Light and abscisic acid pathways antagonistically regulate greening of seedlings by regulating the transcriptional activity of FHY3.
Abstract
The greening of etiolated seedlings is crucial for the growth and survival of plants. After reaching the soil surface and sunlight, etiolated seedlings integrate numerous environmental signals and internal cues to control the initiation and rate of greening thus to improve their survival and adaption. However, the underlying regulatory mechanisms by which light and phytohormones, such as abscisic acid (ABA), coordinately regulate greening of the etiolated seedlings is still unknown. In this study, we showed that Arabidopsis (Arabidopsis thaliana) DE-ETIOLATED1 (DET1), a key negative regulator of photomorphogenesis, positively regulated light-induced greening by repressing ABA responses. Upon irradiating etiolated seedlings with light, DET1 physically interacts with FAR-RED ELONGATED HYPOCOTYL3 (FHY3) and subsequently associates to the promoter region of the FHY3 direct downstream target ABA INSENSITIVE5 (ABI5). Further, DET1 recruits HISTONE DEACETYLASE6 to the locus of the ABI5 promoter and reduces the enrichments of H3K27ac and H3K4me3 modification, thus subsequently repressing ABI5 expression and promoting the greening of etiolated seedlings. This study reveals the physiological and molecular function of DET1 and FHY3 in the greening of seedlings and provides insights into the regulatory mechanism by which plants integrate light and ABA signals to fine-tune early seedling establishment.
Light serves as one of the most important environmental signals to modulate diverse aspects of plant growth and development. In most species, when a seed germinates in dark conditions (e.g. in soil), it exhibits an etiolated development, including an elongated hypocotyl, folded apical hook, and tightly closed cotyledons without chlorophyll biosynthesis. Once the etiolated seedling reaches the soil surface and perceives sunlight, light-induced photomorphogenesis is initiated, including inhibiting the elongation of the hypocotyl, opening the tightly closed cotyledons, and activating chlorophyll production, so the seedling turns green (Gommers and Monte, 2018). In natural conditions, when the etiolated seedlings grow close to or out of the soil surface, the dramatic environmental changes, including reduced moisture, increased temperature, and light intensity, all could affect photomorphogenesis. These external environmental changes mostly act through internal auxin, ethylene, cytokinin, abscisic acid (ABA), and salicylic acid signaling pathways to fine-tune the photomorphogenesis of etiolated seedlings and subsequently increase the acclimatization and survival of plants (Zhong et al., 2009; Guan et al., 2014; Abbas et al., 2015; Riber et al., 2015; Zhang et al., 2016, 2018; Xu et al., 2018; Yang et al., 2018; Huang et al., 2020). Although ABA has been known to play an essential role in seed germination and early seedling establishment (Chen et al., 2008, 2020; Tang et al., 2013; Fernando and Schroeder, 2015), the regulatory mechanisms by which light and ABA coordinately regulate the greening of the etiolated seedlings remain largely unknown.
The Arabidopsis (Arabidopsis thaliana) transcription factor FAR-RED ELONGATED HYPOCOTYL3 (FHY3) was originally identified in the phytochrome A-mediated far-red light signaling pathway (Lin et al., 2007; Wang and Wang, 2015; Ma and Li, 2018). FHY3 also acts as a crucial regulator of multiple cellular processes, including the circadian clock, chloroplast division, chlorophyll biosynthesis, ABA and stress responses, oxidative stress and cell death, shade avoidance responses, leaf senescence, and plant defense (Li et al., 2011; Ouyang et al., 2011; Tang et al., 2012, 2013; Ma et al., 2016, 2017, 2019; Liu et al., 2019, 2020; Tian et al., 2020). FHY3 positively regulates chlorophyll biosynthesis by activating the transcription of HEMB1, which encodes 5-aminolevulinic acid dehydratase, and mutation of FHY3 decreased the accumulation of Pchlide and subsequently promoted the greening of etiolated seedlings (Tang et al., 2012). Moreover, FHY3 positively regulates ABA and stress responses by directly activating the transcription of ABA INSENSITIVE5 (ABI5), which encodes a crucial transcription factor in the ABA signaling pathway (Finkelstein and Lynch, 2000; Tang et al., 2013). Mutation of ABI5 or reduction of its protein abundance promoted seedling greening and early seedling establishment, indicating its important regulatory roles in these processes (Lopez-Molina et al., 2001; Guan et al., 2014). However, the underlying regulatory mechanisms by which FHY3 and its direct target ABI5 mediate light and ABA signals to regulate the greening of etiolated seedlings remain unknown.
Arabidopsis DE-ETIOLATED1 (DET1) is a key negative regulator in photomorphogenesis. In Arabidopsis, the null allele of det1 is lethal, and the weak allele det1-1 exhibits a continuous photomorphogenesis phenotype in darkness (Pepper et al., 1994). DET1 interacts with DAMAGED DNA BINDING PROTEIN1 (DDB1) and CONSTITUTIVE PHOTOMORPHOGENIC10 (COP10), forming the COP10–DET1–DDB1 (CDD) complex and facilitating the protein degradation of the positive regulators of photomorphogenesis (e.g. ELONGATED HYPOCOTYL5), thus repressing photomorphogenesis in darkness (Yanagawa et al., 2004). Meanwhile, DET1 mediates the stabilization of PHYTOCHROME-INTERACTING FACTORS (PIFs, including PIF1, PIF3, PIF4, and PIF5), and thus promotes the elongation of the etiolated seedling (Dong et al., 2014). Besides its key negative role in light signal transduction, the CDD complex negatively regulates ABA responses. The CDD complex interacts with DET1-, DDB1-ASSOCIATED1, which acts as a substrate adaptor to promote the degradation of ABA receptors (e.g. PYRABACTIN RESISTANCE 1-LIKE8), thus, to negatively regulate ABA signal transduction (Irigoyen et al., 2014). Meanwhile, DET1 affects the transcription of a set of dark-to-light transition-related genes by controlling the monoubiquitination of histone 2B or other modifications (Nassrallah et al., 2018). Although DET1 has been shown to play essential roles in light and ABA signaling, the molecular mechanisms by which DET1 integrates light and ABA signals to regulate greening of seedlings remain unknown.
In this study, we found that although det1-1 plants exhibit a constitutive photomorphogenic phenotype, their light-induced greening of seedlings is significantly inhibited, especially in ABA-treated seedlings. Further, we showed that DET1 interacted with FHY3 and repressed its transcriptional activation to ABI5. Finally, we demonstrated that DET1 recruits HISTONE DEACETYLASE6 (HDA6) to the promoter region of ABI5 and subsequently represses the FHY3-mediated transcriptional activation of ABI5 in light- or ABA-treated seedlings. Our finding identified the molecular regulatory mechanisms by which light and ABA coordinately regulate greening of seedlings and ABA responses through DET1, HDA6, and FHY3.
RESULTS
ABA Inhibits Light-Induced Greening of Etiolated Seedlings through DET1
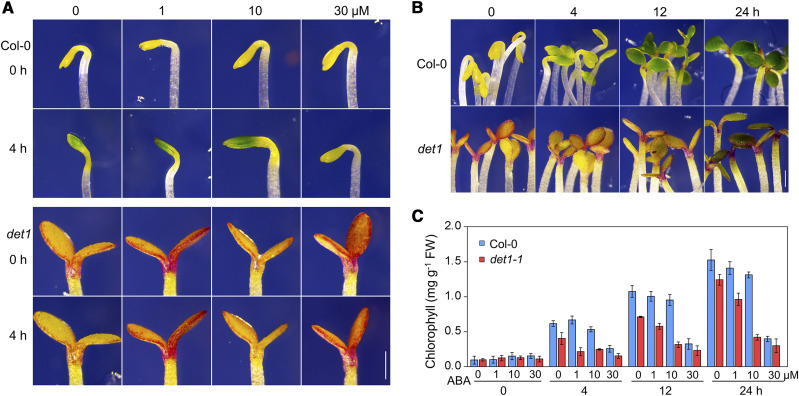
To investigate the relationship between light signaling and the ABA pathway in controlling the greening of etiolated seedlings in Arabidopsis, we measured the greening rates of a set of well-known light-signaling related mutants that were germinated and grown under continuous darkness for 4 d. As shown in Figure 1, A and B, we found that seedlings of the det1-1 mutant (a weak allele) showed a slower greening rate than wild-type Col-0 plants after the seedlings were irradiated with light, although the det1-1 plants exhibited a constitutive photomorphogenic phenotype in darkness. In wild-type Col-0 plants, after being irradiated with light for 4 h, the cotyledons partially turned green (especially the middle region) and the folded apical hook partially opened. The tightly closed cotyledons of Col-0 plants completely opened after being irradiated with light for 12 h, and total chlorophyll contents reached their maximum values after being irradiated with light for 24 h (Fig. 1, B and C). In det1-1 plants, the greening rate was obviously slower than in Col-0 plants (Fig. 1, B and C). Interestingly, light-induced cotyledon greening was slightly decreased in wild-type Col-0 seedlings after being treated with 10 μm of ABA, and strongly decreased after a high concentration (30 μm) of ABA treatment (Fig. 1, A and C). Compared to Col-0 plants, the det1-1 mutant exhibited increased sensitivity to ABA treatment (including 1, 10, and 30 μm; Fig. 1, A and C). All these observations suggest that ABA inhibits the light-induced greening of seedlings, and DET1 plays a negative role in this process.
Figure 1.
ABA inhibits light-induced greening of etiolated seedlings through DET1. A, Effect of ABA on the greening of etiolated Col-0 and det1-1 seedlings. Four-day-old etiolated seedlings were treated without or with 1, 10, or 30 μm of ABA for 4 h under light conditions. Scale bar = 1 mm. B, The greening phenotype of 4-d-old etiolated Col-0 and det1-1 seedlings that were irradiated with light for 0, 4, 12, or 24 h. C, Total chlorophyll contents of the seedlings shown in A and B. Values are means ± sd. FW, Fresh weight.
DET1 Physically Interacts with FHY3 In Vitro and In Vivo
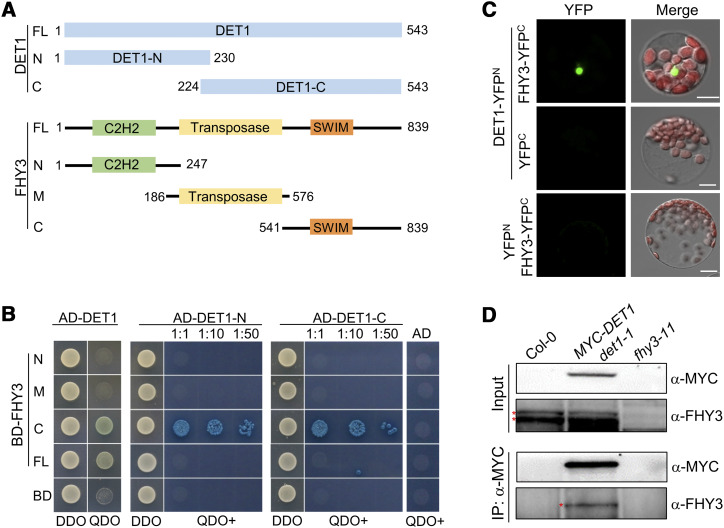
To investigate the regulatory mechanism by which DET1 regulates the greening of seedlings, we performed a yeast two-hybrid screening and found that DET1 interacts with FHY3. As shown in Figure 2, A and B, full-length FHY3 protein weakly interacted with DET1, and the C-terminal region of FHY3 strongly interacted with DET1, indicating that the C-terminal transcriptional activation region of FHY3 is responsible for the physical interaction with DET1 in vitro (Fig. 2, A and B). Given that DET1 forms the CDD complex by interacting with COP10 and DDB1 in planta, we performed a yeast two-hybrid assay and verified that COP10 interacts with the full-length and C-terminal of FHY3 (Supplemental Fig. S1A). These results suggest that DET1 and COP10 physically interact with FHY3 through its C-terminal transcriptional activation domain.
Figure 2.
DET1 physically interacts with FHY3 in vitro and in vivo. A and B, Yeast two-hybrid assays showing DET1 interacts with FHY3 in vitro. The positions of various fragments that used in the yeast two-hybrid assays are shown in A. The various fragments of DET1 and FHY3 were fused to DNA binding domain (BD) or activation domain (AD) of GAL4, respectively. 1:10 and 1:50 indicate the dilutions of yeast cells that were spotted on plates for X-α-gal assays. DDO, Yeast synthetic medium without Trp/Leu; QDO, yeast synthetic medium without Trp/Leu/His/Ade, but with 20 μg mL−1 of X-α-gal and 125 ng mL−1 of AbA; QDO+, QDO medium-plus with 40 μg mL−1 of X-α-gal and 250 ng mL−1 of AbA. C, BiFC assays showing DET1 interacts with FHY3 in Arabidopsis protoplasts. YFPN and YFPC indicate the N- or C-terminal parts of YFP, respectively. Scale bars = 5 μm. D, Co-IP assays showing DET1 interacts with FHY3 in Arabidopsis. Four-day-old various etiolated seedlings were irradiated with light for 30 min and then used to perform Co-IP assays. The fhy3-11 was used as a negative control showing the positions of endogenous FHY3 protein (indicates by asterisk) that detected by anti-FHY3 polyclonal antibodies. Anti-MYC monoclonal antibodies were used to perform the IP and detect the abundance of the MYC-DET1 fusion protein.
To demonstrate the physical interaction between DET1 and FHY3 in vivo, we performed a bimolecular fluorescence complementation (BiFC) assay in Arabidopsis protoplasts. As shown in Figure 2C, coexpressing DET1-YFPN (DET1 fused to the N terminus of yellow fluorescent protein [YFP]) and FHY3-YFPC (FHY3 fused to the C terminus of YFP) produced obvious YFP fluorescence signals in the nucleus, whereas protoplasts harboring DET1-YFPN/YFPC or YFPN/FHY3-YFPC failed to produce any detectable YFP signal. Further, we performed coimmunoprecipitation (Co-IP) assays using 4-d-old seedlings of 35Sp:MYC-DET1 (in the det1-1 background) and 35Sp:FLAG-COP10 (in the cop10-1 background) after being irradiated with light for 30 min (minute). After the total protein immunoprecipitation by anti-MYC (Fig. 2D) or anti-FLAG (Supplemental Fig. S1B) antibodies, specific bands of FHY3 protein were clearly detected in the immunoprecipitated (IP) products of 35Sp:MYC-DET1 and 35Sp:FLAG-COP10 plants, but not in wild-type Col-0 and fhy3-11 plants. All these results indicate that DET1 and COP10 physically interact with FHY3 in vitro and in vivo.
FHY3 Acts Downstream of DET1 in Mediating ABA Responses
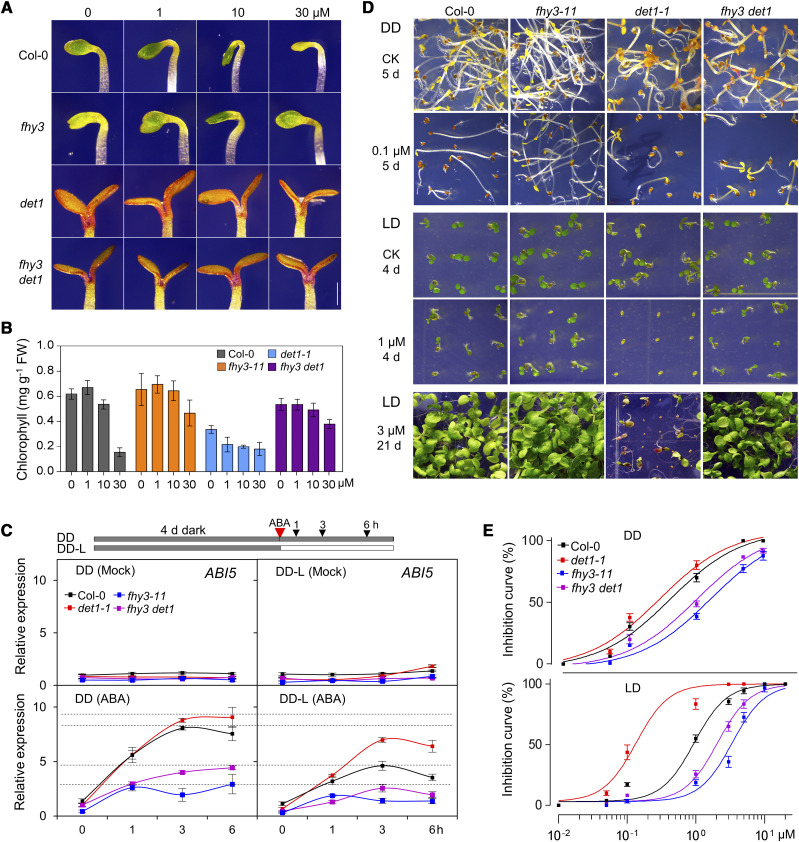
To investigate the genetic relationship between DET1 and FHY3, we crossed the fhy3-11 (null allele) mutant with the det1-1 (weak allele) mutant to generate the fhy3 det1 double mutant in the Col-0 background. To investigate how DET1 and FHY3 mediate the inhibition of greening by ABA, 4-d-old etiolated Col-0, fhy3-11, det1-1, and fhy3 det1 seedlings were treated with different concentrations of ABA after being irradiated with light. Compared to wild-type Col-0 plants, fhy3-11 plants exhibited faster greening rates and higher chlorophyll contents when treated with a high concentration of ABA (30 μm), while det1-1 plants exhibited slower greening rates and much lower chlorophyll contents (Fig. 3, A and B). Under ABA treatment conditions, fhy3 det1 plants showed an intermediate greening response between those of fhy3-11 and det1-1 plants, while closer to fhy3-11, which indicated that FHY3 and DET1 might antagonistically regulate light-induced greening under ABA treatment conditions.
Figure 3.
FHY3 acts downstream of DET1 in mediating ABA responses. A and B, The greening phenotype (A) and chlorophyll contents (B) of 4-d-old various etiolated seedlings treated with 0, 1, 10, or 30 μm of ABA for 4 h under light conditions. FW, Fresh weight. Scale bar = 1 mm. Values are means ± sd. C, RT-qPCR analyses showing the relative expression of ABI5 in various seedlings. Four-day-old various etiolated seedlings were treated without (Mock) or with 30 μm of ABA for the indicated times under DD or after being transferred to light conditions (DD-L). A typical experiment out of three repeated experiments is shown, and means are values ± sd of three technical replicates are presented. D, Phenotypes of various plants germinated and continuously grown on GM without or with 0.1 μm of ABA under DD for 5 d, with 1 μm of ABA under LD for 4 d, or with 3 μm of ABA under LD for 21 d. E, The ABA inhibition response curves of seed germination for various plants under DD (upper) or LD (lower) conditions. ABA inhibition curves were calculated based on the inhibition effect of ABA on germination at the time point that ∼50% wild-type seeds germinated under 1 μm of ABA treatment (at 60 h after imbibition under LD; at 72 h after imbibition under DD, respectively) using the EC50 shift. Values are means ± sd.
To further verify this notion, we measured the expression of the well-studied ABA rapid-response marker genes ABI1, ABI3, ABI4, and ABI5 in seedlings treated without (mock) or with ABA, under constant darkness (DD) or after plants were irradiated with light (DD-L). Without ABA treatment, reverse transcription quantitative PCR (RT-qPCR) analyses showed that their expression levels were not significantly altered in Col-0, det1-1, fhy3-11, and fhy3 det1 plants under DD and DD-L conditions (Fig. 3C; Supplemental Fig. A). After ABA treatment, the expression levels of ABI1, ABI3, ABI4, and ABI5 were obviously increased in Col-0 and det1-1 plants, but only slightly increased in fhy3-11 and fhy3 det1 plants (Fig. 3C; Supplemental Fig. S2A). Interestingly, we noticed that the induction of ABI1, ABI3, ABI4, and ABI5 expression after ABA treatment was much higher in the wild-type Col-0 plants under DD, compared with their expression levels under DD-L, which indicated that ABA-induced expression of these ABI genes was significantly repressed by light (Fig. 3C; Supplemental Fig. S2A).
Further, ABA inhibited seed germination and response curves of Col-0, fhy3-11, det1-1, and fhy3 det1 plants were observed under both DD and long-day (LD) conditions. Compared to wild-type Col-0 plants, det1-1 plants had obviously increased sensitivity to ABA treatment; seed germination and postgerminative growth of det1-1 plants were dramatically inhibited and arrested by a very low concentration of ABA (0.1 μm), while fhy3-11 plants exhibited reduced sensitivity to ABA treatment under both DD and LD conditions (Fig. 3, D and E; Supplemental Fig. S2, B and C). More importantly, although fhy3 det1 plants exhibited intermediate ABA responses between those of fhy3 and det1 plants, the inhibited seed germination and arrested post-germinative growth of det1-1 plants were nearly completely rescued by disruption of FHY3 (Fig. 3, D and E). ABA inhibited seed germination and response curves of Nossen-0 (No-0), fhy3-4 (No-0 ecotype), MYC-DET1 No-0, and MYC-DET1 fhy3-4 plants were further examined. Consistently, under ABA treatment, MYC-DET1 fhy3-4 plants exhibited a very similar (but not additive) seed germination ratio to that of fhy3-4 plants, although a higher percent of germination ratio than No-0 plants was observed in MYC-DET1 No-0 plants (Supplemental Fig. S2, D and E). In addition, with ABA treatment, the root elongation of det1-1 plants was completely inhibited, while fhy3-4, fhy3-4 det1-1, and MYC-DET1 fhy3-4 plants exhibited much longer root lengths than wild-type plants (Supplemental Fig. S2, F and G). We also noticed that, without ABA treatment, the fhy3-4 and fhy3-4 det1-1 plants exhibited longer root lengths than wild-type plants, while the det1-1 mutant exhibited shorter root lengths than wild-type plants. All these results indicate that FHY3 acts downstream of DET1-mediated ABA responses, including gene expression, seed germination, greening of etiolated seedlings, and postgerminative growth.
DET1 Inhibits FHY3 Activation of its Direct Target ABI5 under Light and ABA Treatment Conditions
To reveal the molecular mechanisms by which DET1 regulates ABA responses through FHY3, we first checked the tissue-specific expression pattern of FHY3 and DET1 using FHY3p:GUS and DET1p:GUS reporter lines. GUS staining assays showed that both FHY3 and DET1 were strongly expressed in cotyledons and the radicle (Supplemental Fig. S3A). Further, RT-qPCR and immunoblot assays showed that FHY3 and DET1 shared a similar expression pattern under diurnal and ABA treatment conditions (Supplemental Fig. S3, B–E). Immunoblot assays showed that the abundance of FHY3 protein was not significantly altered in det1-1 plants, while the abundance of MYC-DET1 was slightly increased in MYC-DET1 fhy3-4 plants (Supplemental Fig. S4, A and B).
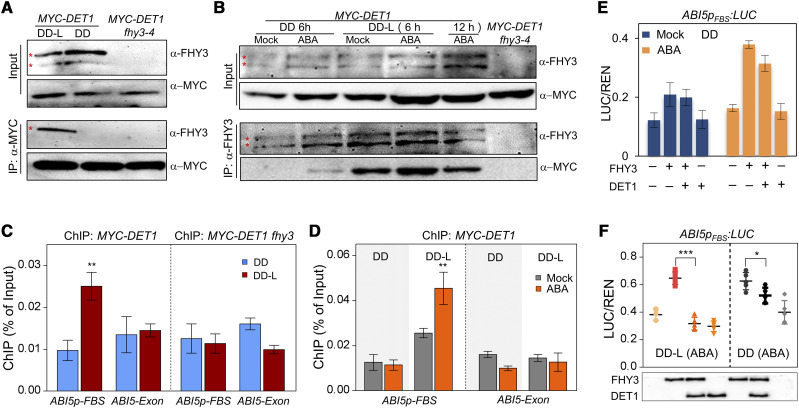
To investigate the regulatory mechanisms of the DET1 and FHY3 interaction during the dark-to-light transition, we performed Co-IP assays in etiolated MYC-DET1 No-0 seedlings. As shown in Figure 4A, after protein immunoprecipitated by anti-MYC monoclonal antibodies, specific FHY3 protein bands were clearly detected in the IP product in MYC-DET1 No-0 seedlings under light conditions (DD-L) but not DD conditions (Fig. 4A), suggesting that the protein interaction between DET1 and FHY3 is induced by light. A Co-IP assay further revealed that ABA treatment significantly enhanced the protein interaction between DET1 and FHY3 under light conditions (Fig. 4B). Meanwhile, the ABA- and FHY3-induced high expression of ABI5 could be obviously reduced by light (Fig. 3C), implying that light promotes the DET1 interaction with FHY3 and subsequently may inhibit the FHY3-mediated transcriptional activation of its direct targets such as ABI5.
Figure 4.
DET1 inhibit the transcriptional activation of ABI5 by FHY3. A and B, Co-IP analyses showing the protein interaction between DET1 and FHY3 dependent on light (A) and enhanced by ABA (B). In A, 4-d-old etiolated MYC-DET1 seedlings (DD) being irradiated with light (DD-L) for 1 h were used to perform Co-IP assays. In B, 4-d-old etiolated MYC-DET1 seedlings treated without (Mock) or with ABA (30 μm) for the indicated times under DD or DD-L conditions were used to perform Co-IP assays. In A and B, MYC-DET1 fhy3 was used as a negative control to indicate the positions (asterisk) of FHY3 protein. C and D, ChIP-qPCR assays showing the association of DET1 with the ABI5 promoter is dependent on FHY3 (C) and enhanced by ABA under light (D). In C, 4-d-old etiolated seedlings (DD) after being irradiated with light (DD-L) for 3 h were used to perform ChIP assays. In D, 4-d-old etiolated seedlings were treated without (Mock) or with ABA (30 μm) for 3 h under DD or DD-L conditions, and then used to perform a ChIP assay. E and F, Transient expression assays showing DET1 represses the transcriptional activation of ABI5 by FHY3. In E, the transformed Arabidopsis protoplasts were first incubated without (Mock) or with 0.3 μm of ABA under darkness for 16 h (DD), then irradiated with light (DD-L) or not for 3 h and used to measure the expression of LUC reporter. In F, the abundances of FHY3-3FLAG and DET1-3FLAG were detected by anti-FLAG antibodies. Data are means of five biological replicates, and error bars represent sd. In C, D, and F, asterisks indicate the statistical significance by Student's t test (*P <0.05, **P <0.01, and ***P <0.001).
To reveal the molecular mechanism by which DET1 affects the FHY3-mediated transcriptional activation of its downstream targets, we first tested whether DET1 associated with the promoter region of FHY3 targets such as ABI5 by interacting with FHY3. To this end, 4-d-old etiolated MYC-DET1 No-0 and MYC-DET1 fhy3-4 seedlings without or with light treatment were used to perform the chromatin immunoprecipitation quantitative PCR (ChIP-qPCR) assays. As shown in Figure 4C, the fragments containing the FHY3/FAR1 binding site (FBS) cis-elements of the ABI5 promoter were specifically enriched in the ChIP products by the MYC-DET1 protein in MYC-DET1 No-0 seedlings after being irradiated with light, but not in MYC-DET1 fhy3-4 seedlings (Fig. 4C). ChIP-qPCR assays in ABA-treated MYC-DET1 No-0 seedlings showed that ABA treatment enhanced the association of DET1 with the ABI5 promoter (Fig. 4D). Meanwhile, ChIP-qPCR assays showed that FHY3 directly bound to the promoter region of ABI5 in both FHY3p:FHY3-YFP and FHY3p:FHY3-YFP det1-1 seedlings (Supplemental Fig. S4C), which suggested that the DNA binding activity of FHY3 was not directly affected by DET1. All these results suggest that the association of DET1 with the promoter region of ABI5 is dependent on FHY3 under light and ABA treatment conditions.
To confirm whether DET1 directly represses the transcriptional activation of ABI5 by FHY3, we performed transient expression assays in yeast cells and Arabidopsis protoplasts. In yeast cells, expression of FHY3 significantly increased the transcription of ABI5pFBS:LacZ, and coexpression of DET1 and FHY3 repressed the transcriptional activation of ABI5pFBS:LacZ by FHY3 (Supplemental Fig. S4D). In Arabidopsis protoplasts, compared with expression of FHY3 alone, coexpression of DET1 and FHY3 repressed the activation of ABI5pFBS:LUC, especially under light and ABA treatment conditions (Fig. 4, E and F; Supplemental Fig. S4E), which is consistent with the specific interaction between DET1 and FHY3 in light (Fig. 4, A and B). All these results suggest that DET1 interacts with FHY3 and subsequently associates to the promoter regions of FHY3 direct targets (such as ABI5) under light or ABA treatment conditions, and then inhibits their expression.
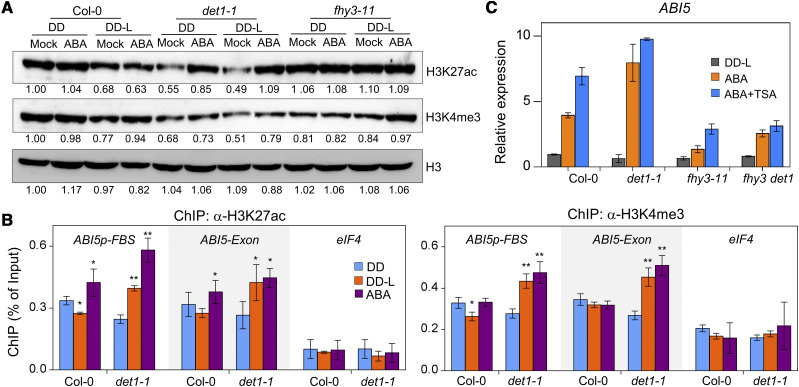
DET1 Mediates H3K27ac and H3K4me3 Modification during the Dark-to-Light Transition and ABA Response
To investigate whether DET1 represses the FHY3-mediated transcriptional activation by influencing histone modifications at the FHY3 target locus, we first measured the global abundances of H3K27ac (acetylation of histone H3lys27) and H3K4me3 (trimethylation of histone H3lys4), two well-studied transcriptional activation-related histone modifications (Santos-Rosa et al., 2002; Katoh et al., 2018), in Col-0, det1-1, fhy3-11, and fhy3 det1 seedlings (Fig. 5A; Supplemental Fig. S5, A and B). In darkness, compared to wild-type Col-0 plants, the global abundances of H3K27ac and H3K4me3 were reduced in det1-1 and fhy3 det1 plants, but were not significantly altered in fhy3-11 plants (Fig. 5A; Supplemental Fig. S5B). Upon irradiating seedlings with light, compared to seedlings grown in darkness, the global abundances of H3K27ac and H3K4me3 were decreased slightly in Col-0 plants, did not significantly change in fhy3-11 plants, and were obviously increased in det1-1 and fhy3 det1 plants (Fig. 5A; Supplemental Fig. S5B). After ABA treatment, compared to untreated plants, the global abundances of H3K27ac and H3K4me3 were significantly increased in det1-1 plants, but not in Col-0 plants and fhy3-11 plants (Fig. 5A). The specific enrichments of H3ac, H3K27ac, and H3K4me3 modifications in the ABI5 promoter and exon regions were further detected in etiolated Col-0 and det1-1 seedlings using ChIP-qPCR. In Col-0 plants, the enrichments of H3ac, H3K27ac, and H3K4me3 in the promoter region of ABI5 were repressed by light and induced by ABA treatment (Fig. 5B; Supplemental Fig. S5C). Compared to wild-type Col-0 plants, the enrichments of H3K27ac and H3K4me3 in the promoter and exon regions of ABI5 were obviously induced in det1-1 plants under both light and ABA treatment conditions (Fig. 5B), consistent with its positive role in the decrease of H3K27ac and H3K4me3 modifications after light or ABA treatment (Fig. 5A). Interestingly, these results revealed that DET1 promoted the accumulation of H3K27ac and H3K4me3 in darkness, while it promoted a decrease of H3K27ac and H3K4me3 after light or ABA treatment.
Figure 5.
DET1 mediates histone modification during the dark-to-light transition and ABA response. A, Immunoblot assays showing the dynamic changes of H3K27ac and H3K4me3 abundance during dark (DD) to light (L) transition and ABA treatment. Four-day-old various etiolated seedlings were treated without (Mock) or with ABA (30 μm) under DD or DD-L for 3 h, and then used to perform immunoblot assays. The abundance of histone H3 was used as a loading control. B, ChIP-qPCR assays showing the association of H3K27ac and H3K4me3 at the promoter and exon regions of ABI5. The exon region of eIF4 (eIF4A1) was used as a negative control for ChIP-qPCR. Asterisks indicate the statistical significance by Student's t test (*P <0.05 and **P <0.01). C, RT-qPCR analysis showing the effect of ABA and TSA treatment on the expression of ABI5. Four-day-old etiolated seedlings were treated with ABA (30 μm) or ABA+TSA (10 μm) for 3 h under light (DD-L), and then used to perform RT-qPCR assays.
The contrasting regulatory roles of DET1 in dark and light suggest that DET1 may specifically interact with HDAs or other histone modification regulators after light or ABA treatment, thus affecting H3K27ac or other modifications. To test this, we performed RT-qPCR assays with Trichostatin A (TSA) treatment, an inhibitor of HDAs (Finnin et al., 1999). In wild-type Col-0 and fhy3-11 plants, TSA treatment obviously increased the induction of ABI5 in response to ABA treatment (Fig. 5C). Compared to wild-type Col-0 plants, TSA treatment only slightly increased the induction of ABI5 in response to ABA treatment in det1-1 and fhy3 det1 plants (Fig. 5C), which suggested that DET1 might inhibit the transcriptional activation of ABI5 through regulating the enrichments of H3K27ac, H3K4me3, or other modifications.
DET1 Recruits HDA6 To Inhibit the Transcriptional Activity of FHY3
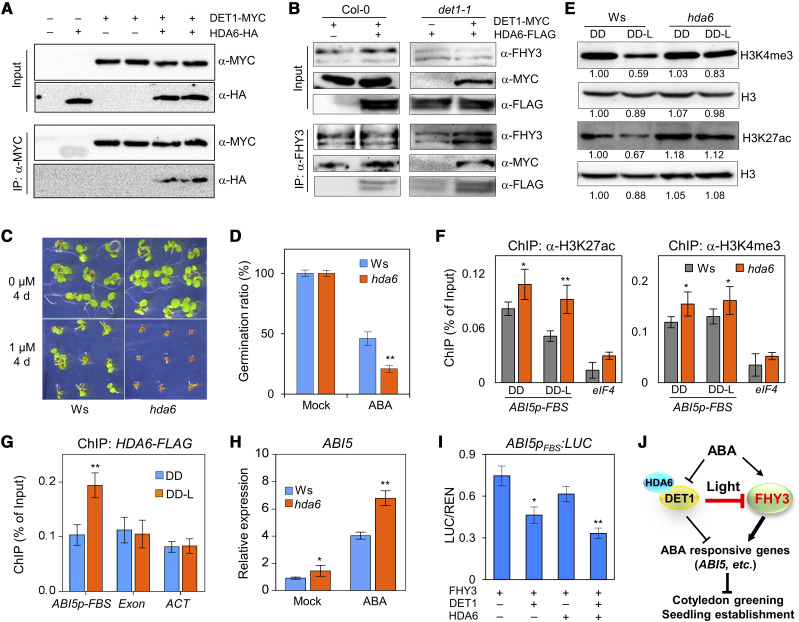
To investigate whether HDAs were involved in the DET1-mediated inhibition of FHY3, we first tested the protein interaction between DET1 and HDA6, an HDA member that has been reported to function in light, ABA, and stress responses (Earley et al., 2006; Tanaka et al., 2008; Tessadori et al., 2009; Chen et al., 2010). As shown in Figure 6A, after being immunoprecipitated by anti-MYC antibodies, specific HDA6-HA protein bands were detected in the samples that coexpressed DET1-MYC and HDA6-HA, which suggested that DET1 interacted with HDA6 in plants (Fig. 6A). To test whether DET1 recruits HDA6 to associate with FHY3, we performed Co-IP assays in the protoplasts of Col-0 and det1-1 plants. After immunoprecipitation by anti-FHY3 antibodies, DET1-MYC and HDA6-FLAG proteins were detected in the protoplasts that coexpressed DET1-MYC and HDA6-FLAG, indicating that FHY3 associated with DET1 and HDA6 in plants (Fig. 6B, left). In det1-1 protoplasts, after being immunoprecipitated with anti-FHY3 antibodies, weak HDA6-FLAG bands could be detected. When coexpressing DET1-MYC and HDA6-FLAG in det1-1 protoplasts, the detected abundance of HDA6-FLAG was significantly increased after being immunoprecipitated with anti-FHY3 antibodies, compared with that in det1-1 protoplasts expressing HDA6-FLAG alone (Fig. 6B), which suggested that DET1 recruited HDA6 to associate with FHY3 in Arabidopsis.
Figure 6.
DET1 recruits HDA6 to inhibit the transcriptional activity of FHY3. A, Co-IP assays in N. benthamiana leaves showing DET1 interacts with HDA6. B, Co-IP assays in protoplasts of wild type (Col-0) and det1-1 showing FHY3 interacts with HDA6 via DET1. C and D, Seed germination phenotypes (C) and ratio (D) of wild-type (Ws) and hda6 plants under 1-μm ABA treatment conditions. Germination ratios were determined from three biological replicates of ∼40 seeds each. E, Immunoblot assays showing the dynamic changes of H3K4me3 and H3K27ac abundance in Ws and hda6 plants during the dark-to-light transition. The abundance of histone H3 was used as a loading control. F, ChIP-qPCR assays showing the effect of HDA6 on the association of H3K4me3 and H3K27ac at ABI5 promoter during the dark-to-light transition. The exon of eIF4 was used as a negative control for ChIP assay. G, ChIP-qPCR assays showing HDA6 associates with the ABI5 promoter after etiolated seedlings irradiated with light (DD-L). The promoter of ACT was used as a negative control. H, RT-qPCR analysis showing the effect of HDA6 on the expression of ABI5. Four-day-old etiolated seedlings were treated with 30 μm of ABA for 3 h under light and then used to perform RT-qPCR assays. I, Transient expression assays in Arabidopsis protoplasts showing HDA6 and DET1 repress the transcriptional activation of ABI5 by FHY3. Data are means of five biological replicates, and error bars represent sd. In F to I, asterisks indicate the statistical significance by Student's t test (*P <0.05 and **P <0.01). J, Working model showing DET1-HDA6 and FHY3 coordinately regulate the effects of light and ABA in the greening of seedling during the dark-to-light transition.
To further investigate whether DET1 could interact with HDA6 to coordinately regulate ABA responses, we examined ABA responses in hda6 plants (an HDA6-RNAi transgenic line in the Wassilewskija ecotype [Ws]; Wu et al., 2008). With a low concentration of ABA (1 μm) treatment, germination of hda6 seeds was completely inhibited (Fig. 6, C and D), similar to det1-1 plants (Fig. 3, D and E). Compared with the Ws control plants, the global abundances of H3K27ac and H3K4me3 were significantly increased when hda6 seedlings were transferred from dark to light (Fig. 6E; Supplemental Fig. S5B). ChIP-qPCR assays further verified that the specific enrichments of H3K27ac and H3K4me3 modifications in the promoter region of ABI5 were significantly higher in hda6 plants, compared with Ws control plants (Fig. 6F). Consistent with these results, the fragments containing the FBS cis-elements in the ABI5 promoter were specifically enriched in the DNA products immunoprecipitated by anti-FLAG of HDA6-FLAG seedlings under DD-L conditions (Fig. 6G). Meanwhile, the expression level of ABI5 was slightly higher in hda6 plants in light (mock), and significantly higher than Ws plants when seedlings were treated with ABA (Fig. 6H). More importantly, when HDA6 was coexpressed with FHY3, the expression level of ABI5pFBS:LUC was slightly repressed, and the repression effect was enhanced when both HDA6 and DET1 were coexpressed with FHY3, suggesting that HDA6 and DET1 have additive roles in this repression process (Fig. 6I). Taken together, our results revealed that DET1 recruits HDA6 to FHY3 target loci and subsequently represses their transcription by reducing the abundance of H3K4me3 and H3K27ac modifications.
DISCUSSION
FHY3 and ABI5 Act Downstream of DET1 in Mediating ABA Responses in Light
As a key repressor of photomorphogenesis and light signal transduction, mutation of DET1 in plants causes a de-etiolated phenotype in darkness, while det1-1 plants exhibit a delayed greening rate after being irradiated with light (Fig. 1). Consistent with this, the det1-1 plants failed to survive after being transferred to light from long-term (such as 7 d) growth in continuous darkness. Besides a delayed greening rate, disruption of DET1 causes very low seed germination, inhibited elongation of primary root, and severely arrested early seedling establishment phenotypes under ABA treatment conditions (Fig. 3, A, B, D, and E; Supplemental Fig. S2). At the transcription level, DET1 repressed the induction of multiple ABI genes (ABI1, ABI3, ABI4, and ABI5), especially ABI5 in the seedlings (Fig. 3C; Supplemental Fig. S2A). Considering the important regulatory roles of these genes (such as ABI5) in ABA responses (Tang et al., 2013; Guan et al., 2014; Hu et al., 2019), the high induction levels of ABI5 or other response genes in det1-1 plants could be one of the major reasons for enhanced ABA response in det1 plants (Figs. 1 and 3; Supplemental Fig. S2). Meanwhile, mutation of ABI5 completely rescues the ABA hypersensitivity in det1-1 plants (Fernando and Schroeder, 2015), which further indicates that ABI5 acts downstream of DET1 in ABA responses. More importantly, ABI5 is one of the direct targets of FHY3, which directly physically interacts with DET1, and disruption of FHY3 in det1-1 plants completely recovers its enhanced ABA responses (Figs. 2 and 3). Additionally, MYC-DET1 fhy3 plants exhibited a similar ABA sensitivity to fhy3 plants, not an additive phenotype (Supplemental Fig. S2, D–G). These results indicate that FHY3 is a major downstream factor of DET1 in mediating ABA response by activating the transcription of ABI5. Therefore, the cascade of DET1-FHY3-ABI5 plays an essential role in ABA response during the dark-to-light transition.
We noticed that the interaction between DET1 and FHY3 only occurred after the seedlings were irradiated with light and enhanced by ABA treatment, which indicated that this protein interaction was induced by light. Interestingly, Co-IP assays showed that DET1 only interacted with the upper-shifted band of FHY3 in plants, suggesting that only modified FHY3 protein interacted with DET1. Phosphatase treatment experiments showed that the upper band contained the phosphorylated form of FHY3 (Supplemental Fig. S6), while the upper band of FHY3 is very stable, not rapidly and significantly changed during the dark-to-light transition or ABA treatment (Fig. 4, A and B; Supplemental Fig. S6). These results indicate that the light-induced interaction between FHY3 and DET1 is not dependent on the phosphorylation of FHY3. In addition, other FHY3 or DET1 interacting proteins related to light signals, such as PIFs or COP1, may interfere with the interaction between FHY3 and DET1. As reported, FHY3 can interact with PIF1 and PIF3 in the dark, and DET1 can interact with PIF1/3/4/5 and COP1 in the dark (Lau and Deng, 2012; Tang et al., 2012; Dong et al., 2014). These FHY3 or DET1 interacting proteins may act as competitors to prevent the protein interaction between DET1 and FHY3 in darkness by an unknown mechanism, while upon being irradiated with light, the competition may decrease and thus allow the interaction between FHY3 and DET1.
In this study, we showed that DET1 repressed the FHY3-mediated transcriptional activation of ABI5 under light and ABA treatment conditions (Figs. 3 and 4). In addition, a recent study has revealed that the induction of ABI5 is mediated by PIF4 in darkness and ABA treatment conditions (Qi et al., 2020). Therefore, in darkness, ABA- or stress-induced high expression of ABI5 could be mediated by both FHY3 and PIF4, while FHY3 is the major transcription factor responsible for the induction of ABI5 upon irradiating etiolated seedlings with light. After the dark-to-light transition, ABA or abiotic stress induced high expression of ABI5 could be further repressed by DET1 through interacting with FHY3, thus, to promote the greening of etiolated seedlings and increase their adaptation to light. Other transcriptional regulators may also be involved in the transcriptional regulation of ABI5 or other ABA response genes during the dark-to-light transition that need to be further explored.
Although we observed that ABA treatment obviously inhibited the greening of etiolated seedlings, the detailed molecular regulatory mechanisms are still largely unknown. Indeed, the transcriptional levels of PROTOCHLOROPHYLLIDE OXIDOREDUCTASEs (PORs), encoding the enzymes that catalyze the conversation from Pchlide to chlorophyllide, were significantly repressed by ABA and dramatically decreased in det1 seedlings (Kusnetsov et al., 1998; Nassrallah et al., 2018), consistent with the delayed greening phenotype (Figs. 1 and 3, A and B). Considering HEMB1, which encodes 5-aminolevulinic acid dehydratase and is a direct target of FHY3, its transcriptional level was not significantly altered in det1-1 plants (Nassrallah et al., 2018), but was obviously reduced in the fhy3 mutant (Tang et al., 2012), which suggested that DET1-FHY3 might also affect the greening of seedlings through directly affecting the expression of chlorophyll-biosynthesis–related genes, such as PROTOCHLOROPHYLLIDE OXIDOREDUCTASEs and HEMB1. In addition to ABA signaling, reactive oxygen species caused by excess accumulation of Pchlide after irradiation by light probably also influence the greening of seedlings during the dark-to-light transition.
DET1 and HDA6 Repress FHY3-Mediated Transcriptional Activation of ABI5
DET1 is an evolutionarily conserved component in plants and animals (Pick et al., 2007; Dubin et al., 2011; Lau and Deng, 2012). Previous studies have revealed that DET1 not only affects the histone modification of Histone H2 by controlling the monoubiquitination of Histone 2B, it also affects histone modification of Histone H3, including H3K4me3, H3K27me3, and H3K36ac by an unknown mechanism (Kang et al., 2015; Nassrallah et al., 2018). In this study, we noticed that the global abundances of H3K27ac and H3K4me3 in det1-1 plants were significantly decreased in darkness, and increased after light or ABA treatment (Fig. 5A; Supplemental Fig. S5, A and B). These results suggest that DET1 plays contrasting regulatory roles in dark and light: In the dark, it promotes the accumulation of H3K27ac and H3K4me3, but after light and ABA treatment, it promotes the decrease of H3K27ac and H3K4me3 (Fig. 5A; Supplemental Fig. S5, A and B). These contrasting regulatory roles of DET1 under dark and light suggest that DET1 may specifically interact with light-regulated histone modification regulators. Subsequently, we showed that DET1 physically interacted with HDA6 (Fig. 6, A and B), an HDA responsible for the deacetylation of H3K27 and affected the trimethylation of H3K4 (Probst et al., 2004; Earley et al., 2006; Tanaka et al., 2008; Tessadori et al., 2009; Chen et al., 2010). Meanwhile, HDA6 plays a negative role in ABA responses (Chen et al., 2010), similar to the physiological role of DET1 in ABA response (Figs. 1 and 3). Disruption of HDA6 produced similar changes of H3K27ac and H3K4me3 as those observed in det1-1 plants (Figs. 5A and 6E). More importantly, DET1 recruits HDA6 to the ABI5 promoter thus coordinately inhibiting the activation of ABI5 by regulating the enrichment of H3K27ac and H3K4me3 modifications (Fig. 6, G–I–I). All these physiological, biochemical, and molecular evidence support the idea that HDA6 functions together with DET1 to coordinately inhibit the transcriptional activity of FHY3.
In this study, we showed that DET1 repressed the transcription of ABI5 by recruiting HDA6 to the promoter region of ABI5 (Fig. 6). Interestingly, we noticed that DET1 could not bind to the exon region of ABI5, but the specific enrichments of H3K27ac and H3K4me3 at the ABI5 exon were significantly increased in det1-1 plants (Fig. 5B). These results suggest that HDA6 or other histone modification regulators that interact with DET1 might also affect the enrichment of H3K27ac and H3K4me3 at the ABI5 promoter and exon regions. Meanwhile, DET1 might influence the stability or function of FHY3 in other mechanisms, although the abundance of FHY3 was not obviously altered in the det1-1 plants (Supplemental Fig. S4A), but this possibility cannot be excluded. Indeed, DET1 may repress the transcription of ABI5 by directly inhibiting the transcription activity of FHY3, because DET1 strongly interacts with the C-terminal region of FHY3 (Fig. 2, A and B) that is responsible for the transcriptional regulation (Lin et al., 2007). Moreover, we noticed that in the transient expression assays, expressed DET1 alone could only slightly reduce the transcription of the LUC reporter driven by the ABI5 promoter (Fig. 4, E and F; Supplemental Fig. S4D), which suggested that DET1 might repress the expression of ABI5 in FHY3-independent manners. Consistently, DET1 has been reported to interact with other transcription factors such as PIF4 that also mediate the transcriptional activation of ABI5 (Dong et al., 2014; Qi et al., 2020). Therefore, DET1 affects the transcription of ABI5 and other aspects of ABA responses through multiple regulatory levels.
Finally, we showed that during the dark-to-light transition, light induced the interaction between DET1 and FHY3, then DET1 associated to the promoters of FHY3 direct targets (such as ABI5), and subsequently recruited HDA6 to the target locus to reduce the enrichments of H3K27ac and H3K4me3 modifications, thus repressing the transcription of ABI5 and other ABA response genes (Fig. 6J). Therefore, our study revealed the molecular mechanisms by which light and ABA coordinately regulate the greening of etiolated seedlings and possibly other physiological processes.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) fhy3-11 (Salk_002711), det1-1 (Chory et al., 1989), 35Sp:MYC-DET1 det1-1 (Saijo et al., 2003), 35Sp:FLAG-COP10 cop10-1 (Yanagawa et al., 2004), and FHY3p:GUS (Lin and Wang, 2004) lines were in the Col-0 ecotype. hda6 (cs24039) is an HDA6-RNAi transgenic line in the Ws ecotype (Wu et al., 2008). fhy3-4 (Wang and Deng, 2002) and FHY3p:FHY3-YFP fhy3-4 plants (Lin et al., 2007) were in the No-0 ecotype. fhy3-4 det1-1, MYC-DET1 fhy3-4, MYC-DET1 No-0, and FHY3p:FHY3-YFP det1-1 plants were generated by genetic crosses. Arabidopsis seeds sown on germination medium (GM) with 1% (w/v) Suc and 0.8% (w/v) agar were incubated at 4°C overnight and transferred to different growth conditions as indicated at 22°C. The light intensity under LD and DD-L conditions was ∼100 μmol m−2 s−1.
Plasmid Construction and Generation of Transgenic Arabidopsis Plants
To generate pCAMBIA1305-35Sp:DET1-3MYC and pCAMBIA1305-35Sp:HDA6-3HA, the coding regions of DET1 and HDA6 were PCR-amplified from complementary DNA (cDNA) of Col-0 plants and inserted into the KpnI- and SalI-digested pCAMBIA1305-3MYC and pCAMBIA1305-3HA, respectively. To generate pPZP211-35Sp:HDA6-3FLAG and pPZP211-35Sp:FHY3-3FLAG, the fragments containing the HDA6 and FHY3 coding regions were inserted into the KpnI- and SalI-digested pPZP211-3FLAG (Ma et al., 2016), respectively. To generate DET1p:GUS, the fragments containing the promoter region of DET1 (1785 bp) were inserted into the SacI- and SalI-digested pPZP211-GUS. The pPZP211-DET1p:GUS and pPZP211-35Sp:HDA6-3FLAG vectors were transformed into Col-0 plants. After transformation, more than 30 independent T1 generation transgenic lines were screened and transplanted in soil. Two representative lines were identified by GUS staining, RT-qPCR, or immunoblot analysis, and further used for functional assays.
ABA Treatments and Greening Assays
Arabidopsis seeds were first incubated at 4°C for 72 h and then transferred to sugar-free GM to perform ABA treatments and greening assays. For ABA-induced inhibition of seed germination, Arabidopsis seeds were transferred to medium with 0, 0.01, 0.05, 0.1, 1, 3, 5, 10, or 20 μm of ABA under LD conditions, or with 0, 0.05, 0.1, 1, 5, or 10 μm of ABA under DD conditions. The seed germination is characterized by radicle protrusion. Germination ratios were measured from three biological replicates of ∼80 seeds each and ABA inhibition curves were calculated based on the inhibition effect of ABA on germination at the time point that ∼50% of wild-type seed germinated under 1 μm of ABA treatment using the EC50 shift in the program Prism 7.0 (https://www.graphpad.com/scientific-software/prism/).
Four-d–old etiolated seedlings were irradiated with light or treated with different concentrations of ABA for the indicated times and then used to perform RT-qPCR, ChIP-qPCR, Co-IP, and immunoblot assays. To measure the total chlorophyll contents, various seedlings were incubated in 95% (v/v) ethanol for 2 d in darkness and then the absorbance at 665 and 649 nm was measured, and the contents of chlorophyll were calculated using the equation (6.63A665 + 18.08A649)/g fresh weight (Tian et al., 2020).
Yeast Assays
To generate the full-length or various fragments of DET1 or FHY3 in pGADT7 and pGBKT7 plasmids, the fragments containing full-length, N-terminal (1–741 bp), transposase (555–1,728 bp), and C-terminal (1,620–2,517 bp) of FHY3 were inserted into the BamHI- and SalI-digested pGBKT7, and full-length (1–1,629 bp), N-terminal (1–690 bp), and C-terminal (669–1,629 bp) fragments of DET1 were inserted into the BamHI- and SalI-digested pGADT7. To perform yeast two-hybrid assays, various pGADT7- and pGBKT7-fusion plasmids were cotransformed into strain Y2H Gold following the procedure described in the Yeast Protocols Handbook (Clontech). DDO indicates yeast synthetic medium without Trp/Leu, QDO indicates yeast synthetic medium without Trp/Leu/His/Ade, but with 20 μg mL−1 of X-α-gal and 125 ng mL−1 of Aureobasidin A (AbA). QDO+ indicates QDO medium with 40 μg mL−1 of X-α-gal and 250 ng mL−1 of AbA.
To generate the pJG-FHY3 plasmid, the fragment containing the full-length FHY3 coding region was inserted into the BamHI- and SalI-digested pJG4-5 vector. The fragment containing FBS cis-elements in the promoter of ABI5 was repeated three times and inserted into the KpnI- and XhoI-digested pLacZi-2μ (Lin et al., 2007) to generate the ABI5pFBS:LacZ. To perform transient expression assays in yeast cells, pGAD-DET1 and pJG-FHY3 plasmids were cotransformed with the ABI5pFBS:LacZ reporter into yeast strain EGY48. After transformation, all cultures were grown on the appropriate yeast synthetic medium. The liquid assay was conducted as described in the Yeast Protocols Handbook (Clontech).
BiFC Assay
Three-week-old wild-type Col-0 plants grown under short-day conditions were used to prepare protoplasts and perform the BiFC assay. The full-length coding regions of DET1 and FHY3 were individually subcloned into the pTOPO vector and further recombined into the pSITE-BIFC-nEYFP (YFPn) or pSITE-BIFC-cEYFP (YFPc) vectors (Martin et al., 2009). The combinations of DET1-YFPN and FHY3-YFPC, DET1-YFPN, and YFPC, YFPN and FHY3-YFPC were cotransformed into Arabidopsis protoplasts. After 16 h of incubation under dim light, the fluorescent signals of YFP were detected by laser scanning confocal microscopy (Zeiss).
Immunoblot and Co-IP Assays
For immunoblot assays, total proteins from various seedlings or protoplasts were extracted in extraction buffer containing 0.5% (w/v) SDS. For Co-IP assays, total proteins were extracted from seedlings, protoplasts, or Nicotiana benthamiana leaves using the IP buffer and further immunoprecipitated using the specific primary antibodies as indicated. The detailed procedures for immunoblot and Co-IP assays have been described in Li et al. (2011). The primary antibodies anti-FHY3 (catalog no. PHY1892A; PhytoAB), anti-MYC (catalog no. 2278S; Cell Signaling Technology), anti-FLAG (catalog no. A8592; Sigma-Aldrich), anti-HA (catalog no. 11583816001; Roche), anti-Actin (catalog no. CW0264M; CWBIO), anti-H3K4me3 (catalog no. 07-473; Millipore), anti-H3K4ac (catalog no. ab176799, Abcam), anti-H3K14ac (catalog no. ab52946; Abcam), anti-H3K27ac (catalog no. ab4729; Abcam), anti-H3ac (K9/K14/K18/K23/K2; catalog no. ab47915; Abcam), and anti-H3 (catalog no. AF0009; Beyotime) were used to perform IP or immunoblot assays. The intensities of each band were quantified using the program ImageJ (https://imagej.nih.gov/ij/). All the immunoblot and Co-IP assays were repeated at least three times.
RT-qPCR and ChIP-qPCR
Total RNA was extracted from various seedlings with an RNA Extraction Kit (Omega), and first-strand cDNA was synthesized with cDNA Synthesis SuperMix (Transgene). qPCR assays were further performed using UltraSYBR Mixture (CWBIO) using a LightCycler 96 PCR machine (Roche). The expression levels of various genes were normalized to the expression level of the internal control gene UBQ1.
Four-day-old etiolated seedlings after light or ABA treatments were used to perform ChIP assays. The primary antibodies anti-MYC, anti-FLAG, anti-GFP, anti-H3K4me3, and anti-H3K27ac were used in the ChIP assays. The detailed procedure for ChIP assays has been described in Ma et al. (2016). The exon region of ABI5 and the promoter region of ACTIN12 (ACT) were used as the negative control for ChIP assays with DET1 and HDA6. The exon region of eIF4A1 (eIF4) and ACT was used as the negative control for ChIP with H3K27ac, H3K4me3, and H3ac. All the RT-qPCR and ChIP-qPCR assays were repeated at least three times. Primers for RT-qPCR and ChIP-qPCR are listed in Supplemental Table S1.
Transient Transcription Dual-Luciferase Assay
To generate the pGreen-0800-ABI5pFBS:LUC plasmid, the fragment containing the FBS cis-elements in the ABI5 promoter region was repeated three times and inserted into the SalI- and KpnI-digested pGreen-0800-35Smini:LUC vector (Chen et al., 2019). The plasmids 35Sp:FHY3-3FLAG, 35Sp:DET1-3FLAG, and 35Sp: HDA6-3FLAG were coexpressed with pGreen-0800-ABI5pFBS:LUC as indicated in the dual-luciferase assays in Arabidopsis protoplasts. The detailed procedure for dual-luciferase assays has been described in Li et al. (2011). For LUC/REN, the expression values of the LUC reporter were normalized to the expression values of REN luciferase driven by the 35S promoter. Data are means of five biological replicates, and error bars represent sd.
Statistical Analysis
To determine statistical significance, we employed Student’s t test. *P < 0.05 was considered to indicate statistical significance, and **P < 0.01 or ***P < 0.001 were considered extremely significant.
Accession Numbers
The Arabidopsis Genome Initiative (https://www.arabidopsis.org) locus identifiers for the genes mentioned in this study are as follows: DET1 (At4g10180), FHY3 (At3g22170), HDA6 (At5g63110), ABI5 (At2g36270), COP10 (At3g13550), ACT (At3g46520), UBQ1 (At3g52590), and eIF4A1 (AT3g13920).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. FHY3 physically interacts with COP10 in vitro and in vivo.
Supplemental Figure S2. FHY3 acts downstream of DET1 in mediating ABA responses.
Supplemental Figure S3. Expression pattern of FHY3 and DET1.
Supplemental Figure S4. Effect of DET1 on the protein abundance and DNA binding activity of FHY3.
Supplemental Figure S5. Influence of DET1 on histone modifications of histone H3 during dark-to-light transition and ABA responses.
Supplemental Figure S6. Phosphatase treatment assays of FHY3 protein in plants.
Supplemental Table S1. List of primers used in this study.
Footnotes
This work was supported by the National Natural Science Foundation of China (grants no. 31670249 and 31870266) and funds of the Shandong “Double Tops” program (to G.L.).
Articles can be viewed without a subscription.
References
- Abbas M, Berckhan S, Rooney DJ, Gibbs DJ, Vicente Conde J, Sousa Correia C, Bassel GW, Marín-de la Rosa N, León J, Alabadí D, et al. (2015) Oxygen sensing coordinates photomorphogenesis to facilitate seedling survival. Curr Biol 25: 1483–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zhang J, Neff MM, Hong SW, Zhang H, Deng XW, Xiong L(2008) Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc Natl Acad Sci USA 105: 4495–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Li GJ, Bressan RA, Song CP, Zhu JK, Zhao Y(2020) Abscisic acid dynamics, signaling, and functions in plants. J Integr Plant Biol 62: 25–54 [DOI] [PubMed] [Google Scholar]
- Chen LT, Luo M, Wang YY, Wu K(2010) Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J Exp Bot 61: 3345–3353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Xu X, Xu D, Zhang H, Zhang C, Li G(2019) WRKY18 and WRKY53 coordinate with HISTONE ACETYLTRANSFERASE1 to regulate rapid responses to sugar. Plant Physiol 180: 2212–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Peto C, Feinbaum R, Pratt L, Ausubel F(1989) Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell 58: 991–999 [DOI] [PubMed] [Google Scholar]
- Dong J, Tang D, Gao Z, Yu R, Li K, He H, Terzaghi W, Deng XW, Chen H(2014) Arabidopsis DE-ETIOLATED1 represses photomorphogenesis by positively regulating phytochrome-interacting factors in the dark. Plant Cell 26: 3630–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin MJ, Kasten S, Nellen W(2011) Characterization of the Dictyostelium homolog of chromatin binding protein DET1 suggests a conserved pathway regulating cell type specification and developmental plasticity. Eukaryot Cell 10: 352–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K, Lawrence RJ, Pontes O, Reuther R, Enciso AJ, Silva M, Neves N, Gross M, Viegas W, Pikaard CS(2006) Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev 20: 1283–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando VC, Schroeder DF(2015) Genetic interactions between DET1 and intermediate genes in Arabidopsis ABA signalling. Plant Sci 239: 166–179 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ(2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP(1999) Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 401: 188–193 [DOI] [PubMed] [Google Scholar]
- Gommers CMM, Monte E(2018) Seedling establishment: A dimmer switch-regulated process between dark and light signaling. Plant Physiol 176: 1061–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan C, Wang X, Feng J, Hong S, Liang Y, Ren B, Zuo J(2014) Cytokinin antagonizes abscisic acid-mediated inhibition of cotyledon greening by promoting the degradation of abscisic acid insensitive5 protein in Arabidopsis. Plant Physiol 164: 1515–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Han X, Yang M, Zhang M, Pan J, Yu D(2019) The transcription factor INDUCER OF CBF EXPRESSION1 interacts with ABSCISIC ACID INSENSITIVE5 and DELLA proteins to fine-tune abscisic acid signaling during seed germination in Arabidopsis. Plant Cell 31: 1520–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Dong Z, Guo P, Zhang X, Qiu Y, Li B, Wang Y, Guo H(2020) Salicylic acid suppresses apical hook formation via NPR1-mediated repression of EIN3 and EIL1 in Arabidopsis. Plant Cell 32: 612–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irigoyen ML, Iniesto E, Rodriguez L, Puga MI, Yanagawa Y, Pick E, Strickland E, Paz-Ares J, Wei N, De Jaeger G, et al. (2014) Targeted degradation of abscisic acid receptors is mediated by the ubiquitin ligase substrate adaptor DDA1 in Arabidopsis. Plant Cell 26: 712–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MY, Yoo SC, Kwon HY, Lee BD, Cho JN, Noh YS, Paek NC(2015) Negative regulatory roles of DE-ETIOLATED1 in flowering time in Arabidopsis. Sci Rep 5: 9728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh N, Kuroda K, Tomikawa J, Ogata-Kawata H, Ozaki R, Ochiai A, Kitade M, Takeda S, Nakabayashi K, Hata K(2018) Reciprocal changes of H3K27ac and H3K27me3 at the promoter regions of the critical genes for endometrial decidualization. Epigenomics 10: 1243–1257 [DOI] [PubMed] [Google Scholar]
- Kusnetsov V, Herrmann RG, Kulaeva ON, Oelmüller R(1998) Cytokinin stimulates and abscisic acid inhibits greening of etiolated Lupinus luteus cotyledons by affecting the expression of the light-sensitive protochlorophyllide oxidoreductase. Mol Gen Genet 259: 21–28 [DOI] [PubMed] [Google Scholar]
- Lau OS, Deng XW(2012) The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci 17: 584–593 [DOI] [PubMed] [Google Scholar]
- Li G, Siddiqui H, Teng Y, Lin R, Wan XY, Li J, Lau OS, Ouyang X, Dai M, Wan J, et al. (2011) Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat Cell Biol 13: 616–622 [DOI] [PubMed] [Google Scholar]
- Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H(2007) Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318: 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Wang H(2004) Arabidopsis FHY3/FAR1 gene family and distinct roles of its members in light control of Arabidopsis development. Plant Physiol 136: 4010–4022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ma M, Li G, Yuan L, Xie Y, Wei H, Ma X, Li Q, Devlin PF, Xu X, et al. (2020) Transcription factors FHY3 and FAR1 regulate light-induced CIRCADIAN CLOCK ASSOCIATED1 gene expression in Arabidopsis. Plant Cell 32: 1464–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wei H, Ma M, Li Q, Kong D, Sun J, Ma X, Wang B, Chen C, Xie Y, et al. (2019) Arabidopsis FHY3 and FAR1 regulate the balance between growth and defense responses under shade conditions. Plant Cell 31: 2089–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH(2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Li G(2018) FAR1-RELATED SEQUENCE (FRS) and FRS-RELATED FACTOR (FRF) family proteins in Arabidopsis growth and development. Front Plant Sci 9: 692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Li Y, Li X, Xu D, Lin X, Liu M, Li G, Qin X(2019) FAR-RED ELONGATED HYPOCOTYLS3 negatively regulates shade avoidance responses in Arabidopsis. Plant Cell Environ 42: 3280–3292 [DOI] [PubMed] [Google Scholar]
- Ma L, Tian T, Lin R, Deng XW, Wang H, Li G(2016) Arabidopsis FHY3 and FAR1 regulate light-induced myo-Inositol biosynthesis and oxidative stress responses by transcriptional activation of MIPS1. Mol Plant 9: 541–557 [DOI] [PubMed] [Google Scholar]
- Ma L, Xue N, Fu X, Zhang H, Li G(2017) Arabidopsis thaliana FAR-RED ELONGATED HYPOCOTYLS3 (FHY3) and FAR-RED-IMPAIRED RESPONSE1 (FAR1) modulate starch synthesis in response to light and sugar. New Phytol 213: 1682–1696 [DOI] [PubMed] [Google Scholar]
- Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM(2009) Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J 59: 150–162 [DOI] [PubMed] [Google Scholar]
- Nassrallah A, Rougée M, Bourbousse C, Drevensek S, Fonseca S, Iniesto E, Ait-Mohamed O, Deton-Cabanillas AF, Zabulon G, Ahmed I, et al. (2018) DET1-mediated degradation of a SAGA-like deubiquitination module controls H2Bub homeostasis. eLife 7: e37892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang X, Li J, Li G, Li B, Chen B, Shen H, Huang X, Mo X, Wan X, Lin R, et al. (2011) Genome-wide binding site analysis of FAR-RED ELONGATED HYPOCOTYL3 reveals its novel function in Arabidopsis development. Plant Cell 23: 2514–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper A, Delaney T, Washburn T, Poole D, Chory J(1994) DET1, a negative regulator of light-mediated development and gene expression in arabidopsis, encodes a novel nuclear-localized protein. Cell 78: 109–116 [DOI] [PubMed] [Google Scholar]
- Pick E, Lau OS, Tsuge T, Menon S, Tong Y, Dohmae N, Plafker SM, Deng XW, Wei N(2007) Mammalian DET1 regulates Cul4A activity and forms stable complexes with E2 ubiquitin-conjugating enzymes. Mol Cell Biol 27: 4708–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AV, Fagard M, Proux F, Mourrain P, Boutet S, Earley K, Lawrence RJ, Pikaard CS, Murfett J, Furner I, et al. (2004) Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell 16: 1021–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L, Liu S, Li C, Fu J, Jing Y, Cheng J, Li H, Zhang D, Wang X, Dong X, et al. (2020) PHYTOCHROME-INTERACTING FACTORS interact with the ABA receptors PYL8 and PYL9 to orchestrate ABA signaling in darkness. Mol Plant 13: 414–430 [DOI] [PubMed] [Google Scholar]
- Riber W, Müller JT, Visser EJ, Sasidharan R, Voesenek LA, Mustroph A(2015) The greening after extended darkness1 is an N-end rule pathway mutant with high tolerance to submergence and starvation. Plant Physiol 167: 1616–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW(2003) The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev 17: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T(2002) Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Kikuchi A, Kamada H(2008) The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol 146: 149–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Ji Q, Huang Y, Jiang Z, Bao M, Wang H, Lin R(2013) FAR-RED ELONGATED HYPOCOTYL3 and FAR-RED IMPAIRED RESPONSE1 transcription factors integrate light and abscisic acid signaling in Arabidopsis. Plant Physiol 163: 857–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Wang W, Chen D, Ji Q, Jing Y, Wang H, Lin R(2012) Transposase-derived proteins FHY3/FAR1 interact with PHYTOCHROME-INTERACTING FACTOR1 to regulate chlorophyll biosynthesis by modulating HEMB1 during deetiolation in Arabidopsis. Plant Cell 24: 1984–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessadori F, van Zanten M, Pavlova P, Clifton R, Pontvianne F, Snoek LB, Millenaar FF, Schulkes RK, van Driel R, Voesenek LA, et al. (2009) Phytochrome B and histone deacetylase 6 control light-induced chromatin compaction in Arabidopsis thaliana. PLoS Genet 5: e1000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Ma L, Liu Y, Xu D, Chen Q, Li G(2020) Arabidopsis FAR-RED ELONGATED HYPOCOTYL3 integrates age and light signals to negatively regulate leaf senescence. Plant Cell 32: 1574–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Deng XW(2002) Arabidopsis FHY3 defines a key phytochrome a signaling component directly interacting with its homologous partner FAR1. EMBO J 21: 1339–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang H(2015) Multifaceted roles of FHY3 and FAR1 in light signaling and beyond. Trends Plant Sci 20: 453–461 [DOI] [PubMed] [Google Scholar]
- Wu K, Zhang L, Zhou C, Yu CW, Chaikam V(2008) HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J Exp Bot 59: 225–234 [DOI] [PubMed] [Google Scholar]
- Xu F, He S, Zhang J, Mao Z, Wang W, Li T, Hua J, Du S, Xu P, Li L, et al. (2018) Photoactivated CRY1 and phyB interact directly with AUX/IAA proteins to inhibit auxin signaling in Arabidopsis. Mol Plant 11: 523–541 [DOI] [PubMed] [Google Scholar]
- Yanagawa Y, Sullivan JA, Komatsu S, Gusmaroli G, Suzuki G, Yin J, Ishibashi T, Saijo Y, Rubio V, Kimura S, et al. (2004) Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev 18: 2172–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Xie F, Jiang Y, Li Z, Huang X, Li L(2018) Phytochrome a negatively regulates the shade avoidance response by increasing auxin/indole acidic acid protein stability. Dev Cell 44: 29–41.e4 [DOI] [PubMed] [Google Scholar]
- Zhang DW, Yuan S, Xu F, Zhu F, Yuan M, Ye HX, Guo HQ, Lv X, Yin Y, Lin HH(2016) Light intensity affects chlorophyll synthesis during greening process by metabolite signal from mitochondrial alternative oxidase in Arabidopsis. Plant Cell Environ 39: 12–25 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang Y, McFarlane HE, Obata T, Richter AS, Lohse M, Grimm B, Persson S, Fernie AR, Giavalisco P(2018) Inhibition of TOR represses nutrient consumption, which improves greening after extended periods of etiolation. Plant Physiol 178: 101–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Zhao M, Shi T, Shi H, An F, Zhao Q, Guo H(2009) EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc Natl Acad Sci USA 106: 21431–21436 [DOI] [PMC free article] [PubMed] [Google Scholar]