In the self-incompatibility response, incompatible pollen stimulates irreversible oxidative protein modifications that affect crucial protein functions.
Abstract
Self-incompatibility (SI) is used by many angiosperms to prevent self-fertilization and inbreeding. In common poppy (Papaver rhoeas), interaction of cognate pollen and pistil S-determinants triggers programmed cell death (PCD) of incompatible pollen. We previously identified that reactive oxygen species (ROS) signal to SI-PCD. ROS-induced oxidative posttranslational modifications (oxPTMs) can regulate protein structure and function. Here, we have identified and mapped oxPTMs triggered by SI in incompatible pollen. Notably, SI-induced pollen had numerous irreversible oxidative modifications, while untreated pollen had virtually none. Our data provide a valuable analysis of the protein targets of ROS in the context of SI-induction and comprise a benchmark because currently there are few reports of irreversible oxPTMs in plants. Strikingly, cytoskeletal proteins and enzymes involved in energy metabolism are a prominent target of ROS. Oxidative modifications to a phosphomimic form of a pyrophosphatase result in a reduction of its activity. Therefore, our results demonstrate irreversible oxidation of pollen proteins during SI and provide evidence that this modification can affect protein function. We suggest that this reduction in cellular activity could lead to PCD.
Angiosperms perform sexual reproduction using pollination, utilizing specific interactions between pollen (male) and pistil (female) tissues. Many angiosperms use self-incompatibility (SI) to prevent self-fertilization and inbreeding. These genetically controlled systems trigger rejection of “self” (incompatible) pollen. Common poppy (Papaver rhoeas) uses a SI system involving the female S-determinant (PrsS) protein, a ligand secreted by the pistil (Foote et al., 1994) and the male S-determinant protein (PrpS; Wheeler et al., 2009). SI also triggers programmed cell death (PCD), involving the activation of a DEVDase/caspase-3-like activity (Bosch and Franklin-Tong, 2007). A MAP kinase, p56, is involved in signaling to SI-PCD (Rudd et al., 2003; Li et al., 2007; Chai et al., 2017). The actin cytoskeleton is an early target of the SI signaling cascade in P. rhoeas pollen (Geitmann et al., 2000; Snowman et al., 2002) beginning with actin depolymerization and later formation of punctate F-actin foci (Geitmann et al., 2000; Snowman et al., 2002; Poulter et al., 2010). SI also triggers increases in reactive oxygen species (ROS) and nitric oxide (NO; Wilkins et al., 2011). Live-cell imaging of ROS in growing P. rhoeas pollen tubes, using chloromethyl- 2′7 ′-dichlorodihydrofluorescein oxidation, showed that SI induces relatively rapid increases in ROS, as early as 2 min after SI in some incompatible pollen tubes. A link between SI-induced ROS and PCD was identified using ROS scavengers, which revealed alleviation of SI-induced events, including formation of actin punctate foci and the activation of a DEVDase/caspase-3-like activity (Wilkins et al., 2011). These data provided evidence that ROS increases are upstream of these key SI markers and are required for SI-PCD (Wilkins et al., 2011) and represented the first steps in understanding ROS signaling in this system.
Exactly how ROS mediate SI-induced events is an important question that needs to be addressed. One possibility is that oxidative posttranslational modifications to proteins (oxPTMs) are involved. These include reversible modifications to Cys (e.g. sulfenylation, disulphide bonds, S-glutathionylation) and Met (Met sulfoxide) as well as a range of irreversible oxPTMs (Møller et al., 2007). In the case of Cys, reversible oxPTMs mediate signaling or changes in protein function (Waszczak et al., 2014, 2015; Akter et al., 2015a). NO produced during SI (Wilkins et al., 2011) also provides the possibility of a role for Cys S-nitrosylation. Although we had previously identified ROS as a signal to SI-PCD (Wilkins et al., 2011), earlier studies did not extend to identifying the protein targets of oxidation.
In this study, we aimed to identify and map oxPTMs on pollen proteins triggered by SI and H2O2 using liquid chromatography tandem mass spectrometry (LC-MS/MS). We analyzed the protein targets of ROS in the context of SI induction and identified and mapped oxidative modifications. Our data reveal that irreversible oxidation is likely an important mechanism involved in SI events in incompatible P. rhoeas pollen and provide a link between irreversible oxPTMs and a ROS-mediated physiological process.
RESULTS
SI Causes Oxidative Modifications to Proteins in Incompatible Pollen
Identifying the nature of the oxPTMs on individual proteins is an important step to understanding how cells interpret oxidative signals and translate them into a response. As we had previously shown that ROS and NO increased during the SI response and played a role in mediating actin alterations and PCD (Wilkins et al., 2011), we wished to examine whether pollen proteins were oxidatively modified after SI. We used LC-MS/MS to examine the extent and type of oxPTMs to pollen proteins during early SI, subjecting P. rhoeas pollen grown in vitro to SI taking samples 12 min after treatment (n = 2). In addition, we exposed P. rhoeas pollen to H2O2 treatments, again taking samples 12 min after treatment (n = 3). We counted the oxPTMs observed after these two treatments, discarding any that also occurred in untreated samples. Likewise, for untreated samples, only oxPTMs identified as uniquely occurring in these samples were counted (n = 3). We compared the SI response with H2O2 treatment to determine which of these modifications were also induced by artificially generated oxidative stress. A number of oxidative modifications were detected following both treatments (Supplemental Tables S1–S6); peptide coverage relating to data in Supplemental Tables S1, S3, and S5 is shown in Supplemental Table S7; annotated spectra for representative examples of each oxidative modification identified are shown in Supplemental Figure S1. As none of the modifications listed were observed in the untreated samples, this provides confidence that they are stimulated by that treatment.
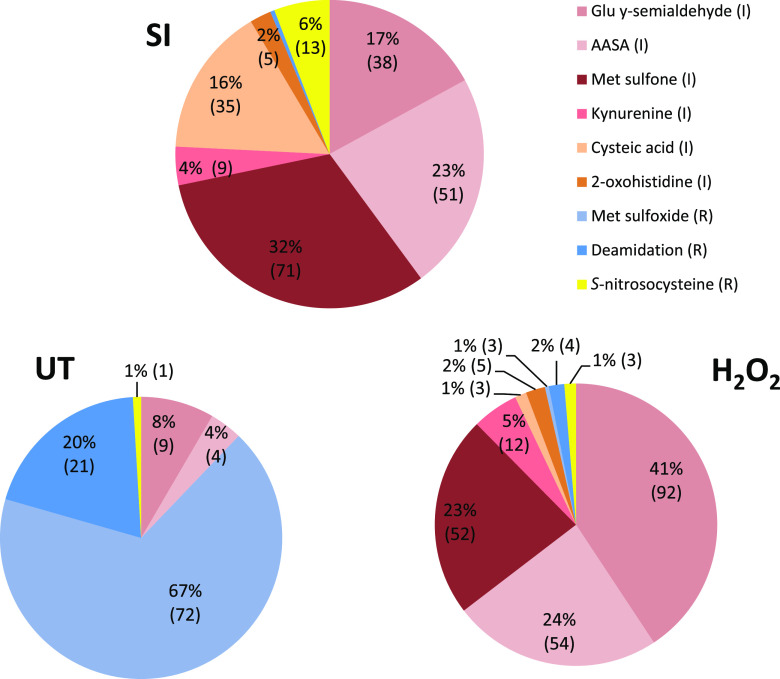
The types of oxidative modifications identified on peptides from SI-induced pollen proteins were quite different from those identified on peptides from untreated pollen (Fig. 1). Notably, we found that the majority (94%) of the oxidatively modified amino acids in the SI sample were irreversibly modified (209/223), compared to only 13/107 (12%) in the untreated pollen sample. Irreversible modifications identified in SI-induced samples included 71 Met to Met sulfone, 51 aminoadipic semialdehyde (AASA) on Lys, 38 Pro to Glu γ-semialdehyde, and 35 Cys to cysteic acid; other modifications were kyneurine on Trp (9) and 2-oxo-His (5; Fig. 1).
Figure 1.
Distribution of types of oxidative modifications of pollen proteins after different treatments. Each unique oxidative modification was identified on a unique peptide for each type of pollen treatment: SI induction (SI), H2O2, or untreated (UT) was categorized according to its type of modification and counted. Irreversible modifications (I) are indicated in red tones, and reversible modifications (R) are indicated in blues. These were represented proportionally in pie charts and are shown as a percentage of total counts, with the actual number of modifications identified in brackets.
Few reports of such irreversible oxidative modifications exist. Cysteines are irreversibly oxidized to Cys sulfonic acid or cysteic acid in response to severe oxidative stress, which generally leads to protein inactivation and degradation (Møller et al., 2007). Modification of Lys to AASA is a carbonylation modification, which is the most common type of irreversible oxidative modification to a protein, which generally inhibits the function of proteins. Together, these data demonstrate that during early SI, many proteins are permanently modified.
By contrast, the majority of the oxPTMs on untreated pollen proteins were of a reversible nature (94 out of 107 modifications identified; Fig. 1). These mainly comprised 72 methionines modified to Met sulfoxide. Untreated samples also had 21 deamidated amino acids. By contrast, the SI-induced pollen had no Met sulfoxide modifications; only one deamidation was identified. H2O2 treatment of pollen also resulted in a majority of irreversible oxidative modifications (Fig. 1), with 218 irreversibly modified amino acids over 155 different peptides; the remaining eight oxPTMs were reversible. Irreversible modifications identified in H2O2-treated samples included 92 Pro to Glu γ-semialdehyde, 54 AASA on Lys, and 52 Met to Met sulfone.
SI pollen proteins had far more oxPTMs than untreated pollen. We identified 181 uniquely modified oxPTM peptides containing 251 different oxidatively modified amino acids in SI-induced pollen (summarized in Supplemental Table S1; full data in Supplemental Table S2), while untreated pollen analyzed in an identical manner side-by-side had 104 uniquely modified peptides with 110 different oxidatively modified amino acids (summarized in Supplemental Table S3; full data in Supplemental Table S4). In H2O2-treated pollen, 262 unique oxPTMs were identified (summarized in Supplemental Table S5; full data in Supplemental Table S6). Notably, proteins that in control conditions contained Met sulfoxide modification often showed increased oxidation to the sulfone form following SI induction and H2O2 treatment (Supplemental Tables S1, S3, and S5).
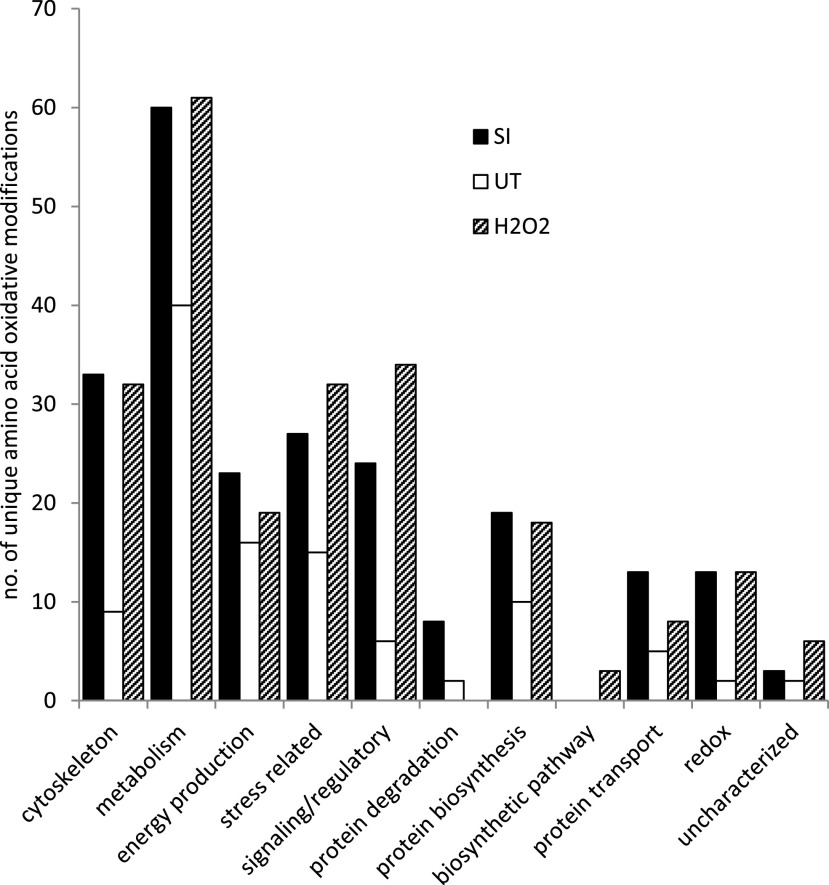
Proteins with oxPTMs were categorized according to their general functions (Fig. 2); any common modifications found between the pollen samples (SI, H2O2, and untreated) were discarded. For all of the functional groups, the SI samples had increased numbers of unique amino acids modified by oxidation compared to untreated pollen. The largest difference in numbers of oxidatively modified amino acids between SI-induced pollen and untreated pollen was found in the general functional grouping of cytoskeleton (33 versus 9), signaling/regulatory (24 versus 6), stress related (27 versus 15), and metabolism (60 versus 40), which together comprised 69% of the modified proteins in SI-induced pollen. However, even in functional groupings where fewer modifications were found in SI pollen proteins, proportionally the difference compared to untreated pollen was large (e.g. for proteins involved in redox, SI had 13 differently modified amino acids, compared to two in untreated). The oxidatively modified proteins identified from the H2O2-treated pollen were also categorized based on their general functions. Like SI treatment, proteins involved in metabolism, signaling/regulation, stress, and cytoskeleton comprised the majority (70%) of those with oxPTMs after H2O2 treatment (Fig. 2). Although the frequency of oxPTMs in the dataset will be influenced by protein abundance, it is striking that cytoskeletal proteins and enzymes involved in energy metabolism represent a prominent target during the SI response (Supplemental Table S1). In relation to energy metabolism, a large proportion of enzymes associated with glycolysis (phosphoglucomutase, pyrophosphate-dependent phosphofructokinase, glyceraldehyde 3-P dehydrogenase, enolase, pyruvate kinase, inorganic pyrophosphatase), organic acid metabolism (aconitase, citrate synthase, citrate lyase, isocitrate dehydrogenase, malate dehydrogenase, phosphoenolpyruvate carboxylase), and ATP synthesis/use (ATP synthase, ATPases) have oxPTMs.
Figure 2.
Distribution of the number of unique oxidative modifications to amino acids on pollen proteins according to function after different treatments. Each unique oxidatively modified amino acid was counted and categorized according to its function for each pollen treatment: SI induction (SI), H2O2, or untreated (UT).
Proteins with oxPTM Common to SI and H2O2 Treatments
To gain a better idea of the overlap between SI- and H2O2-treated samples, we identified peptides with identical oxPTMs in the SI-induced and the H2O2-treated samples, but not in untreated pollen (Table 1; Supplemental Tables S1, S3, and S5). Thirty-two peptides shared 44 oxidatively modified amino acids, with identical modifications found in both SI-induced and H2O2-treated samples. This overlap gives confidence that the modifications triggered in incompatible pollen tubes are authentic ROS-mediated events and that these proteins are rapidly oxidatively modified by ROS formed during SI. There was no overlap between proteins/peptides with S-nitroso-Cys modifications in SI- and H2O2-treated samples, suggesting that those modified during SI might be specific.
Table 1. Overlap between oxidatively modified peptides in SI-induced and H2O2-treated pollen.
Peptides containing the same oxidatively modified amino acids in both SI and H2O2 treated pollen are listed in columns 3 and 4, respectively; those with # have additional oxidative modifications. Modified amino acids are indicated by small bold letters, with the type of oxidative modification indicated by superscript numbers as follows: 1Glu y-semialdehyde (I), 2AASA (I), 3Met sulfone (I), 4kynurenine (I), 5cysteic acid (I), 62-oxo-His (I), 7Met sulfoxide (R), 8carbamidomethyl (R) produced by reaction with iodoacetamide during sample preparation, 9deamidation (R), 10S-nitroso-Cys (R). Most proteins were identified using PANTHER, except a few that were “unclassified” in PANTHER (indicated by *) and identified using a BLAST search.
| Protein Class | Protein | Modified Peptide in SI Pollen | Modified Peptide in H2O2 Pollen |
|---|---|---|---|
| Cytoskeleton | Actin | DLYGNIVLSGGSTMFp1GIADR | DLYGNIVLSGGSTMFp1GIADR |
| Actin | EITALAPSSm3K | EITALAPSSm3K | |
| Actin | YPIEHGIVSNWDDm3EK | YPIEHGIVSNWDDm3EK | |
| Actin | YPIEHGIVTNw4DDMEK | YPIEHGIVTNw4DDm3EK # | |
| Actin | DLYGNIVLSGGTTm3FPGIADR | DLYGNIVLSGGTTm3FPGIADR | |
| Alpha-tubulin | k2LADNc8TGLQGFLVFNAVGGGTGSGLGSLLLER | k2LADNc8TGLQGFLVFNAVGGGTGSGLGSLLLER | |
| Alpha-tubulin | TIQFVDWc4PTGFK | TIQFVDWc4PTGFk2 # | |
| Beta-tubulin | GHYTEGAELIDSVLDVVRk2 | GHYTEGAELIDSVLDVVRk2 | |
| Beta-tubulin | NSSYFVEw4Ip1NNVk2 | NSSYFVEw4Ip1NNVk2 | |
| Beta-tubulin | m3MLTFSVFPSPK | m3MLTFSVFPSPK | |
| Metabolism | GAPDH | VALQRDDVELVAVNDPFITTDYMTYMFk2 | VALQRDDVELVAVNDPFITTDYMTYMFk2 |
| GAPDH | DAp1MFVVGVNEK | DAp1MFVVGVNEK | |
| sPPase | AIGLm3p1MIDQGEKDDK | AIGLm3p1MIDQGEKDDK | |
| sPPase | RSVAAHp1w4h6DLEIGPGAPSVVNAVVEITk2 | RSVAAHp1w4h6DLEIGPGAPSVVNAVVEITk2 | |
| Enolase | KYGQDATNVGDEGGFAPNIQENk2EGLELLK | KYGQDATNVGDEGGFAPNIQENk2EGLELLK | |
| Enolase | SFVSDYPIVSIEDPFDQDDw4Eh6YSk2 | SFVSDYPIVSIEDPFDQDDw4Eh6YSk2 | |
| Stress | HSP70 | NQVAMNp1INTVFDAK | NQVAMNp1INTVFDAK |
| Signaling/regulatory | Elongation Factor 2* | GVQYLNEIKDSVVAGFQwask2 | GVQYLNEIKDSVVAGFQwASk2 |
| Elongation Factor 2* | GVQYLNEIKDSVVAGFQw4Ask2 | GVQYLNEIKDSVVAGFQw4ASk2 | |
| Predicted EF2-like | Nc8DPDGPLm3LYVSK | Nc8DPDGPLm3LYVSK | |
| Predicted EF2-like | LYMEARp1LEDGLAEAIDDGR | LYMEARp1LEDGLAEAIDDGR | |
| Eukaryotic initiation factor 4 | VQVGVFSATMp1PEALEITR | VQVGVFSATMp1PEALEITR | |
| RAB GTPase | LLLIGDSGVGk2 | LLLIGDSGVGk2 | |
| RAB GTPase | FADDSYLESYISTIGVDFk2 | FADDSYLESYISTIGVDFk2 | |
| RAB GDP dissociation inhibitor | NDYYGGESTSLNLIQLWk2 | NDYYGGESTSLNLIQLWk2 | |
| 14-3-3-like protein | QAFDEAISELDTLGEESYk2DSTLIm3QLLR | QAFDEAISELDTLGEESYk2DSTLIm3QLLR | |
| Protein biosynthesis | Met synthase | k2LNLPILPTTTIGSFPQTIELR | k2LNLPILPTTTIGSFPQTIELR |
| Met synthase | GMLTGp1VTILNWSFVR | GMLTGp1VTILNWSFVR | |
| Ser hydroxyl methyl-transferase* | GIELIASENFTSFAVIEALGSALTNk2 | GIELIASENFTSFAVIEALGSALTNk2 | |
| Ser hydroxyl methyl-transferase* | IMGLDLp1SGGHLTHGYYTSGGk2 | IMGLDLp1SGGHLTHGYYTSGGk2 | |
| Redox | SKS (SKU5 similar) | YALNGVSHTDp1ETPLK | YALNGVSHTDp1ETPLKSGKGDGSDAp1LFTLKp1GK # |
| 2-oxoacid dehydrogenase acyltransferase | RTPVSGPKGk2PQALQVk2 | RTPVSGPKGk2PQALQVk2 |
The proteins identified with identical oxPTMs after SI and H2O2 (Table 1) suggest that some key common events are triggered. Actin and tubulin are shared targets, with 10 identical peptides containing 14 shared oxidatively modified amino acids. Other proteins known to be involved in tip growth, e.g. soluble inorganic pyrophosphatases, Rab GTPases, and several elongation factor subunit peptides, were also oxidatively modified in both SI- and H2O2-treated pollen. These modified targets could contribute to inhibition of pollen tube growth. These data further suggest that protein synthesis and energy metabolism is altered by ROS during SI.
Pollen Proteins Are Modified by S-Nitrosylation after SI
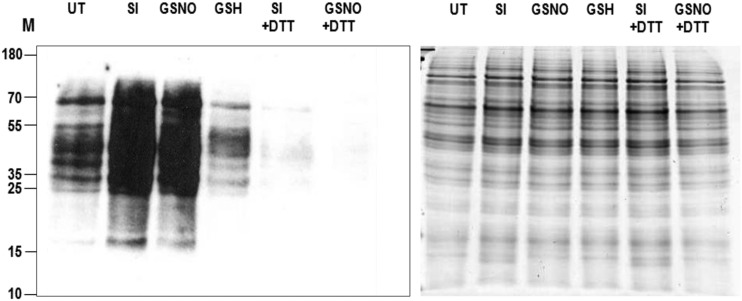
We previously showed that increases in NO were observed after SI induction in incompatible pollen (Wilkins et al., 2011). NO, via S-nitrosoglutathione (GSNO) production, could induce protein S-nitrosylation. Here, we directly examined if SI-stimulated S-nitrosylation by analyzing protein extracts from pollen after SI induction using LC-MS/MS. First, we examined pollen protein extracts for S-nitrosylation using western blotting, treating germinated pollen with GSNO as a comparison. Pollen extracts were selectively labeled for proteins containing an S-nitrosylated Cys using iodoTMTzero, then visualized after western blotting using an anti-TMT antibody. Both SI-induced and GSNO-treated pollen had high levels of S-nitrosylation, whereas little staining of S-nitrosylated proteins was detectable in the untreated pollen (Fig. 3). Addition of the reducing agent dithiothreitol (DTT) during protein extraction resulted in the almost complete loss of staining, verifying that the staining was detecting oxidized proteins. Thus, SI-treated pollen has more S-nitrosylated proteins than untreated pollen. LC-MS/MS identified 13 S-nitroso-Cys modifications in the SI-induced pollen samples (Fig. 1; Supplemental Table S1). In comparison, only one and three S-nitroso-Cys-modified peptides were identified in untreated and H2O2-treated pollen, respectively. This provides good evidence for authentic S-nitrosylation of proteins triggered in pollen by SI.
Figure 3.
Detection of S-nitrosylated proteins from pollen tubes by western blot analysis. Western blot of S-nitrosylated proteins detected with Pierce S-nitrosylation western blot kit. UT, Untreated sample; SI, SI-induced sample; GSNO, addition of NO donor S-nitrosoglutathione; GSH, addition of reducing agent glutathione (GSH); SI+DTT, SI-induced S-nitrosylated proteins reduced by addition of DTT; GSNO+DTT, NO-donor treated S-nitrosylated proteins reduced by addition of DTT. M, Molecular marker (kDa). Right, Coomassie Blue staining of these S-nitrosylated proteins on SDS-PAGE showing equal loading of proteins.
Soluble Inorganic Pyrophosphatases Are Targets of ROS-Mediated Irreversible Modification during SI and H2O2 Treatment
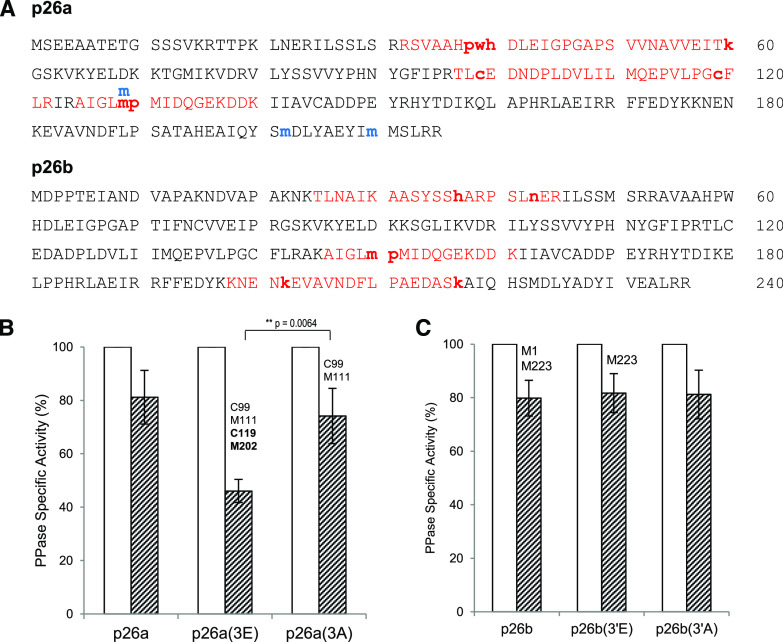
Two proteins that were identified as having oxPTMs after SI induction by LC-MS/MS were the soluble inorganic pyrophosphatases (sPPases) p26.1a/b (referred to here as p26a/p26b). These were previously identified as targets for SI-induced phosphorylation (Rudd et al., 1996; de Graaf et al., 2006; Eaves et al., 2017). Three oxidatively modified peptides from p26a, comprising six oxPTMs, and three from p26b, also comprising six oxPTMs, were identified in SI-induced pollen samples (Supplemental Table S1). Most of the modifications observed in the SI-induced pollen were irreversible; for p26a, Met-129 was irreversibly modified to Met sulfone; Pro-38 and Pro-130 were both irreversibly modified to Glu γ-semialdehyde; Trp-39 was modified to kynurenine, His-40 to 2-oxo-His, and Lys-60 was modified to AASA. Five irreversible oxPTMs were identified on p26b (His-37, 2-oxo-His; Met-150, Met sulfone; Pro-151, Glu γ-semialdehyde; Lys-202 and Lys-217, AASA) and one reversible modification (Asp-43, deamidation; Fig. 4A; Supplemental Table S1). All the modifications identified in the SI-induced samples of p26a were identical to those identified in samples from H2O2-treated pollen, suggesting that they are authentic ROS-stimulated modifications. In untreated pollen, only reversible Met sulfoxide oxPTMs were identified. These data provide good evidence for these p26 sPPases (which play a critical role in modulation of pollen tube growth) as a target of largely irreversible oxidation after SI induction.
Figure 4.
Oxidative modifications identified on the sPPase, p26a/b, and alterations to PPase activity in the p26a(3E) mutant recombinant protein. A, Sequence of the sPPase p26a and p26b from P. rhoeas showing all the peptides (in red) and modifications identified relating to the p26 sPPase from both pollen after SI induction and the recombinant p26 protein. Oxidatively modified amino acids are indicated in bold (small letters); notably all eight were also identified in H2O2-treated samples. Modifications indicated in blue were found in untreated samples. B and C, PPase activities in recombinant p26a (B) and p26b (C) and their phosphomimic/null (3E/A) mutant proteins after treatment with H2O2. Recombinant p26 enzymes were assayed for PPase activity at pH7.2 (white bars) and supplemented with H2O2 (hatched bars). Values for pyrophosphatase (PPase) activity are mean ± sem (n > 3); t test. The phosphomimic protein p26a(3E) was much more sensitive to H2O2 than the wild-type enzyme, displaying significantly lower PPase activity (**P = 0.0064). The oxidative modifications identified on each of these proteins are indicated above the bars; C119 and M202 (indicated in bold typeface) are associated with a drop in sPPase activity.
We examined the possible effects of ROS on p26a/b further, to see if PPase activity might be affected. We had previously made triple phosphomimic mutant recombinant proteins for p26a (S13E, T18E, and S27E, named p26a(3E)) and p26b (T25E, S41E, and S51E, named p26b(3′E)), which mimic the three sites phosphorylated by endogenous pollen kinases during SI and their corresponding phosphonull mutants (p26a(3A) and p26b(3′A)). These phosphomimic mutant proteins exhibited significantly reduced PPase activity in the presence of Ca2+ and/or H2O2 (Eaves et al., 2017). We treated recombinant p26a/b proteins and their mutant forms with H2O2 and then analyzed them for both PPase activity and oxPTMs using LC-MS/MS. The phosphomimic recombinant p26a(3E) protein had reduced PPase activity and contained two unique irreversible oxidative modifications on Cys-119 (cysteic acid) and Met-202 (Met sulfone) that were not found in p26a or the phosphonull p26a(3A) treated with H2O2 (Fig. 4B; Tables 2 and 3; Supplemental Fig. S2; see also Supplemental Tables S8–S11). Two further irreversible oxPTMs were identified (cysteic acid on Cys-99 and Met sulfoxide on Met-111), which were also present on the phosphonull mutant p26a(3A) protein and did not have significantly different PPase activity from the phosphomimic (3E). However, it is plausible that these, when modified in combination with the other oxidized amino acids, Cys-119 and Met-202, may alter function, as Cys-99 and Cys-119 are adjacent to the active site (Cooperman et al., 1992). The phosphomimic protein p26a(3E) was much more sensitive to H2O2 than the wild-type enzyme, displaying significantly lower PPase activity (P = 0.0064; Fig. 4B). By contrast, the phosphonull recombinant p26a(3A) protein did not have significantly different PPase activity from p26a (P = 0.650; Fig. 4B). Irreversible oxidative modifications were also found on the p26b recombinant protein (Met-1 and Met-223, Met sulfone), but no significant alteration in PPase activity was detected in the phosphomimic mutant p26b(3′E) compared to that exhibited by p26b and p26b(3′A) after treatment with H2O2 (not significant, P = 0.852 and 0.966 respectively; Fig. 4C), so these also are unlikely to be involved in modulating PPase activity. These data suggest that the oxidative modifications on the phosphomimic p26a(3E) protein contribute to the reduction in PPase activity.
Table 2. Oxidative modifications identified by LC-MS/MS on the recombinant sPPase protein p26a after H2O2 treatment.
Oxidative modifications identified on the recombinant protein p26a and its phosphomimic mutant p26a(3E) and phosphonull mutant p26a(3A) without and after H2O2 treatment. Bold text indicates irreversible oxPTMs. I, Irreversible; R, reversible. Details of oxidative modifications relating to these experimental data are in Supplemental Tables S8, S9, S10, and S11 and Supplemental Figure S2.
| Residue | Untreated | H2O2-Treated | ||
|---|---|---|---|---|
| p26a | p26a(3E) | p26a(3A) | ||
| C99 | – | – | Cysteic acid (I) | Cysteic acid (I) |
| M111 | – | – | Met sulfone (I) | Met sulfoxide (R) Met sulfone (I) |
| C119 | – | – | Cysteic acid (I) Nitrosyl (R) | – |
| M129 | Met sulfoxide (R) | Met sulfoxide (R) | Met sulfoxide (R) | Met sulfoxide (R) |
| M131 | – | – | Met sulfoxide (R) | Met sulfoxide (R) |
| C145 | – | – | – | Sulfinic acid |
| M202 | – | – | Met sulfoxide (R) Met sulfone (I) | Met sulfoxide (R) |
| M210 | Met sulfoxide (R) | – | Met sulfoxide (R) | Met sulfoxide (R) |
| M211 | Met sulfoxide (R) | – | Met sulfoxide (R) | Met sulfoxide (R) |
Table 3. Oxidative modifications identified by LC-MS/MS on the recombinant sPPase protein p26b after H2O2 treatment.
Oxidative modifications identified on the recombinant proteinp26b and its phosphomimic mutant p26b(3′E) and phosphonull mutant p26b(3′A) without and after H2O2 treatment. Bold text indicates irreversible oxPTMs. I, Irreversible; R, reversible. Details of oxidative modifications relating to these experimental data are in Supplemental Tables S8, S9, S10, and S11 and Supplemental Figure S2
| Residue | Untreated | H2O2-Treated | ||
|---|---|---|---|---|
| p26b | p26b(3′E) | p26b(3′A) | ||
| M1 | Met sulfoxide (R) | Met sulfoxide (R) | Met sulfoxide (R) Met sulfone (I) | Met sulfoxide (R) |
| M150 | – | Met sulfoxide (R) | Met sulfoxide (R) | Met sulfoxide (R) |
| M152 | – | – | Met sufoxide (R) | – |
| M223 | Met sulfoxide (R) | Met sulfoxide (R) Met sulfone (I) | Met sulfoxide (R) Met sulfone (I) | Met sulfoxide (R) Met sulfone (I) |
Cytoskeletal Proteins Are Oxidatively Modified after SI Induction
We identified 30 unique oxidatively modified cytoskeletal protein peptides with 36 different oxidative modifications after SI induction compared to eight peptides with nine different oxPTMs identified in untreated pollen. Notably, these peptides from the SI-induced pollen contained many more irreversible modifications (31/39; Supplemental Table S1) than untreated pollen (1/8; Supplemental Table S3). It is of interest that the H2O2-treated pollen contained 13 identically modified amino acids on actin and tubulin as the SI-induced pollen (Table 1). These data confirm that SI induces a similar ROS response as H2O2 treatment, suggesting these are authentic ROS-mediated events. In addition, three actin binding proteins (ABPs; one profilin and two fimbrins), identified by six different modified peptides containing eight irreversibly modified oxPTMs, were found in the SI-induced sample (Supplemental Table S1). Modification of profilin might alter its affinity for binding to actin filaments or could affect its actin sequestering property. Similarly, modifications to fimbrin could potentially affect its binding to actin and consequently affect actin filament bundling. Thus, oxPTMs to these proteins could potentially impact the organization of the actin cytoskeleton in incompatible pollen. Although previous studies showed that the actin cytoskeleton is a target for ROS signals (Wilkins et al., 2011), these studies were indirect, using ROS scavengers, and we had not previously shown a direct link between increases in H2O2 and formation of actin punctate foci. Having identified many oxPTMs on actin in this study, we examined whether addition of H2O2 might trigger alterations to pollen tube F-actin configuration.
H2O2 Stimulates the Formation of Actin Foci in Pollen Tubes
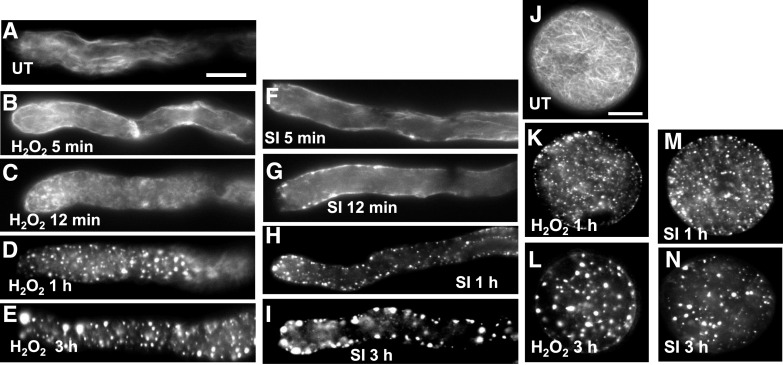
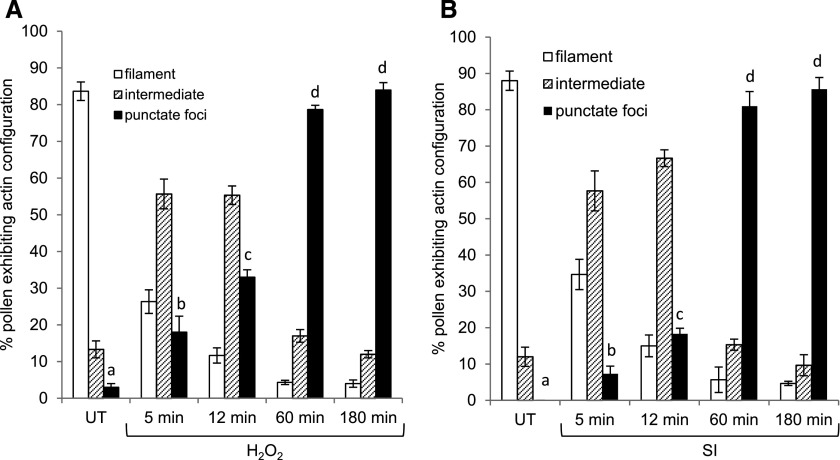
We treated pollen tubes with either H2O2 or recombinant PrsS to induce SI and used rhodamine phalloidin staining to observe the alterations in F-actin configuration. In the untreated pollen tubes (Fig. 5A), F-actin filament bundles were visible. Pollen tubes treated with H2O2 displayed alterations to the F-actin organization as early as 5 min (Fig. 5, B and C); the typical F-actin filament bundles rapidly reduced and the number of pollen tubes displaying punctate actin foci was significantly increased compared to untreated samples (Fig. 6A). After 1 h, small F-actin foci were present (Fig. 5D), and large punctate foci were observed after 3 h of treatment (Fig. 5E); after 1 to 3 h of treatment, ∼ 80% of pollen tubes contained punctate actin foci (Fig. 6A). These alterations triggered by H2O2 appear very similar to those in SI-induced pollen previously observed (Geitmann et al., 2000; Snowman et al., 2002; Poulter et al., 2010; Figs. 5, F–I, and 6B). Nongerminated pollen grains showed a similar response as the pollen tubes (Fig. 5, J–N), showing that these can also respond to ROS. Our data show that ROS can stimulate major changes in actin configuration in pollen that are strikingly similar to those observed during SI. Together with the identification of oxPTMs to actin and associated proteins, this provides further evidence for the involvement of ROS in the formation of SI-stimulated F-actin punctate foci.
Figure 5.
F-actin alterations in pollen are induced by ROS in P. rhoeas pollen tubes. F-actin was visualized with rhodamine-phalloidin using fluorescence microscopy. A, F-actin organization in a representative untreated pollen tube. B to E, H2O2-treated pollen tubes after 5min (B), 12 min (C), 1 h (D), and 3 h (E) of treatment. Alterations were observed as early as 5 to 12 min after treatment. At 1 and 3h large punctate foci of actin were formed. F to I, Pollen tubes at 5 min (F), 12 min (G), 1 h (H), and 3 h (I) after SI induction showed similar alterations to F-actin. J to N, Pollen grains showed similar alterations. J, Untreated pollen grain with F-actin filament bundles; K and L, H2O2-treated pollen grains. M and N, SI-induced pollen grains. Scale bars = 10 μm. SI, SI induction; UT, untreated.
Figure 6.
Quantitation of actin alterations stimulated in P. rhoeas pollen. Pollen tubes were treated with H2O2 (A) or SI induction (SI; B), and samples were fixed at different time points after treatment. F-actin was stained with rhodamine-phalloidin and examined using fluorescence microscopy. The actin configuration was evaluated by placing each pollen tube into one of the three categories according to Snowman et al. (2002): actin filaments only (open bars), foci only (black bars), or intermediate (i.e. filaments and foci; lined bars). Three independent experiments scoring 100 pollen tubes for each treatment expressed as percentage of total. Data are mean ± sem (n = 100). One-way ANOVA followed by Tukey multiple comparison was performed to compare the punctate foci formation across different time points. Different letters represent comparisons where P < 0.05. UT, Untreated.
Increased 20S Proteasomal Activity Is Observed after SI
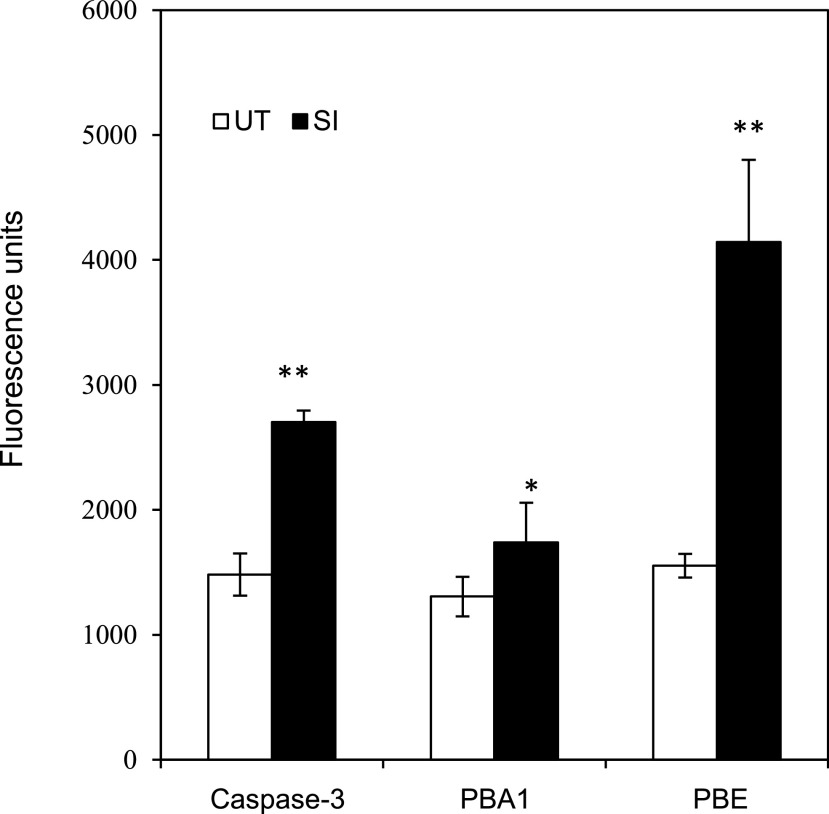
Irreversible oxidation damages proteins. As the 20S proteasome is implicated in removing oxidatively damaged proteins during apoptosis/PCD (Aiken et al., 2011), we investigated whether increased proteasomal activity might be triggered by SI. We characterized the activities of 20S proteasome β-subunits β5 (PBE) and PBA1, during the SI-PCD response, using fluorogenic probes Z-GGL-amc and Ac-nLPnLD-amc as substrates. Five hours after SI, significant increases in both PBA1 and PBE activities were detected (Fig. 7). This provides evidence that the 20S proteasome is activated by SI and could potentially be involved in removal of irreversibly oxidized proteins in incompatible pollen.
Figure 7.
Measurement of various protease activities after SI in P. rhoeas pollen extracts. The 20S proteasomal activities in poppy SI response were measured using fluorogenic peptide substrates in pollen extracts 5 h after SI induction (SI) or in untreated (UT) controls. Caspase-3/DEVDase activity was measured as control. Significant increases of caspase-3/DEVDase, 20S proteasome β-subunit β5 (PBE), and PBA1 subunit activities were observed in the SI extracts (black bars). Mean ± sd, n = 4. *P < 0.05 and **P < 0.01 (Student’s t-test). The actual values of DEVDase, PBA1, and PBE activities are not comparable, because different probes were used.
DISCUSSION
Previously, we showed that SI-induced ROS and NO production are required for pollen tube PCD (Wilkins et al., 2011), but the mechanism was not determined. Both ROS and NO can modify proteins, and we now show that the SI response involves rapid formation of many irreversible oxPTMs and provide evidence that this is linked to altered protein function. Critically, the pattern of oxPTM formation induced by SI overlaps with those induced by exogenous H2O2 and happens sufficiently rapidly (within 12 min) to strongly suggest it is not a consequence of PCD. Irreversible modifications found were Glu y-semialdehyde (from Pro and Arg), aminoadipic acid (AASA from Lys), Met sulfone, kynurenine (from Trp), cysteic acid, and 2-oxo-His. Few reports of rapid irreversible oxidative modifications exist in plants. Moreover, little is known about the functional consequences of these irreversible oxPTMs (Møller et al., 2007; Rinalducci et al., 2008; Jacques et al., 2013, 2015). The reversible modifications Met sulfoxide (Jacques et al., 2013) and S-nitroso-Cys (Astier et al., 2011) were also detected but not sulfenylated cysteines, possibly because our method did not protect these reactive groups during extraction. Protein sulfenylation has been detected in plants following H2O2 treatment by trapping these groups (Waszczak et al., 2014, 2015; Akter et al., 2015b). While it is likely that some of the oxPTMs such as cysteic acid are potentially artifacts formed on unprotected side chains during sample processing, the key point is that more modifications occur in pollen subjected to SI and exogenous H2O2 treatments compared to untreated samples. While Cys sulfenylation and S-nitroso-Cys formation have been implicated as mediators of H2O2 (Smirnoff and Arnaud, 2019) and NO (Astier et al., 2011) signaling, the extent to which irreversible oxPTMs represent damage or have a functional significance is less well understood. Our results provide evidence that rapid production of irreversible oxPTMs is implicated in a physiological response in plants, rather than representing longer-term oxidative damage. However, further work is needed to establish which of these modifications are site specific and to provide a firm link with specific processes occurring during SI. Evidence for a link to inorganic pyrophosphatase and actin is discussed below.
Irreversible modification of proteins is likely to inhibit function, and they can be marked for proteolysis by the proteasome (Grune et al., 1996; Berlett and Stadtman, 1997). Irreversible protein oxidation is particularly detrimental in the cell, as this can render damaged proteins inactive or lead to functional abnormalities. Studies have implicated the 20S proteasome as important for the removal of damaged proteins, as (at least in animal cells) it is more resistant to oxidative stress than the 26S proteasome, maintaining activity even after treatment with moderate to high concentrations of H2O2 (Reinheckel et al., 1998; Aiken et al., 2011; Pajares et al., 2015). Moreover, 20S proteasomes can degrade oxidized proteins in vitro, independent of ubiquitin/ATP (Aiken et al., 2011). We measured a significant increase in 20S proteasomal activity in SI-induced poppy pollen. This is not inconsistent with the idea that protein damage is triggered by SI and that the 20S proteasome may be recruited to degrade oxidatively damaged proteins during the SI response.
It is striking that cytoskeletal proteins and enzymes involved in energy metabolism respond prominently during the SI response. In relation to energy metabolism, a large proportion of enzymes associated with glycolysis, organic acid metabolism, and ATP synthesis/use have oxPTMs. In animal cells, one of the principle targets of protein oxidation is metabolism; evidence suggests that oxidation of a few metabolic enzymes, especially those involved in glycolysis, can dramatically affect the cellular energy status, thereby rapidly inducing cellular dysfunction with a limited number of protein oxidation events. Glyceraldhehyde 3-phosphate dehydrogenase (GAPDH) is one of the best examples of oxidation of a metabolic enzyme having direct control over apoptosis in animal cells (Cecarini et al., 2007; Sirover, 2012; Villa and Ricci, 2016). In yeast (Saccharomyces cerevisiae), oxidative stress inactivates GAPDH, enolase, and aconitase (Cabiscol et al., 2000). Modulation of metabolism resulting in inhibition of glycolysis leads to cell death via ROS-mediated cell death in plants (Kunz et al., 2014). Thus, it is well established that inhibition of glycolysis leads to cell death. It is noteworthy that cytosolic GAPDH from Arabidopsis (Arabidopsis thaliana) was identified as a major H2O2-oxidized protein; reversible Cys oxidation resulted in inhibition of its activity (Hancock et al., 2005; Yang and Zhai, 2017). In plants, there is increasing evidence supporting the idea that plant cytoplasmic GAPDH has alternative, nonmetabolic “moonlighting” functions triggered by oxPTMs of the protein under stress conditions (Zaffagnini et al., 2013). A study using Arabidopsis GAPDH knockout lines displayed accelerated PCD in response to effector-triggered immunity (Henry et al., 2015). Our data provide a mechanistic link between SI, which triggers PCD, and possible protein targets of irreversible oxidation that could result in destruction of metabolism. In animal cells, there is good evidence that during apoptosis, the loss of energy production contributes to the dismantling of the cell. A decrease in ATP content during apoptosis has been shown to be dependent on inhibition of glycolysis, leading to the impairment in the activity of two glycolysis-limiting enzymes, phosphofructokinase and pyruvate kinase (Pradelli et al., 2014). While there is currently limited evidence that the oxPTM modifications observed here specifically cause SI-mediated PCD, the literature suggests that this may be the case, and this possibility should be investigated in future studies.
The sPPase (p26a/p26b) provides an example of an enzyme involved in the SI response (de Graaf et al., 2006; Eaves et al., 2017) that is a target of SI-ROS oxidation, displaying several oxPTMs within a few minutes of SI induction. Previously, we showed that p26a/p26b were phosphorylated following SI, and this reduces PPase enzyme activity (de Graaf et al., 2006); phosphorylation together with Ca2+, ROS, and low pH further inhibited PPase activity (Eaves et al., 2017). Here, we show that H2O2 treatment of the mutant recombinant enzyme p26a(3E) resulted in a reduction in PPase activity. However, we did not observe a differential effect on activity between the different mutant forms of p26, which is expected, as Eaves et al. (2017) showed that PPase activity is reduced and differential only at below pH 7; the PPase assays here were carried out at pH 8.0, which is the optimal activity pH for pyrophosphatases.
Thus, the phosphomimic amino acid substitutions on this enzyme contribute to an increased susceptibility to oxidative modification, resulting in a reduction in PPase activity in vitro. Some of the oxidized residues (Met-111 and Asp-138) are located in regions of the protein that could potentially interfere with the enzyme’s catalytic properties, based on 3D structures of Escherichia coli sPPase (Cooperman et al., 1992). The irreversible modification of Cys residues (Cys-99 and Cys-119) either side of conserved active site residues could affect function. sPPases are enzymes that hydrolyze inorganic pyrophosphate to provide the driving force for many metabolic reactions. Inorganic pyrophosphate is generated during biopolymer synthesis and hydrolyzed to inorganic phosphate; this reaction provides a thermodynamic pull favoring biosynthesis (Kornberg, 1962). In a biological context, phosphorylation of p26a during SI in vivo is rapidly followed by an increase in ROS; this oxidative modification could further reduce PPase activity, which will result in lowering of ATP levels and further impact on cellular energetics. Thus, our data provide insights into a new mechanism whereby PPase activity can be inhibited. Here, we not only show that ROS can contribute to SI by inhibiting a crucial enzyme for biosynthesis, but this provides a significant advance by providing an example of ROS modifying an enzyme to affect its activity. This finding could have implications for many biological systems that involve biosynthesis.
We show that cytoskeletal proteins (both actin and tubulin) and the ABPs fimbrin and profilin are targets of extensive irreversible oxidative modifications. Met residues in actin are commonly oxidized to the irreversible sulfone form, while oxidation of actin methionines has been reported previously (Dalle-Donne et al., 2001). Moreover, oxidation of key Cys residues of actin results in cell death in yeast (Farah and Amberg, 2007). The actin cytoskeleton plays an essential role in pollen tube growth (Gibbon et al., 1999; Vidali et al., 2001) and is implicated in mediating apoptosis in yeast. In yeast, during acute oxidative stress, F-actin forms oxidized actin bodies that sequester actin into immobile, nondynamic structures that regulate the oxidative stress response, playing a pivotal protective role in the decision whether to enter apoptosis (Farah et al., 2011). These oxidized actin bodies appear similar to the highly stable F-actin foci that we observed in SI (Geitmann et al., 2000; Snowman et al., 2002; Poulter et al., 2010) and H2O2-treated pollen (Wilkins et al., 2011). We previously demonstrated that SI-induced ROS and NO production was required for the formation of these distinctive actin structures, which were concomitant with initiation of PCD (Wilkins et al., 2011). Here, we show that H2O2 induces the formation of actin foci. In yeast, it is well established that a decrease in actin dynamics and accumulation of aggregates of stabilized F-actin can induce “actin-mediated apoptosis” involving ROS-mediated apoptosis (Gourlay et al., 2004). The apparent underlying similarities in actin involvement in plant PCD have been commented upon (Franklin-Tong and Gourlay, 2008), and this study reinforces this idea. Together, these data suggest that the oxidation of cytoskeletal proteins observed here may play a key role in SI-PCD in pollen. The role of oxidation in cytoskeletal function in plant cells requires further investigation. Clearly, the cytoskeleton and its associated proteins are an important target during SI, and we have shown that several are oxidatively modified. These modifications may affect cytoskeletal dynamics, as several irreversible modifications occur in the binding domain of actin, which would restrict actin or ABPs to bind with actin and thus might alter actin dynamics.
We identified several S-nitrosylated proteins in the SI-induced pollen samples. The majority of NO affected proteins appear to be modified by S-nitrosylation of the thiol group of a single Cys residue. To date, around 20 different S-nitrosylated proteins have been characterized in detail in plants, and most of them have been reviewed recently with regard to their functional significance in NO signaling (Astier et al., 2011; Lamotte et al., 2015). The identified proteins from plant proteome-wide studies have been shown to take part in major cellular activities, notably primary and secondary metabolism, photosynthesis, protein folding, cellular architecture, and stress responses (Astier et al., 2011). It is thought that NO signaling in plants uses S-nitrosylation of Cys residues of redox-sensitive proteins (Wang et al., 2006; Moreau et al., 2010), which can affect protein activity and so has the potential to be important in regulating cellular events (Lindermayr et al., 2005; Couturier et al., 2013). The phosphomimic mutant recombinant protein p26a(3E) sPPase was not only irreversibly oxidized on Cys-119 to cysteic acid but was also nitrosylated on this site. As this modified protein had significantly reduced PPase activity, it suggests oxidation may play a role.
In conclusion, we have shown that oxidation is an important mechanism triggered by the SI response in P. rhoeas pollen. Here, we have shown that the SI response results in rapid and extensive oxidation of pollen proteins. Strikingly, many of these oxPTMs are irreversible. We provide evidence for increased proteasomal activation, which is consistent with the idea that, following inactivation, oxidized proteins may be removed by the 20S proteasome. The observed oxidative modifications particularly impact enzymes associated with energy production and the cytoskeleton. In some cases (GAPDH and sPPase here), there is evidence that such irreversible modifications inhibit critical core metabolic enzyme activity. These modifications could therefore contribute to the very rapid growth inhibition and PCD following induction of SI. We also show that actin is a target for extensive irreversible oxidation and that oxidation stimulates the formation of stable actin foci in pollen. Actin dynamics have previously been implicated in the decision whether to enter PCD, and this study further suggests that this is the case. Together, our data demonstrate irreversible oxidation of key pollen proteins and suggest that this triggers a catastrophic reduction in cellular activity that could lead to PCD.
MATERIALS AND METHODS
Pollen Tube Growth, SI Induction, and Other Treatments
Common poppy (Papaver rhoeas) pollen was hydrated then grown in vitro in liquid germination medium [0.01% (w/v) H3BO3, 0.01% (w/v) KNO3, 0.01% (w/v) Mg(NO3)20.6H2O, 0.036% (w/v) CaCl2-2H2O, and 13.5% (w/v) Suc] at 25°C for 1 h (Snowman et al., 2002). SI was induced by adding incompatible recombinant S proteins (final concentration 10 μg mL−1) as described previously (Snowman et al., 2002). Samples were taken at 12 min after SI induction. For each SI-induced sample, a noninduced control was prepared by adding only germination medium to the pollen. For H2O2 treatments, germinated pollen tubes were treated with H2O2 (2.5 mm) for 12 min, as this was when ROS increases were detected in incompatible pollen tubes (Wilkins et al., 2011). Two biological replicates were analyzed for the SI treatment and three replicates for the H2O2 treatment and the noninduced control. We did not look for presence of unmodified proteins/peptides, but they were present in all samples (Supplemental Tables S2, S4, and S6). Thus, only modified peptides observed in two replicates were considered for the SI samples. We made comparisons between the treatments and discarded any modifications that were observed in the untreated samples; this gives confidence that they are stimulated by that particular treatment. Thus, we do not comment on the number of unmodified peptides here, but on differences between modifications occurring. Pollen was harvested by centrifuging, resuspended in HEN buffer (250 mm HEPES/pH 7.7, 1 mm EDTA, 0.1 mm neocuproine), homogenized on ice, and clarified by centrifugation. The protein content of the supernatant was determined by Bradford assay (Bradford, 1976), which was stored at −20°C until required.
To generate S-nitrosylated proteins for western blots, germinated pollen was treated with 500 µm NO donor GSNO for ∼30 min. Proteins (60 µg) were extracted as described above except trypsin digests were performed without DTT. Peptides were adjusted to 3 µg µL−1 in HEN buffer. Thiols were blocked on tryptic peptides using 0.2% (w/v) S-methyl methane thiosulfonate and 2.5% (w/v) SDS and incubated for 20 min at 50°C then equilibrated in HEN buffer using Spin 6 columns (Bio-Rad) according to manufacturer’s instructions.
Detection of S-Nitrosylation of Proteins by Western Blot
Protein extracts were prepared as described above and separated by SDS-PAGE. Proteins containing S-nitrosylated Cys were selectively labeled using iodoTMTzero. S-nitrosylated proteins were visualized by western blotting using anti-TMT antibody using a Pierce S-nitrosylation western blot kit according to the manufacturer’s instructions. Fifty millimolars DTT was added to controls during protein extraction.
Sample Preparation for Mass Spectrometry
Sample pollen proteins (60 µg) were run into SDS-PAGE, and gel plugs containing the proteins were digested using trypsin gold (Promega) according to manufacturer’s instructions. Ten millimolars DTT in 100 mm ammonium bicarbonate (pH 8) was added to the protein and incubated for 30 min at 56°C. Samples were cooled to room temperature and alkylated with 50 mm iodoacetamide in the dark for 30 min. Samples were desalted using ZipTipC18 (Merck Millipore). Tips were prewet in 100% (v/v) acetonitrile and rinsed in 2 × 10 µL 0.1% (v/v) trifluoroacetic acid. Samples were loaded according to manufacturer’s instructions. ZipTip were washed with 0.1% (v/v) trifluoroacetic acid (3 × 10 µL) to remove excess salts. Peptides were eluted with 10 µL of 50% (v/v) acetonitrile/0.1% (v/v) trifluoroacetic acid. Samples were dried down to remove the acetonitrile and resuspended in 0.1% (v/v) formic acid solution. Chemicals were from Sigma, Fisher Scientific, and J.T. Baker.
LC-MS/MS Analysis of Peptides
Reversed-phase chromatography was performed to separate tryptic peptides prior the mass spectrometric analysis using an UltiMate 3000 HPLCnano series (Thermo Scientific Dionex) system. Samples were analyzed with two columns, an Acclaim PepMap µ-precolumn cartridge 300 µm inner diameter. × 5 mm, 5 μm, 100 Å, and an analytical column Acclaim PepMap rapid separation liquid chromatography, 75 µm inner diameter × 15 cm, 2 µm, 100 Å (Thermo Scientific Dionex). Mobile phase buffer A was composed of 0.1% (v/v) formic acid in water and mobile phase B 0.1% (v/v) formic acid in acetonitrile. Samples were loaded onto the µ-precolumn, and peptides were eluted onto the analytical column at 300 nL min−1 by increasing the mobile phase B from 3% to 44% over 40 min then to 90% B over 5 min, followed by a 15 min re-equilibration at 3% B. Eluted peptides were converted to gas-phase ions by means of electrospray ionization via a Triversa Nanomate nanospray source (Advion Biosciences) and introduced into a LTQ Orbitrap Velos ETD mass spectrometer (Thermo Fisher Scientific). Survey scans of peptide precursors from 380 to 1600 mass-to-charge ratio (m/z) were performed at 60 K resolution (at 400 m/z) with automatic gain control 1 × 106. Precursor ions with charge state 2 to 4 were analyzed by collision-induced dissociation fragmentation in the ion trap. MS/MS analysis was performed using collision energy 35, automatic gain control 5 × 104, max injection time 100 ms, and isolation width 2 m/z. A dynamic exclusion duration of 30 s was used to select the monoisotopic peaks.
Collision-induced dissociation MS/MS data were searched using the SEQUEST algorithm (Thermo Fisher Scientific). As the complete and annotated genome sequence for P. rhoeas is not currently available, the identifications were limited to peptides identical to those found in other green plants or the few sequences of P. rhoeas submitted to the European Molecular Biology Laboratory by our laboratory, namely Pr-p26.1a, Pr-p26.1b, and MAPK. It was searched for the following modifications: deamidation (N and Q); carbaminomethylation, sulfenic acid, sulfinic acid, cysteic acid, and S-nitro-Cys (C); Met sulfoxide, Met sulfone (M); 2-oxo-His (H); Glu γ-semialdehyde (P); AASA (K); kynurenine (W); and phosphorylation (S, T, and Y). Two missed cleavages were allowed, with precursor mass tolerance of 10 ppm and MS/MS mass tolerance 0.8 D. The false discovery rate was set to 1% at the protein, peptide, and PSM level. The searches for oxidative modifications were conducted as variable modifications; we had to set two different searches, as a maximum of only six modifications could be set for each search. The criteria for “real hit proteins” were accepted as those containing at least two high-confidence peptides. Peptides were analyzed to identify irreversible and reversible oxidative modifications to amino acids. Peptide coverage of beta and gamma ions relating to data in Supplemental Tables S1, S3, and S5 is shown in Supplemental Table S7. Excel files relating to oxidative modifications listed in Supplemental Table S1 (SI treatment), Supplemental Table S3 (untreated samples), and Supplemental Table S5 (H2O2 treatment) are provided as Supplemental Tables S2, S4, and S6, respectively. Supplemental data supporting this research (*.raw files) are uploaded and available under accession number MassIVE MSV000085216, accessible though the publicly available URL ftp://massive.ucsd.edu/MSV000085216/.
For the counts, we disregarded the carbamidomethyl modifications, as these are an artifact of iodoacetamide treatment. However, these modifications are shown in Supplemental Tables S1 to S3 for clarity. We did not count any oxPTMs that were detected in the treated samples if they also occurred in untreated samples, as these were assumed to be modifications that were “normal.” We also counted the number of unique amino acid modifications to oxidatively modified peptides and grouped these according to protein function using the PANTHER classification system, https://www.ncbi.nlm.nih.gov/pubmed/27899595 (Mi et al., 2017); again, any that overlapped with untreated samples were not counted. Where a particular identification (GI number) was not in the PANTHER database, we based the protein identify and protein class according to its classification in NCBI, either directly (the same ID) or through the identification of similar proteins by BLAST searches and/or the PANTHER protein class for a very similar protein. The remaining proteins were labeled “unclassified” but were placed in a functional class if this was obvious from the protein identified. The experiments were all done in replicate and the peptides listed in Supplemental Tables S1 to S3, and figures are all high probability peptide hits, with at least two high-probability peptides identified from each protein, detected in at least two of the n = 2, n = 3 independent biological replicates. Although we did not look for presence of unmodified proteins/peptides, they were present in all samples. However, as none of the modifications listed were observed in the untreated samples, this provides confidence that they are stimulated by that treatment.
p26 Analysis and PPase Assays
Recombinant His-tagged p26 sPPase proteins (p26.1a and p26.1b) and their triple substitution phospho-mutant versions are as follows: phosphomimic with a Glu (E) substitution [p26a/b(3E)] and the corresponding phosphonull with an Ala [A] substitution (p26a/b(3A)) were for p26a (S13E, T18E, and S27E, named p26a(3E)) and p26b (T25E, S41E, and S51E, named p26b(3′E)). They were prepared as described previously (Eaves et al., 2017). The p26 protein was diluted to 10 µm in 50 mm HEPES-KOH, pH 8.0, 50 µm EGTA, 2 mm MgCl2. Two hundred fifty nanogram aliquots were assayed for free phosphate production using a discontinuous PPase assay and 2 mm sodium pyrophosphate as substrate (Fiske and Subbarow, 1925); n > 3 for each assay. The assay buffer was supplemented with 10 mm H2O2 as appropriate. Duplicate assay samples were sent for LC-MS/MS analysis. Excel files relating to oxidative modifications relating to p26 analysis listed in Tables 2 and 3 are provided as Supplemental Tables S8 to S11. Supplemental data supporting this research (*.raw files) are uploaded and available in the MassIVE repository under accession number MassIVE MSV000085216, accessible though the publicly available URL ftp://massive.ucsd.edu/MSV000085216/.
Poppy Pollen Protein Extractions for Proteasome and Caspase Assays
P. rhoeas pollen was collected and snap-frozen in liquid nitrogen. Protein extracts were prepared by grinding pollen using a glass homogenizer in proteasome assay buffer (50 mm Tris-HCl, pH 7.5; 5 mm MgCl2; 250 mm Suc; 1 mm DTT; 0.05 mg mL−1 bovine serum albumin). ATP was freshly added to the buffer to a final concentration of 5 mm before use (Kisselev and Goldberg, 2005). Lysates were sonicated at 10,000 A for 2 × 5 s, incubated on ice for 20 min and centrifuged at 13,200 rpm at 4°C for 20 min. The supernatant was collected and protein concentration was determined by the Bradford assay. Protein extracts were aliquoted and stored at −20°C for use in the proteasome activity assays. Protein samples for caspase activity assay were extracted using caspase extraction buffer (50 mm Na-acetate; 10 mm l-Cys; 10% (v/v) glycerol; 0.1% (w/v) CHAPS; pH 6.0; Bosch and Franklin-Tong, 2007).
Proteasome and Caspase Activity Assays using Fluorogenic Peptide Substrates
Each activity assay (100 μL) contained 10 μg protein lysates and 100 μm of either Z-GGL-amc or Ac-nLPnLD-amc as fluorogenic probes for PBE and PBA1 (both 20S proteasome subunits) activity measurements, respectively. Fluorescence was monitored with the excitation at 380 nm and emission at 460 nm every 10 min over a period of 4 h using a time-resolved fluorescence plate reader (FLUOstar OPTIMA; BMG LABTECH).
Caspase activity was assayed in caspase activity assay buffer (50 mm Na-acetate; 10 mm l-Cys; 10% [v/v] glycerol; 0.1% [w/v] CHAPS; pH 5.0). Each activity assay (100 μL) contained 10 μg protein lysates and 100 μm fluorogenic probes Ac-DEVD-amc. Caspase activity was monitored in the plate reader as described (Bosch and Franklin-Tong, 2007).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Annotated spectra for representative examples of each oxidative modification identified in Supplemental Tables S1, S3, and S5.
Supplemental Figure S2. Annotated spectra for oxidative modifications identified on the p26 recombinant proteins and their mutant variants.
Supplemental Table S1. Summary of oxidative modifications to pollen proteins after SI induction.
Supplemental Table S2. Excel file relating to oxidative modifications listed in Supplemental Table S1.
Supplemental Table S3. Summary of oxidative modifications found to proteins from untreated pollen.
Supplemental Table S4. Excel file relating to oxidative modifications listed in Supplemental Table S3.
Supplemental Table S5. Summary of oxidative modification to pollen proteins after treatment with H2O2.
Supplemental Table S6. Excel file relating to oxidative modifications listed in Supplemental Table S5.
Supplemental Table S7. Peptide coverage of beta and gamma ions relating to data in Supplemental Tables S1, S2, and S3.
Supplemental Table S8. Oxidative modifications in untreated (UT) sPPase p26b.
Supplemental Table S9. Oxidative modifications to p26a after H2O2 treatment.
Supplemental Table S10. Oxidative modifications to the phosphomimic p26 mutant p26b(3E) after H2O2 treatment.
Supplemental Table S11. Oxidative modifications to the phosphonull p26 mutant p26a(3A) after H2O2 treatment.
Footnotes
This work was supported by the Engineering and Physical Sciences Research Council (EPSRC; grant no. EP/L023490/1 to H.J.C.); the Biotechnology and Biological Sciences Research Council (BBSRC; grant nos. BB/I020004/1 and BB/N001311/1 to N.S, grant no. BB/P005489/1 to M.B. and V.E.F.-T., and grant no. BB/G003149/1); a Commonwealth PhD studentship to T.H.; and a PhD studentship from the China Scholarship Council (CSC; to Z.L.). The Advion Triversa Nanomate and Thermo Fisher Orbitrap Velos mass spectrometer used in this research were funded through the Birmingham Science City Translational Medicine: Experimental Medicine Network of Excellence project, with support from Advantage West Midlands (AWM).
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license
References
- Aiken CT, Kaake RM, Wang X, Huang L(2011) Oxidative stress-mediated regulation of proteasome complexes. Mol Cell Proteomics 10: R110.006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akter S, Huang J, Bodra N, De Smet B, Wahni K, Rombaut D, Pauwels J, Gevaert K, Carroll K, Van Breusegem F, et al. (2015a) DYn-2 based identification of Arabidopsis sulfenomes. Mol Cell Proteomics 14: 1183–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akter S, Huang J, Waszczak C, Jacques S, Gevaert K, Van Breusegem F, Messens J(2015b) Cysteines under ROS attack in plants: A proteomics view. J Exp Bot 66: 2935–2944 [DOI] [PubMed] [Google Scholar]
- Astier J, Rasul S, Koen E, Manzoor H, Besson-Bard A, Lamotte O, Jeandroz S, Durner J, Lindermayr C, Wendehenne D(2011) S-nitrosylation: An emerging post-translational protein modification in plants. Plant Sci 181: 527–533 [DOI] [PubMed] [Google Scholar]
- Berlett BS, Stadtman ER(1997) Protein oxidation in aging, disease, and oxidative stress. J Biol Chem 272: 20313–20316 [DOI] [PubMed] [Google Scholar]
- Bosch M, Franklin-Tong VE(2007) Temporal and spatial activation of caspase-like enzymes induced by self-incompatibility in Papaver pollen. Proc Natl Acad Sci USA 104: 18327–18332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM.(1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Cabiscol E, Piulats E, Echave P, Herrero E, Ros J(2000) Oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. J Biol Chem 275: 27393–27398 [DOI] [PubMed] [Google Scholar]
- Cecarini V, Gee J, Fioretti E, Amici M, Angeletti M, Eleuteri AM, Keller JN(2007) Protein oxidation and cellular homeostasis: Emphasis on metabolism. Biochim Biophys Acta 1773: 93–104 [DOI] [PubMed] [Google Scholar]
- Chai L, Tudor RL, Poulter NS, Wilkins KA, Eaves DJ, Franklin CH, Franklin-Tong VE(2017) MAP kinase PrMPK9-1 contributes to the self-incompatibility response. Plant Physiol 174: 1226–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooperman BS, Baykov AA, Lahti R(1992) Evolutionary conservation of the active site of soluble inorganic pyrophosphatase. Trends Biochem Sci 17: 262–266 [DOI] [PubMed] [Google Scholar]
- Couturier J, Chibani K, Jacquot J-P, Rouhier N(2013) Cysteine-based redox regulation and signaling in plants. Front Plant Sci 4: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle-Donne I, Rossi R, Milzani A, Di Simplicio P, Colombo R(2001) The actin cytoskeleton response to oxidants: From small heat shock protein phosphorylation to changes in the redox state of actin itself. Free Radic Biol Med 31: 1624–1632 [DOI] [PubMed] [Google Scholar]
- de Graaf BHJ, Rudd JJ, Wheeler MJ, Perry RM, Bell EM, Osman K, Franklin FCH, Franklin-Tong VE(2006) Self-incompatibility in Papaver targets soluble inorganic pyrophosphatases in pollen. Nature 444: 490–493 [DOI] [PubMed] [Google Scholar]
- Eaves DJ, Haque T, Tudor RL, Barron Y, Zampronio CG, Cotton NPJ, de Graaf BHJ, White SA, Cooper HJ, Franklin FCH, et al. (2017) Identification of phosphorylation sites altering pollen soluble inorganic pyrophosphatase activity. Plant Physiol 173: 1606–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah ME, Amberg DC(2007) Conserved actin cysteine residues are oxidative stress sensors that can regulate cell death in yeast. Mol Biol Cell 18: 1359–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah ME, Sirotkin V, Haarer B, Kakhniashvili D, Amberg DC(2011) Diverse protective roles of the actin cytoskeleton during oxidative stress. Cytoskeleton (Hoboken) 68: 340–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiske CH, Subbarow Y(1925) The colorimetric determination of phophorous. J Biol Chem 66: 375–400 [Google Scholar]
- Foote HCC, Ride JP, Franklin-Tong VE, Walker EA, Lawrence MJ, Franklin FCH(1994) Cloning and expression of a distinctive class of self-incompatibility (S) gene from Papaver rhoeas L. Proc Natl Acad Sci USA 91: 2265–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin-Tong VE, Gourlay CW(2008) A role for actin in regulating apoptosis/programmed cell death: Evidence spanning yeast, plants and animals. Biochem J 413: 389–404 [DOI] [PubMed] [Google Scholar]
- Geitmann A, Snowman BN, Emons AMC, Franklin-Tong VE(2000) Alterations in the actin cytoskeleton of pollen tubes are induced by the self-incompatibility reaction in Papaver rhoeas. Plant Cell 12: 1239–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbon BC, Kovar DR, Staiger CJ(1999) Latrunculin B has different effects on pollen germination and tube growth. Plant Cell 11: 2349–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourlay CW, Carpp LN, Timpson P, Winder SJ, Ayscough KR(2004) A role for the actin cytoskeleton in cell death and aging in yeast. J Cell Biol 164: 803–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grune T, Reinheckel T, Davies KJ(1996) Degradation of oxidized proteins in K562 human hematopoietic cells by proteasome. J Biol Chem 271: 15504–15509 [DOI] [PubMed] [Google Scholar]
- Hancock JT, Henson D, Nyirenda M, Desikan R, Harrison J, Lewis M, Hughes J, Neill SJ(2005) Proteomic identification of glyceraldehyde 3-phosphate dehydrogenase as an inhibitory target of hydrogen peroxide in Arabidopsis. Plant Physiol Biochem 43: 828–835 [DOI] [PubMed] [Google Scholar]
- Henry E, Fung N, Liu J, Drakakaki G, Coaker G(2015) Beyond glycolysis: GAPDHs are multi-functional enzymes involved in regulation of ROS, autophagy, and plant immune responses. PLoS Genet 11: e1005199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques S, Ghesquière B, De Bock P-J, Demol H, Wahni K, Willems P, Messens J, Van Breusegem F, Gevaert K(2015) Protein methionine sulfoxide dynamics in Arabidopsis thaliana under oxidative stress. Mol Cell Proteomics 14: 1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques S, Ghesquière B, Van Breusegem F, Gevaert K(2013) Plant proteins under oxidative attack. Proteomics 13: 932–940 [DOI] [PubMed] [Google Scholar]
- Kisselev AF, Goldberg AL(2005) Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Methods Enzymol 398: 364–378 [DOI] [PubMed] [Google Scholar]
- Kornberg A.(1962) On the metabolic significance of phosphorolytic and pyrophosphorolytic reactions In Kasha M, and Pullman D, eds, Horizons in Biochemistry. Academic Press, New York, pp 251–264 [Google Scholar]
- Kunz S, Pesquet E, Kleczkowski LA(2014) Functional dissection of sugar signals affecting gene expression in Arabidopsis thaliana. PLoS One 9: e100312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamotte O, Bertoldo JB, Besson-Bard A, Rosnoblet C, Aimé S, Hichami S, Terenzi H, Wendehenne D(2015) Protein S-nitrosylation: Specificity and identification strategies in plants. Front Chem 2: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Samaj J, Franklin-Tong VE(2007) A mitogen-activated protein kinase signals to programmed cell death induced by self-incompatibility in Papaver pollen. Plant Physiol 145: 236–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C, Saalbach G, Durner J(2005) Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol 137: 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD(2017) PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45(D1): D183–D189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller IM, Jensen PE, Hansson A(2007) Oxidative modifications to cellular components in plants. Annu Rev Plant Biol 58: 459–481 [DOI] [PubMed] [Google Scholar]
- Moreau M, Lindermayr C, Durner J, Klessig DF(2010) NO synthesis and signaling in plants—where do we stand? Physiol Plant 138: 372–383 [DOI] [PubMed] [Google Scholar]
- Pajares M, Jiménez-Moreno N, Dias IHK, Debelec B, Vucetic M, Fladmark KE, Basaga H, Ribaric S, Milisav I, Cuadrado A(2015) Redox control of protein degradation. Redox Biol 6: 409–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulter NS, Staiger CJ, Rappoport JZ, Franklin-Tong VE(2010) Actin-binding proteins implicated in formation of the punctate actin foci stimulated by the self-incompatibility response in Papaver. Plant Physiol 152: 1274–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradelli LA, Villa E, Zunino B, Marchetti S, Ricci JE(2014) Glucose metabolism is inhibited by caspases upon the induction of apoptosis. Cell Death Dis 5: e1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinheckel T, Sitte N, Ullrich O, Kuckelkorn U, Davies KJ, Grune T(1998) Comparative resistance of the 20S and 26S proteasome to oxidative stress. Biochem J 335: 637–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinalducci S, Murgiano L, Zolla L(2008) Redox proteomics: Basic principles and future perspectives for the detection of protein oxidation in plants. J Exp Bot 59: 3781–3801 [DOI] [PubMed] [Google Scholar]
- Rudd JJ, Franklin F, Lord JM, Franklin-Tong VE(1996) Increased phosphorylation of a 26-kD pollen protein is induced by the self-incompatibility response in Papaver rhoeas. Plant Cell 8: 713–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd JJ, Osman K, Franklin FCH, Franklin-Tong VE(2003) Activation of a putative MAP kinase in pollen is stimulated by the self-incompatibility (SI) response. FEBS Lett 547: 223–227 [DOI] [PubMed] [Google Scholar]
- Sirover MA.(2012) Subcellular dynamics of multifunctional protein regulation: mechanisms of GAPDH intracellular translocation. J Cell Biochem 113: 2193–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnoff N, Arnaud D(2019) Hydrogen peroxide metabolism and functions in plants. New Phytol 221: 1197–1214 [DOI] [PubMed] [Google Scholar]
- Snowman BN, Kovar DR, Shevchenko G, Franklin-Tong VE, Staiger CJ(2002) Signal-mediated depolymerization of actin in pollen during the self-incompatibility response. Plant Cell 14: 2613–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L, McKenna ST, Hepler PK(2001) Actin polymerization is essential for pollen tube growth. Mol Biol Cell 12: 2534–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa E, Ricci J-E(2016) How does metabolism affect cell death in cancer? FEBS J 283: 2653–2660 [DOI] [PubMed] [Google Scholar]
- Wang Y, Yun B-W, Kwon E, Hong JK, Yoon J, Loake GJ(2006) S-nitrosylation: An emerging redox-based post-translational modification in plants. J Exp Bot 57: 1777–1784 [DOI] [PubMed] [Google Scholar]
- Waszczak C, Akter S, Eeckhout D, Persiau G, Wahni K, Bodra N, Van Molle I, De Smet B, Vertommen D, Gevaert K, et al. (2014) Sulfenome mining in Arabidopsis thaliana. Proc Natl Acad Sci USA 111: 11545–11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waszczak C, Akter S, Jacques S, Huang J, Messens J, Van Breusegem F(2015) Oxidative post-translational modifications of cysteine residues in plant signal transduction. J Exp Bot 66: 2923–2934 [DOI] [PubMed] [Google Scholar]
- Wheeler MJ, de Graaf BHJ, Hadjiosif N, Perry RM, Poulter NS, Osman K, Vatovec S, Harper A, Franklin FCH, Franklin-Tong VE(2009) Identification of the pollen self-incompatibility determinant in Papaver rhoeas. Nature 459: 992–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins KA, Bancroft J, Bosch M, Ings J, Smirnoff N, Franklin-Tong VE(2011) Reactive oxygen species and nitric oxide mediate actin reorganization and programmed cell death in the self-incompatibility response of papaver. Plant Physiol 156: 404–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SS, Zhai QH(2017) Cytosolic GAPDH: A key mediator in redox signal transduction in plants. Biol Plant 61: 417–426 [Google Scholar]
- Zaffagnini M, Fermani S, Costa A, Lemaire SD, Trost P(2013) Plant cytoplasmic GAPDH: Redox post-translational modifications and moonlighting properties. Front Plant Sci 4: 450. [DOI] [PMC free article] [PubMed] [Google Scholar]