Abstract
Background
Methotrexate (MTX) is an antifolate agent which is widely used in clinic for treating malignancies, rheumatoid arthritis and ectopic pregnancy. As reported, MTX has side effects on gastrointestinal system, nervous system and reproductive system, while its potential damages on oocyte quality are still unclear. It is known that oocyte quality is essential for healthy conception and the forthcoming embryo development. Thus, this work studied the effects of MTX on the oocyte quality.
Results
We established MTX model mice by single treatment with 5 mg/Kg MTX. Both morphological and molecular biology studies were performed to assess the in-vivo matured oocytes quality and to analyze the related mechanisms. The in-vivo matured oocytes from MTX-treated mice had poor in-vitro fertilization ability, and the resulting embryo formation rates and blastocyst quality were lower than the control group. We found that the in-vivo matured MTX-treated mouse oocytes displayed abnormal transcript expressions for genes of key enzymes in the folate cycles. MTX increased the rate of abnormal chromosome alignment and affected the regulation of chromosome separation via disrupting the spindle morphology and reducing the mRNA expressions of MAD2 and Sgo1. MTX reduced the DNA methylation levels in the in-vivo matured oocytes, and further studies showed that MTX altered the expressions of DNMT1 and DNMT 3b, and may also affect the levels of the methyl donor and its metabolite.
Conclusions
MTX impaired the in-vivo matured mouse oocyte quality by disturbing folate metabolism and affecting chromosome stability and methylation modification.
Keywords: Methotrexate, Oocyte quality, Folate metabolism, Chromosome, Methylation modification
Background
Methotrexate (MTX) is a folate antagonist which is transferred into the cell by the solute carrier family 19 (SLC19A) and competitively inhibits the dihydrofolate reductase (DHFR) activity [1–4]. Thus, MTX reduces catalytic conversion of dihydrofolate (DHF) to tetrahydrofolate (THF), namely seriously disturbing the folate metabolism [1, 2, 5]. The several important enzymes in the folate pathway (including serine hydroxymethyl transferase (SHMT), 5,10-methylene THF reductase (MTHFR), methionine synthase reductase (MTRR), methionine adenosyl transferase (MAT), cystathionine β-synthase (CSB), and so on [6]; as shown in Fig. S1) may also be indirectly inhibited by MTX. Additionally, the folate metabolites (such as purine, thymidine, S-adenosylmethionine (SAM); as shown in Fig. S1), which are essential for the DNA and RNA synthesis and methylation modification [7, 8], may also be affected by the MTX toxicity.
Due to the biological activities of MTX, MTX is widely used in clinic for the treatment of various neoplastic diseases (such as leukemia and lymphoma), various autoimmune and inflammatory disorders (rheumatoid arthritis and psoriasis, for example) [9–12]. For the female patients, MTX also is the common medicament used for the termination of pregnancy, ectopic pregnancy, and other uterine diseases [13, 14]. It is not desirable that MTX causes damages to various organs including stomach, intestine, hematologic system, liver, lung and central nervous system [15–19]. In addition, MTX also has side effects on reproductive system usually with defective oogenesis and spermatogenesis which may result in infertility [20, 21]. Until now, the studies of the association between female fertility and MTX toxicity have been mainly focused on analyzing the effects of MTX on follicle stimulating hormone, antral follicle count, oocyte yield, ovarian reserve, ovarian responsiveness, and so on [22–24]. It is known that oocyte quality is critical for female fertilization and embryonic development. However, the association between MTX toxicity and oocyte quality is still unclear.
The purpose of this study was to explore the effects of MTX on the oocyte quality and its possible mechanisms. We firstly confirmed the effects of MTX on in-vivo matured oocyte quality by analyzing the in-vitro fertilization ability and the resulting embryo formation. Considering the metabolic pathway of MTX, the transcript expressions for genes of the key enzymes in the folate cycles were examined. Chromosome stability is a major factor in developmental potential of oocytes. Therefore, the chromosome alignment and the separation regulation were analyzed in this work. It is known that the correct separation of chromosomes is regulated by spindle formation, spindle assembly checkpoint (SAC) and cohesin dissociation [25, 26]. Mitotic arrest deficient 2 (MAD2) is the main component of SAC, and Shugoshin 1 (Sgo1) plays a key role in protecting cohesion [25–27]. Thus, this work examined the spindle morphology and the mRNA levels of MAD2 and Sgo1 to determine the effects of MTX on chromosome separation. DNA methylation is an important epigenetic modification, which has close link with gene expression and chromatin organization [28, 29]. The folate metabolite, SAM, is required as methyl donor for methyl metabolism [7]. DNA methyltransferases (DNMTs) have the functions of methylating DNA, namely, add a methyl from SAM to the carbon 5 position of CpG dinucleotide [30]. Thus, to determine the MTX-induced changes on DNA methylation modification, we also examined the expressions of DNMTs and the concentrations of SAM and its metabolite, S-adenosylhomocysteine (SAH). Our observations exhibit insights into the origin of MTX toxicity on oocyte quality and developmental competence.
Results
MTX reduced the in-vitro fertilization ability of in-vivo matured oocytes and the embryo development potential
As seen in Table 1, the in-vitro fertilization rate of in-vivo matured oocytes from MTX model mice was 59.9% ± 3.7%, which was much lower than 81.5% ± 2.1% in the control group (p < 0.05). The resulting 2-cell rate, 8-cell rate and blastocyst rate in the MTX group were also greatly reduced (46.9% ± 2.2, 29.9% ± 2.1 and 21.2% ± 3.3% in the MTX group versus 76.7% ± 1.8, 59.4% ± 2.5 and 48.8% ± 3.1% in the control group). Blastocyst quality was assessed by the ratio of ICM cells to total cells. In the control group, the ICM cell number and the total cell number were 14.6 ± 1.7 and 46.1 ± 2.2, the radio of which was 31.7% ± 4.0%. The corresponding estimates in the MTX group were 11.5 ± 1.9, 45.5 ± 2.0 and 25.8% ± 3.9%. There were significant differences in the ICM cell number and the radio between the control and MTX groups.
Table 1.
Effects of MTX on the oocyte fertilization and the fertilized embryo formation
| Fertilization rate (%) | 2-cell rate (%) | 8-cell rate (%) | Blastocyst rate (%) | No. of ICM; No. of total cells; ratio (%) | |
|---|---|---|---|---|---|
| Control group (total number of oocytes, No.168) | 81.5 ± 2.1 | 76.7 ± 1.8 | 59.4 ± 2.5 | 48.8 ± 3.1 |
14.6 ± 1. 7 46.1 ± 2.2 31.7 ± 4.0 |
| MTX group (No.124) | 59.9 ± 3.7* | 46.9 ± 2.2* | 29.9 ± 2.1* | 21.2 ± 3.3* |
11.5 ± 1.9* 45.5 ± 2.0 25.8 ± 3.9* |
* p < 0.05, There is significant difference between the MTX and control groups
MTX reduced the mRNA transcriptional expressions for the key enzymes involved in the folate cycles
There were no significant differences on the total RNA quantity between control (about 0.39 ± 0.03 ng RNA per oocyte) and MTX (about 0.38 ± 0.02 ng RNA per oocyte) groups. Real-time PCR results showed that the mRNA expressions of the key enzymes in the folate cycles, namely, SLC19A, DHFR, SHMT1, SHMT2, MTHFR, MTRR, MAT1A and CSB were significantly reduced in the MTX-treated mouse matured oocytes (0.259 ± 0.086, 0.25 ± 0.069, 0.509 ± 0.095, 0.491 ± 0.069, 0.397 ± 0.061, 0.405 ± 0.043, 0.491 ± 0.112, 0.586 ± 0.095 versus 1.0; Fig. 1).
Fig. 1.
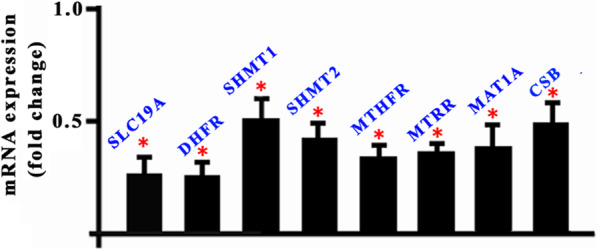
Transcript expressions for the key enzymes of the folate metabolism in the MTX-treated mouse oocytes. * p<0.05
MTX increased the errors in the chromosome alignment
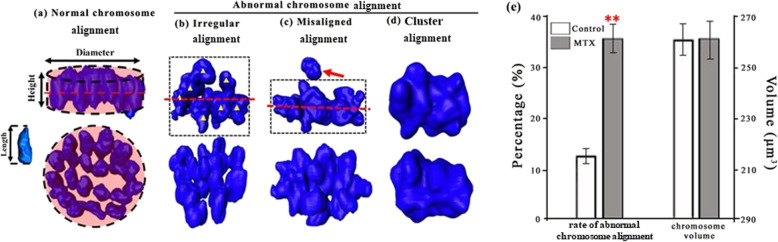
In this work, chromosome alignments at the second metaphase of mouse oocytes (Fig. 2a-d) were divided into normal alignment and abnormal alignment (including irregular alignment, misaligned alignment and cluster alignment). The rate of abnormal alignment in the MTX group was significantly higher than that in the control group (35.0% ± 2.1% versus 12.8% ± 1.2%; Fig. 2e). The oocyte sizes in both MTX and control groups were similar (74.1 ± 5.5 μm versus 75.1 ± 5.0 μm), also, the spatial area occupied by the chromosome fluorescence did not significantly differ (263.2 ± 6.5 μm3 versus 263.7 ± 8.2 μm3; Fig. 2e).
Fig. 2.
Effects of MTX on the chromosome alignment. Chromosomes were visualized using Hoechst 33342, and were reconstructed using Amira 5.2. Chromosome alignments were observed from the directions parallel and perpendicular to the equatorial plate. The dot line indicates the equatorial plate. a Normal chromosome alignment in the in-vivo matured oocyte is rather regular and looks like a disc-shaped plate, namely, all the chromosomes are neatly arranged at the equatorial plate. We used a short circular cylinder as the model of chromosome plate, and the volume and surface area of this cylinder were same to the chromosome pate. The cylinder height (H) and univalent length (L) were measured. The normal alignment had the characteristic of H ≤ 2 L. b The irregular alignment had the characteristic of H>2 L. c The misaligned alignment was that individual univalents were not on the equatorial plate, while others were still along the plate. The red arrow indicates the misaligned chromosome. d The chromosome alignment of all the chromosomes clustered together in a great mass was defined as cluster alignment, and the equatorial plate can’t be found. The rates of abnormal chromosome alignment and chromosome volumes were shown in e. Chromosome volume was estimated by the spatial area occupied by fluorescence, and was calculated using Amira 5.2. The oocyte numbers for analyzing the rates of abnormal chromosome alignment were 384 in the control group, and 275 oocytes in the MTX group. ** p<0.01
MTX affected the regulation of chromosomes separation
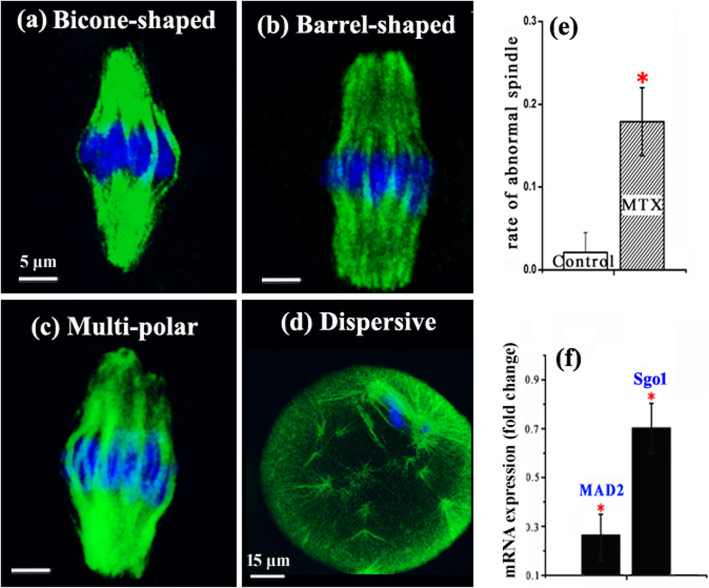
The meiotic spindle morphologies and mRNA levels of MAD2 and Sgo1 were examined to analyze the effects of MTX on centromere-associated regulatory facts in chromosome segregation and spindle assembly checkpoint. In this work, we observed three types of spindles (Fig. 3a-d): bipolar spindle (further divided into bicone-shaped spindle, barrel-shaped spindle), multi-polar spindle and dispersive spindle. Multi-polar spindle and dispersive spindle were defined as abnormal morphology. The rate of abnormal spindles in the MTX group was much higher than that in the control group (17.7% ± 4.2% versus 1.9% ± 2.5%; Fig. 3e). Real-time PCR results showed that the transcript levels of MAD2 and Sgo1 were much lower in the MTX group than the control group (0.262 ± 0.093, 0.697 ± 0.129 versus 1.0; Fig.3f).
Fig. 3.
Effects of MTX on the meiotic spindles and transcripts for MAD2 and Sgo1. Oocytes were labelled with antibodies against β-tubulin (green) and were counterstained with Hoechst 33342 (blue). Different spindle morphologies are shown in a-d. The rates of abnormal spindle are shown in e. mRNA transcript levels of MAD2 and Sgo1 are shown in f. The oocyte numbers for analyzing the rates of abnormal spindle were 384 in the control group, and 275 oocytes in the MTX group. * p<0.05
MTX altered the DNA methylation modification
Fluorescence polarization was used to determine the global DNA methylation level (that is a simple and new method). It was found that the global DNA methylation level in the MTX group was 32.7% ± 3.4%, much lower than 41.2% ± 2.57% in the control group. Besides, we also assessed DNA methylation level by the traditional method of immunostaining for 5MeC, and found the same results that the relative fluorescence intensity of 5MeC in the MTX group was much lower than that in the control group (0.696 ± 0.139 vs 1.0; Supplementary materials).
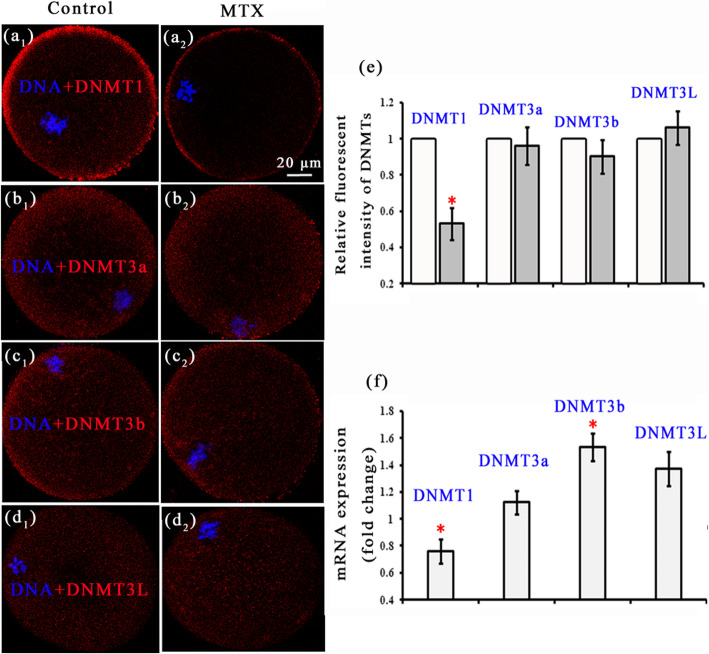
Further, we examined the expressions of DNMTs, and found that in both MTX and control group, DNMT1 in the in-vivo matured oocyte was mainly localized in the cortical region, however, DNMT3a, 3b, 3 L were uniformly distributed in the ooplasm (Fig. 4a-d). The relative fluorescence intensity of DNMT1 in the MTX group was much lower than that in the control group (0.536 ± 0.089 versus 1.0; Fig.4e), while no significant differences were found between the fluorescence intensities of DNMT3a, 3b and 3 L in the MTX and control groups (0.961 ± 0.104, 0.906 ± 0.093, 1.06 ± 0.094 versus 1.0; Fig.4e). Compared with the control group, the mRNA expression for DNMT1 in the MTX group was lower (0.763 ± 0.091 versus 1.0; Fig.4f), while the mRNA expression for DNMT3b was higher (1.536 ± 0.103 versus 1.0; Fig.4f), the others had no obvious differences (1.120 ± 0.086, 1.37 ± 0.127 versus 1.0; Fig.4f).
Fig. 4.
Localizations and transcript expressions of DNMTs. a-d show the localizations of DNMT1, DNMT3a, DNMT3b and DNMT3L. Their relative fluorescent intensities are shown in e. mRNA transcript levels are shown in f. In the control group, 81, 71, 74 and 83 oocytes were used to calculate the relative fluorescent intensities of DNMT1, DNMT3a, DNMT3b and DNMT3L. The corresponding oocytes numbers in the MTX group were 69, 74, 72 and 75. * p<0.05
Since it was not possible to directly analyze the levels of SAM and SAH in the matured oocytes, their concentrations in mouse eyeball blood were measured to obtain some general information on methyl donor availability. The SAM level in the blood was 8.1 ± 1.5 μmol/L in the MTX group, which was not significantly different from 8.2 ± 1.0 μmol/L in the control group. The SAH level in the MTX group was 4.2 ± 0.9 μmol/L, much higher than 1.8 ± 0.7 μmol/L in the control group. The ratios of SAM to SAH were about 1.91 in the MTX group and 4.67 in the control group.
Discussion
MTX, a specific antifolate drug, affects the folate metabolism via competitively inhibiting DHFR activity [1, 2]. Folate pathway includes 2 cycles which are closely related with DNA and RNA synthesis and methylation modification respectively [1, 5]. Both DNA/RNA synthesis and methylation reaction are crucial for oocyte quality and embryonic development [31–33]. However, does MTX affect the oocyte quality? How does MTX affect the oocyte quality ultimately resulting in the female fertility difficulty (including nature pregnancy and artificial insemination)? Up to now, we still lack the related information. This work focused on this issue.
The rate of abnormal chromosome alignment increased with the dosage of MTX (Table S1). When the dosage of MTX was more than or equal to 5 mg/Kg, the rate of abnormal chromosome alignment reached a plateau and kept steady at about 34%. There were no significant differences in the rate of abnormal chromosome alignment between the groups of MTX dosage more than or equal to 5 mg/Kg. The concentrations of intracellular MTX and its metabolite MTX polyglutamates (MTXPG) are crucial for the efficacy of MTX [34]. Due to the saturation of MTX receptor and MTXPG formation, high-dose MTX may not induce higher intracellular concentrations of MTX and MTXPG. According to our dose-response relationship, the MTX dosage of 5 mg/Kg caused the significant chromosome alignment abnormalities, while the higher MTX dosage (> 5 mg/Kg) didn’t induce more chromosome abnormalities. Thus, this work used the administration of single injection of 5 mg/Kg MTX to establish the MTX model mice for further study. Based on the surface area of animal, the dose of 5 mg/kg for mice equals to 0.4 mg/Kg for human. Referring to medication instructions of MTX, the 0.4 mg/Kg MTX for human is a common dose for treating choriocarcinoma, trophoblastic disease, adult lymphoma, osteosarcoma and other diseases.
This work indicated that MTX impaired the oocyte quality, showing the decreased fertilization rate and a decline in the resulting embryo development potential (Table 1). To explore the possible reasons for MTX-induced damages on oocytes, we firstly examined the influences of MTX on the folate cycles and found that the transcripts for genes encoding key enzymes in the folate cycle were reduced in the MTX group compared with the control group (Fig. 1). This is consistent with the reduced transcript expressions in MTX-treated mouse embryos [14]. The low transcripts suggest that there are disturbances in the one-carbon metabolism and in methyl donor availability [35].
Furthermore, we found that MTX increased abnormalities in chromosome alignment and increased chromosome clustering (Fig. 2). Trencsenyi et al. also found differences in chromatin conformation [36]. The correct separation of chromosomes is crucial for gamete formation and embryo development, the errors during separation will result in serious anomalies, for example, aneuploidy [25]. Chromosome separations are regulated by the spindle formation, the establishment of spindle assembly checkpoint (SAC) and cohesin dissociation [25, 26]. This work found that MTX disrupted the spindle formation showing the increased rate of abnormal spindle morphology (Fig. 3). MAD2, a key protein for SAC, plays a significant role in ensuring chromosome arranging neatly and the coming separation [25, 26]. The loss of MAD2 can cause that chromosomes separate in advance before ranking at equatorial plate [37]. This work found that MTX reduced the mRNA transcript expression of MAD2 (Fig. 3). This indicated that MTX may disturb SAC signalling and proper chromosome separation. Shugoshins are known to protect chromosome cohesion [38]. Reduced Sgo1 at centromeres was associated with precocious separation of chromatids and reduced the embryo development potential [27]. The PCR result in this work showed that the amount of Sgo1 mRNA was significantly reduced in the MTX group (Fig. 3). This indicated that MTX may cause the cohesin dissociation errors and the impediment of chromosome separation via affecting the Sgo1 expression. Thus, we speculated that MTX induced a decline in oocyte fertilization and embryo development potential via disturbing the regulation of chromosome separation.
DNA methylation disorders can result in genomic instability and gene expression disruption [28, 29]. DNA methylation levels in the imprinted domains would change during carcinogenesis [39]. Epigenetic modifications had a close link with the developmental capacity of oocytes and embryos [40]. MTX-induced global DNA methylation reduction suggests that MTX may affect the genome stability and the establishment of imprinted gene in oocytes and reduce the oocyte quality.
DNA methylation is performed by DNMTs which add a methyl to the carbon 5 position of CpG dinucleotide from SAM (methyl donor) [30]. Thus, we examined the expressions and transcript levels of DNMTs. Unlike DNMT3 family scattering in the ooplasmic, DNMT1 mainly localized at the cortical region of ooplasmic (Fig. 4). Compared with the control group, the fluorescence intensity and mRNA transcript level of DNMT1 both were reduced in the MTX group (Fig. 4). Many researches have demonstrated that the down-regulation of DNMT1 could induce the hypomethylation [41–43]. The fluorescence intensities and mRNA transcript levels of DNMT3 family basically had no obvious difference between the MTX and control groups, except the transcript of DNMT3b which greatly increased in the MTX group (Fig. 4). The mRNA level of DNMT3b was found to be inconsistent with its protein expression (Fig. 4). This may be due to the MTX-induced changes on regulation of translation from RNA to protein. Sandhu R’s team showed that altered post-transcription regulation of DNMT 3b by microRNAs affected its expression [44]. Our results of DNMTs might suggest that MTX had little effects on the DNMTs localizations but affected the DNMT1 and DNMT3b transcript levels. It was indicated that DNMT1 and DNMT3b co-regulated the DNA methylation [45, 46]. Specifically, depletion of DNMT1 resulted in widespread hypomethylation and DNMT3b knockdown caused that more hypermethylation than hypomethylation events occurred [45]. In this work, the MTX-induced reduction in the global DNA methylation may be due to the co-regulation of DNMT1 and DNMT3b. Further, we measured the concentrations of SAM and its metabolite SAH in the eyeball blood using HPLC as a rough estimate for the oocytes and found that the ratio of SAM to SAH in the blood was decreased in the MTX group. This ratio was a key indicator to assess the methylation level [47]. It is known that blood provides nutrients to the cells and carries away their wastes. The decreased ratio of SAM to SAH in the blood may signify that this ratio in the oocytes was also reduced. This indicated that MTX changed the methyl donor metabolism. Thus, it was reasonable to assume that the low DNA methylation in the MTX group was also related to the reduced ratio of SAM to SAH.
Conclusions
This work is the first to confirm the MTX-induced reduction in the in-vivo matured oocyte quality. We showed that MTX impaired the in-vitro fertilization function and the resulting embryo development potential. Further, our observations suggested that MTX disordered the folate metabolism pathway, increased the instabilities of chromosome alignment, and reduced the global DNA methylation level. In short, MTX had side effects on the in-vivo matured oocyte quality which resulted in the poor female fertility.
Methods
Experimental design
As shown in Fig. S2, the control and MTX model mice were established by treating with normal saline and 5 mg/kg MTX. (The establishment of the effective dosage of MTX referred to Table S1.) This work studied the in-vivo matured oocytes. After retrieving the in-vivo matured oocytes from the control and MTX model mice, the in-vitro fertilization ability and the resulting embryo formation were firstly analyzed to confirm the effects of MTX on the oocyte quality. Then, the action mechanisms of MTX impairing oocytes quality were studied. From morphological point, fluorescence staining and imaging were used to examine the chromosome morphology, DNMT localization and spindle morphology. From the point of molecular biology, real-time PCR was used to examine the transcripts for genes of the key enzymes in the folate pathway, DNMTs, as well as those in the spindle assembly checkpoint and chromosome segregation, MAD2 and Sgo1. Additionally, we quantitatively measured the global DNA methylation using fluorescence polarization and measured the concentrations of SAM and SAH in the blood using high performance liquid chromatography (HPLC). Through the research above, we analyzed the effects of MTX on the in-vivo matured oocyte quality and its possible mechanisms.
Animals
The ICR mice (Chinese Academy of Medical Sciences, Beijing, China) were housed and bred in barrier environment. Food and water were available ad libitum according to the Chinese National Standard (GB 14925–2001).
Oocyte collection
24 h after the treatment with MTX or normal saline, the model mice were treated with 10 IU pregnant mare serum gonadotropin (Bo’ en Pharmaceutical Ltd., Chifeng, China), and after 48 h, were treated with 10 IU human chorionic gonadotropin (Livzon Pharmaceutical Group Inc., Zhuhai, China), then, were euthanized by cervical dislocation 15 h later. Cumulus-oocyte complexes (COCs) were harvested from the oviducts. Cumulus cells were isolated by brief incubation in 0.05% hyaluronidase (Sigma, Louis, MO, USA) at 37 °C for 5 min. This work selected the in-vivo matured oocytes for further study, and these selected oocytes had the features of arresting at the second meiotic metaphase, presenting morphology integrity and good refraction, showing the first polar body.
In-vitro fertilization, embryo culture and morphological examination
ICR male mice were euthanized by cervical dislocation, and their epididymal tails were removed and transferred to HTF droplets. Epididymal tails were punctured to slowly extrude sperm. Then, the sperm suspensions were capacitated in HTF droplets for 90 min. The capacitated sperms were added to the droplets containing COCs and cultured for 6 h. After washing, all the oocytes were transferred to KSOM (Millipore, Massachusetts, USA) droplets and cultured to the blastocyst stage for 4 days. The fertilization and embryo development were recorded by noninvasive imaging technology, full-field optical coherence tomography (FF-OCT), due to the advantages of label-free, three-dimension and high resolution. Fertilization was identified by the appearance of male and female pronuclei. Like our previous work [48], blastomere number was counted by the segmented nuclei which were the dark areas in the images captured by FF-OCT. Additionally, the blastomere localizations were used as a coarse retrieval to distinguish the inner cell mass (ICM) and trophoblast [48].
Immunofluorescence, two-photon fluorescence imaging and fluorescence intensity analysis
The in-vivo matured nude oocytes were fixed in 4% paraformaldehyde in phosphatebuffered saline (PBS) for 30 min. Then, these fixed oocytes were permeabilized with 0.5% Triton-X 100 for 30 min, and were blocked in 2% normal rabbit serum blocking solution for 30 min. For DNMTs staining, they were incubated with anti-DNMT1, anti-DNMT3a, anti-DNMT3b and anti-DNMT3L (Jackson ImmunoResearch Laboratories, West Grove, USA) overnight at 4 °C in darkness. After washing, the oocytes were immersed in a biotinylated goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories, West Grove, USA) for 30 min. Finally, these oocytes were rinsed and reacted with quantum dot 605-streptavidin conjugate (Invitrogen, California, USA) for 1 h. For spindle labeling, the blocked oocytes were incubated with FITC-conjugated anti-β-tubulin antibody (Sigma, Saint Louis, USA) in darkness for 1 h. Chromosomes were counterstained with Hoechst 33342 (2 μg/mL) for 15 min. The fluorescence images were captured by a Bio-Rad MRC 1024 system (Bio-Rad, California, USA) coupled to a Nikon TE300 inverted microscope (Nikon, Tokyo, Japan) with 100× oil objective (Plan Apochromat DIC H, NA 1.4; Nikon, Tokyo, Japan). All the groups experienced the same experimental conditions (namely, the same excitation laser power and wavelength, detector gain). We used software Amira 5.2 to assess the fluorescence intensity which was defined as the average grey value in the three-dimensional spatial region of interest (DNMTs). According to the method of Zhu [49], the fluorescence intensity in the control group was normalized to one, then, the relative fluorescence intensity in the MTX group was obtained.
Analysis of mRNA transcript levels
We used RNeasy Micro Kit (Qiagen, California, USA) to extract the total mRNA from about 35 oocytes. Then, RNA purity was estimated by ultraviolet-visible spectrophotometer and its integrity was detected by gel electrophoresis. Total RNA was reverse transcribed using Sensiscript RT Kit (Qiagen, California, USA). According to the manufacturer’s instructions, reverse transcription was performed using 20 μl reactions containing 2 μl 10× buffer RT, 2 μl dNTP Mix, 2 μl Oligo-dT primer, 1 μl RNase inhibitor, 1 μl Sensiscript Reverse Transcriptase, 7 μl RNase-free water, 7 ng RNA template. Then, the 20 μl mixture was placed in a constant temperature bath at 37 °C for 60 min, and the cDNA was obtained. Each analysis was performed for three times using 15 μl reactions containing 7.5 μl SYBR Green PCR Master Mix (Applied Biosystems, UK), 0.6 μl forward and reverse primers (10 μmol/L), 1.5 μl cDNA, and 5.4 μl RNase free water. The sequences of the forward and reverse primers are shown in Table S2. The relative quantitation calculation was implemented by the 2-△△Ct method. β-actin was used as the internal reference.
Quantitative determination of global DNA methylation
The matured nude oocytes were immersed in Tyrode’s solution (Haling Biotechnology, Shanghai, China) to remove zona pellucid. After washing, these zona-free oocytes were immersed in the lysis buffer (Solarbio, Beijing, China) at 37 °C for 60 min. After sufficient dissociation, the mixed oocyte lysates were centrifuged 10,000 rpm for 2 min, and then saved at − 80 °C.
According to the method of Zhao and Xue [50], a sample of 500 ng DNA was incubated at 37 °C for 12 h in a 50 μl reaction mixture containing 2.5 μl restriction enzyme HpaII (10 U/μg, TaKaRa Biotechnology, Dalian, China), 5 μl 10× Buffer T, 5 μl 0.1% BSA (Solarbio, Beijing, China), 500 ng DNA, and distilled water adding up to 50 μl. Another sample of 500 ng DNA was incubated at the same conditions, but HpaII in the reaction mixture was replaced by MspI (TaKaRa Biotechnology, Dalian, China). After enzyme digestion, 4 μl enzymatic products of HpaII/MspI were placed at 57.8 °C for 60 min in a 15 μl chain-extension reaction mixture containing 1.5 μl 10 × PCR Buffer, 0.63 μl 0.75 U Taq DNA polymerase (TaKaRa Biotechnology, Dalian, China), 0.56 μl TAMRA-dCTP (Applied Biosystems, UK), 4.0 μl restricted DNA and 8.3 μl distilled water. After TAMRA-dCMP incorporating into DNA, the chain-extension reaction products were measured by fluorescence polarization using microplate reader (Molecular SpectraMax Plus). The fluorescence polarization values of HpaII/MspI restriction products were recorded as FPHpaII and FPMspI. The global DNA methylation level of oocytes was calculated by (1- FPHpaII / FPMspI) × 100%.
Quantitative measurement of SAM/SAH concentration in the eyeball blood
The concentration of SAM and SAH were measurement by extracting the mouse eyeball blood. According to the Zhen’s method [51], 100 μl plasma sample were extracted with 10 μl of 35% perchloric acid. Then, the acidified sample was vortexed for 1 min, subsequently, was placed at about 25 °C for 10 min, lastly, were centrifuged with the speed of 15,000 rpm at 4 °C for 10 min. Ultimately, 25 μl supernatant was analyzed using HPLC. HPLC equipment was Agilent 1100. The chromatogram column (Agilent ZORBAX C8, 250 × 4.6 mm, 5 μm) was used for the analysis. The mobile phase consisted of two solvents: solvent 1 was 8 mM octane sulphonic acid and 50 mM NaH2PO4 (PH 3.0); solvent 2 was chromatographic grade methanol. The flow rate was 0.9 mL/min. The column temperature was 35 °C. The UV detection wavelength was set at 450 nm from 0 to 8 min, and 254 nm from 8 to 16 min. The SAM/SAH standards configuring in water to different concentrations were used to trace the radio-labelled peaks.
Statistical analysis
All the experiments were repeated three times. Results were presented as the means ± SEM. Statistical comparisons were performed by One-way ANOVA or chi-square using software SPSS 16.0. p < 0.05 was considered significant.
Supplementary information
Additional file 1 Figure S1. Folate metabolism pathway. The key metabolites and enzymes are listed in the folate cycle. Abbreviations (full name): FR (folate receptor), RFC (reduced folate carrier), DHF (dihydrofolate), DHFR (dihydrofolate reductase), THF (tetrahydrofolate), SHMT (serine hydroxymethyl transferase), MTHFR (5,10-methylene THF reductase), MTRR (methionine synthase reductase), MS (methionine synthetase), MAT (methionine adenosyl transferase), SAM (S-adenosylmethionine), DNMT (DNA methyltransferase), SAH (S-adenosylhomocysteine), SAHH (SAH-hydrolase), Hcy (homocysteine), CSB (cystathionine β-synthase).
Additional file 2 Figure S2. Experimental technology roadmap.
Additional file 3 Table S1. Toxicity assessment of different doses MTX In the control group, mice were treated with normal saline. And the mice in the MTX group were treated with one dose of MTX of 0.5 mg/kg, 5 mg/kg, 10 mg/kg, 20 mg/kg and 50 mg/kg. We analyzed 7 mice in each group. The body weight, the retrieved oocyte number and the rate of chromosome alignment abnormity were used to assess the toxicity of MTX. Specifically, the retrieved oocytes were the in-vivo matured oocytes with the first polar bodies. It was found that there were no obvious differences in the body weight and the retrieved oocytes number between the control group and different doses MTX groups. Compared with the control group, 0.5 mg/Kg MTX had little effects on the chromosome alignment, but the rates of abnormal chromosome alignment in 5 mg/Kg, 10 mg/kg, 20 mg/kg and 50 mg/kg MTX groups were much higher. *p < 0.05. This indicated the administration of single injection of 5 mg/Kg MTX affected oocyte quality showing chromosome instability. Thus, this work used the administration of single injection of 5 mg/Kg MTX to establish the MTX model mice for further study.
Additional file 4 Table S2. Primers used for the real time PCR analysis
Additional file 5: Supplementary Materials. The details of examining the global DNA methylation via immunostaining for 5MeC.
Acknowledgements
Not applicable.
Abbreviations
- MTX
Methotrexate
- SLC19A
Solute carrier family 19
- DHFR
Dihydrofolate reductase
- DHF
Dihydrofolate
- THF
Tetrahydrofolate
- SHMT
Serinehydroxymethyl transferase
- MTHFR
5,10-methylene THF reductase
- MTRR
Methionine synthase reductase
- MAT
Methionine adenosyl transferase
- CSB
Cystathion β-synthase
- SAM
S-adenosylmethionine
- SAC
Spindle assembly checkpoint
- MAD2
Mitotic arrest deficient 2
- Sgo1
Shugoshin 1
- DNMTs
DNA methyltransferases
- SAH
S-adenosylhomocysteine
- HPLC
High performance liquid chromatography
- COCs
Cumulus-oocyte complexes
- FF-OCT
Full-field optical coherence tomography
- ICM
Inner cell mass
- PBS
Phosphatebuffered saline
Authors’ contributions
NT and DYL mainly make contributions to the acquisition, analysis and interpretation of data, and to draft the manuscript. JY and WYM mainly contribute to the conception and design of this study. All the authors read and approved the final manuscript.
Funding
This research was funded by National Nature Science Foundation of China (11804235) and Support plan for young and middle-aged scientific and technological innovation talents of Shenyang City (RC180226). The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study was approved by the Institutional Animal Welfare and Ethics Committee of Peking University (NO. IRBLA2009–020).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ning Tian, Email: tiann517@aliyun.com.
Dan-yu Lv, Email: dy_lu@163.com.
Ji Yu, Email: yuji4268@163.com.
Wan-yun Ma, Email: mawy@mail.tsinghua.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12860-020-00298-7.
References
- 1.Kwong WY, Adamiak SJ, Gwynn A, Singh R, Sinclair KD. Endogenous folates and single-carbon metabolism in the ovarian follicle, oocyte and pre-implantation embryo. Reproduction. 2010;139:705–715. doi: 10.1530/REP-09-0517. [DOI] [PubMed] [Google Scholar]
- 2.Hsieh YC, Tedeschi P, Adebisi LR, et al. Enhanced degradation of dihydrofolate reductase through inhibition of NAD kinase by nicotinamide analogs. Mol Pharmacol. 2013;83:339–353. doi: 10.1124/mol.112.080218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kooistra M, Trasler JM, Baltz JM. Folate transport in mouse cumulus-oocyte complexes and preimplantation embryos. Biol Reprod. 2013;89:63. doi: 10.1095/biolreprod.113.111146. [DOI] [PubMed] [Google Scholar]
- 4.Meredith M, MacNeil AH, Trasler JM, Baltz JM. Growing mouse oocytes transiently activate folate transport via folate receptors as they approach full size. Biol Reprod. 2016;94:125. doi: 10.1095/biolreprod.115.137687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karri S, Vanithakumari G. Effect of methotrexate and leucovorin on female reproductive tract of albino rats. Cell Biochem Funct. 2011;29:1–21. doi: 10.1002/cbf.1711. [DOI] [PubMed] [Google Scholar]
- 6.Ikeda S, Namekawa T, Sugimoto M, Kume S. Expression of methylation pathway enzymes in bovine oocytes and preimplantation embryos. J Exp Zool Ecol Genet Physiol. 2010;313:129–136. doi: 10.1002/jez.581. [DOI] [PubMed] [Google Scholar]
- 7.Menezo Y, Khatchadourian C, Gharib A, Hamidi J, Greenland T, Sarda N. Regulation of S-adenosyl methionine synthesis in the mouse embryo. Life Sci. 1989;44:1601–1609. doi: 10.1016/0024-3205(89)90455-4. [DOI] [PubMed] [Google Scholar]
- 8.O'Neill C. Endogenous folic acid is essential for normal development of preimplantation embryos. Hum Reprod. 1998;13:1312–1316. doi: 10.1093/humrep/13.5.1312. [DOI] [PubMed] [Google Scholar]
- 9.Ramesey LB, Panetta JC, Smith C, Yang WJ, Fan YP, Winick NJ, et al. Genome-wide study of methotrexate clearance replicates SLCO1B1. Blood. 2013;121:898–904. doi: 10.1182/blood-2012-08-452839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoelzer D, Walewski J, Dohner H, et al. Improved outcome of adult Burkitt lymphoma/leukemia with rituximab and chemotherapy: report of a large prospective multicenter trial. Blood. 2014;124:3870–3879. doi: 10.1182/blood-2014-03-563627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoo DH, Hrycaj P, Miranda P, et al. A randomised, double-blind, parallel-group study to demonstrate equivalence in efficacy and safety of CT-P13 compared with innovator infliximab when coadministered with methotrexate in patients with active rheumatoid arthritis: the PLANETRA study. Ann Rheum Dis. 2013;72:1613–1620. doi: 10.1136/annrheumdis-2012-203090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirby B. Drug survival of methotremate in psoriasis. Brit J Dermatol. 2017;177:345–346. doi: 10.1111/bjd.15693. [DOI] [PubMed] [Google Scholar]
- 13.Sholapurkar SL. Methotrexate for ectopic pregnancy: success rates and avoidance of embryopathy. Am J Obstet Gynecol. 2015;212:121–122. doi: 10.1016/j.ajog.2014.09.038. [DOI] [PubMed] [Google Scholar]
- 14.Martin MC, Barbero P, Groisman B, Aguirre MA, Koren G. Methotrexate embryopathy after exposure to low weekly doses in early pregnancy. Reprod Toxicol. 2014;43:26–29. doi: 10.1016/j.reprotox.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 15.Ng C, Xiao YD, Lum BL, Han YH. Quantitative structure-activity relationships of methotrexate and methotrexate analogues transported by the rat multispecific resistance-associated protein 2 (rMrp2) Eur J Pharm Sci. 2005;26:405–413. doi: 10.1016/j.ejps.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 16.Widemann BC, Adamson PC. Understanding and managing methotrexate nephrotoxicity. Oncologist. 2006;11:694–703. doi: 10.1634/theoncologist.11-6-694. [DOI] [PubMed] [Google Scholar]
- 17.Labib R, Hafez H, Badary O. Nigelia sativa oil ameliorates methotrexate-induced liver toxicity. Eur J Cancer. 2011;47:S119. [Google Scholar]
- 18.Ohbayashi M, Shiozawa A, Yasuda M, Kohyama N, Kobayashi Y, Yamamoto T. Involvement of alveolar epithelial cell on methotrexate-induced pulmonary toxicity. J Pharmacol Sci. 2009;109:206P.
- 19.Zhao J, Guan T, Wang J, Xiang Q, Wang M, Wang X, et al. Influence of the antifolate drug methotrexate on the development of murine neural tube defects and genomic instability. J Appl Toxicol. 2013;33:915–923. doi: 10.1002/jat.2769. [DOI] [PubMed] [Google Scholar]
- 20.Bayram M, Ozogul C, Dursun A, Ercan ZS, Isik I, Dilekoz E. Light and electron microscope examination of the effects of methotrexate on the endosalpinx. Eur J Obstet Gynecol Reprod Biol. 2005;120:96–103. doi: 10.1016/j.ejogrb.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 21.Gökçe A, Oktar S, Koc A, Yonden Z. Protective effects of thymoquinone against methotrexate-induced testicular injury. Hum Exp Toxicol. 2011;30:897–903. doi: 10.1177/0960327110382564. [DOI] [PubMed] [Google Scholar]
- 22.Boots CE, Gustofson RL, Feinberg EC. Does methotrexate administration for ectopic pregnancy after in vitro fertilization impact ovarian reserve or ovarian responsiveness? Fertil Steril. 2013;100:1590–1593. doi: 10.1016/j.fertnstert.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Ohannessian A, Loundou A, Courbière B, et al. Ovarian responsiveness in women receiving fertility treatment after methotrexate for ectopic pregnancy: a systematic review and meta-analysis. Hum Reprod. 2014;29:1949–1956. doi: 10.1093/humrep/deu174. [DOI] [PubMed] [Google Scholar]
- 24.McLaren JF, Burney RO, Milki AA, et al. Effect of methotrexate exposure on subsequent fertility in women undergoing controlled ovarian stimulation. Fertil Steril. 2009;92:515–519. doi: 10.1016/j.fertnstert.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 25.Vogt E, Kirsch-Volders M, Parry J, Eichenlaub-Ritter U. Spindle formation, chromosome segregation and the spindle checkpoint in mammalian oocytes and susceptibility to meiotic error. Mutat Res. 2008;651:14–29. doi: 10.1016/j.mrgentox.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 26.Marston AL. Chromosome segregation in budding yeast: sister chromatid cohesion and related mechanisms. Genetics. 2014;196:31–63. doi: 10.1534/genetics.112.145144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin S, Ai JS, Shi LH, Wei L, Yuan J, Ouyang YC, et al. Shugoshin1 may play important roles in separation of homologous chromosomes and sister chromatids during mouse oocyte meiosis. PLoS One. 2008;3:e3516. doi: 10.1371/journal.pone.0003516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato A, Otsu E, Negishi H, Utsunomiya T, Arima T. (2007) Aberrant DNA methylation of imprinted loci in superovulated oocytes. Hum Reprod. 2007; 22: 26–35. [DOI] [PubMed]
- 29.Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 30.Uysal F, Akkoyunlu G, Ozturk S. DNA methyltransferases exhibit dynamic expression during spermatogenesis. Reprod BioMed Online. 2016;33:690–702. doi: 10.1016/j.rbmo.2016.08.022. [DOI] [PubMed] [Google Scholar]
- 31.Gao YY, Chen L, Wang T, Nie ZW, Zhang X, Miao YL. Oocyte aging-induced Neuronatin (NNAT) hypermethylation affects oocyte quality by impairing glucose transport in porcine. Sci Rep. 2016;6:36008. doi: 10.1038/srep36008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaffer B, McGraw S, Xiao SC, Chan D, Trasler J, Chaillet JR. The Dnmt1 intrinsically disordered domain regulates genomic methylation during development. Genetics. 2015;199:533–541. doi: 10.1534/genetics.114.173609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anckaert E, Romero S, Adriaenssens T, Smitz J. Effects of low methyl donor levels in culture medium during mouse follicle culture on oocyte imprinting establishment. Biol Reprod. 2010;83:377–386. doi: 10.1095/biolreprod.109.082164. [DOI] [PubMed] [Google Scholar]
- 34.Galpin AJ, Schuetz JD, Masson E, et al. Differences in folylpolyglutamate synthetase and dihydrofolate reductase expression in human b-lineage versus t-lineage leukemic lymphoblasts: mechanisms for lineage differences in methotrexate polyglutamylation and cytotoxicity. Mol Pharmacol. 1997;52:155–163. doi: 10.1124/mol.52.1.155. [DOI] [PubMed] [Google Scholar]
- 35.Mentch SJ, Locasale JW. One-carbon metabolism and epigenetics: understanding the specificity. Ann N Y Acad Sci. 2016;1363:91–98. doi: 10.1111/nyas.12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trencsenyi G, Bako F, Nagy G, et al. Methotrexate induced apoptotic and necrotic chromatin changes in rat myeloid leukemia cells. Inflamm Res. 2015;64:193–203. doi: 10.1007/s00011-015-0797-x. [DOI] [PubMed] [Google Scholar]
- 37.Homer HA, McDougall A, Levasseur M, Yallop K, Murdoch AP, Herbert M. Mad2 prevents aneuploidy and premature proteolysis of cyclin B and securing during meiosis I in mouse oocytes. Genes Dev. 2005;19:202–207. doi: 10.1101/gad.328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- 39.Bretz CL, Langohr IM, Kim J. Epigenetic response of imprinted domains during carcinogenesis. Clin Epigenetics. 2017;9:90. doi: 10.1186/s13148-017-0393-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bao SQ, Obata Y, Carroll J, Domeki I, Kono T. Epigenetic modifications necessary for normal development are established during oocyte growth in mice. Biol Reprod. 2000;62:616–621. doi: 10.1095/biolreprod62.3.616. [DOI] [PubMed] [Google Scholar]
- 41.Torrisani J, Unterberger A, Tendulkar SR, Shikimi K, Szyf M. Auf1 cell cycle variations define genomic dna methylation by regulation of dnmt1 mrna stability. Mol Cell Biol. 2007;27:395–410. doi: 10.1128/MCB.01236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 43.Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 44.Sandhu R, Rivenbark AG, Coleman WB. Loss of post-transcriptional regulation of DNMT3b by microRNAs: a possible molecular mechanism for the hypermethylation defect observed in a subset of breast cancer cell lines. Int J Oncol. 2012;41:721–732. doi: 10.3892/ijo.2012.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tiedemann R, Choi JH, Robertson K. Acute depletion reveals novel co-regulation of DNA methylation at conserved loci by DNMT1 and DNMT3B. Cancer Res. 2014;74:2305. [Google Scholar]
- 46.Hamatani T, Falco G, Carter MG, Akutsu H, Stagg CA, Sharov AA, Dudekula DB, VanBuren V, Ko MSH. Age-associated alteration of gene expression patterns in mouse oocytes. Hum Mol Genet. 2004;13:2263–2278. doi: 10.1093/hmg/ddh241. [DOI] [PubMed] [Google Scholar]
- 47.Yi P, Melnyk S, Pogribna M, Pogribny IP, Hine RJ, James SJ. Increase in plasma homocysteine associated with parallel increases in plasma S-adenosylhomocysteine and lymphocyte DNA hypomethylation. J Biol Chem. 2000;275:29318–29323. doi: 10.1074/jbc.M002725200. [DOI] [PubMed] [Google Scholar]
- 48.Zheng J, Lu D, Chen T, Wang C, Tian N, Zhao F, et al. Label-free subcellular 3D live imaging of preimplantation mouse embryos with full-field optical coherence tomography. J Biomed Opt. 2012;17:070503. doi: 10.1117/1.JBO.17.7.070503. [DOI] [PubMed] [Google Scholar]
- 49.Zhu CC, Zhang Y, Duan X, Han J, Sun SC. Toxic effects of HT-2 toxin on mouse oocytes and its possible mechanisms. Arch Toxicol. 2016;90:1495–1505. doi: 10.1007/s00204-015-1560-3. [DOI] [PubMed] [Google Scholar]
- 50.Zhao C, Xue H. A simple method for high-throughput quantification of genome-wide DNA methylation by fluorescence polarization. Epigenetics. 2012;7:335–339. doi: 10.4161/epi.19376. [DOI] [PubMed] [Google Scholar]
- 51.Zhen Q, Xu B, Ma L, Tian G, Tang X, Ding M. Simultaneous determination of tryptophan, kynurenine and 5-hydroxytryptamine by HPLC: application in uremic patients undergoing hemodialysis. Clin Biochem. 2011;44:226–230. doi: 10.1016/j.clinbiochem.2010.10.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Figure S1. Folate metabolism pathway. The key metabolites and enzymes are listed in the folate cycle. Abbreviations (full name): FR (folate receptor), RFC (reduced folate carrier), DHF (dihydrofolate), DHFR (dihydrofolate reductase), THF (tetrahydrofolate), SHMT (serine hydroxymethyl transferase), MTHFR (5,10-methylene THF reductase), MTRR (methionine synthase reductase), MS (methionine synthetase), MAT (methionine adenosyl transferase), SAM (S-adenosylmethionine), DNMT (DNA methyltransferase), SAH (S-adenosylhomocysteine), SAHH (SAH-hydrolase), Hcy (homocysteine), CSB (cystathionine β-synthase).
Additional file 2 Figure S2. Experimental technology roadmap.
Additional file 3 Table S1. Toxicity assessment of different doses MTX In the control group, mice were treated with normal saline. And the mice in the MTX group were treated with one dose of MTX of 0.5 mg/kg, 5 mg/kg, 10 mg/kg, 20 mg/kg and 50 mg/kg. We analyzed 7 mice in each group. The body weight, the retrieved oocyte number and the rate of chromosome alignment abnormity were used to assess the toxicity of MTX. Specifically, the retrieved oocytes were the in-vivo matured oocytes with the first polar bodies. It was found that there were no obvious differences in the body weight and the retrieved oocytes number between the control group and different doses MTX groups. Compared with the control group, 0.5 mg/Kg MTX had little effects on the chromosome alignment, but the rates of abnormal chromosome alignment in 5 mg/Kg, 10 mg/kg, 20 mg/kg and 50 mg/kg MTX groups were much higher. *p < 0.05. This indicated the administration of single injection of 5 mg/Kg MTX affected oocyte quality showing chromosome instability. Thus, this work used the administration of single injection of 5 mg/Kg MTX to establish the MTX model mice for further study.
Additional file 4 Table S2. Primers used for the real time PCR analysis
Additional file 5: Supplementary Materials. The details of examining the global DNA methylation via immunostaining for 5MeC.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.