Abstract
CD154 (CD40-ligand) is a critical immune regulator. CD154 expression is tightly regulated and largely restricted to activated CD4 T cells. Using DNase I hypersensitivity site (HSS) mapping, we identified two novel HSS mapping to the human CD154 promoter element and just upstream. Both HSS were activation independent and CD4 T-cell specific. Approximately 350 bp of DNA sequence flanking the upstream HSS site was highly conserved between mouse and man, and was rich in binding sites for GATA and NFAT proteins. Gel shift and chromatin immunoprecipitation assays demonstrated both NFAT1 and the Th2 factor, GATA-3, bound this enhancer element in vitro and in vivo, respectively. A PstI/XbaI 345 bp fragment of this region acted as a transcriptional enhancer of the CD154 promoter in primary human CD4 T cells. Overexpression of repressor of GATA and a dominant negative GATA-3 protein independently inhibited transcription, whereas overexpression of wild-type GATA-3 enhanced transcriptional activity, by this element in primary CD4 T cells. Moreover, more interleukin-4-producing CD4 T cells expressed CD154 following activation than interferon-g-producing CD4 T cells. Thus, we identified a novel T-cell-specific, GATA-3 responsive, CD154 transcriptional enhancer, which may contribute to increased propensity of Th2 cells to express CD154.
Keywords: human, T cells, transcription factors, CD154, GATA-3, NFAT
Introduction
CD154 (CD40 ligand) is a critically important molecule for the immune response. Antibody-producing B lymphocytes depend on the interaction with CD154 expressed on activated CD4 T lymphocytes for their growth, differentiation and antibody formation.1 Because of its multiple potent effects during a normal immune response, CD154 expression by CD4 T cells is tightly regulated, and, normally, CD154 expression is highly regulated at the transcriptional level.2 Conversely, CD154 dysregulation has been shown in multiple diseases, including autoimmune diseases (for example, systemic lupus erythematosus), Alzheimer disease and atherosclerosis, to name a few.3
We previously identified and characterized the human CD154 (hCD154) transcriptional promoter4 and a 3′ enhancer element.5 The promoter is activation dependent and requires the cyclosporin-sensitive nuclear factor of activated Tcells (NFAT) transcription factor for expression. By comparison, the hCD154 3′ enhancer element is dependent on nuclear factor-κB protein engagement for optimal activity. Both the promoter and enhancer display relatively weak transcriptional activity in reporter gene assays in primary human T cells. The observation that the CD154 promoter has weak transcriptional activity as compared with other cytokine genes (for example, interleukin (IL)-2), but has similarly abundant mRNA transcripts,2,5 led us to search for other cis-acting elements that could account for its transcriptional regulation.
To this end, we explored the greater hCD154 gene locus for additional transcription promoting cis-elements by the technique of DNase I hypersensitivity site (HSS) mapping. Herein, we report the identification of a novel CD4 T-cell-specific HSS, which maps just upstream of the hCD154 transcriptional promoter. The immediate surrounding sequence is species conserved and functions in vitro as a transcriptional enhancer. This 5′ enhancer element is bound by NFAT1 and the Th2-specific transcription factor, GATA-3,6 in vitro and in vivo, and GATA-3 positively regulates its transcriptional enhancer activity. Thus, we have identified and partially characterized a novel CD154 transcriptional enhancer element. A better understanding of normal CD154 expression in primary CD4 T cells may ultimately allow for the identification of disease-specific regulatory elements in the greater hCD154 gene locus that contribute to the prolonged and exaggerated expression of CD154 by CD4 T cells of patients with SLE and other autoimmune disorders.
Results
Identification of novel DNase I HSS in and around the hCD154 transcriptional promoter
To date, only the CD154 transcriptional promoter, immediate 3′ enhancer and 3′ UTR mRNA stability element have been characterized as cis-acting CD154 regulatory elements. Together, these elements are relatively weak at driving CD154 expression in vitro. To explore additional CD154 cis-regulatory elements, we analyzed the hCD154 gene locus in Jurkat CD4 T cells by DNase I HSS mapping. We first analyzed a 6.4 kb BamHI fragment, which includes the first two exons of hCD154, and portions of the second intron and upstream flanking region 5′ of the transcription start site (TSS). By increasing the concentration of DNase I, we identified two strong HSS and some minor HSS located between these two sites (Figure 1a). Both sites (HSS 1 and HSS 2) were also present in primary human CD4 T cells both before and after polyclonal T-cell activation (Figure 1b). Interestingly, these sites appeared relatively CD4 T-cell specific, as they were absent in two macrophage and two B cell lines (Figure 1c; and data not shown), and absent in human primary CD8 T cells and primary B cells (data not shown). Fine mapping by Southern blotting revealed that HSS 1 mapped between the two critical NFAT binding sites in the proximal promoter (Figure 2a),4 whereas HSS 2 mapped approximately 1.5 kb upstream of the hCD154 promoter (Figure 2a). Thus, we identified two novel DNase I HSS, which are not dependent on activation, are CD4 T-cell specific and correspond to the heart of the hCD154 transcriptional promoter and to just upstream of the promoter, respectively.2
Figure 1.

Identification of two novel DNase I hypersensitivity sites (HSS) near the human CD154 (hCD154) transcription start site (TSS). Nuclei were isolated from human primary (b) and transformed (Jurkat D1.1; a–c) CD4 T cells, transformed B cells (A2D6; c), and transformed macrophages (U937; c). The nuclei were rested (b, lanes 1 and 2) or stimulated (all other lanes, a–c) for 2 h with phorbol 13-myristate 12-acetate (PMA; 25 ng ml−1) and ionomycin (1.5 μm). Nuclei were then treated with either none (0), or increasing amounts of DNase I (1–7 μg ml−1, as labeled) for 3 min at room temperature (RT). DNA was extracted, BamHI digested, Southern blotted and hybridized with a 280 bp 32-P labeled probe corresponding to the 5′ proximal end (B2 kb upstream of the hCD154 TSS) of a 6.4 kb BamHI fragment surrounding the TSS. MW markers (1 kb ladder; b) and novel HSS are noted. Minimum of N = 3 for each cell type and condition.
Figure 2.

Mapping of the novel hypersensitivity site (HSS) to the CD154 promoter and a sequence conserved region just upstream. (a) Jurkat D1.1 nuclei were digested with 0 (lanes 1–3, 6–8), 2.5 (lane 4) or 3 (lane 5) μg ml−1 of DNase I. Genomic DNA was then digested with nothing (lane 8), BamHI alone (lanes 3–5), or BamHI plus the following: HindIII (lane 1), SnaBI (lane 2), XbaI (lane 6) or PstI (lane 7). HSS 1 (CD154 promoter) and HSS 2 (novel upstream element) are depicted by arrows. Restriction enzymes and associated base pair sizes of fragments are listed to the sides. (b) A graphic depiction (VISTA map) of the degree of DNA sequence conservation (y axis ranges from 50–100% identity) between mouse and human of the regions flanking HSS 1 (CD154 promoter) and HSS 2 (upstream element).
Characterization of a novel 5′ CD154 transcriptional enhancer
DNA sequence alignment using VISTA Map (http://genome.lbl.gov/vista/index.shtml) software revealed that the murine and hCD154 promoters are highly homologous (Figure 2b). Similarly, the sequence flanking HSS 2 just upstream of the promoter is also conserved between species (Figure 2b), whereas regions outside of the HSS are less than 50% homologous. This sequence conservation argues that the DNA immediately surrounding the novel HSS site, HSS 2, may be an important cis-regulatory element, such as a transcriptional modifier (for example, enhancer or silencer). To test this possibility functionally, 345 bp of DNA surrounding HSS 2 were subcloned into a reporter gene directed by the 1.3 kb hCD154 promoter.4 The PstI/XbaI fragment was subcloned in both the forward and reverse orientations at a site separated from the transcriptional promoter by greater than 1 kb in either direction on the plasmid. These plasmids (5′ forward and 5′ reverse) were transiently transfected into primary human peripheral blood CD4 T cells, and the cells were subsequently polyclonally activated for 6 h (only minimal background activity in the absence of stimulation). Luciferase reporter gene activity was compared between each plasmid and equimolar amounts of the parent hCD154 promoter reporter gene. The 345 bp fragment augmented CD154 promoter-driven transcription fivefold over the promoter alone in either orientation (Figure 3a), compared to threefold using the previously published 3′ enhancer.5 Unlike the 3′ enhancer,5 the novel 5′ enhancer did not act heterologously with the IL-2 promoter (Figure 3b). Nevertheless, by acting at a distance from the promoter and by enhancing promoter activity in either orientation, this upstream 345 bp fragment surrounding HSS 2 fulfilled the definition of a transcriptional enhancer element in vitro.8
Figure 3.

DNA surrounding hypersensitivity sites (HSS) 2 upstream of the CD154 promoter functions as a transcriptional enhancer element in primary CD4 T cells. (a) Primary human CD4 T cells were transiently transfected with the following various CD154 promoter driven luciferase reporter genes: (1) the human CD154 (hCD154) promoter alone (CD154-pGL24); (2) 1.3 kb of the CD154 3′ enhancer subcloned into the enhancer site of CD154-pGL2 in the germline (or forward) orientation (3′ UTR5); a PstI/XbaI 345 bp fragment subcloned into the enhancer site of CD154-pGL2 in the forward (3) or reverse (4) orientations. Following a 2-h rest the cells were stimulated for 6 h with phorbol 13-myristate 12-acetate (PMA) and ionomycin, and cell lysates were analyzed for luciferase activity. (b) Primary human CD4 T cells were transfected with a reporter gene consisting of a 0.6 kb of the human interleukin (IL)-2 gene 5′-flanking region (pIL-2) promoter fragment subcloned into the pGL2-Basic plasmid (left bars).7 Similarly, CD4 T cells were transfected with a plasmid containing a PstI/XbaI 345 bp fragment (CD154 5′ enhancer) subcloned at the enhancer site of a pIL-2 plasmid (right bars). The CD4 T cells were then rested (open bars) or stimulated with PMA and ionymycin (filled bars) for 6 h, lysed and assayed for luciferase activity. The results depicted are from one representative experiment of five (a) or four (b) experiments performed.
Transcription factors bound to the novel 5′ CD154 enhancer element
To explore the transcription factors responsible for this enhancer activity, the sequence surrounding HSS 2 was analyzed using the TRANSFAC database.9 Multiple potential binding sites for known T-cell transcription factors were identified, including four NFAT and four GATA-3 sites (Figure 4). Interestingly, this was somewhat analogous to a similarly sized enhancer element10 characterized for the IL-4 gene, which depends on GATA-3 for its expression.6Two composite sites (GATA-3/NFAT and NF-IL-6/GATA-3) in this novel enhancer were studied by electrophoretic mobility shift assay (EMSA) using nuclear extracts from activated Jurkat T cells (Figure 5). Each oligonucleotide composite site demonstrated GATA-3 binding (lanes 1 and 7, bands 1 and 2), similar to GATA-3 binding elements from other genes.11 The lower band remained (band 1), as noted for other GATA-3 sites,12 even under more stringent binding conditions (lane 6). The GATA-3 bands were both inhibited by the presence of either excess cold (unlabeled) self (lanes 2, 3, 8, 9) or the other GATA-3-binding oligonucleotide (lanes 4, 10), demonstrating specific GATA-3 binding to each probe. An additional slower migrating band (Figure 5; lane 1, band 3) was present bound to the GATA-3/NFAT probe but not to the NF-IL-6/GATA-3 probe. A portion of this band supershifted in the presence of anti-NFAT1 antibody, identifying NFAT1 as bound to this probe in vitro (lane 5). In summary, the in vitro binding assays demonstrated GATA-3 binding to both oligonucleotide probes from the 345 bp enhancer element. NFAT1 also bound the GATA-3/NFAT probe, but only GATA-3 bound the NF-IL-6/GATA-3 probe (Figure 5; and data not shown). Similar results were obtained with activated but not resting primary human CD4 T-cell nuclear extracts (data not shown). Therefore, GATA-3 and NFAT1 were likely candidates to bind the 5′ enhancer element in vivo and contribute to transcriptional activity via this cis-element.
Figure 4.

Potential transcription factor binding sites present in the upstream human CD154 (hCD154) enhancer element. The DNA sequence contained within the PstI/XbaI 345 bp fragment approximately 1.5 kb upstream of the hCD154 transcription start site (TSS) was analyzed using the TRANSFAC database. Flanking restriction enzyme sites are highlighted, and potential binding sites for known T-cell-associated transcription factors are noted. Potential GATA-3 binding sites are underlined, nuclear factor of activated T cells (NFAT) sites are underlined, italicized, and bolded, and a NF-IL-6 (C/EBPβ) site is bolded. A recognized potential TATA box is also underlined and bolded.
Figure 5.
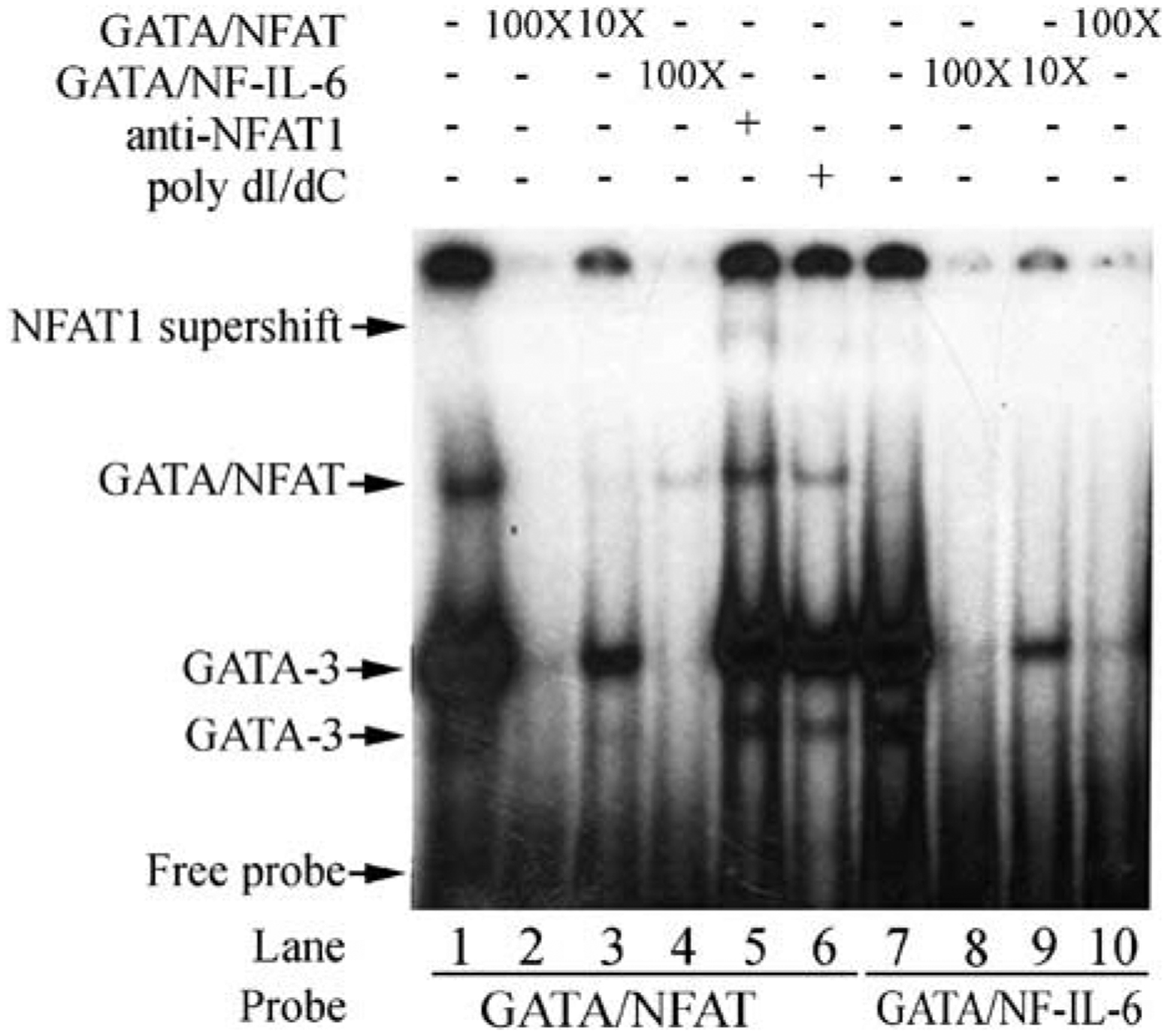
Protein binding to the CD154 5′ enhancer in vitro. Radiolabeled oligonucleotides corresponding to combinatorial binding sites for GATA/NFAT (lanes 1–6) or NF-IL-6/GATA (lanes 7–10) were incubated with nuclear extracts from polyclonally activated Jurkat T cells in the presence or absence of excess self (lanes 2, 3, 8, and 9; 10× or 100×) or cross competing (lanes 4 and 10; 100×) cold competitor oligonucleotides. In lane 5 nuclear extracts were preincubated with anti-NFAT1 antibody. In one lane (lane 6), the reaction was carried out in the presence of poly dI/dC for stringency. Proteins were electrophoresed on Tris-borate-EDTA (TBE) gels and visualized with autoradiography. Antibodies or oligonucleotide competitors are listed above the lanes, and labeled probes at the bottom of lanes. GATA-3 (bands 1 and 2) forms major and minor bands. NFAT or a complex of NFAT/GATA-3 (band 3) shows a single band higher up, and the NFAT1 top arrow (band 4) shows an NFAT1 supershift. Free oligonucleotide is noted at the bottom.
Chromatin immunoprecipitation (ChIP) was used to explore transcription factor binding to this novel 5′ enhancer element in vivo. Jurkat and primary human CD4 T cells were polyclonally activated in vitro for 2 h, and ChIP was carried out using anti-NFAT1 and a panel of commercially available anti-GATA-3 antibodies, which are capable of immunoprecipitation but poor at super-shifting in EMSA. Amplification primers and a fluorescently tagged probe were designed for use using real-time PCR to specifically and quantitatively analyze proteins bound to this 345 bp element in vivo (see Materials and methods). The Jurkat ChIP revealed evidence for both NFAT1 and GATA-3 binding (relative to the isotype control antibody) to the novel CD154 5′ enhancer, as detected by ethidium bromide staining on an agarose gel (data not shown). Real-time PCR analysis also demonstrated a significant shift of the binding curve to the left (that is, fewer cycles required for amplification to reach a threshold level) for the anti-GATA-3 antibody relative to the isotype control (Figure 6a). A summary of the in vivo binding results is presented in Figure 6b for each of the transcription factors studied. All three anti-GATA-3 antibodies demonstrated GATA-3 binding in vivo to the enhancer element in stimulated Jurkat and primary human CD4 T cells (Figure 6b). Similarly, NFAT1 was noted to bind the element in vivo in activated Jurkat T cells, and, particularly, in primary human CD4 T cells (Figure 6b). As a control, primers used to detect the proximal CD154 promoter13 demonstrated NFAT1 but not GATA-3 engagement of the proximal CD154 promoter in vivo (data not shown).13 Thus, both GATA-3 and NFAT1 bind the CD154 5′ enhancer in vivo following T-cell activation.
Figure 6.

GATA-3 and NFAT1 transcription factors bind the CD154 5′ enhancer in vivo following T-cell activation. Jurkat D1.1 (open bars) and primary human CD4 T cells (solid bars) were separately stimulated with phorbol 13-myristate 12-acetate (PMA) and ionomycin for 2 h, and transcription factor binding to the CD154 5′ enhancer was detected by chromatin immunoprecipitation (ChIP) as described.5 Protein cross-linked and sonicated DNA was immunoprecipitated with anti-GATA-3, anti-NFAT1 or isotype control antibodies. Immunoprecipitated DNA was amplified/identified using both CD154 5′ enhancer specific primers and a site-specific internal probe (see Materials and methods for details). Samples were analyzed in duplicate by real-time PCR. (a) A representative real-time plot showing DNA amplified from precipitated material using an anti-GATA-3 antibody (two overlapping left curves) versus an isotype control antibody (two right curves) is shown. Cell cycle numbers required to reach a set threshold value (Ct, horizontal line) are depicted along the x axis. (b) Specific transcription factor binding in vivo is presented as relative binding of the specific anti-transcription factor antibody compared to isotype control. A summary (mean±s.e.m.) of 4–6 experiments per transcription factor binding quantification is depicted for Jurkat D1.1 and primary human CD4 T cells.
GATA-3 is important for the transcriptional activity of the novel CD154 5′ enhancer
We next explored a functional role for GATA-3 in regulating transcription via this novel 5′ enhancer element using freshly isolated human peripheral blood CD4 T cells. Primary CD4 T cells were transiently cotransfected with a GATA-3 mammalian expression vector (or control), along with a CD154 promoter-driven reporter gene or the same reporter gene containing the 5′ enhancer element (see Figure 3). Following polyclonal CD4 T-cell activation, overexpression of GATA-3 augmented CD154-promoter driven transcription in a dose-dependent fashion for the construct containing the 5′ enhancer element, but not for the reporter plasmid with the CD154 promoter alone (Figure 7a). In addition, endogenous GATA-3 was important for enhancer activity in a polyclonal population of primary CD4 T cells, because overexpression of repressor of GATA (ROG) and a dominant negative GATA-3 construct (KRR mutant) independently inhibited transcriptional activity to levels seen using the CD154 promoter alone (Figure 7b). Moreover, overexpression of ROG inhibited activation-triggered endogenous CD154 expression in primary human CD4 T cells by over 50% (Figure 8a, dashed versus dotted line). Thus, GATA-3 is important for optimal CD154 expression in freshly isolated peripheral blood primary human CD4 T cells.
Figure 7.

GATA-3 is important for CD154 5′ enhancer activity. Primary human CD4T cells were transiently transfected with CD154-pGL2 (three left bars)4 or CD154-pGL2 including the 350 bp PstI/XbaI 5′enhancer fragment subcloned into the enhancer site (three right bars) (Figure 3). (a) The cells were cotransfected with increasing amounts of a GATA-3 expression plasmid (hatched and full bars) or control plasmid (open bars) and analyzed for transcriptional activity following 6 h of phorbol 13-myristate 12-acetate (PMA) plus ionomycin stimulation. (b) Similarly, the cells were cotransfected with expression vectors corresponding to repressor of GATA (ROG; hatched bars)14 or a dominant-negative version of GATA-3 (KRR mutant; full bars).15 Results are representative of one of four experiments performed.
Figure 8.

Interleukin (IL)-4-producing primary CD4 T cells have increased propensity for CD154 expression following T-cell activation. (a) Freshly isolated primary human peripheral blood CD4 T cells were transiently transfected with a repressor of GATA (ROG; dashed line) or control (dotted line) expression vector and then polyclonally stimulated for 6 h. Cells were analyzed by flow cytometry for CD154 expression. Isotype control antibody staining is depicted with the solid line. (c) The mean±s.e.m. of the percent maximal CD154 expression (MFI) from control (designated 100%) and ROG-transfected cells from four separate experiments is presented. (b) Peripheral blood human CD4 T cells were primed in vitro as previously described7 and expanded with IL-2 and IL-4. After 14–21 days in culture, the cells were polyclonally activated for 6 h and analyzed for intracellular cytokine staining and surface CD154 expression. CD154 expression is denoted on the x-axis for cells gated on expression of IL-4 (solid line) or interferon-γ (IFNγ; (dashed line). Isotype control antibody staining for CD154 expression is shown with a dotted line. (d) Results are representative of one of four experiments performed, and the means±s.e.m. of the percent CD154 positive for the two subsets are depicted in graphic form.
High propensity for IL-4-producing CD4 T cells to express CD154 following activation
Although GATA-3 is present in low levels in naive CD4 T cells, it is markedly upregulated in IL-4-producing Th2 cells and diminished in interferon-γ (IFNγ)-producing Th1 cells.6 There has been little published data as to differences, or lack thereof, in CD154 expression among different CD4 T-cell subsets. This is, in part, because the vast majority of CD4 T cells express CD154 following polyclonal activation ex vivo.16 In addition, both Th1- and Th2-cloned CD4 T cell lines have been shown to express CD154 following activation.17 However, long-term cloned T cell lines do not always represent the reality of primary CD4 T cells. We, therefore, decided to analyze IL-4- and IFNγ-producing primary human CD4 T cells for their ability to express CD154 following polyclonal T-cell activation.
In order to generate a significant number of both IL-4- and IFNγ-producing primary CD4 T cells, human peripheral blood CD4 T cells were primed in vitro in the presence of IL-2 and IL-4 for 2 weeks (see Materials and methods). The cells were rested without exogenous cytokines for 2 days before polyclonal T-cell activation, and then analyzed 6 h later for surface CD154 expression and intracellular cytokine expression. Interestingly, almost all of the IL-4-producing CD4 T cells expressed high levels of CD154 following activation, whereas only half of the IFNγ-producing CD4 T cells expressed CD154 (Figure 8b). The expression of CD154 was bimodal (all or none) for the IFNγ-producing CD4 T cells (Figure 8b), suggesting that only a subset of these cells was capable of any CD154 expression. It is, therefore, possible that the GATA-3-regulated novel 5′ CD154 enhancer described in this manuscript contributes to the high propensity (490%) of GATA-3-expressing IL-4-producing CD4 T cells to express CD154.
Discussion
Activation-dependent expression of CD154 by CD4 T cells is a critically important event during the immune response. Because of its pleiotropic effects on a wide variety of CD40-expressing partner cells, CD154 is very tightly regulated. This regulation is largely transcriptional, although mRNA stability and posttranslational mechanisms also control CD154 expression. We and others have previously described and characterized the CD154 transcriptional promoter.2,4,13,18–23 In addition, we have identified a 3′ CD154 transcriptional enhancer5 located just downstream of the 3′ UTR mRNA stability element.24,25 In vitro, both the hCD154 transcriptional promoter and 3′ enhancer element are relatively weak, despite the abundant and rapid generation of CD154 mRNA following CD4 T-cell activation.5 This led us to hypothesize that other CD154 cis-transcriptional regulatory elements contribute to CD154 expression.
Using DNase I HSS, we not only identified a novel HSS (HSS 1), which mapped to the hCD154 transcriptional promoter, but another unique HSS (HSS 2) was located just upstream (~1.5 kb) of the TSS (Figure 1a).2 Intriguingly, similar to the DNA present within the CD154 transcriptional promoter, the DNA sequence flanking HSS 2 was highly conserved between mouse and man (Figure 2b), arguing that this region was likely to possess important regulatory function. Recently, other investigators have reported sequence conservation of this stretch of DNA between several species.26 Remarkably analogous to a similarly sized IL-4 transcriptional enhancer, ~345 bp of species conserved sequence surrounding HSS 2 augmented CD154 promoter activity in either orientation and a distance from the promoter (Figure 3a), thus fulfilling in vitro-defined transcriptional enhancer activity.8 Finally, when this region is tested in a longer (1.7 kb) reporter construct that includes the 5′ enhancer contiguous (uninterrupted sequence) with the CD154 proximal promoter, there is approximately a threefold augmentation of transcription relative to the promoter construct (1.2 kb) lacking the upstream 5′ sequence (data not shown). Thus, the enhancer element augments CD154 promoter-driven transcription in the context of the intact 5′-flanking region of the CD154 gene. Roman and colleagues have also recently shown that this region is rich in binding sites for TFE3 and TFEB transcription factors.19
This novel 5′ CD154 transcriptional enhancer, much like the IL-4 3′ enhancer,10 possesses multiple binding sites for NFAT and GATA proteins. In peripheral T cells the only GATA protein present is GATA-3,27 and site-directed disruption of individual GATA-3 sites within the context of reporter genes have minimal to moderate (~50% reduction) inhibitory effects on transcriptional enhancement perhaps because of the redundancy of binding sites in this region (data not shown). Moreover, mutation of a potential GATA binding site in the proximal CD154 promoter has no effect on transcription.28 Nevertheless, both NFAT1 and GATA-3 bound the 5′ enhancer element in vitro and in vivo (Figures 5 and 6), and GATA-3 was found to be important for its transcriptional activity (Figure 7). Moreover, both HSS 1 (promoter) and HSS 2 (5′ enhancer) were present in primary CD4 T cells, and in an activation-independent fashion (Figure 1b). Interestingly, both HSS 1 and HSS 2 were not present in macrophages, transformed and primary B cells, or primary CD8 T cells (Figure 1c; and data not shown), suggesting that the hCD154 promoter and 5′ enhancer both contribute to the highly restricted expression pattern of CD154 by CD4 T cells. Moreover, unlike the hCD154 3′ enhancer,5 the 5′ enhancer does not act heterologously with the IL-2 promoter (Figure 3b), providing another layer of specificity to CD154 expression.
Lastly, the 5′ enhancer may also be important in preferential expression of CD154 among IL-4-producing CD4 T cells. Like the IL-4 3′ enhancer,10 the CD154 5′ enhancer possesses multiple binding sites for the Th2-specific transcription factor, GATA-3 (Figure 4). GATA-3 binding in vivo (Figure 6) also appears necessary for the enhancer activity contributed by this cis-element (Figure 7b) and for optimal CD154 expression in freshly isolated primary human CD4 T cells (Figure 8a). The importance of GATA-3 binding may also help explain the expression of CD154 by almost all IL-4-producing CD4 T cells compared to the all or none expression among subsets of IFNγ-producing CD4 T cells (Figure 8b). It is possible that GATA-3 confers an increased likelihood of CD154 expression via binding the CD154 enhancer, but that this binding is not absolutely required for CD154 expression. Along these lines, we have not noted a difference in CD154 expression in activated freshly isolated splenocytes from wild-type and GATA-3 knock-out animals (data not shown).
One possible scenario depicting the importance of GATA-3 to CD154 expression is that GATA-3 and NFAT1 bind this novel enhancer element together,29 and that NFAT recruits CBP/p300 histone deactylase to help open the locus.30 Teleologically, the expression of CD154 by almost all IL-4-producing CD4 T cells may have evolved in conjunction with IL-4 expression to assist in critical B-cell functions such as immunoglobulin class switching. This expression would still have to be tightly regulated, because continued or prolonged CD154 expression contributes to autoimmunity in the form of SLE and other autoimmune disorders.3,31,32 A better understanding of the cis- and trans-acting factors that participate in CD154 expression should assist in identifying those factors involved in CD154 dysregulation in autoimmune states.
Materials and methods
Transformed cell lines and isolation of various lymphocyte cell types
Primary human CD4 T cells were prepared by negative selection using the Rosette Sep method (Stem Cell Technologies, Vancouver, British Columbia, Canada), as previously described.33 Similarly, primary human CD8 T cell and primary human B cells were isolated by negative selection by analogous approaches. Each specific cell type was more than 90% specific for CD4 T cells, CD8 T cell and B cells, respectively. In vitro primed primary human CD4 T cells were generated as previously described7 with the addition of rIL-4 (10 IU ml−1) in addition to IL-2 (10 IU ml−1). U93734 and THP-135 monocyte/macrophage cell lines were generously provided by Dr Kate Sullivan (Children’s Hospital of Philadelphia, PA, USA). Two different EBV-transformed human B cell lines, A2D6 and A1,36 were graciously supplied by Dr Mariuz Wasik (University of Pennsylvania, Philadelphia, PA, USA), and the Jurkat D1.1 human CD4 thymoma cell line,37 which constitutively expresses CD154, was a gift from Dr Seth Lederman (Columbia University, New York, NY, USA). All transformed cell lines were routinely passed in vitro in tissue culture according to standard protocols. Human peripheral blood was obtained with appropriate IRB approval.
Nuclear preparations and DNase I hypersensitive site mapping
Nuclei from established transformed cell lines or primary human cells were prepared by NP-40 lysis as described by Siebenlist et al.,38 using 0.1–0.3% NP-40 followed by either a sucrose gradient5 or gentle (980 g) centrifugation and subsequent wash in buffer not containing NP-40 after treatment to obtain pure nuclei. Nuclei were gently (by vortex and stirring) resuspended in nuclear isolation buffer (60 mm KCl, 15 mm NaCl, 5 mm MgCl2, 0.1 mm EGTA, 15 mm Tris-HCl (pH 7.0), 0.5 mm dithiothreitol, 0.1 mm AEBSF (4-(2-aminoethyl)-benzene-sulfonylfluoride), 0.3 m sucrose) containing 5% glycerol, and aliquoted into 0.5 ml samples each containing 5–10×106 nuclei. Microgram amounts of DNase I (1 μg μl−1; (Worthington, Lakewood, NJ, USA) were added (in 40 μl volumes containing 5 mm CaCl2, 1 mm MgCl2) to each nuclear sample, with water as a control. Empirically, either twofold dilutions were used (to find the proper range of digestion), or, if in the range of DNase activity, μg ml−1 concentrations were added (for example, 4–6μg ml−1) to successive samples for 3 min at room temperature.39 Reactions were stopped with 50 μl of 5% SDS, 100 mm EDTA. Samples were then digested with 50 μl of 5 mg ml−1 proteinase K at 37 °C overnight, or at 55 °C for 1 h. DNA was subsequently isolated by repeated organic extraction and ethanol precipitation.
DNase I-treated samples were digested with BamHI, and electrophoresed on 1% agarose gels using 10 μg of sample per lane. For fine mapping, double digested (BamHI plus selected restriction endonucleases) fragments of the 6.4 kb DNase-untreated sample were run in parallel lanes. Gels were blotted onto Hybond plus nylon membrane (Pharmacia, Pisctaway, NJ, USA), and Southern blotting was performed overnight in 20×SSC. A 280 bp32P-labeled probe, corresponding to the 5′ end of the 6.4 kb BamHI fragment and containing the promoter and first two exons of the hCD154 gene, was generated by PCR using primers 5′-CTCTGACTTGGGCAATTA-3′ and 5′-CATGGTTTCATTGGGTGATTC-3′. Hybridization was performed in roller bottles using QuickHyb (Strata-gene, La Jolla, CA, USA) solution with 1.25×107 c.p.m. of spun column-purified probe per 10 ml hybridization solution at 68 °C. The final wash of high stringency was with 0.05×SSC and 0.1% SDS at 65 °C for 30 min. Bands were visualized by standard autoradiography as described.40
Reporter plasmid generation and transcription assays
The luciferase containing reporter plasmid pCD154-Luc,4 containing 1.3 kb of the hCD154 proximal promoter inserted into the promoter site of pGL2-Basic (Promega, Madison, WI, USA), was modified by conventional subcloning,5 such that a 345 bp PstI/XbaI fragment from 1.5 kb upstream of the CD154 TSS was inserted into the enhancer site of the plasmid in either the forward or reverse orientations (pCD154–5′F-Luc and pCD154–5′R-luc, respectively). Similarly, the same 345 bp fragment was inserted into pIL-2-Luc, containing 0.6 kb of the hIL-2 promoter upstream of luciferase,7 at the enhancer site in the forward orientation (pIL-2-CD154–5′F-Luc). Multiple independent preparations of each plasmid, to be used in reporter gene assays, were isolated by a commercial affinity column following the manufacturer’s instructions (Qiagen, Chatsworth, CA, USA). The wild-type (WT) (pEF1α-puro-GATA-3)41 and dominant-negative (KRR mutant)15 GATA-3 expression plasmids were generously provided by Dr Gerd Blobel (Children’s Hospital of Philadelphia) and Dr Ginny Shapiro (University of Pennsylvania), respectively. The ROG expression plasmid has been previously described.14
For reporter gene assays, primary human CD4 T cells were transiently cotransfected with luciferase reporter plasmids (see above) along with an expression vector (or control plasmid) using our own published protocol.42 Cells were stimulated in duplicate samples with phorbol 13-myristate 12-acetate (PMA) and ionomycin for 6 h and cell lysates were analyzed for firefly luciferase activity by luminometry as described.7 Transfection efficiency was corrected for by measuring Renilla luciferase activity (Promega) from a cotransfected pRL-null plasmid (Promega) as described,33 and results are presented as means±s.e.m.
Nuclear protein preparation and electrophoretic mobility shift assays
Primary or in vitro primed human CD4, or Jurkat D1.1, T cells were stimulated with PMA (25 ng ml−1) and ionomycin (1.5 μm) for 2 h. Nuclear proteins were extracted by NP-40 detergent lysis43 or dounce homogenization in hypotonic buffer (to isolate nuclei) followed by high salt (1.2 m KCl) extraction and dialysis into binding buffer (10 mm Tris-HCl, pH 7.5, 100 mm KCl, 10% glycerol, 50 ng ml−1 poly(dI-dC), 1.5 mm MgCl2, 0.2 mm EDTA, 1 mm dithiothreitol, and 0.2 mm phenylmethylsulfonyl fluoride), aliquoted and frozen at −70°C. Protein concentration was determined the by Bradford method using a commercial kit (Pierce, Rockford, IL, USA). One μl of 10 μm single-stranded oligonucleotide was labeled with γ32P-ATP using T4 polynucleotide kinase and then annealed (by heating to 95 °C in a heating block and allowing it to cool to room temperature) with an excess of its partner strand. This was then ethanol precipitated with carrier (sonicated salmon sperm DNA), washed extensively with 80% ethanol (by successive centrifugation to remove excess free label), and 5×104 c.p.m. labeled double-stranded (ds) probe was used per reaction. Cold ds inhibitors were made by annealing equal amounts of the partner oligonucleotides. Where indicated, an anti-NFAT1 antibody, N58820 (Transduction Laboratories, Lexington, KY, USA), was preincubated with nuclear extract. The initial reaction with 1–5 μg of nuclear extract was mixed with 1 μl of antibody, cold ds inhibitor (molar amount as indicated), or nothing and incubated at room temperature for 15 min. Then, 32P-labeled ds oligonucleotide was added for another 10-min incubation at room temperature. Ten μl reactions were chilled along with loading dye and run on 6% preformed Tris-borate-EDTA (TBE) gels (Novex; Invitrogen, San Diego, CA, USA) in 0.5×TBE buffer. Gels were dried and bands were identified by autoradiography. Nucleotide sequences for the oligonucleotide probes are the following (NFAT and NF-IL-6 sites are underlined, and GATA sites are boldened on the coding strands):
NF-IL-6-GATA, 5′-TGGCAGATGTAGTGAAAAGCTACATAGATCTGGGCCCAG-3′ (coding strand) and 5′-CTGGGCCCAGATCTATGTAGCTTTTCACTACATCTGCCA-3′ (noncoding strand); GATA-NFAT, 5′-GGCTCTCATGATAATTTTCCTTCAGTGGAACTAAGG-3′ (coding strand) and 5′-CCTTAGTTCCACTGAAGGAAAATTATCATGAGAGCC-3′ (noncoding strand).
Chromatin immunoprecipitation assay
Jurkat D1.1 or primary human CD4 T cells were stimulated for 2 h with phorbol ester and ionomycin. DNA was cross-linked with 1% formaldehyde (Fisher Scientific, Pittsburgh, PA, USA), stopped with 0.125 m (final) glycine, washed with cold phosphate-buffered saline containing protease inhibitor cocktail (Sigma P-8340, St Louis, MO, USA), resuspended in SDS lysis buffer (this and all following reagents from ChIP assay kit no. 17–295; Upstate Biotechnology, Lake Placid, NY, USA) for 30 min on ice with vortexing, passed through a Qiashredder (Qiagen, Valencia, CA, USA) and diluted 1:3 in 10 mM Tris, 1 mM EDTA (TE; to decrease the 1% SDS in lysis buffer to avoid foaming during sonification). Four separate 12-s pulses of sonification, with a Branson 750 Sonifier (setting of 3; Branson Ultrasonics, Danbury, CT, USA) on ice, generated DNA fragments ranging between 300–1000 bp as checked on agarose gel by adding 1% SDS to sonicated and heated (65 °C oven for 4 h) samples. After preclearing divided supernatants (200 μl in 1200 μl of dilution buffer) with 80 μl of immunobead reagent (protein A-agarose/bovine serum albumin (BSA)/ssDNA; or protein G-agarose/BSA/ssDNA) appropriate for the relevant subsequent immunoprecipitating antibody isotype, 10 μl of each test antibody (anti-NFAT1, N58820; HG3–31X mouse mAb (α-GATA-3; Santa Cruz Biotechnology, Santa Cruz, CA, USA); HG3–35X, human specific mouse mAb (α-GATA-3; Santa Cruz Labs); H-48X, a rabbit polyclonal antibody (μ-GATA-3; Santa Cruz Labs)) and their respective (mouse or rabbit) isotype controls, were added to incubate overnight at 4 °C. Appropriate immunobead reagent was added, and beads were gently washed successively (low salt, high salt, LiCl and TE), according to kit instructions. DNA was then eluted with freshly prepared 1% SDS in 0.1 m NaHCO3, reverse cross-linked (65 °C for 4 h in 0.3 m NaCl), treated with proteinase K, extracted with phenol/chloroform, treated with RNase A (40 μg ml−1) and equal amounts of all immunoprecipitated DNA samples were analyzed first by ‘hot start’ PCR (using oligo primers corresponding to 1.5 kb upstream of the hCD154 TSS; CD154ChIP forward 5′-GCCCATTTTGGCTCTCATGA-3′ and CD154ChIP reverse 5′-AGCTTTTCACTACATCTGCCAAGTAG-3′) for 30 cycles, where total accumulated DNA differences versus that of the respective control immunoglublin immunoprecipitate were visualized on agarose gels with ethidium bromide.
To obtain a more critical analysis of immunoprecipitating sample differences from the ChIP experiments above, real-time PCR was done,5,44 which measures curve differences during the log phase of amplification. Primer Express software package that accompanies the Applied Biosystems Model 7700 sequence detector was used to design a Taqman probe, which would cover the region where the transcription factors would be expected to bind the DNA. The 27-mer selected was then synthesized and fluorescent-tag labeled at the 5′ end with 6-fluoro amidite (6-FAM) and at the 3′ end with BlackHole Quencher-1 (BHQ-1; IDT technologies, Coraville, IA, USA) as follows: 5′-FAM-TCAAAGTTCATCACGTGCATCGAAGCC-3BHQ-1/−3′. Then with 900 μm of the above primers (CD154ChIP forward and reverse), 200 μm of the probe was then used in Taqman Universal PCR Master Mix (Roche Diagnostics, Branchberg, NJ, USA) to amplify DNA immunoprecipitating samples above (NFAT1, GATA-3, Oct-1, isotype controls), in triplicate for real-time experiments on the Applied Biosystems Model 7700. Threshold (Ct) values and absorbance curves (ΔRn = y axis, cycle number x = axis) of experimental versus controls were analyzed and compared with SDS software on a MAC computer. Fold induction of the transcription factor specific antibody versus the isotype control at values greater than 1 were considered significant evidence of binding in vivo.
Flow cytometric analyses
For Figure 8a, primary human peripheral blood CD4 T cells were transiently transfected with a ROG expression vector or control (as above) and rested for 2 h before restimulation. The cells were treated with PMA (25 ng ml−1) and ionomycin (1.5 μm) for 6 h before antibody staining for flow cytometry. For Figure 8b, primary human CD4 T cells were primed in vitro for 2–3 weeks in the presence of rIL-2 (50 U ml−1) and rIL-4 (10 ng ml−1) as described above.7 The cells were maintained without exogenous cytokines for 2 days before cytokine analysis. The cells were then stimulated with PMA (25 ng ml−1) and ionomycin (1.5 μm) for 6 h before antibody staining for flow cytometry. Detection of intracellular IL-4 and IFNγ using directly FITC-conjugated reagents (Pharmingen, San Diego, CA, USA) was carried out as previously described.7 Cell surface CD154 was detected with PE-conjugated anti-CD154 mAb (Ancell, Bayport, MN, USA). All negative gates were determined using the appropriate isotype control, fluorochrome-conjugated reagents.
Acknowledgements
We thank Dr Kate Sullivan, Dr Gerd Blobel, Dr Ginny Shapiro, Dr Seth Lederman and Dr Mariuz Wasik for the generous gifts of reagents and cell lines as detailed in the Materials and methods. We also thank Dr Terri Finkel and Dr Gerd Blobel for critical reviews of the article. This work was supported, in part, by grants from the Arthritis Foundation, the Arthritis National Research Foundation, the Dorough Lupus Foundation, the Kahn Foundation for Lupus Research, the American College of Rheumatology Research and Education Foundation and the National Institutes of Health (R01-AR48257, R21-AR49335, P30-HH2815, M01-RR240; RQC).
Footnotes
Conflict of interest
No financial interests to disclose.
References
- 1.Van Kooten C, Banchereau J. CD40-CD40 ligand: a multi-functional receptor-ligand pair. Adv Immunol 1996; 61: 1–77. [DOI] [PubMed] [Google Scholar]
- 2.Cron RQ. CD154 transcriptional regulation in primary human CD4 T cells. Immunol Res 2003; 27: 185–202. [DOI] [PubMed] [Google Scholar]
- 3.Cron RQ. CD154 and lupus. Pediatr Rheumatol Online J 2003; 1: 172–181. [Google Scholar]
- 4.Schubert LA, King G, Cron RQ, Lewis DB, Aruffo A, Hollenbaugh D. The human gp39 promoter. Two distinct nuclear factors of activated T cell protein-binding elements contribute independently to transcriptional activation. J Biol Chem 1995; 270: 29624–29627. [DOI] [PubMed] [Google Scholar]
- 5.Schubert LA, Cron RQ, Cleary AM, Brunner M, Song A, Lu L-S et al. A T cell-specific enhancer of the human CD40 ligand gene. J Biol Chem 2002; 277: 7386–7395. [DOI] [PubMed] [Google Scholar]
- 6.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 1997; 89: 587–596. [DOI] [PubMed] [Google Scholar]
- 7.Cron RQ, Bort SJ, Wang Y, Brunvand MW, Lewis DB. T cell priming enhances IL-4 gene expression by increasing nuclear factor of activated T cells. J Immunol 1999; 162: 860–870. [PubMed] [Google Scholar]
- 8.Blackwood EM, Kadonaga JT. Going the distance: a current view of enhancer action. Science 1998; 281: 61–63. [DOI] [PubMed] [Google Scholar]
- 9.Kel A, Voss N, Jauregui R, Kel-Margoulis O, Wingender E. Beyond microarrays: finding key transcription factors controlling signal transduction pathways. BMC Bioinformatics 2006; 7 (Suppl 2): S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agarwal S, Avni O, Rao A. Cell-type-restricted binding of the transcription factor NFAT to a distal IL-4 enhancer in vivo. Immunity 2000; 12: 643–652. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Shannon MF, Young IG. A role for Ets1, synergizing with AP-1 and GATA-3 in the regulation of IL-5 transcription in mouse Th2 lymphocytes. Int Immunol 2006; 18: 313–323. [DOI] [PubMed] [Google Scholar]
- 12.Kishikawa H, Sun J, Choi A, Miaw SC, Ho IC. The cell type-specific expression of the murine IL-13 gene is regulated by GATA-3. J Immunol 2001; 167: 4414–4420. [DOI] [PubMed] [Google Scholar]
- 13.Cron RQ, Bandyopadhyay R, Genin A, Brunner M, Kersh GJ, Yin J et al. Early growth response-1 is required for CD154 transcription. J Immunol 2006; 176: 811–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miaw SC, Choi A, Yu E, Kishikawa H, Ho IC. ROG, repressor of GATA, regulates the expression of cytokine genes. Immunity 2000; 12: 323–333. [DOI] [PubMed] [Google Scholar]
- 15.Smith VM, Lee PP, Szychowski S, Winoto A. GATA-3 dominant negative mutant. Functional redundancy of the T cell receptor alpha and beta enhancers. J Biol Chem 1995; 270: 1515–1520. [DOI] [PubMed] [Google Scholar]
- 16.Jullien P, Cron RQ, Dabbagh K, Cleary A, Chen L, Tran P et al. Decreased CD154 expression by neonatal CD4+ T cells is due to limitations in both proximal and distal events of T cell activation. Int Immunol 2003; 15: 1461–1472. [DOI] [PubMed] [Google Scholar]
- 17.Roy M, Waldschmidt T, Aruffo A, Ledbetter JA, Noelle RJ. The regulation of the expression of gp39, the CD40 ligand, on normal and cloned CD4+ T cells. J Immunol 1993; 151: 2497–2510. [PubMed] [Google Scholar]
- 18.Tsytsykova AV, Tsitsikov EN, Geha RS. The CD40L promoter contains nuclear factor of activated T cells-binding motifs which require AP-1 binding for activation of transcription. J Biol Chem 1996; 271: 3763–3770. [DOI] [PubMed] [Google Scholar]
- 19.Huan C, Kelly ML, Steele R, Shapira I, Gottesman SR, Roman CA. Transcription factors TFE3 and TFEB are critical for CD40 ligand expression and thymus-dependent humoral immunity. Nat Immunol 2006; 7: 1082–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siddiqa A, Sims-Mourtada JC, Guzman-Rojas L, Rangel R, Guret C, Madrid-Marina V et al. Regulation of CD40 and CD40 ligand by the AT-hook transcription factor AKNA. Nature 2001; 410: 383–387. [DOI] [PubMed] [Google Scholar]
- 21.Lobo FM, Zanjani R, Ho N, Chatila TA, Fuleihan RL. Calcium-dependent activation of TNF family gene expression by Ca2+/calmodulin kinase type IV/Gr and calcineurin. J Immunol 1999; 162: 2057–2063. [PubMed] [Google Scholar]
- 22.Srahna M, Remacle JE, Annamalai K, Pype S, Huylebroeck D, Boogaerts MA et al. NF-kappaB is involved in the regulation of CD154 (CD40 ligand) expression in primary human T cells. Clin Exp Immunol 2001; 125: 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindgren H, Axcrona K, Leanderson T. Regulation of transcriptional activity of the murine CD40 ligand promoter in response to signals through TCR and the costimulatory molecules CD28 and CD2. J Immunol 2001; 166: 4578–4585. [DOI] [PubMed] [Google Scholar]
- 24.Ford GS, Barnhart B, Shone S, Covey LR. Regulation of CD154 (CD40 ligand) mRNA stability during T cell activation. J Immunol 1999; 162: 4037–4044. [PubMed] [Google Scholar]
- 25.Hamilton BJ, Genin A, Cron RQ, Rigby WF. Delineation of a novel pathway that regulates CD154 (CD40 ligand) expression. Mol Cell Biol 2003; 23: 510–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steiper ME, Parikh SJ, Zichello JM. Phylogenetic analysis of the promoter region of the CD40L gene in primates and other mammals. Infect Genet Evol 2008; 8: 406–413. [DOI] [PubMed] [Google Scholar]
- 27.Ho IC, Pai SY. GATA-3—not just for Th2 cells anymore. Cell Mol Immunol 2007; 4: 15–29. [PubMed] [Google Scholar]
- 28.Crist SA, Sprague DL, Ratliff TL. Nuclear factor of activated T cells (NFAT) mediates CD154 expression in megakaryocytes. Blood 2008; 111: 3553–3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell 2002; 109 (Suppl): S67–S79. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Rodriguez C, Rao A. Nuclear factor of activated T cells (NFAT)-dependent transactivation regulated by the coactivators p300/CREB-binding protein (CBP). J Exp Med 1998; 187: 2031–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Datta SK, Kalled SL. CD40-CD40 ligand interaction in autoimmune disease. Arthritis Rheum 1997; 40: 1735–1745. [DOI] [PubMed] [Google Scholar]
- 32.Crow MK, Kirou KA. Regulation of CD40 ligand expression in systemic lupus erythematosus. Curr Opin Rheumatol 2001; 13: 361–369. [DOI] [PubMed] [Google Scholar]
- 33.Cron RQ. HIV-1, NFAT, and cyclosporin: immunosuppresion for the immunosuppressed? DNA Cell Biol 2001; 20: 761–767. [DOI] [PubMed] [Google Scholar]
- 34.Larrick JW, Fischer DG, Anderson SJ, Koren HS. Characterization of a human macrophage-like cell line stimulated in vitro: a model of macrophage functions. J Immunol 1980; 125: 6–12. [PubMed] [Google Scholar]
- 35.Cossu G, Kuo AL, Pessano S, Warren L, Cooper RA. Decreased synthesis of high-molecular-weight glycopeptides in human promyelocytic leukemic cells (HL-60) during phorbol ester-induced macrophage differentiation. Cancer Res 1982; 42: 484–489. [PubMed] [Google Scholar]
- 36.Majewski M, Korecka M, Kossev P, Li S, Goldman J, Moore J et al. The immunosuppressive macrolide RAD inhibits growth of human Epstein-Barr virus-transformed B lymphocytes in vitro and in vivo: a potential approach to prevention and treatment of posttransplant lymphoproliferative disorders. Proc Natl Acad Sci USA 2000; 97: 4285–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yellin MJ, Lee JJ, Chess L, Lederman S. A human CD4- T cell leukemia subclone with contact-dependent helper function. J Immunol 1991; 147: 3389–3395. [PubMed] [Google Scholar]
- 38.Siebenlist U, Franzoso G, Brown K. Structure, regulation and function of NF-kappa B. Annu Rev Cell Biol 1994; 10: 405–455. [DOI] [PubMed] [Google Scholar]
- 39.Cockerill PN. Identification of DNaseI hypersensitive sites within nuclei. Methods Mol Biol 2000; 130: 29–46. [DOI] [PubMed] [Google Scholar]
- 40.Cron RQ, Coligan JE, Bluestone JA. Polymorphisms and diversity of T-cell receptor-gamma proteins expressed in mouse spleen. Immunogenetics 1990; 31: 220–228. [DOI] [PubMed] [Google Scholar]
- 41.Blobel GA, Simon MC, Orkin SH. Rescue of GATA-1-deficient embryonic stem cells by heterologous GATA-binding proteins. Mol Cell Biol 1995; 15: 626–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cron RQ, Schubert LA, Lewis DB, Hughes CC. Consistent transient transfection of DNA into non-transformed human and murine T-lymphocytes. J Immunol Methods 1997; 205: 145–150. [DOI] [PubMed] [Google Scholar]
- 43.Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res 1989; 17: 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Christenson LK, Stouffer RL, Strauss JF III. Quantitative analysis of the hormone-induced hyperacetylation of histone H3 associated with the steroidogenic acute regulatory protein gene promoter. J Biol Chem 2001; 276: 27392–27399. [DOI] [PubMed] [Google Scholar]


