Abstract
Targeted protein degradation by small-molecule degraders represents an emerging mode of action in drug discovery. Proteolysis targeting chimeras (PROTACs) are small molecules that can recruit an E3 ligase and a protein of interest (POI) into proximity, leading to induced ubiquitination and degradation of the POI by the proteasome system. To date, the design and optimization of PROTACs remain empirical due to the complicated mechanism of induced protein degradation. Nevertheless, it is increasingly appreciated that profiling step-by-step along the ubiquitin-proteasome degradation pathway using biochemical and biophysical assays are essential in understanding the structure–activity relationship and facilitating the rational design of PROTACs. This review aims to summarize these assays and to discuss the potential of expanding the toolbox with other new techniques.
Keywords: : biochemical assay, biophysical assay, degraders, PROTAC, protein degradation, protein ubiquitination, ternary complex
Induced degradation of disease-causing proteins by small molecules is becoming one of the most appealing mechanisms of action in drug discovery [1–5]. Protein degradation can be achieved through different strategies including small molecule binders (e.g., fulvestrant), hydrophobic tagging, proteolysis targeting chimeras (PROTACs), lysosome targeting chimeras, autophagy-targeting chimeras and molecular glues [5,6]. PROTACs are heterobifunctional molecules that can bring a target protein into proximity to an E3 ubiquitin ligase and thus induce the ubiquitination and degradation of the target protein. First proposed and demonstrated in a proof-of-concept study by Crews and Deshaies in 2001 [7], PROTAC technology has emerged as a paradigm-shifting approach in drug discovery and has been rapidly progressing toward therapeutic applications [1,3,8–10]. Over the last few years, we have witnessed many proteins that have been successfully degraded by PROTACs, including nuclear receptors (ERRα, AR, PARP1 and RAR) [6,10], protein kinases (AKT, BCR-ABL, FLT3, BTK, ALK, FAK, IRAK4, BRAF, CDK4/6/8/9, RIPK2, PSD-95, CK2, DAPK1, ERK1/2, PI3K, c-Met and p38α/δ) [6,10,11], epigenetic regulators (BRD4/7/9, HDAC6, Sirt2, TRIM24 and Smad3) [6,10], neurodegenerative disease-related proteins (Tau, mutant huntingtin, PSD-95 and α-synuclein) [6,10], regulatory proteins (CRABP-I/II, TACC3, AHR, FKBP12, MDM2 and X-protein) [6,10], anti-apoptotic proteins (MCL-1, BCL-2 and BCL-XL) [12–15], virus-related protein (NS3/4A) [16] and transcription factors (STAT3 and BCL-6) [6,10,17]. The advancement of two PROTAC molecules, ARV-110 and ARV-471, into clinical trials by Arvinas Inc. (CT, USA) [18,19] is a milestone in the field of PROTAC-mediated target protein degradation, propelling a fast-growing PROTAC field in the coming years.
Despite the rapid progress in this area, PROTAC discovery is still a laborious and largely empirical process. Thus, it is urgent to have a set of techniques and assays that can not only characterize and rank these hard-to-obtain compounds rapidly and efficiently, but also provide valuable feedback to guide medicinal chemists during the lead optimization process. The review article by Hughes and Ciulli has already stressed the importance of characterizing the ternary complex in guiding the design of chemical degraders [20]. Daniels et al. have briefly discussed historical and current technologies for monitoring and deciphering protein degradation pathways inside cells with an emphasis on nano-bioluminescence resonance energy transfer (Nano-BRET®) technology [21]. The goals of this review are to summarize currently available techniques and assays for the screening of PROTAC degraders, to compare and contrast the pros and cons of each method, as well as to propose potential new assays that could be introduced for the characterization of PROTAC degraders.
Pathway for PROTACs to degrade target proteins: from cell penetration to target degradation
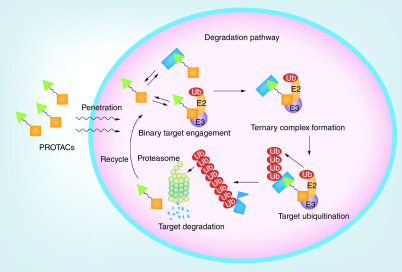
To induce protein degradation in cells, a PROTAC degrader has to go through a complicated process called the degradation pathway (Figure 1) [21]. The degrader must have reasonable chemical stability, solubility and cellular permeability to enter the cells. Once inside the cell, the degrader should be able to engage with either the specific E3 ligase or the target protein to form the corresponding binary complex, followed by the involvement of the third binding partner to form the ternary complex. Several properties of the ternary complex determine whether ubiquitins can be covalently connected to one or more lysine residues of the target protein. The poly-ubiquitinated target protein would then be recognized and recruited by proteasomes, resulting in the degradation of the target protein. When the degradation of a target protein is faster than its expression, the net intracellular target protein level would decrease, leading to a downstream pharmacological effect. Tracking the performance of a degrader along the degradation pathway can provide valuable insights into the mechanism of action of the degrader and offer a clear structure–activity relationship (SAR) to guide further structural modifications.
Figure 1. . Degradation pathway for proteolysis targeting chimeras.
PROTAC: Proteolysis targeting chimera.
Cell penetration
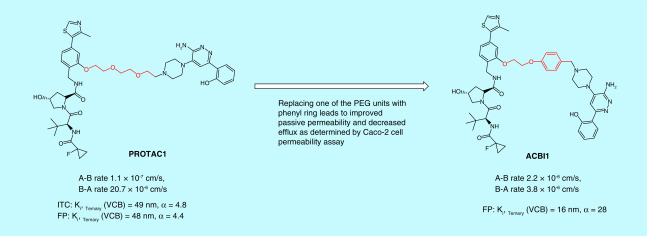
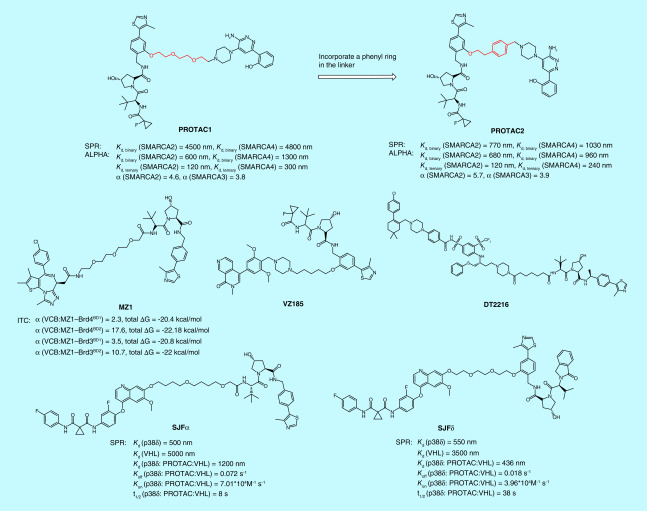
The bivalent nature of PROTACs, assembled by connecting two small-molecule ligands through a linker unit, makes these molecules large in size and against the Lipinski’s ‘rule of five’, [22]. Nevertheless, studies over the past few years have shown that these unusual looking molecules could have surprisingly normal pharmaceutical properties [18]. Chemical stability, solubility and cell membrane permeability are the first three barriers that PROTAC degraders have to overcome. Several structural modification strategies have been applied to improve the physicochemical properties of PROTACs. For example, basic nitrogen-containing groups such as a pyridinyl or a piperazinyl group were empirically introduced in the linker to increase the solubility of PROTACs [23], while an amide bond was avoided in an attempt to improve permeability [24]. Technically, stability, solubility and permeability of PROTAC degraders can be determined with the same methods commonly used for small molecule drugs. For example, a Caco-2 cell permeability assay was used to evaluate the permeability of the PROTACs targeting SMARCA2/4 for degradation. With this assay, Farnaby et al. demonstrated that replacing one of the polyethylene glycol (PEG) units in the linker of PROTAC1 with a phenyl ring dramatically improved its passive permeability and decreased efflux (Figure 2) [25].
Figure 2. . An example of determining proteolysis targeting chimera cell permeability by using Caco-2 cells.
α: Cooperativity; A-B rate: Permeability from apical side to basal side; B-A rate: Permeability from basal side to apical side; FP: Fluorescence polarization; ITC: Isothermal titration calorimetry; PEG: Polyethylene glycol; VCB: von Hippel–Lindau–ElonginC–ElonginB complex.
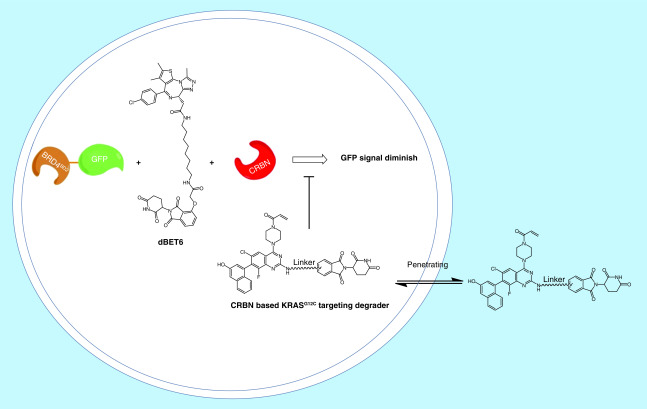
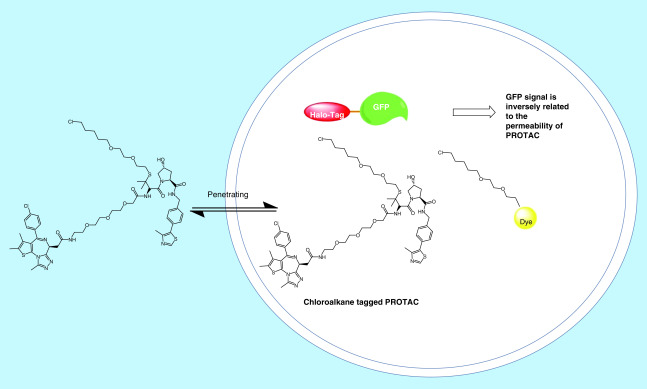
However, current methods to assess cell permeability of small molecules rely on liquid chromatography–mass spectrometry (LC–MS) to determine their apparent permeability. Low permeable PROTACs could have the concentrations that fall below the detection limit of LC–MS. To provide a quantitative ranking of cell permeability for PROTAC degraders, Zeng et al. utilized a competitive CRBN engagement assay for the evaluation of cell permeability [26]. They co-treated BRD4BD2-GFP reporter cells with dBET6, a CRBN-based PROTAC targeting BRD4 and a CRBN-based KRASG12C targeting PROTAC, which allows the displacement of dBET6 from CRBN and thus rescue BRD4BD2-GFP levels (Figure 3). The permeability of KRASG12C targeting PROTAC correlates with their ability to displace dBET6 from CRBN. Although this assay provided a quantitative method to rank the permeability of PROTACs, the permeability data obtained from this assay is a combination of permeability and CRBN engagement. More recently, a chloroalkane penetration assay was developed by Foley et al. (Figure 4) [27]. This assay uses a cell line that stably expresses a HaloTag–GFP fusion protein that is anchored to the outer mitochondrial membrane facing the cytosol. Chloroalkane-tagged molecules of interest can be trapped covalently inside the cell if they are cell permeable. Following a washing step, chloroalkane-tagged dye is added to react with any remaining HaloTag–GFP fusion proteins. Flow cytometry is then employed to quantify the resulting fluorescence intensity, which is inversely proportional to the permeability of the chloroalkane-tagged molecule. With the chloroalkane penetration assay, the authors obtained structure–permeability relationships of the degraders. However, the major limitation of this assay is that it requires adding a chloroalkane tag on the PROTACs, which may not be achievable for compounds that do not have a good derivatization site. Moreover, attaching the same chloroalkane tag to different PROTACs may not equally influence the molecules’ cell permeability. Therefore, the permeability data obtained from the chloroalkane-tagged derivatives may not directly represent the permeability of their parent compounds.
Figure 3. . Competitive cereblon engagement assay for the measuring of cell permeability.
CRBN: Cereblon.
Figure 4. . Chloroalkane penetration assay for the measuring of cell permeability.
PROTAC: Proteolysis targeting chimera.
Binary/ternary binding affinities & ternary complex
Compared with traditional protein inhibitors that mainly rely on binary interactions between small-molecule ligands and target proteins, PROTAC degraders work by involving more complicated three-body binding equilibria. Besides determining binary protein–ligand interaction, characterizing protein–protein interaction (PPI), ternary complex formation, ternary complex stability and cooperativity are also important in elucidating the SAR. The SAR obtained from cell-free biochemical assays can provide valuable feedback to rational lead optimization. Increasing evidence has also shown that ternary complex formation, ternary complex stability and cooperativity of PPI are more predictive of a PROTAC’s degradation activity than its binary interactions [11,28,29]. A ternary complex can be characterized by several different assays. Common biochemical and biophysical assays used to profile ternary complex formation, population, stability, binding affinities, cooperativity or kinetics are discussed below:
Fluorescence polarization assay
The fluorescence polarization (FP) assay can, in principle, be used to quantitatively analyze the binding of any small soluble fluorescent molecule (or any molecules that compete with it) to a protein. It is based on the principle that the degree of FP of a fluorescent molecule is proportional to its rotational relaxation time (ρ) which can be described by the Stokes equation:
Where ρ is the rotation relaxation time, defined as the time taken by the fluorescent molecule to rotate 68.5°; V is molecular volume including hydration; η represents viscosity; R and T are the gas constant and thermodynamic temperatures, respectively.
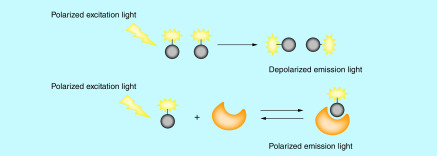
Interaction of a large molecule (protein) with a fluorescent ligand (tracer) can change the effective molecular volume of the fluorescent ligand, and thus alter its rotational relaxation time and eventually its polarization that can be detected through plane-polarized light (Figure 5). As such, the binding affinity between a fluorescent ligand and a protein can be monitored using plane-polarized light in a nondestructive and separation-free manner.
Figure 5. . Illustration of the mechanism of the fluorescence polarization assay.
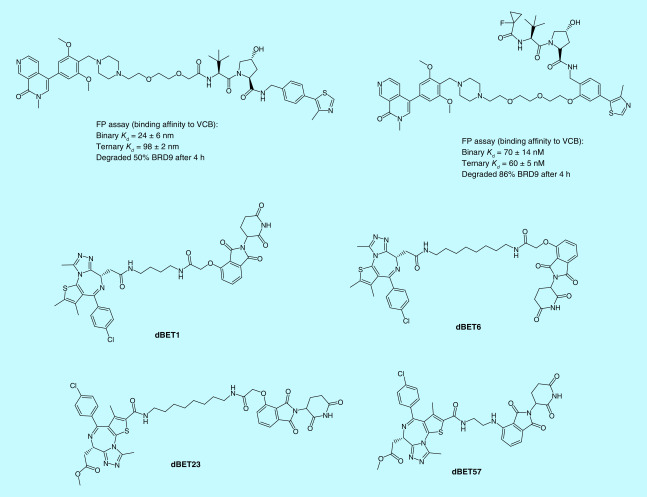
The FP assay is an economical and homogeneous assay that can provide rapid screening for a large number of compounds. As such, the FP assay has been extensively used in high-throughput screening (HTS) programs in drug discovery. In the PROTAC field, the FP assay provides a powerful tool to determine binary binding affinities, ternary binding affinities and cooperativities. The von Hippel-Lindau (VHL) E3 ligase is one of the most commonly hijacked E3 ligases by PROTAC degraders. The development of the competitive FP assay is based on the binding between the FAM-labeled HIF-1α peptide and VCB (VHL–ElonginC–ElonginB) protein complex provides an effective way to screen VHL ligand-based degraders or VHL ligands for their binding affinities to VHL [30–33]. Zoppi et al. developed VHL-based PROTACs targeting BRD7/9 for degradation. Two of the PROTACs were built with the same BRD7/9 ligand and VHL ligand, but with different derivatization sites at the VHL ligand. Using the FP assay, the authors show that the PROTAC with a higher ternary binding affinity to VCB, albeit with lower binary binding affinity, is more potent in degrading BRD9 (Figure 6) [34], indicating that the ternary binding affinity of a PROTAC is more predictive for the ability of the PROTAC to degrade its target protein. The FP assay was also used by Fischer’s group to characterize their BRD4 degraders that recruit CRBN. Negative cooperativities of their PROTAC degraders to DDB1ΔB-CRBN were revealed as the binding affinities for all the PROTAC degraders (dBET1, dBET6, dBET23 and dBET57) (Figure 6) tested showed decreased binding affinity to DDB1ΔB-CRBN in the presence of BRD4BD1 or BRD4BD2 versus in the absence of BRD4BD1 or BRD4BD2 [35]. Their study provided the first evidence that PROTACs can induce protein degradation despite the negative cooperativity. Using the FAM-labeled small molecular VHL ligand as a fluorescent probe, Wang’s group was able to determine the binary binding affinities of their newly designed VHL ligands to the VCB complex through the competitive FP assay [23].
Figure 6. . Examples of proteolysis targeting chimeras characterized by the fluorescence polarization assay.
FP: Fluorescence polarization.
The FP assay is one of the most accessible assays that can be easily set up with various protocols available [36,37]. The fluorescent probe is the key for the FP assay, which can be synthesized by a wide range of commercial services and academic core facilities or can be prepared in-house by attaching a fluorophore to a ligand. Various fluorophores are available, with fluorescein and rhodamine being used most commonly. Fluorescent probes are preferred to be as small as possible to maximize the molecular weight difference between a fluorescent probe and its binding protein. The binding affinity of a fluorescent probe to its target protein is also important, because the higher the binding affinity of a fluorescent probe, the wider the range of inhibitor potencies that can be resolved [38]. However, the affinity of a fluorescent probe will be considered too high if the instrument cannot detect the fluorescent signal when it is lower than twice the Kd [36,38]. The application of the FP assay for characterizing the binary binding affinity of PROTACs to their target protein or E3 ligase is the same as that of a small-molecule ligand. However, when repurposed for ternary binding affinity or cooperativity testing, the FP assay requires saturating the PROTAC with one binding protein first, then titrating into the other protein as a binary complex. As such, a large amount of the proteins are required, making it infeasible for HTS.
Amplified luminescent proximity homogeneous assay
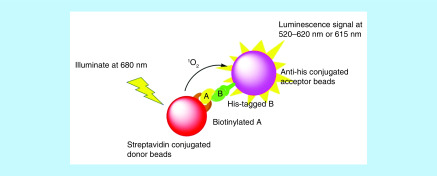
Amplified luminescent proximity homogeneous assay (ALPHA) technology is a bead-based proximity assay that can be used to study interactions between molecules in a microplate format. In this assay, one binding partner (A) is attached to the donor bead, the other binding partner (B) is conjugated to the acceptor bead. The donor beads are coated with photosensitizer that can convert ambient O2 to singlet O2 (1O2) once illuminated at 680 nm. The singlet O2 has a half-life of 4 μs, allowing it to travel about 200 nm in the solution. If acceptor beads are within this distance, thioxene derivatives coated on the acceptor beads will accept the energy from singlet O2 and emit light at 520–620 nm (ALPHAScreen) or at 615 nm (ALPHALISA). The chance that acceptor beads are in proximity to donor beads would be increased significantly when binding partners A and B interact, and the tighter the binding, the higher the luminescence signal produced by the acceptor beads. As such, the interaction between A and B can be quantified simply by detecting the luminescence signal (Figure 7). A competitor that competes with A in binding to B would decrease the luminescence signal in a concentration-dependent manner, providing a competitive assay for the screening of inhibitors. PerkinElmer® (MA, USA) has made ALPHA very practical and easy by commercializing numerous ALPHA reagents. Meanwhile, the high signal-to-background ratio, high dynamic range, high sensitivity and wash-free procedure associated with the ALPHA makes this technology suitable for HTS application, allowing for the discovery of hits from a big library of compounds [39–41]. In addition to the application of screening enzyme or protein inhibitors [42,43], the ALPHA has also been utilized to quantify the concentrations of proteins [44,45], DNAs [46] or small molecules [47], and to capture PPI [48,49], as well as to characterize ternary complexes formed between a target protein, a PROTAC degrader and an E3 ligase [25,29,34,50,51]. Detecting ternary complex formation is one of the most ueful applications of the ALPHA for PROTAC degraders. By titrating a PROTAC degrader to its target protein and E3 ligase, a bell-shaped curve can be produced when plotting ALPHA signals against the concentrations of the PROTAC. The height of the bell-shaped curve reflects the relative population of the ternary complex, allowing scientists to rank PROTAC degraders according to their ability to form ternary complexes. Thus far, the ternary complexes that have been profiled by the ALPHA include BRD4:MZ1:VCB [29], BRD4:dBET1:CRBN-DDB1 [50], BRD4:JQ-1-IMiD PROTAC (or JQ-1-VHL PROTAC):CRBN-DDB1 (or VCB) [51], BRD9:VZ185:VCB [34], BCL-XL:DT-2216:VCB [12] and p38α:SJFα:VCB (Figure 8) [11]. The ternary complex formation data obtained from the ALPHA helped authors to explain some observations during the development of PROTAC degraders. For example, using the ALPHA ternary complex formation assay, Smith et al. demonstrated that SJFα can selectively degrade p38α because their ALPHAs revealed that only SJFα, but not SJFδ, can form ternary complexes with p38α and VCB complex [11]. Besides ternary complex formation, the ALPHA can also be repurposed to determine ternary complex affinity and cooperativity. Farnaby et al. developed an ALPHA as an orthogonal method of surface plasmon resonance (SPR) assay, to determine the binary binding affinities, ternary binding affinities, as well as cooperativities of their PROTAC molecules (PROTAC1 and PROTAC2) (Figure 8) to SMARCA2 and SMARCA4, respectively [25]. Although Kd values obtained from the ALPHA were much lower than those resulted from the SPR assay, the two assays gave the same conclusion that PROTAC2 had stronger binding affinities to SMARCA2/4 than PROTAC1.
Figure 7. . Illustration of the amplified luminescent proximity homogeneous assay.
Figure 8. . Examples of proteolysis targeting chimeras that have been characterized with amplified luminescent proximity homogeneous assay, time resolved-fluorescence energy transfer, isothermal titration calorimetry or surface plasma resonance assays.
α: Cooperativity; ΔG: Gibbs free energy; ALPHA: Amplified luminescent proximity homogeneous assay; ITC: Isothermal titration calorimetry; SPR: Surface plasma resonance; TR-FRET: Time-resolved fluorescence energy transfer; VHL: von Hippel–Lindau.
With properly tagged proteins and a variety of commercially available beads to choose from, the ALPHA can be easily set up by following protocols from Promega (WI, USA). Although being wash-free is a big advantage of the ALPHA over conventional ELISA, the ALPHA involves multiple incubation steps, and the order of adding reagents may need to be optimized as one reagent may interfere with the binding of another. While the FP assay uses a fluorescence signal, the ALPHA uses a chemiluminescence signal, thus it has a higher dynamic range and is more sensitive due to the low background interference. However, the ALPHA relies on singlet oxygen to transfer energy from donor beads to acceptor beads. Singlet oxygen is a reactive oxgen species that can be quenched by certain chemicals, leading to false results. In addition, conjugating binding partners introduce steric hindrance, which could potentially interfere with the binding. At last, the donor beads of the ALPHA are coated with a light-sensitive photosensitizer. As such, special care needs to be taken to handle them.
Time-resolved fluorescence energy transfer assay
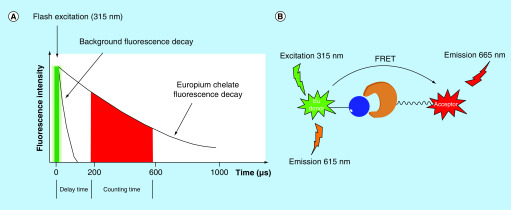
Similar to the ALPHA, the time-resolved fluorescence energy transfer (TR-FRET) assay is also a proximity-based technology. It combines time-resolved fluorimetry with Förster resonance energy transfer (FRET), resulting in an assay that allows for very sensitive detection of binding/dissociation events in a homogeneous format. While the ALPHA uses a singlet oxygen transferred chemiluminescence signal to measure binding between two partners, the TR-FRET assay utilizes long-lived fluorophores combined with time-gated fluorescence intensity measurements to quantitate molecular association or dissociation events. In this assay, one binding partner is labeled with a donor fluorophore (usually a europium or terbium chelate/cryptate); the other binding partner is conjugated to a corresponding acceptor fluorophore. Europium and terbium are both lanthanides that have a long fluorescence life-time, large Stokes shift and a narrow emission peak. With these properties, europium and terbium can emit the fluorescent signals after the delay time when background fluorescence has decayed (Figure 9A). In addition, the acceptor fluorophores used in the TR-FRET assay are chosen to have excitation wavelengths that are overlapped with the emission wavelengths of their paired donor fluorophores, allowing the energy transfer from donor fluorophores to acceptor fluorophores when they are in proximity (Figure 9B). By detecting the emitted fluorescence signal of both the acceptor fluorophore and the donor fluorophore, the TR-FRET signal can be determined through a ratiometric or delta method, which is used to quantify the binding events of two binding partners that are conjugated to donor and acceptor fluorophores, respectively.
Figure 9. . Principle of the time-resolved fluorescence energy transfer assay.
(A) Time-gated measurement of fluorescence intensity; (B) proximity-based energy transfer.
FRET: Förster resonance energy transfer.
By designing the TR-FRET assay in different formats, this assay can be applied to perform HTS on small-molecule libraries [52–54], to quantify protein concentrations [55–59], to screen PPI targeting small molecules [60], as well as to characterize the ternary complex for PROTACs [35,61]. Similar to the ALPHA, the ternary complex formation assay performed with TR-FRET also results in a bell-shaped curve, whose peak height reflects the relative population of the ternary complex. Using the TR-FRET assay, Radoslaw et al. showed that dBET23 (Figure 6), a CRBN-based PROTAC targeting BRD4 for degradation, mediated ternary complex population was affected by the interaction between BRD4BD1 and CRBN, because mutations on the key residues of BRD4BD1 that mediated the interaction between BRD4BD1 and CRBN resulted in changes in the peak height [35]. By labeling BTK with donor fluorophore terbium, and CRBN with acceptor fluorophore XLA665, Zorba et al. developed a TR-FRET-based ternary complex formation assay for their PROTACs and found that changes in the linker length of PROTACs led to changes in the peak height of the bell-shaped curve, and thus established a relationship between the linker length of the PROTACs and their ternary complex formation capability [61].
Both the ALPHA and the TR-FRET assays are proximity-based assays, making them alternatives to each other. Compared with the ALPHA that captures binding partner to beads, the TR-FRET assay labels binding partner with fluorescent molecules, which are much smaller than beads, to provide more entropic freedom. Choosing an appropriate fluorescent pair is the key to a TR-FRET assay. There are vendors (e.g., Cisbio, Molecular Devices, Invitrogen and PerkinElmer) that provide commercialized fluorescent pairs, making the assay readily accessible. In addition, instead of monitoring signals from acceptor beads alone in the ALPHA, the TR-FRET assay detects the emitted fluorescent signal from both donor and acceptor fluorophores, allowing a ratiometric correction for liquid dispensing errors, reducing assay variability and improving data quality. The TR-FRET assay is also advantageous over the FP assay in offering higher dynamic range and sensitivity, although both assays rely on fluorescent signals. However, like all other assays, the TR-FRET assay does not come without limitations. The major one is the interference from quenching and fluorescent compounds. The nonspecific effect of an interfering compound on only the donor or only the acceptor signal will affect the ratio calculation and could lead to an incorrect conclusion [62].
Isothermal titration calorimetry
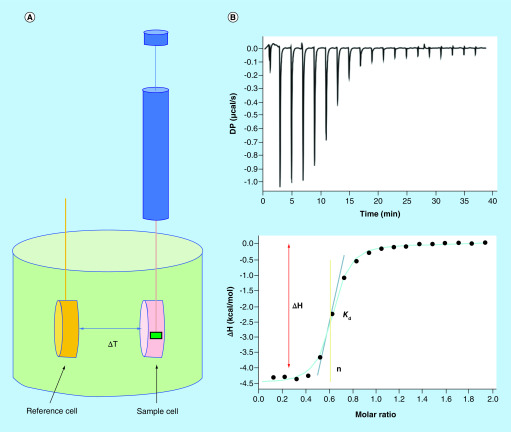
Isothermal titration calorimetry (ITC) has been the gold standard for direct binding measurement. It measures the generation or consumption of heat following the titration of a ligand solution into a protein solution (or the reverse) to determine parameters including stoichiometry, Kd. changes in enthalpy (ΔH), changes in entropy (ΔS) and the heat capacity change (ΔCP). In each ITC experiment, stepwise injections of one reagent into a calorimetric cell containing the second reagent are performed and the exothermic or endothermic process of each injection is monitored. Analysis of the reaction mixture heat as a function of the analyte concentration provides a complete thermodynamic characterization of a binding event (Figure 10) [63].
Figure 10. . Basics of an isothermal titration calorimetry experiment.
(A) The titrant is loaded into the syringe and injected into the sample cell containing analytes and the temperature difference between the reference cell and the sample cells are recorded. (B) Representative data obtained from an ITC experiment.
ΔH: Enthalpy change; ΔT: Temperature difference; ITC: Isothermal titration calorimetry; n: Stoichiometry.
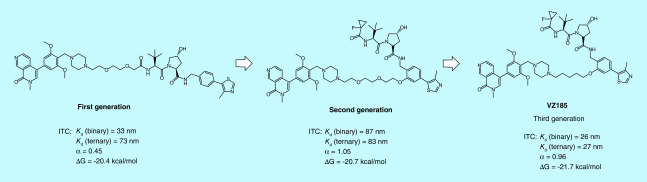
ITC is superior to any other biophysical techniques in that it can determine all binding parameters including stoichiometry (n), Kd. ΔH and ΔS in a single experiment. When in combination with structural information, ITC data can provide deeper insights into the mechanisms of binding, the binding driving force and the structure–function relationships [64]. ITC is also a true label-free biophysical method that allows the binding affinities to be determined without the need to tag or immobilize the binding partner, which is advantageous as tagging or immobilizing an analyte is sometimes technically difficult and may even interfere with the binding. In addition, the size of a molecule is not an issue for the ITC assay, whereas the sensitivity of other biophysical techniques such as SPR and bio-layer interferometry (BLI) is largely dependent on the molecular weight of their analytes. ITC has been increasingly [65] used in determining protein–protein [66,67], protein–nucleic acid [68–70] and protein–small molecule interactions [71,72]. In recent years, ITC has also been applied in profiling the thermodynamic parameters and cooperativities of PROTAC molecules, providing invaluable insights into the interplay between a PROTAC molecule and its target protein and E3 ligase [25,29,34,73,74]. Using the ITC assay, Zoppi et al. solved the thermodynamic parameters of a VHL-based PROTAC targeting BRD9 for degradation and found that it had negative cooperativities to both VCB complex and BRD9, consistent with the weak and partial degradation of BRD9 [34]. Improving the ability of ternary complex formation and cooperativity by changing the derivatization site, linker length and linker composition led to the second and the third generations of PROTACs that are more potent in degrading BRD9 than the initially developed BRD9 PROTACs (Figure 11) [34]. ITC-guided PROTAC optimization was also presented by Farnaby et al. Cooperative recognition of their first PROTAC by VCB complex and SMARCA2BD determined by ITC titration prompted them to further investigate this compound and eventually led to a more cooperative, more potent, and more cell membrane permeable PROTAC degrader ACBI1 (Figure 2) [25]. In addition, the Gibbs free energy (ΔG) value obtained from the ITC assay can be used to evaluate the stability of the ternary complex. MZ1 is a VHL-based PROTAC targeting bromodomain- and extraterminal (BET) family proteins for degradation. The ITC assay revealed that MZ1 is isoform-specific (prefers BD2 domain) because VCB:MZ1:BRD4BD2 (or BRD3BD2) has not only higher cooperativities but also lower ΔG value than VCB:MZ1:BRD4BD1 (or BRD3BD1) (Figure 8). The lower ΔG value indicates that VCB:MZ1:BRD4BD2 (or BRD3BD2) is a more stable ternary complex [29].
Figure 11. . Example of isothermal titration calorimetry-guided proteolysis targeting chimera modification.
α: Cooperativity; ΔG: Gibbs free energy; ITC: Isothermal titration calorimetry; Kd. Dissociation constant.
The sensitivity and accuracy of ITC instruments are improving with time. Two of the most commonly used, highly sensitive isothermal titration calorimeters are ITC-200 from Malvern Instrument (previously MicroCal, Malvern, UK) and Nano-ITC from TA Instrument (DE, USA). However, even the most advanced ITC instrument requires at least 200 μl of sample limited by the sensitivity of heat measurement, which is a major disadvantage of the ITC assay when precious samples are being used. Application of ITC for determining binary binding affinities of PROTACs is pretty straightforward, and protocols for the determination of binary binding affinities are available [75,76]. However, like the FP and ALPHA assays, ITC also requires saturating the PROTAC with one of the binding proteins when performing ternary binding experiments, which can be a challenge when a large amount of a high-purity protein is hard to obtain. ITC is also demanding in aqueous solubility of the analytes as the concentration of the titrant has to be >10 × Kd. which can be problematic for PROTACs with low solubility and proteins that aggregate at high concentrations. Furthermore, with each titration experiment taking about 40 min, ITC is a low-throughput assay. Thus, ITC is usually a secondary assay for compound screening.
SPR/BLI assay
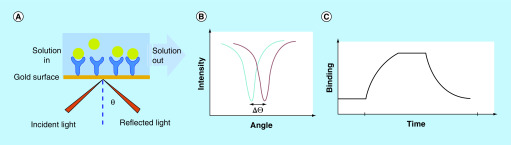
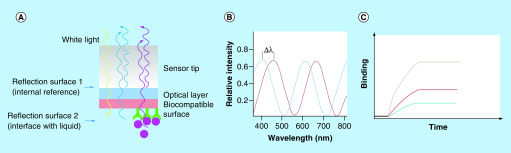
Both SPR and BLI are biophysical techniques that detect binding events through a spectroscopic method; the resulting sensorgrams show the kinetics of binding in real-time. The SPR assay is based on the changes in the refractive index of the medium directly in contact with the sensor chip surface, whereas BLI detects the changes in the interference pattern on the biosensor tip surface. In the SPR assay, an incident light shines on the gold film (where targets are immobilized) of the sensor chip and the reflected light intensity is monitored. When the frequency of the incident photon equals to the vibration frequency of electrons on the gold surface, the electrons of the gold surface absorb the photon energy and resonance occurs with a plasmonic wave generated on the surface of the gold, a phenomenon known as SPR. This phenomenon results in a dip in the reflection intensity. The shape and location of the dip carry the information of the sensor chip surface. As the ligand solution flows across the sensor chip, the binding of ligands to the gold film changes its reflective index, which changes the angle of the incident light required for SPR. The reflective angles also change accordingly, resulting in shifts in the SPR dip. These angle changes or SPR dip shifts are defined as SPR response signals. A sensorgram can then be obtained by plotting SPR response signals against time during the interaction between an analyte and a target (Figure 12). During a BLI experiment, however, white light shines through the optical fiber tip, and can be reflected back from two biosensor tip layers, a layer of immobilized targets on the tip, and an internal reference layer. Binding of ligands from solution to the immobilized targets changes the thickness of the tip surface, resulting in an interference pattern change of the tip, which is recorded as wavelength shift (Δλ). Δλ is a direct measure of the thickness of the tip surface. Plotting Δλ over time during the interaction between targets and a ligand affords a sensorgram (Figure 13).
Figure 12. . Illustration of surface plasma resonance experiment.
⊖: Reflective angle; Δ⊖: Reflective angle change.
Figure 13. . Illustration of bio-layer interferometry experiment.
Δλ: Wavelength shift.
Both SPR and BLI are label-free biophysical methods for determining interactions between ligands and targets, although they both require immobilization of one binding partner on the sensor chips/tips. The immobilization strategies for SPR and BLI are the same, but the availability of the corresponding modified sensor chips/tips is instrument dependent. The two broad approaches for performing the immobilization are chemical coupling and capture. Chemical coupling strategy results in covalent bonds between the ligands and the sensor chips/tips surface, while the capture strategy takes advantage of the strong affinity between the chemistry of the sensor chips/tips and the tag of the ligand. For example, a nickel-NTA chip can be used for the capture of His-tagged proteins, and a streptavidin or NeutrAvidin chip can be used for the capture of biotinylated molecules.
The biggest difference between SPR and BLI, however, is that SPR uses microfluidic systems for continuous registration of the signal, while BLI moves the ‘dip-and-read’ sensor tips among wells of an open microwell plate. Without the use of a microfluidic system, BLI offers several advantages over SPR: it is possible to test biological liquids (cell lysate, plasma and serum) with BLI; BLI can test up to 16 samples in one run, and thus provides a higher throughput; sample recovery is also possible in BLI assay. However, because of a different principle of theory employed, BLI has a lower sensitivity than SPR, especially for detecting interaction between small molecules and macromolecule targets. Nevertheless, BLI and SPR are superior to the other technologies discussed previously in that they can determine binding kinetics and binding mechanisms in real-time without the use of fluorescent probes. In the past two decades we have witnessed an increasing application of SPR technology in every aspect of pharmaceutical analyses, from fragment-based drug screening [77,78] and HTS [79] to pharmacokinetic drug profiling [80], early absorption, distribution, metabolism, excretion and toxicity studies [81] and quality control [82]. In recent years, biosensor technology also finds its application in PROTACs, not only for binary binding determination [17,61,83] but also for ternary complex characterization [11,84]. In this aspect, SPR is more advantageous than ITC and other assays, as it not only provides ternary binding affinity (Kd) but also kinetics (Kon. Koff and t1/2) of a ternary complex. Roy et al. developed an SPR-based assay to measure dissociation kinetics of PROTAC ternary complexes and found that ternary complex dissociative half-life correlates well with intracellular target degradation, suggesting that the kinetic parameter drives effective target degradation [84]. In a study published by Smith et al., PROTAC SJFδ and PROTAC SJFα are two foretinib-based PROTACs recruiting VHL using two different linkage vectors. These two PROTACs were found to degrade p38 in an isoform-selective manner. An SPR kinetics study revealed that SJFδ had a higher ternary binding affinity (p38δ:SJFδ:VHL) and a longer ternary complex dissociation than those of SJFα (p38δ:SJFα:VHL) (Figure 8), indicating that p38δ:SJFδ:VHL is the more favorable and more stable, and higher populated ternary complex than p38δ:SJFα:VHL, which explains the selective degradation of p38δ by SJFδ [11].
The requirement for immobilization of the binding partner is a major limitation for these two biosensor technologies, as immobilizing a molecule on to a surface may interfere with its binding properties by obstructing the binding site or by restricting entropic freedom. In addition, sensitivity is always a concern when small molecules are tested on a BLI or SPR instrument, so immobilizing PROTACs, instead of proteins, to sensor tips/chips can be a method of choice to improve sensitivity. However, chemical modification of small molecules is required to facilitate immobilization, which in turn may change the binding affinities of the parent molecules. Mass transport limitation is another issue because the maximal Kon or the minimum Koff that can be determined by SPR/BLI is limited by the diffusion rate. Moreover, although these biosensor technologies can provide informative data, the expense of sensor chips/tips and instruments is much higher than other technologies.
Nano-BRET assay
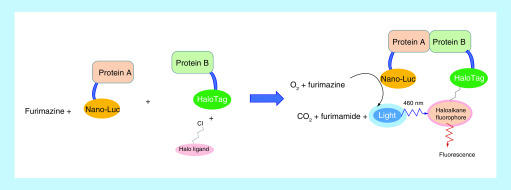
BRET is a biophysical technology that relies on energy transfer from a donor luciferase to an acceptor fluorophore to monitor proximity and interaction within a live cell. Since its first description more than 20 years ago, BRET technology has been evolving over time with the development of several new substrates and energy donor/acceptor couples (e.g., Renilla luciferase donor + coelenterazine [substrate] + yellow fluorescent protein acceptor and Renilla luciferase donor + bisdeoxycoelenterazine [substrate] + green fluorescent protein acceptor) [85,86]. Nano-BRET is an optimized BRET technology using Nanoluc luciferase coupled with its substrate furimazine as the donor system [87]. The optimum properties of the Nanoluc and furimazine combination, such as high physical stability, high luminescence signal and small size, make Nano-BRET advantageous over previous BRET technologies and the combination of Nanoluc with various fluorescent protein acceptors greatly expands the application of BRET technology [85,88–90]. The recent introduction of the Halo-Tag system as an alternative to fluorescent proteins offers an opportunity for multiplexing as fluorophores can be chosen as needed as long as chloroalkane–fluorophore conjugates are available [91]. In this new Nano-BRET system, one binding partner (e.g., protein A) is fused to Nanoluc and the other binding partner (e.g., protein B) is fused to Halo-Tag. The substrate of Nanoluc (furimazine) and ligand of Halo-Tag (chloroalkane–fluorophore conjugate) are added separately to the cells. Once in the cell, furimazine can be converted to furimamide by luciferase in the presence of oxygen and emit a luminescence signal at around 460 nm, whereas the chloroalkane–fluorophore conjugate attaches to Halo-Tag covalently and can be excited at 460 nm and emit light at around 618 nm. Interaction between protein A and B brings luciferase and Halo-Tag in proximity, allowing the luminescence signal to be transferred to a fluorophore (Figure 14). With proper modification and adaptation, Nano-BRET technology can be applied to almost every step along the degradation pathway of PROTAC within live cells [92]: from PROTAC target engagement, ternary complex formation [12,91,93] and target ubiquitination to target degradation and target protein level detection [34,93]. Using the HiBiT-BET protein complemented with Large BiT (LgBiT) as an energy donor and Halo-Tag fused to an E3 ligase component or ubiquitin as the respective energy acceptor, Krinstin et al. developed a set of Nano-BRET assays to determine ternary complex formation, target ubiquitination and protein degradation in a live-cell platform [93]. Nano-BRET can also be used to explain the mechanism of action of PROTAC molecules. Recently, Khan et al. demonstrated that DT2216, a VHL-based PROTAC, can only selectively degrade BCL-XL although its warhead binds to both BCL-2 and BCL-XL. The ALPHA disclosed that both BCL-XL:DT2216:VCB and BCL-2:DT2216:VCB complexes were formed and appeared to be stable in vitro. However, a further ternary complex formation assay using Nano-BRET revealed that DT2216 can only induce energy transfer between HiBiT–BCL-XL and HaloTag–VHL but not between HiBiT–BCL-2 and HaloTag–VHL, indicating that the lack of BCL-2 degradation is due at least in part to the absence of the formation of intracellular BCL-2:DT2216:VCB ternary complex [12]. This study implies that a ternary complex assay carried out within live cells is more predictive of target degradation induced by PROTACs.
Figure 14. . Illustration of nano-bioluminescence resonance energy transfer technology combined with a Halo-Tag system.
Nano-BRET is a relatively new technology developed and commercialized by Promega (WI, USA). It offers several advantages over other methods in profiling PROTAC molecules: Nano-BRET assay is carried out within live cells, and thus the results are based on physiological conditions; Nano-BRET uses endogenous protein in live cells, thus avoiding protein expression and purification process, which is favorable for target proteins that are hard to obtain or are in a large protein complex; the Nano-BRET assay can kinetically monitor target engagement and ternary complex formation, as well as target ubiquitination and degradation in real-time, providing more insights into the mechanism of action of PROTAC molecules; Nano-BRET is versatile as it can be combined with other technologies such as CRISPR/Cas9 or can be adapted to fulfill one’s needs.
Others
Assays other than the aforementioned that have also been used to characterize ternary complexes include size exclusive chromatography (SEC) [94,95], crystallography [25,35,74], co-immunoprecipitation (Co-IP) [83,96], mass spectrometry (MS) [61,97] and the NanoBiT® assay [95].
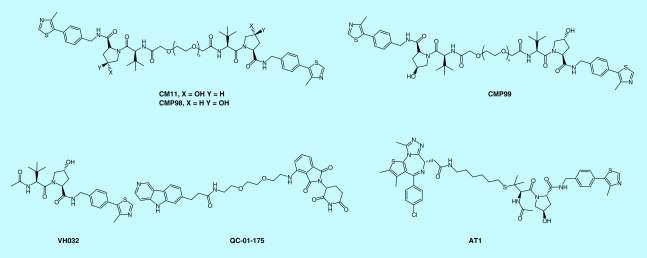
The SEC ternary complex formation assay is based on the size difference between the binary complex and the ternary complex. By detecting the elution volume and elution time of proteins following PROTAC treatment, one can compare the relative ternary complex formation among different PROTAC molecules. Using the SEC assay, Maniaci et al. found that their homo-PROTAC CM11 (trans-trans) can form a much higher population of a ternary complex with VCB complex than its cis-trans control compound CMP99, while its cis-cis control compound CM98 or VHL ligand VHL032 cannot (Figure 15) [94].
Figure 15. . Examples of compounds mentioned in size exclusive chromatography, co-immunoprecipitation and mass spectrometry.
Crystallography is a challenging yet very useful biophysical technique. The ternary complex crystal structures of PROTAC degraders can provide valuable insights into how they bind to their target protein and E3 ligase, which can provide better guidance on the rational design and optimization of new degraders. The first ternary complex crystal structure for PROTACs is VHL:MZ1:BRD4BD2, solved by Ciulli’s group [29], and has been used to guide the design of macrocyclic PROTACs [74]. Ternary complex crystal structure of VCB:PROTAC1:SMARCA2BD was also reported by Ciulli’s group. Their work first demonstrated the structure-guided approach for PROTAC modification [25]. Multiple crystal structures of ternary complexes involving CRBN and BRD4 published by Fisher’s group have demonstrated that plasticity in binding confers selectivity in ligand-induced protein degradation [35].
Co-IP can be used to detect PPI in live cells by using target protein-specific antibodies to indirectly capture proteins that are bound to a specific target protein. The enhanced capture in the presence of a PROTAC is an indication of ternary complex formation. The advantage of co-IP over other ternary complex formation assays is that it is most related to physiological conditions. QC-01-175 (Figure 15) is a CRBN-based PROTAC targeting Tau for degradation, Silva et al. demonstrated that the presence of QC-01-175 could help both DDB1 to pull down Tau protein or Tau protein to pull down DDB1, indicating CRBN–QC-01-175–Tau ternary complex formed inside the cell [83]. Although co-IP detects ternary complex formation for a PROTAC in physiological conditions, it relies on a high-quality antibody and the wash step of this assay may diminish weak PPI, leading to a false negative result.
MS is another label-free technique that can provide insights into PROTAC degrader-mediated PPI [61,97]. According to the study by Beveridge et al., native MS can preferentially reveal the E3–PROTAC–POI ternary complex in competition experiments with multiple substrate proteins present, thereby suggesting that it is not only an ideal HTS strategy for the development of new PROTACs [97] but also a valuable tool to dissect the mechanism of actions, selectivity and specificity of PROTACs. A native electrospray ionization mass spectrometry (ESI–MS) experiment of VCB and AT1 (Figure 15) or MZ1 (Figure 8) in the presence of equal molar amounts of BRD4BD2, BRD3BD2 and BRD4BD1 revealed that both PROTAC AT1 and MZ1 prefer to form a ternary complex with BRD4BD2 and VCB than with BRD3BD2 or BRD4BD1 and VCB, consistent with the data obtained from ITC and SPR experiments [97].
NanoBiT is a protein-fragment complementation assay that can detect PPI and ternary complex formation in live cells. In this assay, one binding partner is fused to a LgBiT (18 kDa) subunit, while the other binding partner is fused to a small BiT (SmBiT; 11 amino acid peptide). SmBiT is a complementary peptide of LgBiT that is designed to have a very low binding affinity to LgBiT. However, once SmBiT and LgBiT interact, they form an active enzyme that can generate a bright luminescent signal in the presence of a substrate. Interaction between LgBiT-fused and SmBiT-fused binding partners brings these two complementary proteins together, resulting in a luminescent signal as a measurement of PPI between binding partner proteins. With this assay, PROTAC induced ternary complex (BCR-ABL:PROTAC:VHL ternary complex) was detected in HEK293 cells [95]. Nevertheless, NanoBiT cannot detect PPI that is weaker than the interaction between LgBiT and SmBiT.
The thermal shift assay (TSA), also known as differential scanning calorimetry (DSF), studies thermal stabilization of proteins upon ligand binding. Given its capability to screen ligands that occupy the nonactive sites of proteins, DSF assay could be the potential assay to offer nonfunctional binding ligands for PROTAC design. By applying the TSA in a cellular formate, the cellular TSA (CETSA) allows studies of target engagement of PROTACs in a cellular context. The CETSA involves treatment of cells with a PROTAC of interest, heating to denature and precipitate proteins, cell lysis and the separation of cell debris and aggregates from the soluble protein fraction. Whereas unbound proteins denature and precipitate as temperatures elevate, ligand-bound proteins remain in solution which can be quantified by either western blot or by ALPHA [98]. The increase of the denaturing temperature of the target proteins/E3 ligases in the presence of PROTACs is an indication of targets/E3 ligases engagement. Although its application for ternary complex formation is yet to be developed, Smith et al. has used CETSA to compare the binary target engagement of their PROTACs SJFα and SJFδ [11]. According to their data, target protein p38δ denatures at higher temperatures in the SJFδ-treated cells, suggesting that SJFδ has a stronger target engagement than SJFα.
Microscale thermophoresis is a relatively new technology that detects the interaction between a ligand and target protein based on motion changes of fluorescently labeled proteins along a microscopic temperature gradient upon binding. Although it has not yet been applied for PROTAC degraders, microscale thermophoresis has been used to illustrate the mechanism of Diels–Alderase ribozyme in catalyzing Diels–Alder reaction between two free small substrates (N-pentylmaleimide and anthracene-9-carboxylic acid), demonstrating its capability in carrying out a ternary complex binding assay [99]. This technology also offers advantages that other assays cannot match, such as high sensitivity, low sample volumes and is independent of molecular size.
While x-ray crystallography is a powerful biophysical technique to solve the crystal structures of ternary complexes formed between PROTACs and their E3 ligases and target proteins, requiring the crystallization of samples has limited its major application, especially when the dynamics of the target hamper crystallization. Cryogenic electron microscopy (cryo-EM) is emerging as a complementary technique to x-ray crystallography to provide 3D structures of macromolecular complexes. With numerous cryo-EM structures being solved in the past few years accompanied by the technical improvements in image acquisition and image processing, cryo-EM is moving to the forefront of drug discovery [100]. The application of cryo-EM to solve the ternary complex structure of DCAF15–DDB1–DDA1–indisulam–RBM39 [101] indicates its potential use to solve the ternary complex structures induced by PROTACs as well. The several advantages over crystallography technique offered by cryo-EM, such as access to larger and/or more complex biological systems, analysis of samples in solution and potentially providing insights into the states of the macromolecule that are closer to those that are more biologically relevant [102] will make cryo-EM a powerful and essential technique for PROTAC development in the future.
Protein ubiquitination assay
Poly-ubiquitination is often the defining step in triggering target protein degradation. Carried out by the ubiquitin proteasome system, protein ubiquitination involves concerted actions of E1, E2 and E3 enzymes to attach ubiquitins to lysine residues of a target protein and subsequent ubiquitin chain elongation. The ubiquitinated proteins can be recognized, recruited and degraded by the 26S proteasome, a very large multicatalytic protease complex that breaks down ubiquitinated proteins to small peptides [103].
Ubiquitination assays can be done both in a cell-free system and in live cells using either electrophoresis-based or nonelectrophoresis-based methods. Using an in vitro ubiquitination assay coupled with western blot, Zeng et al. revealed that the inability of the KRASG12C degrader to degrade endogenous KRASG12C is likely attributed to the lack of induction of KRASG12C poly-ubiquitination [26]. Substrate protein ubiquitination can also be detected in vitro with nonelectrophoresis-based methods such as FP [104], ALPHA [105] and TR-FRET assays [106]. Nevertheless, all these in vitro ubiquitination assays require additives such as ATP and E1, E2 and E3 enzymes, which is quite complicated in setting up the assay conditions and are not physiologically related. Ubiquitination assays carried out in live cells, however, allows for ubiquitination detection under native conditions by taking advantage of their own ubiquitination machinery. Immunoprecipitation of substrate proteins followed by ubiquitin immunoblotting is the simplest and one of the most commonly used methods to detect protein ubiquitination and it is likely to capture the ubiquitinated target protein specifically given that a good antibody against the substrate protein works in IP [107]. Alternatively, the tagged versions of a target protein and ubiquitin can be expressed in cells to facilitate the detection [11]. For example, to demonstrate the selective ubiquitination of p38α by PROTAC SJFα, HeLa cell was transfected with HA-Ubiquitin and FLAG-p38α. Followed with FLAG immunoprecipitation and immunoblotting HA (Ub), Smith et al. managed to detect ubiquitinations of FLAG-p38α in PROTAC SJFα-treated cells, but the ubiquitination of p38α was barely detected in SJFδ-treated cells [11]. More recently, NanoBRET technology was applied to monitor intracellular BET protein ubiquitination induced by PROTAC degraders [93]. In this NanoBRET-based assay, HiBiT-BET protein, its complementary protein LgBiT, and the luminescence substrate furimazine were used as an energy donor system, while the Halo-Tag fused ubiquitin and haloalkane fluorophore were employed as the respective energy acceptor system. A PROTAC degrader that can induce the ubiquitination of BET protein can bring HiBiT-BET protein in proximity to HaloTag-fused ubiquitin. With the involvement of LgBiT, furimazine and haloalkane fluorophore, an energy transfer from donor to acceptor can be expected, resulting in a fluorescence signal that reflects the intensity of ubiquitination [93].
It has been well-established that different ubiquitination sites and ubiquitination types can determine the fates of an ubiquitinated target protein [108]. MS is a powerful technology that can not only be used in detecting total protein ubiquitination but also allows identification of ubiquitination sites and ubiquitination types [109–113]. For example, by coupling an in vitro ubiquitination assay with MS analysis, Gadd et al. identified K346 on BRD4BD2 and several sites on the other BET bromodomains as the ubiquitination sites induced by MZ1 [29]. MS was also used by Khan et al. in identifying K87 as the only site on BCL-XL that can be ubiquitinated through DT2216 treatment [12]. Using MS-based proteomic analysis, Ottis et al. found that different E3 ligases can induce different ubiquitin linkage types following PROTAC treatment. Their study revealed that K48 and K11 are the major ubiquitin linkage types induced by VHL and CRBN, whereas βTrCP and parkin predominantly induce K48 and K6 ubiquitin linkages, respectively [114].
A label-free technology named nanopore makes real-time detection of protein ubiquitination possible [115], yet it has not been applied to PROTAC degraders. In contrast to current ubiquitination assays that only detect end-point ubiquitination, the nanopore is capable of monitoring the E1–E2–E3 ubiquitination cascade kinetically. The TUBE-ALPHALISA and the TUBE-DELFIA (dissociation-enhanced lanthanide fluoroimmunoassay) are also two of the assays that can potentially be applied for quantifying ubiquitinated proteins in cell lysates following PROTAC treatment. The ease of operation and plate-based format of these two assays make high-throughput ubiquitination screening possible [116].
Target degradation
Protein degradation, also called proteolysis, is a process that results in the hydrolysis of one or more peptide bonds in a protein. The ubiquitin-proteasome pathway (UPP) and autophagy are two main pathways and machineries that mediate degradation of intracellular proteins [117], while extracellular proteins and some cell surface proteins are taken up by endocytosis and are degraded within lysosomes [103]. Protein degradation induced by PROTAC degraders is anticipated through UPP. Given the unpredictable nature of PROTAC degraders in inducing target protein degradation [84], it is of great importance to introduce reliable degradation assays to ultimately evaluate if the designed PROTAC is a target protein degrader. Methods that have been used to detect intracellular protein levels post PROTAC treatment include western blot [118,119], capillary-based immunoassay [61,120], fluorescence or luminescence-based reporter assays [26,93] and MS based-proteomics [12,17,25,28,61].
Immunoassays
Western blot is the most frequently used method to measure the relative target protein levels in cells. However, the western blot assay relies on specific and high-quality antibodies for protein detection and is not suitable to accurately quantify the protein levels. In addition, the low sensitivity and multistep procedures of western blot could bring artifacts to the results. A capillary electrophoresis immunoassay is simpler than western blot with less sample consumption, simpler procedures and a shorter analysis time [121]. It has been applied to determine the levels of BTK [61] and pirin protein [120] after cells treated with respective PROTACs. However, similar to western blot, this method also depends on the specific interaction between an antibody and an antigen for protein detection, presenting as a semiquantitative technique for protein detection. Other antibody-based methods that have been developed to quantitate protein levels include ELISA and ALPHA. Both ELISA and ALPHA provide highly sensitive protein level detection with the ALPHA being wash-free, having a larger dynamic range and allowing protein detection homogeneously [122]. However, neither the ELISA nor the ALPHA have been applied to PROTAC degraders for protein degradation.
Reporter assays
Fluorescence- or luminescence-based reporter assays represent a rapid and sensitive method to measure protein degradation in situ. For example, by generating GFP-KRASG12C reporter cells, Zeng et al. were able to detect the degradation of GFP-KRASG12C induced by their KRAS targeting PROTAC degraders through the strength of the fluorescence signal [26]. Similarly, the development of a luciferase-based HiBiT tagging system offered another reporter assay to monitor protein degradation. In this assay, a target protein is fused with HiBiT using CRISPR/Cas9 editing in cell lines stably expressing LgBiT which complements with HiBiT to form the luminescent NanoBiT luciferase. Post PROTAC treatment, degradation of a target protein is reflected in the loss of luminescence signal [93]. With the capability of monitoring protein degradation kinetically, this system has been used to characterize the protein degradation profiles of BRD2, BRD3 and BRD4 induced by PROTAC MZ1 (Figure 8) [93], as well as the protein degradation profiles of BRD7 and BRD9 induced by PROTAC VZ185 (Figure 8) [34]. One big challenge in these fluorescence- and luminescence-based assays is background interference, which can potentially decrease the dynamic range and sensitivity of these assays. However, once properly set up, these reporter assays can offer robust phenotype-based HTS methods for PROTACs.
Mass spectrometry
MS analysis is a method that offers sensitive protein detection and quantification without relying on antibodies or tags. Various proteomic approaches are available to understand the mechanism of action of PROTAC degraders [123]. The quantitative approach uses synthetic stable isotope-labeled proteins which can precisely mimic their endogenous counterparts as the internal standards to quantify the corresponding target protein [124]. A global proteomic analysis study is widely used to examine the abundance change of proteins post treatment of with PROTAC degraders, to validate the degradation selectivity of the PROTAC degrader, and to reveal any off-target effects [12,17,25,28,61,125]. For example, using proteome dynamics profiling, Savitski et al. identified TREX complex adaptor FYTTD1 and SOAT1 as the off-targets of JQ1-VHL PROTAC. The activities of these off-targets are probably responsible for the observed molecular and phenotypic responses [125]. By offering an antibody- and tag-free protein quantitative method, MS makes intact protein detection possible. Moreover, with its capability to investigate the causes and consequences of protein degradation in biological systems [125], we anticipate its increasing application to the study of PROTAC degraders.
Protein degradation-based phenotypic screening
One of the big challenges for PROTAC development is that only a few E3 ligases have ligands. Since protein degradation is a phenotypic outcome of a PROTAC degrader, protein degradation-based phenotypic screening has been applied to identify engaged E3 ligases by a PROTAC. By screening electrophile-containing compounds that can react with cysteine(s) on E3 ligase, followed by incorporating these electrophilic probes into heterobifunctional compounds consisting of a reversible target ligand, E3 ligases that are capable of supporting targeted protein degradation can be identified with these covalent PROTACs. E3 ligases that are discovered via this method include RNF114 [126], DCAF16 [127] and RNF4 [128].
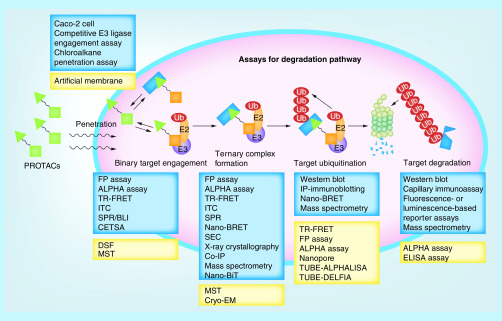
Figure 16. . Assays (in the blue frame) and potential assays (in the yellow frame) that can be applied for the degradation pathway of proteolysis targeting chimeras degraders.
ALPHA: Amplified luminescent proximity homogeneous assay; BLI: Bio-layer interferometry; CETSA: Cellular thermal shift assay; Co-IP: Co-immunoprecipitation; Cryo-EM: Cryogenic electron microscopy; DSF: Differential scanning fluorimetry; FP: Fluorescence polarization; ITC: Isothermal titration calorimetry; MST: Microscale thermophoresis; Nano-BiT: NanoLuc binary technology; Nano-BRET: Nano-bioluminescence resonance energy transfer; PROTAC: Proteolysis targeting chimera; FP: Fluorescence polarization; SPR: Surface plasma resonance; TR-FRET: Time-resolved fluorescence energy transfer; TUBE-ALPHALISA: Tandem ubiquitin-binding entities-amplified luminescent proximity homogeneous and enzyme-linked immunosorbent assay; TUBE-DELFIA: Tandem ubiquitin-binding entities-dissociation-enhanced lanthanide fluorescent immunoassay.
Future perspective
Targeting protein for degradation is an emerging mode of action in drug discovery [5] and promises to shift the drug discovery paradigm. Ubiquitin proteasome system-based PROTAC degraders are appealing modalities in inducing target protein degradation with their compound library expanding at an ever-increasing speed. Accompanied by the fast-expanding compound library is the development of all kinds of biochemical and biophysical assays. The suite of biophysical and biochemical methods that are now available represents a powerful toolbox for future PROTACs development. These assays played a significant role in helping us to understand the mechanism of PROTAC-induced protein degradation, particularly in understanding kinetics and thermodynamics of ternary complex formation, ternary complex stability and cooperativity and other factors that contribute to effective protein degradation. However, much remains to be understood: why some proteins cannot be degraded by PROTACs, how to predict the degradability of a target protein, how to match different E3 ligases and a target protein in the most efficient way, how to predict which lysine residue on a target protein is the most feasible for ubiquitination, to which level and what kind of ubiquitination is required for specific protein degradation and which step is the rate-limiting step along the degradation pathway for a specific protein? To answer these questions, new assays and new technologies need to be developed or new theories may need to be proposed. Vice versa, answers to these questions will help to identify the best assay formats and lead to better degraders. Expanding the current biochemical and biophysical assay toolbox with new technologies and new assays can potentially provide us with new insights into the mechanism of action of PROTACs and offer new methods for the screening and development of new compounds.
Executive summary.
Proteolysis targeting chimera (PROTAC) degraders are emerging drug discovery modalities and they can achieve target protein degradation in four major steps: penetrating into cells, ternary complex formation, target ubiquitination and target degradation.
Characterizing PROTAC degraders along each step of the protein degradation pathway with biochemical and biophysical assays provides valuable information for a better understanding of the mechanism of action and structure–activity relationship, which is essential for further structural modifications of the degraders.
Assays for cell permeability: Caco-2 cell assay, competitive E3 engagement assay and chloroalkane penetrating assay.
Assays for ternary complex formation and characterization: fluorescence polarization (FP) assay, amplified luminescent proximity homogeneous assay (ALPHA), time-resolved fluorescence energy transfer (TR-FRET), isothermal titration calorimetry (ITC), surface plasmon resonance/bio-layer interferometry (SPR/BLI), nano-bioluminescence resonance energy transfer (Nano-BRET®), size exclusive chromatography (SEC), crystallography, mass spectrometry (MS) and co-immunoprecipitation and NanoBiT.
Assays for target protein ubiquitination: in vitro enzyme assay, western blot, FP, time-resolved fluorescence energy transfer, amplified luminescent proximity homogeneous assay, immunoprecipitation/western blot, Nano-BRET and immunoprecipitation/MS.
Assays for target protein degradation: western blot, capillary immunoassay, MS, Nano-BRET and fluorescence-based reporter assays.
Assays and potential assays that can be applied to PROTAC degraders are summarized in Figure 16.
Choosing the right assay for characterizing a PROTAC is essential to provide feedback to medicinal chemists for optimization and to understanding the mechanism of action of a PROTAC molecule. For example, if a PROTAC does not induce protein degradation as detected by western blot, albeit with reasonable binary/ternary binding affinity, the permeability assay, intracellular ternary complex assay and ubiquitination assay of the PROTAC might uncover some clues. Cross-validation among different assays is also important as no assay is perfect. While the FP assay can be easily set up, the range of affinity that can be tested is limited. Although cell-based assays are more physiologically related, the structure–activity relationship obtained from these assays may be complicated by the the differences of compound permeability. A label-free method such as isothermal titration calorimetry is attractive, but it is highly demanding in terms of a sample’s quality and quantity.
Expanding the current biophysical and biochemical assay toolbox by introducing new technologies or new assays can potentially provide new insights into the mode of action of PROTAC degraders.
Acknowledgments
The authors thank JS Wiegand for editing and proofreading the manuscript.
Footnotes
Financial & competing interests disclosure
This work was supported in part by NIH grants R01CA211963, R01CA219836, R01CA242003 and R21 CA223371. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Open access
This work is licensed under the Attribution-NonCommercial-NoDerivatives 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.An S, Fu L. Small-molecule PROTACs: an emerging and promising approach for the development of targeted therapy drugs. EBioMedicine 36, 553–562 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciulli A, Farnaby W. Protein degradation for drug discovery. Drug Discov. Today Technol. 31, 1–3 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Lai AC, Crews CM. Induced protein degradation: an emerging drug discovery paradigm. Nat. Rev. Drug Discov. 16(2), 101–114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schapira M, Calabrese MF, Bullock AN, Crews CM. Targeted protein degradation: expanding the toolbox. Nat. Rev. Drug Discov. 1–15 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Valeur E, Narjes F, Ottmann CT, Plowright A. Emerging modes-of-action in drug discovery. MedChemComm 10(9), 1550–1568 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun X, Gao H, Yang Y. et al. PROTACs: great opportunities for academia and industry. Signal Transduct. Target Ther. 4(1), 1–33 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Summarized the majority of the targets that have been targeted by proteolysis targeting chimera (PROTACs) so far.
- 7.Sakamoto KM, Kim KB, Kumagai A, Mercurio F, Crews CM, Deshaies RJ. PROTACs: chimeric molecules that target proteins to the Skp1–Cullin-F box complex for ubiquitination and degradation. Proc. Natl Acad. Sci. USA 98(15), 8554–8559 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neklesa TK, Winkler JD, Crews CM. Targeted protein degradation by PROTACs. Pharmacol. Ther. 174, 138–144 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Toure M, Crews CM. Small-molecule PROTACS: new approaches to protein degradation. Angew. Chem. Int. Ed. Engl. 55(6), 1966–1973 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Zou Y, Ma D, Wang Y. The PROTAC technology in drug development. Cell Biochem. Funct. 37(1), 21–30 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith BE, Wang SL, Jaime-Figueroa S. et al. Differential PROTAC substrate specificity dictated by orientation of recruited E3 ligase. Nat. Commun. 10(1), 131 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; • To explain the selective degradation demonstrated by their PROTAC degraders, the authors characterized the ternary complex formed among POI (protein of interest):PROTAC:VHL (von Hippel–Lindau E3 ligase) and found that ternary complex kinetics is the driving force for target degradation.
- 12.Khan S, Zhang X, Lv D. et al. A selective BCL-X L PROTAC degrader achieves safe and potent antitumor activity. Nat. Med. 25(12), 1938–1947 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papatzimas JW, Gorobets E, Maity R. et al. From inhibition to degradation: targeting the antiapoptotic protein myeloid cell leukemia 1 (MCL1). J. Med. Chem. 62(11), 5522–5540 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Wang Z, He N, Guo Z. et al. Proteolysis targeting chimeras for the selective degradation of Mcl-1/Bcl-2 derived from nonselective target binding ligands. J. Med. Chem. 62(17), 8152–8163 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Thummuri D, He Y. et al. Utilizing PROTAC technology to address the on-target platelet toxicity associated with inhibition of BCL-XL. Chem. Commun. 55(98), 14765–14768 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Wispelaere M, Du G, Donovan KA. et al. Small molecule degraders of the hepatitis C virus protease reduce susceptibility to resistance mutations. Nat. Commun. 10(1), 1–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bai L, Zhou H, Xu R. et al. A potent and selective small-molecule degrader of STAT3 achieves complete tumor regression in vivo. Cancer Cell 36(5), 498–511.e17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mullard A. First targeted protein degrader hits the clinic. Nat. Rev. Drug Discov. 18, 237–239 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Clinical Trials Arena (2019). https://www.clinicaltrialsarena.com/news/arvinas-cancer-drugs-phase1-results/
- 20.Hughes SJ, Ciulli A. Molecular recognition of ternary complexes: a new dimension in the structure-guided design of chemical degraders. Essays Biochem. 61(5), 505–516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daniels DL, Riching KM, Urh M. Monitoring and deciphering protein degradation pathways inside cells. Drug Discov. Today Technol. 31, 61–68 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Edmondson SD, Yang B, Fallan C. Proteolysis targeting chimeras (PROTACs) in ‘beyond rule-of-five’ chemical space: recent progress and future challenges. Bioorg. Med. Chem Lett. 29(13), 1555–1564 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Han X, Wang C, Qin C. et al. Discovery of ARD-69 as a highly potent proteolysis targeting chimera (PROTAC) degrader of androgen receptor (AR) for the treatment of prostate cancer. J. Med. Chem. 62(2), 941–964 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Cromm PM, Samarasinghe KTG, Hines J, Crews CM. Addressing kinase-independent functions of FAK via PROTAC-mediated degradation. J. Am. Chem. Soc. 140(49), 17019–17026 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Farnaby W, Koegl M, Roy MJ. et al. BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design. Nat. Chem. Biol. 15(7), 672 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Provided the first example of using nonfunctional binding ligand as a warhead for PROTAC design. It also demonstrated the application of structure-guided approach for PROTAC degrader modification.
- 26.Zeng M, Xiong Y, Safaee N. et al. Exploring targeted degradation strategy for oncogenic KRASG12C. Cell Chem. Biol. 27(1), 19.e6–31.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Foley CA, Potjewyd F, Lamb KN, James LI, Frye SV. Assessing the cell permeability of bivalent chemical degraders using the chloroalkane penetration assay. ACS Chem. Biol. 15(1), 290–295 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bondeson DP, Smith BE, Burslem GM. et al. Lessons in PROTAC design from selective degradation with a promiscuous warhead. Cell Chem. Biol. 25(1), 78–87.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gadd MS, Testa A, Lucas X. et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat. Chem. Biol. 13(5), 514–521 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buckley DL, Gustafson JL, van Molle I. et al. Small-molecule inhibitors of the interaction between the E3 Ligase VHL and HIF1α. Angew. Chem. Int. Ed. Engl. 51(46), 11463–11467 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buckley DL, van Molle I, Gareiss PC. et al. Targeting the von Hippel–Lindau E3 ubiquitin ligase using small molecules to disrupt the VHL/HIF-1α interaction. J. Am. Chem. Soc. 134(10), 4465–4468 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Reported the generation of the first small molecule targeting VHL E3 ligase through rational design. The biochemical and biophysical methods used include isothermal titration calorimetry, NMR, fluorescence polarization (FP) and crystallography.
- 32.Crew AP, Raina K, Dong H. et al. Identification and characterization of von Hippel–Lindau-recruiting proteolysis targeting chimeras (PROTACs) of TANK-binding kinase 1. J. Med. Chem. 61(2), 583–598 (2018). [DOI] [PubMed] [Google Scholar]
- 33.van Molle I, Thomann A, Buckley DL. et al. Dissecting fragment-based lead discovery at the von Hippel–Lindau protein: hypoxia inducible factor 1α protein–protein interface. Chem. Biol. 19(10), 1300–1312 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zoppi V, Hughes SJ, Maniaci C. et al. Iterative design and optimization of initially inactive proteolysis targeting chimeras (PROTACs) identify VZ185 as a potent, fast, and selective von hippel–lindau (VHL) based dual degrader probe of BRD9 and BRD7. J. Med. Chem. 62(2), 699–726 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nowak RP, DeAngelo SL, Buckley D. et al. Plasticity in binding confers selectivity in ligand induced protein degradation. Nat. Chem. Biol. 14(7), 706–714 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moerke NJ. Fluorescence polarization (FP) assays for monitoring peptide-protein or nucleic acid-protein binding. Curr. Protoc. Chem. Biol. 1(1), 1–15 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Rossi AM, Taylor CW. Analysis of protein–ligand interactions by fluorescence polarization. Nat. Protoc. 6(3), 365–387 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang X. Fluorescence polarization competition assay: the range of resolvable inhibitor potency is limited by the affinity of the fluorescent ligand. J. Biomol. Screen. 8(1), 34–38 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Wagner T, Greschik H, Burgahn T. et al. Identification of a small-molecule ligand of the epigenetic reader protein spindlin1 via a versatile screening platform. Nucleic Acids Res. 44(9), e88–e88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warner G, Illy C, Pedro L, Roby P, Bossé R. AlphaScreen kinase HTS platforms. Curr. Med. Chem. 11(6), 721–730 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Yasgar A, Jadhav A, Simeonov A, Coussens NP. AlphaScreen-based assays: ultra-high-throughput screening for small-molecule inhibitors of challenging enzymes and protein–protein interactions. Methods Mol. Biol. 1439, 77–98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guenat S, Rouleau N, Bielmann C. et al. Homogeneous and nonradioactive high-throughput screening platform for the characterization of kinase inhibitors in cell lysates. J. Biomol. Screen. 11(8), 1015–1026 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Rouleau N, Turcotte S, Mondou M-H, Roby P, Bossé R. Development of a versatile platform for nuclear receptor screening using AlphaScreen. J. Biomol. Screen. 8(2), 191–197 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Cauchon E, Liu S, Percival MD. et al. Development of a homogeneous immunoassay for the detection of angiotensin I in plasma using AlphaLISA acceptor beads technology. Anal. Biochem. 388(1), 134–139 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Poulsen F, Jensen KB. A luminescent oxygen channeling immunoassay for the determination of insulin in human plasma. J. Biomol. Screen. 12(2), 240–247 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Patel R, Pollner R, de Keczer S. et al. Quantification of DNA using the luminescent oxygen channeling assay. Clin. Chem. 46(9), 1471–1477 (2000). [PubMed] [Google Scholar]
- 47.Huang X, Schmidt TA, Shortt C. et al. A competitive alphascreen assay for detection of hyaluronan. Glycobiology 28(3), 137–147 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Prashek J, Bouyain S, Fu M, Li Y, Berkes D, Yao X. Interaction between the pH and START domains of ceramide transfer protein competes with phosphatidylinositol 4-phosphate binding by the PH domain. J. Biol. Chem. 292(34), 14217–14228 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsirkone VG, Blokken J, Wit FD. et al. N-terminal half of transportin SR2 interacts with HIV integrase. J. Biol. Chem. 292(23), 9699–9710 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winter GE, Buckley DL, Paulk J. et al. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348(6241), 1376–1381 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wurz RP, Dellamaggiore K, Dou H. et al. A “click chemistry platform” for the rapid synthesis of bispecific molecules for inducing protein degradation. J. Med. Chem. 61(2), 453–461 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Hermanson SB, Carlson CB, Riddle SM. et al. Screening for novel LRRK2 inhibitors using a high-throughput TR-FRET cellular assay for LRRK2 Ser935 phosphorylation. PLoS ONE 7(8), e43580 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin W, Goktug AN, Wu J, Currier DG, Chen T. High-throughput screening identifies 1,4,5-substituted 1,2,3-triazole analogs as potent and specific antagonists of pregnane X receptor. Assay Drug Dev. Technol. 15(8), 383–394 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiong J, Pecchi VG, Qui M. et al. Development of a time-resolved fluorescence resonance energy transfer ultrahigh-throughput screening assay for targeting the NSD3 and MYC interaction. Assay Drug Dev. Technol. 16(2), 96–106 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bidinosti M, Shimshek DR, Mollenhauer B. et al. Novel one-step immunoassays to quantify α-synuclein: applications for biomarker development and high-throughput screening. J. Biol. Chem. 287(40), 33691–33705 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cui X, Liang Q, Liang Y, Lu M, Ding Y, Lu B. TR-FRET assays of huntingtin protein fragments reveal temperature and polyQ length-dependent conformational changes. Sci. Rep. 4, 5601 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liang Y, Yao Y, Lu M, Hou J, Yu S, Lu B. TR-FRET assays for endogenous huntingtin protein level in mouse cells. J. Huntingtons Dis. 3(3), 253–259 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Schutzius G, Bleckmann D, Kapps-Fouthier S, di Giorgio F, Gerhartz B, Weiss A. A quantitative homogeneous assay for fragile X mental retardation 1 protein. J. Neurodev. Disord. 5(1), 8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gaborit N, Larbouret C, Vallaghe J. et al. Time-resolved fluorescence resonance energy transfer (TR-FRET) to analyze the disruption of EGFR/HER2 dimers. J. Biol. Chem. 286(13), 11337–11345 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stroik DR, Yuen SL, Janicek KA. et al. Targeting protein–protein interactions for therapeutic discovery via FRET-based high-throughput screening in living cells. Sci. Rep. 8(1), 1–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zorba A, Nguyen C, Xu Y. et al. Delineating the role of cooperativity in the design of potent PROTACs for BTK. Proc. Natl Acad. Sci. USA 115(31), E7285–E7292 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Imbert P-E, Unterreiner V, Siebert D, Gubler H, Parker C, Gabriel D. Recommendations for the reduction of compound artifacts in time-resolved fluorescence resonance energy transfer assays. Assay Drug Dev. Technol. 5(3), 363–372 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Velazquez-Campoy A, Leavitt SA, Freire E. Characterization of protein–protein interactions by isothermal titration calorimetry. Methods Mol. Biol. 261, 35–54 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Freire E. Do enthalpy and entropy distinguish first in class from best in class? Drug Discov. Today 13(19), 869–874 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Falconer RJ. Applications of isothermal titration calorimetry – the research and technical developments from 2011 to 2015. J. Mol. Recognit. 29(10), 504–515 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Bulatov E, Martin EM, Chatterjee S. et al. Biophysical studies on interactions and assembly of full-size E3 ubiquitin ligase suppressor of cytokine signaling 2 (SOCS2)-elongin bc-cullin 5-ring box protein 2 (RBX2). J. Biol. Chem. 290(7), 4178–4191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chabot PR, Raiola L, Lussier-Price M. et al. Structural and functional characterization of a complex between the acidic transactivation domain of EBNA2 and the Tfb1/p62 Subunit of TFIIH. PLoS Pathog. 10(3), e1004042 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cai S, Yan J, Xiong H, Liu Y, Peng D, Liu Z. Investigations on the interface of nucleic acid aptamers and binding targets. Analyst 143(22), 5317–5338 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Seo M, Lei L, Egli M. Label-free electrophoretic mobility shift assay (EMSA) for measuring dissociation constants of protein-RNA complexes. Curr. Protoc. Nucleic Acid Chem. 76(1), e70 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Z, Oni O, Liu J. New insights into a classic aptamer: binding sites, cooperativity and more sensitive adenosine detection. Nucleic Acids Res. 45(13), 7593–7601 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gal M, Bloch I, Shechter N, Romanenko O, Shir OM. Efficient isothermal titration calorimetry technique identifies direct interaction of small molecule inhibitors with the target protein. Comb. Chem. High Throughput Screen. 19(1), 4–13 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Myrianthopoulos V, Gaboriaud-Kolar N, Tallant C. et al. Discovery and optimization of a selective ligand for the switch/sucrose nonfermenting-related bromodomains of polybromo protein-1 by the use of virtual screening and hydration analysis. J. Med. Chem. 59(19), 8787–8803 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lu M, Liu T, Jiao Q. et al. Discovery of a Keap1-dependent peptide PROTAC to knockdown Tau by ubiquitination-proteasome degradation pathway. Eur. J. Med. Chem. 146, 251–259 (2018). [DOI] [PubMed] [Google Scholar]
- 74.Testa A, Hughes SJ, Lucas X, Wright JE, Ciulli A. Structure-based design of a macrocyclic PROTAC. Angew. Chem. Int. Ed. Engl. 59(4), 1727–1734 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Velázquez-Campoy A, Ohtaka H, Nezami A, Muzammil S, Freire E. Isothermal titration calorimetry. Curr. Protoc. Cell Biol. 23(1), 17.8.1–17.8.24 (2004). [DOI] [PubMed] [Google Scholar]
- 76.Saponaro A. Isothermal titration calorimetry: a biophysical method to characterize the interaction between label-free biomolecules in solution. Bio Protoc. 8(15), (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Alvarado C, Stahl E, Koessel K. et al. Development of a fragment-based screening assay for the focal adhesion targeting domain using SPR and NMR. Molecules 24(18), 3352 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huber S, Casagrande F, Hug MN. et al. SPR-based fragment screening with neurotensin receptor 1 generates novel small molecule ligands. PLoS ONE 12(5), e0175842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Giannetti AM. From experimental design to validated hits a comprehensive walk-through of fragment lead identification using surface plasmon resonance. Methods Enzymol. 493, 169–218 (2011). [DOI] [PubMed] [Google Scholar]
- 80.Abdiche YN, Myszka DG. Probing the mechanism of drug/lipid membrane interactions using Biacore. Anal. Biochem. 328(2), 233–243 (2004). [DOI] [PubMed] [Google Scholar]
- 81.Frostell-Karlsson Å, Remaeus A, Roos H. et al. Biosensor analysis of the interaction between immobilized human serum albumin and drug compounds for prediction of human serum albumin binding levels. J. Med. Chem. 43(10), 1986–1992 (2000). [DOI] [PubMed] [Google Scholar]
- 82.Petz M. Recent applications of surface plasmon resonance biosensors for analyzing residues and contaminants in food. Monatsh Chem. 140(8), 953–964 (2009). [Google Scholar]
- 83.Silva MC, Ferguson FM, Cai Q. et al. Targeted degradation of aberrant tau in frontotemporal dementia patient-derived neuronal cell models. eLife 8, e45457 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Roy MJ, Winkler S, Hughes SJ. et al. SPR-measured kinetics of PROTAC ternary complexes influence target degradation rate. ACS Chem. Biol. 14(3), 361–368 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Surface plasmon resonance assay was developed to profile PROTAC ternary complexes kinetically.
- 85.Dale NC, Johnstone EKM, White CW, Pfleger KDG. NanoBRET: the bright future of proximity-aased assays. Front. Bioeng. Biotechnol. 7, 56 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bacart J, Corbel C, Jockers R, Bach S, Couturier C. The BRET technology and its application to screening assays. Biotechnol. J. 3(3), 311–324 (2008). [DOI] [PubMed] [Google Scholar]
- 87.Hall MP, Unch J, Binkowski BF. et al. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol. 7(11), 1848–1857 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eyre NS, Aloia AL, Joyce MA, Chulanetra M, Tyrrell DL, Beard MR. Sensitive luminescent reporter viruses reveal appreciable release of hepatitis C virus NS5A protein into the extracellular environment. Virology 507, 20–31 (2017). [DOI] [PubMed] [Google Scholar]
- 89.Kim J, Grailhe R. Nanoluciferase signal brightness using furimazine substrates opens bioluminescence resonance energy transfer to widefield microscopy. Cytometry Part A 89(8), 742–746 (2016). [DOI] [PubMed] [Google Scholar]
- 90.Yoshida T, Kakizuka A, Imamura H. BTeam, a novel BRET-based biosensor for the accurate quantification of ATP concentration within living cells. Sci. Rep. 6(1), 1–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Machleidt T, Woodroofe CC, Schwinn MK. et al. NanoBRET – a novel BRET platform for the analysis of protein–protein interactions. ACS Chem. Biol. 10(8), 1797–1804 (2015). [DOI] [PubMed] [Google Scholar]
- 92.Daniels DL, Riching KM, Urh M. Monitoring and deciphering protein degradation pathways inside cells. Drug Discov. Today Technol. 31, 61–68 (2019). [DOI] [PubMed] [Google Scholar]
- 93.Riching KM, Mahan S, Corona CR. et al. Quantitative live-cell kinetic degradation and mechanistic profiling of PROTAC mode of action. ACS Chem. Biol. 13(9), 2758–2770 (2018). [DOI] [PubMed] [Google Scholar]; • Nano-bioluminescence resonance energy transfer (Nano-BRET®) assay was developed as a technology to characterize PROTAC degraders in live cells.
- 94.Maniaci C, Hughes SJ, Testa A. et al. Homo-PROTACs: bivalent small-molecule dimerizers of the VHL E3 ubiquitin ligase to induce self-degradation. Nat. Commun. 8(1), 830 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhao Q, Ren C, Liu L. et al. Discovery of SIAIS178 as an effective BCR-ABL degrader by recruiting von Hippel–Lindau (VHL) E3 ubiquitin ligase. J. Med. Chem. 62(20), 9281–9298 (2019). [DOI] [PubMed] [Google Scholar]
- 96.Simonetta KR, Taygerly J, Boyle K. et al. Prospective discovery of small molecule enhancers of an E3 ligase–substrate interaction. Nat. Commun. 10(1), 1402 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Beveridge R, Kessler D, Rumpel K, Ettmayer P, Meinhart A, Clausen T. Native mass spectrometry can effectively predict PROTAC efficacy. bioRxiv 851980 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jafari R, Almqvist H, Axelsson H. et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat. Protoc. 9(9), 2100–2122 (2014). [DOI] [PubMed] [Google Scholar]
- 99.Gaffarogullari EC, Krause A, Balbo J, Herten D-P, Jäschke A. Microscale thermophoresis provides insights into mechanism and thermodynamics of ribozyme catalysis. RNA Biol. 10(12), 1815–1821 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Renaud J-P, Chari A, Ciferri C. et al. Cryo-EM in drug discovery: achievements, limitations and prospects. Nat. Rev. Drug Discov. 17(7), 471–492 (2018). [DOI] [PubMed] [Google Scholar]
- 101.Bussiere DE, Xie L, Srinivas H. et al. Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex. Nat. Chem. Biol. 16(1), 15–23 (2020). [DOI] [PubMed] [Google Scholar]
- 102.Scapin G, Potter CS, Carragher B. Cryo-EM for small molecules discovery, design, understanding, and application. Cell Chem. Biol. 25(11), 1318–1325 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin–proteasome pathway in normal and disease states. J. Am. Soc. Nephrol. 17(7), 1807–1819 (2006). [DOI] [PubMed] [Google Scholar]
- 104.Mot AC, Prell E, Klecker M. et al. Real-time detection of N-end rule-mediated ubiquitination via fluorescently labeled substrate probes. New Phytologist 217(2), 613–624 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kus B, Gajadhar A, Stanger K. et al. A high throughput screen to identify substrates for the ubiquitin ligase Rsp5. J. Biol. Chem. 280(33), 29470–29478 (2005). [DOI] [PubMed] [Google Scholar]
- 106.Madiraju C, Welsh K, Cuddy MP. et al. TR-FRET-based high-throughput screening assay for identification of UBC13 inhibitors. J. Biomol. Screen. 17(2), 163–176 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sigismund S, Polo S. Strategies to detect endogenous ubiquitination of a target mammalian protein. Method Mol. Biol. 1449, 143–151 (2016). [DOI] [PubMed] [Google Scholar]
- 108.Grice GL, Nathan JA. The recognition of ubiquitinated proteins by the proteasome. Cell. Mol. Life Sci. 73(18), 3497–3506 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Heap RE, Gant MS, Lamoliatte F, Peltier J, Trost M. Mass spectrometry techniques for studying the ubiquitin system. Biochem. Soc. Trans. 45(5), 1137–1148 (2017). [DOI] [PubMed] [Google Scholar]
- 110.Ordureau A, Münch C, Harper JW. Quantifying ubiquitin signaling. Mol. Cell 58(4), 660–676 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Udeshi ND, Mertins P, Svinkina T, Carr SA. Large-scale identification of ubiquitination sites by mass spectrometry. Nat. Protoc. 8(10), 1950–1960 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xu G, Jaffrey SR. Proteomic identification of protein ubiquitination events. Biotechnol. Genet. Eng. Rev. 29, 73–109 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yoshida Y, Saeki Y, Murakami A. et al. A comprehensive method for detecting ubiquitinated substrates using TR-TUBE. Proc. Natl Acad. Sci. USA 112(15), 4630–4635 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ottis P, Toure M, Cromm PM, Ko E, Gustafson JL, Crews CM. Assessing different E3 ligases for small molecule induced protein ubiquitination and degradation. ACS Chem. Biol. 12(10), 2570–2578 (2017). [DOI] [PubMed] [Google Scholar]
- 115.Wloka C, van Meervelt V, van Gelder D. et al. Label-free and real-time detection of protein ubiquitination with a biological nanopore. ACS Nano 11(5), 4387–4394 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mata-Cantero L, Cid C, Gomez-Lorenzo MG. et al. Development of two novel high-throughput assays to quantify ubiquitylated proteins in cell lysates: application to screening of new anti-malarials. Malar. J. 14, 200 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Varshavsky A. Proteolysis. : Encyclopedia of Genetics. Brenner S, Miller JH (). Academic Press, NY, USA, 1573–1575 (2001). [Google Scholar]
- 118.Lai AC, Toure M, Hellerschmied D. et al. Modular PROTAC design for the degradation of oncogenic BCR-ABL. Angew. Chem. Int. Ed. Engl. 55(2), 807–810 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lu J, Qian Y, Altieri M. et al. Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem. Biol. 22(6), 755–763 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chessum NEA, Sharp SY, Caldwell JJ. et al. Demonstrating in-cell target engagement using a pirin protein degradation probe (CCT367766). J. Med. Chem. 61(3), 918–933 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yeung WSB, Luo GA, Wang QG, Ou JP. Capillary electrophoresis-based immunoassay. J. Chromatogr. B Analyt. Technol. Biomed Life Sci. 797(1), 217–228 (2003). [DOI] [PubMed] [Google Scholar]
- 122.Eglen RM, Reisine T, Roby P. et al. The use of AlphaScreen technology in HTS: current status. Curr. Chem. Genomics 1, 2–10 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Grandi P, Bantscheff M. Advanced proteomics approaches to unravel protein homeostasis. Drug Discov. Today Technol. 31, 99–108 (2019). [DOI] [PubMed] [Google Scholar]
- 124.Pan S, Aebersold R, Chen R. et al. Mass spectrometry based targeted protein quantification: methods and applications. J. Proteome Res. 8(2), 787–797 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Savitski MM, Zinn N, Faelth-Savitski M. et al. Multiplexed proteome dynamics profiling reveals mechanisms controlling protein homeostasis. Cell 173(1), 260–274.e25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Spradlin JN, Hu X, Ward CC. et al. Harnessing the anti-cancer natural product nimbolide for targeted protein degradation. Nat. Chem. Biol. 15(7), 747–755 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang X, Crowley VM, Wucherpfennig TG, Dix MM, Cravatt BF. Electrophilic PROTACs that degrade nuclear proteins by engaging DCAF16. Nat. Chem. Biol. 15(7), 737–746 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ward CC, Kleinman JI, Brittain SM. et al. Covalent ligand screening uncovers a RNF4 E3 ligase recruiter for targeted protein degradation applications. ACS Chem. Biol. 14(11), 2430–2440 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]