3H12A/MCPIP1 and CDKN1A/p21 seems to be a common characteristic of CD4+ T cells and monocytes subpopulations from different HIV-positive groups.
Fig. 5.
CDKN1A/p21 and ZC3H12A/MCPIP1 mRNA levels are positively correlated in total, HLA-DR− and HIV-specific CD4+ T cells and primary monocytes. The ZC3H12A/MCPIP1 and isoform of CDKN1A/p21 (probe ILMN_1784602) normalized expression correlations were performed with data from previous studies in a, b total CD4+ T cells (GSE18233, GSE28128), c HLA-DR- CD4+ T cells (GSE23879), d HIV-specific CD4+ T cells and e, f primary monocytes (GSE52900, GSE18464) considering all groups (EC, VC, NC-TP, NC-LTNP, ART-suppressed, ART-non-suppressed, and HIV-negative subjects). The points’ colors indicate the patient group, accordingly to the legend. Correlation coefficients (Spearman’s ρ) are shown in the bottom right corner of each graph. P-values < 0.05 were considered statistically significant
Discussion
In this study, we described a positive correlation of ZC3H12A/MCPIP1 and CDKN1A/p21 mRNA expression in PBMC, CD4+ T cells and monocytes from HIV-infected and-uninfected subjects and we further detected that the mRNA levels of ZC3H12A/MCPIP1 and CDKN1A/p21 were significantly increased in PBMC of HIC compared to cells of HIV-negative and ART-suppressed individuals. We also observed higher MCPIP1 and p21 protein levels in PBMC and CD4+ T cells from HIC when compared with HIV-negative subjects.
Analyses of mRNA expression data in PBMC, CD4+ T cells and primary monocytes obtained from different cohorts of HIV–infected subjects described here and in previous studies clearly support a coordinated expression of ZC3H12A/MCPIP1 and at least one isoform of CDKN1A/p21, consistent with findings on human carcinoma cell line [32]. Of note, the ZC3H12A/MCPIP1 and CDKN1A/p21 mRNA expression seems to be much better correlated in HLA-DR− CD4+ T cells than in total CD4+ T cells, indicating that activation of CD4+ T cells might partially disrupt that correlation. Consistent with this hypothesis, we observed that levels of CDKN1A/p21 mRNA, but not ZC3H12A/MCPIP1, were positively correlated with CD4+ T cell activation in HIC. We speculate that expression of CDKN1A/p21 in CD4+ T cells could be modulated by ZC3H12A/MCPIP1 and by stimuli that activate the TCR. Because ZC3H12A/MCPIP1 acts as a broad suppressor of the biogenesis pathway of both cellular [39] and viral miRNA [40], it might enhance the expression of CDKN1A/p21 by downregulating miRNAs that target its mRNA, like let-7c miRNA [41].
While elevated mRNA expression of CDKN1A/p21 in PBMC and CD4+ T cells of HIC had already been previously described in others cohorts [5, 21, 24–26], this is the first study to show simultaneous upregulation of ZC3H12A/MCPIP1 in HIC. The increased transcripts levels were mirrored by the protein products of these genes in PBMC, in accordance with previous studies [21, 42]. One limitation of our study is the impossibility of assigning which cell(s) population(s) has increased expression of ZC3H12A/MCPIP1 and CDKN1A/p21 in our cohort of HIC as the expression profile of these multifunctional proteins and RF may vary between different PBMC sub-populations [8]. Another limitation of our study is that the PBMC composition may vary among individuals depending on HIV-infection status, virologic control and immune deregulation, thus making comparison between different groups of HIV-positive and -negative individuals more difficult to interpret. Future studies comparing the expression levels of these multifunctional proteins in specific cell types might better decipher the mechanisms of host factors regulation in the setting of natural control of HIV-1 infection.
The reanalysis of previously reported expression profiles in specific PBMC subpopulations provide some conflicting results. Consistent with our study, we detect a modest, but significant, upregulation of both ZC3H12A/MCPIP1 and CDKN1A/p21 in CD4+ T cells of HIC compared to HIV-negative individuals from different cohorts [33, 34] as well as in HIV-specific CD4+ T cells of HIC respect to non-controllers and ART-suppressed individuals [36]. We also detected significant upregulation of CDKN1A/p21, but not ZC3H12A/MCPIP1, in HLA-DR− CD4+ T cells from HIC respect to ART-suppressed individuals [33]. Finally, we failed to detect significant upregulation of these genes in total CD4+ T cells from HIC respect to HIV-infected control groups [34, 35]. These findings support that expression levels of ZC3H12A/MCPIP1 and CDKN1A/p21 might greatly vary among distinct PBMC sub-populations and HIV-infected individuals [43] and that the upregulation of those multifunctional proteins might not be a host signature common to all HIC.
Increased expression of some host RF has been previously observed in CD4+ T cells (i.e., SAMHD1, SLFN11 and IFITM1) [5, 7, 8] and PBMC (i.e., Mx1, Mx2, Tetherin and SLFN11) [6, 9] from some HIC. Although a few HIC from our cohort displayed mRNA levels of SAMHD1 and/or SLFN11 well above the normal range (Additional file 1: Figure S1), none of those RF were significantly upregulated in PBMC of our HIC cohort, with the only exception of IFITM1. Most ISGs are upregulated in the chronic phase of HIV-1 infection in viremic untreated patients [34, 44, 45] and their expression is positively correlated with the percentage of activated T cells and negatively correlated with CD4+ T cell counts [34, 44–47]. It is possible that residual or low-level viremia detected in HIC might not be enough to induce a generalized upregulation of ISGs during chronic infection [34]. In addition, ZC3H12A/MCPIP1 [48, 49] and CDKN1A/p21 [16] negatively regulate the NF-κB cascade and their upregulation may also contribute to limit the chronic upregulation of ISGs in our HIC cohort. While most RF are mainly induced by IFN type I, IFITM1 can also be induced by IFN type II [50] indicating that another pathway may have stimulated its expression in our HIC cohort.
The particular set of host RF and multifunctional proteins that are selectively upregulated clearly vary across disparate HIC cohorts. While one study detected upregulation of SAMHD1 mRNA in PBMCs from HIC compared to HIV-negative individuals [7], other study failed to detect significant differences in SAMHD1 expression between those groups [51]. Contrasting findings could be observed even among studies that analyzed specific cell types. Some works detected increased expression of CDKN1A/p21 [21, 24] or IFITM1 [8] in CD4+ T cells of HIC compared to HIV-negative individuals, but other study failed to detected significant upregulation of those proteins in CD4+ T cells of HIC [5]. Thus, cell types are not the only source of variation across studies of HIC. We propose that the genetic and immunologic heterogeneity of HIC individuals and HIV-infected control groups combined with the reduced size of most HIC cohorts might explain the contrasting findings across studies [5–9, 51].
Selective upregulation of ZC3H12A/MCPIP1 and CDKN1A/p21 in CD4+ T, macrophages and/or dendritic cells may have a synergistic antiviral effect and directly limit HIV-1 replication by (1) reducing the reverse transcription and chromosomal integration of HIV-1 in quiescent cells and thus limiting the size of the latent proviral reservoir [18–20, 52–54]; (2) restricting HIV-1 LTR transcription [48, 49, 55, 56]; and (3) degrading viral mRNA and miRNA [28, 39, 40, 57]. Whether upregulation of these multifunctional proteins directly contributes to reduce the susceptibility of CD4+ T cells and/or macrophages to HIV-1 infection in vivo, however, has been more difficult to prove. Chen et al. [21] demonstrate that elevated levels of CDKN1A/p21 in CD4+ T cells from HIC functionally contribute to the resistance of cells to HIV-1 infection in vitro. Saez-Cirion et al. [24] also detected high levels of CDKN1A/p21 in CD4+ T cells from HIC ex vivo, but found no evidence of a direct role of this protein in the reduced susceptibility of CD4+ T cells and macrophages to HIV-1 infection in vitro. Future studies of the susceptibility of PBMC of HIC to HIV-1 infection in vitro should analyze the potential role of both CDKN1A/p21 and ZC3H12A/MCPIP1 simultaneously.
Selective upregulation of ZC3H12A/MCPIP1 and CDKN1A/p21 may also play a crucial role in the control of HIV-1 infection by indirect mechanisms. HIV-1 preferentially infects the HIV-specific CD4+ memory T cells in vivo [58] and reduces their life span [59], compromising the generation of effective immune responses to the virus. Notably, our analysis of mRNA expression data in HIV-specific CD4+ T cells from a previous study [36] revealed increased levels of both ZC3H12A/MCPIP1 and CDKN1A/p21 transcripts in cells from HIC respect to other HIV-infected control groups. This may limit the seeding and size of the viral reservoir and further assist to preserve the specific CD4+ T response against HIV-1 in HIC. Because CDKN1A/p21 and ZC3H12A/MCPIP1 are negative regulators of the proliferation of activated/memory T cells [10, 13, 14, 29] and of macrophage-mediated inflammatory responses [15–17, 60, 61], upregulation of these proteins may also indirectly limit HIV-1 replication and further prevent CD4+ T cells decline by reducing chronic IFN-I signaling, generalized inflammation and over-activation of the immune system [10, 13–17, 60–63].
Our results confirm previous observations that levels of CDKN1A/p21 mRNA are positively correlated with CD4+ T cell activation in HIC [5] and further support a positive correlation between CDKN1A/p21 and ZC3H12A/MCPIP1 mRNA with monocyte activation in EC. Because induction of ZC3H12A/MCPIP1 mRNA upon CD4+ T cell activation is more ephemeral (< 12 h) than upon stimulation of macrophages (≥ 24 h) [60, 61, 64, 65], a direct correlation between ZC3H12A/MCPIP1 mRNA and CD4+ T cell activation could be more difficult to be detected. Increased expression of CDKN1A/p21 and ZC3H12A/MCPIP1 associated with T cell and/or monocyte activation may be a distinctive homoeostatic response to limit the deleterious effects of aberrant chronic immune activation and inflammation-driven by HIV-1 infection. Notably, similar correlations were not observed in other HIV-infected subjects in this or in previous studies [21], suggesting that this homoeostatic response could be a unique characteristic of some HIC/EC.
Transcript levels of RF here analyzed were not significantly correlated with T cell activation or sCD14 in HIC, with the only exception of a negative correlation between APOBEC3G mRNA and sCD14 levels in EC (r = − 0.73. P = 0.006; data not shown). Surprisingly, transcripts levels of APOBEC3G were also negatively correlated with ZC3H12A/MCPIP1 and CDKN1A/p21 mRNA levels in EC. One possible explanation for these negative correlations lies in the interaction of APOBEC3G, ZC3H12A/MCPIP1, and CDKN1A/p21 with the product of an important monocyte differentiation gene, the Kruppel-like factor 4 (KLF4). The expression of KLF4 in human macrophages is induced after IFN-γ, LPS, or TNF-α stimulus [66], mediating the proinflammatory signaling and the direct transcriptional regulation of CD14 in vitro [67] and also inducing expression of both ZC3H12A/MCPIP1 [68] and CDKN1A/p21 [69, 70]. Interestingly, APOBEC3G binds to the 3′-UTR of KLF4 mRNA and results in the reduction of its expression [71]. Thus, lower levels of APOBEC3G mRNA may be associated with an upregulation of KLF4 that in turn induce higher levels of sCD14, ZC3H12A/MCPIP1 and CDKN1A/p21 mRNA in EC.
Conclusions
In summary, our data confirm the high levels of CDKN1A/p21 expression and show for the first-time the concurrent upregulation of ZC3H12A/MCPIP1 in HIC, suggesting a possible synergistic effect of both innate host multifunctional proteins on natural suppression of HIV-1 replication in vivo. Moreover, we found a positive correlation between CDKN1A/p21 and ZC3H12A/MCPIP1 transcripts in PBMC, CD4+ T cells, and monocytes from different HIV-infected cohorts as well as positive correlation between expression of those multifunctional proteins and activation of CD4+ T cells and/or macrophages. These findings point that pharmacological manipulation of CDKN1A/p21 and ZC3H12A/MCPIP1 may open novel therapeutic perspectives to reduce HIV-1 replication and to attenuate HIV-associated inflammation and immune activation in vivo.
Methods
Study subjects
We analyzed a cohort of 21 HIC subjects followed-up at the Instituto Nacional de Infectologia Evandro Chagas (INI) in Rio de Janeiro, Brazil. All HIC maintained RNA VL of < 2000 copies/ml without antiretroviral therapy for at least 5 years and were subdivided in two sub-groups: EC (n = 13) when most (≥ 70%) plasma VL determinations were below the limit of detection (LOD), and VC (n = 8) when most (≥ 70%) VL determinations were > LOD and < 2000 copies/ml. The limit of detection of plasma VL determinations varied over the follow-up period in according to the Brazilian Ministry of Health guidelines, with methodologies being updated overtime to improve sensitivity: Nuclisens HIV-1 RNA QT assay (Organon Teknika, Durham, NC, limit of detection: 80 copies/mL) from 1999 to 2007; the Versant HIV-1 3.0 RNA assay (bDNA 3.0, Siemens, Tarrytown, NY, limit of detection: 50 copies/mL) from 2007 to 2013; and the Abbott RealTime HIV-1 assay (Abbott Laboratories, Wiesbaden, Germany, limit of detection: 40 copies/mL) from 2013 to until today. Virological and immunological characteristics of these subjects were described in detail in previous studies [72, 73]. Two groups of ART-suppressed subjects (ART, n = 8) and healthy HIV-1-uninfected subjects (NEG, n = 10) were used as controls.
mRNA gene-expression analysis
Total RNA was extracted from 1 × 107 PBMC using RNeasy mini kit (Qiagen, Hilden, North Rhine-Westphalia, Germany) in which buffer RLT was supplemented with β-mercaptoethanol and displaced on-column DNase treatment using a Qiagen RNase-Free DNase Set (Qiagen, Hilden, North Rhine-Westphalia, Germany) according to manufacturer’s instruction. Total RNA yield and quality were determined using NanoDrop® 8000 spectrophotometer and an Agilent® 2100 Bioanalyzer. Only samples with an RNA integrity number (RIN) greater than 8.0 were used. Purified RNA (1 μg) was reverse-transcribed to cDNA using RT2 First Strand Kit (Qiagen, Hilden, North Rhine-Westphalia, Germany). The cDNA was mixed with RT2SYBR Green/ROX qPCR Master Mix (Qiagen, Hilden, North Rhine-Westphalia, Germany) and the mixture was added into customized RT2RNA PCR Array (Qiagen, Hilden, North Rhine-Westphalia, Germany) to measure the mRNA expression of 10 cellular target genes (APOBEC3G, SAMHD1, Tetherin, Mx1, Mx2, SLFN11, IFITM1, IFITM3, ZC3H12A/MCPIP1, and CDKN1A/p21) besides three housekeeping genes (GAPDH, β-actin, and RNase-P), according to manufacturer’s instructions. Values of the crossing point at the maximum of the second derivative of the four-parameters fitted sigmoid curve second derivative, Cp, was determined for each sample. The efficiency of each amplification reaction was calculated as the ratio between the fluorescence of the cycle of quantification and fluorescence of the cycle immediately preceding that. Genes used in the normalization among samples were selected by the geNorm method [74]. Data were expressed as fold-changes in mRNA abundance calculated as the normalized gene expression in any test sample divided by the mean normalized gene expression in the control HIV-negative group.
Western blotting
We selected a subset of the eight cryopreserved PBMC samples (two elite controller, two viremic controllers and four healthy HIV-1-uninfected subjects) chosen based on a range of mRNA expression and availability of the biorepository for quantification of protein expression in PBMC and CD4+ T cells. CD4+ T cells were enriched from cryopreserved PBMCs using the EasySep Human CD4+ T Cell enrichment magnetic kit (StemCell Technologies, Vancouver, BC, Canada), according to the manufacturer’s instructions. The whole cell lysates were prepared from the 2 × 106 PBMC and CD4+ T cells and lysed with RIPA buffer (Tris–HCl 50 mM, pH 8.0, 150 mM NaCl, 1% NP40, 0.1% SDS, 0.5% Sodium Deoxycholate, 1 mM PMSF, and protease inhibitor cocktail). Supernatants were recovered after 14,000 rpm centrifugation (10 min, 4 °C) and protein concentrations were determined according to the Lowry method (Bio-Rad DC protein assay kit). 30 µg of proteins from each sample were resolved in 10% SDS-PAGE, and proteins were electrotransferred to polyvinylidene difluoride (PVDF) membranes. Unspecific binding sites were blocked with 5% skimmed milk in Tris-buffered saline containing 0.1% Tween 20 (TBST) for 2h under agitation, before the probing with anti-p21 antibody (Cell Signaling Technology, Danvers, Massachusetts, EUA) overnight at 4° C. Unbound primary antibody was washed out with TBST before incubation with secondary anti- rabbit antibody (Cell Signaling Technology, Danvers, Massachusetts, EUA) for 1h. After that, the membranes were washed 3× with TBST and 2x with Tris-buffered saline (TBS) 1x before development by Enhanced Chemiluminescence (Thermo Scientific SuperSignal West Dura ECL) on Hyperfilm (Amersham Hyperfilm ECL). The second probing proceeded with anti-MCPIP1 antibody (Proteintech, Rosemont, IL 60,018, USA) incubation for 1.5h at room temperature (accordingly to the manufacturer), followed by the same steps of washing and incubation with secondary anti-rabbit antibody before ECL development. Finally, the membranes were probed and developed (Amersham ECL Prime) for β-Actin (Cell Signaling Technology, Danvers, Massachusetts, EUA), and the immunoblots were photodocumented for image analysis in the ImageJ free software. The expression of p21 and MCPIP1 proteins were normalized with β-Actin for subsequent analysis.
Reanalysis of publicly available microarray datasets
We searched ZC3H12A/MCPIP1 and CDKN1A/p21 gene expression data from published studies that evaluated transcriptome in different cell populations of HIV-infected individuals through the Gene Expression Browser (GXB)—http://hiv.gxbsidra.org/dm3/geneBrowser/list. Raw expression profile data, with special focus on the genes of interest, ZC3H12A and CDKN1A, were obtained from six studies [33–38] through the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo). In studies that used the Illumina microarray platform, the ZC3H12A gene was represented by one probe (ILMN_1672295) while the CDKN1A gene was represented by two probes (ILMN_1784602 and ILMN_1787212). In the study that used the Affymetrix Human Clariom D Assay, the ZC3H12A (TC0100007832.hg.1) and CDKN1A (TC0600007847.hg.1) genes were both represented by one probe. Two datasets (GSE18233 and GSE23879) reported expression profiles in CD4+ T cells from EC, ART-suppressed and HIV-negative subjects (Additional file 1: Tables S3 and S4). The third dataset (GSE28128) reported expression profiles in CD4+ T and CD8+ T cells from EC, NC-LTNP and NC-TP subjects (Additional file 1: Table S5). The fourth dataset (GSE129872/GSE128296) reported expression profiles in HIV-specific CD4+ T cells from EC, NC-TP and ART-suppressed subjects (Additional file 1: Table S6). The fifth dataset (GSE52900) reported expression profiles in monocytes from ART-suppressed, ART-non-suppressed and HIV-negative subjects (Additional file 1: Table S7). The sixth dataset (GSE18464) reported expression profiles in monocytes from High and Low -viral loads and HIV-negative subjects (Additional file 1: Table S8). Raw expression profile data from these six studies were normalized and transformed to log2 using the limma R-package [75], and correlations were performed for both genes.
T cell and monocyte activation analyses
We used data of T cell and monocyte activation obtained in a previous study conducted by our group including these patients [73], in which plasma levels of soluble CD14 (sCD14) were determined by ELISA-sCD14 Quantikine assay (R&D Systems Minneapolis, MN) according to the manufacturer’s protocol and surface expression of combined HLA-DR and CD38 on CD4+ and CD8+ T cells was analyzed by flow cytometry.
Data analyses
The comparisons of mean log-fold changes (log-FC) in mRNA abundance were performed by either t-tests or one-way ANOVA nonparametric permutation tests (B = 1000 permutations), followed by pair-wise comparisons with Holm-Bonferroni adjustment [76], for two or more groups respectively. The differential expression analysis from studies containing public data from EC individuals was performed with the limma R-package [75]. LogFC and FDR values were obtained for both genes (two/three probes) for all comparisons. Spearman coefficient was used for correlation analyses. A first-order log-Normal multiple regression analysis was fitted to model p21 gene expression as a function of MCPIP1 gene expression, CD4+ T cell activation (HLA-DR+CD38+), and HIC groups (EC and VC). The threshold for statistical significance was set to P < 0.05. Data were analyzed with R software (version 3.5.2) [77].
Supplementary information
Additional file 1. Extended Materials with information about the cohorts of HIV-infected and uninfected subjects (Tables S2–S8) used in this study, and supplementary results (Figures S1–S11 and Table S1) regarding ZC3H12A/MCPIP1 and CDKN1A/p21 mRNA and protein expression levels, classical restriction factors mRNA expression and immune activation.
Acknowledgements
The authors thank the patients, who participated in the study, as well as all the technical staff involved in the clinical follow-up of these patients. We also thank Ms. Marilia Alves Figueira de Melo for the excellent technical support in RNA quantification and integrity analyses and the Plataforma de PCR em Tempo–RJ (RPT09A)–FIOCRUZ and Plataforma de Sequenciamento de Ácidos Nucléicos de Nova Geração–RJ (RPT01J)–FIOCRUZ. We are grateful for support from the Coordination for the Improvement of Higher Education Personnel (CAPES).
Abbreviations
- HIV
Human immunodeficiency virus
- HIC
HIV controllers
- EC
Elite controllers
- VC
Viremic controllers
- PBMC
Peripheral blood mononuclear cells
- VL
Viral loads
- LOD
Limit of detection
- RF
Restriction factors
- APOBEC3G
Apolipoprotein B mRNA-Editing enzyme, Catalytic polypeptide-like
- BST2
Bone Stromal Tumor protein 2
- SAMHD1
Sterile Alpha Motif domain and HD domain-containing protein 1 (SAMHD1)
- Mx
Myxovirus resistance protein
- IFITM
Interferon-inducible transmembrane family proteins
- SLFN11
Schlafen 11
- ART
Antiretroviral
- NEG
Healthy HIV-1-uninfected individuals
- CDKN1A
Cyclin-dependent kinase (CDK) inhibitor p21
- MCPIP1
Monocyte chemotactic protein–induced protein 1
- MALT1
Mucosa-associated lymphoid-tissue lymphoma-translocation 1
- Caki-1 cells
Human renal carcinoma cell line
- KLF4
Kruppel-like factor 4
- RIN
RNA integrity number
- NC-TP
Untreated non-controllers typical progressors
- NC-LTNP
Untreated non-controllers long-term nonprogressors
Author’s contributions
GB and TMLS conceived and designed the study and supervised the experiments. SSDA conducted experiments and analyzed the data together with MR-A, ED, LS, HN and GB. FH performed the CD4+ T cell and monocyte activation assays and helped SSDA to sorting assay CD4+ T cells. ED collaborated with mRNA gene-expression analysis. LS and SSDA performed the western blots assays. BH, BG, and VGV conducted patient recruitment and follow-up. FH, ED, and MGM provided intellectual input for results interpretations. SSDA, GB and MR-A wrote the first draft. All authors read and approved the final manuscript.
Funding
This work was supported by the Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro–FAPERJ (Grant number E-26/110.123/2014) and the Conselho Nacional de Desenvolvimento Científico e Tecnológico–CNPq (Grant Number 401220/2016-8). SSDA had her PhD funded by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro–FAPERJ. Currently, SSDA is financed by Postdoctoral fellowship from the “Pós-Doutorado Nota 10 (PDR 10)” by Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro–FAPERJ, but during the execution of this work she also was financed by the “Pós-Doutorado Júnior (PDJ)” by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
Availability of data and materials
The datasets during and/or analyzed during the current study available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study was carried out in accordance with the recommendations of the ethical committee of Instituto Nacional de Infectologia Evandro Chagas (INI-Fiocruz) that approved the study protocol (CAAE 1717.0.000.009-07). All subjects gave written informed consent in accordance with the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Suwellen S. D. de Azevedo, Email: suwellen@ioc.fiocruz.br, Email: suwellendias@gmail.com
Marcelo Ribeiro-Alves, Email: marcelo.ribeiro@ini.fiocruz.br.
Fernanda H. Côrtes, Email: fheloise@ioc.fiocruz.br
Edson Delatorre, Email: edsondelatorre@gmail.com.
Lucia Spangenberg, Email: lucia83@gmail.com.
Hugo Naya, Email: blakngu@gmail.com.
Leonardo N. Seito, Email: leonardo.seito@far.fiocruz.br
Brenda Hoagland, Email: brenda.fiocruz@gmail.com.
Beatriz Grinsztejn, Email: beatriz.grinsztejn@gmail.com.
Valdilea G. Veloso, Email: valdilea.veloso@gmail.com
Mariza G. Morgado, Email: mmorgado@ioc.fiocruz.br
Thiago Moreno L. Souza, Email: souzatml@gmail.com
Gonzalo Bello, Email: gbellobr@gmail.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12977-020-00522-4.
References
- 1.Walker BD, Yu XG. Unravelling the mechanisms of durable control of HIV-1. Nat Rev Immunol. 2013;13:487–498. doi: 10.1038/nri3478. [DOI] [PubMed] [Google Scholar]
- 2.Harris RS, Hultquist JF, Evans DT. The restriction factors of human immunodeficiency virus. J Biol Chem. 2012;287:40875–40883. doi: 10.1074/jbc.R112.416925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colomer-Lluch M, Ruiz A, Moris A, Prado JG. Restriction factors: from intrinsic viral restriction to shaping cellular immunity against HIV-1. Front Immunol. 2018;9:2876. doi: 10.3389/fimmu.2018.02876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doyle T, Goujon C, Malim MH. HIV-1 and interferons: who’s interfering with whom? Nat Rev Microbiol. 2015;13:403–413. doi: 10.1038/nrmicro3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdel-Mohsen M, Raposo RAS, Deng X, Li M, Liegler T, Sinclair E, et al. Expression profile of host restriction factors in HIV-1 elite controllers. Retrovirology. 2013;10:1–13. doi: 10.1186/1742-4690-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krishnan S, Wilson EMP, Sheikh V, Rupert A, Mendoza D, Yang J, et al. Evidence for innate immune system activation in HIV type 1-infected elite controllers. J Infect Dis. 2014;209:931–939. doi: 10.1093/infdis/jit581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riveira-Muñoz E, Ruiz A, Pauls E, Permanyer M, Badia R, Mothe B, et al. Increased expression of SAMHD1 in a subset of HIV-1 elite controllers. J Antimicrob Chemother. 2014;69:3057–3060. doi: 10.1093/jac/dku276. [DOI] [PubMed] [Google Scholar]
- 8.Canoui E, Noël N, Lécuroux C, Boufassa F, Sáez-Cirión A, Bourgeois C, et al. Strong ifitm1 expression in CD4 T cells in HIV controllers is correlated with immune activation. J Acquir Immune Defic Syndr. 2017;74:e56–e59. doi: 10.1097/QAI.0000000000001166. [DOI] [PubMed] [Google Scholar]
- 9.Van Hecke C, Trypsteen W, Malatinkova E, De Spiegelaere W, Vervisch K, Rutsaert S, et al. Early treated HIV-1 positive individuals demonstrate similar restriction factor expression profile as long-term non-progressors. EBioMed. 2019;41:1–12. doi: 10.1016/j.ebiom.2019.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khanna AK, Plummer M, Nilakantan V, Pieper GM. Recombinant p21 protein inhibits lymphocyte proliferation and transcription factors. J. Immunol. 2005;174:7610–7617. doi: 10.4049/jimmunol.174.12.7610. [DOI] [PubMed] [Google Scholar]
- 11.Khanna AK. Reciprocal role of cyclins and cyclin kinase inhibitor p21WAF1/CIP1 on lymphocyte proliferation, allo-immune activation and inflammation. BMC Immunol. 2005;6:22. doi: 10.1186/1471-2172-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balomenos D, Martín-Caballero J, García MI, Prieto I, Flores JM, Serrano M, et al. The cell cycle inhibitor p21 controls T-cell proliferation and sex-linked lupus development. Nat Med. 2000;6:171–176. doi: 10.1038/72272. [DOI] [PubMed] [Google Scholar]
- 13.Arias CF, Ballesteros-Tato A, Garcia MI, Martin-Caballero J, Flores JM, Martinez-A C, et al. p21CIP1/WAF1 controls proliferation of activated/memory T cells and affects homeostasis and memory T cell responses. J. Immunol. 2007;178:2296–2306. doi: 10.4049/jimmunol.178.4.2296. [DOI] [PubMed] [Google Scholar]
- 14.Santiago-Raber M-L, Lawson BR, Dummer W, Barnhouse M, Koundouris S, Wilson CB, et al. Role of cyclin kinase inhibitor p21 in systemic autoimmunity. J. Immunol. 2001;167:4067–4074. doi: 10.4049/jimmunol.167.7.4067. [DOI] [PubMed] [Google Scholar]
- 15.Lloberas J, Celada A. p21 waf1/CIP1, a CDK inhibitor and a negative feedback system that controls macrophage activation. Eur J Immunol. 2009;39:691–694. doi: 10.1002/eji.200939262. [DOI] [PubMed] [Google Scholar]
- 16.Trakala M, Arias CF, García MI, Moreno-Ortiz MC, Tsilingiri K, Fernández PJ, et al. Regulation of macrophage activation and septic shock susceptibility via p21(WAF1/CIP1) Eur J Immunol. 2009;39:810–819. doi: 10.1002/eji.200838676. [DOI] [PubMed] [Google Scholar]
- 17.Scatizzi JC, Mavers M, Hutcheson J, Young B, Shi B, Pope RM, et al. The CDK domain of p21 is a suppressor of IL-1β-mediated inflammation in activated macrophages. Eur J Immunol. 2009;39:820–825. doi: 10.1002/eji.200838683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, Scadden DT, Crumpacker CS. Primitive hematopoietic cells resist HIV-1 infection via p21Waf1/Cip1/Sdi1. J Clin Invest. 2007;117:473–481. doi: 10.1172/JCI28971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bergamaschi A, David A, Le Rouzic E, Nisole S, Barre-Sinoussi F, Pancino G. The CDK inhibitor p21Cip1/WAF1 is induced by Fc R activation and restricts the replication of human immunodeficiency virus type 1 and related primate lentiviruses in human macrophages. J Virol. 2009;83:12253–12265. doi: 10.1128/JVI.01395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valle-Casuso JC, Allouch A, David A, Lenzi GM, Studdard L, Barré-Sinoussi F, et al. p21 Restricts HIV-1 in monocyte-derived dendritic cells through the reduction of deoxynucleoside triphosphate biosynthesis and regulation of SAMHD1 antiviral activity. Kirchhoff F, editor. J Virol. 2017;91:1–18. doi: 10.1128/JVI.01324-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H, Li C, Huang J, Cung T, Seiss K, Beamon J, et al. CD4 + T cells from elite controllers resist HIV-1 infection by selective upregulation of p21. J Clin Invest. 2011;121:1549–1560. doi: 10.1172/JCI44539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elahi S, Weiss RH, Merani S. Atorvastatin restricts HIV replication in CD4 + T cells by upregulation of p21. AIDS. 2016;30:171–183. doi: 10.1097/QAD.0000000000000917. [DOI] [PubMed] [Google Scholar]
- 23.Elahi S, Niki T, Hirashima M, Horton H. Galectin-9 binding to Tim-3 renders activated human CD4 + T cells less susceptible to HIV-1 infection. Blood. 2012;119:4192–4204. doi: 10.1182/blood-2011-11-389585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saez-Cirion A, Hamimi C, Bergamaschi A, David A, Versmisse P, Melard A, et al. Restriction of HIV-1 replication in macrophages and CD4 + T cells from HIV controllers. Blood. 2011;118:955–964. doi: 10.1182/blood-2010-12-327106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moosa Y, Tanko RF, Ramsuran V, Singh R, Madzivhandila M, Yende-Zuma N, et al. Case report: mechanisms of HIV elite control in two African women. BMC Infect Dis. 2018;18:1–7. doi: 10.1186/s12879-018-2961-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madlala P, Van de Velde P, Van Remoortel B, Vets S, Van Wijngaerden E, Van Laethem K, et al. Analysis of ex vivo HIV-1 infection in a controller-discordant couple. J Virus Erad. 2018;4:170–173. [PMC free article] [PubMed] [Google Scholar]
- 27.Fu M, Blackshear PJ. RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins. Nat Rev Immunol. 2017;17:130–143. doi: 10.1038/nri.2016.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu S, Qiu C, Miao R, Zhou J, Lee A, Liu B, et al. MCPIP1 restricts HIV infection and is rapidly degraded in activated CD4 + T cells. Proc Natl Acad Sci USA. 2013;110:19083–19088. doi: 10.1073/pnas.1316208110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uehata T, Iwasaki H, Vandenbon A, Matsushita K, Hernandez-cuellar E, Kuniyoshi K, et al. Malt1-induced cleavage of regnase-1 in CD4 + helper T cells regulates immune activation. Cell. 2013;153:1036–1049. doi: 10.1016/j.cell.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 30.Jeltsch KM, Hu D, Brenner S, Zöller J, Heinz GA, Nagel D, et al. Cleavage of roquin and regnase-1 by the paracaspase MALT1 releases their cooperatively repressed targets to promote TH17 differentiation. Nat Immunol. 2014;15:1079–1089. doi: 10.1038/ni.3008. [DOI] [PubMed] [Google Scholar]
- 31.Jura J, Skalniak L, Koj A. Monocyte chemotactic protein-1-induced protein-1 (MCPIP1) is a novel multifunctional modulator of inflammatory reactions. Biochim Biophys Acta. 2012;1823:1905–1913. doi: 10.1016/j.bbamcr.2012.06.029. [DOI] [PubMed] [Google Scholar]
- 32.Lichawska-Cieslar A, Pietrzycka R, Ligeza J, Kulecka M, Paziewska A, Kalita A, et al. RNA sequencing reveals widespread transcriptome changes in a renal carcinoma cell line. Oncotarget. 2018;9:8597–8613. doi: 10.18632/oncotarget.24269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vigneault F, Woods M, Buzon MJ, Li C, Pereyra F, Crosby SD, et al. Transcriptional profiling of CD4 T cells identifies distinct subgroups of HIV-1 elite controllers. J Virol. 2011;85:3015–3019. doi: 10.1128/JVI.01846-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rotger M, Dang KK, Fellay J, Heinzen EL, Feng S, Descombes P, et al. Genome-wide mRNA expression correlates of viral control in CD4 + T-cells from HIV-1-infected individuals. Emerman M, editor. PLoS Pathog. 2010;6:e1000781. doi: 10.1371/journal.ppat.1000781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rotger M, Dalmau J, Rauch A, McLaren P, Bosinger SE, Martinez R, et al. Comparative transcriptomics of extreme phenotypes of human HIV-1 infection and SIV infection in sooty mangabey and rhesus macaque. J Clin Invest. 2011;121:2391–2400. doi: 10.1172/JCI45235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morou A, Brunet-Ratnasingham E, Dubé M, Charlebois R, Mercier E, Darko S, et al. Altered differentiation is central to HIV-specific CD4 + T cell dysfunction in progressive disease. Nat Immunol. 2019;20:1059–1070. doi: 10.1038/s41590-019-0418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu JQ, Sassé TR, Saksena MM, Saksena NK. Transcriptome analysis of primary monocytes from HIV-positive patients with differential responses to antiretroviral therapy. Virol J. 2013;10:1–16. doi: 10.1186/1743-422X-10-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rempel H, Sun B, Calosing C, Pillai SK, Pulliam L. Interferon-α drives monocyte gene expression in chronic unsuppressed HIV-1 infection. AIDS. 2010;24:1415–1423. doi: 10.1097/QAD.0b013e32833ac623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki HI, Arase M, Matsuyama H, Choi YL, Ueno T, Mano H, et al. MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Mol Cell. 2011;44:424–436. doi: 10.1016/j.molcel.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 40.Happel C, Ramalingam D, Ziegelbauer JM. Virus-mediated alterations in miRNA factors and degradation of viral miRNAs by MCPIP1. Sugden B, editor. PLoS Biol. 2016;14:e2000998. doi: 10.1371/journal.pbio.2000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farberov L, Herzig E, Modai S, Isakov O, Hizi A, Shomron N. MicroRNA-mediated regulation of p21 and TASK1 cellular restriction factors enhances HIV-1 infection. J Cell Sci. 2015;128:1607–1616. doi: 10.1242/jcs.167817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin R-J, Chu J-S, Chien H-L, Tseng C-H, Ko P-C, Mei Y-Y, et al. MCPIP1 suppresses hepatitis C virus replication and negatively regulates virus-induced proinflammatory cytokine responses. J Immunol. 2014;193:4159–4168. doi: 10.4049/jimmunol.1400337. [DOI] [PubMed] [Google Scholar]
- 43.De Pablo A, Bogoi R, Bejarano I, Toro C, Valencia E, Moreno V, et al. Short communication: p21/CDKN1A expression shows broad interindividual diversity in a subset of HIV-1 elite controllers. AIDS Res Hum Retroviruses. 2016;32:1–5. doi: 10.1089/AID.2015.0137. [DOI] [PubMed] [Google Scholar]
- 44.Sedaghat AR, German J, Teslovich TM, Cofrancesco J, Jie CC, Talbot CC, et al. Chronic CD4 + T-cell activation and depletion in human immunodeficiency virus type 1 infection: type I interferon-mediated disruption of T-cell dynamics. J Virol. 2008;82:1870–1883. doi: 10.1128/JVI.02228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hyrcza MD, Kovacs C, Loutfy M, Halpenny R, Heisler L, Yang S, et al. Distinct transcriptional profiles in ex vivo CD4 + and CD8 + T cells are established early in human immunodeficiency virus type 1 infection and are characterized by a chronic interferon response as well as extensive transcriptional changes in CD8 + T cells. J Virol. 2007;81:3477–3486. doi: 10.1128/JVI.01552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hardy GAD, Sieg S, Rodriguez B, Anthony D, Asaad R, Jiang W, et al. Interferon-α Is the primary plasma type-I IFN in HIV-1 infection and correlates with immune activation and disease markers. Gray CM, editor. PLoS One. 2013;8:e56527. doi: 10.1371/journal.pone.0056527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fernandez S, Tanaskovic S, Helbig K, Rajasuriar R, Kramski M, Murray JM, et al. CD4 + T-cell deficiency in HIV patients responding to antiretroviral therapy is associated with increased expression of interferon-stimulated genes in CD4 + T cells. J Infect Dis. 2011;204:1927–1935. doi: 10.1093/infdis/jir659. [DOI] [PubMed] [Google Scholar]
- 48.Skalniak L, Mizgalska D, Zarebski A, Wyrzykowska P, Koj A, Jura J. Regulatory feedback loop between NF-κB and MCP-1-induced protein 1 RNase. FEBS J. 2009;276:5892–5905. doi: 10.1111/j.1742-4658.2009.07273.x. [DOI] [PubMed] [Google Scholar]
- 49.Liang J, Saad Y, Lei T, Wang J, Qi D, Yang Q, et al. MCP-induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF-κB signaling. J Exp Med. 2010;207:2959–2973. doi: 10.1084/jem.20092641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol. 2013;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buchanan EL, Mcalexander MA, Witwer KW. SAMHD1 expression in blood cells of HIV-1 elite suppressors and viraemic progressors. J Antimicrob Chemother. 2015;70:954–956. doi: 10.1093/jac/dku428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pauls E, Ruiz A, Riveira-Munoz E, Permanyer M, Badia R, Clotet B, et al. p21 regulates the HIV-1 restriction factor SAMHD1. Proc Natl Acad Sci. 2014;111:E1322–E1324. doi: 10.1073/pnas.1322059111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Allouch A, David A, Amie SM, Lahouassa H, Chartier L, Margottin-Goguet F, et al. p21-mediated RNR2 repression restricts HIV-1 replication in macrophages by inhibiting dNTP biosynthesis pathway. Proc Natl Acad Sci. 2013;110:E3997–E4006. doi: 10.1073/pnas.1306719110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leng J, Ho H-P, Buzon MJ, Pereyra F, Walker BD, Yu XG, et al. A cell-intrinsic inhibitor of HIV-1 reverse transcription in CD4 + T cells from elite controllers. Cell Host Microbe. 2014;15:717–728. doi: 10.1016/j.chom.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang D, de la Fuente C, Deng L, Wang L, Zilberman I, Eadie C, et al. Inhibition of human immunodeficiency virus type 1 transcription by chemical cyclin-dependent kinase inhibitors. J Virol. 2001;75:7266–7279. doi: 10.1128/JVI.75.16.7266-7279.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumari N, Iordanskiy S, Kovalskyy D, Breuer D, Niu X, Lin X, et al. Phenyl-1-Pyridin-2yl-Ethanone-based iron chelators increase IκB-α expression, modulate CDK2 and CDK9 activities, and inhibit HIV-1 transcription. Antimicrob Agents Chemother. 2014;58:6558–6571. doi: 10.1128/AAC.02918-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin R-J, Chien H-L, Lin S-Y, Chang B-L, Yu H-P, Tang W-C, et al. MCPIP1 ribonuclease exhibits broad-spectrum antiviral effects through viral RNA binding and degradation. Nucleic Acids Res. 2013;41:3314–3326. doi: 10.1093/nar/gkt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, et al. HIV preferentially infects HIV-specific CD4 + T cells. Nature. 2002;417:95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- 59.Brenchley JM, Ruff LE, Casazza JP, Koup RA, Price DA, Douek DC. Preferential infection shortens the life span of human immunodeficiency virus-specific CD4 + T cells in vivo. J Virol. 2006;80:6801–6809. doi: 10.1128/JVI.00070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liang J, Song W, Tromp G, Kolattukudy PE, Fu M. Genome-wide survey and expression profiling of CCCH-zinc finger family reveals a functional module in macrophage activation. PLoS One. 2008;3:e2880. doi: 10.1371/journal.pone.0002880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matsushita K, Takeuchi O, Standley DM, Kumagai Y, Kawagoe T, Miyake T, et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009;458:1185–1190. doi: 10.1038/nature07924. [DOI] [PubMed] [Google Scholar]
- 62.Li Y, Huang X, Huang S, He H, Lei T, Saaoud F, et al. Central role of myeloid MCPIP1 in protecting against LPS-induced inflammation and lung injury. Signal Transduct Target Ther. 2017;2:17066. doi: 10.1038/sigtrans.2017.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uehata T, Takeuchi O. Regnase-1 is an endoribonuclease essential for the maintenance of immune homeostasis. J Interf Cytokine Res. 2017;37:220–229. doi: 10.1089/jir.2017.0001. [DOI] [PubMed] [Google Scholar]
- 64.Liang J, Wang J, Azfer A, Song W, Tromp G, Kolattukudy PE, et al. A novel CCCH-zinc finger protein family regulates proinflammatory activation of macrophages. J Biol Chem. 2008;283:6337–6346. doi: 10.1074/jbc.M707861200. [DOI] [PubMed] [Google Scholar]
- 65.Li M, Cao W, Liu H, Zhang W, Liu X, Cai Z, et al. MCPIP1 down-regulates IL-2 expression through an ARE-independent pathway. PLoS ONE. 2012;7:e49841. doi: 10.1371/journal.pone.0049841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feinberg MW, Cao Z, Wara AK, Lebedeva MA, Senbanerjee S, Jain MK. Kruppel-like factor 4 is a mediator of proinflammatory signaling in macrophages. J Biol Chem. 2005;280:38247–38258. doi: 10.1074/jbc.M509378200. [DOI] [PubMed] [Google Scholar]
- 67.Feinberg MW, Wara AK, Cao Z, Lebedeva MA, Rosenbauer F, Iwasaki H, et al. The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. EMBO J. 2007;26:4138–4148. doi: 10.1038/sj.emboj.7601824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kapoor N, Niu J, Saad Y, Kumar S, Sirakova T, Becerra E, et al. Transcription factors STAT6 and KLF4 implement macrophage polarization via the dual catalytic powers of MCPIP. J Immunol. 2015;194:6011–6023. doi: 10.4049/jimmunol.1402797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yoon HS, Chen X, Yang VW. Kruppel-like factor 4 mediates p53-dependent G1/S cell cycle arrest in response to DNA damage. J Biol Chem. 2003;278:2101–2105. doi: 10.1074/jbc.M211027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang W, Geiman DE, Shields JM, Dang DT, Mahatan CS, Kaestner KH, et al. The gut-enriched Kruppel-like factor (Kruppel-like factor 4) mediates the transactivating effect of p53 on the p21(WAF1)/(Cip)1 promoter. J Biol Chem. 2000;275:18391–18398. doi: 10.1074/jbc.C000062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garg A, Kaul D, Chauhan N. APOBEC3G governs to ensure cellular oncogenic transformation. Blood Cells Mol Dis. 2015;55:248–254. doi: 10.1016/j.bcmd.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 72.Azevedo SSD, Caetano DG, Côrtes FH, Teixeira SLM, Santos Silva K, Hoagland B, et al. Highly divergent patterns of genetic diversity and evolution in proviral quasispecies from HIV controllers. Retrovirol BioMed Central. 2017;14:1–13. doi: 10.1186/s12977-017-0354-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Côrtes FH, de Paula HHS, Bello G, Ribeiro-Alves M, de Azevedo SSD, Caetano DG, et al. Plasmatic levels of IL-18, IP-10, and activated CD8 + T cells are potential biomarkers to identify HIV-1 elite controllers with a true functional cure profile. Front Immunol. 2018;9:1–11. doi: 10.3389/fimmu.2018.01576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Basso D, Pesarin F, Salmaso L, Solari A. Nonparametric One-Way ANOVA. Permut. Tests Stoch. Ordering ANOVA. New York: Springer; 2009. pp. 105–132. [Google Scholar]
- 77.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, editor. Vienna. 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Extended Materials with information about the cohorts of HIV-infected and uninfected subjects (Tables S2–S8) used in this study, and supplementary results (Figures S1–S11 and Table S1) regarding ZC3H12A/MCPIP1 and CDKN1A/p21 mRNA and protein expression levels, classical restriction factors mRNA expression and immune activation.
Data Availability Statement
The datasets during and/or analyzed during the current study available from the corresponding author on reasonable request.