Abstract
Studies over the past three years have substantially expanded the involvements of eukaryotic initiation factor 3 (eIF3) in messenger RNA (mRNA) translation. It now appears that this multi-subunit complex is involved in every possible form of mRNA translation, controlling every step of protein synthesis from initiation to elongation, termination, and quality control in positive as well as negative fashion. Through the study of eIF3, we are beginning to appreciate protein synthesis as a highly integrated process coordinating protein production with protein folding, subcellular targeting, and degradation. At the same time, eIF3 subunits appear to have specific functions that probably vary between different tissues and individual cells. Considering the broad functions of eIF3 in protein homeostasis, it comes as little surprise that eIF3 is increasingly implicated in major human diseases and first attempts at therapeutically targeting eIF3 have been undertaken. Much remains to be learned, however, about subunit- and tissue-specific functions of eIF3 in protein synthesis and disease and their regulation by environmental conditions and post-translational modifications.
Keywords: mRNA translation, translation initiation factor, eIF3, protein homeostasis, cancer
Introduction
Protein synthesis is a complex process accomplished by the ribosome and its associated factors that encompasses several highly coordinated steps, including the initiation of messenger RNA (mRNA) translation, translation elongation, and termination as well as folding, targeting, and quality control of the nascent polypeptide. The canonical reactions of mRNA m7G cap-dependent translation have been studied for decades and have been fully reconstituted from purified components in vitro (Sonenberg and Hinnebusch, 2009). In canonical initiation, mRNA is recruited to the ribosome through eukaryotic initiation factor 4E (eIF4E)-mediated cap binding. eIF4E interacts with eIF4G to form eIF4F, which binds to eIF3 to attract the 43S pre-initiation complex consisting of the 40S ribosome loaded with methionyl-tRNA and additional eIFs. The 40S ribosome then scans the mRNA from its 5′-end to identify the start codon. Upon start codon recognition, the 60S ribosomal subunit joins to produce an actively translating 80S ribosome, whereas all eIFs are thought to be ejected.
Although all eukaryotic mRNAs possess an m7G cap, disabling the cap-binding protein eIF4E through various strategies has surprisingly little impact on mRNA translation, affecting as few as ~200 mRNAs (Hsieh et al., 2012; Thoreen et al., 2012; Morita et al., 2013; Truitt et al., 2015). This suggests the existence of additional cap-binding proteins as well as cap-independent means of initiating translation. These alternative pathways might recruit ribosomes to mRNA that would then scan from the 5′-end for a start codon essentially in the same way as established for canonical eIF4E-dependent initiation (Shatsky et al., 2018). Recent evidence has implicated the multi-subunit eIF3 complex in several non-canonical modes of initiation that are discussed in this review.
eIF3 function is particularly well studied in the budding yeast Saccharomyces cerevisiae (Hinnebusch, 2017). Whereas the core functions of eIF3 in scanning and AUG recognition are well conserved between budding yeast and higher eukaryotes, S. cerevisiae eIF3 with five subunits (eIF3a, eIF3b, eIF3c, eIF3g, and eIF3i) is considerably smaller than the 12-subunit mammalian complex and thus misses several components, including eIF3d, eIF3l, and eIF3h, which have recently been attributed crucial functions. Knockdown studies in human cells have raised the possibility that a yeast core-like complex may perform essential translation initiation functions also in mammalian cells (Wagner et al., 2014). A potential yet unproven implication of this model is that non-essential mammalian eIF3 subunits have evolved to carry out specific translation functions beyond basic initiation. A thirteens subunit, eIF3j, transiently associates with holo-eIF3 but is not considered a bona fide subunit.
Recent reviews have provided comprehensive summaries of the roles of eIF3 in canonical translation initiation, including scanning, start codon recognition, and termination uniquely informed by structural insights obtained by X-ray crystallography and cryo-electron microscopy (Cate, 2017; Hinnebusch, 2017; Valášek et al., 2017). In addition, the sometimes conflicting involvements of eIF3 subunits in human cancer have been reviewed recently (Hershey, 2015; Sesen et al., 2017; Gomes-Duarte et al., 2018). In the present article, we are focusing on recent progress regarding canonical and non-canonical roles of eIF3 in translation initiation, elongation, and repression and describe novel findings implicating eIF3 in diseases other than cancer.
New non-canonical mechanisms of translation initiation involving eIF3
Methyladenosine modification as an eIF3-recruiting mRNA modification
While all mRNAs carry an m7G cap, several thousand mRNAs are also modified by N6 methylation of adenosines (m6A). Dynamic m6A modification is enriched at the stop codon and the 3′-untranslated regions (UTRs) and also occurs in other regions of the mRNA, including the 5′-UTRs (Meyer et al., 2015; Tong et al., 2018). m6A is known to broadly impact on mRNA metabolism, including splicing, nuclear export, translation, and mRNA decay. With respect to translation, it was shown that m6A can recruit eIF3 to induce 48S initiation complex formation independent of eIF4E cap binding (Meyer et al., 2015). Direct binding of eIF3 to m6A appears to occur within 5′-UTRs, especially under heat shock conditions when cap-dependent initiation is blocked and m6A relocalizes from 3′-UTRs to 5′-UTRs. Importantly, while cap-independent, such m6A-mediated initiation mechanism does not operate as a cellular counterpart to viral internal ribosome entry sites (IRESs), because, unlike viral IRESs, it requires an accessible mRNA 5′-end (Shatsky et al., 2018).
The m6A modification is added to mRNA by a family of methyltransferases (e.g. METTL3; ‘writers’), interpreted by a family of m6A-binding YTH domain proteins (e.g. YTHDF1, YTHDF2, YTHDF3; ‘readers’), and removed by several demethylases (e.g. FTO; ‘erasers’). Readers of the YTHDF group have been implicated in m6A-dependent translation initiation. YTHDF1 was found to bind to ~3600 m6A-containing mRNAs and increase their ribosome occupancy (Wang et al., 2015b). The same study showed that artificial, m6A-independent, tethering of YTHDF1 to 3′-UTRs stimulated translation of a luciferase encoding reporter mRNA. Mechanistically, YTHDF1 binds to eIF3 in an RNA-independent and possibly direct manner, although it remains formally unproven that YTHDF1-mediated translation depends on interaction with eIF3. Unlike with direct eIF3–m6A binding, eIF3 recruitment via YTHDF1 is thought to occur through the m6A modification within the 3′-UTR. Thus, eIF3−YTHDF1 interaction would lead to formation of the closed mRNA loop known to promote translation (Tarun et al., 1997).
Interestingly, while sharing ~50% of mRNA targets, YTHDF1 and its related YTHDF2 impart differential control. Whereas initial YTHDF1 binding promotes mRNA translation, subsequent YTHDF2 binding triggers mRNA decay (Wang et al., 2015b), thus suggesting a temporally ordered scheme of control of gene expression by m6A reader proteins. Under heat stress conditions, YTHDF2 was shown to play a different role, promoting the translation of HSP70 and possibly other heat-induced transcripts depending on m6A modification localized to the 5′-UTR, although this function was not linked to eIF3 (Zhou et al., 2015). Rather, YTHDF2, in this context, is thought to protect m6A-modified mRNA from demethylation, thus promoting cap-independent translation, possibly through direct recruitment of eIF3 to m6A as described above (Meyer et al., 2015).
Another m6A reader, YTHDF3, promotes translation apparently by serving as a specificity determinant of YTHDF1 interacting with m6A in the 3′-UTR (Shi et al., 2017). Whereas YTHDF3 does not interact with eIF3 directly, it appears to join the YTHDF1−eIF3 complex through binding to YTHDF1. On the other hand, YTHDF3 binding to m6A, apparently in a YTHDF1-independent fashion, was shown to mediate the translation of circular mRNAs (Yang et al., 2017). In this context, YTHDF3 functions in association with eIF4G2 and eIF3. Another factor that has recently been implicated in initiation mediated by m6A via the 5′-UTR is ABCF1, although its possible relationship to YTHDF readers and eIF3 remains unknown (Coots et al., 2017).
In addition to m6A readers, the m6A writer METTL3 has been implicated in translation initiation through eIF3. METTL3 binding to m6A-modified 3′-UTRs reinforces the closed loop structure mediated by eIF4G−poly-A-binding protein (PABP) interactions. This occurs through direct binding of METTL3 to eIF3h. Notably, this is independent of the catalytic methyltransferase activity of METTL3 and does not require YTH domain readers (Lin et al., 2016). The METTL3−eIF3 mechanism augments the translation of thousands of mRNAs, a subset of which is involved in tumor progression and apoptosis (Choe et al., 2018). Consequently, knockdown of METTL3 inhibits cell migration and tumor growth in mice, whereas overexpression of METTL3 can transform mouse fibroblasts. The newly discovered role of METTL3 in boosting translation through eIF3h potentially complicates the conclusions drawn from previous studies using METTL3 knockdown to experimentally manipulate m6A levels without awareness that METTL3 has an additional function (Meyer et al., 2015; Wang et al., 2015b; Coots et al., 2017).
Direct cap binding of eIF3 subunits
Using PAR-CLIP, eIF3 was shown to bind to the 5′-UTRs of hundreds of mRNAs with many of the binding sites overlapping with m6A-modified sites (Lee et al., 2015; Meyer et al., 2015). Although only two of these binding sites, located in the c-JUN and BTG1 transcripts, have been studied in detail, they can either activate or suppress translation (Lee et al., 2015). The stimulatory motif in the c-JUN 5′-UTR functions independent of the eIF4F but nevertheless requires the m7G cap. These disparate observations were reconciled when it was found that eIF3 subunit d has a cryptic cap-binding activity that becomes activated upon binding to the structured motif within the c-JUN 5′-UTR (Lee et al., 2016). It thus appears that dual binding of eIF3 to the 5′-cap and the structured motif recruits the 40S ribosome to activate translation. However, whether eIF3 controls the synthesis of endogenous c-JUN protein through this mechanism remains to be demonstrated.
Another subunit of eIF3, eIF3l, was also shown to have cap-binding activity (Kumar et al., 2016). Since eIF3l and eIF3k are not required for cell growth and 48S complex formation in vitro (Smith et al., 2013; Wagner et al., 2016) and were recently shown to inhibit mRNA recruitment to the 43S pre-initiation complex (Herrmannová et al., 2019), the function of eIF3l’s cap-binding activity is currently unknown.
eIF3−PABP interaction in IRES-mediated initiation
eIF3 was also implicated in translation initiation at the IRES of the XIAP mRNA (Thakor et al., 2017). eIF3 interacts directly with the XIAP IRES in a fashion that is facilitated by PABP and results in 40S recruitment. The eIF3−PABP interaction is thought to promote closed loop formation. Since eIF4A and eIF2α are not required, XIAP IRES initiation is proposed to be driven by direct eIF3-mediated recruitment of the ribosome to the vicinity of the start codon, similar to what is observed for several viral IRESs.
Common themes emerging
The above studies have substantially diversified the function of eIF3 in translation. It now appears that eIF3 is a central mediator of translation under both normal and stress conditions. Several common themes have emerged from these studies.
m6A exerts distinct location-dependent modes of regulation
m6A modification located in the 5′-UTR is stress-inducible and mediates cap-independent translation under stress conditions through direct recruitment of eIF3 to a limited set of stress response mRNAs (Meyer et al., 2015; Coots et al., 2017). In contrast, m6A modification located in the 3′-UTR appears to be constitutive and to provide a general boost to cap-dependent translation by promoting closed loop formation via interaction of eIF3 with m6A reader and writer proteins (Wang et al., 2015b; Lin et al., 2016; Shi et al., 2017; Choe et al., 2018).
eIF3 is involved in IRES- and non-IRES-mediated initiation
Cap-independent initiation through binding of eIF3 to m6A located in the 5′-UTR was shown to require 5′-end-dependent scanning and was thus classified as non-IRES-mediated (Meyer et al., 2015; Coots et al., 2017). In contrast, m6A−YTHDF3−eIF3-mediated translation of circular RNA is by definition 5′-end-independent and thus IRES-like (Yang et al., 2017). Likewise, initiation mediated by eIF3 cap binding or eIF3−PABP interactions proceeds independently of eIF4A and is thus scanning-independent (Lee et al., 2016; Thakor et al., 2017).
eIF3 mediates translation under stress conditions
Cap-independent initiation mediated by eIF3 may be particularly useful in stress conditions when canonical cap-dependent initiation is blocked. For example, amino acid starvation inactivates eIF4E (Bar-Peled and Sabatini, 2014). More generally, however, stress conditions such as heat shock and endoplasmic reticulum (ER) stress lead to inactivation of eIF2 (Wek, 2018). During chronic ER stress with low eIF2 activity, translation initiation switches to an eIF3-dependent, but eIF2-independent, mechanism (Guan et al., 2017). However, translation of heat shock protein 70 mRNA (HSP70, HSPA1A) mediated by direct binding of eIF3 to m6A in the 5′-UTR requires eIF2 (Meyer et al., 2015). In addition, binding of eIF2 to HSP70 (HSPA1A) mRNA is increased by heat stress (Coots et al., 2017). Thus, despite reduced availability of eIF2 for ternary complex formation under heat stress, residual eIF2 appears to be preferentially reserved for delivering methionyl-tRNA for m6A-mediated translation, at least in the case of HSPA1A mRNA. Whether other cap-independent mRNAs also use eIF2 during stress conditions or whether other factors deliver methionyl-tRNA—for example eIF5B (Guan et al., 2017)—remains to be established.
eIF3 and translational repression
Whereas the eIF3 holo-complex typically activates mRNA translation in a reconstituted in vitro system, it was shown to act as a repressor in some instances. eIF3 PAR-CLIP studies identified a structured motif in the mRNA encoding BTG1 as a negative regulatory element (Lee et al., 2015). While eIF3 does not appear to bind to this element directly, removal of the motif strongly derepresses the translation of luciferase reporters in vitro and in vivo. Direct binding of eIF3 to a sequence motif in the 5′-UTR was recently implicated in translational suppression of the mRNA encoding ferritin light chain (FTL; Pulos-Holmes et al., 2019). The eIF3-binding motif contains two single nucleotide polymorphisms associated with hyperferritinemia. Significantly, the disease-causing alleles abolish eIF3 binding and derepress FTL translation. However, it still remains unclear whether disabling eIF3 function will derepress the synthesis of BTG1 and FTL proteins in vivo and whether this is mediated through the structured eIF3-binding motifs.
eIF3 was also described as a repressor of nanos1 mRNA in developing Xenopus embryos (Aguero et al., 2017). In fertilized embryos where Nanos1 protein is synthesized, the repression is relieved by accumulation of the Dnd1 helicase and its binding to eIF3f. Dnd1 appears to interact with eIF3f to neutralize the repressive effect of holo-eIF3. Consistent with this idea and previous reports (Shi et al., 2006), ectopic eIF3f inhibits nanos1 translation in vitro and in vivo. However, these dominant effects contrast with clear inhibition of protein synthesis observed upon knockdown of eIF3f in human HeLa cells (Wagner et al., 2016) and the dramatic effects of eIF3f deletion on protein synthesis and viability in the fission yeast Schizosaccharomyces pombe (Zhou et al., 2005).
Despite these uncertainties, many global datasets have documented positive as well as negative effects of individual eIF3 subunits on mRNA translation (Kim et al., 2007; Grzmil et al., 2010; Shah et al., 2016). It is thus likely that intricate subunit-selective mechanisms regulate eIF3 function in stimulation as well as repression of mRNA translation.
eIF3 in translation elongation and quality control
Based on in vitro studies, it has long been believed that binding of eIF3 to the 40S ribosome inhibits 60S subunit joining and that eIF3 is therefore ejected from the 40S prior to subunit joining (Peterson et al., 1979; Trachsel and Staehelin, 1979). However, proteomic evidence has accumulated supporting the idea that eIF3 can form stable interactions with 80S ribosomes and translation elongation factors in various organisms (Guerrero et al., 2008; Sha et al., 2009; Reading et al., 2013; Wang et al., 2015a). eIF3−80S complexes also contain additional factors involved in protein quality control, including chaperones and the 26S proteasome to form a particle dubbed the ‘translasome’ (Sha et al., 2009).
Recent cryo-EM structures of the 48S complex are consistent with the possibility that eIF3 remains bound to the ribosome after subunit joining (Llácer et al., 2015, 2018; Simonetti et al., 2016; Eliseev et al., 2018). Whereas eIF3 is bound to the solvent exposed side of the 40S ribosome within the 43S pre-initiation complex, the eIF3b–eIF3g–eIF3i module appears to transiently move to the inter-subunit side during scanning only to move back to the solvent exposed side upon AUG recognition. Thus, there is no obvious steric hindrance that would prevent eIF3 to remain attached to the 80S ribosome.
Clear evidence that eIF3 remains associated with the 80S ribosome at least during translation of the first 5–10 codons has been provided recently. This was demonstrated for translation of upstream open reading frames (uORFs) of the budding yeast GCN4 mRNA (Mohammad et al., 2017). The data showed that eIF3 remains associated with 80S ribosomes during translation of uORFs and that this facilitates reinitiation at downstream uORFs. This raises the intriguing possibility that eIF3 remains associated with 80S ribosomes independently of uORF translation. Indeed, several recent reports, which are still pending peer review, have applied selective ribosome profiling to demonstrate physical interaction of eIF3 with translating ribosomes proximal to the start codon of the coding ORF (Bohlen et al., 2019; Wagner et al., 2019; Lin et al., 2019). In other work, the possibility was invoked that eIF3−80S ribosome interactions may mediate early translational slowdown, thus giving rise to the ‘5′-ramp’ of ribosome density widely observed within the first ~100 codons of S. cerevisiae mRNAs (Weinberg et al., 2016). However, the 5′-ramp is not typically observed in studies of human cells (Ingolia et al., 2011; Shalgi et al., 2013), and the wider applicability of this proposed function of eIF3−80S interactions to eukaryotic gene expression remains questionable.
In the context of the translasome, eIF3−80S ribosome interactions may have to be considered more broadly with respect to the fate of the nascent chains emerging from translating ribosomes. Genetic loss-of-function screening has firmly implicated eIF3 in protein quality control (Pegoraro et al., 2012). Given the important role of early elongation speed in determining the fate of nascent membrane proteins (Kramer et al., 2009; Acosta-Sampson et al., 2017), it is not inconceivable that eIF3’s potential role in controlling elongation is linked to its role in protein quality control. These connections invoke a scenario in which eIF3 and its associated proteins cooperate in an integrated fashion to orchestrate the folding and targeting of nascent chains to appropriate locales. If proper folding/targeting fails, eIF3’s association with the proteasome and ubiquitylating enzymes may trigger ubiquitin-mediated co-translational degradation (Turner, 2000; Medicherla and Goldberg, 2008; Duttler et al., 2013; Wang et al., 2013).
eIF3 in diseases: cancer and beyond
With respect to the anabolic nature of cancer cell metabolism, mechanisms controlling protein synthesis have been at the center of attention for many years. The cancer-promoting activity of eIF4E, which acts downstream of mTORC1 signaling, has been well documented (Siddiqui and Sonenberg, 2015). Likewise, eIF3 has been placed at the cross-roads of mTOR and S6K signaling (Holz et al., 2005). Since then, many subunits of eIF3 have been implicated in cancer in various positive as well as negative ways as discussed in detail by Hershey (2015). In addition, eIF3 was shown to control the translation of proliferation-related mRNAs (Lee et al., 2015). Nevertheless, no unified mechanism has arisen as to the oncogenic or tumor suppressive roles of eIF3. These roles are likely multifaceted with different subunits playing distinct roles in different tissues. Although a preponderance of evidence suggests a positive role of eIF3 in cancer, rigorous genetic evidence for tissue-specific oncogenic or tumor suppressive functions of individual eIF3 subunits remains outstanding. Conditional eIF3 mutant mice are beginning to be established that will enable such testing (Zeng et al., 2013; Sadato et al., 2018).
A recent study has suggested a possible involvement of eIF3 in cystic fibrosis. Integration of genetic and proteomic screening data implicated eIF3a as well as some other eIF3 subunits in the synthesis and folding of the cystic fibrosis transmembrane conductance regulator (CFTR; Hutt et al., 2018). Counterintuitively, knockdown of eIF3a, which probably caused partial impairment of the eIF3 holo-complex, led to increased production of functional CTFR containing disease-causing mutations. The study proposed that the functional rescue of mutant CFTR by eIF3 attenuation is due to decreased translation initiation, thus reducing the load of nascent peptides. This in turn would free chaperone capacity for the folding of mutant CTFR. However, the study does not provide data sufficient to rule out alternative scenarios consistent with previous protein interaction studies according to which knockdown of eIF3 function would slow down translation elongation and prevent the recruitment of the proteasome to the ribosome (see above paragraphs). Both events may promote the expression of mutant CFTR: Reduced translation elongation rate may provide additional time for folding during synthesis, whereas compromised proteasome recruitment may decelerate co-translational degradation of slowly folding mutant protein. Like the proteasome inhibitor MG132, knockdown of eIF3a strongly inhibits the proteasomal degradation of mutant CFTR (Hutt et al., 2018). In fact, eIF3a knockdown is considerably more efficient than MG132, consistent with the above notion that impairment of eIF3 may promote CFTR expression in multiple proteasome-dependent and independent ways. Regardless, these recent findings nominated eIF3 as a potential drug target in cystic fibrosis.
Finally, a recent study implicated eIF3 in neurodegenerative diseases marked by expansion of simple DNA repeat sequences (Ayhan et al., 2018). The repeats, which are sometimes intronic, can give rise to sense and anti-sense transcripts that are translated via repeat-associated non-ATG (RAN) translation (Zu et al., 2011). RAN translation results in homopolymeric proteins, e.g. poly-Gln, poly-Ser, poly-Ala, or poly-Gly-Pro. Accumulation of such homopolymeric proteins is thought to lead to neurotoxic aggregates. A search for RAN transactivating factors that promote initiation independently of a start codon led to the demonstration that eIF3f specifically stimulates RAN translation but not ATG-dependent translation. Although the exact mechanism is still unclear, the authors proposed that eIF3f may promote RAN through an IRES-like pathway (Ayhan et al., 2018). Whether this initiation function requires eIF3f within the eIF3 holo-complex is presently unknown.
Exploring eIF3 as a therapeutic target
Targeting of eIF3 with small molecules and other approaches is in its infancy. Both in cancer and cystic fibrosis, inhibition of eIF3 function would appear desirable. However, eIF3 does not have any intrinsic enzymatic activities amenable to inhibitor development. A screen for chemicals able to disrupt the interaction of the recombinant eIF3 octameric core complex with the hepatitis C virus (HCV) IRES identified two crude extracts isolated from marine actinobacteria (Zhu et al., 2017). The fractions also modestly inhibited cap-dependent and IRES-mediated translation in vitro as assessed by reporter assay. Assuming a molecular mass of 400 for a single active ingredient present in the crude fractions, inhibition of the eIF3−HCV IRES interaction in an electrophoretic mobility shift assay requires ~100 μM compound. The potency of these reagents is thus limited, in addition to their molecular structure remaining unknown. Nevertheless, these studies provide initial proof-of-concept for small molecule targeting of eIF3. Interestingly, the same study also identified compound HP-3 as a promoter of the eIF3−HCV IRES interaction and a stimulator of IRES-selective translation in vitro and in vivo. Mechanistically, HP-3 was shown to bind to the HCV IRES and alter its structure, apparently in ways that facilitate 40S recruitment. Nevertheless, the utility of a compound stimulating HCV IRES activity remains presently unclear.
Based on the findings that eIF3f inhibits protein synthesis and is downregulated in multiple cancers, including pancreatic adenocarcinoma, melanoma, and breast cancer among others, cell penetrating eIF3f protein was studied for potential anti-cancer activity. Nanomolar doses of either full-length eIF3f or a truncation mutant encompassing the MPN domain were effectively internalized by cells and induced apoptosis, apparently in a cancer cell selective fashion (Marchione et al., 2015). Whereas the mechanism of induction of apoptosis by eIF3f remains uncharacterized and the protein delivery approach meets with several practical challenges, an eIF3f-based protein drug may potentially be developed in the future. Once again, these studies provide early proof-of-concept for potentially targeting eIF3 function through biologics.
Concluding remarks
Whereas the findings summarized above have implicated eIF3 in all known steps of protein synthesis, targeting, and quality control (Figure 1), much remains to be learned. For one, we still know very little about the biochemistry of the eIF3−m6A interaction. Which subunit(s) of eIF3 bind to m6A and YTH domain reader proteins? Likewise, there is very little mechanistic understanding of how eIF3 inhibits the translation of certain mRNAs. In addition, it is unknown whether eIF3 has a role in translation elongation beyond uORF translation in select yeast mRNAs. If eIF3 has a more general role in translation elongation, is this role confined to certain regions of the mRNA (such as the 5′-ramp, for example) or does it extend along the entire mRNA length? Furthermore, what is the structure of the apparent eIF3−80S complex? Many questions also remain regarding the potential oncogenic or tumor suppressive functions of eIF3 and its individual subunits. Are these linked to mRNA-selective translation or to unexplored moonlighting functions of eIF3 subunits outside of holo-eIF3? Likewise, several eIF3 subunits were shown to localize to the nucleus, but their potential nuclear functions remain entirely elusive. Considering these important unresolved questions, eIF3 promises to remain an exciting multifunctional protein complex with many more unexpected roles in cellular physiology and pathophysiology ready to be unearthed in years to come.
Figure 1.
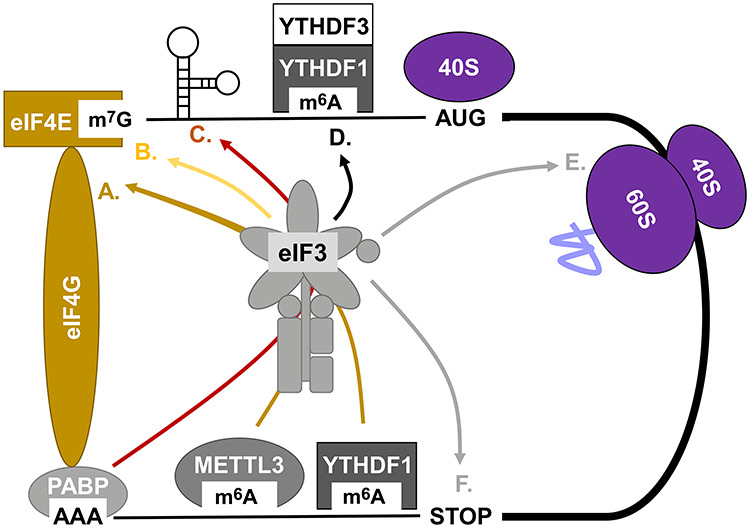
Multiple roles of eIF3 in canonical and non-canonical translation. (A) Promotion of closed loop formation through 3′-UTR m6A binding of writer and reader proteins and eIF3 recruitment. (B) Direct cap and stem loop binding of eIF3 via eIF3d. (C) Promotion of closed loop formation through PABP-facilitated recruitment of eIF3 to cellular IRES elements. (D) Direct or YTH reader-mediated recruitment of eIF3 to 5′-UTR m6A sites to promote cap-independent but scanning-dependent initiation. (E) Potential role of eIF3 in translation elongation through direct interaction with translating 80S ribosomes. (F) The role of eIF3 in translational read-through and termination/ribosome recycling (not discussed here).
Funding
D.A.W.’s lab at Xiamen University is funded through grants 81773771 and 31770813 from the National Science Foundation of China and the 1000 Talent Program.
Conflict of interest: none declared.
References
- Acosta-Sampson L., Döring K., Lin Y., et al. (2017). Role for ribosome-associated complex and stress-seventy subfamily B (RAC-Ssb) in integral membrane protein translation. J. Biol. Chem. 292, 19610–19627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguero T., Jin Z., Chorghade S., et al. (2017). Maternal dead-end 1 promotes translation of nanos1 through binding the eIF3 complex. Development 144, 3755–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayhan F., Perez B.A., Shorrock H.K., et al. (2018). SCA8 RAN polySer protein preferentially accumulates in white matter regions and is regulated by eIF3F. EMBO J. 37, e99023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L., and Sabatini D.M. (2014). Regulation of mTORC1 by amino acids. Trends Cell Biol. 24, 400–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlen J., Fenzl K., Kramer G., et al. (2019). Selective 40S footprinting reveals that scanning ribosomes remain cap-tethered in human cells. BioRxiv, doi: 10.1101/806364. [DOI] [PubMed] [Google Scholar]
- Cate J.H.D. (2017). Human eIF3: from ‘blobology’ to biological insight. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, pii: 20160176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe J., Lin S., Zhang W., et al. (2018). mRNA circularization by METTL3–eIF3h enhances translation and promotes oncogenesis. Nature 561, 556–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coots R.A., Liu X.-M., Mao Y., et al. (2017). m6A facilitates eIF4F-independent mRNA translation. Mol. Cell 68, 504–514.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duttler S., Pechmann S., and Frydman J. (2013). Principles of cotranslational ubiquitination and quality control at the ribosome. Mol. Cell 50, 379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliseev B., Yeramala L., Leitner A., et al. (2018). Structure of a human cap-dependent 48S translation pre-initiation complex. Nucleic Acids Res. 46, 2678–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes-Duarte A., Lacerda R., Menezes J., et al. (2018). eIF3: a factor for human health and disease. RNA Biol. 15, 24–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzmil M., Rzymski T., Milani M., et al. (2010). An oncogenic role of eIF3e/INT6 in human breast cancer. Oncogene 29, 4080–4089. [DOI] [PubMed] [Google Scholar]
- Guan B.-J., Hoef V., Jobava R., et al. (2017). A unique ISR program determines cellular responses to chronic stress. Mol. Cell 68, 885–900.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero C., Milenković T., Pržulj N., et al. (2008). Characterization of the proteasome interaction network using a QTAX-based tag-team strategy and protein interaction network analysis. Proc. Natl Acad. Sci. USA 105, 13333–13338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmannová A., Prilepskaja T., Wagner S., et al. (2019). Adapted formaldehyde gradient cross-linking protocol implicates human eIF3d and eIF3c, k and l subunits in the 43S and 48S pre-initiation complex assembly, respectively. Nucleic Acids Res. 48, 1969–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey J.W.B. (2015). The role of eIF3 and its individual subunits in cancer. Biochim. Biophys. Acta 1849, 792–800. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A.G. (2017). Structural insights into the mechanism of scanning and start codon recognition in eukaryotic translation initiation. Trends Biochem. Sci. 42, 589–611. [DOI] [PubMed] [Google Scholar]
- Holz M.K., Ballif B.A., Gygi S.P., et al. (2005). mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 123, 569–580. [DOI] [PubMed] [Google Scholar]
- Hsieh A.C., Liu Y., Edlind M.P., et al. (2012). The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485, 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutt D.M., Loguercio S., Roth D.M., et al. (2018). Correcting the F508del-CFTR variant by modulating eukaryotic translation initiation factor 3-mediated translation initiation. J. Biol. Chem. 293, 13477–13495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N.T., Lareau L.F., and Weissman J.S. (2011). Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.H., Cai X., Vaughn J.N., et al. (2007). On the functions of the h subunit of eukaryotic initiation factor 3 in late stages of translation initiation. Genome Biol. 8, R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer G., Boehringer D., Ban N., et al. (2009). The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol. 16, 589–597. [DOI] [PubMed] [Google Scholar]
- Kumar P., Hellen C.U.T., and Pestova T.V. (2016). Toward the mechanism of eIF4F-mediated ribosomal attachment to mammalian capped mRNAs. Genes Dev. 30, 1573–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.S.Y., Kranzusch P.J., and Cate J.H.D. (2015). eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature 522, 111–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.S.Y., Kranzusch P.J., Doudna J.A., et al. (2016). eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 536, 96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Choe J., Du P., et al. (2016). The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell 62, 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Li F., Huang L., et al. (2019). eIF3 associates with 80S ribosomes to promote translation elongation, mitochondrial homeostasis, and muscle health. BioRxiv, doi: 10.1101/651240. [DOI] [PubMed] [Google Scholar]
- Llácer J.L., Hussain T., Marler L., et al. (2015). Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol. Cell 59, 399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llácer J.L., Hussain T., Saini A.K., et al. (2018). Translational initiation factor eIF5 replaces eIF1 on the 40S ribosomal subunit to promote start-codon recognition. eLife 7, e39273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchione R., Laurin D., Liguori L., et al. (2015). MD11-mediated delivery of recombinant eIF3f induces melanoma and colorectal carcinoma cell death. Mol. Ther. Methods Clin. Dev. 2, 14056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medicherla B., and Goldberg A.L. (2008). Heat shock and oxygen radicals stimulate ubiquitin-dependent degradation mainly of newly synthesized proteins. J. Cell Biol. 182, 663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K.D., Patil D.P., Zhou J., et al. (2015). 5′ UTR m6A promotes cap-independent translation. Cell 163, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad M.P., Munzarová Pondělíčková V., Zeman J., et al. (2017). In vivo evidence that eIF3 stays bound to ribosomes elongating and terminating on short upstream ORFs to promote reinitiation. Nucleic Acids Res. 45, 2658–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M., Gravel S.-P., Chénard V., et al. (2013). mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 18, 698–711. [DOI] [PubMed] [Google Scholar]
- Pegoraro G., Voss T.C., Martin S.E., et al. (2012). Identification of mammalian protein quality control factors by high-throughput cellular imaging. PLoS One 7, e31684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson D.T., Merrick W.C., and Safer B. (1979). Binding and release of radiolabeled eukaryotic initiation factors 2 and 3 during 80 S initiation complex formation. J. Biol. Chem. 254, 2509–2516. [PubMed] [Google Scholar]
- Pulos-Holmes M.C., Srole D.N., Juarez M.G., et al. (2019). Repression of ferritin light chain translation by human eIF3. eLife 8, e48193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reading B.J., Williams V.N., Chapman R.W., et al. (2013). Dynamics of the striped bass (Morone saxatilis) ovary proteome reveal a complex network of the translasome. J. Proteome Res. 12, 1691–1699. [DOI] [PubMed] [Google Scholar]
- Sadato D., Ono T., Gotoh-Saito S., et al. (2018). Eukaryotic translation initiation factor 3 (eIF3) subunit e is essential for embryonic development and cell proliferation. FEBS Open Bio 8, 1188–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesen J., Casaos J., Scotland S.J., et al. (2017). The bad, the good and eIF3e/INT6. Front. Biosci. 22, 1–20. [DOI] [PubMed] [Google Scholar]
- Sha Z., Brill L.M., Cabrera R., et al. (2009). The eIF3 interactome reveals the translasome, a supercomplex linking protein synthesis and degradation machineries. Mol. Cell 36, 141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M., Su D., Scheliga J.S., et al. (2016). A transcript-specific eIF3 complex mediates global translational control of energy metabolism. Cell Rep. 16, 1891–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalgi R., Hurt J.A., Krykbaeva I., et al. (2013). Widespread regulation of translation by elongation pausing in heat shock. Mol. Cell 49, 439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatsky I.N., Terenin I.M., Smirnova V.V., et al. (2018). Cap-independent translation: what’s in a name? Trends Biochem. Sci. 43, 882–895. [DOI] [PubMed] [Google Scholar]
- Shi H., Wang X., Lu Z., et al. (2017). YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 27, 315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Kahle A., Hershey J.W.B., et al. (2006). Decreased expression of eukaryotic initiation factor 3f deregulates translation and apoptosis in tumor cells. Oncogene 25, 4923–4936. [DOI] [PubMed] [Google Scholar]
- Siddiqui N., and Sonenberg N. (2015). Signalling to eIF4E in cancer. Biochem. Soc. Trans. 43, 763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti A., Brito Querido J., Myasnikov A.G., et al. (2016). eIF3 peripheral subunits rearrangement after mRNA binding and start-codon recognition. Mol. Cell 63, 206–217. [DOI] [PubMed] [Google Scholar]
- Smith M.D., Gu Y., Querol-Audí J., et al. (2013). Human-like eukaryotic translation initiation factor 3 from Neurospora crassa. PLoS One 8, e78715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N., and Hinnebusch A.G. (2009). Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun S.Z., Wells S.E., Deardorff J.A., et al. (1997). Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc. Natl Acad. Sci. USA 94, 9046–9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakor N., Smith M.D., Roberts L., et al. (2017). Cellular mRNA recruits the ribosome via eIF3–PABP bridge to initiate internal translation. RNA Biol. 14, 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen C.C., Chantranupong L., Keys H.R., et al. (2012). A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J., Flavell R.A., and Li H.-B. (2018). RNA m6A modification and its function in diseases. Front. Med. 12, 481–489. [DOI] [PubMed] [Google Scholar]
- Trachsel H., and Staehelin T. (1979). Initiation of mammalian protein synthesis the multiple functions of the initiation factor eIF-3. Biochim. Biophys. Acta 565, 305–314. [DOI] [PubMed] [Google Scholar]
- Truitt M.L., Conn C.S., Shi Z., et al. (2015). Differential requirements for eIF4E dose in normal development and cancer. Cell 162, 59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner G.C. (2000). Detecting and measuring cotranslational protein degradation in vivo. Science 289, 2117–2120. [DOI] [PubMed] [Google Scholar]
- Valášek L.S., Zeman J., Wagner S., et al. (2017). Embraced by eIF3: structural and functional insights into the roles of eIF3 across the translation cycle. Nucleic Acids Res. 45, 10948–10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S., Herrmannova A., Hronova V., et al. (2019). Selective translation complex profiling reveals staged initiation and co-translational assembly of initiation factor complexes. BioRxiv, doi: 10.1101/806125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S., Herrmannová A., Malík R., et al. (2014). Functional and biochemical characterization of human eukaryotic translation initiation factor 3 in living cells. Mol. Cell. Biol. 34, 3041–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S., Herrmannová A., Šikrová D., et al. (2016). Human eIF3b and eIF3a serve as the nucleation core for the assembly of eIF3 into two interconnected modules: the yeast-like core and the octamer. Nucleic Acids Res. 44, 10772–10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Durfee L.A., and Huibregtse J.M. (2013). A cotranslational ubiquitination pathway for quality control of misfolded proteins. Mol. Cell 50, 368–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Delahunty C., Fritz-Wolf K., et al. (2015a). Characterization of the 26S proteasome network in Plasmodium falciparum. Sci. Rep. 5, 17818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhao B.S., Roundtree I.A., et al. (2015b). N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg D.E., Shah P., Eichhorn S.W., et al. (2016). Improved ribosome-footprint and mRNA measurements provide insights into dynamics and regulation of yeast translation. Cell Rep. 14, 1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek R.C. (2018). Role of eIF2α kinases in translational control and adaptation to cellular stress. Cold Spring Harb. Perspect. Biol. 10, a032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Fan X., Mao M., et al. (2017). Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 27, 626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L., Wan Y., Li D., et al. (2013). The m-subunit of murine translation initiation factor eIF3 maintains the integrity of the eIF3 complex and is required for embryonic development, homeostasis, and organ size control. J. Biol. Chem. 288, 30087–30093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Arslan F., Wee S., et al. (2005). PCI proteins eIF3e and eIF3m define distinct translation initiation factor 3 complexes. BMC Biol. 3, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wan J., Gao X., et al. (2015). Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 526, 591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Huang P., Yang N., et al. (2017). Establishment and application of a high throughput screening system targeting the interaction between HCV internal ribosome entry site and human eukaryotic translation initiation factor 3. Front. Microbiol. 8, 977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T., Gibbens B., Doty N.S., et al. (2011). Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl Acad. Sci. USA 108, 260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]


