Abstract
Background
There are many situations of abnormal metabolism influencing liver graft function. This study aims to provide data for the development of liver function recovery after liver transplantation by dynamically analyzing metabolites of bile acids pathway in serum.
Material/Methods
A comprehensive metabolomics profiling of serum of 9 liver transplantation patients before transplantation, on the 1st, 3rd, and 7th days after liver transplantation, and healthy individuals were performed by ultra-performance liquid chromatography-mass spectrometry (UPLC-MS). Multivariate data and dynamic analysis were used to search for biomarkers between the metabolomics profiles present in perioperative liver transplantation and normal controls.
Results
Thirty-three differential endogenous metabolites were screened by the threshold of variable importance in the projection (VIP) from an orthogonal partial least square discriminant analysis (OPLS-DA) greater than 1.0, q-value <0.05, and fold change (FC) ≤0.8 or ≥1.2 between the preoperative group and the normal controls in negative mode. The metabolite intensities of taurocholic acid, taurochenodeoxycholic acid, chenodeoxycholic acid glycine conjugate, and glycocholic acid pre-transplantation were significantly higher than those of normal controls. The average metabolite intensities of taurocholic acid and taurochenodesoxycholic acid on the first day after liver transplantation were lower than those observed pre-transplantation. The average metabolite intensities on day 3 after liver transplantation showed a sudden increase and then decreased after 7 postoperative days. The average metabolite intensities of glycocholic acid and chenodeoxycholic acid glycine conjugate showed an increasing trend on the 1st, 3rd, and 7th days after liver transplantation.
Conclusions
Use of taurocholic acid and taurochenodeoxycholic acid-related bile secretion, liver regeneration, and de novo bile acid synthesis may help clinical evaluation and provide data for the development of liver function recovery after liver transplantation.
MeSH Keywords: Liver Transplantation, Metabolomics, Perioperative Period
Background
As a new research area built upon successive genomics and proteomics development, metabolomics studies the collection of all metabolites from cells, tissue, or organs [1]. A series of different chemical types of molecules contain peptides, carbohydrates, lipids, nucleic acids, and catabolic products of exogenous compounds [2]. Metabolomics often responds to the genetic, disease, and environmental impacts of the final stages in the body or “downstream” of genes and proteins. Unlike genetics and proteomics, the metabolomics reaction is phenotypic changes, revealing more functional changes. Some researchers have used metabolomics as potential biomarkers to investigate therapeutic responses [3,4].
Primary liver cancer (PHC) is one of the 5 most common cancers in the world, and it is also one of the most common malignant tumors in China [5,6]. Liver transplantation is the most effective treatment for PHC [7]. Preoperative and postoperative liver transplantation has different situations, which are assessed with many indexes [8]. Some studies have shown that bile acids can be used as a tool to assess the quality and function of the donor liver early after transplantation [9]. However, the relationship between the metabolites of the bile acid pathway and perioperative liver transplantation recipients has not been reported. Analysis of the metabolites of the bile acid pathway in perioperative liver transplant recipients will help improve our understanding of liver function recovery in patients after liver transplantation. This study was performed by ultra-performance liquid chromatography-mass spectrometry (UPLC-MS) combined with pattern recognition analysis multiplatform next-generation sequencing, which were used to analyze the amount of metabolites produced and the pathways involved [10].
Material and Methods
Patients
Serum samples were collected from 9 patients with primary hepatic carcinoma. The patients were recruited from the Nephrology Department at Guilin No. 181 Hospital (Guilin No. 181 Hospital officially changed its name to Guilin No. 924 Hospital), Guangxi, China. All of the patients had undergone transplantation surgery without recurrence of acute rejection after surgery, and without recurrence of hepatocellular cancer after liver transplantation. Serum samples were collected from patients at various time points: preoperative patients (Pre), 1-day postoperative patients (P1), 3-day postoperative patients (P3), and 7-day postoperative patients (P7). The transplanted livers were obtained from brain-dead donors. The donors did not have a history of liver disease, a medical history of cancer in the last 10 years, hepatitis B surface antigen (HBsAg), hepatitis C virus, or human immunodeficiency virus (HIV) antibodies. Fat infiltration (macrovesicular steatosis >40%), any fibrosis and atherosclerosis of the hepatic artery observed post-transplantation of the liver, and donors after cardiac death were also excluded [11]. Serum samples were also collected from 9 healthy individuals as normal controls (NC) from the Physical Center of the Guilin No. 181 Hospital. The clinical characteristics of patients and healthy individuals were extracted from the hospital database and are summarized in Table 1. Aspartate aminotransferase (AST) and alanine aminotransferase (ALT) <50 IU/L were considered normal; therefore, the liver function of patients gradually recovered to normal. All of the peripheral serum samples were obtained after receiving informed consent from the participating subjects. This study was performed according to the Hospital Ethics Committee, which abides by the Helsinki Declaration on ethical principles for medical research involving human subjects.
Table 1.
Characteristics of the patients and the normal controls.
| Characteristic | NC | Patients | |||
|---|---|---|---|---|---|
| Pre | P1 | P3 | P7 | ||
| Peripheral samples(n) | 9 | 9 | 9 | 9 | 9 |
| Age (years) | 43±12 | 46±15 | 46±15 | 46±15 | 46±15 |
| Sex: Male/Female (n/n) | 8/1 | 8/1 | 8/1 | 8/1 | 8/1 |
| ALT (IU/L) | 23.0±8.0 | 72.0±16.0 | 286.0±52.0 | 212.0±43.0 | 84.0±31.0 |
| AST (IU/L) | 16.0±6.8 | 54.1±21.9 | 257.0 ±58.0 | 181.4±46.2 | 56.3±25.7 |
| TBIL (μmol/L) | 12.4±4.2 | 13.9±3.8 | 38.9±11.1 | 35.4±8.6 | 19.5±9.8 |
Serum collection and storage
Whole-blood samples were taken from a peripheral vein between 7: 00 and 9: 00 AM. Serum samples were collected from 9 patients and 9 healthy individuals between 2014 and 2015. The serum samples used for metabolite detection were collected from patients either in the Emergency Department or during physical examination. Every sample was centrifuged at 3500×g for 5 min, and 2 mL of the supernatant was transferred into clean tubes and frozen at −80°C until UPLC-MS/MS analysis. Each 250 μL serum sample was mixed with 750 μL of cold methanol and vortexed for 1 min. After centrifugation at 20 000×g for 20 min, 90 μL of supernatant was transferred into clean tubes and freeze-dried. The dried samples were reconstituted with 90 μL of 10% aqueous methanol.
UPLC-MS/MS analysis
The chromatographic separation was performed on a Waters Acquity™ 2777C ultra-performance liquid chromatography (UPLC) system (Waters, U.K.) and a SYNAPT G2 XS QTOF spectrometer (Waters, U.K.). The sample was loaded onto an Acquity UPLC HSS T3 (1.8 μm) (Waters, U.K.) column held at 30°C. The UPLC mobile phase consisted of water (solution A) and methanol (solution B), with a splitless flow rate of 0.4 mL/min and an injection volume of 10 μL. The following gradient elution program was used: 0–2 min, 100%–100% A; 2–12 min, 100% A-0% A; 12–14 min, 0% A-0% A; and 14–15 min, 0% A-100% A.
Mass spectrometry
A quadrupole time-of-flight (Q-TOF) instrument (Waters Corp., Milford, USA) was used to carry out the mass spectrometry with electrospray ionization (ESI), which had a capillary voltage of 1000 V in positive mode and 2000 V in negative mode and a sampling cone voltage of 40 V in both modes. The desolvation temperature was set at 350°C, and the cone gas flow was set at 50 L h–1. The source temperature was set at 120°C. Mass spectrometry (MS) data range was from 100 to 1200 m/z. The instrument performance and data quality were monitored by analyzing all samples with a rigorous quality assurance strategy [12].
Statistical analyses
All the UPLC-MS raw files were converted using MarkerLynx software (Version 4.1 Waters, U.K.) for the compound statistics principal component analysis (PCA), the orthogonal partial least square discriminant analysis (OPLS-DA) [13,14], and the correlation analysis. Variables with a threshold of variable importance in the projection (VIP) from the OPLS-DA greater than 1.0, a q-value <0.05, and fold change (FC) ≤0.8 or ≥1.2 were considered to be differential ions. The Q-value means that the p-value was assessed using a t test and corrected using the false discovery rate (FDR). Differential ions were identified using Progenesis QI (version 2.1). Progenesis QI was used to obtain the mass-to-charge ratio, the retention time, and the ion area of metabolites by extracting the peak.
Identification of biomarkers and metabolic pathway analysis
The accurate MS fragments of the metabolites were matched from the Human Metabolome Database (http://www.hmdb.ca/) for identifying the metabolites. The Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/pathway.html) was used for pathway analysis, which visualizes the metabolomics.
Results
Multivariate statistical analysis of the metabolite profiles
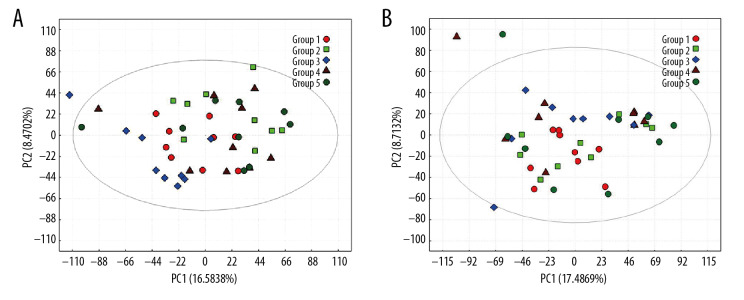
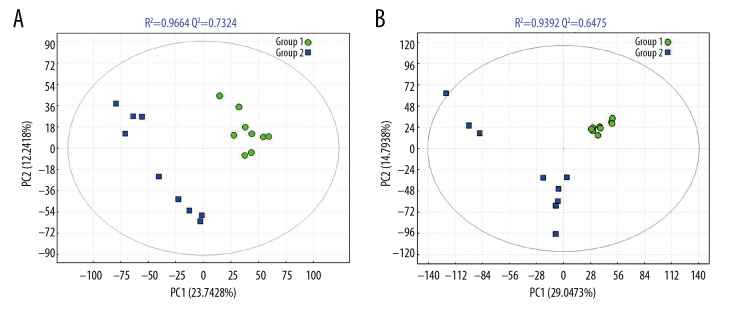
The PCA score map was used for multivariate data analysis to observe the clustering between perioperative liver transplant patients and the control groups (Figure 1A, 1B), which suggested a clear separation between the perioperative liver transplantation patients and healthy individuals, with a few overlaps. The OPLS-DA score plot of preoperative patients and healthy controls showed remarkable group separation, with no sample overlapping for negative and positive patients (Figure 2A, 2B). When the R2 and Q2 values of the model parameters are higher, the OPLS-DA model is more reliable. A more rigorous model validation procedure was applied by class permutation testing, and the results strongly confirmed model consistency and reliability. The model parameters were permutation tested 1000 times. The p-value of Pre vs. NC was 0.012 for the model of positive ions and 0.003 for the model of negative ions; p<0.01 indicates the difference between the 2 groups was extremely significantly. Therefore, we chose negative ions of Pre vs. NC to select differential makers. Metabolite identification was subsequently performed to yield 33 annotated endogenous metabolites; detailed information is depicted in Table 2. The 4 ions belong to the bile acid pathway among the metabolites.
Figure 1.
PCA scores for metabolic pattern to visualize group clustering between perioperative liver transplantation and NC samples. PCA score plots of serum samples collected from NC (group 1), Pre (group 2), P1 (group 3), P3 (group 4), and P7 (group 5) groups in negative ion mode (A) and positive ion mode (B). The sample clusters of negative ion mode were tighter than those of positive ion mode, and no extreme outliers were observed.
Figure 2.
The PLS-DA models of NC (group 1) and Pre (group 2) in negative ion mode (A) and positive ion mode (B). The PLS-DA score plot for the negative ion mode has a clearer separation than that for the positive ion mode between Pre and NC samples.
Table 2.
Potential metabolite makers of negative ions from Pre vs. NC.
| Metabolite name | m/z | Change | VIP | Fold change | q-value | Related pathway |
|---|---|---|---|---|---|---|
| Threonic acid | 135.0296 | ↑ | 2.920519 | 6.722945 | 0.002249 | Ascorbate and aldarate metabolism |
| Threonic acid | 135.0299 | ↑ | 3.101811 | 8.680666 | 0.002504 | Ascorbate and aldarate metabolism |
| Phosphoribosyl formamidocarboxamide | 365.0538 | ↑ | 1.429341 | 1.667861 | 0.015919 | Purine metabolism; Metabolic pathways; Biosynthesis of secondary metabolites |
| 8(R)-Hydroperoxylinoleic acid | 311.2199 | ↑ | 1.985069 | 3.354787 | 0.049389 | Linoleic acid metabolism |
| 13-L-Hydroperoxylinoleic acid | 311.2199 | ↑ | 1.985069 | 3.354787 | 0.049389 | Linoleic acid metabolism; Metabolic pathways |
| 9(S)-HPODE | 311.2199 | ↑ | 1.985069 | 3.354787 | 0.049389 | Linoleic acid metabolism; Metabolic pathways |
| Taurocholic acid | 514.2828 | ↑ | 3.725514 | 121.6349 | 0.025012 | Primary bile acid biosynthesis; Secondary bile acid biosynthesis; Taurine and hypotaurine metabolism; Metabolic pathways; Bile secretion |
| 8-HETE | 319.2261 | ↓ | 3.539139 | 0.104499 | 0.007557 | Arachidonic acid metabolism; Metabolic pathways; PPAR signaling pathway |
| 8,9-Epoxyeicosatrienoic acid | 319.2261 | ↓ | 3.539139 | 0.104499 | 0.007557 | Arachidonic acid metabolism; Metabolic pathways; Vascular smooth muscle contraction |
| 19(S)-HETE | 319.2261 | ↓ | 3.539139 | 0.104499 | 0.007557 | Arachidonic acid metabolism; Metabolic pathways |
| 11(R)-HETE | 319.2261 | ↓ | 3.539139 | 0.104499 | 0.007557 | Arachidonic acid metabolism |
| 15(S)-HETE | 319.2261 | ↓ | 3.539139 | 0.104499 | 0.007557 | Arachidonic acid metabolism |
| 20-Hydroxyeicosatetraenoic acid | 319.2261 | ↓ | 3.539139 | 0.104499 | 0.007557 | Arachidonic acid metabolism; Metabolic pathways; Vascular smooth muscle contraction |
| 5,6-Epoxy-8,11,14-eicosatrienoic acid | 319.2261 | ↓ | 3.539139 | 0.104499 | 0.007557 | Arachidonic acid metabolism; Metabolic pathways; Vascular smooth muscle contraction |
| 16(R)-HETE | 319.2261 | ↓ | 3.539139 | 0.104499 | 0.007557 | Arachidonic acid metabolism |
| 14,15-Epoxy-5,8,11-eicosatrienoic acid | 319.2261 | ↓ | 3.539139 | 0.104499 | 0.007557 | Arachidonic acid metabolism; Metabolic pathways; Vascular smooth muscle contraction |
| 11,12-Epoxyeicosatrienoic acid | 319.2261 | ↓ | 3.539139 | 0.104499 | 0.007557 | Arachidonic acid metabolism; Metabolic pathways; Vascular smooth muscle contraction |
| 5-HETE | 319.2261 | ↓ | 3.539139 | 0.104499 | 0.007557 | Arachidonic acid metabolism |
| Docosapentaenoic acid | 329.2469 | ↓ | 1.976434 | 0.413162 | 0.008974 | Biosynthesis of unsaturated fatty acids |
| Arachidonic acid | 303.2320 | ↓ | 2.263972 | 0.331697 | 0.000831 | Arachidonic acid metabolism; Linoleic acid metabolism; Biosynthesis of unsaturated fatty acids; Metabolic pathways; Vascular smooth muscle contraction; Fc epsilon RI signaling pathway; Fc gamma R-mediated phagocytosis; Long-term depression; Phototransduction – fly; GnRH signaling pathway; Leishmaniasis; Amoebiasis |
| Taurochenodeoxycholic acid | 498.2879 | ↑ | 2.958113 | 33.07833 | 0.044239 | Primary bile acid biosynthesis; Secondary bile acid biosynthesis; Bile secretion |
| 9,10-Epoxyoctadecenoic acid | 295.2257 | ↓ | 1.591986 | 0.55364 | 0.02713 | Linoleic acid metabolism; Metabolic pathways |
| Alpha-dimorphecolic acid | 295.2257 | ↓ | 1.591986 | 0.55364 | 0.02713 | Linoleic acid metabolism; PPAR signaling pathway |
| 12,13-EpOME | 295.2257 | ↓ | 1.591986 | 0.55364 | 0.02713 | Linoleic acid metabolism; Metabolic pathways |
| 13S-hydroxyoctadecadienoic acid | 295.2257 | ↓ | 1.591986 | 0.55364 | 0.02713 | Linoleic acid metabolism; PPAR signaling pathway |
| Chenodeoxycholic acid glycine conjugate | 448.3056 | ↑ | 2.59988 | 6.271501 | 0.010639 | Primary bile acid biosynthesis; Secondary bile acid biosynthesis; Bile secretion |
| Glycocholic acid | 464.3005 | ↑ | 2.925803 | 9.113965 | 0.015133 | Primary bile acid biosynthesis; Secondary bile acid biosynthesis; Metabolic pathways; Bile secretion |
| 9,12,13-TriHOME | 329.2304 | ↑ | 2.3531 | 3.288932 | 0.006036 | Linoleic acid metabolism |
| 9,10,13-TriHOME | 329.2304 | ↑ | 2.3531 | 3.288932 | 0.006036 | Linoleic acid metabolism |
| Adipate semialdehyde | 129.0544 | ↓ | 1.13189 | 0.720488 | 0.025688 | Caprolactam degradation; Metabolic pathways; Microbial metabolism in diverse environments |
| Ketoleucine | 129.0544 | ↓ | 1.13189 | 0.720488 | 0.025688 | Valine, leucine and isoleucine degradation; Valine, leucine and isoleucine biosynthesis; Glucosinolate biosynthesis; Metabolic pathways; Biosynthesis of secondary metabolites |
| Phosphoribosyl formamidocarboxamide | 401.0251 | ↑ | 1.649845 | 1.751775 | 0.003328 | Purine metabolism; Metabolic pathways; Biosynthesis of secondary metabolites |
| Trans-Cinnamic acid | 147.0438 | ↑ | 1.2355 | 1.388167 | 0.001396 | Phenylalanine metabolism; Phenylpropanoid biosynthesis; Metabolic pathways; Biosynthesis of secondary metabolites; Microbial metabolism in diverse environments |
The markers were chosen on the basis of VIP from the OPLS-DA greater than 1.0, q-value <0.05, and fold change ≤0.8 or ≥1.2. “↑” and “↓” represent the compound is up- and down-regulated, respectively, in preoperative patients compared with the normal control.
Changes in the metabolites of the bile acid pathway
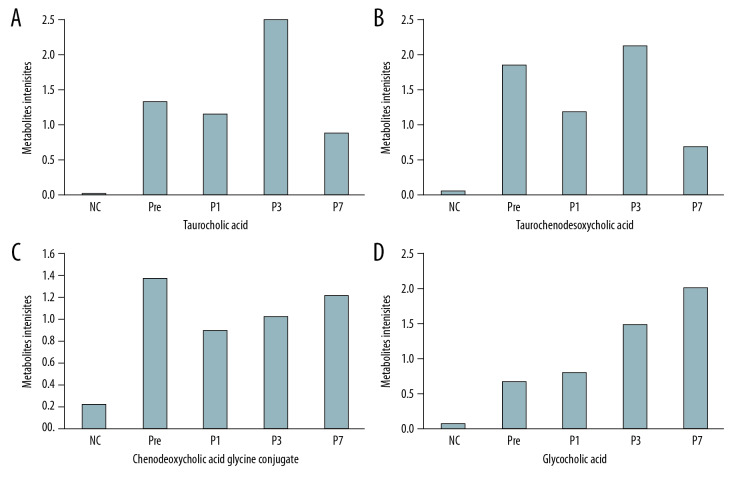
Four differential metabolites from the Pre patient group were distinguished from those of the controls (VIP from the OPLS-DA greater than 1.0, q-value <0.05 and fold change ≤0.8 or ≥1.2). Overall, these metabolite intensities displayed dynamic changes between control and perioperative liver transplantation samples, including taurocholic acid (Figure 3A), taurochenodeoxycholic acid (Figure 3B), chenodeoxycholic acid glycine conjugate (Figure 3C), and glycocholic acid (Figure 3D). The 4 types of metabolites had varying degrees of up- and down-regulation before and after transplantation, which indicates that the metabolites of the bile acid pathway play an important role in perioperative liver transplantation. The results showed that the metabolite intensities of taurocholic acid (VIP=3.7255, FC=121.63, q=0.0250), taurochenodeoxycholic acid (VIP=2.9581, FC=33.078, q=0.0442), chenodeoxycholic acid glycine conjugate (VIP=2.5999, FC=6.2715, q=0.0106), and glycocholic acid (VIP=2.9258, FC=9.1140 q=0.0151) were significantly higher in pre-transplantation patients than in normal controls. The average metabolite intensities of taurocholic acid, taurochenodeoxycholic acid, and chenodeoxycholic acid glycine conjugate at the first day after liver transplantation were lower than those detected pre-transplantation. The average metabolite intensities of taurocholic acid and taurochenodeoxycholic acid on day 3 after liver transplantation had a sudden increase and then decreased after 7 postoperative days. The average metabolite intensity on day 7 after liver transplantation tended to approach NC. The average metabolite intensities of chenodeoxycholic acid glycine conjugate and glycocholic acid showed an increasing trend on the 1st, 3rd, and 7th days after liver transplantation. The average ionic intensities of glycocholic acid on postoperative day 7 were significantly higher than those of NC (VIP=2.9072, FC=27.001, q=0.0384). The results of the differential comparison between groups are listed in Table 3.
Figure 3.
(A–D) Intensities of the metabolites of the bile acid pathway in the control and perioperative liver transplantation groups.
Table 3.
The differential comparison for ionic strength of metabolites.
| Groups | Taurocholic acid | Taurochenodeoxycholic acid | Chenodeoxycholic acid glycine conjugate | Glycocholic acid | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VIP | Fold change | q-value | VIP | fold change | q-value | VIP | fold change | q-value | VIP | fold change | q-value | |
| Pre vs. NC | 3.7255* | 121.63* | 0.0250* | 2.9581* | 33.078* | 0.0442* | 2.5999* | 6.2715* | 0.0106* | 2.9258* | 9.1140* | 0.0151* |
| P1 vs. Pre | 0.1995 | 0.8706 | 0.58367 | 0.9535 | 0.6365 | 0.3168 | 1.4928 | 0.6484 | 0.1004 | 0.9049 | 1.1897 | 0.3272 |
| P3 vs. P1 | 1.0553 | 2.1777 | 0.5727 | 0.3641 | 1.8009 | 0.7085 | 0.5288 | 1.1475 | 0.6667 | 1.7125 | 1.8460 | 0.3533 |
| P7 vs. P3 | 0.6284 | 0.3481 | 0.9997 | 0.1090 | 0.32701 | 0.9997 | 0.7702 | 1.1803 | 0.9997 | 0.0039 | 1.3490 | 0.9999 |
| P7 vs. NC | 3.7079* | 80.275* | 0.0243* | 1.9267 | 12.399 | 0.1930 | 1.2458 | 5.5076 | 0.2866 | 2.9072* | 27.001* | 0.0384* |
VIP from the OPLS-DA greater than 1.0, q-value <0.05 and fold change ≤0.8 or ≥1.2 indicates the difference between the 2 groups was significantly.
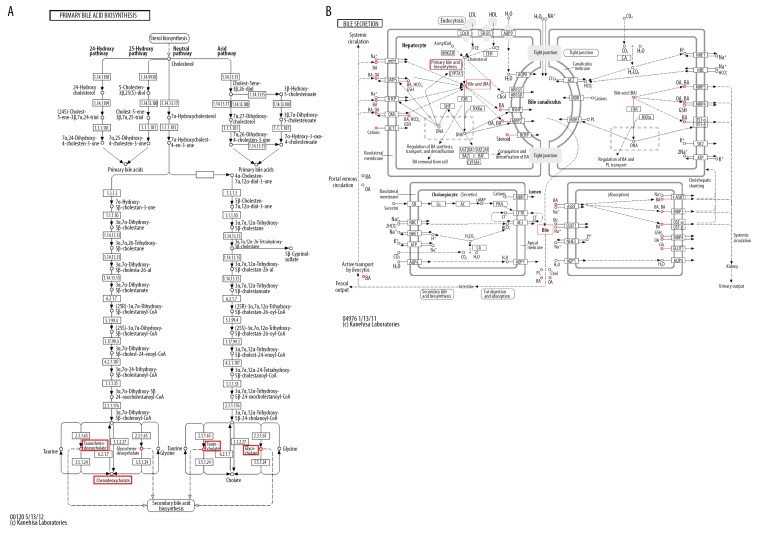
Metabolic network visualization for bile acid pathway
Figure 4A shows the significantly enriched metabolites of pathways for primary and secondary bile acid biosynthesis in the perioperative liver transplantation samples. Among these metabolites, taurocholic acid, taurochenodeoxycholic acid, chenodeoxycholic acid glycine conjugate, and glycocholic acid were involved in both primary and secondary bile acid biosynthesis. Primary and secondary bile acid biosynthesis were involved in bile secretion (Figure 4B). Finally, bile acids were produced through synthetic circulation. Taurocholic acid, taurochenodeoxycholic acid, chenodeoxycholic acid glycine conjugate, and glycocholic acid are metabolites of circulation. The results showed that these metabolites circulate through bile acid pathways by active transport in the liver, and recovered bile acids are secreted through bile [15,16].
Figure 4.
KEGG pathway analysis: summary of the metabolites in the bile acid pathway shown in the red box and described in this article. (A) The primary and secondary bile acid biosynthesis pathways. (B) The bile secretion pathway.
Discussion
Endogenous metabolites are involved in human metabolite pathways and are associated with the development of some diseases [17]. It is important that some methods detect the metabolites of these pathways to find diagnostic biomarkers. Although many studies have reported metabolite biomarkers for liver disease and liver transplantation [10,18–22], which also included bile acids [23], the dynamic analysis value of the metabolites of the bile acid pathway for monitoring perioperative liver transplantation remains largely unknown.
Liquid chromatograph mass spectrometer (LC-MS)-based metabolomics, which process large datasets and classifies sample groups as well as the nature of these metabolites, could be an advanced tool to find metabolites [10,24,25]. In this study, we used UPLC-MS/MS to conduct differential metabolite profiling between Pre and NC samples. The PCA revealed a significant separation between the perioperative liver transplantation and control samples. According to the PLS-DA model based on the R2 and Q2 values, we chose negative ions of Pre vs. NC to select 33 differentially expressed endogenous metabolites, which were chosen according to VIP from OPLS-DA greater than 1.0, q-value <0.05, and fold change ≤0.8 or ≥1.2. Patients who usually undergo liver transplantation for primary liver cancer will be observed for whether they have transplant rejection after surgery, whether hepatocarcinoma recurs, and whether liver function is restored. In this study, we found that patients had no transplant rejection and recurrence of hepatocellular carcinoma after surgery; therefore, we chose to discuss the liver function with dynamic changes after surgery. The bile acids in transplant recipients provide information on the pattern of recovery of bile secretion immediately after graft reperfusion and may be early indicators of the development of primary graft dysfunction [18]. We assume that metabolites of the bile acid pathway could help to further potentiate the role of bile acid analysis as a tool to assess graft function during perioperative liver transplantation. Therefore, we chose 4 out of 33 metabolites in the bile acid pathway to dynamically analyze the metabolic level between NC and perioperative liver transplantation. The KEGG analysis for visualizing the metabolic network of the bile acid pathway showed that UPLC-MS/MS is reliable for detecting metabolites of the bile acid pathway.
Previous reports have suggested that the delayed recovery of hepatic bile secretion observed in donor livers may hamper graft bile flow recovery in recipients, which is an early sign of proper liver function [9]. A recent study also suggested that taurocholic acid and taurochenodeoxycholic acid are significantly increased after transplantation and that this effect might be linked to liver regeneration and de novo bile acid synthesis [26]. In the present study, we also found that taurocholic acid and taurochenodeoxycholic acid were increased on postoperative day 3 compared with postoperative day 1 and showed a decrease on postoperative day 7. This result showed that increased taurocholic acid and taurochenodeoxycholic acid were involved in liver regeneration and de novo bile acid biosynthesis. The metabolic levels after 7 postoperative days were gradually becoming closer to those of NC, which may suggest that grafts recover normal function by normal bile secretion through the bile acid pathway. KEGG pathway analysis also showed that taurocholic acid and taurochenodeoxycholic acid are involved in the biosynthesis of primary bile acid biosynthesis in the bile acid pathway. Although chenodeoxycholic acid glycine conjugate and glycocholic acid are also involved in the biosynthesis of primary bile acid biosynthesis in the bile acid pathway, their expression levels are not similar to those of taurocholic acid and taurochenodeoxycholic acid, which first increased and then stabilized. This effect may be because the number of specimens was small, which could introduce errors into the experimental results. There may also be other alternative pathways that affect chenodeoxycholic acid glycine conjugate and glycocholic acid expression. Therefore, we will consider lengthening the follow-up time and increasing the collection of specimens in future related studies. We also plan to study the regulatory networks of metabolomics, gene expression, and proteins in perioperative liver transplantation.
Conclusions
Taurocholic acid and taurochenodeoxycholic acid may be important metabolites to use in evaluating liver function recovery after liver transplantation. Our research provides a new concept for studying dynamic changes in the metabolites of metabolic pathways to identify biomarkers during perioperative liver transplantation.
Abbreviations
- UPLC-MS
ultra-performance liquid chromatography – mass spectrometry
- VIP
variable importance in the projection
- OPLS-DA
orthogonal partial least square discriminant analysis
- FC
fold change
- PHC,primary hepatic carcinoma; Pre
preoperative patients
- P1
1-day postoperative patients
- P3
3-day postoperative patients
- P7
7-day postoperative patients
- HbsAg
hepatitis B surface antigen
- NC
normal controls
- AST,aspartate aminotransferase; ALT
alanine aminotransferase
- UPLC
ultra-performance liquid chromatography
- MS
mass spectrometry
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LC-MS
liquid chromatograph mass spectrometer
Footnotes
Source of support: The authors gratefully acknowledge financial support received from the Science and Technology Planning Project of Guangdong Province, China (No. 2016A050503009), and the Natural Science Foundation of Guangxi Province (2017GXNSFAA198185)
References
- 1.Fiehn O. Metabolomics – the link between genotypes and phenotypes. Plant Mol Biol. 2002;48:155–71. [PubMed] [Google Scholar]
- 2.Saghatelian A, Cravatt BF. Global strategies to integrate the proteome and metabolome. Curr Opin Chem Biol. 2005;9:62–68. doi: 10.1016/j.cbpa.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Mapstone M, Cheema AK, Fiandaca MS, et al. Plasma phospholipids identify antecedent memory impairment in older adults. Nat Med. 2014;20:415–18. doi: 10.1038/nm.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin W, Zhang J, Liu Y, et al. Studies on diagnostic biomarkers and therapeutic mechanism of Alzheimer’s disease through metabolomics and hippocampal proteomics. Eur J Pharm Sci. 2017;105:119–26. doi: 10.1016/j.ejps.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Wallis M, David AG. Role of liver transplantation for hepatobiliary malignant disorders. Lancet Oncol. 2004;5:480–88. doi: 10.1016/S1470-2045(04)01527-X. [DOI] [PubMed] [Google Scholar]
- 6.Chao-Xu Y. [Clinical study progression of oxaliplatin for primary hepatic carcinoma]. Chinese Clinical Oncology. 2010 [in Chinese] [Google Scholar]
- 7.Sui W, Gan Q, Liu F, et al. The differentially expressed circular ribonucleic acids of primary hepatic carcinoma following liver transplantation as new diagnostic biomarkers for primary hepatic carcinoma. Tumor Biol. 2018;40:101042831876692. doi: 10.1177/1010428318766928. [DOI] [PubMed] [Google Scholar]
- 8.Singh S, Nasa V, Tandon M. Perioperative monitoring in liver transplant patients. J Clin Exp Hepatol. 2012;2:271–78. doi: 10.1016/j.jceh.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melendez HV, Rela M, Setchell KDR, et al. Bile acids analysis: A tool to assess graft function in human liver transplantation. Transpl Int. 2004;17:286–92. doi: 10.1007/s00147-004-0719-2. [DOI] [PubMed] [Google Scholar]
- 10.Liang Q, Wang C, Li BB, Zhang AH. Metabolomics of alcoholic liver disease: A clinical discovery study. RSC Advances. 2015;5(98):80381–87. [Google Scholar]
- 11.Marsman WA, Wiesner RH, Rodriguez L, et al. Use of fatty donor liver is associated with diminished early patient and graft survival. Transplantation. 1996;62:1246–51. doi: 10.1097/00007890-199611150-00011. [DOI] [PubMed] [Google Scholar]
- 12.Quintás G, Portillo N, Garcíacañaveras JC, et al. Chemometric approaches to improve PLSDA model outcome for predicting human non-alcoholic fatty liver disease using UPLC-MS as a metabolic profiling tool. Metabolomics. 2012;8:86–98. [Google Scholar]
- 13.Barker M, Rayens W. Partial least squares for discrimination. J Chemometr. 2003;17:166–73. [Google Scholar]
- 14.Westerhuis JA, Hoefsloot HCJ, Smit S, et al. Assessment of PLSDA cross validation. Metabolomics. 2008;4:81–89. [Google Scholar]
- 15.Hagenbuch B, Meier PJ. Sinusoidal (basolateral) bile salt uptake systems of hepatocytes. Semin Liver Dis. 1996;16:129–36. doi: 10.1055/s-2007-1007226. [DOI] [PubMed] [Google Scholar]
- 16.Agellon LB, Torchia EC. Intracellular transport of bile acids. Biochim Biophys Acta. 2000;1486:198–209. doi: 10.1016/s1388-1981(00)00057-3. [DOI] [PubMed] [Google Scholar]
- 17.Gregg C, Joseph G. The role of endogenous metabolite alterations in neuropsychiatric disease. ACS Chem Neurosci. 2018;9:2101–13. doi: 10.1021/acschemneuro.8b00145. [DOI] [PubMed] [Google Scholar]
- 18.Perera MT, Higdon R, Richards DA, et al. Biomarker differences between cadaveric grafts used in human orthotopic liver transplantation as identified by coulometric electrochemical array detection (CEAD) metabolomics. OMICS. 2014;18:767–77. doi: 10.1089/omi.2014.0094. [DOI] [PubMed] [Google Scholar]
- 19.Cortes M, Pareja E, Garcíacañaveras JC, et al. Metabolomics discloses donor liver biomarkers associated with early allograft dysfunction. J Hepatol. 2014;61:564–74. doi: 10.1016/j.jhep.2014.04.023. [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Wang W, Lv S, et al. Metabonomics study of liver cancer based on ultra performance liquid chromatography coupled to mass spectrometry with HILIC and RPLC separations. Anal Chim Acta. 2009;650:3–9. doi: 10.1016/j.aca.2009.03.039. [DOI] [PubMed] [Google Scholar]
- 21.Citores MJ, Lucena JL, Fuente SDL, et al. Serum biomarkers and risk of hepatocellular carcinoma recurrence after liver transplantation. World J Hepatol. 2019;11:50–64. doi: 10.4254/wjh.v11.i1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golobschwarzl N, Krassnig S, Toeglhofer AM, et al. New liver cancer biomarkers: PI3K/AKT/mTOR pathway members and eukaryotic translation initiation factors. Eur J Cancer. 2018;56:56–70. doi: 10.1016/j.ejca.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 23.Ferraris R, Colombatti G, Fiorentini MT, et al. Diagnostic value of serum bile acids and routine liver function tests in hepatobiliary diseases. Dig Dis Sci. 1983;28:129–36. doi: 10.1007/BF01315142. [DOI] [PubMed] [Google Scholar]
- 24.Zhang A, Sun H, Han Y, et al. Urinary metabolic biomarker and pathway study of hepatitis B virus infected patients based on UPLC-MS system. PLoS One. 2013;8:e64381. doi: 10.1371/journal.pone.0064381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang TJ, Larson MG, Vasan RS, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–53. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Legido-Quigley C, Mcdermott L, Vilca-Melendez H, et al. Bile UPLC-MS fingerprinting and bile acid fluxes during human liver transplantation. Electrophoresis. 2011;32:2063–70. doi: 10.1002/elps.201000512. [DOI] [PubMed] [Google Scholar]